Abstract
Genetic relationships among 132 strains of Vibrio vulnificus (clinical, environmental, and diseased-eel isolates from different geographic origins, as well as seawater and shellfish isolates from the western Mediterranean coast, including reference strains) were analyzed by random amplified polymorphic DNA (RAPD) PCR. Results were validated by ribotyping. For ribotyping, DNAs were digested with KpnI and hybridized with an oligonucleotide probe complementary to a highly conserved sequence in the 23S rRNA gene. Random amplification of DNA was performed with M13 and T3 universal primers. The comparison between ribotyping and RAPD PCR revealed an overall agreement regarding the high level of homogeneity of diseased-eel isolates in contrast to the genetic heterogeneity of Mediterranean isolates. The latter suggests the existence of autochthonous clones present in Mediterranean coastal waters. Both techniques have revealed a genetic proximity among Spanish fish farm isolates and a close relationship between four Spanish eel farm isolates and some Mediterranean isolates. Whereas the differentiation within diseased-eel isolates was only possible by ribotyping, RAPD PCR was able to differentiate phenotypically atypical isolates of V. vulnificus. On the basis of our results, RAPD PCR is proposed as a better technique than ribotyping for rapid typing in the routine analysis of new V. vulnificus isolates.
Vibrio vulnificus is an autochthonous marine and estuarine bacterium which is able to colonize surfaces and internal organs of invertebrate and vertebrate marine animals (21). It may cause a rapid, invasive, and highly lethal disease associated with oyster consumption (17, 18), especially in individuals who have a preexisting chronic illness or who are immunocompromised, and may cause fatal wound infections through exposure to seawater even in people without preexisting disease (13, 22). Its isolation and its role in human infections were first reported in the United States, Japan, and Taiwan (17, 20, 32), but since 1992 reports from other geographical areas including Australia, Brazil, and Europe are increasing (4, 10, 15, 22, 25, 31, 40, 41). Infection occurs either after consumption of contaminated seafood, mainly oysters, or through wounds exposed to seawater (13, 22). V. vulnificus, being a human pathogen, includes some strains, originally defined by Tison et al. (39) as biotype 2, that constitute a serologically homogeneous group recently defined as serovar E by Biosca et al. (9) and that are responsible for severe epizootics in eel farms (3, 7). The strains pathogenic to humans are serologically and genetically diverse and have been isolated from seawater and shellfish, whereas serovar E strains have been isolated only from diseased eels or from people who handle eels (3, 40).
Epidemiological reports have revealed that, in most cases, the incidence of infections by V. vulnificus is related to the consumption of raw oysters (17, 18, 27). In Spain, clinical reports of infections caused by V. vulnificus are very scarce (16, 31), in accordance with the low incidence of this species in the Mediterranean Sea (4, 5). Temperature and salinity have proven to be the two most relevant physicochemical parameters correlated with the distribution of this species (23, 32). In the Mediterranean, summer temperatures are always above 20°C, clearly favorable for this species. Winter temperatures are never below 10°C, and therefore the presence of viable but nonculturable cells in response to low temperatures is not expected. In contrast, salinity values (around 35‰) are much above the optimal values described for this species (23). Until very recently, the only V. vulnificus isolates at the Spanish Mediterranean coast corresponded to serovar E strains recovered from diseased eels at an eel farm with an intensive culture system placed close to the sea, although not directly connected to it, and to a few non-serovar E strains recovered from healthy eels or tank water at the same farm (3, 7, 8). This farm presently uses recirculated freshwater, but at the time of the first outbreaks (1989 to 1990) it used well water of 1.7‰ salinity. In a very recent study, and after a specific search for this species at several sea sites, a search combining culture methods for isolation and identification by PCR using specific primers (4), we were able to recover non-serovar E strains from seawater and shellfish for the first time in our coastal waters.
The Mediterranean isolates together with fish farm, clinical, and environmental isolates from different geographic origins (132 strains) were subjected to ribotyping and randomly amplified polymorphic DNA (RAPD) PCR. Both techniques have been widely used for the typing of bacteria in epidemiological and ecological studies, and their advantages and disadvantages are well known. Ribotyping offers highly reproducible patterns and has already been used for intraspecific differentiation of V. vulnificus from different origins (2, 6, 9, 19, 38). We have used this technique successfully in a previous study of the fish farm isolates, together with strains from other origins (2). RAPD PCR is less laborious and time-consuming than other DNA-based techniques and seems to be the fastest genetic typing method that could be employed for a rapid identification, although it is less reproducible among different laboratories. It has also been widely used as a typing technique for both gram-positive and gram-negative bacteria (11, 26, 27, 30), and more specifically to differentiate pathogenic Vibrio species, including V. vulnificus (6, 19, 42).
In the present study we have used both techniques on all our serovar E and non-serovar E isolates in order to (i) analyze the genetic diversity of the Mediterranean isolates by ribotyping and RAPD PCR, by comparing the obtained profiles with the ones shown by V. vulnificus strains from different origins, (ii) determine the genetic proximity of the V. vulnificus strains isolated at the Spanish fish farm, including serovar E and non-serovar E strains, over several years, and (iii) evaluate the usefulness of RAPD as a typing technique for our isolates, by comparing the results with the ones obtained by ribotyping.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 132 strains of V. vulnificus, including diseased-eel, clinical, and environmental isolates from different origins, were used in this study. These and their sources, when known, are listed in Table 1. Strains were grown in nutrient agar (Oxoid), supplemented with 0.5% NaCl (wt/vol). Clinical strains were incubated at 37°C, and environmental isolates were incubated at 25°C.
TABLE 1.
Strains used in this study
| Straina | Source (date [mo-yr]) | Country | Serovar Ee | Ribotypef | RAPD PCR resultg with primer:
|
|
|---|---|---|---|---|---|---|
| M13 | T3 | |||||
| ATCC 27562T | Human wound infection | USAi | − | 1 | 4 | —h |
| C7184b | Human blood | USA | − | 2 | 5 | 5 |
| UMH1b | Fatal wound infection | USA | − | 3 | 1 | — |
| Vv1b | Fatal wound infection | USA | − | 4 | 5 | 1 |
| 374b | Septicemia case | USA | − | 5 | 5 | 5 |
| L180b | Septicemia case | USA | − | NDj | — | 7 |
| TW1 | Tank water fish farm (10-90) | Spain | − | 6 | 1 | 5 |
| LMG 12095 | Eel | Belgium | − | 6 | 8 | 7 |
| M631 | Eel (9-90) | Spain | − | 7 | 2 | 2 |
| D10 | Sea bream fish farm (7-90) | Spain | − | 8 | 6 | 7 |
| M76 | Eel (7-90) | Spain | − | 8 | 6 | 7 |
| E109 | Eel (7-90) | Spain | − | 8 | 6 | 7 |
| E110 | Eel (7-90) | Spain | − | 8 | 6 | 7 |
| M40 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M47 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M63 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M79 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M89 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M90 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M324 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M328 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M335 | Eel (3-90) | Spain | − | 9 | 1 | 3 |
| M626 | Eel (9-90) | Spain | − | 9 | 1 | 1 |
| M647 | Eel (9-90) | Spain | − | 9 | 2 | 1 |
| E112 | Eel (9-90) | Spain | − | 9 | 2 | |
| E113 | Eel (9-90) | Spain | − | 9 | 2 | |
| E114 | Eel (9-90) | Spain | − | 9 | 1 | 1 |
| P3 | Tank water fish farm (6-97) | Spain | − | 10 | 1 | 4 |
| P4 | Tank water fish farm (6-97) | Spain | − | 10 | 1 | 4 |
| P5 | Tank water fish farm (6-97) | Spain | − | 10 | 1 | 4 |
| P6 | Tank water fish farm (6-97) | Spain | − | 10 | 1 | 4 |
| 143c | Eel | Belgium | − | 11 | 4 | 7 |
| LMG 12090c | Eel | Belgium | − | 11 | 4 | 7 |
| 147c | Eel | Belgium | − | 11 | 4 | 7 |
| 149c | Eel | Belgium | − | 11 | 4 | 7 |
| LMG 12092 | Eel | Belgium | − | 11 | 4 | 7 |
| 155c | Eel | Belgium | − | 11 | 4 | 7 |
| 161c | Eel | Belgium | − | 11 | 4 | 7 |
| 169c | Eel | Belgium | − | 11 | — | — |
| 160c | Eel | Belgium | − | 12 | 8 | 6 |
| 167c | Glass eel | Belgium | − | 13 | 9 | 1 |
| 248 | Eel | Belgium | − | 13 | 9 | 2 |
| 249c | Eel | Belgium | − | 13 | 9 | 2 |
| 170c | Human blood | Belgium | − | 14 | — | — |
| LMG 13637 | Shrimp | Thailand | − | 15 | 8 | 6 |
| VIB 537d | Shrimp | Thailand | − | 15 | — | 6 |
| VIB 521d | Unknown | Unknown | − | 16 | — | 7 |
| VIB 535d | Eel | Sweden | − | 17 | — | — |
| VIB 536d | Eel | Sweden | − | 17 | — | 7 |
| C1 | Seawater (4-96) | Spain | − | 18 | 7 | 7 |
| C2 | Seawater (4-96) | Spain | − | 18 | 7 | 7 |
| C32 | Seawater (12-96) | Spain | − | 18 | 7 | 7 |
| C33 | Seawater (12-96) | Spain | − | 18 | 7 | 7 |
| C36 | Seawater (5-97) | Spain | − | 18 | 6 | 7 |
| C37 | Seawater (5-97) | Spain | − | 18 | 6 | 7 |
| C38 | Seawater (5-97) | Spain | − | 18 | 6 | 7 |
| C39 | Seawater (5-97) | Spain | − | 18 | 6 | 7 |
| C3 | Seawater (4-96) | Spain | − | 19 | 1 | 4 |
| C4–C25 | Wedge shell (5-96) | Spain | − | 20 | 1 | 7 |
| C26 | Seawater (5-96) | Spain | − | 21 | 11 | 12 |
| C27 | Seawater (5-96) | Spain | − | 22 | 11 | — |
| C28 | Seawater (5-96) | Spain | − | 22 | — | — |
| C29 | Seawater (5-96) | Spain | − | 23 | 13 | 12 |
| C30 | Seawater (5-96) | Spain | − | 23 | 13 | 12 |
| C31 | Seawater (5-96) | Spain | − | 24 | 7 | 7 |
| C34 | Seawater (5-97) | Spain | − | 25 | 12 | 9 |
| C35 | Seawater (5-97) | Spain | − | 25 | 12 | 9 |
| C40 | Wedge shell (5-97) | Spain | − | 26 | — | — |
| C41 | Seawater (6-97) | Spain | − | 27 | 10 | 11 |
| C42 | Seawater (6-97) | Spain | − | 28 | 10 | 10 |
| C43 | Common littleneck (6-97) | Spain | − | ND | 10 | 11 |
| C44 | Common littleneck (8-97) | Spain | − | 29 | 10 | — |
| C45 | Wedge shell (8-97) | Spain | − | 30 | 10 | 10 |
| C46 | Wedge shell (8-97) | Spain | − | 31 | 5 | 7 |
| C47 | Wedge shell (8-97) | Spain | − | 31 | 5 | 7 |
| ATCC 33149 | Diseased eel | Japan | + | 32 | 3 | 8 |
| NCIMB 2136 | Diseased eel | Japan | + | 32 | 3 | 8 |
| NCIMB 2137 | Diseased eel | Japan | + | 32 | 3 | 8 |
| E4 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E12 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E22 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E24 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E32 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E37 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E39 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E40 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E52 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E56 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E58 | Diseased eel (89) | Spain | + | 32 | 3 | 8 |
| E64 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| E80 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| E86 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| E92 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| M206 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| M338 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| E103 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| E105 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| E106 | Diseased eel (90) | Spain | + | 32 | 3 | 8 |
| ER1 | Diseased eel (92) | Spain | + | 32 | 3 | 8 |
| R1 | Diseased eel (94) | Spain | + | 32 | 3 | 8 |
| R2 | Diseased eel (94) | Spain | + | 32 | 3 | 8 |
| VIB 525d | Diseased eel | Sweden | + | 32 | 3 | 8 |
| VIB 526d | Diseased eel | Sweden | + | 32 | 3 | 8 |
| VIB 527d | Diseased eel | Sweden | + | 32 | 3 | 8 |
| VIB 524d | Diseased eel | Norway | + | 32 | 3 | 8 |
| 171c | Diseased eel | Belgium | + | 32 | — | 7 |
| VIB 523d | Diseased eel | Sweden | + | 32 | — | 7 |
| NCIMB 2138 | Diseased eel | Japan | + | 33 | 3 | 8 |
| LMG 13638 | Shrimp | Unknown | + | 33 | 3 | 8 |
| A1 | Diseased eel (97) | Spain | + | 33 | 3 | 8 |
| VIB 522d | Human blood | Unknown | + | 34 | 3 | 8 |
ATCC, American Type Culture Collection, Manassas, Va.; NCIMB, National Collection of Industrial and Marine Bacteria, Aberdeen, Scotland; LMG, Laboratorium voor Microbiologie, Rijksuniversiteit, Ghent, Belgium. Strains in boldface correspond to the new V. vulnificus isolates (C1 to C47 are Mediterranean isolates; P3 to P6 and A1 are new eel farm isolates).
Supplied by J. D. Oliver, University of North Carolina, Charlotte.
Supplied by L. Grisez, Katholieke Universiteit, Leuven, Belgium.
Supplied by L. Verdonck, Laboratorium voor Microbiologie, Rijksuniversiteit, Ghent, Belgium.
Ascription to serovar E was performed by slide agglutination with serum anti-E39 (diseased-eel isolate). +, serovar E; −, not serovar E.
Ribotype designations are each preceded by RT (e.g., RT 1).
Clusters obtained by RAPD PCR after the UPGMA analysis.
—, ungrouped strain.
USA, United States.
ND, not determined.
DNA isolation.
Chromosomal DNA was extracted by the guanidinium thiocyanate method of Pitcher et al. (34) and further purified by RNase and proteinase K treatments (only for ribotyping) as described by Sambrook et al. (35).
Ribotyping.
Ribotypes were obtained as described in a preliminary study by Aznar et al. (6), where different enzymes and probes were tested to determine the combination of KpnI digests and 23S rRNA directed probe (1038) yielding the best band discrimination. In accordance with the methods of Aznar et al. (6), 5 μg of chromosomal DNA was digested with endonuclease KpnI (GIBCO BRL) as recommended by the manufacturer and the DNA restriction fragments were separated by electrophoresis in 0.8% (wt/vol) agarose gels with TAE (Tris-acetate-EDTA) buffer. DNA was transferred to a noncharged nylon membrane (Qiabrane; Qiagen) under vacuum (Vacu-AID System; Hybaid). After transfer, DNA was hybridized with the 18-mer universal 1038 probe, complementary to a highly conserved region of eubacterial 23S rRNA genes that were digoxigenin labeled. Hybrid detection was performed by chemiluminescence (DIG Luminescent Detection Kit; Boehringer Mannheim). The hybridization temperature was 48°C. Band patterns displayed on X-OMAT 5 (Kodak) films were recorded with a video camera (Gel Station; Technologia para Diagnóstico e Investigación, Madrid, Spain) and stored as TIFF files.
RAPD PCR.
A new database, based on the RAPD PCR fingerprintings of the 131 strains included in this study, was created. Universal primers M13 (5′GAAACAGCTATGACCATG3′) and T3 (5′ATTAACCCTCACTAAAGG3′) used in this study, as well as the PCR conditions, were described previously by Aznar et al. (6). PCR was conducted in a total volume of 50 μl containing 5 μM universal primer, 0.03 U of Taq polymerase (SuperTherm), 5 μl of Taq reaction buffer, 10 μl of 25 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, and 100 ng of template DNA. Reaction mixtures were overlaid with 30 μl of mineral oil (Sigma, St. Louis, Mo.) and subjected to one cycle (OMNIGENE, St. Louis, Mo.) of 94°C for 5 min, 40°C for 5 min, and 72°C for 5 min. This was followed by 33 cycles of 94°C for 20 s, 48°C for 30 s, and 72°C for 45 s. The amplification products were electrophoresed on a 1.6% agarose gel, stained with ethidium bromide, and photographed under UV light. Gel images were recorded with a video camera (Gel Station; TDI) and stored as TIFF files.
Banding pattern analysis.
Digitized images were converted, normalized, analyzed, and combined with the software package Gel Compar, version 4.0 (Applied Maths, Kortrijk, Belgium). In order to normalize the banding patterns, molecular weight markers were included every four to six tracks. The levels of similarity between pairs of traces were computed by using the Pearson product-moment correlation coefficient for RAPD PCR and the Dice similarity coefficient (SD) for ribotyping (37). Data were clustered by using the unweighted pair group method by arithmetic averaging (UPGMA) algorithm (37).
RESULTS
Ribotyping.
A total of 132 V. vulnificus strains, including 80 isolates previously analyzed (2), 47 Mediterranean non-serovar-E isolates (4), and 5 new isolates recovered at the Spanish eel farm during a recent outbreak, were analyzed in the present work. For the eel farm isolates, one (A1) was recovered from a diseased eel and belonged to serovar E and four (P3 to P6) were non-serovar E isolates. In all, 34 ribotypes were distinguished among the strains analyzed. They have been designated RT 1 to RT 34 (Table 1). The Mediterranean isolates exhibited 14 ribotypes different from the ones already recorded in our database (2). The new isolates from the fish farm displayed two different ribotypes: RT 33, shown by strain A1, which corresponded to an already-described serovar E ribotype, and RT 10 (shared by the rest of strains), which was not previously recorded.
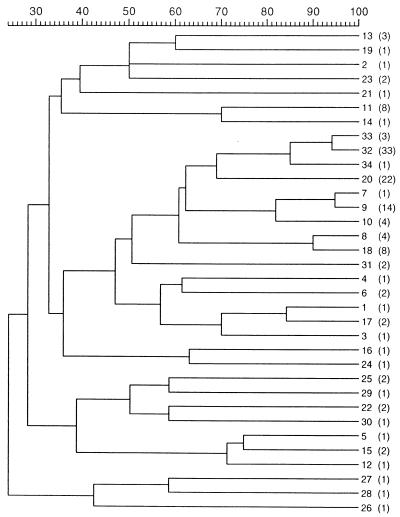
Figure 1 shows the dendrogram obtained with the 34 different profiles, after the UPGMA clustering. Despite the high level of diversity observed, the inclusion of new ribotypes in the cluster analysis did not perturb the previous groupings (2). Indeed, RT 32 to RT 34 comprised all serovar E strains, irrespective of geographic origin or outbreak. None of the 19 seawater or 28 bivalve isolates belonged either to serovar E or to RT 32 to RT 34. Non-serovar E strains isolated at the Spanish eel farm between 1990 and 1997 exhibited RT 6 to RT 10. RT 10, which was displayed by strains isolated in 1997, clustered with RT 7 and RT 9 (isolated in 1990) at 82% similarity. Eight seawater isolates from a place close to the fish farm, recovered in three different samplings over one year, showed a new ribotype (RT 18), which clustered with that of four eel farm isolates (RT 8). The shellfish isolates showed five new ribotypes, RT 20, RT 26, and RT 29 to RT 31, and 22 strains shared the same ribotype (RT 20), which was the closest neighbor to serovar E ribotypes. The rest of new ribotypes were quite diverse as demonstrated by the low levels of similarity among them and could not be related to geographical origins or sources.
FIG. 1.
Dendrogram based on UPGMA cluster analysis of the 34 different ribotypes obtained in this study. The scale measures the percentage of similarity. Numbers next to the dendrogram are ribotype designations, and numbers of strains sharing the same ribotype are shown in parentheses.
RAPD PCR.
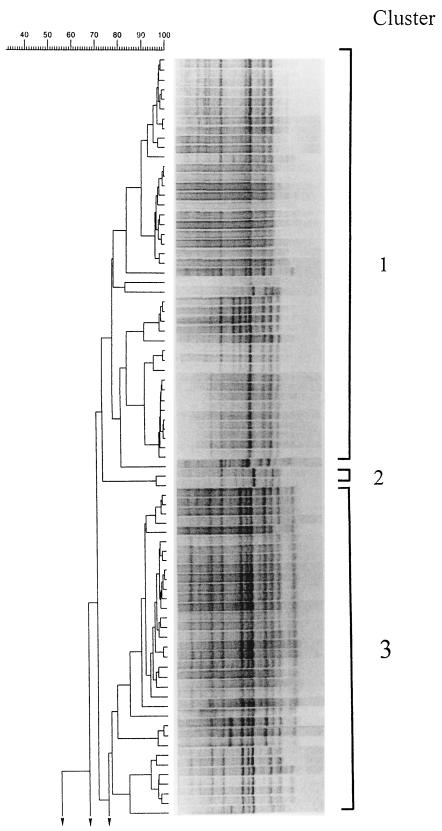
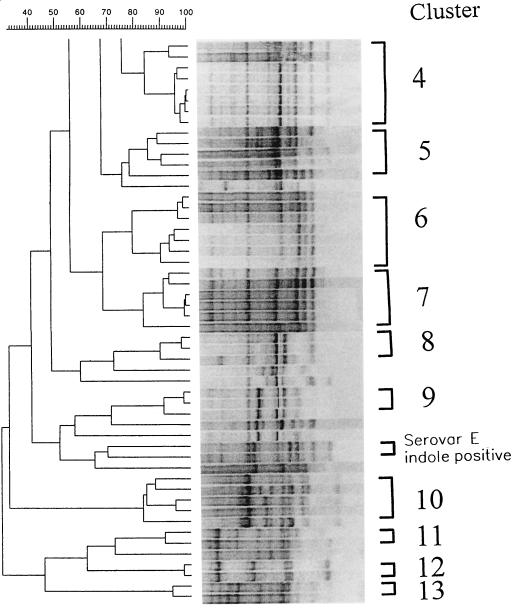
RAPD PCR with primers M13 and T3 rendered reproducible profiles consisting of 7 to 14 bands ranging from 162 to 3,800 bp. Results obtained with both primers allowed the differentiation of the isolates at the intraspecific level. Figure 2 shows the 13 clusters defined at the 77% similarity level by RAPD patterns obtained with primer M13 and the 10 strains which remained ungrouped. Cluster 1 includes mainly Spanish eel farm and shellfish isolates and a U.S. clinical isolate (strain UMH1). Cluster 3 includes all serovar E isolates, with the exception of strains 171 and VIB 523 (both positive for indole production), which grouped apart. Cluster 4 includes Belgium eel isolates together with the type strain (ATCC 27562). Cluster 5 includes clinical and shellfish strains. Clusters 6 and 7 include Spanish seawater and eel farm isolates. Clusters 8 and 9 include eel and shellfish isolates from different origins. Cluster 10 includes Spanish shellfish and seawater isolates. Clusters 11, 12, and 13 include Spanish seawater isolates which exhibited atypical responses for some phenotypic traits.
FIG. 2.
RAPD PCR patterns of V. vulnificus. The dendrogram was derived from an UPGMA cluster analysis of the RAPD PCR profiles of 131 V. vulnificus strains. The tracks show the processed band patterns after conversion, normalization, and subtraction of the background.
A cluster analysis of RAPD profiles obtained with primer T3 yielded 12 clusters at the 85% similarity level, and seven strains remained ungrouped. As with primer M13, all serovar E isolates except strains 171 and VIB 523 grouped together (cluster 8). The rest of the clusters included clinical and environmental isolates, and the phenotypically atypical strains formed several small clusters (Table 1).
Comparison of ribotyping and RAPD PCR results.
The comparison of the two fingerprinting techniques was performed by determining the location of all the strains with the same ribotype in the RAPD PCR dendrogram. For serovar E, a genetically homogeneous group, results with both techniques were in agreement. Ribotyping produced three very closely related profiles (RT 32 to RT 34), which were included in clearly defined clusters by RAPD analysis (cluster 3 with primer M13 or cluster 8 with primer T3). The only two serovar E strains that were not included in any of the two clusters corresponded to atypical biogroup 2 isolates, according to the criteria of Tison et al. (39), now serovar E according to Biosca et al. (9). These isolates shared the main serovar E ribotype (RT 32) but clustered quite far from the rest of the serovar E isolates with both primers by RAPD PCR. Isolates from healthy eels or tank water at the Spanish eel farm that showed closely related ribotypes (RT 7, RT 9, and RT 10) belonged to the same cluster (cluster 1), as determined with primer M13, or to closely related ones (clusters 1 to 4), as determined with T3. RT 8, exhibited by four isolates from healthy eels at the Spanish eel farm (strains D10, M76, E109, and E110), clustered with RT 18 (main ribotype exhibited by Spanish seawater isolates) (Table 1); by RAPD PCR with primer M13 these strains were included in cluster 6 together with Spanish seawater isolates belonging to RT 18. When primer T3 was used, these strains were placed in cluster 7 together with strains from other origins, including the same Spanish seawater isolates (Table 1). Shellfish strains showing RT 20, which clustered close to serovar E and non-serovar E Spanish eel farm ribotypes (Fig. 1), were also determined to be related to these strains by RAPD PCR. In general, the correspondence between results obtained by ribotyping and RAPD analysis was better when primer M13 was employed to generate the profiles.
DISCUSSION
In recent years several studies have been performed in order to analyze the intraspecific diversity of V. vulnificus. This characterization has been carried out by different techniques including molecular typing methods such as lipopolysaccharide typing, total protein profiles (sodium dodecyl sulfate-polyacrylamide gel electrophoresis), and nucleic acid-based methods: DNA sequencing, plasmid profiling, ribotyping, pulsed-field gel electrophoresis, RAPD PCR, and amplified-fragment length polymorphisms (AFLP). Several serotypes could be differentiated based on lipopolysaccharide or capsular antigens (9, 28, 36), although many isolates remained untypeable. Total protein profiles revealed a high level of homogeneity within the species. This is not useful for intraspecific differentiation (3). Plasmid profiles appeared to be good markers for serovar E isolates (8), but their value for the rest of the species is limited (14). Isolates related by common geographic origin or by serotype could be grouped by ribotyping (2, 9, 38) or RAPD profiles (6). Pulsed-field gel electrophoresis and AFLP allowed a finer differentiation of isolates from the same origin, revealing a high level of intraspecific diversity (2, 12). Recently, some of these typing techniques have been combined in order to obtain a better picture of the species diversity (3, 19, 38), although different protocols employed for the same technique by different authors have led to different results (6, 19).
In this work we have searched for the genetic relationships among clinical, environmental, and diseased-eel isolates of V. vulnificus, including a high number of strains from an eel farm that has suffered several infections by this bacterium and seawater strains from the Spanish Mediterranean coast. We had previously used ribotyping for 80 V. vulnificus strains (2) and RAPD analysis for 21 V. vulnificus strains and 5 strains of other Vibrio species (6). Both were found to be good techniques for intraspecific differentiation, with RAPD analysis being more rapid and simpler to perform. The 52 new isolates yielded 15 new ribotypes in addition to the 19 already obtained in our previous study (2). Most of the new isolates used in the present work represent the first seawater and shellfish isolates of V. vulnificus recovered so far on the west coast of the Mediterranean (4). The rest (five strains) are recent isolates from the same eel farm where the Spanish diseased-eel isolates analyzed were recovered (2).
Among the new eel farm isolates, strain A1 was recovered from a diseased eel during an outbreak that occurred in July 1997 and was assigned to serovar E by slide agglutination (unpublished data). It displays RT 33, which was previously assigned to two non-Spanish isolates and which is one of the ribotypes displayed by serovar E isolates. All strains isolated from diseased eels during epizootics that occurred from 1989 to 1994 in the Spanish eel farm shared RT 32. This ribotype is also exhibited by three of four Japanese reference strains (ATCC 33149, NCIMB 2136, and NCIMB 2137), originally isolated from diseased eels, and by diseased-eel isolates from other European countries as well. The fact that the strain isolated during the last outbreak at the Spanish farm exhibits the same ribotype as the fourth Japanese reference strain (NCIMB 2138) could be explained by the existence of a few original clones that have become widely distributed and that are responsible for outbreaks in several countries. This finding, together with the lack of reports on the isolation of serovar E strains from seawater samples, even in the vicinity of the fish farm that has suffered several outbreaks caused by this serovar, supports the hypothesis of animal-to-animal transmission (eel to eel or eel to man) and casts doubts on the hypothesis of water transmission of the disease, a hypothesis based only on a laboratory study (1). The existence of one serovar E strain isolated from human blood would support the animal-to-animal transmission hypothesis, since human infections related to the handling of eels have been reported (13, 40, 41).
In addition to strain A1, some non-serovar E strains (P3 to P6) were isolated from tank water during the last outbreak at the Spanish fish farm. All of them shared RT 10, which is very similar to other eel farm ribotypes (RT 7 and RT 9) described for strains isolated 7 years ago, revealing the close relationship among fish farm isolates. As expected, diversity among seawater isolates was higher and the ribotypes obtained did not cluster, but eight strains isolated in three different samplings over a year showed the same ribotype (RT 18). Interestingly, they were recovered from a sampling site located at the seaside close to the fish farm and clustered at 90% similarity with an eel farm ribotype (RT 8). The diversity in ribotypes found in the seawater isolates suggests the existence of autochthonous clones of V. vulnificus present on the west coast of the Mediterranean.
A V. vulnificus RAPD profile database has been created with the 131 strains, and the results of this technique have been compared with the ones obtained by ribotyping. In all, a good correspondence between the two typing techniques was observed. Of the two primers used, M13 allowed a more accurate definition of clusters and a better correspondence with the results obtained by ribotyping. Both primers differentiated serovar E strains as a tight group, clearly separated from the rest of strains. Furthermore, with both primers the serovar E strain recovered from the last outbreak (A1) clustered with the rest of the strains of serovar E isolated several years before with a very high level of similarity, in accordance with the results obtained by ribotyping. Nevertheless, its differentiation from the rest of the strains of serovar E was only possible through ribotyping. The close relationship between some eel farm isolates (displaying RT 8) and seawater isolates obtained at the sampling site located nearest to the fish farm (displaying RT 18) was identified as well. The genetic proximity among the Spanish fish farm isolates, irrespective of the source (eel or tank water), detected by ribotyping analysis was also detected by RAPD PCR with both primers. Phenotypically atypical strains clustered separately on the basis of RAPD analysis, whereas they were not differentiated by ribotyping. In this sense, it is noteworthy that the two atypical (indole-positive) serovar E strains (171 and VIB 523) did not cluster with the rest of strains belonging to this serovar when RAPD PCR was used, whereas they displayed the main ribotype (RT 32). This result is in accordance with the ones obtained by AFLP fingerprinting in a previous work (2) and reveals the usefulness of RAPD analysis for intraspecific differentiation of V. vulnificus, as found in other studies (6, 42). Nevertheless, other authors did not find a good correspondence between the two techniques: RAPD PCR was found to be less discriminatory (19). In any case, reports on comparisons among techniques have to be taken with caution, and only when exactly the same protocols are employed are results comparable. The existence of large databases for the different techniques that allow the comparison of new isolates is also highly desirable.
The results obtained in this study have demonstrated that both ribotyping and RAPD PCR are good typing techniques for V. vulnificus at the intraspecific level; they have allowed us to differentiate among V. vulnificus strains from different types of samples and geographic origins. They have revealed a high level of genomic diversity among seawater isolates from the western Mediterranean coast; these isolates seem to constitute an autochthonous population, some members of which are closely related to environmental strains recovered from a fish farm in the same area. By contrast, the high level of homogeneity among diseased-eel isolates, irrespective of geographic origin, was confirmed by both techniques. In a previous work we had reported the usefulness of ribotyping for intraspecific differentiation of V. vulnificus biotypes, which has allowed in this study the differentiation of ribotypes for diseased-eel isolates from the same eel farm. The RAPD PCR technique has been compared with and validated by ribotyping; it yielded similar overall results and was especially useful for the differentiation of phenotypically atypical strains. On the basis of our experience, and taking into account the results of previous studies, we propose the use of RAPD PCR for rapid typing in the routine analysis of new isolates of V. vulnificus and ribotyping or AFLP for a finer discrimination within diseased-eel isolates.
ACKNOWLEDGMENTS
This work has been supported by “Comisión Interministerial de Ciencia y Tecnología” grant AGF95-0264. C.R.A. is the recipient of a Ph.D. fellowship from the Ministerio de Educación y Ciencia.
REFERENCES
- 1.Amaro C, Biosca E G, Fouz B, Alcaide E, Esteve C. Evidence that water transmits Vibrio vulnificus biotype 2 infections to eels. Appl Environ Microbiol. 1995;61:1133–1137. doi: 10.1128/aem.61.3.1133-1137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias C R, Verdonck L, Swings J, Garay E, Aznar R. Intraspecific differentiation of Vibrio vulnificus biotypes by amplified fragment length polymorphism and ribotyping. Appl Environ Microbiol. 1997;63:2600–2606. doi: 10.1128/aem.63.7.2600-2606.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias C R, Verdonck L, Swings J, Aznar R, Garay E. A polyphasic approach to study the intraspecific diversity amongst Vibrio vulnificus isolates. Syst Appl Microbiol. 1997;20:622–633. [Google Scholar]
- 4.Arias C R, Aznar R, Pujalte M J, Garay E. A comparison of strategies for the detection and recovery of Vibrio vulnificus from marine samples of the Western Mediterranean coast. Syst Appl Microbiol. 1998;21:128–134. doi: 10.1016/S0723-2020(98)80016-7. [DOI] [PubMed] [Google Scholar]
- 5.Arias, C. R., M. C. Macián, R. Aznar, E. Garay, and M. J. Pujalte. Low incidence of V. vulnificus among Vibrio isolates from seawater and shellfish of the Western Mediterranean coast. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 6.Aznar R, Ludwig W, Schleifer K-H. Ribotyping and randomly amplified polymorphic DNA analysis of Vibrio vulnificus biotypes. Syst Appl Microbiol. 1993;16:303–309. [Google Scholar]
- 7.Biosca E, Amaro C, Esteve C, Alcaide E, Garay E. First record of Vibrio vulnificus biotype 2 from diseased European eel, Anguilla anguilla L. J Fish Dis. 1991;14:103–109. [Google Scholar]
- 8.Biosca E, Oliver J, Amaro C. Phenotypic characterization of Vibrio vulnificus biotype 2, a lipopolysaccharide-based homogeneous O serogroup within Vibrio vulnificus. Appl Environ Microbiol. 1996;62:918–927. doi: 10.1128/aem.62.3.918-927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biosca E G, Amaro C, Larsen J L, Pedersen K. Phenotypic and genotypic characterization of Vibrio vulnificus: proposal for the substitution of the subspecific taxon biotype for serovar. Appl Environ Microbiol. 1997;63:1460–1466. doi: 10.1128/aem.63.4.1460-1466.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bock T, Christensen N, Eriksen N H R, Winter S, Rygaard H, Jørgensen F. The first fatal case of Vibrio vulnificus in Denmark. APMIS. 1994;102:874–876. [PubMed] [Google Scholar]
- 11.Brousseau R, Saint-Onge A, Préfontaine G, Masson L, Cabana J. Arbitrary primer polymerase chain reaction, a powerful method to identify Bacillus thuringiensis serovars and strains. Appl Environ Microbiol. 1993;59:114–119. doi: 10.1128/aem.59.1.114-119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchrieser C, Gangar V V, Murpree R L, Tamplin M L, Kaspar C W. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl Environ Microbiol. 1995;61:1163–1168. doi: 10.1128/aem.61.3.1163-1168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalsgaard A, Frimodt-Møller N, Brunn B, Høi L, Larsen J L. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur J Microbiol Infect Dis. 1996;15:227–232. doi: 10.1007/BF01591359. [DOI] [PubMed] [Google Scholar]
- 14.Davidson L S, Oliver J D. Plasmid carriage in Vibrio vulnificus and other lactose-fermenting marine vibrios. Appl Environ Microbiol. 1986;51:211–213. doi: 10.1128/aem.52.1.211-213.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh H K, Bowen T E. Halophilic vibrios from human tissue infections on the Pacific coast of Australia. Pathology. 1980;12:397–402. doi: 10.3109/00313028009077102. [DOI] [PubMed] [Google Scholar]
- 16.Hernáez J, González F, Provencio M, Romera M A, Portero M F, Pérez-Maestu R, Masa-Vázquez C, Martínez de Letona J. Septicemia por Vibrio vulnificus. Rev Esp Microbiol Clin. 1991;6:144–145. [Google Scholar]
- 17.Hlady W G, Mullen R C, Hopkins R S. Vibrio vulnificus from raw oysters leading cause of reported deaths from foodborne illness in Florida. J Florida Med Assoc. 1993;80:2–4. [PubMed] [Google Scholar]
- 18.Hlady W G, Klontz K C. The epidemiology of Vibrio infections in Florida, 1991–1993. J Infect Dis. 1996;173:1176–1183. doi: 10.1093/infdis/173.5.1176. [DOI] [PubMed] [Google Scholar]
- 19.Høi L, Dalsgaard A, Larsen J L, Warner J M, Oliver J D. Comparison of ribotyping and randomly amplified polymorphic DNA PCR for characterization of Vibrio vulnificus. Appl Environ Microbiol. 1997;63:1674–1678. doi: 10.1128/aem.63.5.1674-1678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hor L I, Goo C T, Wan L. Isolation and characterization of Vibrio vulnificus inhabiting the marine environment of the Southwestern area of Taiwan. J Biomed Sci. 1995;2:384–389. doi: 10.1007/BF02255226. [DOI] [PubMed] [Google Scholar]
- 21.Horré R, Marklein G, Schaal K P. Vibrio vulnificus, an emerging human pathogen. Zentbl Bakteriol. 1996;284:273–284. doi: 10.1016/s0934-8840(96)80103-4. [DOI] [PubMed] [Google Scholar]
- 22.Hoyer J, Engelmann E, Liehr R-M, Distler A, Hahn H, Shimada T. Septic shock due to Vibrio vulnificus serogroup O4 wound infection acquired from the Baltic Sea. Eur J Clin Microbiol Infect Dis. 1995;14:1016–1018. doi: 10.1007/BF01691388. [DOI] [PubMed] [Google Scholar]
- 23.Kaspar C W, Tamplin M L. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl Environ Microbiol. 1993;59:2425–2429. doi: 10.1128/aem.59.8.2425-2429.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klontz K C, Lieb S, Schreiber M, Janowski H T, Baldy L M, Gunn R A. Syndromes of Vibrio vulnificus infections: clinical and epidemiological features in Florida cases, 1981–1987. Ann Intern Med. 1988;109:318–323. doi: 10.7326/0003-4819-109-4-318. [DOI] [PubMed] [Google Scholar]
- 25.Landgraf M, Leme K B P, Garcia-Moreno M L. Occurrence of emerging pathogenic Vibrio spp. in seafood consumed in São Paulo city, Brazil. Rev Microbiol. 1996;27:126–130. [Google Scholar]
- 26.Lawrence L M, Harvey J, Gilmour A. Development of a random amplification of polymorphic DNA typing method for Lysteria monocytogenes. Appl Environ Microbiol. 1993;59:3117–3119. doi: 10.1128/aem.59.9.3117-3119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linton C J, Smart A D, Leeming J P, Jalal H, Telenti A, Bodmer T, Millar M R. Comparison of random amplified polymorphic DNA with restriction fragment length polymorphism as epidemiological typing methods for Mycobacterium tuberculosis. J Clin Pathol. 1995;48:M133–M135. doi: 10.1136/mp.48.3.m133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin S J, Siebeling R J. Identification of Vibrio vulnificus O serovars with antilipopolysaccharide monoclonal antibody. J Clin Microbiol. 1991;29:1684–1688. doi: 10.1128/jcm.29.8.1684-1688.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascola L, Torney M, Dassey D. Vibrio vulnificus infections associated with eating raw oysters. JAMA. 1996;276:937–938. [PubMed] [Google Scholar]
- 30.Mazurier S, van de Giessen A, Heuvelman K, Wearnars K. RAPD analysis of Campylobacter isolates: DNA fingerprinting without the need to purify DNA. Lett Appl Microbiol. 1992;14:260–262. doi: 10.1111/j.1472-765x.1992.tb00700.x. [DOI] [PubMed] [Google Scholar]
- 31.Melhus Å, Holmdahl T, Tjernberg I. First documented case of bacteremia with Vibrio vulnificus in Sweden. Scand J Infect Dis. 1995;27:81–82. doi: 10.3109/00365549509018980. [DOI] [PubMed] [Google Scholar]
- 32.Oliver J D, Warner R A, Cleland D R. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl Environ Microbiol. 1983;45:985–998. doi: 10.1128/aem.45.3.985-998.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Moreno M M O, Romera G, Pous G, Jardí A M, Zaragoza J, Buj J I, Pérez F J. Bacteremia por Vibrio vulnificus en paciente con úlcera cutánea expuesta a agua de mar. Enferm Infecc Microbiol Clin. 1996;14:66–67. [PubMed] [Google Scholar]
- 34.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidinium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Simonson J, Siebeling R J. Immunogenicity of Vibrio vulnificus capsular polysaccharides and polysaccharide-protein conjugates. Infect Immun. 1993;61:2053–2058. doi: 10.1128/iai.61.5.2053-2058.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sneath P H A, Sokal R R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 38.Tamplin M L, Jackson J K, Buchrieser C, Murphfee R L, Portier K M, Gangar V, Miller L G, Kaspar C W. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl Environ Microbiol. 1996;62:3572–3580. doi: 10.1128/aem.62.10.3572-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tison D L, Nishibushi M, Greenwood J D, Seidler R J. Vibrio vulnificus biotype 2: new biogroup pathogenic for eels. Appl Environ Microbiol. 1982;44:640–646. doi: 10.1128/aem.44.3.640-646.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veenstra J, Rietra P J, Stoutenbeek C P, Coster J M, de Gier H H, Dirks-Go S. Infection by an indole-negative variant of Vibrio vulnificus transmitted by eels. J Infect Dis. 1992;166:209–210. doi: 10.1093/infdis/166.1.209. [DOI] [PubMed] [Google Scholar]
- 41.Veenstra J, Rietra P J G M, Coster J M, Slaats E, Dirks-Go S. Seasonal variations in the occurrence of Vibrio vulnificus along the Dutch coast. Epidemiol Infect. 1994;112:285–290. doi: 10.1017/s0950268800057691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J-J, Hor L-I, Shiau S-L. Differentiation of Vibrio vulnificus strains by an arbitrarily primed polymerase chain reaction. Chin J Microbiol Immunol. 1995;28:70–78. [PubMed] [Google Scholar]