Abstract
A previously identified insert expressing an endopeptidase from a Lactobacillus helveticus CNRZ32 genomic library was characterized. Nucleotide sequence analysis revealed an open reading frame of 1,941 bp encoding a putative protein of 71.2 kDa which contained a zinc-protease motif. Protein homology searches revealed that this enzyme has 40% similarity with endopeptidase O (PepO) from Lactococcus lactis P8-2-47. Northern hybridization revealed that pepO is monocistronic and is expressed throughout the growth phase. CNRZ32 derivatives lacking PepO activity were constructed via gene replacement. Enzyme assays revealed that the PepO mutant had significantly reduced endopeptidase activity when compared to CNRZ32 with two of the three substrates examined. Growth studies indicated that PepO has no detectable effect on growth rate or acid production by Lactobacillus helveticus CNRZ32 in amino acid defined or skim milk medium.
Proteolytic enzymes of lactic acid bacteria (LAB) contribute to their ability to obtain essential amino acids from milk and to development of flavor in bacterium-ripened cheese varieties (e.g., Cheddar or Gouda). LAB are multiple-amino-acid auxotrophs and therefore must obtain essential amino acids from the growth medium. The quantities of free amino acids and small peptides present in milk are not sufficient to support the growth of LAB to a high cell density (16). Therefore, LAB require a proteolytic system to obtain essential amino acids from caseins, the primary proteins present in milk. Among LAB, the proteolytic system of Lactococcus bacteria and the relationship of specific components to the ability of these organisms to obtain essential amino acids from caseins are the best characterized. Caseins are hydrolyzed by the lactococcal cell envelope-associated proteinase to produce peptides which are transported into the cell by an oligopeptide transport system. Once inside the lactococcal cell, these peptides are hydrolyzed by a variety of endopeptidases, aminopeptidases, and di- and tripeptidases to yield free amino acids (16).
To date, two distinct endopeptidases from lactococci, designated PepO and PepF, have been reported (21, 28, 31). It was found that growth in milk of lactococcal strains lacking either PepO or PepF was indistinguishable from growth of the wild-type strain. However, two highly related enzymes designated PepO2 (12a) and PepF2 (22) have been identified. These results suggest that the milk growth studies may not accurately reflect the importance of PepO and PepF.
While the proteolytic systems of lactobacilli are not as well characterized as those of lactococci, the results obtained to date suggest that their proteolytic systems are similar (14, 25). We are interested in Lactobacillus helveticus CNRZ32 due to the demonstrated ability of this strain to accelerate cheese ripening and reduce bitterness when used as an adjunct culture (4, 5). We have focused on proteolytic enzymes from this organism because they are believed to be responsible for its beneficial attributes. Recently, we have focused on endopeptidases from this organism since this class of peptidases is the most poorly characterized in lactobacilli (8). To date, a thiol-dependent endopeptidase from Lactobacillus helveticus CNRZ32 has been purified and characterized and the gene encoding this enzyme has been characterized (12). The present report describes the characterization of a metalloendopeptidase gene (pepO) of CNRZ32 and the construction and characterization of derivatives lacking PepO. The use of CNRZ32 derivatives deficient in endopeptidase(s) to clarify their role in cheese flavor development is currently under way.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Lactobacillus helveticus CNRZ32 (15) and JLS200 (CNRZ32 lacking X-prolyl-dipeptidyl aminopeptidase activity, PepX [32]) and their derivatives were grown in MRS broth (Difco Laboratories, Detroit, Mich.) (10) at 37°C. Lactococcus lactis LM0230 was obtained from L. L. McKay (University of Minnesota, St. Paul) and propagated at 30°C in M17-glucose broth (Difco Laboratories) (29). Escherichia coli DH5α (Gibco-BRL Life Technologies Inc., Gaithersburg, Md.) and DPWC and BW26 (Gold Biotechnology Inc., St. Louis, Mo.) were grown in LB broth (24) at 37°C with aeration. Agar plates were prepared by adding 1.5% (wt/vol) granulated agar (Difco Laboratories) to liquid media. The concentration of antibiotics added to liquid media or agar plates for selection of plasmids in E. coli were as follows: pKF1 (12), 1.0 mg of erythromycin per ml; pMOB (Gold Biotechnology), 100 μg of ampicillin or 100 μg of carbenicillin per ml; pSA3 (9), 12.5 mg of tetracycline or 100 μg of chloramphenicol per ml. To select for pSA3 or pTRKL2 (23) in Lactobacillus helveticus or Lactococcus lactis, 5 μg of erythromycin per ml was used. All antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.).
Molecular cloning.
Plasmid isolation from E. coli and chromosomal DNA isolation from Lactobacillus helveticus were performed as described by Sambrook et al. (24). Plasmid isolation from lactococci was conducted as described by Anderson and McKay (2). Restriction enzymes and T4 DNA ligase were purchased from Gibco-BRL Life Technologies Inc. and were used as recommended by the manufacturer. Electroporations were conducted with a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.). Electroporation of E. coli was performed as recommended by the manufacturer (Bio-Rad). Electroporations of Lactobacillus helveticus CNRZ32 and JLS200 were performed essentially as described by Bhowmik and Steele (7). The only differences were the following: (i) instead of electroporation buffer, cells were washed with ice-cold, sterile, double-distilled water; and (ii) 50 mM proline was added to the electroporation buffer. Electroporation of Lactococcus lactis LM0230 was performed as described by Holo and Nes (13). Subcloning of pKF1 (12)-derived fragments into pMOB was conducted essentially as described by Sambrook et al. (24).
DNA sequencing and sequence analysis.
PCR and DNA sequencing reactions were performed in a Perkin-Elmer (Norwalk, Conn.) model 480 thermal cycler. DNA sequencing reactions were conducted with the Prism Ready Reaction DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.). DNA templates were purified by using the modified alkaline lysis-polyethylene glycol precipitation procedure recommended by Applied Biosystems, Inc. Additional primers were designed by using the Affinity program supplied by Ransom Hill Bioscience, Inc. (Ramona, Calif.) and were synthesized by using GIBCO-BRL (Grand Island, N.Y.) Custom Primers. Nested sets of Tn1000 insertions were generated by using the Tn1000 kit (Gold Biotechnology, Inc.). Vector- and transposon-specific primers supplied with the Tn1000 kit were used for mapping of the Tn1000 insertion sites by PCR. DNA sequencing was conducted with the primers supplied with the Tn1000 kit and with synthesized primers. DNA sequence determination was conducted by the Nucleic Acid and Protein Facility of the University of Wisconsin—Madison Biotechnology Center, by using an ABI model 370/3 automated sequencer. Sequences were analyzed by use of the Genetics Computer Group (Madison, Wis.) sequence analysis package. Protein homology searches were performed by using the BLAST network service (1).
Construction of LAB derivatives.
A fragment of pKF1 insert was cloned into pSA3. A 381-bp internal in-frame deletion was introduced by digestion with restriction enzymes AflII and BbsI (see Fig. 1), filling-in with Klenow fragment, and ligation to yield pSUW50. The deletion was confirmed by restriction endonuclease digestions and DNA sequencing. Construction of Lactobacillus helveticus CNRZ32 PepO-deficient derivatives was conducted via gene replacement with pSUW50 (6). Identification of the mutants was accomplished by performing PCR with pepO internal primers and Southern hybridization with digoxigenin-labeled DNA probes generated by PCR from the same set of primers (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). The nucleotide sequences of the primers were 5′CCGAATGGTTGTCTAAAGCA3′ (YC-1) and 5′CCAGCATCCAGCCTTTAATTTC3′ (YC-2).
FIG. 1.
Nucleotide sequence and deduced amino acid sequence of pepO from Lactobacillus helveticus CNRZ32. A putative Shine-Dalgarno sequence is shown in boldface type and labeled rbs. The −10 and −35 regions are indicated as well as the zinc-metalloprotease motif. The two horizontal arrows indicate the putative transcriptional terminator. The 5′ end of the pepO mRNA is marked with a vertical arrow. Relevant restriction endonuclease sites are shown in boldface type and labeled. The nucleotides between the ≫≪ symbols were deleted in the PepO− derivatives.
A SmaI-SalI fragment containing a complete copy of pepO from pKF1 was subcloned into pTRKL2 to yield pSUW51. A Lactococcus lactis LM0230 derivative containing pSUW51 was constructed by electroporation and confirmed by plasmid analysis.
Enzyme assays.
Overnight cultures were harvested by centrifugation at 3,840 × g, and the pellet was washed and suspended in 50 mM Na2HPO4 (pH 8.0; Sigma). Cell extracts from Lactobacillus helveticus and Lactococcus lactis were obtained by alternately vortexing the samples with glass beads and then cooling them on ice, for 1 min each, with six repetitions. The protein content of cell extracts was determined by the method of Lowry et al. (18) with the Sigma Total Protein Kit by using bovine serum albumin (Sigma) as the standard. Substrates previously used to screen for endopeptidase activity (12), N-benzoyl-Phe-Val-Arg-p-nitroanilide (pNA), N-benzoyl-Pro-Phe-Arg-pNA, and N-benzoyl-Val-Gly-Arg-pNA (Sigma), were employed at final concentrations of 0.1, 0.5, and 0.5 mM, respectively. Enzyme assays were conducted with cell extracts normalized to 30 μg of protein/ml in 50 mM Na2HPO4 (pH 8.0) and preequilibrated at 37°C. Reactions were initiated by the addition of substrate. Reactions were stopped by the addition of 200 μl of 30% acetic acid to 800-μl reaction mixtures. Absorbance at 410 nm was determined by using a Beckman (Fullerton, Calif.) DU-65 spectrophotometer. Reaction rates were verified to be linear under these conditions and were quantified on the basis of release of pNA (extinction coefficient of 8.8 mM−1 cm−1 at 410 nm) (11). Enzyme assays were performed in duplicate on two independently prepared cell extracts. Endopeptidase activity was expressed as micromoles of pNA released per minute per milligram of protein. The endopeptidase activities of Lactobacillus helveticus CNRZ32 and Lactococcus lactis LM0230 were normalized to 100%. The relative endopeptidase activities of CNRZ32 PepO− and LM0230(pSUW51) were calculated relative to those of their parental strains.
Growth studies.
Growth studies were performed in double-steamed, pasteurized skim milk medium (pasteurized skim milk was steamed for 20 min, held at 42°C for 1 h, and then steamed for another 20 min) and amino acid defined medium (salts were prepared according to the ingredients of MRS broth, with the supplement of complete amino acids and glucose as the carbon source) (8a). Cultures propagated in MRS broth at 42°C to exponential phase were washed and resuspended in 0.85% NaCl to the original volume. A 0.1% inoculation (initial cell density, approximately 1.0 × 106 cells/ml) was made into both media, and the cultures were incubated at 42°C. Samples for pH and absorbance determinations were taken at 1-h intervals. The pH was determined with a pH meter (model 410A; Orion Research, Boston, Mass.) with an Ingold puncture-type pH probe (LoT406-M6-DXK-S7/25; Mettler-Toledo, Urdorf, Switzerland). The cell density in amino acid defined medium was determined by monitoring absorbance at 600 nm. The cell density in skim milk medium was determined by monitoring the absorbance at 600 nm of clarified samples (8a). Briefly, 0.5 ml of skim milk culture was mixed with 0.5 ml of 2 M borate–200 mM EDTA (pH 8.0) and incubated at 55°C for 10 min. The cells were then harvested by centrifugation and washed once with 1.0 ml of 2 M borate–200 mM EDTA (pH 8.0). The cell pellet was washed twice with 100 mM BisTris buffer (bis[2-hydroxyethyl]iminotris[hydroxymethyl]methane) (pH 6.5), and the absorbance at 600 nm was determined from dilutions (if necessary) in the 0.03 to 0.30 linear range. All sampling was performed in triplicate from duplicate growth curves.
Southern hybridization.
A 774-bp internal pepO fragment (nucleotides 98 to 871) was used to synthesize a digoxigenin-labeled probe by PCR with the YC-1 and YC-2 primers and the Genius system (Boehringer Mannheim). Southern hybridizations were performed by the procedure supplied by the manufacturer.
RNA methods.
Total RNA was isolated by using the RNeasy kit (Qiagen). A 1,548-bp internal pepO fragment (nucleotides 387 to 1934) was amplified and used to synthesize a digoxigenin-labeled probe with the Genius system (Boehringer Mannheim) for Northern hybridization. The nucleotide sequences of the two primers were 5′CTTCATGGGTCCATATGCC3′ and 5′GTAATTCTATCTTCAGGATC3′. RNA molecular weight markers, solutions, and reagents used in Northern hybridization and chemiluminescent detection were purchased from Boehringer Mannheim. Northern hybridizations were performed by the procedure supplied by the manufacturer. Mapping of the 5′ end of the pepO transcript was conducted by using the 5′ rapid amplification of cDNA (5′ RACE) kit (version 2.0; Gibco-BRL). The nucleotide sequences of the three primers used for 5′ RACE were 5′CTGTATTTTTCATGTCAGCATC3′ (YC-3), 5′CTCCGGCAGAAGTTTG3′ (YC-4), and 5′TTTGATCTGCAGG3′ (YC-5). First-strand cDNA synthesis was performed with primer YC-3. Nested amplification of first-strand cDNA was carried out with primer YC-4 and the anchor primer supplied by the kit. Sequencing reactions were conducted with primer YC-5 by using the nested amplification product as the template.
Nucleotide sequence accession number.
The sequence for pepO has been submitted to GenBank and assigned accession no. AF019410.
RESULTS
Subcloning of pKF1.
Previously, an endopeptidase-positive clone, designated pKF1, was identified in a Lactobacillus helveticus CNRZ32 genomic library constructed in E. coli DH5α (12). A restriction endonuclease map revealed an insert size of 5.7 kb (data not shown). All attempts to subclone the entire insert into pMOB were unsuccessful. Three pKF1 fragments were subcloned separately into pMOB. Enzyme assays indicated that none of the subcloned fragments expressed endopeptidase activity (data not shown).
Tn1000 mutagenesis and DNA sequence analysis.
Sequence analysis was employed to identify the pKF1 region which confers endopeptidase activity. Insertion sites of Tn1000 within the three subcloned fragments were determined by PCR. The junctions of the subcloned fragments were sequenced by using synthesized primers and pKF1 as the template. The nucleic acid sequence of pepO is shown in Fig. 1. An open reading frame (ORF) of 1,941 bp which encodes a putative protein of 647 amino acids was identified. This protein has 40% sequence similarity to PepO from Lactococcus lactis P8-2-47 (20). There is a Shine-Dalgarno sequence (AAGGAG; ΔG = −12.8 kcal) (26) 6 bases upstream from the putative start codon AUG; in addition, a putative transcriptional terminator (ΔG = −22.4 kcal) (30) was identified 16 bases downstream of the putative stop codon UAA. Additionally, a search of the PROSITE Dictionary of Protein Sites and Patterns (3) with the deduced amino acid sequence identified a zinc-protease motif (His-Glu-Xxx-Xxx-His). No signal sequence was detected from the hydrophilicity plot (17).
CNRZ32 pepO-negative derivatives.
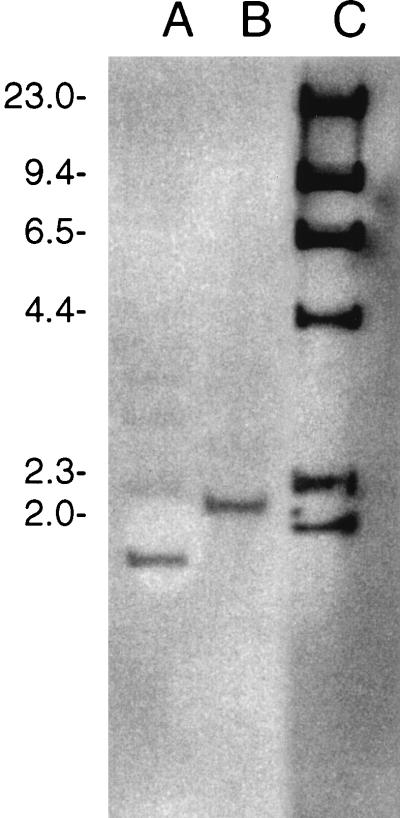
Two CNRZ32PepO-negative derivatives were constructed: a CNRZ32 pepO single mutant and a CNRZ32 pepX pepO double mutant. The CNRZ32 pepX pepO mutant was constructed from JLS200. Results from both PCR (data not shown) and Southern hybridization (Fig. 2) confirmed that an approximately 400-bp deletion had been introduced into the chromosomal pepO gene. Southern hybridization (Fig. 2) with total chromosomal DNA digested with PstI detected single bands which hybridized with the pepO probe in both CNRZ32 (2.2 kb) and its pepO mutant (1.8 kb).
FIG. 2.
Detection of Lactobacillus helveticus CNRZ32 pepO mutants by Southern hybridization. Lanes: A, CNRZ32 pepO; B, CNRZ32 wild type; C, digoxigenin-labeled λ DNA/HindIII molecular weight markers (sizes of markers, in thousands, are shown on the left).
Growth characteristics.
The growth and acidification rates for the CNRZ32 wild type, five pepO mutants, two pepO+ revertants, two pepX pepO mutants, and two pepX pepO+ revertants in both amino acid defined medium and skim milk medium were compared (data not shown). No differences among the CNRZ32 wild type, pepO mutants, or the pepO+ revertants were observed. Similarly, no differences among the pepX, pepX pepO mutants, or pepX pepO+ revertants were observed.
Enzyme assay.
The pepO mutant examined had 79 and >94% lower activities than that of CNRZ32 with N-benzoyl-Phe-Val-Arg-pNA and N-benzoyl-Val-Gly-Arg-pNA, respectively (Table 1). The introduction of the CNRZ32 pepO into Lactococcus lactis LM0230 on the low-copy-number vector pTRKL2 (6 to 9 copies/cell) did not result in a significant increase in endopeptidase activity (data not shown).
TABLE 1.
Endopeptidase activities of Lactobacillus helveticus CNRZ32 and a CNRZ32 pepO mutant
| Strain | Endopeptidase activity on substrate of:
|
|||||
|---|---|---|---|---|---|---|
|
N-benzoyl-Phe-Val-Arg-pNA
|
N-benzoyl-Pro-Phe-Arg-pNA
|
N-benzoyl-Val-Gly-Arg-pNA
|
||||
| Means ± SDa | RA (%)b | Mean ± SD | RA (%) | Mean ± SD | RA (%) | |
| CNRZ32 | 0.39 ± 0.01 | 100 | 0.081 ± 0.001 | 100 | 0.085 ± 0.004 | 100 |
| CNRZ32 pepO | 0.083 ± 0.001 | 21 | 0.081 ± 0.003 | 100 | BQLc | <6 |
Mean of specific activity (micromoles of pNA minute−1 milligram of protein−1) ± standard deviation.
RA, relative activity.
BQL, below quantifiable level (0.01 μmol of pNA min−1 mg of protein−1).
mRNA analysis.
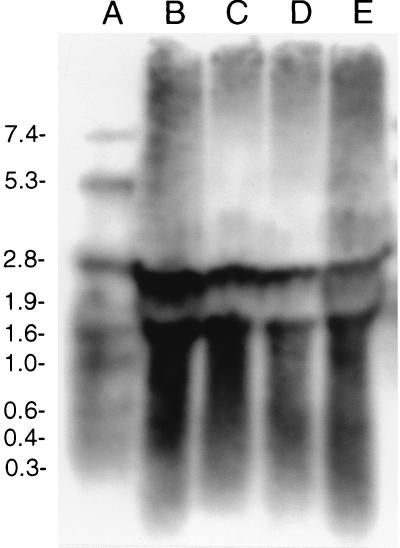
Two transcripts were detected in CNRZ32 throughout the growth phase (Fig. 3). One transcript had a calculated size of 2.2 kb, which corresponds to the size of the ORF of pepO. The other transcript had a calculated size of 1.5 kb. Similar analysis with a CNRZ32 pepO mutant detected transcripts of 1.8 and 1.5 kb (data not shown).
FIG. 3.
Detection of the Lactobacillus helveticus CNRZ32 pepO transcript during growth in MRS broth by using Northern hybridization. Lanes: A, digoxigenin-labeled RNA molecular weight markers; B to E, CNRZ32 total RNA isolated from early, mid-log, late log, and stationary phases (optical densities at 600 nm of 0.6, 1.2, 2.0, and 3.5), respectively. Sizes of molecular weight markers, in thousands, are shown on the left.
Sequencing of the nested amplification product from 5′ RACE revealed that the transcription start site was located 41 nucleotides upstream of the start codon (Fig. 1).
DISCUSSION
In this study, a previously identified endopeptidase-expressing clone (12) was characterized and determined to contain a 5.7-kb insert. Nucleic acid sequencing identified a 1,941-bp pepO ORF with an upstream AT-rich sequence which might serve as the putative −10 and −35 promoter regions. Data which suggest that the CNRZ32 pepO gene is monocistronic include (i) the putative promoter region and the putative terminator, and (ii) results from Northern hybridization. The high level of protein sequence homology of the CNRZ32 PepO to the PepO from Lactococcus lactis P8-2-47 suggests an ancestral association between these two enzymes. In addition, a metalloprotease motif (His-Glu-Xxx-Xxx-His) identified from the deduced PepO sequence was also present in strain P8-2-47 (20). The lack of a signal sequence suggests an intracellular location for PepO. The lactococcal PepO is also believed to be located intracellularly (27, 31).
In contrast to the lactococcal pepO (20, 28, 31), nucleic acid sequence analysis of 2.82 kb upstream and 0.55 kb downstream of the CNRZ32 pepO gene suggests that this gene is not associated with oligopeptide transport genes. A second copy of pepO in lactococcal strains, designated pepO2, has been reported recently (12a). The presence of single bands in Southern hybridizations with pepO as the probe in both CNRZ32 and its pepO derivatives indicates that there is only one copy of pepO in CNRZ32. Additionally, the >94% reduction in hydrolysis of N-benzoyl-Val-Gly-Arg-pNA by the pepO mutant suggests that there is a single copy of pepO in CNRZ32 and that this substrate could function to selectively quantify PepO activity.
To evaluate the physiological role of PepO, studies were conducted to compare the growth and acidification rates of the CNRZ32 wild type and pepO, pepO pepX, and pepX mutants in both amino acid defined medium and skim milk medium. The results revealed that the pepO and pepO pepX strains did not differ significantly in their growth or acidification rates from those of the wild type and the pepX mutant, respectively. This is similar to the results reported by Mierau et al. (19) for PepO in lactococci. These results suggest one or more of the following: (i) that PepO is not involved in the hydrolysis of milk-derived peptides, (ii) that other peptidases possess overlapping specificities with PepO, and (iii) that alternative milk-derived peptides can be utilized to obtain essential amino acids. Further investigation is required to determine what role, if any, PepO has in the development of cheese flavor.
ACKNOWLEDGMENTS
This project was supported by the Center for Dairy Research through funding from the National Dairy Promotion and Research Board and the College of Agricultural and Life Science at the University of Wisconsin—Madison.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A. The PROSITE dictionary of sites and patterns in proteins, its current status. Nucleic Acids Res. 1993;21:3097–3103. doi: 10.1093/nar/21.13.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartels H J, Johnson M E, Olson N F. Accelerated ripening of Gouda cheese. 1. Effect of heat-shocked thermophilic lactobacilli and streptococci on proteolysis and flavor development. Milchwissenschaft. 1987;42:83–88. [Google Scholar]
- 5.Bartels H J, Johnson M E, Olson N F. Accelerated ripening of Gouda cheese. 1. Effect of freeze-shocked Lactobacillus helveticus on proteolysis and flavor development. Milchwissenschaft. 1987;42:139–144. [Google Scholar]
- 6.Bhowmik T, Fernández L, Steele J L. Gene replacement in Lactobacillus helveticus. J Bacteriol. 1993;175:6341–6344. doi: 10.1128/jb.175.19.6341-6344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhowmik T, Steele J L. Development of an electroporation procedure for gene disruption in Lactobacillus helveticus CNRZ32. J Gen Microbiol. 1993;139:1433–1439. [Google Scholar]
- 8.Bockelmann W, Hoppe-Seyler T, Heller K J. Purification and characterization of an endopeptidase from Lactobacillus delbrueckii subsp. bulgaricus B14. Int Dairy J. 1996;6:1167–1180. [Google Scholar]
- 8a.Christensen, J. E., and J. L. Steele. Unpublished data.
- 9.Dao M L, Ferretti J J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985;49:115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMan J, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 11.Erlanger B F, Kowkowsky N, Cohen W. The preparation and properties of two new chromogenic substrates of trypsin. Arch Biochem Biophys. 1961;95:271–278. doi: 10.1016/0003-9861(61)90145-x. [DOI] [PubMed] [Google Scholar]
- 12.Fenster K M, Parkin K L, Steele J L. Characterization of a thiol-dependent endopeptidase from Lactobacillus helveticus CNRZ32. J Bacteriol. 1997;179:2529–2533. doi: 10.1128/jb.179.8.2529-2533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Hellendoorn, M. A., I. Mierau, M. van der Horst, G. Venema, and J. Kok. 1996. Abstracts of FEMS 5th Symposium on Lactic Acid Bacteria 1996, abstr. K13.
- 13.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalid N M, Marth E H. Lactobacilli—their enzymes and role in ripening and spoilage of cheese: a review. J Dairy Sci. 1990;73:2669–2684. [Google Scholar]
- 15.Khalid N M, Marth E H. Purification and partial characterization of a prolyldipeptidyl aminopeptidase from Lactobacillus helveticus CNRZ32. Appl Environ Microbiol. 1990;56:381–388. doi: 10.1128/aem.56.2.381-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunji E R S, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 17.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N H, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:263–275. [PubMed] [Google Scholar]
- 19.Mierau I, Kunji E R S, Leenhouts K J, Hellendoorn M A, Haandrikman A J, Poolman B, Konings W N, Venema G, Kok J. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J Bacteriol. 1996;178:2794–2803. doi: 10.1128/jb.178.10.2794-2803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mierau I, Tan P S T, Haandrikman A J, Kok J, Leenhouts K J, Konings W N, Venema G. Cloning and sequencing of the gene for a lactococcal endopeptidase, an enzyme with sequence similarity to mammalian enkephalinase. J Bacteriol. 1993;175:2087–2096. doi: 10.1128/jb.175.7.2087-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monnet V, Nardi M, Chopin A, Chopin M, Gripon J. Biochemical and genetic characterization of PepF, an oligopeptidase from Lactococcus lactis. J Biol Chem. 1994;269:32070–32076. [PubMed] [Google Scholar]
- 22.Nardi M, Renault P, Monnet V. Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis. J Bacteriol. 1997;179:4164–4171. doi: 10.1128/jb.179.13.4164-4171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Sullivan D J, Klaenhammer T R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Sasaki M, Bosman B W, Tan P S T. Immunological and electrophoretic study of the proteolytic enzymes from various Lactococcus and Lactobacillus strains. J Dairy Res. 1995;62:611–620. doi: 10.1017/s0022029900031344. [DOI] [PubMed] [Google Scholar]
- 26.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan P S T, Chapot-Chartier M-P, Pos K M, Rousseau M, Boquien C-Y, Gripon J-C, Konings W N. Localization of peptidases in lactococci. Appl Environ Microbiol. 1992;58:285–290. doi: 10.1128/aem.58.1.285-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan P S T, Pos K M, Konings W N. Purification and characterization of an endopeptidase from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1991;57:3593–3599. doi: 10.1128/aem.57.12.3593-3599.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinoco I J, Borer P N, Dengler B, Levine M D. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 31.Tynkkynen S, Buist G, Kunji E, Kok J, Poolman B, Venema G, Haandrikman A. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J Bacteriol. 1993;175:7523–7532. doi: 10.1128/jb.175.23.7523-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yüksel G, Steele J L. DNA sequence analysis, expression, distribution, and physiological role of the Xaa-prolyldipeptidyl aminopeptidase gene from Lactobacillus helveticus CNRZ32. Appl Microbiol Biotechnol. 1995;44:766–773. doi: 10.1007/BF00178616. [DOI] [PubMed] [Google Scholar]