Abstract
Examining the frequency and distribution of hybrids across contact zones provide insights into the factors mediating hybridization. In this study, we examined the effect of habitat and climate on hybridization patterns for three phenotypically, genetically, and ecologically distinct groups of the Canada jay (Perisoreus canadensis) in a secondary contact zone in western North America. Additionally, we tested whether the frequency of hybridization involving the three groups (referred to as Boreal, Pacific and Rocky Mountain morphotypes) is similar across the hybrid zones or whether some pairs have hybridized more frequently than others. We reanalyzed microsatellite, mtDNA and plumage data, and new microsatellite and plumage data for 526 individuals to identify putative genetic and phenotypic hybrids. The genetically and phenotypically distinct groups are associated with different habitats and occupy distinct climate niches across the contact zone. Most putative genetic hybrids (86%) had Rocky Mountain ancestry. Hybrids were observed most commonly in intermediate climate niches and in habitats where Engelmann spruce (Picea engelmannii) overlaps broadly with boreal and subalpine tree species. Our finding that hybrids occupy intermediate climate niches relative to parental morphotypes matches patterns for other plant and animal species found in this region. This study demonstrates how habitat and climate influence hybridization patterns in areas of secondary contact and adds to the growing body of research on tri-species hybrid zones.
Subject terms: Genetic variation, Biogeography
Introduction
Hybridization occurs across diverse taxa, although it is much more common in plant (25%) than animal (10%) species (Rieseberg 1997; Mallet 2005; Boecklen 2017). Examining patterns of hybridization in areas where species or divergent populations come into contact can provide important insights into factors influencing reproductive isolation (Hewitt 1988; Taylor et al. 2015; Billerman et al. 2016). Furthermore, hybridization has significant ecological and evolutionary consequences (Gompert et al. 2017) including the origin of new species (Soltis and Soltis 2009; Toews et al. 2011), collapse or extinction of species complexes (Taylor et al. 2006; Seehausen et al. 2008; Behm et al. 2010), and adaptive introgression (Borge et al. 2005; Chhatre et al. 2018).
Hybrid zones are both complex and variable. The shape of the zone, phylogenetic relatedness, degree of range overlap, level of pre- and post-zygotic isolation, natural history, dispersal, selection, and fitness all influence hybridization (Borge et al. 2005; Lemmon and Lemmon 2010; McKenzie et al. 2015; Kovach et al. 2015; Moran et al. 2019; Irwin 2020). Hybrid zones can be quite variable across time as well as space (Wielstra 2019). Whereas some hybrid zones have remained stable with respect to the location of contact and the frequency of hybrids (Taylor et al. 2014; Wang et al. 2019; Aguillon and Rohwer 2022), others are dynamic in space and time (Billerman et al. 2016, 2019). Studying hybrid zones is especially important in the context of anthropogenic and climate changes and provides important insights into the evolutionary and ecological repercussions of hybridization on biodiversity (Taylor et al. 2015; Ottenburghs 2021).
Whereas the majority of studies have focused on hybridization between two species or populations, a growing body of research has examined hybridization between three species or genetic lineages. Examples of tri-species hybrid complexes are fewer for animal species (Crow et al. 2007; Grossen et al. 2016; Natola and Burg 2018; Grant and Grant 2020; Ottenburghs 2021; Natola et al. 2022) compared to plants. Studies of oak (Quercus sp.), spruce (Picea sp.), and poplar (Populus sp.) species, for example, have provided insights into the dynamics of tri-species hybridization and the frequency and consistency of hybridization in these systems (Chhatre et al. 2018; Cronk and Suarez-Gonzalez 2018; Haselhorst et al. 2019; Cannon and Petit 2020). The term syngameon is often used to describe species complexes where distinct populations or species hybridize frequently in the absence of reproductive isolation (Boecklen 2017; Cannon and Petit 2020). Hybridization in these systems is uneven and often dominated by one species (Grant 1981; Boecklen 2017).
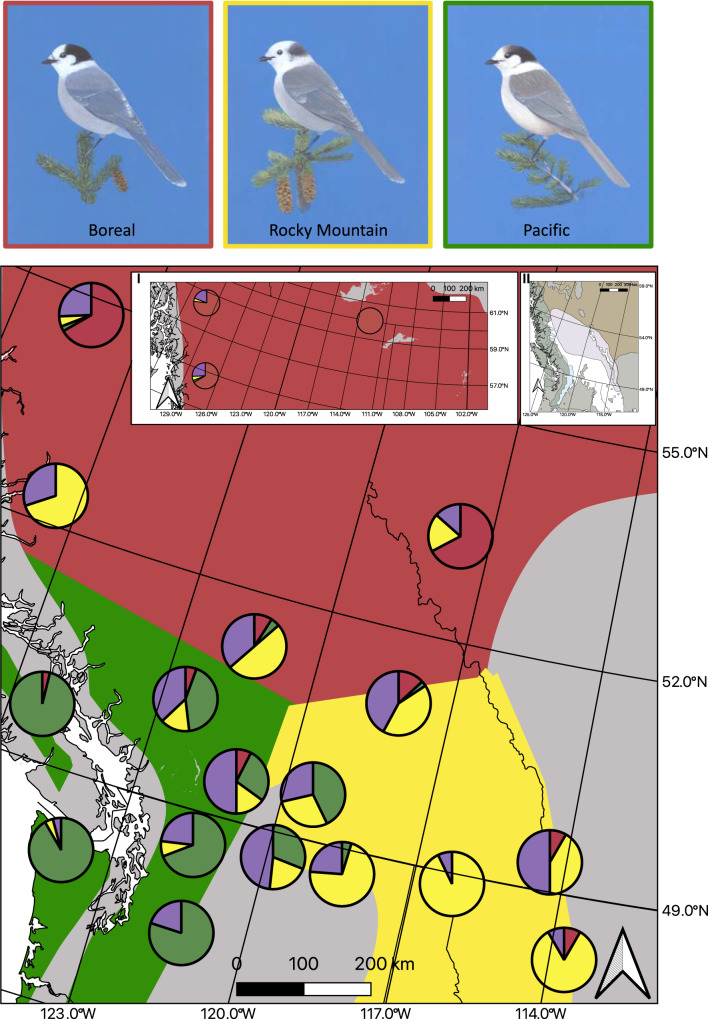
Canada jays (Perisoreus canadensis) are resident, food-storing passerines of North America with a wide distribution across boreal and subalpine habitats (Fig. 1; Strickland and Ouellet 2020). They show high levels of phenotypic and genetic variation (van Els et al. 2012; Dohms et al. 2017; Graham et al. 2021), and consist of three phenotypic groups (hereafter referred to as Boreal, Rocky Mountain, and Pacific morphotypes) that are genetically and ecologically distinct from each other (Strickland and Ouellet 2020). In its transcontinental range from Alaska to Newfoundland, the Boreal morphotype is strongly associated with white spruce (Picea glauca) and black spruce (P. mariana). The Rocky Mountain morphotype occurs from southeastern British Columbia to northern New Mexico and eastern Arizona in the Rocky Mountains and is strongly associated with Engelmann spruce (P. engelmannii). Finally, the Pacific morphotype is not necessarily associated with spruce at all. Although it occurs from northern California to southwestern British Columbia in high-elevation forests that may contain Engelmann spruce and also in coastal Sitka spruce (P. sitchensis) in northern California, it is absent from seemingly similar coastal stands in British Columbia, breeding instead, on Vancouver Island for example, at elevations above 850 m in spruce-less subalpine forests characterized by mountain hemlock (Tsuga mertensiana), Pacific silver fir (Abies amabilis), and yellow cedar (Callitropsis nootkatensis; Quarrell et al. 2022).
Fig. 1. Sketches show the plumage differences of the three Canada jay (Perisoreus canadensis) morphotypes (Boreal, Rocky Mountain, and Pacific) and the key tree species they are associated with (illustrations by Howard Coneybeare).
Boreal morphotypes are strongly associated with white spruce (Picea glauca), Rocky Mountain morphotypes are associated with Engelmann spruce (P. engelmanni), and Pacific morphotypes are not associated with any particular spruce species but are found in coniferous forests of the western subalpine. Map shows the distributions of the three Canada jay morphotypes across the contact in southwestern Canada and northwestern United States: Boreal (red), Rocky Mountain (yellow), and Pacific (green). Inset I) shows populations from further north in northern British Columbia and the Northwest Territories Canada. Pie charts show the nuclear genetic group that individuals were assigned to with STRUCTURE and GENODIVE (as found in Fig. 3): Boreal (red), Rocky Mountain (yellow), Pacific (green), and putative hybrids (purple). For information on the individuals included in each group, please see Table S1. Inset II) Map showing the distribution of white spruce (Picea glauca; light brown), Engelmann spruce (P. engelmanni; white), and western subalpine (dark gray), and the areas of overlap between white and Engelmann spruce (light purple), and western subalpine and Engelmann spruce (light blue) across the contact zone.
All three morphotypes come into secondary contact in south-central British Columbia and north-central Washington, where they show high levels of nuclear and mitochondrial introgression (Graham et al. 2021; Fig. 1). This region is also an active contact zone for many other plant and animal species (Brunsfeld et al. 2000; Swenson and Howard 2005; Toews and Irwin 2008; Gugger et al. 2010; Chavez et al. 2011; De La Torre et al. 2014; De La Torre et al. 2015; Natola and Burg 2018; Haselhorst et al. 2019), including three or more species of spruce, poplar (Populus sp.), and sapsuckers (Sphyrapicus sp.; Seneviratne et al. 2012; De La Torre et al. 2014; De La Torre et al. 2015; Grossen et al. 2016; Natola and Burg 2018; Billerman et al. 2019; Haselhorst et al. 2019). Thus, multiple tri-lineage contact zones occur in this phylogeographically complex region of northwestern North America (Brunsfeld et al. 2000).
Secondary contact of the Boreal and Rocky Mountain morphotypes (Fig. 1) mirrors, and was presumably facilitated by, post-glacial contact between white and Engelmann spruce, two closely related tree species with which the morphotypes are believed to have co-diverged in ice-free refugia during the Last Glacial Maximum (Hamilton et al. 2015). Engelmann and white spruce now co-occur in a ~376,000 km2 area (Fig. 1) in western North America, with the former species occupying a high-elevation ring around the entire contact zone. The exact distribution is exceedingly complex and is further complicated by widespread hybridization of these two spruce species, leading to intermediate forms commonly known as “interior spruce” (Haselhorst et al. 2019). At the western edge of the Engelmann-white spruce zone, spruce-dominated forests abruptly give way to the spruce-less subalpine zone of the Coast Mountains dominated by Pacific silver fir, yellow cedar, and mountain hemlock. The transition from forests dominated by Engelmann spruce to the spruce-less subalpine zone coincides with the beginning of the distribution range for the Pacific Canada jay morphotype (Fig. 1). The contact zone between Engelmann spruce and subalpine forest species is narrower, with Engelmann spruce co-occurring with mountain hemlock and Pacific silver in ~64,000 km2 region west of the Cascade Range.
We combined genetic, phenotypic, habitat, and climate data to examine factors influencing hybridization patterns among Canada jay morphotypes in the secondary contact zone. We genotyped individuals at twelve variable microsatellite markers to identify putative genetic hybrids and examined plumage characteristics to identify putative phenotypic hybrids. Given the nature of this contact zone where all three morphotypes can co-occur, we examined the ancestry for each putative genetic hybrid to determine whether the frequency of hybridization among morphotypes is similar across the contact zone or whether any of the three morphotype pairs hybridize more frequently. Next, we analyzed habitat and climate data to determine the extent to which putative phenotypic and genetic hybrids are associated with specific habitats, and whether morphotypes and putative hybrids occupy separate climate niches within the contact zone. Finally, we examined the influence of Engelmann spruce on hybridization for Canada jay morphotypes. Previous research suggests that the northern expansion of Engelmann spruce from glacial refugia and subsequent secondary contact with boreal and subalpine forest species has resulted in a greater frequency of hybridization among morphotypes (van Els et al. 2012; Dohms et al. 2017; Graham et al. 2021). To examine this last hypothesis, we compared genetic patterns in areas where Engelmann spruce occurs in sympatry with boreal and subalpine forests and areas where Engelmann spruce does not have such associations.
Methods
Phenotypic assignment
Canada jays exhibit high phenotypic diversity across their range. The Pacific morphotype is distinguished from the other two morphotypes by its paler breast, conspicuous white-shafted feathers on its back, and its darker nuchal patch that extends forward to, and sometimes past, the eyes (Fig. 1). The Rocky Mountain and Boreal morphotypes share many similar plumage traits including a gray, unstreaked back, white tipped secondaries and rectrices, and a white throat and collar that contrast with the variably gray breast and belly. They are, however, diagnosable based on crown differences –the Rocky Mountain morphotype has a pale-headed appearance due to a reduced nuchal patch that does not reach as far forward as the eye, whereas the Boreal morphotype has a darker nuchal patch that extends to its eye. Within the hybrid zone, phenotypic hybrids (hereafter referred to as intergrades) are also present. These individuals are characterized by admixture of diagnostic traits from two or more morphotypes, with those typical of the Rocky Mountain morphotypes being most commonly observed. Images of intergrade morphotypes are found in Graham et al. (2021).
All individuals sampled for this study (N = 526, Supplementary Table S1) were assigned to a morphotype by an observer (DS, JW, CC, and BAG); of these assignments, 410 were from museum specimens and 116 were from live birds trapped and released in the field; 309 of the birds were previously assigned to a phenotype in Graham et al. (2021), while the remaining 217 were assigned a phenotype for the first time in this study. We validated our ability to assign individuals to the correct morphotype by examining 194 adult specimens in collections housed at the Beaty Museum of Biodiversity, Canadian Museum of Nature, Royal Alberta Museum, Royal British Columbia Museum, and Royal Ontario Museum (177 of the 194 adult samples used for phenotypic analysis were included in our microsatellite genetic analysis). We assigned each specimen to one of the three morphotypes or identified it as an intergrade if it had plumage characteristics from two or more morphs, and collected measurements for eight plumage traits: (a) extent of crown plumage, (b) whether the dark crown plumage reached the eye, (c) neck-breast demarcation, (d) colouration of back plumage relative to breast plumage color, (e) presence of streaked feather shafts on the back, (f) presence of white tips on retrices, (g) presence of white tips on secondary coverts, and (h) presence of white edging on primaries (Fig. 1). All adult plumage traits were scored on a continuous scale as outlined in Graham et al. (2021). All variables conformed to normality and we used discriminant function analysis to test the accuracy of observer assignment. Discriminant function analysis was conducted in SPSS (version 23.0, SPSS Inc., Chicago IL) using the leave-one-out classification approach. We tested all variables for intercorrelations and found that none of the variable intercorrelations exceeded 0.7; thus, we included all the variables in the analyses. Following cross-validation, 93.4% of the birds were classified to the same morphotype assigned by the observer, demonstrating that we could assign birds accurately to the correct morphotype.
Genetic analyses
We analyzed 526 blood, feather, tissue or toe pad samples of Canada jays from Alberta, British Columbia, Northwest Territories, Idaho, Montana, and Washington; the majority of areas sampled are within or adjacent to the contact zone among the three morphotypes (Fig. 1). Samples were collected between 1890 and 2019 and included 309 samples used in a previous study (Graham et al. 2021) plus 217 new samples that were included to enhance the coverage of samples analyzed within the contact zone (Table S1).
All 309 samples included in the mtDNA control region dataset analyzed in this study were previously genotyped in Graham et al. (2021). The 309 mtDNA samples were included in this study because they fell within the contact zone studied or were adjacent to it and were reanalyzed to assess clade assignment. For 190 of these samples sequenced by Graham et al. (2021), we analyzed a 506 bp fragment of the mtDNA control region (CR) to examine genetic differentiation between morphotypes (see below). The remaining 119 samples genotyped by Graham et al. (2021) used primers that targeted fixed nucleotide differences between each clade were not included in the analyses of mtDNA genetic differentiation between morphotypes. Due to the age of the samples (some were over 100 years old), we had difficulties amplifying them with the mtDNA CR primers. As a result, we did not add any new mtDNA CR samples to this dataset.
For this study, we used microsatellite genotype data from 309 samples in Graham et al. (2021) and generated new genotypes for 217 new samples. All 526 samples were genotyped at 12 microsatellite loci using PCR protocols in Dohms et al. (2017) and Graham et al. (2021); the loci included Apco 30, Apco 37, Apco 40, Apco 41, Apco 91 (Stenzler and Fitzpatrick 2002); LTML 8 (McDonald and Potts 1994); Lox1 (Bensch et al. 1997); MJG1 (Li et al. 1997); Pdo 5 (Griffith et al. 1999); AIAAAAG13 (Delaney and Wayne 2005); Ck.2 A5A (Tarr and Fleisher 1998); and PJGATA2 (Busch et al. 2009). We tested for deviations from Hardy-Weinberg equilibrium and linkage disequilibrium with Genepop 4.7 (Rousset 2008). For these analyses, the 526 samples were grouped into twenty-two distinct populations based on the geographic sampling location (see Table S1 for population information). We corrected for all pairwise comparisons using the sequential Bonferroni method (Rice 1989). All individuals included in the analyses were genotyped at a minimum of six of the 12 loci, although most samples were genotyped for at least ten loci.
We examined population differentiation between morphotypes using both microsatellite and mtDNA markers. We calculated pairwise FST values (microsatellites) using GENODIVE 3.04 (Meirmans and Van Tienderen 2004) and pairwise θST values (mtDNA) with the R package haplotypes (Aktas 2020). We created a haplotype network in TCS (Clement et al. 2002) to visualize the distribution of phenotypes within each of the mtDNA clades using the 190 individuals that we had mtDNA control region sequence data for.
We validated the power of our microsatellite data to detect putative genetic hybrids with the program HYBRIDLAB 1.0 (Nielsen et al. 2006), following an approach similar to the one implemented by Costa et al. (2020). We generated 100 parental Boreal, Rocky Mountain, and Pacific genotypes and then ran multiple simulations to produce genotypes for putative Boreal x Rocky Mountain, Boreal x Pacific, and Rocky Mountain x Pacific F1 hybrids. We generated the simulated parental genotypes by selecting 61 individuals (approximately 20 from each morphotype) from allopatric populations that were genetically and morphologically distinct. We used the Bayesian clustering program STRUCTURE V2.3.4 (Pritchard et al. 2000) to quantify ancestry (Q) for each of the individuals from parental and hybrid populations using the following settings: admixture model with correlated alleles and no loc-priors (where population origin is not used to assist with population assignment), with a burn in of 50,000 chains followed by 100,000 steps for five iterations. For these simulations, we chose K = 3 to test the ability and power of STRUCTURE to assign individuals from each of the three parental populations to a distinct genetic cluster. Parental populations showed high assignment to their respective genetic cluster (Qmean = 0.86–0.91), demonstrating that our genetic data had considerable power to detect putative genetic hybrids. From these results, we chose a threshold Q value of 0.70 as 296 of the 300 simulated parental genotypes had Q values that exceeded this threshold. Of the 300 F1 hybrid genotypes we simulated, 292 fell below this 0.70 threshold (1 Boreal x Pacific, and 7 Pacific x Rocky Mountain F1 hybrids were above this threshold; range 0.70–0.79). Further, we chose to use this threshold to account for processes like homoplasy, where alleles may arise in populations via convergent evolution.
To examine population genetic structure, we used the Bayesian clustering program STRUCTURE V2.3.4 (Pritchard et al. 2000). Previous analyses of western populations revealed K = 3 as the optimal K (Graham et al. 2021), corresponding to the three mtDNA clades (van Els et al. 2012; Dohms et al. 2017). Based on these previous results, we examined population genetic structure at K = 3 within the contact zone. We used the admixture model with correlated alleles and no loc-priors, with a burn in of 50,000 chains followed by 100,000 steps for ten iterations. As stated above, we assigned individuals to a given cluster if their ancestry value was ≥ 0.7.
We calculated hybrid indices in GENODIVE 3.04 (Meirmans and Van Tienderen 2004) using the method outlined by Buerkle (2005) to complement our results with STRUCTURE. For those individuals with Q < 0.7 (N = 257 out of 526), we used hybrid indices to better determine the ancestry for each putative genetic hybrid. This method uses maximum likelihood estimates to calculate hybrid scores. Given that only two parental populations can be used in GENODIVE, we conducted three separate runs with the three possible parental combinations: Boreal and Rocky Mountain birds; Boreal and Pacific birds; and Rocky Mountain and Pacific birds. We chose 61 individuals (26 Boreal, 21 Rocky Mountain, and 14 Pacific) to act as reference individuals for each parental population; these individuals were selected from allopatric populations adjacent to the contact zone with Q > 0.9 to their parental population. We assigned individuals the hybrid index score with the least negative likelihood score following the three sets of analyses. Hybrid indices range from zero to one and we assigned an individual to one of the parental populations if the hybrid index score was <0.2 or >0.8 or as a putative genetic hybrid if their hybrid index score fell between 0.2 and 0.8. We considered an individual to be a hybrid if identified as a hybrid using both STRUCTURE and hybrid index scores.
Habitat and environment analyses
To quantify habitat, we used the ranges of five key tree species that Canada jay morphotypes are associated with across their respective distributions, and that are distributed within, or occur at the edge of, the contact zone (Strickland and Ouellet 2020): Engelmann spruce, black spruce, white spruce, Pacific silver fir, and mountain hemlock. We downloaded shape files for each tree species from the Conservation Biology Institute Data Basin (https://databasin.org) and plotted their distributions across the contact zone in QGIS 3.10 (QGIS.org). We then used the point-sampling tool in QGIS to determine which tree species were present in the area from which each of the 526 Canada jay samples were collected and assigned individuals to a designated forest type based on the presence/absence of these five tree species (Fig. 1). We assigned individuals to western subalpine forests if mountain hemlock and/or Pacific silver fir were present; western subalpine x Engelmann spruce forest if one or both of the subalpine forest tree species were found in the presence of Engelmann spruce; Engelmann spruce forest if only Engelmann spruce was present; Engelmann spruce x boreal forest if Engelmann spruce co-occurred with black and/or white spruce; and boreal forest if only black and/or white spruce were present and Engelmann spruce was absent. We had no samples from areas where boreal and subalpine forest species overlap, so we were unable to analyze this designated habitat type. This analysis assumes homogeneity of forest types within the study region, and based upon previous studies that have characterized the distribution of spruce species within our study region (De La Torre et al. 2014; De La Torre et al. 2014; De La Torre et al. 2015; Hamilton et al. 2015; Hasselhorst et al. 2019), we feel confident that the shapefiles used in this study accurately depict the distribution of these species. Furthermore, the strong associations between Rocky Mountain and Boreal morphotypes with their respective spruce species and the absence of a relationship between spruce and Pacific morphotypes (Strickland and Ouellet 2020) indicate that the assumptions made for this analysis are robust.
We also examined climate variation across the contact zone to complement our habitat analyses. For each individual, we collected data from the world Bioclim dataset (version 2.5, Hijmans et al. 2005) for elevation and nineteen environmental variables. We tested for intercorrelations among the twenty variables using a Spearman’s rank correlation and removed all variables with intercorrelations that exceeded 0.7 following the approach of Ruegg et al. (2006). Eight environmental variables remained: elevation (m), annual mean temperature (°C), isothermality, mean temperature during the driest quarter (°C), mean annual precipitation (mm), mean precipitation during the driest month (mm), precipitation seasonality, and mean precipitation during the warmest quarter (mm). Using these eight variables, we performed a Principal Component Analysis (PCA) to create an environmental index following the approach used by Bell and Irian (2019). We performed the PCA in SPSS (version 23.0, SPSS Inc., Chicago, IL), used a direct oblimin rotation as it allows for correlations among components, and retained the first principal component (Eigenvalue = 2.9) which explained 35.8% of the variance for environmental characteristics across the contact zone (see Table S2). Five environmental variables summarizing temperature and precipitation variation were associated with the first principal component: mean temperature during the driest quarter, annual mean temperature, isothermality, annual mean precipitation, and mean precipitation during the driest quarter. To visualize the environmental distribution of morphotype, and genetic variation, we plotted the frequency of each phenotype, mtDNA, microsatellite, and hybrid group along this environmental gradient.
Correlates of genetic and phenotypic patterns with habitat and environment
We used several multivariate approaches to examine the relationship between genetic and phenotypic patterns with environment and habitat variation. First, we used distance-based redundancy analysis (dbRDA) to examine how habitat and environment influence genetic and phenotypic distance; dbRDA is a multiple regression method that has been used in a number of ecological and evolutionary studies (Potvin and Clegg 2015), and provides greater power for detecting linear relationships (Fortin and Legendre 2010). For this analysis, we examined the effect of habitat, plumage classification, environmental variation, and geographic distance on genetic distance. For genetic distance, we calculated Cavalli-Sforza and Edwards chord distance between all 526 individuals in our microsatellite dataset using GENODIVE (V 3.0, Meirmans and Van Tienderen 2004). For our explanatory variables, we used forest type (western subalpine forest, western subalpine x Engelmann spruce forest, Engelmann spruce forest, boreal x Engelmann spruce forest, and boreal forest) as our habitat variable, the first principal component from our PCA as our environmental index measurement, morphotype (Boreal, Rocky Mountain, Pacific, and intergrade) as our plumage measurement, and Euclidean distance between geographic sampling points as our geographic distance measurement.
Next, we used correspondence analysis to examine the relationship between habitat (i.e., forest tree species), genetic variation, and plumage. Correspondence analysis is a multivariate ordination method that uses contingency tables to examine the relationships among variables (van Dam et al. 2021). We counted the number of occurrences for each genetic and phenotypic category in the presence of each forest type and examined genetic patterns for both mtDNA and microsatellite data. For the mtDNA analysis, the three categories coincided with the three mtDNA clades (Pacific, Rocky Mountain, and Boreal) present in our study. For the microsatellite analysis, we had six categories of genotypes based on our hybrid index assignment that included the same three groups plus hybrids between them (Rocky Mountain × Boreal hybrid, Rocky Mountain × Pacific hybrid, and Pacific × Boreal hybrid). For our phenotypic analysis, we had four categories (Rocky Mountain, Boreal, Pacific, and intergrade) based on the morphotype assignment of each specimen.
Given our previous hypothesis that secondary contact among Canada jay morphotypes occurred following post-Pleistocene expansion of Engelmann spruce (Graham et al. 2021), we examined the influence of Engelmann spruce on hybridization. We compared the mean hybrid index for Boreal and Pacific morphotypes between forests where Engelmann spruce was absent and present using a Mann–Whitney test, similar to the approach of Ortego et al. (2014). We conducted a similar analysis for Rocky Mountain morphotypes, whereby the two comparison groups included forests where only Engelmann spruce was found and forests where Engelmann spruce overlapped with either boreal or subalpine tree species. All analyses were conducted in Past software version 3.0. (Hammer et al. 2001).
Finally, we used a Kruskal–Wallis test to determine if the differences between the environments for each of the different phenotypic and genetic groups are significant. We examined mtDNA, microsatellite, and morphotype groups separately using the same three, six, and four categories, respectively, as described above for our first retained principal component from our PCA. All analyses were conducted in Past 3.0, and we used Bonferroni post-hoc tests to account for multiple comparisons.
Results
Phenotypic analyses
Overall, we identified 185 Pacific, 159 Rocky Mountain, 151 Boreal, and 31 intergrade morphotypes across the contact zone. Thus, of the 526 individuals examined, less than 6% were identified as phenotypic intergrades based on our assessments.
Genetic analyses
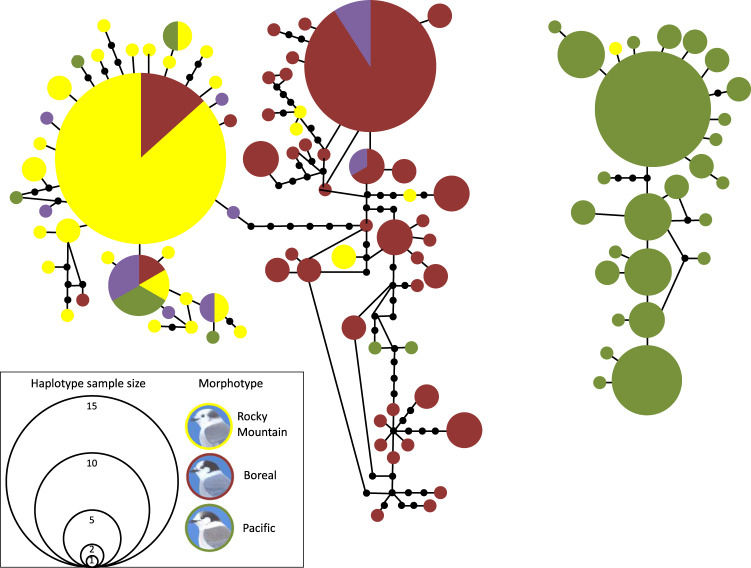
The statistical parsimony network revealed that the three morphotypes are primarily associated with a distinct mtDNA clade (Fig. 2). Intergrade morphotypes are primarily associated with the Intermountain West clade (hereafter referred to as Rocky Mountain haplotypes) previously identified in Dohms et al. (2017), although two individuals had Boreal haplotypes. Of the 89 Boreal morphotypes that we genotyped at mtDNA, 83% had Boreal haplotypes while the remaining 17% had Rocky Mountain haplotypes. Among 76 Rocky Mountain morphotypes, 93.4% of individuals had Rocky Mountain haplotypes while 5.3 and 1.3% had Boreal and Pacific haplotypes, respectively. For the 129 Pacific morphotypes, 73.6% had Pacific haplotypes while 22.5% had Rocky Mountain and 3.9% had Boreal haplotypes.
Fig. 2. Statistical parsimony network showing the relationship between the three Canada jay (Perisoreus canadensis) morphotypes and putative phenotypic hybrids (intergrades) for 190 individuals using a 506 bp fragment of the mtDNA control region.
Colors correspond with the morphotype (Boreal = red; Rocky Mountain = yellow; Pacific = green; intergrade = purple) assigned to each individual, while small black circles represent inferred haplotypes. Canada jay morphotype illustrations by Howard Coneybeare.
Nine of 242 loci by population comparisons (3.7%) showed departures from Hardy-Weinberg equilibrium following sequential Bonferroni corrections (p < 0.0002); five of these nine loci × population departures occurred in populations with known admixture between nuclear genetic clusters and mitochondrial clades (Graham et al. 2021). No locus pair comparisons showed evidence of linkage disequilibrium. Based on these patterns, we included all 12 microsatellite loci for genetic analyses.
Genetic patterns among morphotypes
The three morphotypes are genetically distinct from each other based on pairwise ΦST (mtDNA) and FST (microsatellites) comparisons (Table 1). The greatest genetic differences were between Pacific morphotypes and both Boreal and Rocky Mountain morphotypes. Genetic differences between Boreal and Rocky Mountain morphotypes are relatively smaller. Intergrade morphotypes are genetically distinct from all three morphotypes based on microsatellite markers, but are only genetically distinct from Boreal and Pacific morphotypes based on mtDNA.
Table 1.
Pairwise comparisons (microsatellite: FST/p-values below diagonal and mtDNA: θST/p-values above diagonal) for the four Canada jay (Perisoreus canadensis) morphotypes.
| Boreal morphotype | Rocky Mountain morphotype | Pacific morphotype | Intergrade morphotype | |
|---|---|---|---|---|
| Boreal morphotype | – | 0.37/ < 0.001 | 0.73/ < 0.001 | 0.34/ < 0.001 |
| Rocky Mountain morphotype | 0.02/ < 0.001 | – | 0.68/ < 0.001 | 0.00/0.86 |
| Pacific morphotype | 0.10/ < 0.001 | 0.09/ < 0.001 | – | 0.71/ < 0.001 |
| intergrade morphotype | 0.04/ < 0.001 | 0.03/ < 0.001 | 0.04/ < 0.001 | – |
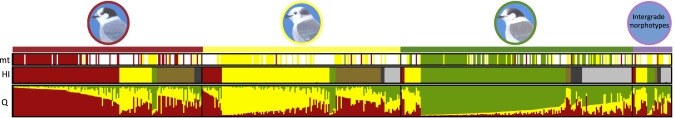
STRUCTURE at K = 3 assigned 269 individuals to one of the three genetic clusters (Fig. 3); the remaining 257 individuals were identified as admixed (individuals with Q values < 0.7 to one of the three genetic clusters). Of the 257 individuals that showed admixture for STRUCTURE, 154 individuals were identified as putative hybrids based on hybrid index scores using GENODIVE. Boreal × Rocky Mountain hybrids (N = 72) and Rocky Mountain × Pacific hybrids (N = 61) were more common than Boreal × Pacific hybrids (N = 21) across the contact zone. The remaining 103 were assigned to one of the three genetic clusters. Putative genetic hybrids were most common at the edges of the contact zone between the three morphotypes (Fig. 1). Additionally, individuals with genetic x phenotype mismatches were also found at the edges of the contact zone. Combining STRUCTURE and GENODIVE results, 56% of Boreal, 54% of Rocky Mountain and 63% of Pacific morphotypes showed a match between phenotype and genotype. The most common mismatches for phenotype and genotype were Boreal morphotypes with Rocky Mountain ancestry (N = 26), while we found relatively fewer Rocky Mountain morphs with Boreal ancestry (N = 16) or Pacific ancestry (N = 5), Pacific morphotypes with Rocky Mountain (N = 13) or Boreal (N = 3) ancestry, or Boreal morphotypes with Pacific ancestry (N = 4) (Fig. 3). Forty-two percent of intergrade morphotypes were identified as putative genetic hybrids (i.e., those individuals that both STRUCTURE and GENODIVE identified as hybrids), while 24% of Boreal, 33% of Rocky Mountain, and 29% of Pacific morphotypes were identified as putative genetic hybrids. Overall, putative Boreal × Rocky Mountain hybrids (46.7%) were more common than Pacific × Rocky Mountain hybrids (39.6%), while Boreal × Pacific hybrids (13.6%) were relatively less common.
Fig. 3. Histogram showing genetic assignment of the three Canada jay (Perisoreus canadensis) morphotypes (Boreal, Rocky Mountain, and Pacific) and putative phenotypic hybrids (referred to as intergrade morphotypes) using mtDNA (mt), GENODIVE (HI) and Structure (Q).
For microsatellite data, individuals were first assigned to a genetic cluster (red = Boreal, yellow = Rocky Mountain, Pacific = green) with STRUCTURE. Following this STRUCTURE run, individuals with admixed ancestry (i.e., those individuals with a Q < 0.70) were assigned to a genetic group or identified as a putative genetic hybrid type (olive = Boreal × Rocky Mountain, light gray = Pacific × Rocky Mountain, dark gray = Pacific × Boreal) with GENODIVE. For mtDNA the colors correspond with the three mtDNA clades (red = Boreal, yellow = Rocky Mountain, Pacific = green, white = not genotyped for mtDNA).
Plumage and genetic patterns across forest types
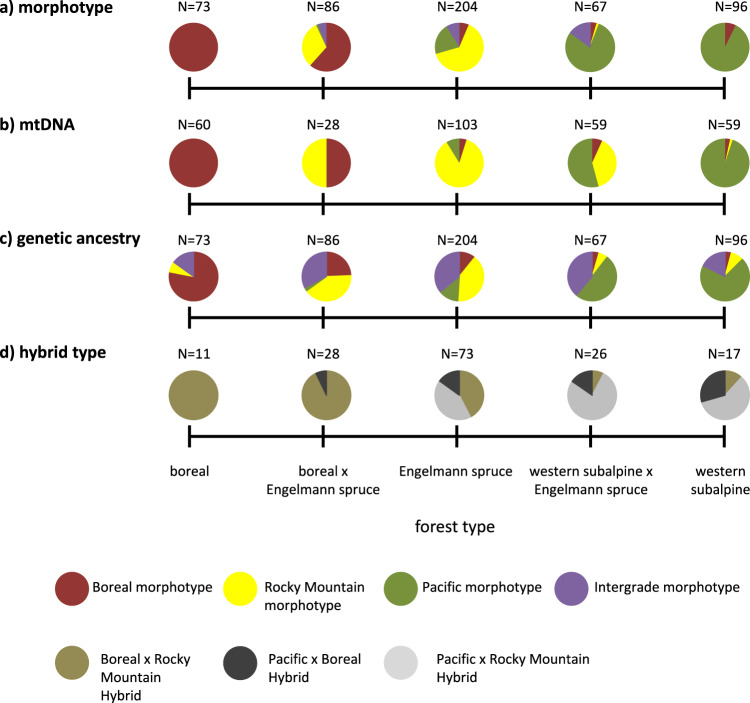
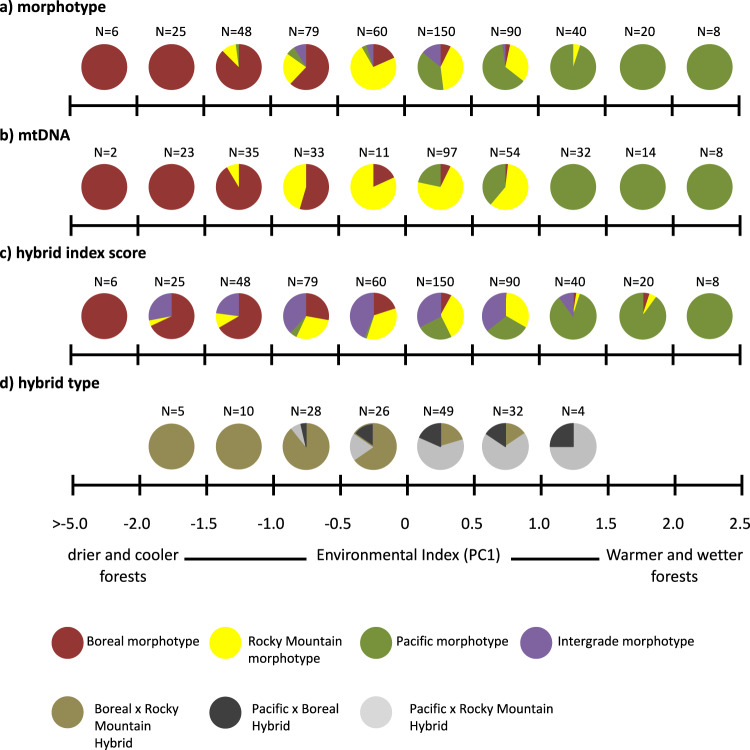
Boreal morphotypes were the only morphotype found in boreal forests and were the most frequent morphotype in Boreal × Engelmann spruce forests (Fig. 4a). Rocky Mountain morphotypes were the most frequent morphotype found in Engelmann spruce forests, while Pacific morphotypes were most frequent in western subalpine and western subalpine × Engelmann spruce forests. Finally, intergrade morphotypes were only found in forests where Engelmann spruce was present.
Fig. 4. Summary figure showing the distribution of Canada jay (Perisoreus canadensis) phenotypes and genotypes across forest types.
a Phenotypic patterns based on morphotype assignment by observers; b MtDNA clade; c Genotype assignment of microsatellite data based on hybrid index scores; d Distribution of putative hybrids across forest type (olive = Boreal × Rocky Mountain, dark gray = Pacific × Boreal, light gray = Pacific × Rocky Mountain). Colors in a-c represent Boreal (red), Rocky Mountain (yellow), Pacific (green), and intergrades (purple).
Boreal mtDNA haplotypes were detected for all of the 60 individuals genotyped from boreal forest habitats (Fig. 4b). In boreal × Engelmann spruce forests, both Boreal (14 of 28; 50%) and Rocky Mountain (14 of 28; 50%) mtDNA haplotypes were detected with equal frequency. The majority of the 103 individuals in Engelmann spruce forests had Rocky Mountain mtDNA haplotypes (86.4%), while relatively few individuals in that habitat had Pacific (8.7%) or Boreal (4.9%) mtDNA haplotypes. Pacific haplotypes (31 of 59; 52.5%) were more frequent than both Rocky Mountain (24 of 59; 40.7%) and Boreal (4 of 59; 6.8%) mtDNA haplotypes in western subalpine × Engelmann spruce forests. In the western subalpine forests, 94.9% of the 59 individuals had Pacific mtDNA haplotypes although both Boreal (3.4%) and Rocky Mountain (1.7%) mtDNA haplotypes were also found in this forest type.
Genetic ancestry (based on microsatellite genotyping) varied among forest types (Fig. 4c). Of the 73 birds genotyped from boreal forests, 57 (78%) were assigned to the Boreal genetic cluster. In boreal × Engelmann spruce forests, 36% and 24% of individuals were assigned to the Rocky Mountain and Boreal genetic clusters, respectively. Most (30%) of the remaining individuals in that habitat type were Boreal x Rocky Mountain hybrids. In Engelmann spruce forests, a greater proportion of individuals were assigned to the Rocky Mountain (40%) than the Pacific (13%) or Boreal (11%) genetic clusters. In both western subalpine × Engelmann spruce and western subalpine forests, 63% of individuals (N = 163) were assigned to the Pacific genetic cluster. The number of putative genetic hybrids was greatest in Engelmann spruce forests (N = 204; Fig. 4c, d), where both putative Pacific × Rocky Mountain, and Boreal × Rocky Mountain hybrids were found with similar frequency (N = 32 and 30, respectively). Fewer putative hybrids were found outside of these forests, with the fewest putative hybrids found in western subalpine (N = 17) and boreal (N = 11) forests.
Forest type accounted for genetic differentiation (adjusted r2 = 6.1%; p < 0.001; Table 1) based on dbRDA and partial-dbRDA models that examined microsatellite genetic distance across forest types. Forest type explained a comparable portion of the variance relative to morphotype (adjusted r2 = 6.9%; p < 0.001) and a greater portion of the variance relative to environmental variation (adjusted r2 = 4.0%; p < 0.001) or geographic distance (r2 = 3.6%; p < 0.001). When we controlled for geographic distance, forest type (adjusted r2 = 2.9%; p < 0.001) or environment (adjusted r2 = 1.6%; p < 0.001) accounted for a relatively small portion of genetic variation.
Hybridization varied across habitat type and correspondence analysis revealed strong associations between morphotype (axis 1: r = 0.65; axis 2: r = 0.43; Fig. S1), mtDNA genotype (axis 1: r = 0.73; axis 2: r = 0.53), and microsatellite genotypes (axis 1: r = 0.60; axis 2: r = 0.26) with the five forest types examined. Boreal morphotypes and genotypes showed a strong association with boreal forests across all three analyses, while Rocky Mountain morphotypes and genotypes showed a strong association with Engelmann spruce forests. By comparison, Pacific morphotypes and genotypes were closely associated with western subalpine forests. Finally, intergrade morphotypes were associated with forests where Engelmann spruce was present; a similar pattern was observed for putative genetic hybrids, although Boreal × Rocky Mountain hybrids were most strongly associated with boreal × Engelmann spruce forests. By comparison Pacific × Rocky Mountain hybrids were more closely associated with western subalpine x Engelmann spruce forests and Engelmann spruce, while Boreal × Pacific hybrids were not closely associated with any one forest type (Table 2).
Table 2.
Summary table for redundancy and partial-redundancy models examining the relationship between genetic distance with morphotype, forest type, environmental index, and geographic distance for Canada jay (Perisoreus canadensis) morphotypes.
| Variable | F | p | inertia | %r2 | %adj r2 |
|---|---|---|---|---|---|
| Morphotype | 14.09 | 0.001 | 15.45 | 7.5 | 6.9 |
| Forest type | 9.55 | 0.001 | 14.08 | 6.8 | 6.1 |
| Environmental index | 23.08 | 0.001 | 8.70 | 4.2 | 4.0 |
| Geographic distance | 20.72 | 0.001 | 7.84 | 3.8 | 3.6 |
| Forest type | geographic distance | 5.15 | 0.001 | 7.57 | 3.6 | 2.9 |
| Environmental index | geographic distance | 9.65 | 0.001 | 3.60 | 1.7 | 1.6 |
Pseudo F-score (F), p-value (p), amount of variation in the model (inertia), proportion of variation explained by the variable(s) (%r2); and proportion of variation explained by the variable(s) following adjustments for the number of predictors in the model (adj%r2) are shown. All models are significant at p < 0.05.
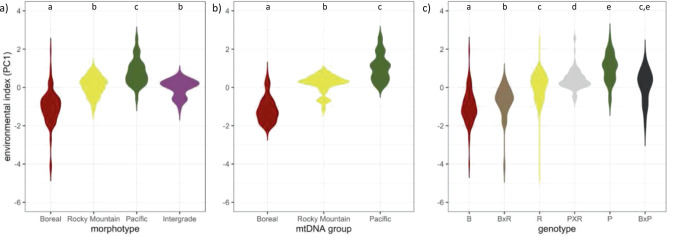
Canada jay plumage, mtDNA and nuclear ancestry, and hybridization patterns change along an ecological gradient (Fig. 5a–d). Boreal and Pacific plumage, mtDNA, and genetic groups occupy the most divergent habitats (cooler drier forests versus warmer wetter forests, respectively). The Rocky Mountain morphotype and genetic group occupies intermediate forests along this environmental gradient. Putative genetic hybrids are found across this environmental gradient but are more frequent in intermediate habitats. Across the environmental gradient, Pacific × Rocky Mountain hybrids replace Boreal × Rocky Mountain hybrids as environmental conditions become warmer and wetter, whereas Boreal × Pacific hybrids occur less frequently and are found across the environmental gradient. Overall, the three putative hybrid genotypes occupy different environments than Boreal, Rocky Mountain, and Pacific morphotypes. The environments occupied by the three morphotypes (χ2df=3 = 295.7, p < 0.001; Fig. 6a) and their corresponding mtDNA (χ2df=2 = 197.7, p < 0.001; Fig. 6b) and microsatellite genetic (χ2df=5 = 254.3, p < 0.001; Fig. 6c) groups are ecologically distinct. The three hybrid types also occupy distinct environments, although the habitats where Boreal × Pacific hybrids are found were more similar to the habitats where Rocky Mountain jays are found.
Fig. 5. Summary figure showing the distribution of Canada jay (Perisoreus canadensis) phenotypes and genotypes across an environmental gradient.
a Phenotypic patterns based on morphotype assignment by observers; b MtDNA clade distribution across forest type; c Genotype assignment of microsatellite data based on hybrid index scores; d Distribution of putative hybrids across forest type (olive = Boreal × Rocky Mountain, dark gray = Pacific × Boreal, light gray = Pacific × Rocky Mountain). Colors across a–d represent Boreal (red), Rocky Mountain (yellow), Pacific (green), and intergrade morphotypes (purple). For the mtDNA patterns, Rocky Mountain refers to the Intermountain West clade described in Dohms et al. (2017).
Fig. 6. Violin plots examining the relationship between environmental variation and phenotypic and genetic variation.
a morphotypes, b mtDNA lineages, and c microsatellite genotypes in Canada jays (Perisoreus canadensis). Plots with different letters indicate comparisons that were significantly different at p < 0.05 following least squares differences tests.
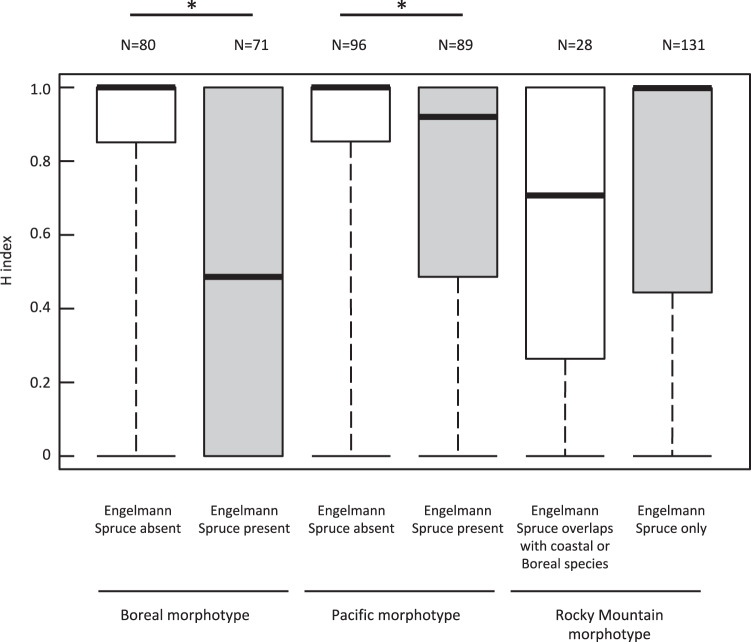
Comparisons of hybrid indices in the presence and absence of Engelmann spruce (Fig. 7) support results from the correspondence analysis (Fig. S1). Boreal (0.84 ± 0.07; mean ± 95% CI) and Pacific (0.84 ± 0.06) morphotypes had higher hybrid index scores and there were fewer hybrids in the absence of Engelmann spruce (Boreal: 0.49 ± 0.09; z = 5.8, p < 0.001; Pacific: 0.71 ± 0.07; z = 3.1, p < 0.002), and had lower hybrid index scores indicating that putative genetic hybrids are more likely to be found in areas where Engelmann spruce is found. Although Rocky Mountain morphotypes had lower hybrid index scores (0.63 ± 0.15) in areas where Engelmann spruce overlaps with western subalpine or boreal forest tree species than in areas where only Engelmann spruce was present (0.70 ± 0.06), hybrid indices were not significantly different between areas (zRocky Mountain = −1.0, p = 0.33).
Fig. 7. Box plots of hybrid index scores for Canada jay (Perisoreus canadensis) Boreal and Pacific morphotypes in the presence and absence of Engelmann spruce.
For Rocky Mountain morphotypes which are associated with Engelmann spruce, we compared hybrid indices in forests where only Engelmann spruce was found and where Engelmann spruce overlapped with coastal and boreal tree species. Dark lines represent the median and the bottom and top of box represent the 25th and 75th quartiles, respectively, while the whiskers extend to the highest and lowest values. Asterisk above paired plots show comparisons significant at p < 0.05.
Discussion
Across the Canada jay contact zone, hybridization is asymmetric and appears to be associated with habitat and geography. Hybridization occurs less frequently between Boreal and Pacific morphotypes (13.6%) than between Rocky Mountain and Boreal morphotypes (46.7%) and Pacific and Rocky Mountain (39.6%) morphotypes. Boreal and Pacific morphotypes occupy the most ecologically distinct habitats, and there is relatively little overlap in habitat as the range limits of Boreal and Pacific morphotypes are separated by Engelmann spruce habitat that is associated with the Rocky Mountain morphotype. The distribution of Engelmann spruce, however, overlaps more extensively with the ranges of western subalpine and boreal forests, thus forming areas with mosaic habitat. The greater habitat overlap appears to facilitate hybridization between Rocky Mountain and both Boreal and Pacific morphotypes. It is well established that sympatry increases the potential for hybridization (Jasso-Martínez et al. 2018; Tea et al. 2020), which explains why a greater proportion of putative hybrids had Rocky Mountain phenotypes and ancestry. Overall, the ecological differences observed among morphotypes and genetic groups across the contact zone reveal how variable environmental conditions can be within contact zones (Taylor et al. 2015).
We used two different methods (STRUCTURE and hybrid indexes) to detect hybrids. Overall STRUCTURE detected a greater proportion of hybrids than hybrid index scores did. One potential reason is that the efficiency of STRUCTURE in detecting hybrids decreases when FST between parental populations is relatively low, and few loci are used (Vaha and Primmer 2006). Notably in our study, pairwise FST comparisons were relatively low between morphotypes and we used relatively few loci, six to twelve, which may have decreased the efficiency of STRUCTURE to detect hybrids. Although the number of loci and type of loci used may affect the efficiency of these approaches to detect hybrids, our previous assessment of genetic patterns in western Canada jay populations revealed a high frequency of cytonuclear discordance (Graham et al. 2021), indicating high rates of recent and historical gene flow among morphotypes, especially in British Columbia and northern Washington (the focal areas of this study). Additionally, our analyses show high levels of admixture among intergrade morphotypes; only 24% of intergrade morphotypes had Q > 0.7 to a single cluster. Therefore, we feel confident in our ability to detect hybridization and introgression for this study system, and the high number of hybrids detected is indicative of recent and historic gene flow present in this system.
Our finding that hybridization in Canada jays increases in transitional habitats and environments outside of the climate niches occupied by the three morphotypes is not surprising given the results of other studies (Billerman et al. 2016; Wood et al. 2016; Chhatre et al. 2018). The distribution of intermediate morphotypes, mtDNA introgression, admixture and putative genetic hybrids all show congruent patterns indicating that habitat and climate influence hybridization in this species. Furthermore, the continuum of hybrids across the contact zone suggests ongoing gene flow and incomplete reproductive isolation among Canada jay morphotypes. The genetic structure and distribution of hybrids within this contact zone, where hybridization is asymmetric and dominated by one morphotype, share many characteristics with other natural and simulated syngameous systems (De La Torre et al. 2014; Ortego et al. 2014; De La Torre et al. 2015; Hamilton et al. 2015; Boecklen 2017; Chhatre et al. 2018; Haselhorst et al. 2019). For example, Engelmann and white spruce constitute one of the best-documented syngameous systems and hybridize extensively (De La Torre et al. 2015; Hamilton et al. 2015) in the same geographic area where Canada jays hybridize. Previous studies have shown that hybridization increases as spatial and temporal environmental differences between the habitats occupied by parental populations decreases (Swenson and Howard 2005; Seehausen et al. 2008; Ortego et al. 2014; Grabenstein and Taylor 2018). Thus, higher hybridization is expected in transitional areas between the habitats of the two parental populations. Our findings match this expectation because the three morphotypes live in different habitats, and hybridization among them increases in intermediate environments.
The complexity of Canada jay genetic patterns within the contact zone examined in this study reflects the complex natural history of Picea spp. Spruce species hybridize extensively in this region despite occupying distinct climatic niches and being ecologically, genetically, and morphologically distinct outside of the contact zone (De La Torre et al. 2014; De La Torre et al. 2014; De La Torre et al. 2015; Haselhorst et al. 2019). Despite these differences, hybrids form stable populations in areas with intermediate climates relative to parental species, and hybridization has been ongoing for the last 21,000 years. The fact that spruce hybridization also occurs along a climatic gradient and that the Canada jay and spruce hybrid zones overlap may explain why we see such a strong association between hybridization, habitat, and climate for Canada jays. It is also likely that some historical hybridization has occurred with Canada jay morphotypes given the high levels of mtDNA introgression and admixture within the contact zone. The congruent patterns between Canada jays and spruce are not surprising given that the genetic structure of many forest-dependent vertebrate species reflects the genetic patterns of the tree species they depend on (Arboghast and Kenagy 2001; Chavez et al. 2011; Graham and Burg 2012; Adams and Burg 2015).
The prevalence of hybridization among Canada jay morphotypes may bring into question our previous assessment that the Pacific morphotype be elevated to species status (Graham et al. 2021). Many plant and animal species remain separate species despite weak reproductive isolation and extensive hybridization in areas of secondary contact (Crow et al. 2007; Taylor et al. 2014; Chhatre et al. 2018; Natola and Burg 2018; Haselhorst et al. 2019; Grant and Grant 2020). Among avian species hybrid zones are typically narrow (130 ± 44 km; Slager et al. 2020), although a recent study discovered a broad hybrid zone (>900 km) between two North American crow species (genus: Corvus; Slager et al. 2020). Reproductive isolation remains incomplete among Canada Jay morphotypes in areas of contact, but there is minimal evidence that Pacific morphotypes hybridize outside of the contact zone (Graham et al. 2021). Further, coalescence times indicate that secondary contact among morphotypes is recent (Dohms et al. 2017), as is the case with other boreal species (following the LGM ~ 20 kya; Weir and Schluter 2004). The reduced levels of gene flow between Pacific morphotypes and other morphotypes combined with ecological, behavioral, and plumage differences (Strickland and Ouellet 2020; Graham et al. 2021) all suggest that the elevation of Pacific morphotype to species status should be considered.
We found a greater proportion of putative genetic hybrids (N = 154) than putative phenotypic hybrids (N = 31) across the contact zone. The discrepancy between genetic and phenotypic patterns is not surprising for several reasons. First, neutral genetic markers are not always strong predictors of plumage colouration as these markers often represent a small portion of the genome. Recent studies have found a strong association between regulatory regions of genes and plumage variation, and gene expression within these regions affects plumage colouration (Funk and Taylor 2019; De Zwaan et al. 2021). Previous analyses of Canada jay genetic patterns have reported the discrepancy between phenotypic and genetic patterns (Graham et al. 2021), especially for the genetically diverse Rocky Mountain morphotype, which is paraphyletic (van Els et al. 2012; Dohms et al. 2017) and exhibits high levels of cytonuclear discordance (Graham et al. 2021). Our study suggests high levels of gene flow among morphotypes and a strong association between habitat and genetic patterns. These results need to be interpreted conservatively, however, given the restricted number of markers used in this study. Given how extensive hybridization appears to be among the different morphotypes and that hybridization occurs over a broad area, future studies of this contact zone should incorporate next-generation sequencing tools. Multiple studies have shown that genomic studies using single nucleotide polymorphisms (SNP) are better able to detect hybridization patterns than microsatellites because they analyze a greater proportion of the genome (Väli et al. 2010; reviewed in Ottenburghs 2021). Further, using a greater number of markers will allow for the detection of later generation backcrosses (Ottenburghs 2021), something that we were unable to do within this study. Despite these caveats, our study represents a first attempt to examine the relationship between habitat and genetic patterns within this complex contact zone and can be used to identify important geographic areas that should be examined with a genomic dataset.
We studied the role of climate and habitat in hybridization among Canada jay morphotypes in western North America. Overall, our study provides greater insights into the complexity of genetic patterns for this species complex and adds to the growing body of studies on tri-lineage hybrid zones. Hybrid zones are naturally complex and variable, and our research emphasizes the importance of habitat and environmental conditions in hybridization. Finally, our study provides another example of a syngameon system for animals, which to date are relatively rare compared to plant systems.
Supplementary information
Habitat and climate influence hybridization among three genetically distinct Canada jay (Perisoreus canadensis) morphotypes in an avian hybrid zone complex
Acknowledgements
We thank all of the field assistants and graduate students that assisted with field sample collection, and Quinn McCallum and Fei Ying who helped with data collection for the museum samples. We thank the Burke Museum, American Museum of Natural History, Royal Saskatchewan Museum, Royal Alberta Museum, Beaty Biodiversity Museum, Royal British Columbia Museum, Royal Ontario Museum and Canadian Museum of Nature for tissue and specimen loans.
Author contributions
BAG collected samples from the field, collected data from museum skins, genotyped samples, analyzed all the data, and wrote and edited the manuscript. IS collected data from museum skins and edited the manuscript. CC collected samples from the field, collected data from museum skins, and edited the manuscript. DS collected data from museum skins and edited the manuscript. JW collected data from museum skins and edited the manuscript. HC drew the sketches of Canada Jay and tree species featured in the figures, collected data from museum skins, and edited the paper. KMD collected samples from the field, genotyped samples, and edited the manuscript. TMB collected samples from the field, collected data from museum skins, assisted with the writing of the manuscript, edited the manuscript, and was responsible for securing funding for the project.
Funding
This work was funded by NSERC Discovery and Alberta Innovates grants (TMB). All methods and procedures were approved by the University of Lethbridge Animal Welfare Committee (#1901).
Data availability
All data have been archived in the Dryad Digital Repository 10.5061/dryad.vmcvdnd05.
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor: Rui Faria
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-023-00652-3.
References
- Adams RV, Burg TM. Gene flow of a forest-dependent bird across a fragmented landscape. PLoS One. 2015;10:1–22. doi: 10.1371/journal.pone.0140938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguillon S, Rohwer V. Revisiting a classic hybrid zone: movement of the northern flicker hybrid zone in contemporary times. Evolution. 2022;76:1082–1090. doi: 10.1111/evo.14474. [DOI] [PubMed] [Google Scholar]
- Aktas C (2020) Manipulating DNA sequences and estimating unambiguous haplotype network with statistical parsimony. https://cran.r-project.org.
- Arboghast BS, Kenagy G. Phylogeography as an integrative comparative phylogeography approach to historical biogeography. J Biogeogr. 2001;28:819–825. [Google Scholar]
- Behm JE, Ives AR, Boughman JW. Breakdown in postmating isolation and the collapse of a species pair through hybridization. Am Nat. 2010;175:11–26. doi: 10.1086/648559. [DOI] [PubMed] [Google Scholar]
- Bell RC, Irian CG. Phenotypic and genetic divergence in reed frogs across a mosaic hybrid zone on São Tomé Island. Biol J Linn Soc. 2019;128:672–680. [Google Scholar]
- Bensch S, Price T, Kohn J. Isolation and characterization of microsatellite loci in a Phyloscopus warbler. Mol Ecol. 1997;6:91–92. doi: 10.1046/j.1365-294x.1997.00150.x. [DOI] [PubMed] [Google Scholar]
- Billerman SM, Cicero C, Bowie RCK, Carling MD. Phenotypic and genetic introgression across a moving woodpecker hybrid zone. Mol Ecol. 2019;28:1692–1708. doi: 10.1111/mec.15043. [DOI] [PubMed] [Google Scholar]
- Billerman SM, Murphy MA, Carling MD. Changing climate mediates sapsucker (Aves: Sphyrapicus) hybrid zone movement. Ecol Evol. 2016;6:7976–7990. doi: 10.1002/ece3.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boecklen WJ. Topology of syngameons. Ecol Evol. 2017;7:10486–10491. doi: 10.1002/ece3.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borge T, Lindroos K, Nádvorník P, Syvänen AC, Sætre GP. Amount of introgression in flycatcher hybrid zones reflects regional differences in pre and post-zygotic barriers to gene exchange. J Evol Biol. 2005;18:1416–1424. doi: 10.1111/j.1420-9101.2005.00964.x. [DOI] [PubMed] [Google Scholar]
- Brunsfeld SJ, Sullivan J, Soltis DE, Soltis PS. Comparative phylogeography of north-western North America: a synthesis. In: Silvertown J, Antonovics J, editors. Integrating Ecology and Evolution in a Spatial Context. Oxford: Blackwell Science; 2000. [Google Scholar]
- Buerkle CA. Maximum-likelihood estimation of a hybrid index based on molecular markers. Mol Ecol Notes. 2005;5:684–687. [Google Scholar]
- Busch JD, Benford R, Pearson T, Palmer E, Balda RP, Keim P. Development of polymorphic tetranucleotide microsatellites for pinyon jays (Gymnorhinus cyanocephalus) Conserv Genet. 2009;10:689–691. [Google Scholar]
- Cannon CH, Petit RJ. The oak syngameon: more than the sum of its parts. New Phytol. 2020;226:978–983. doi: 10.1111/nph.16091. [DOI] [PubMed] [Google Scholar]
- Chavez AS, Saltzberg CJ, Kenagy GJ. Genetic and phenotypic variation across a hybrid zone between ecologically divergent tree squirrels (Tamiasciurus) Mol Ecol. 2011;20:3350–3366. doi: 10.1111/j.1365-294X.2011.05184.x. [DOI] [PubMed] [Google Scholar]
- Chhatre VE, Evans LM, DiFazio SP, Keller SR. Adaptive introgression and maintenance of a trispecies hybrid complex in range-edge populations of Populus. Mol Ecol. 2018;27:4820–4838. doi: 10.1111/mec.14820. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene geaneologies. Mol Ecol. 2002;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Costa D, Sotelo G, Kaliantzopoulou A, Carvalho J, Butlin R, Hollander J, Faria R. Hybridization patterns between two marine snails, Littorina fabalis and L. obtusa. Ecol Evol. 2020;10:1058–1079. doi: 10.1002/ece3.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk QC, Suarez-Gonzalez A. The role of interspecific hybridization in adaptive potential at range margins. Mol Ecol. 2018;27:4653–4656. doi: 10.1111/mec.14927. [DOI] [PubMed] [Google Scholar]
- Crow KD, Munehara H, Kanamoto Z, Balanov A, Antonenko D, Bernardi G. Maintenance of species boundaries despite rampant hybridization between three species of reef fishes (Hexagrammidae): Implications for the role of selection. Biol J Linn Soc. 2007;91:135–147. [Google Scholar]
- Delaney KS, Wayne RK. Adaptive units for conservation: population distinction and historic extinctions in the island scrub-jay. Conserv Biol. 2005;19:523–533. [Google Scholar]
- De La Torre AR, Ingvarsson PK, Aitken SN. Genetic architecture and genomic patterns of gene flow between hybridizing species of Picea. Heredity. 2015;115:153–164. doi: 10.1038/hdy.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Roberts DR, Aitken SN. Genome-wide admixture and ecological niche modelling reveal the maintenance of species boundaries despite long history of interspecific gene flow. Mol Ecol. 2014;23:2046–2059. doi: 10.1111/mec.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Wang T, Jaquish B, Aitken SN. Adaptation and exogenous selection in a Picea glauca × Picea engelmannii hybrid zone: Implications for forest management under climate change. New Phytol. 2014;201:687–699. doi: 10.1111/nph.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zwaan DR, Mackenzie J, Mikklesen E, Wood C, Wang S (2021) Opposing dominance within a color gene block underpins hybrid plumage signal discordance. [DOI] [PMC free article] [PubMed]
- Dohms KM, Graham BA, Burg TM. Multilocus genetic analyses and spatial modeling reveal complex population structure and history in a widespread resident North American passerine (Perisoreus canadensis) Ecol Evol. 2017;7:9869–9889. doi: 10.1002/ece3.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk ER, Taylor SA. High-throughput sequencing is revealing genetic associations with avian plumage color. The Auk. 2019;136:1–7. [Google Scholar]
- Gompert Z, Mandeville EG, Buerkle CA. Analysis of population genomic data from hybrid zones. Annu Rev Ecol Evol Syst. 2017;48:207–229. [Google Scholar]
- Grabenstein KC, Taylor SA. Breaking barriers: causes, consequences, and experimental utility of human-mediated hybridization. Trends Ecol Evol. 2018;33:198–212. doi: 10.1016/j.tree.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Graham BA, Burg TM. Molecular markers provide insights into contemporary and historic gene flow for a non-migratory species. J Avian Biol. 2012;43:198–214. [Google Scholar]
- Graham BA, Cicero C, Strickland D, Woods JG, Coneybeare H, Dohms KM, et al. Cryptic genetic diversity and cytonuclear discordance characterize contact among Canada jay (Perisoreus canadensis) morphotypes in western North America. Biol J Linn Soc. 2021;132:725–740. [Google Scholar]
- Grant V (1981) The Syngameon. In: Plant Speciation. New York, NY, Columbia University Press, p 234–241
- Grant PR, Grant BR. Triad hybridization via a conduit species. Proc Natl Acad Sci USA. 2020;117:7888–7896. doi: 10.1073/pnas.2000388117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith SC, Stewart IRK, Dawson DA, Owens IPF, Burke T. Contrasting patterns of extra-pair paternity in mainland and island populations of the house sparrow (Passer domesticus): Is there an ‘island effect’? Biol J Linn Soc. 1999;68:303–316. [Google Scholar]
- Grossen C, Seneviratne SS, Croll D, Irwin DE. Strong reproductive isolation and narrow genomic tracts of differentiation among three woodpecker species in secondary contact. Mol Ecol. 2016;25:4247–4266. doi: 10.1111/mec.13751. [DOI] [PubMed] [Google Scholar]
- Gugger PF, Sugita S, Cavender-Bares J. Phylogeography of Douglas-fir based on mitochondrial and chloroplast DNA sequences: testing hypotheses from the fossil record. Mol Ecol. 2010;19:1877–1897. doi: 10.1111/j.1365-294X.2010.04622.x. [DOI] [PubMed] [Google Scholar]
- Hamilton JA, De la Torre AR, Aitken SN. Fine-scale environmental variation contributes to introgression in a three-species spruce hybrid complex. Tree Genet Genomes. 2015;11:817. [Google Scholar]
- Hammer Ø, Harper DA, Ryan PD. Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- Haselhorst MSH, Parchman TL, Buerkle CA. Genetic evidence for species cohesion, substructure and hybrids in spruce. Mol Ecol. 2019;28:2029–2045. doi: 10.1111/mec.15056. [DOI] [PubMed] [Google Scholar]
- Hewitt G. Hybrid zones-natural laboratories for evolutionary studies. Trends Ecol Evol. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- Irwin DE. Assortative mating in hybrid zones is remarkably Ineffective in promoting speciation. Evolution. 2020;195:E150–E167. doi: 10.1086/708529. [DOI] [PubMed] [Google Scholar]
- Jasso-Martínez JM, Machkour-M’Rabet S, Vila R, Rodríguez-Arnaiz R, Castañeda-Sortibrán AN. Molecular evidence of hybridization in sympatric populations of the Enantia jethys complex (Lepidoptera: Pieridae) PLoS One. 2018;13:1–23. doi: 10.1371/journal.pone.0197116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach RP, Muhlfeld CC, Boyer MC, Lowe WH, Allendorf FW, Luikart G. Dispersal and selection mediate hybridization between a native and invasive species. Proc R Soc B. 2015;282:20142454. doi: 10.1098/rspb.2014.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon EM, Lemmon AR. Reinforcement in chorus frogs: lifetime fitness estimates including intrinsic natural selection and sexual selection against hybrids. Evolution. 2010;64:1748–1761. doi: 10.1111/j.1558-5646.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- Li SH, Huang Y, Brown J. Isolation of tetranucleotide microsatellites from the Mexican jay Aphelcoma ultramarina. Mol Ecol. 1997;6:499–501. doi: 10.1046/j.1365-294x.1997.00215.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- McDonald DB, Potts WK. Cooperative display and relatedness among males in a lek-mating bird. Science. 1994;266:1030–1032. doi: 10.1126/science.7973654. [DOI] [PubMed] [Google Scholar]
- McKenzie JL, Dhillon RS, Schulte PM, McKenzie JL. Evidence for a bimodal distribution of hybrid indices in a hybrid zone with high admixture. R Soc Open Sci. 2015;2:150285. doi: 10.1098/rsos.150285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–794. [Google Scholar]
- Moran PA, Hunt J, Mitchell C, Ritchie MG, Bailey NW. Behavioural mechanisms of sexual isolation involving multiple modalities and their inheritance. J Evol Biol. 2019;32:243–258. doi: 10.1111/jeb.13408. [DOI] [PubMed] [Google Scholar]
- Natola L, Burg TM. Population genetics and speciation of yellow-bellied, red-naped, and red-breasted sapsuckers (Sphyrapicus varius, S. nuchalis, and S. ruber) J Hered. 2018;109:663–674. doi: 10.1093/jhered/esy034. [DOI] [PubMed] [Google Scholar]
- Natola L, Seneviratne S, Irwin D. Population genomics of an emergent tri-species hybrid zone. Mol Ecol. 2022;31:5356–5367. doi: 10.1111/mec.16650. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Bach L, Kotlicki P. HYBRIDALB (version 1.0): a program for generating simulated hybrids from population samples. Mol Ecol Notes. 2006;6:971–973. [Google Scholar]
- Ortego J, Gugger PF, Riordan EC, Sork VL. Influence of climatic niche suitability and geographical overlap on hybridization patterns among southern Californian oaks. J Biogeogr. 2014;41:1895–1908. [Google Scholar]
- Ottenburghs J. The genic view of hybridization in the Anthropocene. Evol Appl. 2021;14:2342–2360. doi: 10.1111/eva.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin DA, Clegg SM. The relative roles of cultural drift and acoustic adaptation in shaping syllable repertoires of island bird populations change with time since colonization. Evolution. 2015;69:368–380. doi: 10.1111/evo.12573. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrell NJ, Strickland D, Norris DR. Investigating factors that set the lower elevational limit of Canada jays (Perisoreus canadensis) on Vancouver Island, British Columbia, Canada. Can J Zool. 2022;100:64–76. [Google Scholar]
- Rice W. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Hybrid origins of plant species. Annu Rev Ecol Syst. 1997;28:359–389. [Google Scholar]
- Rousset F. GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Ruegg K, Slabbekoorn H, Clegg S, Smith TB. Divergence in mating signals correlates with ecological variation in the migratory songbird, Swainson’s thrush (Catharus ustulatus) Mol Ecol. 2006;15:3147–3156. doi: 10.1111/j.1365-294X.2006.03011.x. [DOI] [PubMed] [Google Scholar]
- Seehausen O, Takimoto G, Roy D, Jokela J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol Ecol. 2008;17:30–44. doi: 10.1111/j.1365-294X.2007.03529.x. [DOI] [PubMed] [Google Scholar]
- Seneviratne SS, Toews DPL, Brelsford A, Irwin DE. Concordance of genetic and phenotypic characters across a sapsucker hybrid zone. J Avian Biol. 2012;43:119–130. [Google Scholar]
- Slager DL, Epperley KL, Ha RR, Rowher S, Woods C, Van Hemert C, Klicka J. Cryptic and extensive hybridization between ancient lineages of American crows. Mol Ecol. 2020;29:959–969. doi: 10.1111/mec.15377. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Stenzler LM, Fitzpatrick JW. Isolation of microsatellite loci in the Florida scrub-jay Aphelcoma coerulescens. Mol Ecol Notes. 2002;2:547–550. [Google Scholar]
- Strickland DS, Ouellet H (2020) Canada jay (Perisoreus canadensis), version 1.0. In Birds of the World.
- Swenson NG, Howard DJ. Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am Nat. 2005;166:581–591. doi: 10.1086/491688. [DOI] [PubMed] [Google Scholar]
- Tarr CL, Fleisher RC. Primers for polymorphic GT microsatellites isolated from the Mariana crow, Corvus kabaryi. Mol Ecol. 1998;7:252–255. [PubMed] [Google Scholar]
- Taylor EB, Boughman JW, Groenenboom M, Sniatynski M, Schluter D, Gow JL. Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol. 2006;15:343–55. doi: 10.1111/j.1365-294X.2005.02794.x. [DOI] [PubMed] [Google Scholar]
- Taylor SA, Larson EL, Harrison RG. Hybrid zones: windows on climate change. Trends Ecol Evol. 2015;30:398–406. doi: 10.1016/j.tree.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, White TA, Hochachka WM, Ferretti V, Curry RL, Lovette I. Climate-mediated movement of an avian hybrid zone. Curr Biol. 2014;24:671–676. doi: 10.1016/j.cub.2014.01.069. [DOI] [PubMed] [Google Scholar]
- Tea YK, Hobbs JPA, Vitelli F, DiBattista JD, Ho SYW, Lo N. Angels in disguise: sympatric hybridization in the marine angelfishes is widespread and occurs between deeply divergent lineages. Proc Biol Sci. 2020;287:20201459. doi: 10.1098/rspb.2020.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews DPL, Iewin DE. Cryptic specieation in a holarctic passerine revelaed by genetic and bioacoustic analyses. Mol Ecol. 2008;17:2691–2705. doi: 10.1111/j.1365-294X.2008.03769.x. [DOI] [PubMed] [Google Scholar]
- Toews DPL, Brelsford A, Irwin DE. Hybridization between Townsend’s Dendroica townsendi and black-throated green warblers D. virens in an avian suture zone. J Avian Biol. 2011;42:434–446. [Google Scholar]
- Vaha JP, Primmer CR. Efficiency of model based Bayesian methods for detecting hybrid individuals under different hybridization scenarios with different number of loci. Mol. Ecol. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Väli U, Saag P, Dombrovsky V, Meyburg BU, Maciorowski G, Mizera T, Treinys R, Fagerberg S. Microsatellite and single nucleotide polymorphisms in avian hybrid identification: a comparative case study. J. Avian Biol. 2010;41:34–49. [Google Scholar]
- van Dam A, Dekker M, Morales-Castilla I, Rodríguez M, Wichmann D, Baudena M. Correspondence analysis, spectral clustering and graph embedding: Applications to ecology and economic complexity. Sci Rep. 2021;11:8926. doi: 10.1038/s41598-021-87971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Els P, Cicero C, Klicka J. High latitudes and high genetic diversity: Phylogeography of a widespread boreal bird, the gray jay (Perisoreus canadensis) Mol Phylogenet Evol. 2012;63:456–465. doi: 10.1016/j.ympev.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Wang S, Rohwer S, Delmore K, Irwin DE. Cross-decades stability of an avian hybrid zone. J Evol Biol. 2019;32:1242–1251. doi: 10.1111/jeb.13524. [DOI] [PubMed] [Google Scholar]
- Weir JT, Schluter D. Ice sheets promote speciation in birds. Proc Roy Soc B Biol Sci. 2004;271:1881–1885. doi: 10.1098/rspb.2004.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielstra B. Historical hybrid movement: more pervasive than appreciated. J Biogeography. 2019;46:1300–1305. [Google Scholar]
- Wood EM, Barker Swarthout SE, Hochachka WM, Larkin JL, Rohrbaugh RW, Rosenberg KV, et al. Intermediate habitat associations by hybrids may facilitate genetic introgression in a songbird. J Avian Biol. 2016;47:508–520. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Habitat and climate influence hybridization among three genetically distinct Canada jay (Perisoreus canadensis) morphotypes in an avian hybrid zone complex
Data Availability Statement
All data have been archived in the Dryad Digital Repository 10.5061/dryad.vmcvdnd05.