Abstract
The phenotypic and genotypic adaptation of a freshwater sedimentary microbial community to elevated (22 to 217 μg g [dry weight] of sediment−1) levels of polycyclic aromatic hydrocarbons (PAHs) was determined by using an integrated biomolecular approach. Central to the approach was the use of phospholipid fatty acid (PLFA) profiles to characterize the microbial community structure and nucleic acid analysis to quantify the frequency of degradative genes. The study site was the Little Scioto River, a highly impacted, channelized riverine system located in central Ohio. This study site is a unique lotic system, with all sampling stations having similar flow and sediment characteristics both upstream and downstream from the source of contamination. These characteristics allowed for the specific analysis of PAH impact on the microbial community. PAH concentrations in impacted sediments ranged from 22 to 217 μg g (dry weight) of sediment−1, while PAH concentrations in ambient sediments ranged from below detection levels to 1.5 μg g (dry weight) of sediment−1. Total microbial biomass measured by phospholipid phosphate (PLP) analysis ranged from 95 to 345 nmol of PLP g (dry weight) of sediment−1. Nucleic acid analysis showed the presence of PAH-degradative genes at all sites, although observed frequencies were typically higher at contaminated sites. Principal component analysis of PLFA profiles indicated that moderate to high PAH concentrations altered microbial community structure and that seasonal changes were comparable in magnitude to the effects of PAH pollution. These data indicate that this community responded to PAH contamination at both the phenotypic and the genotypic level.
The prevalence of organic pollution within the environment and the major role of microorganisms in its decomposition point to the need for increased understanding of how microbial communities are affected by and interact with these compounds. Knowledge derived primarily from laboratory culture-based studies needs to be expanded and tested under environmental conditions in order to further the application of microbial degradative potential in bioremediation. Recent studies into bioremediation suggest that natural attenuation or enhancement of natural intrinsic degradative potential may serve as a more cost-effective and less disruptive method for remediating organic pollution in the environment (28, 40). Fundamental research in community dynamics and microbial ecology at the biomolecular level (both genotypic and phenotypic) is needed to better understand these natural intrinsic processes of bioremediation in contaminated sites and how they may be enhanced (33). Previously, it was difficult to quantify the nature of phenotypic and genotypic changes in microbial communities, but recent developments in biomolecular analysis of microbial communities allow for their quantitative description (1, 11, 12, 29, 34, 39, 41). These analyses also provide the tools necessary for the assessment of pollutant impact on ecosystems and the means for tracking potential degradative consortia in the environment.
The Little Scioto River is a channelized riverine system located in central Ohio that was contaminated by chronic illegal discharge of creosote from a nearby wood processing plant. This site is no longer active (discharge ended in 1977) and allows for the comprehensive study of in situ microbial communities experiencing polycyclic aromatic hydrocarbon (PAH) stress. Creosote is a complex mixture made up of ∼200 different compounds that are classified into three broad groups: PAHs, phenolics, and nitrogen-, oxygen-, and sulfur-containing aromatic compounds. PAHs are major constituents of creosote (∼85% by weight) and are common environmental pollutants due to their wide use in wood preservatives (27, 31). They constitute a class of hazardous organic chemicals that pose potential health risks to many forms of life due to their toxic, carcinogenic, and mutagenic effects. These compounds are introduced into aquatic environments from a multitude of sources, with the sediments serving as the major repository of deposition (7).
Microorganisms capable of degrading PAHs are commonly isolated from contaminated soils and sediments (17, 18, 42), but their role within microbial communities is not well known. Similarly, the catabolic genes (many times plasmid borne) are commonly isolated from these environments (21, 35), but again their distribution within microbial communities is little understood. In this study, we examined the phenotypic and genotypic responses of the Little Scioto River sedimentary community to PAH contamination. To do so, we determined PAH concentration, PAH-degradative potential, microbial community structure, and degradative gene frequency in sediments from both ambient and PAH-contaminated sediments.
MATERIALS AND METHODS
Study site.
Six study stations were established on the Little Scioto River proximal to recognizable geographic features or state and county roads. Sampling stations were designated as either ambient (≤1.56 μg g [dry weight] of sediment−1) or contaminated, based on PAH concentration in the sediments. Stations were labeled from north to south in the direction of river flow as stations A to F. Stations A and B are located upstream from the source of contamination and are designated ambient stations; stations C to F were established downstream from the source of contamination and serve as PAH-contaminated stations. This study location is unique and serves as a natural extension of culture-based laboratory studies because at all study stations the river is 100% pool and glide with no fast current or riffles; sediments are predominantly muck, silt, and sand with detrital material (sticks, branches, and leaves); and tree cover is moderate to sparse along both banks (30). The station similarities are the result of the river being channelized by the Army Corps of Engineers in the early 1900s. The Ohio American Water Company measured temperature, pH, and water depth daily near station B. Water temperature ranged from 5 to 23°C seasonally; pH remained relatively constant at 7.1; and water depth, dependent on season, ranged from 1 to 2 m except during episodic floods.
Sampling scheme.
Sediments from stations A and E were sampled in December 1994. In April 1995 stations A to E were sampled, and in July 1995 all six stations were sampled. Not all stations were sampled at all times because we added stations based on PAH concentrations measured during previous samplings. Sediments were collected by using 10-cm-diameter push cores and were sampled in triplicate. The 0- to 1-cm horizon of each core was subsampled with a 5-ml syringe with the cannula end removed. Sediments for total microbial biomass, phospholipid fatty acid (PLFA) profiles, and PAH analysis were preserved on site by placing them in a modified dichloromethane-methanol-phosphate buffer extraction mixture (4, 10, 13). On July 1995 the remaining top 0 to 5 cm of the sediment cores was used to determine 14C-radiolabeled PAH metabolism and the degradative gene frequency of the sedimentary microbial community. These samples were placed on ice and transported to the lab for further processing.
Metabolism of 14C-radiolabeled substrates.
Biodegradation potential was measured on the basis of 14CO2 evolution from 14C-labeled naphthalene and anthracene (34). Two-gram subsamples from samples of the top 5 cm of sediment were slurried with 1 ml of sterile deionized H2O in 40-ml vials (Pierce, Rockford, Ill.). An 8-ml vial containing 0.5 ml of a 0.5 N NaOH solution was inserted into each 40-ml vial to serve as a 14CO2 trap. Either 160,000 cpm of [1-14C]naphthalene (specific activity, 8.9 mCi/mmol; Sigma, St. Louis, Mo.) or 110,000 cpm of [UL-14C]anthracene (specific activity, 15.0 mCi/mmol; Sigma) was added to each experimental vial. The mineralization vials were grouped into triplicates, with two vials serving as experimental samples and one vial serving as an abiotic degradation control. The abiotic control was treated with 0.5 ml of 2 N H2SO4 at the beginning of the incubation. All vials were sealed with Teflon-lined septa and screw caps and incubated at 25°C and 100 rpm on a rotary shaker. Naphthalene incubations were terminated at 0, 2, 6, 12, and 24 h by the addition of 0.5 ml of 2 N H2SO4. Anthracene incubations were terminated at 0, 24, 48, 96, and 168 h. After addition of the acid, the samples were allowed to shake at least 1 h prior to removal of the NaOH trap solution, which was added to 10 ml of Ready Safe scintillation cocktail (Beckman, Fullerton, Calif.) and 1 ml of sterile distilled H2O. The amount of 14CO2 evolved during the assays was determined by liquid scintillation counting.
PAH, total microbial biomass, and PLFA analyses.
Sediments were extracted in the dichloromethane-methanol-buffer mixture for at least 48 h, the extraction mixture was partitioned into aqueous and organic fractions through the addition of dichloromethane and water, and the organic fraction (which contains the lipids and lipophilic pollutants) was recovered. The organic fraction was subsampled for total microbial biomass. Total microbial biomass was determined by the phospholipid phosphate (PLP) method (13). The remaining lipid was fractionated by differential elution from silicic acid columns, using SPE technology and a vacuum manifold (10). The PAH fraction was recovered in hexane, and the phospholipid fraction was recovered in methanol. Individual PAHs were quantified by gas chromatography (GC)-mass spectroscopy (MS) (Hewlett-Packard 5890 Series II Plus interfaced with a Hewlett-Packard 5972 Mass Selective Detector) with individual response factors generated from known quantitative standards. A deuterium-labeled external standard (phenanthrene-d10) was added to each sample at the time of extraction for determination of PAH recovery efficiency. PAHs were identified by relative retention times, coelution with known standards, and comparison of their mass spectra with published spectra and/or spectra of known standards (10). Microbial community structure was determined by PLFA analysis according to the procedures of White (43) as modified by Findlay (11), with functional groups assigned as specified by Findlay et al. (14). PLFAs were analyzed as fatty acid methyl esters (FAMEs). FAMEs were quantified by GC, with identifications based on relative retention times, coelution with known standards, and MS analysis. PLFA analysis provides information on microbial community structure by relating the complex mixture of FAMEs back to the organisms present in the samples (41).
DNA extraction from sediments, nucleic acid hybridization and quantification, and probe preparation.
DNA was extracted from 10- to 18-g samples of sediment according to the method of Ogram et al. (29), including in situ cell lysis by ballistic disintegration in a Bead-Beater (BioSpec, Bartlesville, Okla.) (24, 39). Nucleic acid extracts were hybridized with the 32P-radiolabeled catabolic gene probes alkB (alkane hydroxylase), nahA (naphthalene dioxygenase), nahH (2,3-catechol dioxygenase), and todC1/C2 (toluene dioxygenase) as previously described (20, 36, 37, 39). The two non-PAH gene probes were used because of the high potential for introduction of nonaromatic and monoaromatic compounds into the Little Scioto River (25, 44). A Universal 16S ribosomal DNA (rDNA) oligonucleotide probe located at position 1392 on the Escherichia coli map (38) was used to determine total microbial abundance. After chloroform-phenol extraction, portions of DNA from all samples were vacuum blotted onto nylon membranes (0.2-μm mesh size; ICN Biomedical, Costa Mesa, Calif.) by using a slot blot apparatus (Bio-Rad, Hercules, Calif.), and the nucleic acids were heat fixed. The slot blots were prehybridized for at least 1 h at 65°C (8, 39), then the 32P-radiolabeled gene probe of interest was added, and the mixture was allowed to hybridize overnight. Single-stranded catabolic gene probes were generated by asymmetric PCR with the addition of [α-32P]dCTP (ICN Biomedical) in place of the unlabeled dCTP present in the PCR kit (Perkin-Elmer, Foster City, Calif.) (39). PCR-generated probes were purified by using NucTrap push columns (Stratagene, La Jolla, Calif.). Probes with a specific activity of at least 105 cpm/μl, as determined by scintillation counting of 5 μl of purified probe in 10 ml of Beckman Ready Safe scintillation cocktail, were added to the slot blots. The 32P-radiolabeled 16S rDNA oligonucleotide probe was end labeled by using a 5′-terminus labeling kit (Boehringer Mannheim, Indianapolis, Ind.). The membranes were hybridized at 42°C in hybridization buffer and washed, and the concentration of bound probe was determined by autoradiography and digital imaging (39). Cell numbers were determined by using the algorithms of Applegate et al. (1), modified for use with the universal 16S rDNA probe (39).
Statistics.
Weight percent fatty acid data for use in principal component analysis were log transformed [ln (x + 1)] prior to analysis. Statistical analyses were performed with Systat for Macintosh version 5.2.1. Analysis of variance of the frequency of gene occurrence indicated significant heteroscedasticity (i.e., the error variance was not constant in all cases), which was not removed by transformation of the data. Subsequent statistical analyses were performed on grouped data (ambient versus contaminated), using a Kruskal-Wallis one-way analysis of variance (nonparametric).
RESULTS
PAHs.
PAH concentrations in ambient Little Scioto River sediments were variable over the sampling period and ranged from below detection (<10 pg g [dry weight] of sediment−1) to 1.56 μg g (dry weight) of sediment−1 (Table 1). In contrast, PAH concentrations at the contaminated stations ranged from 22 to 217 μg g (dry weight) of sediment−1. Fourteen individual PAHs, ranging in size from 2-bromonaphthalene to benzo[g,h,i]perylene, were identified from the sediments (Table 2). The identified compounds represented 63.5% of the total PAH recovered from the sediments. Neither naphthalene nor acenaphthalene was detected, indicating the aged nature of the contamination. The high standard deviations observed for the PAH concentrations are likely the result of heterogeneous distribution of PAHs in the sediments, again due to the aged nature of this study site, as the techniques used have been demonstrated to be both accurate and precise (10).
TABLE 1.
PAH concentration and biodegradation potential (percent mineralization) of Little Scioto River sediments
| Site | Mean PAH concn (μg g [dry wt] of sediment−1) ± SD (n = 3)a
|
Mean % mineralization ± SD (n = 2)
|
|||
|---|---|---|---|---|---|
| December | April | July | Naphthalene (48 h) | Anthracene (168 h) | |
| A | 1.56 ± 0.41 | 0.24 ± 0.41 | — | 6.01 ± 5.95 | 2.54 ± 1.29 |
| B | NS | 0.43 ± 0.74 | — | NDb | ND |
| C | NS | 105.51 ± 70.12 | 29.46 ± 7.02 | ND | ND |
| D | NS | 215.36 ± 79.46 | 175.17 ± 67.75 | ND | ND |
| E | 154.88 ± 65.86 | 216.83 ± 36.01 | 100.04 ± 81.12 | 8.31 ± 7.19 | 10.02 ± 9.02 |
| F | NS | NS | 21.82 ± 9.74 | ND | ND |
NS, not sampled; —, below detection limit.
ND, not determined.
TABLE 2.
Little Scioto River PAHs
| Retention time (min) | Compound | Mean PAH concna (μg g [dry wt] of sediment−1) ± SD (n = 3) |
|---|---|---|
| 6.52 | Naphthalene | ND |
| 9.59 | Acenaphthalene | ND |
| 9.95 | 2-Bromonaphthalene | 2.21 ± 2.62 |
| 10.04 | Acenaphthene | 0.68 ± 0.30 |
| 11.61 | Fluorene | 2.31 ± 1.91 |
| 14.73 | Phenanthrene | 13.35 ± 15.21 |
| 14.97 | Anthracene | 11.59 ± 11.51 |
| 19.11 | Fluoranthene | 8.85 ± 5.90 |
| 19.96 | Pyrene | 20.18 ± 17.76 |
| 24.57 | Benzo[a]anthracene | 8.99 ± 6.51 |
| 24.74 | Chrysene | 13.55 ± 8.74 |
| 28.45 | Benzo[a]fluoranthene | 8.85 ± 4.26 |
| 29.50 | Benzo[a]pyrene | 3.13 ± 1.49 |
| 33.00 | Idenol[1,2,3-cd]pyrene | 2.48 ± 1.76 |
| 33.11 | Dibenzo[a,h]anthracene | 1.26 ± 0.21 |
| 33.82 | Benzo[g,h,i]perylene | 2.59 ± 1.05 |
| Total identified | 100.04 ± 67.35 | |
| Total unidentifiedb | 36.01 ± 24.24 | |
| % Identified | 63.54 |
Station E, July 1995. ND, not detected.
Assuming GC-MS response equal to closest retention time for identified PAHs.
Metabolism of 14C-radiolabeled naphthalene and anthracene.
Mineralization experiments with radiolabeled PAHs using ambient (station A) and contaminated (station E) sediments indicated that mineralization occurred at both stations in the Little Scioto River (Table 1). Microbial degradation of naphthalene was observed 6 h after inoculation of sediments from the station E. Initiation of mineralization in station A sediments was not observed until 12 h. After the onset of mineralization, the values for percent naphthalene mineralized per hour were similar for sediments from both stations. Microbial degradation of anthracene was first observed 24 h after inoculation of sediments from station E. The initiation of anthracene mineralization in the station A sediments was not observed until 96 h. After 168 h, station E sediments had mineralized 10% of the added anthracene whereas ambient sediments had mineralized only 2.5%.
Total microbial biomass.
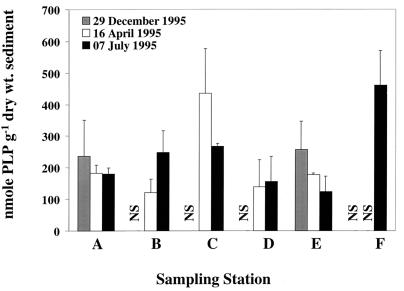
The ranges of total microbial biomass measured as PLP were similar at the two ambient stations and four contaminated stations. At the ambient stations (A and B), biomass ranged from 120 to 247 nmol of PLP g (dry weight) of sediment−1, and the contaminated stations (C to F) had biomass measurements that ranged from 109 to 460 nmol of PLP g (dry weight) of sediment−1 (Fig. 1). Stations with intermediate PAH concentration (stations C and F) had the highest total microbial biomass (435 and 460 nmol of PLP g [dry weight] of sediment−1, respectively). Using the conversion factor of 50 nmol of PLP to 2.0 × 109 cells (3, 9), estimated cell numbers ranged from 4.8 × 109 to 1.84 × 1010 cells g (dry weight)−1.
FIG. 1.
Total microbial biomass for Little Scioto River sediments. Bars represent means (n = 3), and error bars represent plus 1 standard deviation. NS, not sampled.
Sedimentary microbial community structure.
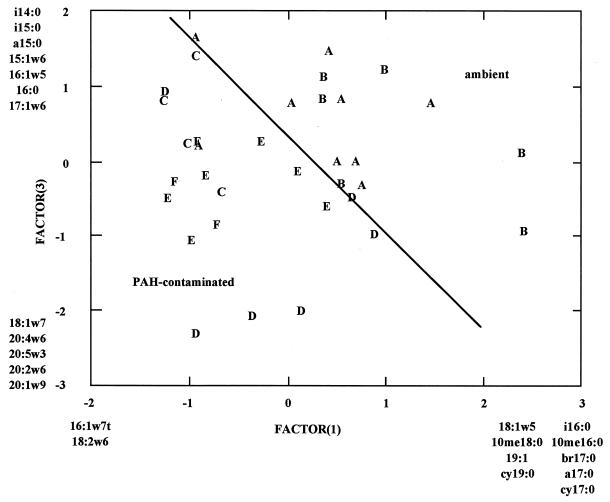
Principal component analysis of PLFA profiles revealed three significant components of variation within the data set describing community structure. The data matrix used in the analysis was composed of the weight percent of 39 PLFAs from 34 replicate samples. Principal components 1, 2, and 3 accounted for 18, 21, and 16%, respectively, of the variance within the data set. The combination of principal components 1 and 3 mapped the majority of samples (14 of 15 [93%]) from ambient stations into one group and the majority of samples (18 of 19 [95%]) from contaminated stations into another group (Fig. 2). The pattern was somewhat complex in that samples from ambient stations showed positive eigenvalues for principal component 1, principal component 3, or both, while samples from contaminated stations showed negative eigenvalues for principal component 1, principal component 3, or both. Samples with positive eigenvalues for principal component 1 were enriched in fatty acids i16:0, 10me16:0, br17:0, a17:0, cy17:0, 18:1ω5, 10me18:0, 19:1, and cy19:0, while samples with negative eigenvalues were enriched in fatty acids 16:1ω7t and 18:2ω6. Fatty acids i16:0, 10me16:0, br17:0, a17:0, cy17:0, 18:1ω5, 10me18:0, 19:1, and cy19:0 are indicative of bacteria, in particular gram-positive and anaerobic gram-negative bacteria. The fatty acid 18:2ω6 has a wide phylogenetic distribution but is more common in eukaryotic organisms, and the fatty acid 16:1ω7t is commonly associated with metabolic stress in aerobic gram-negative bacteria. Samples with positive eigenvalues for principal component 3 were enriched in fatty acids i14:0, i15:0, a15:0, 15:1ω6, 16:1ω5, 16:0, and 17:1ω6, while samples with negative eigenvalues were enriched in fatty acids 18:1ω7, 20:4ω6, 20:5ω3, 20:2ω6, and 20:1ω9. Fatty acids i14:0, i15:0, a15:0, 15:1ω6, i16:0, 16:1ω5, 16:0, and 17:1ω6 are indicative of Bacillus-type organisms (i14:0, i15:0, a15:0, and i16:0) or anaerobic gram-negative bacteria, while fatty acids 20:4ω6, 20:5ω3, 20:2ω6, and 20:1ω9 indicate the presence of heterotrophic microeukaryotes. The fatty acid 18:1ω7 is indicative of aerobic gram-negative bacteria.
FIG. 2.
Principal component analysis of the Little Scioto River sediment PLFA profiles, factor 1 by factor 3. Letters indicate individual samples and represent sampling stations (A to F). Fatty acids listed at the extremes of the principal components are disproportionately abundant in samples that map to those extremes.
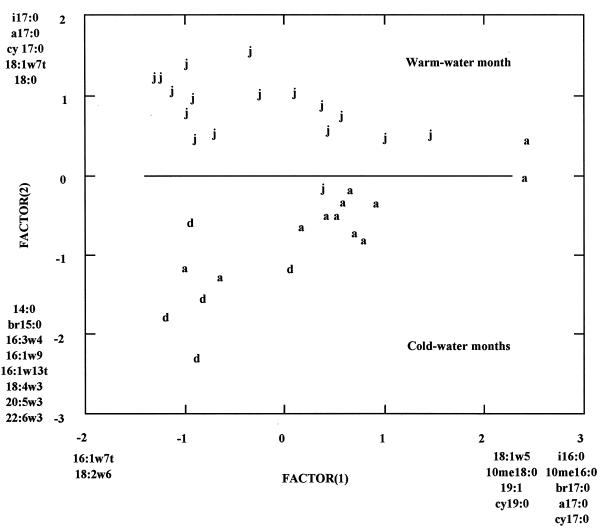
Graphic analysis indicated that principal component 2 mapped the samples into two distinct groups. These two groups were the majority of samples (16 of 17 [94%]) taken during July 1995 and the majority of samples (16 of 17 [94%]) taken during December 1994 and April 1995 (Fig. 3). Fatty acids i17:0, a17:0, cy17:0, 18:1ω7t, and 18:0 were disproportionately more abundant in samples taken during July 1995, while 14:0, br15:0, 16:3ω4, 16:1ω9, 16:1ω13t, 18:4ω3, 20:5ω3, and 22:6ω3 were disproportionately more abundant in December 1994 and April 1995. Fatty acids i17:0, a17:0, cy17:0, 18:1ω7t, and 18:0 are associated with bacteria; i17:0, a17:0, and cy17:0 are indicative of anaerobic bacteria, and 18:1ω7t is indicative of aerobic, gram-negative bacteria under metabolic stress (41). In contrast, 16:3ω4, 16:1ω9, 16:1ω13t, 18:4ω3, 20:5ω3, and 22:6ω3 are considered marker fatty acids for eukaryotic phototrophs (12). These changes in PLFA profiles indicate that phototrophic eukaryotes were more abundant and anaerobic bacteria were less abundant in all sediments during December and April, while the converse was true of all sediments during July.
FIG. 3.
Principal component analysis of the Little Scioto River sediment PLFA profiles, factor 1 by factor 2. Letters indicate individual samples and represent sampling dates (d, December; a, April; j, July). Fatty acids listed at the extremes of the principal components are disproportionately abundant in samples that map to those extremes.
Sedimentary microbial community molecular analysis.
Total microbial abundance, measured with a Universal 16S rDNA oligonucleotide probe, ranged from 0.73 × 109 to 3.03 × 109 cells g (dry weight) of sediment−1. Nucleic acid-degradative gene analysis indicated that degradative gene sequences (nahA, nahH, todC1/C2, and alkB) were present in the microbial community at all stations on the Little Scioto River with the exception that nahH was not detected at station E. Because of the limited sampling for nucleic acid analysis, these data are summarized here. The frequency of bacterial populations containing the alkB gene sequences ranged from 11 to 13% in sediments from the ambient stations and 8 to 66% in sediments from the contaminated stations; todC1/C2 gene sequences ranged from 10 to 18% in sediments from the ambient stations and 2 to 56% in sediments from the contaminated stations; nahH gene sequences ranged from 1 to 3% in sediments from the ambient stations and 0 to 22% in sediments from the contaminated stations; and nahA gene sequences ranged from 6 to 9% in sediments from the ambient stations and 5 to 37% in sediments from the contaminated stations. Nonparametric statistical analysis of the frequency of occurrence of degradative genes indicated that nahA and alkB gene sequences occurred with significantly greater frequency (P = 0.038) in contaminated sediments than in ambient sediments.
DISCUSSION
Prior to our initial characterization of the sedimentary microbial community in the Little Scioto River, we predicted three possible responses of the microbial community to increased concentrations of PAHs. We hypothesized that (i) the PAH-degradative potential would increase, (ii) the microbial community structure would be altered, and (iii) the frequency of degradative gene sequences in the community would increase. Our findings allow an initial assessment of each of these predictions.
Mineralization data from the ambient station A and the contaminated station E site indicated that the capacity to biodegrade low-molecular-weight PAHs was present in both ambient and contaminated sediments. The percent naphthalene mineralized initially was greater in the contaminated sediments but within 48 h became statistically similar for sediments from both stations. In contrast, the percent anthracene mineralized was greater for contaminated sediments and remained so throughout the incubation period. The solubility of individual PAHs and the ability of microorganisms to degrade them is inversely proportional to molecular weight and the number of benzene rings contained (2, 7). This relationship and the age of the spill likely explain the absence of naphthalene and high concentrations of anthracene in the sediments (Table 2). Madsen et al. (26) found that mineralization activity was greater in urban subsurface sediments chronically exposed to PAHs than in previously unexposed sediments. The similarity in mineralization of naphthalene by ambient and PAH-contaminated sediments is likely due to the absence of chronic exposure to naphthalene. The increased mineralization of anthracene in PAH-contaminated sediments is likely due to their persistence in the environment.
As expected, chronic exposure to PAHs altered microbial community structure, but quite interestingly, principal component analysis indicated that seasonal variations explained a similar percentage of the variation in community structure (Fig. 2 and 3). Sediments from cold-water months showed a consistent pattern of increased relative abundance of phototrophic eukaryotes. In contrast, samples taken during periods of warmer water temperatures showed increased relative abundance of anaerobic prokaryotes. Findlay and Watling (15) observed similar shifts in sedimentary microbial community structure at a pristine shallow-water marine site. Findlay et al. (16) have also explored the relative strength of natural versus anthropogenic causes in inducing changes in microbial community structure by comparing the patterns of seasonal change in pristine and organically enriched sites. Sites experiencing a two- to threefold increase in benthic carbon flux exhibited a marked change in community structure (an increase in the abundance of Beggiatoa-type organisms), yet seasonal variation was the major component of variation in community structure and followed a pattern similar to that observed in pristine sediments and by us in Little Scioto River sediments. The finding that seasonal changes in community structure are a major component of variation in three surface sediment systems (pristine, organically enriched, and PAH contaminated) strengthens the case that this variation should be considered in attempts to use changes in microbial community structure to detect anthropogenic stresses and when one is evaluating natural or intrinsic bioremediation.
PAH-contaminated sediments were enriched in two groups of fatty acids: those associated with heterotrophic eukaryotes (18:2ω6, 20:4ω6, 20:5ω3, 20:2ω6, and 20:1ω9) and some associated with aerobic, gram-negative bacteria (16:1ω7t and 18:1ω7c), while ambient sediments were enriched in fatty acids associated with either Bacillus-type organisms, gram-positive bacteria, or anaerobic gram-negative bacteria. The increase in aerobic, gram-negative bacteria in PAH-contaminated sediments is not unexpected, as it is well known that low-molecular-weight PAHs are rapidly degraded under aerobic conditions (2, 7). The increase in heterotrophic microeukaryotes suggests that these organisms are also responding to the contamination, which in turn suggests the development of a food web in which degradative bacteria serve as a food source for predatory microeukaryotes. Ghiorse et al. (19) and Carman et al. (6) have documented development of such food webs in PAH-contaminated groundwater and estuarine sediments, respectively.
Microbial biomass levels were similar at all stations, with the exception of increased biomass at station C on 16 April 1995 and station F on 7 July 1995. The similarity in biomass abundance indicates that the observed changes in community structure reflect replacement of organisms rather than the addition of novel organisms to the community. Both estimates of abundance (16S rDNA and total phospholipid) showed general agreement, although estimates derived from PLP analysis were generally fourfold greater. This is likely due to the differences in sampling technique—PLP was determined on the top 1 cm of sediment, while DNA was extracted from the top 5 cm of sediment.
Nucleic acid hybridization indicated that hydrocarbon-degradative gene sequences were present at all stations, and the genes nahA and alkB occurred at greater frequencies in PAH-contaminated sediments. The occurrence of degradative gene sequences in ambient sediments is not wholly unexpected, considering the episodic presence of trace amounts of PAH in these sediments. The presence of this biodegradative potential suggests that only trace levels of PAH are necessary to maintain degradative sequences within the microbial community at some baseline percentage. The proportion of the community containing degradative gene sequences increased approximately 200% in response to PAH contamination. The PAH-degradative genes assayed for in this study are commonly isolated from plasmids (20, 24, 44). In general, catabolic plasmids are widespread in nature, have been shown to increase in communities in response to pollutant stress, and are known to transfer between members of microbial communities (5, 22, 35). The catabolic plasmids containing the degradative genes probed for in this study have been found exclusively (to the best of our knowledge) in gram-negative bacteria, in particular Pseudomonas spp. (20, 23, 32, 35–37). PLFA analysis indicated that aerobic, gram-negative bacteria (the component of the bacterial community that responded positively to PAH contamination) increased in biomass some 15 to 30%. The increased frequency of degradative genes and increased biomass of gram-negative bacteria in PAH-contaminated sediments suggest that gram-negative bacteria containing catabolic plasmids are responsible, at least in part, for the increased degradative potential observed in these sediments. The difference in magnitude of the changes in gene frequency compared to biomass suggests that the community has the capacity to respond to PAH contamination either genotypically, by transferring degradative gene sequences between members of the community, or through replacement of nondegradative populations.
ACKNOWLEDGMENTS
This study was funded in part by the Sigma Xi National Research Society and the Graduate School and Department of Microbiology at Miami University.
We thank Jim Smoot and Stacy Wilson for sampling assistance.
REFERENCES
- 1.Applegate B M, Matrabutham U, Sanserivino J, Sayler G S. Biodegradation genes as marker genes in microbial ecosystems. In: Akkermans A D L, Elsas J D, Bruijn F J, editors. Molecular microbial ecology manual. Norwell, Mass: Kluwer Academic Publishers; 1996. pp. 6.1.8–6.1.14. [Google Scholar]
- 2.Atlas R M. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol Rev. 1981;45:180–209. doi: 10.1128/mr.45.1.180-209.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balkwill D L, Leach F R, Wilson J T, McNabb J F, White D C. Equivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate, direct counts in subsurface aquifer sediments. Microbiol Ecol. 1989;16:73–84. doi: 10.1007/BF02097406. [DOI] [PubMed] [Google Scholar]
- 4.Bligh E G, Dyer W J. A rapid method of lipid extraction and purification. Can J Biochem Physiol. 1959;35:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 5.Burton N F, Day M J, Bull A T. Distribution of plasmids in clean and polluted sites in a South Wales river. Appl Environ Microbiol. 1982;44:1026–1029. doi: 10.1128/aem.44.5.1026-1029.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman K R, Fleeger J W, Means J C, Pomarico S M, McMillin D J. Experimental investigation of the effects of polynuclear aromatic hydrocarbons on an estuarine sediment food web. Mar Environ Res. 1995;40:289–318. [Google Scholar]
- 7.Cerniglia C E. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- 8.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbs F C, Findlay R H. Analysis of microbial lipids to determine biomass and to detect the response of sedimentary microorganisms to disturbance. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbiology. Boca Raton, Fla: Lewis Publications; 1993. pp. 347–358. [Google Scholar]
- 10.Fang J, Findlay R H. The use of a classic lipid extraction method for simultaneous recovery of organic pollutants and microbial lipids from sediments. J Microbiol Methods. 1996;27:63–71. [Google Scholar]
- 11.Findlay R H. The use of phospholipid fatty acids to determine microbial community structure. In: Akkermans A D L, Elsas J D, Bruijn F J, editors. Molecular microbial ecology manual. Norwell, Mass: Kluwer Academic Publishers; 1996. pp. 4.1.4/1–4.1.4/17. [Google Scholar]
- 12.Findlay R H, Dobbs F C. Quantitative description of microbial communities using lipid analysis. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbiology. Boca Raton, Fla: Lewis Publications; 1993. pp. 271–284. [Google Scholar]
- 13.Findlay R H, King G M, Watling L. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl Environ Microbiol. 1989;55:2888–2893. doi: 10.1128/aem.55.11.2888-2893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Findlay R H, Trexler M B, Guckert J B, White D C. Response of a benthic microbial community to biotic disturbance. Mar Ecol Prog Ser. 1990;61:135–148. [Google Scholar]
- 15.Findlay R H, Watling L. Seasonal variation in the structure of a marine benthic community. Microb Ecol. 1988;36:23–30. doi: 10.1007/s002489900089. [DOI] [PubMed] [Google Scholar]
- 16.Findlay R H, Watling L, Mayer L M. Environmental impact of salmon net-pen culture on marine benthic communities in Maine: a case study. Estuaries. 1995;18:145–179. [Google Scholar]
- 17.Foght J M, Fedorak P M, Westlake D W S. Mineralization of 14C-hexadecane and 14C-phenanthrene in crude oil: specificity among bacterial isolates. Can J Microbiol. 1989;36:169–175. doi: 10.1139/m90-030. [DOI] [PubMed] [Google Scholar]
- 18.Geiselbrecht A D, Herwig R P, Deming J W, Staley J T. Enumeration and phylogenetic analysis of polycyclic aromatic hydrocarbon-degrading marine bacteria from Puget Sound sediments. Appl Environ Microbiol. 1996;62:3344–3349. doi: 10.1128/aem.62.9.3344-3349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghiorse W C, Herrick J B, Sandoli R L, Madsen E L. Natural selection of PAH-degrading bacterial guilds at coal-tar disposal sites. Environ Health Perspect. 1995;103:107–111. doi: 10.1289/ehp.95103s4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosal W C, Balkwill D L. Nucleotide sequence and expression of gene nahH of plasmid NAH7 and homology with gene xylE of TOL pWWO. Gene. 1987;55:19–28. doi: 10.1016/0378-1119(87)90244-7. [DOI] [PubMed] [Google Scholar]
- 21.Gunsalus I C, Yen K M. Pathogenicity and ecology of bacterial plasmids. In: Levy S B, editor. Molecular biology. New York, N.Y: Plenum Press; 1981. pp. 499–509. [Google Scholar]
- 22.Hada H S, Sizemore R K. Incidence of plasmids in marine Vibrio spp. isolated from an oil field in the northwestern Gulf of Mexico. Appl Environ Microbiol. 1981;41:199–202. doi: 10.1128/aem.41.1.199-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrick J B, Stuart Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston W H, Stapleton R D, Sayler G S. Direct extraction of microbial DNA from soils and sediments. In: Akkermans A D L, Elsas J D, Bruijn F J, editors. Molecular microbial ecology manual. Norwell, Mass: Kluwer Academic Publishers; 1996. pp. 1.3.2/1–1.3.2/9. [Google Scholar]
- 25.Kok M, Oldenhuis R, van der Linden M P G, Raatjes P, Kingma J, van Lelyveld P H, Witholt B. The Pseudomonas oleovorans alkane hydroxylase gene, sequence and expression. J Biol Chem. 1989;264:5435–5441. [PubMed] [Google Scholar]
- 26.Madsen E L, Winding A, Malachowsky K, Thomas C T, Ghiose W C. Contrasts between subsurface microbial communities and their metabolic adaptation to polycyclic aromatic hydrocarbons at a forested and urban coal-tar disposal site. Microb Ecol. 1992;24:199–213. doi: 10.1007/BF00174455. [DOI] [PubMed] [Google Scholar]
- 27.Mueller J G, Chapman P J, Pritchard P H. Creosote-contaminated sites. Environ Sci Technol. 1989;23:1197–1201. [Google Scholar]
- 28.National Research Council. In situ bioremediation: when does it work? Washington, D.C: National Academy Press; 1993. [Google Scholar]
- 29.Ogram A, Sayler G S, Barklay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 30.Ohio Environmental Protection Agency. Bottom sediment evaluation of the Little Scioto River. Columbus, Ohio: Division of Water Quality Planning and Assessment; 1992. [Google Scholar]
- 31.Priddle M W, MacQuarrie K T B. Dissolution of creosote in groundwater: an experimental and modeling investigation. J Contam Hydrol. 1994;15:27–56. [Google Scholar]
- 32.Resnick S M, Lee K, Gibson D T. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Indust Microbiol Biotechnol. 1996;17:5–6. [Google Scholar]
- 33.Rittmann B E. In situ bioremediation. Park Ridge, N.J: Noyes Publications; 1994. [Google Scholar]
- 34.Sanseverino J, Werner C, Flemming J, Applegate B, King J M H, Sayler G S. Molecular diagnostics of polycyclic aromatic hydrocarbon biodegradation in manufactured gas plant soils. Biodegradation. 1993;4:303–321. doi: 10.1007/BF00695976. [DOI] [PubMed] [Google Scholar]
- 35.Sayler G S, Hooper S W, Layton A C, King J M H. Catabolic plasmids of environmental and ecological significance. Microb Ecol. 1990;19:1–20. doi: 10.1007/BF02015050. [DOI] [PubMed] [Google Scholar]
- 36.Simon M J, Osslaund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 37.Singer R T, Finnerty W R. Genetics of PAH-degrading microorganisms. In: Atlas R M, editor. Petroleum microbiology. New York, N.Y: Macmillan Publishing; 1984. pp. 299–354. [Google Scholar]
- 38.Stahl D A, Flescher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stapleton R D, Ripp S, Jimenez L, Cheol-Koh S, Flemming J T, Gregory I R, Sayler G S. Nucleic acid analytical approaches in bioremediation. Site assessment and characterization. J Microbiol Methods. 1998;32:165–178. [Google Scholar]
- 40.U.S. Environmental Protection Agency. Understanding bioremediation: a guidebook for citizens. EPA/540/2-91/002. U.S. Washington, D.C: Environmental Protection Agency; 1991. [Google Scholar]
- 41.Vestal J R, White D C. Lipid analysis in microbial ecology. BioScience. 1989;39:535–541. [PubMed] [Google Scholar]
- 42.West P A, Okpokwasili G C, Brayton P R, Grimes D J, Colwell R R. Numerical taxonomy of phenanthrene-degrading bacteria isolated from the Chesapeake Bay. Appl Environ Microbiol. 1984;48:988–993. doi: 10.1128/aem.48.5.988-993.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White D C. Analysis of microorganisms in terms of quantity and activity in natural environments. In: Slater J H, Whittenbury R, Wimpenny J W T, editors. Microbes in their natural environments. Cambridge, England: Cambridge University Press; 1983. pp. 1–30. [Google Scholar]
- 44.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]