Abstract
Simple Summary
Theileria orientalis Ikeda poses significant challenges to the cattle industry, encompassing economic repercussions, trade-related issues, and constraints regarding control and treatment measures. This parasite can precipitate a range of adverse effects in cattle, including diminished milk production, weight loss, anemia, and, in severe instances, cattle fatalities. These adverse impacts directly translate into economic burdens for farmers and the broader cattle industry. The financial costs associated with treating afflicted animals and implementing control strategies can be substantial. Additionally, occurrences of Theileria orientalis Ikeda in regions where it is not endemic can lead to trade restrictions and impediments affecting both cattle and cattle products. This can curtail the international movement of cattle and influence global trade in livestock. Furthermore, there is a dearth of effective treatment options for infected cattle, and severe cases frequently culminate in fatalities. This lack of viable treatment exacerbates the difficulties in disease management. Addressing the presence of Theileria orientalis Ikeda in the USA necessitates comprehensive efforts to understand the prevalence and distribution of the pathogen. This study utilizing PCR followed by DNA sequencing identified two cases of Theileria orientalis Ikeda-positive cattle, marking the first report of this virulent genotype in Alabama, USA.
Abstract
Theileria orientalis Ikeda genotype, a parasite causing a disease in cattle that leads to significant economic challenges in Asia, New Zealand, and Australia, has been identified in seven U.S. States since 2017. Two previously validated PCR tests for Theileria followed by DNA sequencing were performed to test blood samples collected from 219 cattle in Alabama, USA, during the period of 2022–2023. Bidirectional Sanger sequencing revealed that the MPSP gene sequences (639–660 bp) from two cattle in Lee and Mobile Counties of Alabama exhibited a 100% match with those of recognized T. orientalis Ikeda strains, and showed similarities ranging from 76% to 88% with ten other T. orientalis genotypes. A high copy number of T. orientalis Ikeda was detected in the blood of infected cattle (ALP-1: 1.7 × 105 and 1.3 × 106/mL whole blood, six months apart; ALP-2: 7.1 × 106/mL whole blood). Although the confirmed competent vector for T. orientalis Ikeda, Haemaphysalis longicornis tick, has not yet been identified in Alabama, the persistent nature of T. orientalis Ikeda infection and the detection of a high pathogen burden in seemingly healthy cattle in this study suggest that other tick species, as well as shared needles and dehorning procedures, could facilitate pathogen transmission within the herd. Continued investigations are necessary for the surveillance of T. orientalis Ikeda and Haemaphysalis longicornis ticks in Alabama and other U.S. states, along with assessing the pathogenicity of T. orientalis Ikeda infections in cattle.
Keywords: Theileria orientalis Ikeda, cattle, Alabama, Haemaphysalis longicornis tick
1. Background
Theileria orientalis is a hemoprotozoan parasite belonging to the phylum Apicomplexa [1]. The infection of cattle with this parasite can lead to various adverse effects, including anemia, weakness, reduced milk production, diminished weight gain, abortion, and occasionally, death [2,3,4,5,6]. Researchers have identified eleven genotypes of T. orientalis (Sivakumar et al., 2014), with the most frequently encountered genotypes being Buffeli, Chitose, and Ikeda [1,6,7,8]. While T. orientalis Buffeli is typically clinically benign, Chitose and Ikeda tend to be more virulent, resulting in increased morbidity and mortality in affected cattle [2,8].
Although sporadic and limited, outbreaks of T. orientalis Buffeli were reported in the United States (U.S.) before 2017 [9,10,11]. Recently, T. orientalis Ikeda has become endemic in Virginia [5,6,11,12], and has also been detected in seven U.S. states to date [12].
To assess the T. orientalis status in cattle, sheep, and goats in Alabama, USA, we conducted PCR assays followed by DNA sequencing analysis on existing convenience samples of whole blood and blood samples obtained from farms in Alabama.
2. Materials and Methods
2.1. Whole Blood Samples
Samples for analysis came from two sources. Whole-blood samples in EDTA from cattle (n = 72) presenting to the Auburn University Large Animal Teaching hospital between September 2022 and August 2023 for regular physical examination and the diagnosis and treatment of diseases where whole blood was submitted to the Clinical Pathology Laboratory were used in the study. Aliquots of the whole-blood samples were used for routine complete blood counts and biochemical profiles. Additionally, 147 bovine blood samples from three cattle farms associated with Auburn University College of Veterinary Medicine located in Marion County (n = 50), Lee County (n = 33), and Tallapoosa County (n = 64) of Alabama were also used to screen Theileria in this study. The protocol for the collection of bovine samples was reviewed and approved by the Institutional Animal Care and Use Committee of Auburn University (IACUC #2022-5112).
Blood samples were sent to Auburn at ambient temperature. The High-Pure PCR Template Preparation Kit (Roche Molecular Biochemicals, Indianapolis, IN, USA) was used to extract DNA from 400 µL aliquots as published [13,14]. The extracted DNA was eluted in 200 µL elution buffer, and was preserved at −80 °C until PCR was performed in this study.
2.2. Detection of Theileria and Anaplasma DNA by PCRs
Two previously validated quantitative PCRs, an MPSP gene-based Theileria TaqMan PCR [15] and a FRET-PCR targeting the rRNA of Theileria [14], were used to detect Theileria DNA in this study. The primers and probes as well as the primers to sequence the MPSP gene are shown in Table 1.
Table 1.
Primers and probes used in this study.
| PCR and the Target Gene | Oligonucleotides | References |
|---|---|---|
| TaqMan-PCR; MPSP gene of all T. orientalis genotypes | MPSP-F: 5′-GCAAACAAGGATTTGCACGC-3′ | Bogema et al., 2015 [15] |
| MPSP-R: 5′-TGTGAGACTCAATGCGCCTAGA | ||
| Pr-U: 5′-FAM-TCGACAAGTTCTCACCAC-MGB-NFQ-3′ | ||
| FRET-PCR; rRNA for all Theileria spp. | F: 5′-TAGTGACAAGAAATAACAATACGGGGCTT-3′ | Yang et al., 2014 [14] |
| R: 5′- CAGCAGAAATTCAACTACGAGCTTTTTAACT-3′ | ||
| Pr-1: 5′-CCAATTGATACTCTGGAAGAGGTTT-(6-FAM)-3′ | ||
| Pr-2: 5′-(LCRed640)-AATTCCCATCATTCCAATTACAAGAC-phosphate-3′ | ||
| Sequencing primers for Ikeda genotype | RTF-I: ATTGGTAGACGGAAAATGGAAGAAGG | Bogema et al., 2015 [15] |
| RTR-I: GAGACTCAATGCGCCTAGAGATAATAGA |
All PCR reactions were performed on a Roche Light Cycler 480 II Thermocycler as described [15]. In brief, 10 µL of the extracted DNA was added to a 10 µL reaction mixture containing 5× PCR FRET buffer, 400 µM dNTP (Roche Diagnostics GmbH, Mannheim, Germany), 0.34 units of Platinum Taq DNA Polymerase (Invitrogen, Boston, MA, USA), 1 µM of each forward and reverse primer (Integrated DNA Technologies, Coralville, IA, USA). Thermal cycling consisted of 18 high-stringency step-down cycles followed by 25 relaxed-stringency fluorescence acquisition cycles. The 18 high-stringency step-down thermal cycles were 6 × 15 s @ 95 °C, 60 s @ 75 °C; 9 × 15 s @ 95 °C, 60 s @ 73 °C; and 3 × 15 s @ 95 °C, 30 s @ 71 °C, 30 s @ 72 °C; this was followed by the relaxed-stringency fluorescence acquisition cycling of 25 × 15 s @ 95 °C, 8 s @ 58 °C; this is was followed by fluorescence acquisition, 30 s @ 65 °C, and 30 s @ 72 °C.
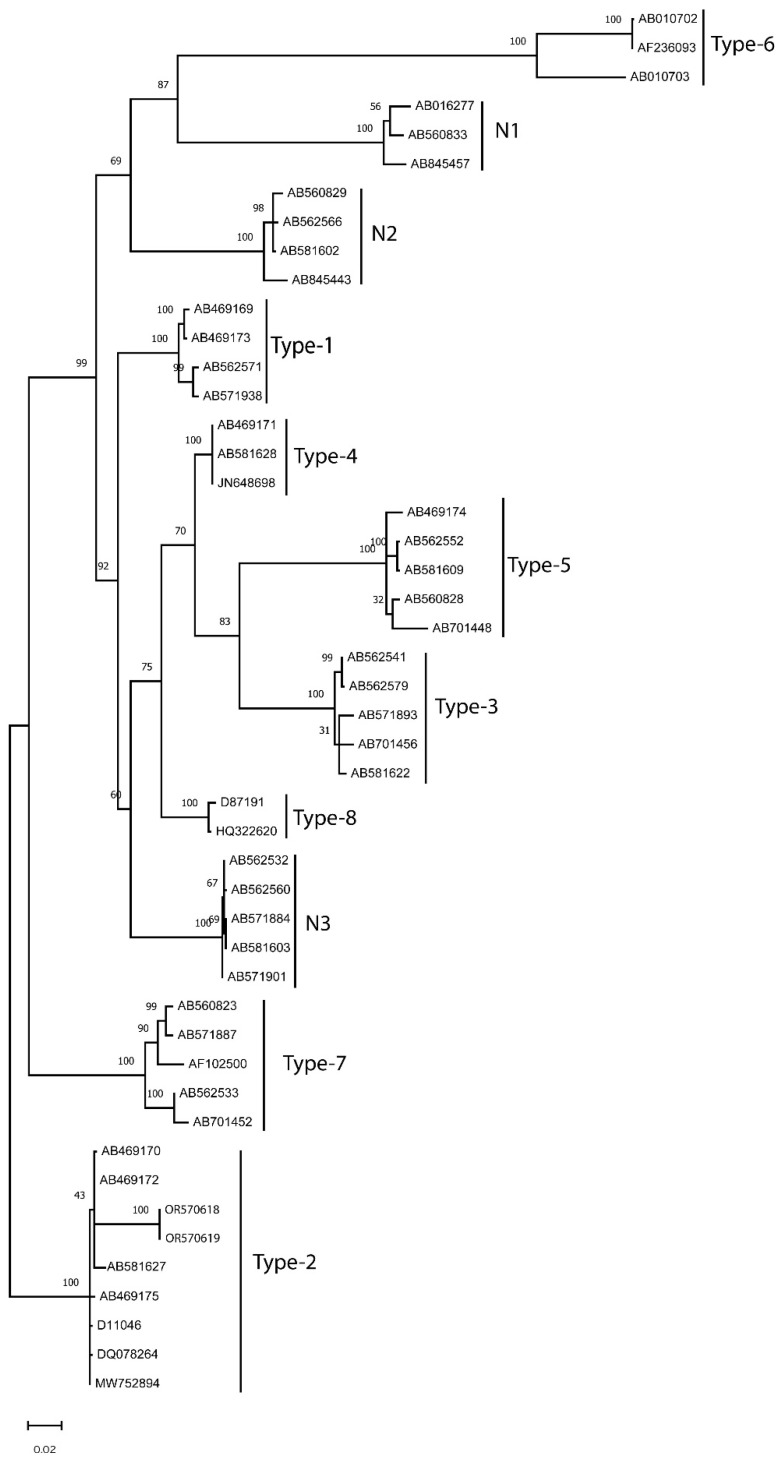
The products of Theileria-positive PCRs were sent to ELIM Biopharmaceuticals (Hayward, CA, USA) for Bidirectional Sanger sequencing. The nucleotide sequences were submitted to NCBI to obtain GenBank Accession numbers (OR570618, OR570619), and a phylogenetic tree was generated to compare the nucleotide sequences of Theileria identified in this study with those of all recognized 11 T. orientalis genotypes (Figure 1).
Figure 1.
Phylogenetic tree displaying major piroplasm surface protein (MPSP) gene sequences for Theileria orientalis. The 739–760-bp nucleotide sequences of the MPSP gene, representing 11 genotypes of T. orientalis, were concatenated and aligned using CLUSTALW. A maximum likelihood phylogenetic tree was generated employing a p-distance model. Bootstrap values are expressed as percentages based on 1000 replications, and the bar represents evolutionary distances as 0.05 changes per nucleotide position. The genotypes of T. orientalis, along with their GenBank accession numbers, are provided. Notably, the sequences of T. orientalis Ikeda identified in this study (ALP-1: OR570618; ALP-2: OR570619) exhibited 100% identity with those of recognized T. orientalis Ikeda strains, while demonstrating 76–88% similarity with other T. orientalis genotypes.
The Theileria-positive samples identified in this study were submitted to the Molecular diagnostic laboratory at Auburn University College of Veterinary Medicine for the detection of Anaplasma DNA following the published protocol [16].
3. Results
Both the MPSP-based TaqMan PCR and rRNA-targeting FRET-PCR, followed by DNA sequencing, successfully identified T. orientalis DNA in 2 out of 89 convenience blood samples. However, none of the 147 bovine blood samples from three different farms tested positive for T. orientalis. Notably, the two T. orientalis-positive samples were found to be free of Anaplasma spp.
The MPSP serves as an antigenic marker for Theileria spp. and is widely employed for genotyping the 11 distinct T. orientalis groups [1,17]. The 739–760 bp nucleotide sequences of the MPSP gene, representing these 11 T. orientalis genotypes, were concatenated and aligned using CLUSTALW. Subsequently, a maximum likelihood phylogenetic tree was constructed. The sequences from T. orientalis identified in this study (ALP-1, ALP-2) displayed a 100% match with those from recognized T. orientalis Ikeda strains (Figure 1). Furthermore, they exhibited similarities ranging from 76% to 88% with other T. orientalis genotypes (Type 1: 86–87%; Type-3: 82–83%; Type-4: 87%; Type-5: 80–81%; Type-6: 76%; Type 7: 88%; Type-8: 86%; N1: 79%; N2: 83–84%; N3: 86–87%) (Figure 1). Although the internal segment of the small subunit gene was not amplified in the Theileria-positive samples in this study, the phylogenetic trees, based on the long MPSP fragment, unequivocally confirmed the presence of T. orientalis Ikeda DNA in two cattle from Alabama.
In Lee County, Alabama, a one-year-old female heifer (ALP-1 with an accession number OR570618) was found to carry T. orientalis Ikeda DNA. This heifer exhibited normal red blood cell (RBC) and hemoglobin (HGB) counts but displayed elevated monocyte counts (2.3/µL) and white blood cell counts (2.1 × 103/µL). Additionally, increased levels of glucose (313 mg/dL) were observed, alongside low iron levels (25 µg/dL). This heifer was being treated for an acute free gas bloat resulting from grain overload. She recovered from this episode of ruminal acidosis following treatment and has been a healthy member of the resident herd since discharge. The initial test revealed the presence of 1.7 × 105 T. orientalis Ikeda/mL in whole blood, and a follow-up test six months later showed the increased count of 1.3 × 106/mL in whole blood.
A four-month-old male calf from Mobile County, Alabama, presented with fever, swollen lymph nodes, anorexia, pyrexia, cough, dyspnea, lethargy, tachypnea, polyuria, and diarrhea. Bloodwork, including chemistry and a complete blood count, revealed no significant abnormalities except for decreased fibrinogen levels, indicating recovery from a previous inflammatory event. A urinalysis was also performed, revealing dilute urine but no signs of infection or inflammation of the bladder. This calf tested positive for T. orientalis Ikeda (ALP-2 with an accession number OR570619), with a copy number of 7.1 × 106/mL in whole blood.
4. Discussion
T. orientalis Ikeda is a protozoan parasite of cattle that has significant implications for the livestock industry. In this study, we reported for the first time the identification of this virulent genotype Ikeda in the cattle from Alabama, USA. Persistent infection with different variants of T. orientalis is a common occurrence [7,18], and the immune mechanisms responsible for disease resistance are not yet fully understood. In this study, a follow-up test conducted six months later revealed an increased count of 1.3 × 106/mL of T. orientalis Ikeda in the whole blood of one animal, compared to the initial count of 1.7 × 105 T. orientalis Ikeda/mL whole blood, with both cattle appearing to be in relatively good health, and no evidence of anemia or fever being observed. This supports the notion that T. orientalis can lead to chronic and persistent infections, emphasizing the need for further research into the pathogenicity of T. orientalis Ikeda infections in ruminants.
An experimental transmission trial conducted by the USDA’s Agricultural Research Service (ARS) in collaboration with the Virginia Tech Animal Laboratory Services (ViTALS) laboratory has confirmed the vector competence of the Haemaphysalis longicornis tick (ALHT: Asian long-horn tick) for T. orientalis Ikeda [17]. ALHT can feed on a wide range of wildlife and domestic species, including birds, white-tailed deer, companion animals, livestock, equines, and humans. As of September 2023, ALHT has been identified in 18 states [19]. Once introduced into suitable habitats, ALHT populations can proliferate rapidly, partly due to the exhibition of multiple genetic types, including both parthenogenetic and bisexual reproduction [20,21].
Although ALHT has not yet been identified in Alabama, given the persistent nature of T. orientalis Ikeda infection and the detection of a high burden of T. orientalis Ikeda in apparently healthy cattle in this study, it is plausible that other tick species, as well as shared needles and dehorning procedures, could facilitate the spread of the pathogen within the herd. Telionis et al. reported an 8.7% (172/1980) prevalence of genotype Ikeda in Virginia Market Cattle, 2018–2020 [6]. It is likely that T. orientalis Ikeda will be identified in many other states and become established throughout the USA.
5. Conclusions
In conclusion, PCR tests followed by DNA sequencing confirmed the presence of T. orientalis Ikeda DNA in the blood of two cattle in Alabama, USA. Continued research in epidemiology, vector ecology and vaccine development will contribute to a better understanding of Theileria orientalis Ikeda and help develop more effective strategies for its control and prevention, benefiting both the cattle industry and animal health.
Author Contributions
Conceptualization, C.W. and P.H.W.; methodology, N.I., S.B., S.F., C.A., J.W.S. and A.M.; formal analysis, N.I., S.B. and C.W.; writing—original draft preparation, C.W.; writing—review and editing, C.W., N.I., S.B. and P.H.W.; All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care and Use Committee of Auburn University (IACUC #2022-5112 approved on 25 October 2022).
Informed Consent Statement
Informed consent was waived as this study utilized convenient and anonymous diagnostic samples.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was supported, in part, by the Molecular Diagnostic Laboratory at Department of Pathobiology Auburn University.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sivakumar T., Hayashida K., Sugimoto C., Yokoyama N. Evolution and genetic diversity of Theileria. Infect. Genet. Evol. 2014;27:250–263. doi: 10.1016/j.meegid.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Watts J.G., Playford M.C., Hickey K.L. Theileria orientalis: A review. N. Z. Vet. J. 2016;64:3–9. doi: 10.1080/00480169.2015.1064792. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence K.E., Forsyth S.F., Vaatstra B.L., McFadden A., Pulford D.J., Govindaraju K., Pomroy W.E. Clinical haematology and biochemistry profiles of cattle naturally infected with Theileria orientalis Ikeda type in New Zealand. N. Z. Vet. J. 2018;66:21–29. doi: 10.1080/00480169.2017.1391142. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence K.E., Gedye K., Pomroy W.E. A longitudinal study of the effect of Theileria orientalis Ikeda type infection on three New Zealand dairy farms naturally infected at pasture. Vet. Parasitol. 2019;276:108977. doi: 10.1016/j.vetpar.2019.108977. [DOI] [PubMed] [Google Scholar]
- 5.Oakes V.J., Todd S.M., Carbonello A.A., Michalak P., Lahmers K.K. Coinfection of cattle in Virginia with Theileria orientalis Ikeda genotype and Anaplasma marginale. J. Vet. Diagn. Investig. 2022;34:36–41. doi: 10.1177/10406387211057627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telionis A., Lahmers K., Todd M., Carbonello A., Broaddus C.C., Bissett C.J., Hungerford L.L. Distribution of Theileria orientalis in Virginia Market Cattle, 2018–2020. Pathogens. 2022;11:1353. doi: 10.3390/pathogens11111353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eamens G.J., Bailey G., Jenkins C., Gonsalves J.R. Significance of Theileria orientalis types in individual affected beef herds in New South Wales based on clinical, smear and PCR findings. Vet. Parasitol. 2013;196:96–105. doi: 10.1016/j.vetpar.2012.12.059. [DOI] [PubMed] [Google Scholar]
- 8.Eamens G.J., Gonsalves J.R., Jenkins C., Collins D., Bailey G. Theileria orientalis MPSP types in Australian cattle herds associated with outbreaks of clinical disease and their association with clinical pathology findings. Vet. Parasitol. 2013;191:209–217. doi: 10.1016/j.vetpar.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Uilenberg G., Perié N.M., Spanjer A.A., Franssen F.F. Theileria orientalis, a cosmopolitan blood parasite of cattle: Demonstration of the schizont stage. Res. Vet. Sci. 1985;38:352–360. doi: 10.1016/S0034-5288(18)31808-3. [DOI] [PubMed] [Google Scholar]
- 10.Stockham S.L., Kjemtrup A.M., Conrad P.A., Schmidt D.A., Scott M.A., Robinson T.W., Tyler J.W., Johnson G.C., Carson C.A., Cuddihee P. Theileriosis in a Missouri beef herd caused by Theileria buffeli: Case report, herd investigation, ultrastructure, phylogenetic analysis, and experimental transmission. Vet. Pathol. 2000;37:11–21. doi: 10.1354/vp.37-1-11. [DOI] [PubMed] [Google Scholar]
- 11.Thompson A.T., White S., Shaw D., Egizi A., Lahmers K., Ruder M.G., Yabsley M.J. Theileria orientalis Ikeda in host-seeking Haemaphysalis longicornis in Virginia, U.S.A. Ticks Tick Borne Dis. 2020;11:101450. doi: 10.1016/j.ttbdis.2020.101450. [DOI] [PubMed] [Google Scholar]
- 12.Lokting B. A New Tick-Borne Disease Is Killing Cattle in the US. MIT Technology Review 2022. [(accessed on 15 September 2023)]. Available online: https://www.technologyreview.com/2022/11/17/1063352/new-tick-borne-disease-killing-cattle-in-us/
- 13.Barua S., Newbolt C.H., Ditchkoff S.S., Johnson C., Zohdy S., Smith R., Wang C. Absence of SARS-CoV-2 in a captive white-tailed deer population in Alabama, USA. Emerg. Microbes Infect. 2022;11:1707–1710. doi: 10.1080/22221751.2022.2090282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Mao Y., Kelly P., Yang Z., Luan L., Zhang J., Li J., El-Mahallawy H.S., Wang C. A pan-Theileria FRET-qPCR survey for Theileria spp. in ruminants from nine provinces of China. Parasites Vectors. 2014;7:413. doi: 10.1186/1756-3305-7-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogema D.R., Deutscher A.T., Fell S., Collins D., Eamens G.J., Jenkins C. Development and validation of a quantitative PCR assay using multiplexed hydrolysis probes for detection and quantification of Theileria orientalis isolates and differentiation of clinically relevant subtypes. J. Clin. Microbiol. 2015;53:941–950. doi: 10.1128/JCM.03387-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu H., Kelly P.J., Zhang J., Luo Q., Yang Y., Mao Y., Yang Z., Li J., Wu H., Wang C. Molecular detection of Anaplasma spp. and Ehrlichia spp. in ruminants from twelve provinces of China. Can. J. Infect. Dis. Med. Microbiol. 2016;2016:9183861. doi: 10.1155/2016/9183861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinkel K.D., Herndon D.R., Noh S.M., Lahmers K.K., Todd S.M., Ueti M.W., Scoles G.A., Mason K.L., Fry L.M. A U.S. isolate of Theileria orientalis, Ikeda genotype, is transmitted to cattle by the invasive Asian longhorned tick, Haemaphysalis longicornis. Parasites Vectors. 2021;14:157. doi: 10.1186/s13071-021-04659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yam J., Bogema D.R., Jenkins C. Oriental theileriosis. Ticks Tick Borne Pathog. 2019:1–31. doi: 10.5772/intechopen.81198. [DOI] [Google Scholar]
- 19.USDA National Haemaphysalis longicornis (Asian Longhorned Tick) Situation Report. [(accessed on 15 September 2023)];2023 Available online: https://www.aphis.usda.gov/animal_health/animal_diseases/tick/downloads/longhorned-tick-sitrep.pdf.
- 20.Hutcheson H.J., Dergousoff S.J., Lindsay L.R. Haemaphysalis longicornis: A tick of considerable veterinary importance, now established in North America. Can. Vet. J. 2019;6:27–28. doi: 10.17269/s41997-018-0152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heath A.C.G. Biology, ecology and distribution of the tick, Haemaphysalis longicornis Neumann (Acari: Ix odidae) in New Zealand. N. Z. Vet. J. 2016;64:10–20. doi: 10.1080/00480169.2015.1035769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.