Abstract
Bacterial aggregates from a chemolithoautotrophic, nitrifying fluidized bed reactor were investigated with microsensors and rRNA-based molecular techniques. The microprofiles of O2, NH4+, NO2−, and NO3− demonstrated the occurrence of complete nitrification in the outer 125 μm of the aggregates. The ammonia oxidizers were identified as members of the Nitrosospira group by fluorescence in situ hybridization (FISH). No ammonia- or nitrite-oxidizing bacteria of the genus Nitrosomonas or Nitrobacter, respectively, could be detected by FISH. To identify the nitrite oxidizers, a 16S ribosomal DNA clone library was constructed and screened by denaturing gradient gel electrophoresis and selected clones were sequenced. The organisms represented by these sequences formed two phylogenetically distinct clusters affiliated with the nitrite oxidizer Nitrospira moscoviensis. 16S rRNA-targeted oligonucleotide probes were designed for in situ detection of these organisms. FISH analysis showed that the dominant populations of Nitrospira spp. and Nitrosospira spp. formed separate, dense clusters which were in contact with each other and occurred throughout the aggregate. A second, smaller, morphologically and genetically different population of Nitrospira spp. was restricted to the outer nitrifying zones.
Lithoautotrophic nitrification, the sequential transformation of NH4+ via NO2− to NO3−, is typically catalyzed by two phylogenetically distinct groups of bacteria, i.e., the ammonia-oxidizing bacteria and the nitrite-oxidizing bacteria. All characterized freshwater ammonia oxidizers belong to a coherent group within the β subclass of the class Proteobacteria, comprising the genera Nitrosomonas (formerly Nitrosococcus mobilis and Nitrosomonas), and Nitrosospira (formerly Nitrosospira, Nitrosovibrio, and Nitrosolobus) (14, 30). For a long time, all nitrite oxidizers isolated from freshwater habitats belonged to the genus Nitrobacter within the α subclass of Proteobacteria (7). Recently, Nitrospira moscoviensis, a member of an independent phylum (13), was isolated from a corroded iron pipe in Moscow.
Although nitrification is the central aerobic process of microbial nitrogen cycling, only limited knowledge about the ecological relevance of its various protagonists is available. Medium selectivity and the ability of some Nitrosomonas strains to outcompete other ammonia oxidizers in liquid cultures (6) made Nitrosomonas europaea the most commonly isolated and best investigated ammonia oxidizer. However, there is increasing evidence of dominance by other species or genera in particular environments (24). This situation might be similar for the Nitrobacter spp., the best-investigated nitrite oxidizers. The introduction of molecular techniques into microbial ecology has enabled the detection and reliable quantification of natural populations of nitrifiers. Based on comparative 16S rRNA sequence analysis, PCR primers (15, 20, 31, 35) and oligonucleotide probes (16, 21, 33) specific for ammonia oxidizers from the β subclass of the class Proteobacteria have been developed. Application of specific PCR in combination with the construction of clone libraries (28) or denaturing gradient gel electrophoresis (DGGE) (18) revealed a high genetic diversity within the ammonia oxidizers of the β subclass of Proteobacteria and a wide distribution of Nitrosospira-like sequences in natural samples like lake water, sediments, or soils. On the other hand, quantitative hybridizations with oligonucleotide probes showed that in ammonium-rich systems like activated sludge (33) or biofilm reactors (21, 26), the dominant ammonia oxidizers were members of the genus Nitrosomonas.
DNA from the genus Nitrobacter has been amplified from soil by using specific PCR primers (12), and oligonucleotide probes have been designed for use in quantitative hybridizations (34). However, with the exception of two biofilm reactors (21, 26), no Nitrobacter spp. could be detected in activated sludge, eutrophic and oligotrophic biofilms, river water (21, 34), or aquaria (16) by using molecular techniques. It was therefore suggested that other nitrite oxidizers are present and active in these systems. Consistent with this hypothesis, Hovanec et al. recently reported Nitrospira spp. to be associated with nitrite oxidation in freshwater aquaria (17).
In this study, the nitrifying community of a chemolithoautotrophic fluidized bed reactor was investigated. The reactor, originally operated with high ammonia concentrations (11), was for this study operated with low ammonia concentrations, so that it rather resembled a natural freshwater habitat than a wastewater treatment system (24). The construction of a 16S rRNA clone library from directly isolated DNA was combined with fluorescence in situ hybridization (4). Microsensor measurements to relate the localization of the nitrifying populations to functional zones within the biofilm were performed.
MATERIALS AND METHODS
Nitrifying aggregates.
The lab-scale fluidized bed reactor used as a source for nitrifying aggregates has been described in detail before (11). However, the recirculation rate was increased from 1 to 1.8 ml/s, resulting in ammonium limitation in the reactor instead of oxygen limitation; i.e., ammonium was depleted 50 cm above the inlet but oxygen was still present. At the time of sampling, the reactor had been running for more than 8 months under constant conditions. The temperature was kept at 30°C, and the pH was adjusted to 8 in the aeration tank. Chemical parameters at the sampling point, 30 cm above the inlet, were as follows: [O2], 150 μM; [NH4+], 40 μM; [NO2−], 6.7 μM; [NO3−], 5.5 mM; pH 7.7. No organic carbon source was added to the reactor. The nitrifying aggregates were irregularly shaped conglomerates with diameters of 1 to 2 mm and consisted of spheroid subunits with ca. 50-μm diameters.
Microsensor measurements.
Clark-type O2 microsensors with internal references and guard cathodes were prepared and calibrated as described previously (25). Tip diameters were <10 μm, and stirring sensitivities were <2%.
Liquid ion-exchanging membrane (LIX) microsensors for NH4+, NO2−, and NO3− with solidified tips and protein coatings were prepared as described by de Beer et al. (10). The tip diameters were 5 μm for NH4+ and NO3− microsensors and 15 μm for NO2− microsensors. Calibration was done in a dilution series of NH4+, NO2−, and NO3− in the medium used for the measurements.
Aggregates were attached to insect needles, placed in a flow cell, and perfused with medium as described previously (11). [O2], [NH4+], [NO2−], pH, and temperature in the medium were adjusted to in situ conditions; [NO3−] was 100 μM. After 15 min of incubation, sufficient time to reach steady state, microprofiles were recorded by moving the sensors with a motor-driven micromanipulator at depth steps of 25 μm from the bulk liquid into the aggregate. The position relative to the aggregate surface was determined with a dissection microscope.
Chemical analysis.
[NH4+], [NO2−], and [NO3−] in the reactor column were determined colorimetrically (Spectroquant; Merck, Darmstadt, Germany). [O2] and pH were measured by an O2 microsensor and a pH electrode (Radiometer, Copenhagen, Denmark), respectively, which were positioned in the reactor at the sampling point.
DNA extraction.
Aggregate samples were homogenized by vigorous vortexing and subjected to three freeze-thaw cycles. An aliquot of 200 μl was suspended in 500 μl of AE buffer (20 mM sodium acetate [pH 5.5], 1 mM EDTA), and 1 ml of hot phenol-chloroform-isoamyl alcohol (25:24:1) and 100 μl of 25% (wt/vol) sodium dodecyl sulfate (SDS) were added. The mixture was shaken at 60°C for 30 min and then chilled on ice. The subsequent hot phenol extraction was done as described in the protocol of Wawer et al. (36). Nucleic acids were resuspended in distilled, sterile water, and their quality was checked by agarose gel electrophoresis (1%, wt/vol).
Amplification and cloning of 16S rDNA.
Almost-full-length bacterial 16S rRNA gene (rDNA) fragments were amplified from extracted DNA by the PCR. According to Muyzer et al. (23), primer pair GM3F (Escherichia coli 16S ribosomal DNA positions 8 to 24 [9])-GM4R (E. coli positions 1492 to 1507) was used. One microliter of the PCR product was directly ligated into the pGEM-T vector cloning system (Promega, Mannheim, Germany) and transformed into competent cells (high-efficiency E. coli JM109 [Promega]) as described in the manufacturer’s instructions.
DGGE screening of the gene library.
A 550-bp-long 16S rDNA fragment for DGGE analysis was directly amplified from each 16S rDNA clone by using 1 μl of cell suspension in the PCR. PCR and DGGE were performed as described by Muyzer et al. (22). For DGGE, a gradient of 35 to 65% denaturant at 60°C and an electrophoresis time of 17 h at 100 V were used.
16S rDNA sequencing.
Plasmids were extracted and purified from clones of interest with the Wizard Plus Minipreps DNA purification system (Promega) in accordance with the manufacturer’s instructions. Sequencing of the 16S rDNA inserts was done with a LICOR automated sequencer by MWG-Biotech, Ebersberg, Germany. Partial sequences (ca. 900 bases) and almost full sequences (ca. 1,500 bases) were determined on one strand.
Data analysis.
Sequences were added to the 16S rRNA database of the Technical University Munich by use of the program package ARB (29). The tool ARB_EDIT was used for sequence alignment. The 16S rRNA-based phylogenetic tree was calculated by using distance matrix, maximum-parsimony, and maximum-likelihood analyses, and the topologies of the resulting trees were compared. Branchings not supported by all three methods are displayed as multifurcations. Only sequences that were at least 90% complete were used for tree reconstruction. Partial sequences were added by using maximum parsimony without changing the overall topology of the tree.
Based on the newly retrieved Nitrospira-like sequences, oligonucleotide probes were designed. Their specificity was evaluated by using the ARB tool PROBE_FUNCTIONS and the rRNA database of the Technical University Munich (version 3/97).
Fixation and sectioning of aggregates.
For in situ hybridization, aggregates were fixed in 4% paraformaldehyde for 1 h at 4°C, washed in phosphate-buffered saline, and stored in a 1:1 mixture of phosphate-buffered saline and 96% ethanol at −20°C (2). Prior to the cryosectioning, aggregates were embedded in OCT compound (Tissue-Tek II; Miles, Elkhart, Ind.) overnight and subsequently frozen in a cryomicrotome (Mikrom, Walldorf, Germany) at −35°C. Semithick cryosections (5 to 20 μm) were cut at −17°C, and the single sections were placed on poly-l-lysine (0.01% solution; Sigma, Deisenhofen, Germany)-coated microscopic slides. After air drying overnight to allow optimal attachment of the sections to the slides, the OCT compound was removed by dissolving it in a drop of distilled water and carefully dipping the slide in distilled water. Again the slides were allowed to air dry. Finally, the sections were dehydrated in an ethanol dilution series (50, 70, and 96%) and stored at room temperature.
Oligonucleotide probes.
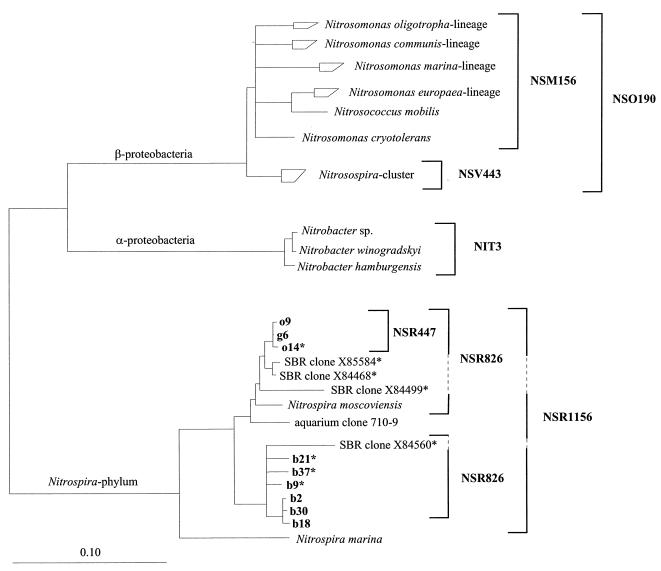
The following rRNA-targeted oligonucleotides were used (probe nomenclature according to Alm et al. [1] is given in parentheses): (i) EUB (S-D-Bact-0338-a-A-18); (ii) ALF1b (S-Sc-aProt-0019-a-A-17), BET42a (L-Sc-bProt-1027-a-A-17), and GAM42a (L-Sc-gProt-1027-a-A-17); (iii) NSO190 (S-*-Ntros-0190-a-A-19); (iv) NSV443 (S-*-Ntros-0443-a-A-19); (v) NSM156 (S-*-Nsom-0156-a-A-19); (vi) NIT3 (S-G-Nbac-1035-a-A-18); (vii) NSR1156 (S-*-Nspir-1156-a-A-18); (viii) NSR826 (S-*-Nspir-0826-a-A-18); and (ix) NSR447 (S-*-Nspir-0447-a-A-18). Oligonucleotides were synthesized and fluorescently labeled with a hydrophilic sulfoindocyanine dye, CY3 or CY5, or with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS) at the 5′ end by Interactiva Biotechnologie GmbH (Ulm, Germany). All probe sequences, their hybridization conditions, and references are given in Table 1. The respective target organisms of probes specific for nitrifying bacteria are indicated in the phylogenetic tree (see Fig. 2).
TABLE 1.
Oligonucleotide probes
| Probe | Specificity | Probe sequence (5′-3′) | Target sitea (rRNA positions) | % FAb | NaCl (mM)c | Reference |
|---|---|---|---|---|---|---|
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 16S (338–355) | 20 | 225 | 3 |
| ALF1b | α Subclass of the class Proteobacteria, several members of the δ subclass of Proteobacteria, genus Nitrospira, most spirochetes | CGTTCGYTCTGAGCCAG | 16S (19–35) | 20 | 225 | 19 |
| BET42a | β Subclass of Proteobacteria | GCCTTCCCACTTCGTTT | 23S (1027–1043) | 35d | 80 | 19 |
| GAM42a | γ Subclass of Proteobacteria | GCCTTCCCACATCGTTT | 23S (1027–1043) | 35e | 80 | 19 |
| NSO190 | Ammonia-oxidizing β-subclass Proteobacteria | CGATCCCCTGCTTTTCTCC | 16S (190–208) | 55 | 20 | 21 |
| NSV443 | Nitrosospira spp. | CCGTGACCGTTTCGTTCCG | 16S (444–462) | 30 | 112 | 21 |
| NSM156 | Nitrosomonas spp. | TATTAGCACATCTTTCGAT | 16S (653–670) | 40 | 56 | 21 |
| NIT3 | Nitrobacter spp. | CCTGTGCTCCATGCTCCG | 16S (1035–1048) | 40f | 56 | 34 |
| NSR826 | Freshwater Nitrospira spp. | GTAACCCGCCGACACTTA | 16S (826–843) | 20 | 225 | This study |
| NSR1156 | Freshwater Nitrospira spp. | CCCGTTCTCCTGGGCAGT | 16S (1156–1173) | 30 | 112 | This study |
| NSR447 | Clones g6, o9, and o14 | GGTTTCCCGTTCCATCTT | 16S (447–464) | 30 | 112 | This study |
E. coli numbering (9).
Percentage formamide in the hybridization buffer.
Millimolar concentration of sodium chloride in the washing buffer.
Used with an equimolar amount of unlabeled competitor oligonucleotide GAM42.
Used with an equimolar amount of unlabeled competitor oligonucleotide BET42a.
Used with an equimolar amount of unlabeled competitor oligonucleotide CNIT3 (34).
FIG. 2.
Phylogenetic tree inferred from comparative analysis of 16S rRNA sequences. The tree is based on the results of maximum-likelihood analysis of sequences of >1,400 nucleotides. Partial sequences (marked by an asterisk) were added by maximum-parsimony analysis without changing the tree topology. Target organisms of probes used in this study are indicated by brackets; SBR clones are not sequenced in the probe target regions. The bar represents 0.1 estimated change per nucleotide.
In situ hybridization.
All hybridizations were performed as described by Manz et al. (19) at 46°C for 90 min in hybridization buffer containing 0.9 M NaCl, formamide at the percentage shown in Table 1, 20 mM Tris-HCl (pH 7.4), and 0.01% SDS. The probe concentration was 5 ng/μl. Hybridization was followed by a stringent washing step at 48°C for 10 min in washing buffer containing 20 mM Tris-HCl (pH 7.4), NaCl at the concentrations listed in Table 1, and 0.01% SDS. Washing buffer was removed by rinsing the slides with distilled water. The slides were air dried, stained with 4,6-diamidino-2-phenylindole (DAPI; 1 μg/ml) for 10 min in the dark on ice, and finally rinsed again with distilled water. The slides were mounted in Vectashield (Vector Laboratories Inc., Burlingame, Calif.) to avoid bleaching and examined with a Zeiss Axioplan epifluorescence microscope equipped with filter sets CY3-HQ and CY5-HQ (Chroma Technology Corp., Brattleboro, Vt.) and 01 and 09 (Carl Zeiss, Jena, Germany). A Zeiss LSM 510 confocal laser scanning microscope, equipped with an Ar ion laser (488 nm) and two HeNe lasers (543 nm, 633 nm), was used to record optical sections as described by Wagner et al. (32).
Nucleotide sequence accession numbers.
16S rDNA sequence data obtained in this study will appear in the EMBL, GenBank, and DDBJ databases under accession no. AJ224038 to AJ224047.
RESULTS
Microsensor measurements.
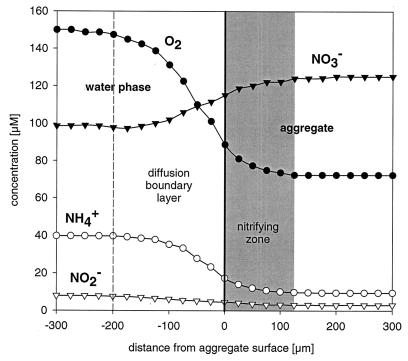
All investigated aggregates showed consumption of O2, NH4+, and NO2− and production of NO3− (Fig. 1). The consumed NH4+ was completely converted to NO3− without an intermediate NO2− peak, and even NO2− present in the bulk water was consumed. This indicates that nitrite oxidation was at least as fast as ammonia oxidation. The active nitrifying zone was restricted to the outer 125 μm of the aggregates, although O2 and NH4+ were both still present in the center of the aggregates at concentrations of about 75 and 10 μM, respectively.
FIG. 1.
Typical microprofiles of [O2], [NH4+], [NO2−], and [NO3−] in nitrifying aggregates. The gray area marks the zone with nitrifying activity.
FISH analysis.
Phase-contrast light microscopy of the aggregate sections showed dense bacterial clusters, void spaces, and an irregular surface. If excited with blue light (488 nm), cells and extracellular material exhibited strong green autofluorescence, which made it difficult to reliably detect FLUOS-labeled oligonucleotide probes. Therefore, we exclusively applied CY3- or CY5-labeled probes for fluorescence in situ hybridization (FISH) analysis.
In situ hybridization with probe EUB proved that the ribosome content of most cells was sufficient for single-cell detection and that probes penetrated most, if not all, cell clusters. Only a few DAPI-stained cells in the central regions of aggregates were not detectable by FISH. Application of probes ALF1b, BET42a, and GAM42a indicated that the population consisted mainly of approximately equal fractions of ALF1b- and BET42a-positive cells. No cells hybridizing with probe GAM42a were detected. ALF1b-positive cells had diameters of below 0.6 μm and formed clusters 3 to 10 μm in size which were situated close to BET42a-positive cells that had diameters of ca. 1 μm and formed clusters 5 to 20 μm in size. Taken together, both populations made up >90% of all cells in the nitrifying zone as well as in the central region of the aggregates. Virtually all cells detected by probe BET42a also hybridized with probes NSO190 and NSV443 (Fig. 3), but no cells could be detected with probe NSM156. Thereby, the ammonia-oxidizing cells were identified as members of the genus Nitrosospira. In contrast, none of the cells detected by probe ALF1b hybridized with probe NIT3, which was specific for the nitrite-oxidizing bacteria of the α subclass of Proteobacteria of the genus Nitrobacter.
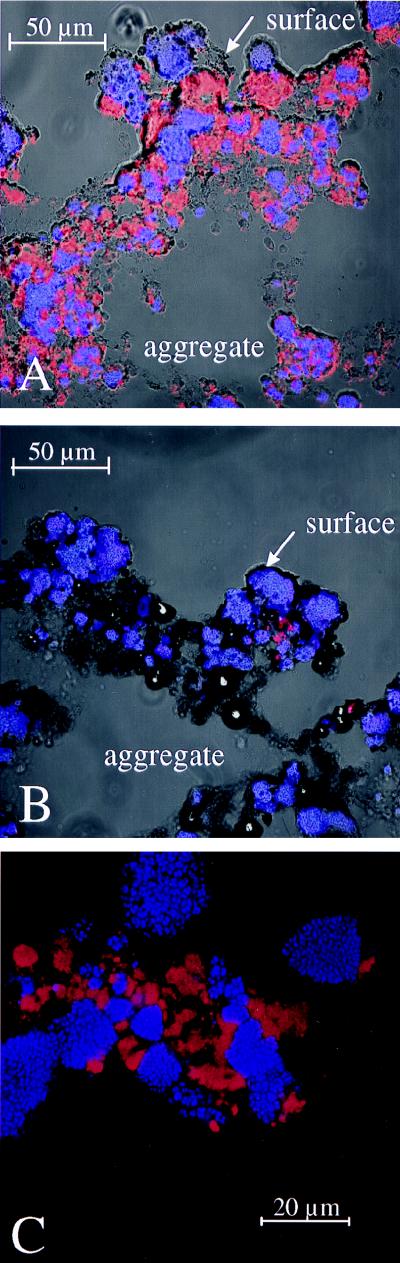
FIG. 3.
In situ identification of nitrifying bacteria in aggregate sections demonstrating their spatial distribution. Images are composed of overlays of phase-contrast microscopic images and two confocal microscopic images. (A) Simultaneous in situ hybridization with CY5-labeled probe NSV443 and CY3-labeled probe NSR1156. Cells of Nitrosospira spp. are shown in blue; cells of Nitrospira-like bacteria are red. (B) Simultaneous in situ hybridization with CY5-labeled probe NSV443 and CY3-labeled probe NSR447. Cells of Nitrosospira spp. are blue; a smaller subpopulation of Nitrospira spp. is visualized in red. (C) Simultaneous in situ hybridization with CY5-labeled probe NSV443 and CY3-labeled probe NSR1156 displaying the morphology and close association of ammonia- and nitrite-oxidizing bacteria.
Clone library.
A 16S rDNA clone library was constructed from aggregate samples to identify the unknown nitrite oxidizers. Fifty-five clones were selected at random. To avoid redundant sequencing, PCR-amplified rDNA fragments of all clones were analyzed by DGGE. Ten distinct categories of bands could be identified, and one representative of each category was selected for comparative sequence analysis. The 16S rRNA sequences of 9 of the 10 clones were affiliated with those of the nitrite oxidizers of the genus Nitrospira (13). Three clone sequences (g6, o9, and o14) were closely related to Nitrospira moscoviensis (96% similarity), whereas the other six (b2, b9, b18, b21, b30, and b37) formed a distinct branch within the genus Nitrospira but were still more closely related to Nitrospira moscoviensis (92% similarity) than to Nitrospira marina (87% similarity). Table 2 shows the 16S rRNA similarity values for the five almost-full-length sequences determined in this study, and Fig. 2 illustrates their affiliation in a phylogenetic tree. The remaining partial sequence (g14) was related to the 16S rRNA gene of a Hyphomonas sp. of the α subclass of Proteobacteria. No sequences affiliated with the genus Nitrobacter or with any known ammonia-oxidizing bacterium were retrieved.
TABLE 2.
Similarity matrix of clones affiliated with Nitrospira spp.
| Species or clone | % Similarity to:
|
||||||
|---|---|---|---|---|---|---|---|
| N. mosco- viensis | N. marina | b2 | b18 | b30 | g6 | o9 | |
| N. moscoviensis | |||||||
| N. marina | 89.3 | ||||||
| b2 | 92.8 | 87.8 | |||||
| b18 | 92.1 | 87.5 | 97.1 | ||||
| b30 | 92.8 | 88.2 | 98.9 | 97.5 | |||
| g6 | 95.8 | 87.6 | 91.7 | 92.6 | 91.6 | ||
| o9 | 95.8 | 87.6 | 90.1 | 92.3 | 90.1 | 99.5 | |
| Clone 710-9 (17) | 94.7 | 87.7 | 90.4 | 90.1 | 90.3 | 92.3 | 89.9 |
Probe design and in situ detection of Nitrospira.
Based on the retrieved sequences, two probes specific for all clones that included Nitrospira moscoviensis, NSR1156 and NSR826 (with one mismatch to clone b18), were developed. Another probe, NSR447, was designed to be complementary to clones g6, o9, and o14 only (Fig. 2). Hybridization conditions of the probes were evaluated in situ by using increasing concentrations of formamide in the hybridization buffer. Probes NSR1156 and NSR447 showed good signal and specificity in buffer with 30% formamide; probe NSR826 did so in buffer with 20% formamide. When applied to the aggregate sections, probes NSR1156 and NSR826 hybridized to virtually all ALF1b-positive cells. A 16S rRNA sequence database check revealed that, indeed, all Nitrospira spp. have the target sequence for probe ALF1b. The nitrifying community active in the fluidized bed reactor was shown to consist of members of the genera Nitrosospira and Nitrospira (Fig. 3A). Only a minor fraction of all NSR1156- and NSR826-positive cells also hybridized with probe NSR447. These cells were slightly bigger, formed smaller clusters than the major part of the Nitrospira sp. cells, and were located exclusively in the nitrifying part of the aggregates (Fig. 3B). Single-cell resolution of the very small Nitrospira spp. was almost impossible even with confocal laser scanning microscopy (Fig. 3C).
DISCUSSION
Microprofiles.
The in situ activity measurements clearly indicate that both ammonia and nitrite oxidizers were active within a 125-μm zone at the aggregate surface (Fig. 1). In contrast, no activity was measured in the inner part of the aggregates, although dense populations of nitrifiers could be detected (Fig. 3) and oxygen and ammonium were present. One possible explanation is that the ammonium concentration in this zone (10 μM) was substantially below the half-saturation constant (Ks) for ammonia oxidation. Indeed, the lowest Ks values reported so far for ammonia-oxidizing bacteria are 30 to 75 μM NH4+ plus NH3 (at pH 7.8) for strains of the Nitrosomonas oligotropha group, isolated from the River Elbe (27). No such data are currently available for members of the genus Nitrosospira.
Ammonia oxidizers.
By using FISH with a set of specific probes, it was shown that members of the genus Nitrosospira (in its extended definition [14] also including the former genera Nitrosolobus and Nitrosovibrio) were the only ammonia-oxidizing bacteria in the aggregates. As previously shown for members of the genus Nitrosomonas in biofilms (21, 26), Nitrosospira spp. also tended to form monospecies clusters in close association with clusters of nitrite oxidizers. Cluster formation could be disadvantageous due to the longer diffusion path of substrates. However, since the nitrifying bacteria rely on CO2 fixation by ribulose bisphosphate carboxylase (RubisCO), it might be a mechanism for increasing the efficacy of CO2 fixation by lowering the [O2] in the clusters. Cluster formation might also provide protection against hazardous environmental factors, or it might just be an ecologically neutral result of cell division. In our study, the clustering of Nitrosospira sp. was probably the reason that no 16S rDNA clone sequences of this genus were obtained. Intact clusters could still be detected after the DNA isolation procedure, indicating resistance against the DNA extraction protocol based on freeze-thawing and hot phenol and SDS extraction. This potential for formation of highly resistant clusters should be considered in all attempts to monitor ammonia-oxidizing bacteria with methods based on DNA extraction.
The in situ detection of Nitrosospira spp. in this ammonium-poor system is in agreement with observations that in natural systems that are low in ammonium, Nitrosospira spp. can be detected rather than Nitrosomonas spp. (15, 18, 28). However, these studies depend on DNA extraction and PCR while true quantitative in situ analyses of these habitats are missing. In contrast, Nitrosomonas spp., i.e., members of the Nitrosomonas europaea lineage, are more abundant in activated sludge, biofilms, and enrichments with millimolar concentrations of ammonium (21, 26, 28, 33). The main adaptations to the different niches are probably via the ammonia oxidation kinetics and the growth rate. In that respect, Nitrosospira and Nitrosomonas seem to be typical K and r strategists, respectively (5). However, the isolation of Nitrosomonas oligotropha and related strains with remarkably low Ks values (27) already indicates limits for such a generalizing statement.
Nitrite oxidizers.
Nitrospira-like bacteria were identified in the nitrifying aggregates by a combination of comparative 16S rRNA sequence analysis and FISH. Their localization within the nitrifying zone and their association with the ammonia-oxidizing Nitrosospira species lend strong evidence that they are responsible for nitrite oxidation in our system. Two genetically different populations were distinguished in this study. The main population, which was more distantly related to Nitrospira moscoviensis, also occurred in the inactive center of the aggregates. The second, smaller population had a higher level of 16S rRNA similarity to Nitrospira moscoviensis. It was restricted to the active nitrifying zone. The genotypic differences obviously coincide with different physiological adaptations leading to these distinct spatial distributions. The exact description of the differing physiological properties of the new nitrite oxidizers, however, will require their isolation. A directed isolation can be monitored by the oligonucleotide probes developed in this study.
The detection of Nitrospira spp. also yields independent support for a recent molecular study reporting that Nitrospira-like bacteria were the main nitrite-oxidizing population in freshwater aquaria (17). Before, Nitrospira-like 16S rDNA sequences had also been isolated from activated sludge (8), but, at that time, reference sequences of cultured Nitrospira spp. with proven nitrite-oxidizing capability were not available. Therefore, it was impossible to link the genotype with a phenotype. This clearly shows the importance of cultivation of bacteria. Considering the rare in situ detection of Nitrobacter species in freshwater systems (16, 21, 34), Nitrospira spp. might be of more general importance for nitrite oxidation. The molecular detection of Nitrospira-like bacteria in habitats as diverse as activated sludge, freshwater aquaria, and an oligotrophic fluidized bed reactor and the initial isolation of Nitrospira moscoviensis from a corroded water pipe may indicate a wide distribution in nitrifying systems.
Conclusion.
Nitrosospira and Nitrospira spp. were identified as main components of nitrifying aggregates. Their activity and spatial distribution could be shown by the combination of microsensor measurements and in situ hybridization. Whether the combination of these two genera is unique to the investigated system or is of general relevance in more oligotrophic freshwater habitats must be evaluated in the future.
ACKNOWLEDGMENTS
This work was supported by the Körber Stiftung, the Max-Planck-Gesellschaft, and the DFG (SFB411-98).
We thank G. Eickert, A. Eggers, and V. Hübner for preparation of oxygen microelectrodes and S. P. P. Ottengraf and J. C. van den Heuvel (Laboratory for Chemical Technology, University of Amsterdam, Amsterdam, The Netherlands) for access to the fluidized bed reactor. G. Muyzer is gratefully acknowledged for introducing DGGE, and F. O. Glöckner and W. Schönhuber are thanked for help with ARB.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews J H, Harris R F. r- and K-selection and microbial ecology. Adv Microb Ecol. 1986;9:99–147. [Google Scholar]
- 6.Belser L W. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 7.Bock E, Koops H-P. The genus Nitrobacter and related genera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 2302–2309. [Google Scholar]
- 8.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structure of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.de Beer D, Schramm A, Santegoeds C M, Kühl M. A nitrite microsensor for profiling environmental biofilms. Appl Environ Microbiol. 1997;63:973–977. doi: 10.1128/aem.63.3.973-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Beer D, van den Heuvel J C, Ottengraf S P P. Microelectrode measurements of the activity distribution in nitrifying bacterial aggregates. Appl Environ Microbiol. 1993;59:573–579. doi: 10.1128/aem.59.2.573-579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degrange V, Bardin R. Detection and counting of Nitrobacter populations in soil by PCR. Appl Environ Microbiol. 1995;61:2093–2098. doi: 10.1128/aem.61.6.2093-2098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov., and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 14.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 15.Hiorns W D, Hastings R C, Head I M, McCarthy A J, Saunders J R, Pickup R W, Hall G H. Amplification of 16S ribosomal RNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of nitrosospiras in the environment. Microbiology. 1995;141:2793–2800. doi: 10.1099/13500872-141-11-2793. [DOI] [PubMed] [Google Scholar]
- 16.Hovanec T A, DeLong E F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hovanec T A, Taylor L T, Blakis A, Delong E F. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol. 1998;64:258–264. doi: 10.1128/aem.64.1.258-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalchuk G A, Stephen J R, de Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 20.McCaig A E, Embley T M, Prosser J I. Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol Lett. 1994;120:363–368. doi: 10.1111/j.1574-6968.1994.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 21.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C M, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual, supplement 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 23.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 24.Prosser J I. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 25.Revsbech N P. An oxygen microelectrode with a guard cathode. Limnol Oceanogr. 1989;34:474–478. [Google Scholar]
- 26.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R, Schleifer K-H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stehr G, Böttcher B, Dittberner P, Rath G, Koops H-P. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol Ecol. 1995;17:177–186. [Google Scholar]
- 28.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, M. Lenke, T. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. ARB: a software environment for sequence data. http://www.mikro.biologie.tu-muenchen.de. Department of Microbiology, Technische Universität München, Munich, Germany.
- 30.Teske A, Alm E, Regan J M, Toze S, Rittman B E, Stahl D A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voytek M A, Ward B B. Detection of ammonia-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol. 1995;61:1444–1450. doi: 10.1128/aem.61.4.1444-1450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner M, Assmus B, Hartmann A, Hutzler P, Amann R. In situ analysis of microbial consortia in activated sludge using fluorescently labelled, rRNA-targeted oligonucleotide probes and confocal scanning laser microscopy. J Microsc. 1994;176:181–187. doi: 10.1111/j.1365-2818.1994.tb03513.x. [DOI] [PubMed] [Google Scholar]
- 33.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 34.Wagner M, Rath G, Koops H-P, Flood J, Amann R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol. 1996;34:237–244. [Google Scholar]
- 35.Ward B B, Voytek M A, Witzel K-P. Phylogenetic diversity of natural populations of ammonia oxidizers investigated by specific PCR amplification. Microb Ecol. 1997;33:87–96. doi: 10.1007/s002489900011. [DOI] [PubMed] [Google Scholar]
- 36.Wawer C, Jetten M S M, Muyzer G. Genetic diversity and expression of the [NiFe] hydrogenase large-subunit gene of Desulfovibrio spp. in environmental samples. Appl Environ Microbiol. 1997;63:4360–4369. doi: 10.1128/aem.63.11.4360-4369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]