Abstract
Pluralibacter gergoviae is a member of the Enterobacteriaceae family that has been reported sporadically. Although P. gergoviae strains exhibiting multidrug-resistant profiles have been identified an in-depth genomic analysis focusing on antimicrobial resistance (AMR) has been lacking, and was therefore performed in this study. Forty-eight P. gergoviae strains, isolated from humans, animals, foods, and the environment during 1970–2023, were analyzed. A large number of single-nucleotide polymorphisms were found, indicating a highly diverse population. Whilst P. gergoviae strains were found to be circulating at the One Health interface, only human and environmental strains exhibited multidrug resistance genotypes. Sixty-one different antimicrobial resistance genes (ARGs) were identified, highlighting genes encoding mobile colistin resistance, carbapenemases, and extended-spectrum β-lactamases. Worryingly, the co-occurrence of mcr-9.1, blaKPC-2, blaCTX-M-9, and blaSHV-12, as well as mcr-10.1, blaNDM-5, and blaSHV-7, was detected. Plasmid sequences were identified as carrying clinically important ARGs, evidencing IncX3 plasmids harboring blaKPC-2, blaNDM-5, or blaSHV-12 genes. Virulence genotyping underlined P. gergoviae as being a low-virulence species. In this regard, P. gergoviae is emerging as a new multidrug-resistant species belonging to the Enterobacteriaceae family. Therefore, continuous epidemiological genomic surveillance of P. gergoviae is required.
Keywords: Enterobacteriaceae, genomic analysis, multidrug resistance, plasmid-mediated quinolone resistance, gene transfer, One Health
1. Introduction
Antimicrobial resistance (AMR) is a growing global health concern that affects several sectors. In clinical settings, AMR reduces the effectiveness of antimicrobials and leads to longer hospital stays. Consequently, there is an increase in mortality rates [1]. Furthermore, healthcare-associated infections caused by MDR pathogens are on the rise, which is concerning [2]. In the environment, antimicrobial-resistant strains are circulating especially in water bodies and agricultural soils, impacting human and animal populations [3]. These strains may be reaching food products and leading to difficult-to-treat foodborne infections [4]. In this context, AMR is a multifactorial problem that requires a One Health approach [5].
Pluralibacter gergoviae, formerly Enterobacter gergoviae, was first described in the 1980s in human clinical specimens and the environment in France and the United States [6,7]. Subsequently, this species was identified as contaminating cosmetic formulations, colonizing the human oral cavity, and causing nosocomial outbreaks [8,9,10,11]. From the 2010s, multidrug-resistant (MDR) P. gergoviae strains, harboring clinically important antimicrobial resistance genes (ARGs) and highlighting carbapenemases, began to emerge in clinical settings [11,12,13]. Whilst MDR P. gergoviae strains have been described, an in-depth genomic analysis is lacking. Therefore, this study aimed to perform a genomic characterization of P. gergoviae strains circulating at the One Health interface and provide genomic insights into AMR.
2. Materials and Methods
Bacterial Genomes and Genomic Analysis
All P. gergoviae genomes (NCBI:txid61647) available at the GenBank® database were downloaded on 1 August 2023. The derived, contaminated, and duplicated genomes were removed. Resistome, virulome, and plasmid replicons were identified using ResFinder v.4.1, VirulenceFinder v.2.0, and PlasmidFinder v.2.1, respectively. Single-nucleotide polymorphism (SNP) counts were determined using CSI Phylogeny v.1.4, with P. gergoviae strain FDAARGOS_186 (BioSample: SAMN05004741) as a reference. Antimicrobial susceptibility was predicted using ResFinder v.4.1 and the strains were classified as MDR according to Magiorakos et al. [14]. All aforementioned analyses were performed using tdefault settings at the Center for Genomic Epidemiology (https://www.genomicepidemiology.org/) (accessed on 2 August 2023). The SNP-based phylogenetic tree was visualized and edited using iTOL v.6 (https://itol.embl.de/) (accessed on 10 August 2023). The plasmid contigs were predicted using mlplasmids v.2.1.0 [15]. The insertion sequence elements were identified using ISfinder [16]. The plasmid sequences were manually curated and visualized using Geneious Prime® v.2023.0.4. Comparative plasmid sequence analysis was carried out using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (accessed on 21 August 2023).
3. Results
3.1. Distribution of P. gergoviae Strains
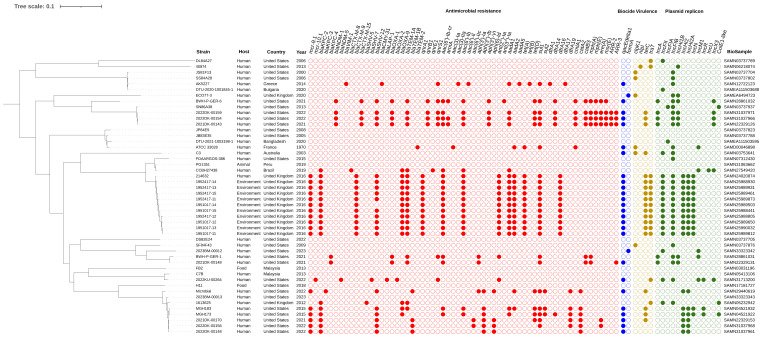
Forty-eight P. gergoviae strains were included in this study. These strains were isolated from humans (abscesses, blood, bronchoalveolar lavage, dentine caries, drainage, endotracheal and tracheal aspirate, feces, gastrojejunostomy tubes, nasopharyngeal swabs, peri-bile duct tissue, right shin wound cultures, tracheostomy sites, and urine), an animal (dog, urine), foods (fish balls and ground beef), or the environment (wastewater), evidencing their circulation at the One Health interface. These strains were obtained from 1970 onwards in countries belonging to the Australian (Australia), American (Brazil, Peru, and the United States), Asian (Bangladesh and Malaysia), and European (Bulgaria, Greece, France, and the United Kingdom) continents (Figure 1).
Figure 1.
The SNP-based phylogenetic tree of P. gergoviae strains distributed worldwide. Strain, host, country, year, and BioSample data were retrieved from the GenBank® database. Red, blue, yellow, and green painted circles represent positive antimicrobial resistance genes, biocide resistance genes, virulence genes, and plasmid replicons, respectively. The tree was rooted at the midpoint.
3.2. SNPs Data
SNP differences among all P. gergoviae genomes ranged from 27 to 33,993, whereas among human and environmental strains SNP differences ranged from 80 to 33,993 and 27 to 1073, respectively. Between food strains (FB2 and H11), differences of 22,871 SNPs were found. Overall, P. gergoviae genomes were grouped according to country or host, but divergences in these characteristics were also observed. The largest clade grouped human and environmental strains (214632, 1951017-11 to -15, and 1952417-11 to -15) from the United Kingdom. Interestingly, human and animal strains (FDAARGOS-386, PG1351, and CCBH27438) from the American continent (Brazil, Peru, and the United States) were clustered. In this regard, the finding of a large number of SNPs indicates a highly diverse population with a range of genetic adaptations that may be related to genetic exchanges or recombination events.
3.3. ARG-Producing Strains
A great diversity of ARGs were found to relate to resistance to polymyxins (mcr-9.1 and mcr-10.1), β-lactams (blaKPC-2, blaKPC-3, blaKPC-4, blaNDM-1, blaNDM-5, blaVIM-1, blaCTX-M-8, blaCTX-M-9, blaCTX-M-15, blaSHV-5, blaSHV-7, blaSHV-12, blaCMY-31, blaLAP-1, blaOXA-1, blaOXA-2, blaOXA-9, blaTEM-1A, blaTEM-1B, and blaTEM-2), fluoroquinolones [qnrA1, qnrB1, qnrE1, qnrS1, and aac(6′)-Ib-cr], aminoglycosides [armA, aac(3)-Ia, aac(3)-IIa, aac(6′)-Ib, aac(6′)-Il, aac(6′)-IIc, aph(3′)-Ia, aph(3″)-Ib, aph(3′)-VI, aph(6)-Id, ant(2″)-Ia, ant(3″)-Ia, aadA1, aadA2, and aadA5], tetracyclines [tet(A), tet(B), and tet(D)], folate pathway antagonists (sul1, sul2, dfrA1, dfrA14, dfrA16, dfrA17, and dfrA19), amphenicols (cmlA1, catA2, catB3), macrolides [mph(A), mph(E), ere(A), and msr(E)], and rifamycins (ARR-2 and ARR-3). Some human-derived strains, and all of those from foods and animals, did not contain ARGs. Furthermore, biocide resistance genes were also detected, including qacEΔ1 (quaternary ammonium compound) in 54.1% of strains and formA (aldehyde) in 2023BM-00012 and ECO77-3 strains (Figure 1).
Most strains (58%) carried at least four ARGs. Among them, strains isolated from humans and the environment in Greece, the United Kingdom, and the United States harbored ten or more ARGs related mainly to polymyxins, β-lactams, fluoroquinolones, aminoglycosides, tetracyclines, and folate pathway antagonists. The presence of genes that encode mobile colistin resistance (mcr-9.1 and mcr-10.1), carbapenemases (blaKPC, blaNDM, and blaVIM), extended-spectrum β-lactamases (ESBLs; blaCTX-M and blaSHV), and plasmid-mediated quinolone resistance [PMQR; qnr and aac(6′)-Ib-cr] is highlighted. Interestingly, the coexistence of the aforementioned clinically relevant ARGs was identified in human and environmental strains from the United States and the United Kingdom, respectively. In addition, three human strains (2022DK-00159, 2022DK-00154, and 2021DK-00143) presented more than 20 ARGs (Figure 1). These characteristics contribute to the emergence of multidrug-resistant strains and the spread of clinically important ARGs worldwide.
3.4. WGS-Predicted Antimicrobial Susceptibility
Corroborating with the resistome, it was found that most strains (58%) were predicted to be MDR since they presented resistance to one or more antimicrobial agents in three or more antimicrobial categories. Although MDR strains presented resistance to critically important antimicrobials [e.g., polymyxins, penicillins + β-lactamase inhibitors, extended-spectrum cephalosporins (3rd and 4th generations), monobactams, carbapenems, fluoroquinolones, and/or aminoglycosides], they were susceptible mainly to minocycline, tigecycline, and/or fosfomycin. MDR strains were obtained exclusively from humans and the environment, while pan-susceptible strains were recovered from humans, animals, and foods (Table 1).
Table 1.
WGS-predicted phenotype based on plasmid-mediated ARGs of P. gergoviae genomes.
| Strain 1 | Antimicrobial Agent 2,3 | Pattern 4 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COL | AMP | AMC | PTZ | CFO | CAZ | CPM | ATM | MPM | CIP | STP | GEN | AMI | TET | MIN | TGC | SXT | CHL | FOS | ||
| 1613625 * | S | R | R | R | R | R | R | R | R | S | S | S | S | S | S | S | S | S | S | MDR |
| 2021DK-00143 * | S | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | R | S | MDR |
| 2021DK-00148 * | S | R | R | R | R | R | R | R | R | R | S | S | S | S | S | S | R | R | S | MDR |
| 2021DK-00170 * | R | R | R | R | R | R | R | R | R | S | R | R | S | R | S | S | R | R | S | MDR |
| 2022DK-00148 * | R | R | R | R | R | R | R | R | R | S | R | S | S | S | S | S | S | R | S | MDR |
| 2022DK-00154 * | S | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | R | S | MDR |
| 2022DK-00156 * | R | R | R | R | R | R | R | R | R | S | R | R | S | S | S | S | R | R | S | MDR |
| 2022DK-00159 * | S | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | R | S | MDR |
| 2022KU-00264 * | R | R | R | R | R | R | R | R | R | R | S | R | S | S | S | S | S | S | S | MDR |
| 2023BM-00012 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| 2023BM-00013 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| 214632 * | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 40874 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| AK5227 * | S | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | S | S | MDR |
| ATCC 33028 * | S | R | S | S | S | S | S | S | S | S | R | R | S | R | S | S | R | R | S | MDR |
| BWH-P-GER-1 * | S | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | R | S | MDR |
| BWH-P-GER-6 * | S | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | R | S | MDR |
| C3 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| C7B * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| CCBH27438 * | S | R | R | R | R | R | R | R | R | R | R | S | R | S | S | S | S | S | S | MDR |
| DL84A27 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| DS82E24 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| DTU-2020-1001845-1 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| DTU-2021-1003198-1 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| ECO77-3 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| FDAARGOS-386 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| JB83E35 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| JP84E9 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| JS81F13 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| MGH173 * | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | R | R | S | MDR |
| MGH183 * | R | R | R | R | R | R | R | R | R | R | R | S | R | R | S | S | R | S | S | MDR |
| Microbial * | R | R | R | R | R | R | R | R | R | S | R | S | R | R | S | S | R | R | S | MDR |
| SF84F43 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| SN86A38 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| SS84A28 * | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| PG1351 ^ | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| FB2 $ | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| H11 $ | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | – |
| 1951017-11 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1951017-12 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1951017-13 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1951017-14 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1951017-15 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1952417-11 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1952417-12 # | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | S | R | S | S | MDR |
| 1952417-13 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1952417-14 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
| 1952417-15 # | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | S | MDR |
1 *, strains from humans; ^, strain from dog; $, strains from foods; #, strains from the environment. 2 S in green color, susceptible; R in red color, resistant. 3 Antimicrobial categories as follows: polymyxins (COL, colistin), penicillins (AMP, ampicillin), penicillins + β-lactamase inhibitors (AMC, amoxicillin-clavulanic acid), antipseudomal penicillins + β-lactamase inhibitors (PTZ, piperacillin-tazobactam), cephamycins (CFO, cefoxitin), extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins (CAZ, ceftazidime; CPM, cefepime); monobactams (ATM, aztreonam); carbapenems (MPM, meropenem); fluoroquinolones (CIP, ciprofloxacin); aminoglycosides (STP, streptomycin; GEN, gentamicin; AMI, amikacin); tetracyclines (TET, tetracycline; MIN, minocycline); glycylcyclines (TGC, tigecycline); folate pathway antagonists (SXT, trimethoprim-sulfamethoxazole); phenicols (CHL, chloramphenicol); and phosphonic acids (FOS, fosfomycin). 4 Classification as MDR according to Magiorakos et al. [9]; MDR, multidrug-resistant; –, non-MDR.
3.5. Plasmid-Mediated Clinically Important ARGs
Various plasmid replicons were detected, including IncA, IncFII, IncFIA, IncFIB, IncHI1B, IncHI2, IncHI2A, IncN, IncM1, IncR, IncU, IncX3, and Col(pHAD28) (Figure 1). Plasmidome analysis revealed that all mcr, bla, and PMQR genes were housed on plasmid contigs. Furthermore, the association between ARGs and Inc groups was identified in some strains as follows: IncHI2/HI2A plasmid contigs (>140 kb) were identified as harboring the mcr-9.1 gene embedded on IS903B-mcr-9.1-IS1-ΔTn3 structure in the 1951017-13 and 1952417-15 strains; an IncFII(K9) fragment (85 kb) was detected as carrying the mcr-10.1 gene located on the xerC-mcr-10.1-ISEc36 structure in the 2022KU-00264 strain; an IncHI1B/FIB plasmid contig (>200 kb) was found to be harboring the blaNDM-1 gene, flanked by intact and truncated ISAba125 elements in the BWH-P-GER-6 strain; an IncA plasmid contig (>200 kb) was detected as housing the blaKPC-4 gene encoded within a truncated Tn4401 element in the BWH-P-GER-1 strain; and IncF plasmid contigs (>100 kb) were identified as harboring the blaKPC-2 gene with Tn4401a and Tn4401b isoforms in the 1613525 and 2022DK-00156 strains, respectively.
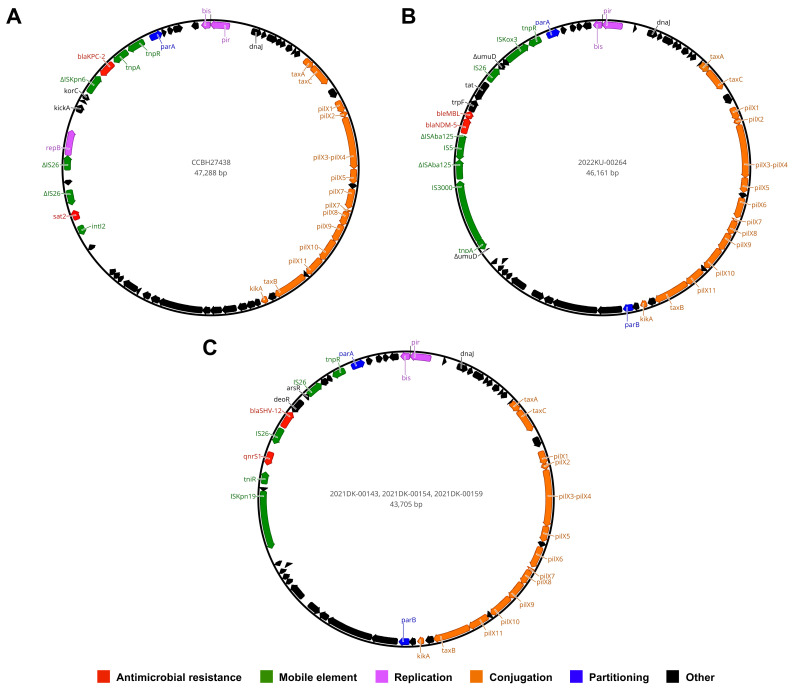
In-depth genomic analysis revealed complete sequences of IncX3 plasmids ranging from 43 to 47 kb in length, carrying genes related mainly to antimicrobial resistance, mobile elements, replication, conjugation, and partitioning. An IncX3-IncU plasmid harboring the blaKPC-2 gene on a non-Tn4401 (NTEKPC-Ic) was identified in CCBH27438 (Figure 2A), while an IncX3 plasmid housing the blaNDM-5 gene located on the ΔumuD genetic structure containing IS3000, ΔISAba125, IS5, bleMBL, trpF, tat, and IS26 (Figure 2B) was detected in the 2022KU-00264 strain. IncX3 plasmids from the 2021DK-00143, 2021DK-00154, and 2021DK-00159 strains harbored the blaSHV-12 gene flanked by IS26 and the qnrS1 gene located proximally to tniR and ISKpn19 (Figure 2C). The presence of the aforementioned ARGs and Inc groups in different plasmid contigs was also detected in other strains, supporting the conclusion that these same plasmids may also be circulating among P. gergoviae strains. BLASTn analysis showed that partial or complete plasmid sequences presented high query coverage (>99%) and nucleotide identity (>99%), with other sequences from Enterobacteriaceae (e.g., Salmonella enterica, Klebsiella pneumoniae, Enterobacter spp., and Citrobacter freundii) distributed worldwide.
Figure 2.
Circular maps of IncX3 plasmids harboring clinically relevant ARGs in P. gergoviae strains. (A) IncX3-IncU plasmid carrying the blaKPC-2 gene in CCBH27438 strain. (B) IncX3 plasmid housing the blaNDM-5 gene in 2022KU-00264 strain. (C) IncX3 plasmid carrying blaSHV-12 and qnrS1 genes in 2021DK-00143, 2021DK-00154, and 2021DK-00159 strains.
3.6. Virulence Genotyping
Virulence genes that encode a heat shock survival AAA family ATPase (clpK1), a lipoprotein NlpI precursor (nlpI), an outer-membrane protein complement resistance (traT), and a tellurium ion resistance protein (terC) were identified among the P. gergoviae genomes. Most strains presented only one of the virulence genes found; however, some of them harbored two genes. Strain C3 from Australia was isolated from a patient with cystic fibrosis and presented clpK1 and terC genes, while environmental strains from the United Kingdom carried terC and traT genes (Figure 1).
4. Discussion
In this study, we present a first comparative genomic analysis of P. gergoviae distributed worldwide. Overall, P. gergoviae strains were recovered from humans, animals, foods, and the environment, but MDR strains were isolated only from humans and the environment, which may be related to the limited number of food and animal strains analyzed. In this context, it can be said that this species has been circulating at the One Health interface and that MDR strains may also emerge within it. Therefore, AMR is a multifaceted challenge that requires coordinated global activities to reduce the spread of MDR bacteria [17]. Following the emergence of P. gergoviae strains as MDR, high-resolution molecular typing methods, including multilocus sequence typing, should be encouraged in order to improve population structure analysis, strain differentiation, international comparisons, and longitudinal studies.
Species belonging to the Enterobacteriaceae family, including E. coli and K. pneumoniae, are known to be potentially pathogenic and MDR [18]. In general, AMR studies are focused on them, while in other species, mainly newer ones, little is known about the AMR’s genetic background. Recently, Raoultella species have gained notoriety for being MDR and shedding clinically important ARGs [19,20]. In this regard, P. gergoviae emerges as a new species with these traits since it harbors an arsenal of plasmid-mediated ARGs. As enterobacteria are known for their ability to exchange genetic materials [21], P. gergoviae also demonstrates the potential to act as a disseminator of ARG-bearing plasmids. Furthermore, P. gergoviae strains also carried biocide resistance genes, contributing to the co-selection of AMR and their persistence in clinical settings [22].
Among known AMR-related plasmid families in Enterobacteriaceae, IncA, IncF, IncH, and IncN stand out for harboring various ARGs. These conjugative plasmids have been reported especially in medically important bacteria [23], and now also in P. gergoviae. Curiously, the first report of AMR-related plasmids in P. gergoviae was published in the United States in 2022. In this report, large IncA and IncHI1B/FIB plasmids carrying blaKPC-4 and blaNDM-1, respectively, were identified as referring to BWH-P-GER-1 and BWH-P-GER-6 strains [24]. Furthermore, IncX3 plasmids have great conjugation ability and high stability, and have been identified as carrying ESBL- or carbapenemase-encoding genes in P. gergoviae strains, reinforcing the importance of P. gergoviae in the dissemination of ARGs within Enterobacteriaceae [25].
Recently, single- and multireplicon plasmids carrying mcr-9 or mcr-10 have been identified in different sources. Consequently, mcr-positive and carbapenem-resistant Enterobacteriaceae strains have emerged worldwide [26,27] and were also described in this study. In addition, the co-occurrence of ESBL- and carbapenemase-encoding genes in P. gergoviae strains has been detected, demonstrating a similar genotype to other medically important enterobacteria [28]. Based on the P. gergoviae resistome, it is possible to predict antimicrobial susceptibility and highlight the multidrug resistance phenotype. Although some mcr genes do not confer resistance to polymyxins in certain bacterial species [29], good correlation between the genotypic and phenotypic resistance for other antimicrobials was documented [30], including in clinical P. gergoviae strains [24]. Furthermore, antimicrobial susceptibility may vary substantially from predictions and may even be underestimated since mutational mechanisms were not investigated in this study, suggesting that MDR strains could still be classified as extensively drug-resistant or pandrug-resistant.
Previous studies have reported that P. gergoviae causes opportunistic infections and even nosocomial outbreaks [11,31,32]; however, virulence genotyping strengthened that P. gergoviae is a low-virulence bacterium [24], although virulence databases are still limited in the case of this species. Indeed, the role of virulence genes in the pathogenicity of this species needs to be further investigated. Worryingly, P. gergoviae-derived infections, especially those caused by carbapenem-resistant strains, can be difficult to treat, representing a significant healthcare challenge and serious public health concern [33]. As described in high-risk clones of E. coli and K. pneumoniae [34], the convergence of virulence and AMR did not occur in P. gergoviae. In addition, no species- and host-specific genes related to AMR or virulence were identified, evidencing an acquired and diverse genetic background.
5. Conclusions
In summary, this study provides the first genomic overview of P. gergoviae strains circulating at the One Health interface. Comparative analysis revealed a highly diverse population that harbored an arsenal of ARGs. Conjugative plasmids were identified as carrying clinically important ARGs embedded in conserved genetic contexts, evidencing their capture of other medically important bacteria. In this sense, P. gergoviae is emerging as a new multidrug-resistant species belonging to the Enterobacteriaceae family. Therefore, more studies investigating the genetic background of AMR in non-clinically relevant enterobacteria should be encouraged and continuous epidemiological genomic surveillance of P. gergoviae should be required.
Acknowledgments
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; grant no. 150712/2022-7 and 304905/2022-4) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)—Finance code 001 for fellowships.
Author Contributions
Conceptualization, formal analysis, investigation, data curation, writing—original draft, writing—review & editing, J.P.R.F. Conceptualization, supervision, project administration, writing—review & editing, E.G.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [grant no. 2021/01655-7].
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Dadgostar P. Antimicrobial Resistance: Implications and Costs. Infect. Drug. Resist. 2019;12:3903–3910. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golli A.L., Cristea O.M., Zlatian O., Glodeanu A.D., Balasoiu A.T., Ionescu M., Popa S. Prevalence of Multidrug-Resistant Pathogens Causing Bloodstream Infections in an Intensive Care Unit. Infect. Drug. Resist. 2022;15:5981–5992. doi: 10.2147/IDR.S383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsson D.G.J., Flach C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022;20:257–269. doi: 10.1038/s41579-021-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samtiya M., Matthews K.R., Dhewa T., Puniya A.K. Antimicrobial Resistance in the Food Chain: Trends, Mechanisms, Pathways, and Possible Regulation Strategies. Foods. 2022;11:2966. doi: 10.3390/foods11192966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velazquez-Meza M.E., Galarde-López M., Carrillo-Quiróz B., Alpuche-Aranda C.M. Antimicrobial resistance: One Health approach. Vet. World. 2022;15:743–749. doi: 10.14202/vetworld.2022.743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner D.J., Richard C., Steigerwalt A.G., Asbury M.A., Mandel M. Enterobacter gergoviae sp. nov.: A New Species of Enterobacteriaceae Found in Clinical Specimens and the Environment. Syst. Appl. Microbiol. 1980;30:1–6. doi: 10.1099/00207713-30-1-1. [DOI] [Google Scholar]
- 7.Brady C., Cleenwerck I., Venter S., Coutinho T., De Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst. Appl. Microbiol. 2013;36:309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Davin-Regli A., Chollet R., Bredin J., Chevalier J., Lepine F., Pagès J.M. Enterobacter gergoviae and the prevalence of efflux in parabens resistance. J. Antimicrob. Chemother. 2006;57:757–760. doi: 10.1093/jac/dkl023. [DOI] [PubMed] [Google Scholar]
- 9.Leão-Vasconcelos L.S., Lima A.B., Costa Dde M., Rocha-Vilefort L.O., Oliveira A.C., Gonçalves N.F., Vieira J.D., Prado-Palos M.A. Enterobacteriaceae isolates from the oral cavity of workers in a Brazilian oncology hospital. Rev. Inst. Med. Trop. Sao Paulo. 2015;57:121–127. doi: 10.1590/S0036-46652015000200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganeswire R., Thong K.L., Puthucheary S.D. Nosocomial outbreak of Enterobacter gergoviae bacteraemia in a neonatal intensive care unit. J. Hosp. Infect. 2003;53:292–296. doi: 10.1053/jhin.2002.1371. [DOI] [PubMed] [Google Scholar]
- 11.Freire M.P., de Oliveira Garcia D., Cury A.P., Spadão F., Di Gioia T.S.R., Francisco G.R., Bueno M.F.C., Tomaz M., de Paula F.J., de Faro L.B., et al. Outbreak of IMP-producing carbapenem-resistant Enterobacter gergoviae among kidney transplant recipients. J. Antimicrob. Chemother. 2016;71:2577–2585. doi: 10.1093/jac/dkw165. [DOI] [PubMed] [Google Scholar]
- 12.Satlin M.J., Jenkins S.G., Chen L., Helfgott D., Feldman E.J., Kreiswirth B.N., Schuetz A.N. Septic shock caused by Klebsiella pneumoniae carbapenemase-producing Enterobacter gergoviae in a neutropenic patient with leukemia. J. Clin. Microbiol. 2013;51:2794–2796. doi: 10.1128/JCM.00004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida A.C.S., de Castro K.K.A., Fehlberg L.C.C., Gales A.C., Vilela M.A., de Morais M.M.C. Carbapenem-resistant Enterobacter gergoviae harbouring blaKPC-2 in Brazil. Int. J. Antimicrob. Agents. 2014;44:369–370. doi: 10.1016/j.ijantimicag.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 15.Arredondo-Alonso S., Rogers M.R.C., Braat J.C., Verschuuren T.D., Top J., Corander J., Willems R.J.L., Schürch A.C. mlplasmids: A user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb. Genom. 2018;4:e000224. doi: 10.1099/mgen.0.000249. Erratum in Microb Genom. 2019, 5, e000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prestinaci F., Pezzotti P., Pantosti A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health. 2015;109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch J.P., 3rd, Clark N.M., Zhanel G.G. Escalating antimicrobial resistance among Enterobacteriaceae: Focus on carbapenemases. Expert Opin. Pharmacother. 2021;22:1455–1473. doi: 10.1080/14656566.2021.1904891. Erratum in Expert Opin. Pharmacother. 2021, 22, i–ii. [DOI] [PubMed] [Google Scholar]
- 19.Zou H., Berglund B., Wang S., Zhou Z., Gu C., Zhao L., Meng C., Li X. Emergence of blaNDM-1, blaNDM-5, blaKPC-2 and blaIMP-4 carrying plasmids in Raoultella spp. in the environment. Environ. Pollut. 2022;306:119437. doi: 10.1016/j.envpol.2022.119437. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q., Wang X., Chen C., Zhao L., Ma J., Dong K. Analysis of NDM-1 and IMP-8 carbapenemase producing Raoultella planticola clinical isolates. Acta Microbiol. Immunol. Hung. 2023;70:193–198. doi: 10.1556/030.2023.02078. [DOI] [PubMed] [Google Scholar]
- 21.El Salabi A., Walsh T.R., Chouchani C. Extended spectrum β-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gram-negative bacteria. Crit. Rev. Microbiol. 2013;39:113–122. doi: 10.3109/1040841X.2012.691870. [DOI] [PubMed] [Google Scholar]
- 22.Verdial C., Serrano I., Tavares L., Gil S., Oliveira M. Mechanisms of Antibiotic and Biocide Resistance That Contribute to Pseudomonas aeruginosa Persistence in the Hospital Environment. Biomedicines. 2023;11:1221. doi: 10.3390/biomedicines11041221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozwandowicz M., Brouwer M.S.M., Fischer J., Wagenaar J.A., Gonzalez-Zorn B., Guerra B., Mevius D.J., Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 24.Weiss Z.F., Hoffmann M., Seetharaman S., Taffner S., Allerd M., Luo Y., Pearson Z., Baker M.A., Klompas M., Bry L., et al. Nosocomial Pluralibacter gergoviae Isolates Expressing NDM and KPC Carbapenemases Characterized Using Whole-Genome Sequencing. Antimicrob. Agents Chemother. 2022;66:e0109322. doi: 10.1128/aac.01093-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X., Chen R., Wang Q., Li C., Ge H., Qiao J., Li Y. Global prevalence, characteristics, and future prospects of IncX3 plasmids: A review. Front. Microbiol. 2022;13:979558. doi: 10.3389/fmicb.2022.979558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macesic N., Blakeway L.V., Stewart J.D., Hawkey J., Wyres K.L., Judd L.M., Wick R.R., Jenney A.W., Holt K.E., Peleg A.Y. Silent spread of mobile colistin resistance gene mcr-9.1 on IncHI2 ‘superplasmids’ in clinical carbapenem-resistant Enterobacterales. Clin. Microbiol. Infect. 2021;27:1856.e7–1856.e13. doi: 10.1016/j.cmi.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H., Wang S., Wu Y., Dong N., Ju X., Cai C., Li R., Li Y., Liu C., Lu J., et al. Carriage of the mcr-9 and mcr-10 genes in clinical strains of the Enterobacter cloacae complex in China: A prevalence and molecular epidemiology study. Int. J. Antimicrob. Agents. 2022;60:106645. doi: 10.1016/j.ijantimicag.2022.106645. [DOI] [PubMed] [Google Scholar]
- 28.Marsh J.W., Mustapha M.M., Griffith M.P., Evans D.R., Ezeonwuka C., Pasculle A.W., Shutt K.A., Sundermann A., Ayres A.M., Shields R.K., et al. Evolution of Outbreak-Causing Carbapenem-Resistant Klebsiella pneumoniae ST258 at a Tertiary Care Hospital over 8 Years. mBio. 2019;10:e01945-19. doi: 10.1128/mBio.01945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyson G.H., Li C., Hsu C.H., Ayers S., Borenstein S., Mukherjee S., Tran T.T., McDermott P.F., Zhao S. The mcr-9 Gene of Salmonella and Escherichia coli Is Not Associated with Colistin Resistance in the United States. Antimicrob. Agents Chemother. 2020;64:e00573-20. doi: 10.1128/AAC.00573-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruppé E., Cherkaoui A., Charretier Y., Girard M., Schicklin S., Lazarevic V., Schrenzel J. From genotype to antibiotic susceptibility phenotype in the order Enterobacterales: A clinical perspective. Clin. Microbiol. Infect. 2020;26:643.e1–643.e7. doi: 10.1016/j.cmi.2019.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Rashid A.M.A., Lim C.T.S. Enterobacter gergoviae peritonitis in a patient on chronic ambulatory peritoneal dialysis-first reported case. Malaysian J. Med. Health Sci. 2017;13:67–69. [Google Scholar]
- 32.Khashei R., Edalati Sarvestani F., Malekzadegan Y., Motamedifar M. The first report of Enterobacter gergoviae carrying blaNDM-1 in Iran. Iran J. Basic Med. Sci. 2020;23:1184–1190. doi: 10.22038/ijbms.2020.41225.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suay-García B., Pérez-Gracia M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics. 2019;8:122. doi: 10.3390/antibiotics8030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bueris V., Sellera F.P., Fuga B., Sano E., Carvalho M.P.N., Couto S.C.F., Moura Q., Lincopan N. Convergence of virulence and resistance in international clones of WHO critical priority Enterobacterales isolated from Marine Bivalves. Sci. Rep. 2022;12:5707. doi: 10.1038/s41598-022-09598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.