Abstract
Using a constant-depth film fermentor, we have grown a six-membered biofilm community with a bacterial composition similar to that found in supragingival dental plaque. Cryosectioning revealed the distribution of bacteria throughout the biofilm. Exposure to 0.2% chlorhexidine for up to 5 min had little effect on biofilm viability.
Most of the bacteria comprising the normal microflora of the oral cavity of humans are present as biofilms (dental plaques) on tooth surfaces. Unless these biofilms are repeatedly removed they may initiate some of the most prevalent infectious diseases of humans—caries and the periodontal diseases. Because of the difficulty of ensuring adequate removal of plaque by mechanical means, there is increasing interest in the use of antimicrobial agents to supplement these mechanical approaches to the control of dental plaque (12, 17). Unfortunately, most laboratory studies of the potential effectiveness of such agents have employed methods which are appropriate for systemic infections (e.g., determination of MICs) but not for diseases in which the causative organisms are present as biofilms (18, 19). It has been well established that the susceptibility of extraoral bacteria to antimicrobial agents is much lower when they comprise a biofilm than when they are in an aqueous suspension (3). However, few studies have investigated the susceptibility of multispecies biofilms of oral (or, indeed, nonoral) bacteria to antimicrobial agents. The purposes of the present study were to grow multispecies biofilms comprised of bacteria normally found in supragingival dental plaque and to determine their bacterial compositions and their susceptibilities to chlorhexidine gluconate (CHG).
The organisms used in the study were Streptococcus sanguis NCTC 10904, Streptococcus mutans NCTC 10449, Streptococcus oralis NCTC 11427, Actinomyces naeslundii NCTC 10951, Neisseria subflava ATCC A1078, and Veillonella dispar NCTC 11831. The inoculum was prepared by growing confluent cultures of each organism on blood agar plates and transferring them aseptically into 10 ml of nutrient broth (Oxoid Ltd., Basingstoke, United Kingdom [UK]) containing 10% glycerol (BDH Chemicals, Poole, UK). Aliquots (1.0 ml) of the resulting mixed suspension were stored at −70°C.
A constant-depth film fermentor (CDFF) fed with a complex medium containing hog gastric mucin (CM) (13) was used to produce biofilms in a manner similar to that described previously (20). The biofilms were grown on bovine enamel discs (5-mm diameter) recessed to a depth of 300 μm. The CDFF was inoculated with the mixed bacterial suspension in 5 ml of CM which was aseptically added to the rotating pans via the sample port. After 1 h, the CDFF was connected to a reservoir of CM (flow rate of 0.72 liter per day—the mean resting flow rate of saliva in humans [7, 10]).
Sampling pans were removed from the CDFF daily, and the number of viable bacteria per biofilm was determined as follows. The enamel discs were gently removed and vortex mixed for 60 s in 10 ml of CM. Serial dilutions were prepared in CM, and 20-μl aliquots of each dilution were plated in quadruplicate onto a range of selective media and Wilkins-Chalgren agar (Oxoid) containing 5% unlysed horse blood. The selective media consisted of mitis salivarius agar (Difco Laboratories, Detroit, Mich.) for streptococci, Veillonella agar (Difco) for V. dispar, cadmium fluoride-acriflavine-tellurite agar (22) for A. naeslundii, and Thayer-Martin agar (Oxoid) for N. subflava. Plates were incubated anaerobically for 4 days at 37°C, except for those containing Thayer-Martin agar (aerobic, 4 days, 37°C). Colonies growing on each of the plates were Gram stained to confirm their identities and counted; the different streptococcal species were distinguished on the basis of colonial morphology.
The cryosectioning methodology was based on the work of Kinniment (8). The fermentor pan was placed in 5 ml of a 25% dextran solution, a cryoprotectant (1), in CM for 2 h at 37°C before being removed and placed in 5 ml of 8% formaldehyde for 20 min. Discs were removed and placed onto a thin layer of OCT embedding compound (Raymond A. Lamb, London, UK) on a cryostat chuck. This was then placed in a −70°C freezer for 20 min until the OCT compound and the biofilm were frozen. Additional OCT compound was then placed over the top of the biofilm so that the entire sample was embedded. Horizontal sections (30 μm thick) were then cut from the biofilm, and each sample was placed in phosphate-buffered saline with cooled forceps. In preliminary studies, the ability of the BacLight live/dead viability kit (Molecular Probes Inc., Eugene, Oreg.) to distinguish between viable and nonviable cells of each of the six organisms was ascertained before and after heat killing (10 min, 100°C). The viability stain protocol was carried out as described previously (14). Sample pans were removed, and the susceptibility of the biofilms to exposure to 0.2% (wt/vol) CHG for 1, 5, and 60 min was determined as described previously (13). The numbers of surviving organisms were determined by viable-cell counting as described above. Biofilms for electron microscopy were prepared as follows. Samples were fixed in 3% glutaraldehyde in 0.1 M sodium cacodylate buffer at 4°C overnight. The specimens were postfixed in 1% osmium tetroxide at 4°C for 2 h before being dehydrated in a graded series of alcohol (20 to 100%; 15-min application time). The biofilms were embedded in fresh Araldite CY212 (Agar Scientific, Stanstead, United Kingdom), and 90- to 100-nm sections were cut. The sections were stained with lead citrate and uranyl acetate and viewed with a JEOL 100 CX transmission electron microscope.
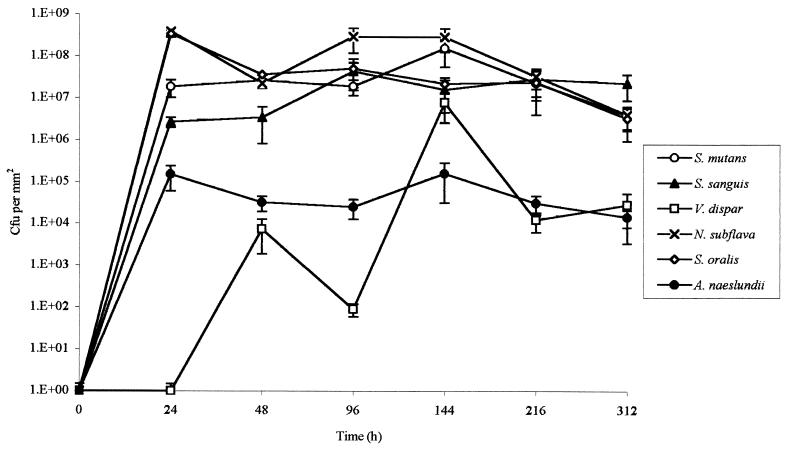
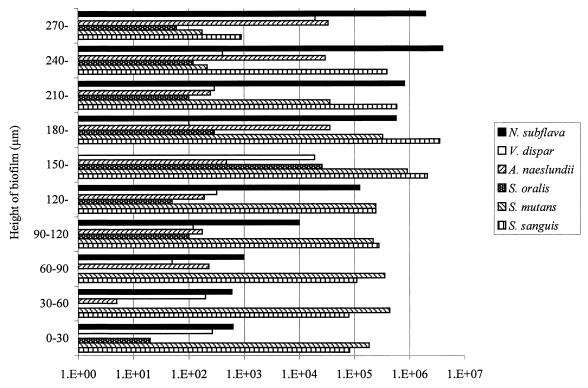
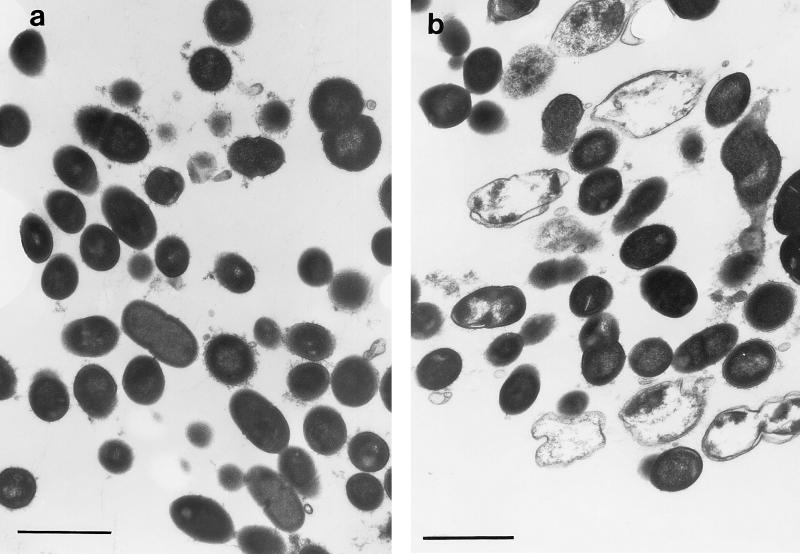
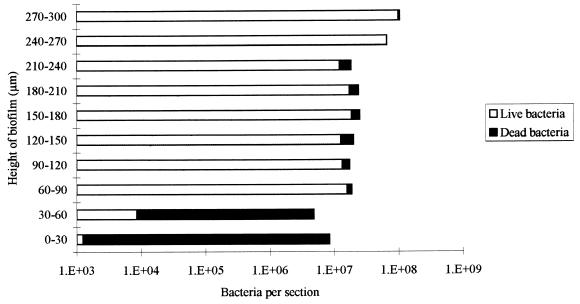
Growth of the multispecies biofilms in the CDFF is shown in Fig. 1, which is based on data from a typical run. The total viable count of the biofilms reached a maximum after approximately 24 h, and the viable-cell density at this point was 7 × 108 CFU/mm2. Table 1 shows the total viable count and the proportions of each species in the biofilms after 216 h in three separate runs. From this it can be seen that streptococci constituted the predominant organisms in the biofilms. N. subflava was the next most prevalent organism, while A. naeslundii and V. dispar generally comprised smaller proportions of the total viable count. The distribution of the various bacterial species throughout the biofilm as revealed by viable-cell counting of 30-μm sections through the biofilm is shown in Fig. 2, which shows the results from a representative biofilm. From this it can be seen that the proportion of the obligate aerobe N. subflava in each of the sections decreased, in a fairly regular manner, with biofilm depth. In the section from the biofilm-air interface (i.e., a biofilm height of 270 to 300 μm) this organism comprised 97.5% of the total viable count, whereas at the base of the film it accounted for only 0.002% of the viable count. Only in the three uppermost sections (i.e., a biofilm height of 210 to 300 μm) of the biofilm did it account for more than 50% of the total viable count. In contrast, no general trend could be discerned for the obligate anaerobe V. dispar, which comprised only a small proportion of the viable organisms in each section of the biofilm and was distributed fairly uniformly throughout the biofilm. There was no trend towards increasing proportions of the organism in the deeper layers of the biofilm. The proportion of V. dispar in this particular biofilm was lower than those generally found in other biofilms, but the distribution of the organism within the other biofilms examined followed a pattern similar to that described above. The distribution of A. naeslundii throughout the biofilm was similar to that found for V. dispar. Two of the three streptococcal species (S. sanguis and S. mutans) collectively comprised the dominant organisms in each section except in the uppermost 60 μm of the biofilm, where N. subflava predominated S. oralis generally comprised a lower proportion of the biofilms than the other streptococci. Transmission electron micrographs of sections through the biofilms revealed high proportions of gram-negative cocci (presumably Neisseria spp.) at the biofilm-air surface (Fig. 3a), while gram-positive cocci were distributed throughout. “Ghost” cells could be observed at the base of the biofilm (Fig. 3b) together with a predominance of gram-positive cocci. The results of vital staining of the biofilm are shown in Fig. 4, from which it can be seen that the uppermost 240 μm of the biofilm consisted mainly of live bacteria while the two innermost 30-μm sections contained mainly dead cells.
FIG. 1.
Growth of individual species comprising a multispecies biofilm formed on bovine enamel discs with CM as the sole nutrient source. Error bars represent standard deviations (n = 4).
TABLE 1.
Total viable counts and proportions of each species at 216 h in biofilmsa
| Organism | Run 1
|
Run 2
|
Run 3
|
Mean % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CFU/mm2 | SD | % of total CFU | CFU/mm2 | SD | % of total CFU | CFU/mm2 | SD | % of total CFU | ||
| N. subflava | 3.4 × 107 | 1.2 × 107 | 25.7 | 7.8 × 107 | 5.9 × 107 | 45.3 | 3.4 × 107 | 1.2 × 107 | 16.4 | 29.1 |
| A. naeslundii | 7.1 × 106 | 1.1 × 106 | 5.4 | 4.1 × 106 | 1.5 × 106 | 2.4 | 7.1 × 107 | 1.1 × 107 | 34.2 | 14 |
| V. dispar | 3.5 × 106 | 1.5 × 106 | 2.6 | 3.1 × 107 | 9.2 × 106 | 18 | 3.5 × 106 | 1.5 × 106 | 1.7 | 7.4 |
| S. sanguis | 4 × 107 | 1 × 107 | 30.3 | 2 × 107 | 5.2 × 106 | 11.6 | 4 × 107 | 1 × 107 | 19.3 | 20.4 |
| S. mutans | 4.8 × 107 | 1.5 × 107 | 36 | 1.8 × 107 | 6.1 × 106 | 10.4 | 4.8 × 107 | 1.5 × 107 | 23 | 23.1 |
| S. oralis | 1.1 × 105 | 3.7 × 104 | 0.1 | 2.2 × 107 | 9.3 × 106 | 12.6 | 1.1 × 107 | 3.7 × 106 | 5.5 | 6.1 |
The biofilms were formed during three separate runs (the number of biofilms per run was 4).
FIG. 2.
Viable counts of constituent bacteria in 30-μm sections of the 300-μm-thick biofilms. Numbers on the x axis are CFU per square millimeter.
FIG. 3.
Transmission electron micrographs of a transverse section of biofilm (216 h) at the biofilm-air interface (a) and at the biofilm-substratum interface (b). Bars represent 2 μm.
FIG. 4.
Relative proportions of live and dead bacteria in 30-μm sections of a 300-μm-thick biofilm at 216 h.
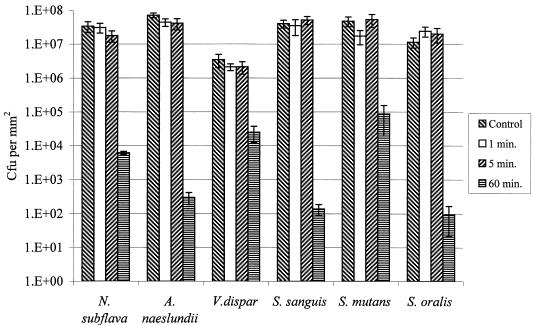
The effect on the viabilities of the various species comprising biofilms is shown in Fig. 5. Exposure to 0.2% CHG for 1 or 5 min had no statistically significant effect on the viability of any of the six species in the biofilms. However, a 60-min exposure resulted in significant kills of all of the organisms. S. sanguis was the most susceptible, with approximately a 5-log10-unit reduction, whereas V. dispar was the least susceptible, with a 2-log10-unit reduction. Both S. oralis and A. naeslundii displayed susceptibilities similar to that of S. sanguis, whereas S. mutans and N. subflava had susceptibilities intermediate between those of S. sanguis and V. dispar.
FIG. 5.
Response of individual species comprising multispecies biofilms to various periods of exposure to 0.2% CHG. Error bars represent standard deviations (n = 4).
Despite the tremendous interest being shown in the use of antimicrobial agents for the control and treatment of dental-plaque-related diseases, few studies of the antimicrobial susceptibility of biofilms of oral bacteria have been published (19). In contrast, the susceptibility of biofilms of nonoral bacteria to antimicrobial agents has received considerable attention (4), and many studies have confirmed that the sessile forms of these bacteria are considerably less susceptible to antibiotics and antiseptics than their corresponding planktonic forms (3). Meaningful laboratory evaluation of agents for use in the prevention and treatment of plaque-related diseases requires the use of models which mimic more closely the situation in the oral cavity. We have employed the CDFF for this purpose and have used it to determine the susceptibility of monospecies biofilms of oral bacteria, and microcosm dental plaques, to antimicrobial agents (14). In this study we have used the CDFF to investigate the composition and antimicrobial susceptibility of defined, multispecies biofilms of oral bacteria. The predominant organisms comprising the biofilms were found to be streptococci, with lower proportions (although still substantial) of N. subflava, A. naeslundii, and V. dispar. The biofilms, therefore, were similar in composition to supragingival dental plaques found in vivo, bearing in mind the enormous variation in composition encountered from site to site within an individual and among the same sites in different individuals (10, 11). Despite the fact that the inoculum in each case was derived from the same source, repeat runs, not surprisingly, resulted in biofilms with different compositions. Such variations are most likely to be the result of unavoidable slight differences between runs in terms of the relative proportions of bacteria in the inoculum, composition of the CM, and flow rates, etc. Such between-run variations are not surprising and have been reported for other attempts to model mixed communities of oral bacteria (6, 9). Vital staining of biofilm sections revealed a high proportion of dead cells at the base of the biofilm, and the total viable count was also much lower at the biofilm base than at the biofilm-air interface. Electron microscopy of these biofilms revealed that the basal layers contained high proportions of ghosts, implying a preponderance of dead, or compromised, cells. Such observations have been described for in vivo dental plaques, a layer of ghost cells being present at the plaque-enamel interface (16). The distribution of the various species within the biofilms, however, revealed some surprises. It might have been predicted that the only anaerobic species in the community, V. dispar, would have been found mainly in the basal layers, where anaerobic conditions would be likely to prevail. However, while this organism was found at the bases of the biofilms, it was also abundant in the upper layers, i.e., at the biofilm-air interface. As was expected, the only obligate aerobe, N. subflava, also predominated in these layers and comprised 97.5 and 90.5% of the viable count in the two uppermost layers. Other studies have reported similar findings (15), and the distribution of this organism is most likely to be related to the establishment of a decreasing oxygen gradient through these deep biofilms due to diffusion limitations coupled with oxygen utilization in the upper layers (21). Active consumption of oxygen by N. subflava may have provided suitable atmospheric conditions and redox potentials to enable survival of V. dispar in the upper layers of the biofilms. The protection afforded by N. subflava to oral anaerobes in biofilms has been demonstrated previously by Bradshaw et al. (2).
The well-known refractory reaction of biofilms to antimicrobial agents was evident in this study, where exposure of the biofilms to chlorhexidine concentrations as high as 0.2% (the maximum concentration used in mouthwashes) for 1 min had no significant effect on bacterial viability. Substantial reductions in bacterial viability resulted only from a 60-min exposure to the antimicrobial agent, illustrating that the effectiveness of an agent for use in the control of plaque-related diseases will depend very much on its substantivity, i.e., its ability to be retained in the oral cavity following the application of a mouthwash or a toothpaste (5). Interest in the use of antimicrobial agents for the treatment of plaque-related diseases, such as caries and periodontitis, has generated a need for laboratory models for the evaluation of agents effective against oral bacterial biofilms, and the CDFF has proved to be useful in this respect. Most studies, however, have employed single-species biofilms of oral bacteria rather than the complex communities characteristic of supragingival plaques. The results of this study have demonstrated that the CDFF can be used to grow multispecies biofilms under conditions similar to those prevailing in the oral cavity, resulting in biofilms in which the bacterial composition, and the spatial distribution of the various species within these biofilms, was similar to that found in supragingival dental plaques. Such biofilms were found to be refractory to chlorhexidine, a commonly used oral antiseptic. As well as being useful in studies of the susceptibility of multispecies biofilms to antimicrobial agents, this model can also be used to evaluate antiplaque agents and to investigate community interactions in such biofilms.
Acknowledgments
This work was supported by SmithKline Beecham Consumer Healthcare.
REFERENCES
- 1.Ashwood-Smith M J, Warby C. Studies on the molecular weight and cryoprotective properties of polyvinylpyrrolidone and dextran with bacteria and erythrocytes. Cryobiology. 1971;8:453–464. doi: 10.1016/0011-2240(71)90036-8. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw D J, Homer K A, Marsh P D, Beighton D. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology. 1994;140:3407–3412. doi: 10.1099/13500872-140-12-3407. [DOI] [PubMed] [Google Scholar]
- 3.Brown M R, Allison W D G, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother. 1988;22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- 4.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. . (Review.) [DOI] [PubMed] [Google Scholar]
- 5.Cummins D, Creeth J E. Delivery of antiplaque agents from dentifrices, gels and mouthwash. J Dent Res. 1992;71:1439–1449. doi: 10.1177/00220345920710071601. [DOI] [PubMed] [Google Scholar]
- 6.Donoghue H D, Perrons C J. Establishment of defined mixed bacterial plaques on teeth in a laboratory microcosm (model mouth) Microb Ecol Health Dis. 1988;1:193–200. [Google Scholar]
- 7.Guyton A C. Human physiology and mechanisms of disease. Philadelphia, Pa: Saunders Co.; 1992. p. 486. [Google Scholar]
- 8.Kinniment S L. Cryosectioning of biofilm. In: Wimpenny J, Nichols W, Stickler D, Lappin-Scott H, editors. Bacterial biofilms and their control in medicine and industry. Cardiff, Wales, United Kingdom: Bioline; 1994. pp. 53–56. [Google Scholar]
- 9.Kinniment S L, Wimpenny J W, Adams D, Marsh P D. Development of a steady-state oral microbial biofilm community using the constant-depth film fermenter. Microbiology. 1996;142:631–638. doi: 10.1099/13500872-142-3-631. [DOI] [PubMed] [Google Scholar]
- 10.Lamb J F. Essentials of physiology. Oxford, United Kingdom: Blackwell Scientific; 1991. p. 93. [Google Scholar]
- 11.Marsh P D, Martin M. Oral microbiology. London, United Kingdom: Chapman & Hall; 1992. p. 70. [Google Scholar]
- 12.Marsh P D, Bradshaw D J. Microbiological effects of new agents in dentifrices for plaque control. Int Dent J. 1993;43:399–406. [PubMed] [Google Scholar]
- 13.Pratten, J., K. Wills, P. Barnett, and M. Wilson. In vitro studies of the effect of antiseptic-containing mouthwashes on the formation and viability of Streptococcus sanguis biofilms. J. App. Microbiol., in press. [DOI] [PubMed]
- 14.Pratten, J., A. W. Smith, and M. Wilson. Response of single species biofilms and microcosm dental plaques to pulsing with chlorhexidine. J. Antimicrob. Chemother., in press. [DOI] [PubMed]
- 15.Ritz H L. Fluorescent antibody staining of Neisseria, Streptococcus and Veillonella in frozen sections of human dental plaque. Arch Oral Biol. 1969;14:1073–1083. doi: 10.1016/0003-9969(69)90077-6. [DOI] [PubMed] [Google Scholar]
- 16.Schroeder H E. The structure and relationship of plaque to the hard and soft tissues: electron microscopic interpretation. Int Dent J. 1970;20:353–381. [PubMed] [Google Scholar]
- 17.Stanley A, Wilson M, Newman H N. The in vitro effects of chlorhexidine on subgingival plaque bacteria. J Clin Periodontol. 1989;16:259–264. doi: 10.1111/j.1600-051x.1989.tb01651.x. [DOI] [PubMed] [Google Scholar]
- 18.Thrower Y, Pinney R J, Wilson M. Susceptibilities of Actinobacillus actinomycetemcomitans biofilms to oral antiseptics. J Med Microbiol. 1997;46:425–429. doi: 10.1099/00222615-46-5-425. [DOI] [PubMed] [Google Scholar]
- 19.Wilson M. Susceptibility of oral bacterial biofilms to antimicrobial agents. J Med Microbiol. 1996;44:79–87. doi: 10.1099/00222615-44-2-79. [DOI] [PubMed] [Google Scholar]
- 20.Wilson M, Kpendema H, Noar J, Hunt N, Morden N. Corrosion of intra-oral magnets in the presence and absence of Streptococcus sanguis biofilms. Biomaterials. 1995;16:721–725. doi: 10.1016/0142-9612(95)99701-m. [DOI] [PubMed] [Google Scholar]
- 21.Wimpenny J W, Peters A, Scourfield M A. Modelling spatial gradients. In: Characklis W G, Wilderer P A, editors. Structure and function of biofilms. Chichester, United Kingdom: Wiley and Sons; 1989. pp. 111–127. [Google Scholar]
- 22.Zylber L J, Jordan H V. Development of a selective medium for detection and enumeration of Actinomyces viscosus and Actinomyces naeslundii in dental plaque. J Clin Microbiol. 1982;15:253–259. doi: 10.1128/jcm.15.2.253-259.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]