Abstract
After years of only low-resolution and partial assemblies, the entire human pre-initiation complex (PIC), including the large and flexible Mediator and TFIID complexes, has come into focus. Five recent papers from three different research groups have transformed our understanding of transcription initiation by RNA polymerase II.
Since the discovery of RNA polymerase II (pol II)1, structural data lagged behind the biochemical and cellular studies that established fundamental aspects of pol II function and its regulation by associated factors. Among myriad pol II-associated factors, a core set is required to direct it to transcription start sites on genomic DNA and to control pol II activity once bound to these sites. This core set of factors is known as the pre-initiation complex (PIC), comprised of TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, pol II, and Mediator2. The PIC is approximately 4.0 MDa in size and its Mediator, TFIID, and TFIIH components are both large and structurally dynamic. This presents many challenges for structural biologists, but these challenges have been overcome by the Cramer, He, and Xu labs, who report high-resolution structures of the complete human PIC for the first time3–7.
While important prior work established PIC structural models in yeast8–10, pol II transcription is more complex in humans, as human genomes possess diverse combinations of core promoter sequence motifs, and Mediator and TFIID contain subunits and domains not present in yeast. The recent structural elucidation of human PICs reveals that yeast PIC structures served as a good basic model, but structural differences were evident in each study. The most extensive structural changes between yeast and human Mediator-containing PICs were characterized by Chen and co-workers6. This may result from their use of PICs containing TFIID instead of TBP (as in Abdella et al.3 and Rengachari et al.7), and their analysis of a conformationally distinct Mediator complex.
Because the structure of the human Mediator complex remained poorly defined, each of the three new cryo-EM studies of Mediator-containing human PICs yielded new high-resolution data for Mediator that are in agreement with a recent structure of the mouse Mediator complex11, with a notable exception (see below). As expected from yeast and mouse Mediator structural data, human MED14 and MED17 are major structural scaffolds. Data from the Xu lab, however, revealed that the metazoan-specific MED26 subunit provides another potential structural hub within human Mediator because it interacts with up to nine other Mediator subunits, based upon crosslinking-mass spectrometry data6.
The Xu lab6 also reports a Mediator conformational state that is markedly distinct from reported structures from Abdella et al.3 and Rengachari et al7. This alternate structure, called MEDB because of a bent conformation for the “tail” region of Mediator, showed large-scale re-organization of the tail, with rotation and movements of 25Å - 55Å among domains within the MED16, MED23, and MED24 subunits. Furthermore, portions of the tail subunits MED25 and MED16 were disordered in the MEDB conformational state, such that they could not be resolved by cryo-EM. By contrast, MED16 and a portion of MED25 were structured in the MEDE structural state (E = extended tail domain). Structural data from the He lab3 represent the MEDE structural state, and is consistent with data from Rengachari et al.7, although the latter study did not include the “distal” tail subunits MED15, MED16, or MED23 – 25.
The Xu lab separately determined the complete MEDE structure, and linked the MEDB structural state to expression of isoform-1 of MED16. MEDE was found to contain MED16 isoform-2. The structural changes between MEDB and MEDE center around the MED16 subunit, but the differences between MED16 isoform-1 and MED16 isoform-2 are minor. MED16 is an 877-residue protein (isoform-1) and isoform-2 (841 residues) is identical except for about 40 residues at the C-terminus. Further verification is needed, but if the structural differences between MEDB and MEDE derive entirely from the distinct MED16 isoforms, this would represent an intriguing example of an alternatively spliced isoform that impacts the structure, and presumably the function, of Mediator.
Cryo-EM structures of Mediator-containing PICs published by Abdella et al., Chen et al., and Rengachari et al. are generally consistent3,6,7, despite the different purification protocols and PIC assembly methods used for sample preparation. Some differences that were apparent likely reflect differences in PIC composition (see below). Each lab reported a similar location and orientation of Mediator relative to pol II and the rest of the PIC (Fig. 1). Mediator interactions with the TFIIH-associated CDK7 kinase module (called the CAK, for CDK-activating kinase) were shown to involve the MED6 subunit and the hook domain, with each binding on opposite sides of the CAK. Each study showed similar, but not identical, Mediator-pol II structural interfaces with the pol II stalk. Mediator-pol II interactions were also observed with the pol II dock domain within RPB1, the pol II RPB3/11 subunits, RPB86, and with the C-terminal domain (CTD) of the largest pol II subunit, RPB13,6. Mediator interactions with the B-ribbon of TFIIB were also observed7, which connects directly to the pol II active site.
Figure 1: Comparison of recent PIC and Mediator-PIC structures.
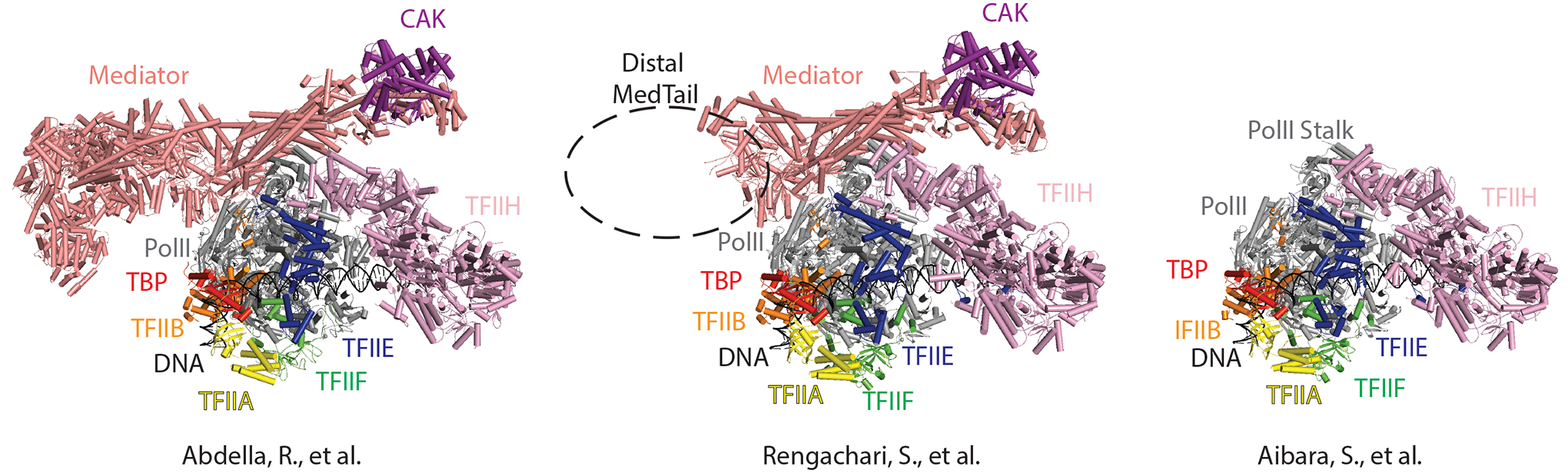
The left structure was reported by Abdella et al. (PDB 7LBM), the middle by Rengachari et al. (PDB 7NVR), and the right by Aibara et al. (PDB 7NVY). Labels are shaded according to the color of the factor. The dotted oval represents the missing Mediator “tail” subunits in the Rengachari et al. study. The most recently published TFIID-containing Mediator-PIC structure from Chen et al.6 was not yet available for comparison here.
The pol II CTD is intrinsically disordered and consists of 52 heptad repeats of the general consensus sequence YSPTSPS. Because of its disordered nature, structural data for the CTD is sparse. The He/Tjian and Xu labs were each able to resolve and model two CTD fragments into their Mediator-PIC cryo-EM structures, and the He/Tjian team was able to determine the directionality (i.e. N-terminus to C-terminus) of the repetitive CTD sequence. As shown in Figure 2, the pol II CTD binds Mediator and CTD heptad repeats C-terminal to the Mediator binding site are then stably positioned to bind the CDK7 active site for phosphorylation. The Mediator-dependent juxtaposition of the pol II CTD and the CAK domain of TFIIH provides a straightforward mechanism by which Mediator promotes pol II CTD phosphorylation by CDK7. Separately, the He/Tjian group showed that the CDK7 kinase was in an active conformation in the PIC3. CTD phosphorylation is fundamentally important to regulate RNA processing (e.g. capping, splicing, poly-adenylation), which ensures that pol II mRNA transcripts are stable and can be translated into proteins. Mediator is known to bind the unphosphorylated CTD, whereas CTD phosphorylation blocks Mediator binding12. TFIIH undergoes structural changes upon binding the PIC (Fig. 3A), including a Mediator-dependent stabilization of the CAK.
Figure 2: Model of the Pol II CTD path to Mediator and CDK7.
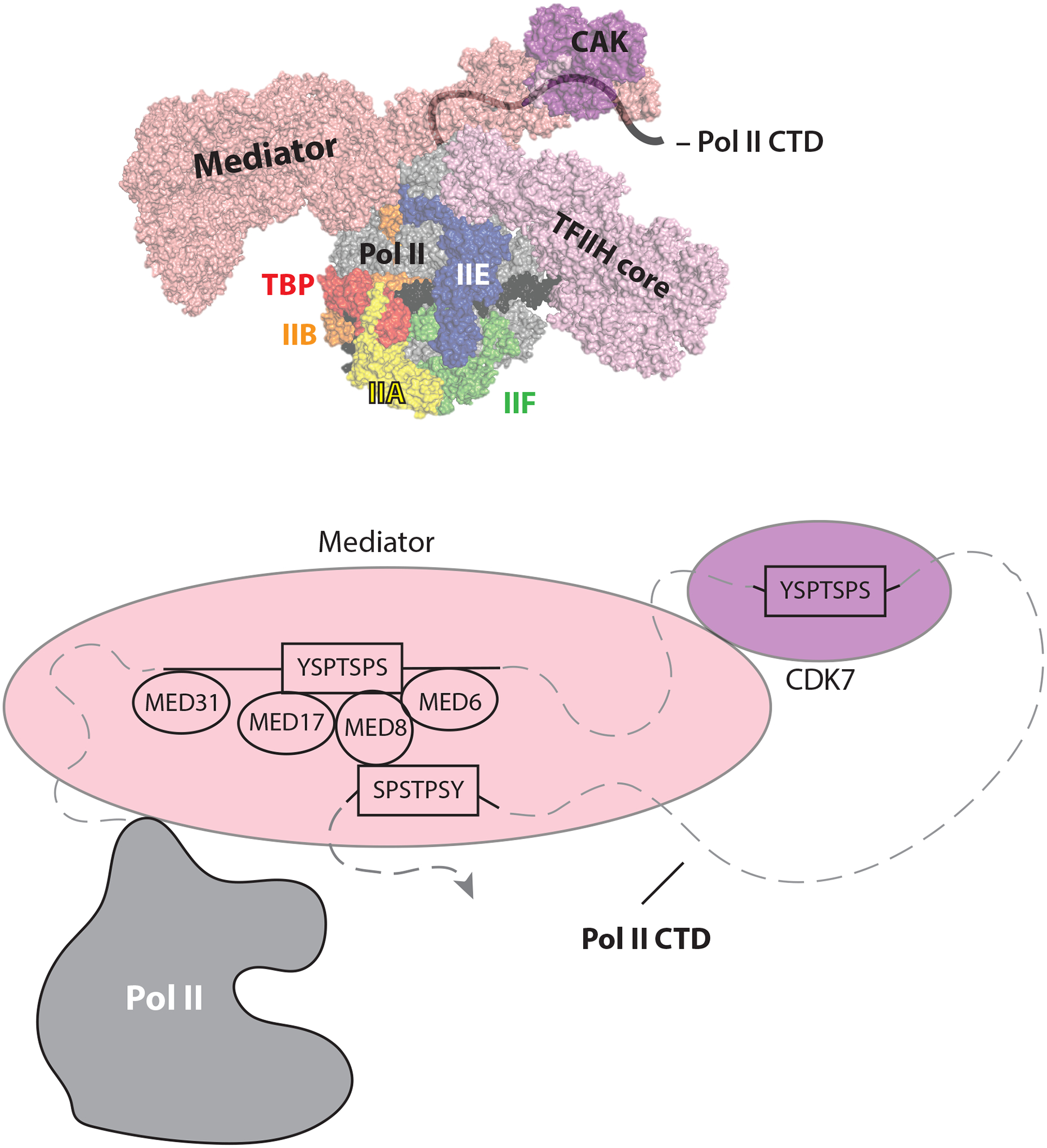
The top panel shows an overview of the Mediator-PIC with an approximate path of the Pol II CTD drawn in. The orientation and color scheme is identical to Abdella et al. (PDB 7LBM) shown in Figure 1. The bottom panel shows a schematic of the pol II CTD peptides mapped onto the Med-PIC structure, based upon data from Adella et al.3 and Chen et al6. It is not clear which of the 52 pol II CTD heptad repeats (YSPTSPS consensus) bind Mediator and CDK7 in the PIC complexes. The Mediator subunits that interact with the CTD are also indicated. Structured and modeled portions of the CTD are represented by solid lines and unstructured portions are represented with dashed lines.
Figure 3: Selected structural details important for pol II transcription initiation.
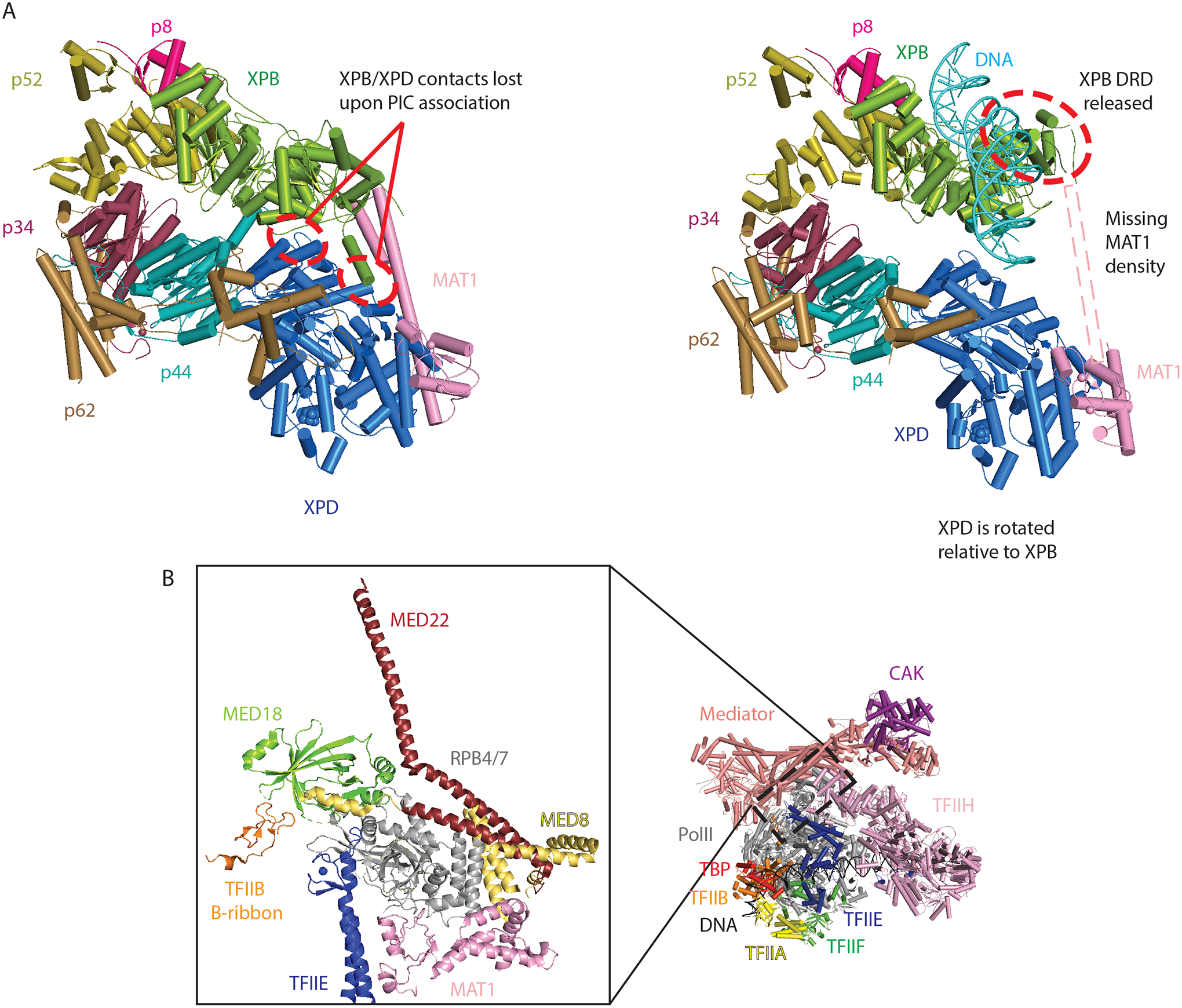
A) Comparison of free core TFIIH25 (left, PDB 6NMI) and PIC-bound TFIIH4 (right, PDB 7NVY) highlight structural transitions that occur upon DNA binding. DRD: damage recognition domain. B) A zoomed-in view of structural interactions centered on the pol II stalk (RPB4/7; PDB 7NVR), which undergoes structural changes upon promoter opening. Labels are colored according to the colors for each domain.
Perhaps the biggest gap in our understanding of PIC structure involves the 1.3 MDa TFIID complex. TFIID initiates PIC assembly by recognizing promoter DNA elements downstream of the transcription start site via its TAF1, TAF2, and TAF7 subunits, followed by structural re-organization to deposit the TATA-binding protein (TBP) at a DNA sequence upstream of the transcription start site5,13. The two recent papers from Chen and co-workers5,6 provide key insights about how TFIID orchestrates pol II transcription initiation. Here, we focus on the Mediator-bound PIC work, which includes TFIID6. TFIID was shown to bind TFIIH through two different sites. First, the TFIID subunit TAF1 contacts XPB and promotes its interaction with the pol II jaw (RPB5 subunit); second, the TFIID subunit TAF2 binds the p8 and p52 subunits of TFIIH. These interactions appear to direct the p8 and p52 subunits to move in a concerted fashion, together with XPB, and these movements are coordinated with a structural shift in the pol II stalk that coincides with structural changes in the E-ribbon domain of TFIIE and the Mediator subunit MED8 around the pol II stalk6. Collectively, this TFIID-induced re-organization alters Mediator orientation with respect to pol II, and establishes a new contact between the Mediator hook domain and XPB. As a result, XPB becomes an interaction hub, simultaneously contacting downstream promoter DNA, pol II, TFIID, and Mediator. This XPB contact network likely sets up subsequent structural transitions required for transcription initiation and pol II promoter escape.
In an article that accompanies their TBP-based Mediator-bound PIC structure7, the Cramer group determined high-resolution PIC structures with both closed and open promoter DNA4. Although these PIC complexes lacked TFIID or Mediator, the data reveal structural changes that are likely essential for remodeling PIC structure to allow pol II to “escape” the promoter and elongate a transcript. The open promoter complex was generated simply by addition of the ATP analog ADP•BeF3, which bound the TFIIH XPB subunit that possesses ATPase and DNA translocase activity14. Unlike ATP, ADP•BeF3 cannot be hydrolyzed by XPB, yet its binding was sufficient to partially open the DNA template near the transcription start site4. This suggests that a single ATP hydrolysis/translocation step is sufficient to open human pol II promoters, in agreement with recent biochemical results15. Interestingly, structural data for the open complex revealed that the pol II clamp closes, and this coincides with a shift in position of the pol II stalk and disruption of MAT1 interaction with the stalk, releasing the MAT1 contact.
Many interesting new hypotheses have been raised by this outstanding set of papers3–7. Below, we briefly touch upon a few that pull together some common themes among the studies, and speculate about potential implications for pol II transcription.
Abdella et al. and Chen et al. each propose a CTD tracking or “gating” mechanism by which Mediator can bind and release the unphosphorylated CTD to enable iterative CDK7-catalyzed CTD phosphorylation. This mechanism is supported by the observation of 2 different CTD-bound structural states in the TBP- vs. TFIID-containing PICs. With TFIID-containing PICs, the Mediator head and middle modules undergo a more substantial structural shift that forms a “sandwich” around the CTD, whereas these structural transitions are incomplete in the TBP-based PICs. The distinct CTD-bound structures may represent functional intermediates that provide a means to thread the CTD through the Mediator “gateway” to CDK7. Two different Mediator-bound segments (i.e. non-sequential; Fig. 2) of the 52-heptad repeat CTD sequence were resolved by Chen et al.6, and they reasonably speculate that threading may involve transient release of one or both segments. Alternatively, the multi-repeat CTD binding represented by the “sandwiched” Mediator structural state may simply act to localize and properly orient the large, disordered CTD for efficient phosphorylation by CDK7. The Mediator-CTD binding affinity (KD, measured with yeast Mediator) is about 1 nM, with off rates of approximately 8.0E−5/second8, which corresponds to a residence time of 3+ hours. Consistent with these results, Chen et al.6 showed that Mediator engulfs the CTD to form the “sandwiched” interaction, through structural changes centered around the hook and knob region. This effectively buries at least four CTD repeats in a hydrophobic environment, which is typical for high-affinity, stable protein-protein interactions.
Regardless of the precise mechanism(s) by which Mediator promotes pol II CTD phosphorylation, it is apparent that Mediator releases the CTD at some point during transcription initiation. How might this happen? Some possible clues are provided by the open promoter PIC structure from Aibara et al.4, in which the pol II clamp and stalk undergo conformational changes. The pol II stalk is a tightly packed interaction hub in the Mediator-containing PIC (Fig. 3B), and structural shifts at the stalk could trigger conformational changes that promote opening of the “CTD sandwich” formed by the Mediator hook and knob domains. Coincident with this, structural shifts in XPB are required to translocate DNA to form the open complex4. Data from Chen et al.6 show that XPB is another interaction hub in the PIC, and conformational changes in XPB could affect not only DNA structure, but also impact TFIID and Mediator structure as well.
An important yet challenging future direction is to assess whether distinct PIC structural intermediates are formed during activated transcription; that is, transcription controlled by sequence-specific, DNA-binding transcription factors (TFs). TFs were not explicitly included or resolved in the present PIC structures, but this may reflect structural disorder. For the PIC factors, potential distinct TF-dependent activation mechanisms may be especially sensitive to Mediator, because it is stably bound by many different TFs and because TFIID may be less frequently targeted16. Mediator also appears to alter its structural state upon TF binding17, but this remains controversial because high-resolution data are generally lacking18.
In cells, accumulating evidence supports TF-dependent “transcriptional bursting”, in which multiple pol II complexes initiate multiple rounds of transcription from the same promoter19. This mechanism requires rapid pol II promoter escape, promoter-proximal pause release, and re-initiation20,21, which implies that Mediator-CTD interactions will be transient and XPB-dependent promoter opening accelerated. Structural data from Rengachari et al.7 revealed an interaction between the MED18 Mediator subunit and the B-ribbon of TFIIB (Fig. 3B). Adjacent motifs in TFIIB reside in the pol II active site and help stabilize the open complex via interactions with single-stranded DNA. Prior work suggested that transcriptional bursting was Mediator dependent22. We speculate that one way for Mediator to promote rapid pol II re-initiation would be to act through TFIIB to maintain an open, “pre-melted” DNA template at the transcription start site.
Finally, we note that substantial portions of the TFIID- and Mediator-containing PICs remain unresolved due to conformational flexibility or intrinsic disorder; such unstructured regions may become ordered upon binding other proteins (e.g. TFs) or may help regulate PIC partitioning into molecular condensates23. Crosslinking-mass spectrometry data from Chen et al.6 suggest that disordered regions (i.e. not resolved in these studies) mediate additional protein-protein contacts within Mediator. For example, MED26 formed crosslinks with nine different subunits, two of which (MED6 and MED8) are separated from the structured C-terminal MED26 residues; similarly, MED1 formed crosslinks to MED29, which is distant from the structured N-terminal MED1 domain. These data likely reflect stable structural states, given that mass spectrometry detection requires perhaps 100 million crosslinking events between the same two residues, which is not likely with random sampling. As noted by Abdella et al. and Chen et al., many sites bound by TFs remain disordered in the PIC structures (e.g. MED1, MED15, MED25), and unresolved regions on MED26 interact with pol II elongation factors24.
These “everything at once” structural insights have answered many long-standing questions in the transcription field. Although important new questions will continue to drive the field forward, future research will benefit from this more solid foundation, based upon a far more detailed and accurate understanding of pol II transcription initiation.
References
- 1.Roeder RG 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat Struct Mol Biol 26, 783–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schier AC & Taatjes DJ Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34, 465–488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdella R et al. Structure of the human Mediator-bound transcription preinitiation complex. Science 372, 52–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aibara S, Schilbach S & Cramer P Structures of mammalian RNA polymerase II pre-initiation complexes. Nature (2021). [DOI] [PubMed] [Google Scholar]
- 5.Chen X et al. Structural insights into preinitiation complex assembly on core promoters. Science 372 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Chen X et al. Structures of the human Mediator and Mediator-bound preinitiation complex. Science (2021). [DOI] [PubMed] [Google Scholar]
- 7.Rengachari S, Schilbach S, Aibara S, Dienemann C & Cramer P Structure of human Mediator-RNA polymerase II pre-initiation complex. Nature (2021). [DOI] [PubMed] [Google Scholar]
- 8.Robinson PJ et al. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 166, 1411–1422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schilbach S et al. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai KL et al. Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H et al. Structure of mammalian Mediator complex reveals Tail module architecture and interaction with a conserved core. Nat Commun 12, 1355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sogaard TM & Svejstrup JQ Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem 282, 14113–20 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Patel AB et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fishburn J, Tomko E, Galburt E & Hahn S Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A 112, 3961–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomko EJ et al. The Role of XPB/Ssl2 dsDNA Translocase Processivity in Transcription Start-site Scanning. J Mol Biol, 166813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi MJ et al. A high-resolution protein architecture of the budding yeast genome. Nature 592, 309–314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taatjes DJ, Naar AM, Andel F, Nogales E & Tjian R Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058–1062 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Henley MJ et al. Unexpected specificity within dynamic transcriptional protein-protein complexes. Proc Natl Acad Sci U S A 117, 27346–27353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez J & Larson DR Transcription in Living Cells: Molecular Mechanisms of Bursting. Annu Rev Biochem 89, 189–212 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Gressel S et al. CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife 6, e29736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao W & Zeitlinger J Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet 49, 1045–1051 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Tantale K et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat Commun 7, 12248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabari BR, Dall’Agnese A & Young RA Biomolecular Condensates in the Nucleus. Trends Biochem Sci 45, 961–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi H et al. Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greber BJ, Toso DB, Fang J & Nogales E The complete structure of the human TFIIH core complex. Elife 8, e44771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


