Abstract
Expression of Phanerochaete chrysosporium genes encoding ligninolytic enzymes was assessed in wood. Poly(A) RNA was extracted from colonized wood chips by magnetic capture, and specific transcripts were quantified by competitive reverse transcriptase PCR. mRNA levels varied substantially among lignin peroxidase genes, and transcript patterns were dramatically different from those in previous studies with defined media.
Lignin depolymerization is catalyzed by extracellular enzymes of white rot basidiomycetes such as Phanerochaete chrysosporium. Major components of this system include lignin peroxidases (LiPs), manganese-dependent lignin peroxidases (MnPs), and a peroxide-generating enzyme, glyoxal oxidase (GLOX) (for review, see references 6, 11, and 17). Under nutrient limitation in defined media, multiple peroxidase and GLOX isozymes are secreted.
The peroxidases of P. chrysosporium are encoded by families of structurally related genes. Ten LiP genes, designated lipA through lipJ, have been characterized and shown to be distributed on three linkage groups (reviewed in reference 12). The three known MnP genes (mnp genes) are unlinked to each other or to any LiP genes (reference 28 and unpublished data). In contrast, GLOX is encoded by a single gene (glx) with two alleles (20, 23). The precise roles and interactions of these genes in lignin degradation and in commercial processes such as biomechanical pulping (for review, see reference 24) are poorly understood.
Numerous studies have demonstrated differential regulation of LiP and MnP genes in response to culture conditions. Northern blots showed lipD transcripts dominating in carbon-starved cultures (18) and in defined media supplemented with balled-mill straw (19). In contrast, lipA transcripts were relatively more abundant in nitrogen-limited media (18). Nuclease protection assays identified lipE as the major transcript in both carbon- and nitrogen-starved cultures (30). Quantitative reverse transcriptase-mediated PCR (RT-PCR) techniques largely confirmed Northern blots and also showed dramatic upregulation of lipC and lipJ under nitrogen starvation (31). All LiP gene transcripts except lipF were detected in anthracene-contaminated soil cultures (3). The MnP genes of P. chrysosporium exhibit complex regulation by nutrient limitation (15, 29), Mn concentration (7, 9, 14), culture agitation, heat shock (8), H2O2 concentration, and other chemical stresses (25). mnp3 appears not to be regulated by Mn. In contrast, mnp1 and mnp2 respond strongly to Mn and are differentially regulated in response to culture agitation (15). The three MnP genes are coordinately transcribed in soil cultures (4). Nothing is known of the regulation of P. chrysosporium peroxidase genes in woody tissue, the natural substrate.
To assess transcript levels of all known LiP, MnP, and GLOX genes in P. chrysosporium-colonized wood, 2.5 kg of aspen wood chips was steam sterilized and inoculated by standard biomechanical pulping methods (1) (reviewed in reference 2). Poly(A) RNA was extracted from 10-g samples as described elsewhere (32), with minor modifications. Specifically, the initial extract buffer was squeezed through Miracloth (Calbiochem, Inc., La Jolla, Calif.) filters, and following incubation with Dynabeads oligo(dT)25 (Dynal, Great Neck, N.Y.), the hybridization buffer was twice extracted with a model MPC-1 magnetic concentrator. Poly(A) RNA levels were too low to accurately quantify (<1 μg/10 g), but yields were adequate for a minimum of 600 separate RT-PCRs. The competitive RT-PCR protocol was adapted from the work of Gilliland et al. (16) with gene-specific primers (Table 1). Competitive templates, in the form of full-length genomic subclones, were added to 50-μl PCR mixtures as 10-fold serial dilutions ranging from 10 ng to 0.1 fg. Preliminary experiments quantifying lipA, lipC, lipE, and lipF transcripts with various amounts of poly(A) template in RT-PCRs showed no evidence for RT inhibition (10).
TABLE 1.
Competitive PCR primers
| Genea | 5′ primer | 3′ primer |
|---|---|---|
| lipA | TCCATCGCAATTTCGCCC | ACACGGTTGATGATTTGG |
| lipB | GCTATTGCCATCTCTCCT | ACACGAGCGATGATCTGG |
| lipC | GCCATCGCTATCTCTCCC | ACACGGTCGATGATTTGG |
| lipD | TCCATCGCTATCTCGCCC | ATGCGAGCGAGAACCTGA |
| lipE | TCCATCGCCATCTCGCCC | ACGCGGGCGATGATCTGG |
| lipF | TGCCCTTGAGTCTCAAGG | ACGCGAGAGATGATCTGG |
| lipG | TCGATCGCCATCTCGCCC | ACACGCTCGATGAGCTGG |
| lipH | GCAATTGCCATCTCGCCC | ACACGGTTAATGAGCTGG |
| lipI | TCTATCGCTATCTCTCCC | ACACGGCTGATGATTTGA |
| lipJ | GCCATCGCGATCTCTCCC | ATCCGAGCCAGGATCTGA |
| mnp1 | CCGACGGCACCCGCGTCAGC | CGAGCGGGAGCGGCGACGCC |
| mnp2 | CAGACGGTACCCGCGTCACC | AGTGGGAGCGGCGACATCAC |
| mnp3 | CCGACGGTACCAAGGTCAAC | AGCGGCAGCGGCGACGCGAC |
| glx | TCACACCTTCGCTCTACACG | TATTTACTCCAGGGTCGGCG |
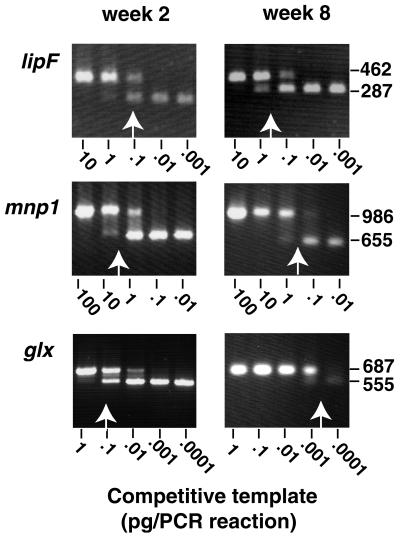
PCR products were size fractionated on 1.5% agarose gels and ethidium bromide stained, and the image was recorded with a Foto/Analyst digital camera (Fotodyne, Inc., Hartland, Wis.). The image was digitized with NIH Image software (version 1.61). Linear regressions were determined by plotting ratios of genomic competitor to cDNA target against the concentration of competitive template. Adjusting for length differences, equivalence points were determined on linear regressions where the ratios were 1.5 for lip genes, 1.3 for mnp genes, and 1.19 for glx. Results were expressed in picograms of cDNA (Fig. 1). Independent analysis for lipC and mnp2 transcript levels in separate wood chip cultures varied less than 12%.
FIG. 1.
Competitive PCRs comparing transcripts of three genes in samples collected after 2 and 8 weeks of incubation. Vertical arrows represent approximate equivalence points. Digitized images were acquired and labeled with Adobe Photoshop 3.0 and Illustrator 7.0, respectively. Numbers at right indicate molecular size in base pairs.
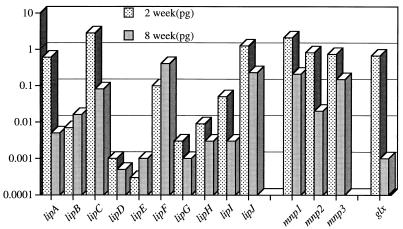
Differences in transcript levels ranged up to 10,000 fold (Fig. 2), and transcript patterns in aspen were unlike the patterns previously observed in defined media or in soil cultures. Transcripts of lipF, absent in soil cultures, were abundant. lipD and lipE, major transcripts in soil and defined media, ranked lowest among LiP gene transcripts in aspen. Transcripts of lipI, represented by a single functional allele (lipI1) in dikaryotic strain BKM-F-1767, were at high levels relative to those in defined media (31). (The alternative allele, lipI2, is transcriptionally inactive due to insertion of a repetitive element [13].) Relative to lip genes, the mnp genes showed less difference in transcript levels.
FIG. 2.
Comparison of transcript levels of all known peroxidase and GLOX genes in aspen wood chip cultures after incubation for 2 or 8 weeks. Results are expressed as picograms of cDNA. The nomenclature for lip and mnp gene designations follows the work of Gaskell et al. (12) and Gettemy et al. (15), respectively.
The identification of glx transcripts in wood was consistent with a close physiological connection between extracellular peroxidases and GLOX (21, 22). The simultaneous detection of lip and glx transcripts was reported in defined media (20, 23) and in soil cultures (3).
Transcript levels generally declined by 8 weeks of incubation, although it is unclear whether transcription was reduced or whether mRNA was partially degraded. lipA and glx transcripts decreased more than 100-fold, while lipB, lipE, and lipF transcripts increased 3- to 4-fold. Temporal shifts in transcription have been observed in defined media (3, 5, 25).
No clear relationship between genomic organization and transcription emerges from these and previous results. Within the two LiP gene clusters (lipA, lipB, lipC, lipE and lipI, lipG, lipH, lipJ), no patterns are evident. The unlinked LiP genes, lipD and lipF, show patterns very different from those of one another and from those of most other lip genes. In comparing all genes on all substrates, lipD and lipE transcript patterns are most alike, although the lipD transcript levels are consistently 5- to 10-fold higher than those of lipE.
The substantial differences in transcript levels probably reflect enzyme activity in aspen, as has been demonstrated in defined media (5, 26, 27) and in soil cultures (3, 4). Thus, genes previously shown to be highly expressed under a wide range of cultural conditions, such as lipD and lipE (18, 19, 30), are unlikely to play a major role in biopulping performance on aspen. It remains to be determined if these genes are differentially regulated under other conditions (e.g., wood species, temperature, and moisture).
Acknowledgments
This research was supported by grant DE-FG02-87ER13712 from the U.S. Department of Energy to D.C. B.J.H.J. was supported by grants from the Foundation for Research Development (South Africa) and Mondi Kraft (South Africa).
REFERENCES
- 1.Akhtar, M. April 1997. U.S. Patent 5,620,564.
- 2.Akhtar M, Blanchette R, Myers G, Kirk T K. An overview of biomechanical pulping research. In: Young R, Akhtar M, editors. Environmentally friendly technologies for the pulp and paper industry. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 309–340. [Google Scholar]
- 3.Bogan B, Schoenike B, Lamar R, Cullen D. Expression of lip genes during growth in soil and oxidation of anthracene by Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:3697–3703. doi: 10.1128/aem.62.10.3697-3703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogan B, Schoenike B, Lamar R, Cullen D. Expression of mnp genes during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:2381–2386. doi: 10.1128/aem.62.7.2381-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boominathan K, D’Souza T M, Naidu P S, Dosoretz C, Reddy C A. Temporal expression of the major lignin peroxidase genes of Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:3946–3950. doi: 10.1128/aem.59.11.3946-3950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broda P, Birch P, Brooks P, Sims P. Lignocellulose degradation by Phanerochaete chrysosporium: gene families and gene expression for a complex process. Mol Microbiol. 1996;19:923–932. doi: 10.1046/j.1365-2958.1996.474966.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown J, Alic M, Gold M. Manganese peroxidase gene transcription in Phanerochaete chrysosporium: activation by manganese. J Bacteriol. 1991;173:4101–4106. doi: 10.1128/jb.173.13.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J, Li D, Alic M, Gold M. Heat shock induction of manganese peroxidase gene transcription in Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:4295–4299. doi: 10.1128/aem.59.12.4295-4299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown J A, Glenn J K, Gold M H. Manganese regulates expression of manganese peroxidase by Phanerochaete chrysosporium. J Bacteriol. 1990;172:3125–3130. doi: 10.1128/jb.172.6.3125-3130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler D, Wagnon C A, Bolton H. Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl Environ Microbiol. 1998;64:669–677. doi: 10.1128/aem.64.2.669-677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen D. Recent advances on the molecular genetics of ligninolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- 12.Gaskell J, Stewart P, Kersten P, Covert S, Reiser J, Cullen D. Establishment of genetic linkage by allele-specific polymerase chain reaction: application to the lignin peroxidase gene family of Phanerochaete chrysosporium. Bio/Technology. 1994;12:1372–1375. doi: 10.1038/nbt1294-1372. [DOI] [PubMed] [Google Scholar]
- 13.Gaskell J, Vanden Wymelenberg A, Cullen D. Structure, inheritance, and transcriptional effects of pce1, an insertional element within Phanerochaete chrysosporium lignin peroxidase gene lipI. Proc Natl Acad Sci USA. 1995;92:7465–7469. doi: 10.1073/pnas.92.16.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gettemy J M, Li D, Alic M, Gold M H. Truncated-gene reporter system for studying the regulation of manganese peroxidase expression. Curr Genet. 1997;31:519–524. doi: 10.1007/s002940050239. [DOI] [PubMed] [Google Scholar]
- 15.Gettemy J M, Ma B, Alic M, Gold M H. Reverse transcription-PCR analysis of the regulation of the manganese peroxidase gene family. Appl Environ Microbiol. 1998;64:569–574. doi: 10.1128/aem.64.2.569-574.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilliland G, Perrin S, Bunn H. Competitive PCR for quantitation of mRNA. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols. New York, N.Y: Academic Press, Inc.; 1990. pp. 60–69. [Google Scholar]
- 17.Gold M, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holzbaur E, Tien M. Structure and regulation of a lignin peroxidase gene from Phanerochaete chrysosporium. Biochem Biophys Res Commun. 1988;155:626–633. doi: 10.1016/s0006-291x(88)80541-2. [DOI] [PubMed] [Google Scholar]
- 19.James C M, Felipe M S S, Sims P F G, Broda P. Expression of a single lignin peroxidase-encoding gene in Phanerochaete chrysosporium strain ME446. Gene. 1992;114:217–222. doi: 10.1016/0378-1119(92)90577-c. [DOI] [PubMed] [Google Scholar]
- 20.Kersten P, Cullen D. Cloning and characterization of a cDNA encoding glyoxal oxidase, a peroxide-producing enzyme from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Proc Natl Acad Sci USA. 1993;90:7411–7413. doi: 10.1073/pnas.90.15.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kersten P J. Glyoxal oxidase of Phanerochaete chrysosporium; its characterization and activation by lignin peroxidase. Proc Natl Acad Sci USA. 1990;87:2936–2940. doi: 10.1073/pnas.87.8.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kersten P J, Kirk T K. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J Bacteriol. 1987;169:2195–2201. doi: 10.1128/jb.169.5.2195-2201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kersten P J, Witek C, Vanden Wymelenberg A, Cullen D. Phanerochaete chrysosporium glyoxal oxidase is encoded by two allelic variants: structure, genomic organization, and heterologous expression of glx1 and glx2. J Bacteriol. 1995;177:6106–6110. doi: 10.1128/jb.177.21.6106-6110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirk T K, Cullen D. Enzymology and molecular genetics of wood degradation by white-rot fungi. In: Young R A, Akhtar M, editors. Environmentally friendly technologies for the pulp and paper industry. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 273–308. [Google Scholar]
- 25.Li D, Alic M, Brown J, Gold M H. Regulation of manganese peroxidase gene transcription by hydrogen peroxide, chemical stress, and molecular oxygen. Appl Environ Microbiol. 1995;61:341–345. doi: 10.1128/aem.61.1.341-345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Alic M, Gold M. Nitrogen regulation of lignin peroxidase gene transcription. Appl Environ Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moukha S, Wosten H, Mylius E, Asther M, Wessels J. Spatial and temporal accumulation of mRNAs encoding two common lignin peroxidases in Phanerochaete chrysosporium. J Bacteriol. 1993;175:3672–3678. doi: 10.1128/jb.175.11.3672-3678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orth A, Rzhetskaya M, Cullen D, Tien M. Characterization of a cDNA encoding a manganese peroxidase from Phanerochaete chrysosporium: genomic organization of lignin and manganese peroxidase genes. Gene. 1994;148:161–165. doi: 10.1016/0378-1119(94)90251-8. [DOI] [PubMed] [Google Scholar]
- 29.Pease E, Tien M. Heterogeneity and regulation of manganese peroxidases from Phanerochaete chrysosporium. J Bacteriol. 1992;174:3532–3540. doi: 10.1128/jb.174.11.3532-3540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiser J, Walther I, Fraefel C, Fiechter A. Methods to investigate the expression of lignin peroxidase genes by the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:2897–2903. doi: 10.1128/aem.59.9.2897-2903.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart P, Kersten P, Vanden Wymelenberg A, Gaskell J, Cullen D. The lignin peroxidase gene family of Phanerochaete chrysosporium: complex regulation by carbon and nitrogen limitation and the identification of a second dimorphic chromosome. J Bacteriol. 1992;174:5036–5042. doi: 10.1128/jb.174.15.5036-5042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallim M A, Janse B J H, Gaskell J, Pizzirani-Kleiner A, Cullen D. Phanerochaete chrysosporium cellobiohydrolase and cellobiose dehydrogenase transcripts in wood. Appl Environ Microbiol. 1998;64:1924–1928. doi: 10.1128/aem.64.5.1924-1928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]