Abstract
Gastric cancer (GC) has emerged as a significant issue in public health all worldwide as a result of its high mortality rate and dismal prognosis. AT-rich interactive domain 1 A (ARID1A) is a vital component of the switch/sucrose-non-fermentable (SWI/SNF) chromatin remodeling complex, and ARID1A mutations occur in various tumors, leading to protein loss and decreased expression; it then affects the tumor biological behavior or prognosis. More significantly, ARID1A mutations will likely be biological markers for immune checkpoint blockade (ICB) treatment and selective targeted therapy. To provide theoretical support for future research on the stratification of individuals with gastric cancer with ARID1A as a biomarker to achieve precision therapy, we have focused on the clinical significance, predictive value, underlying mechanisms, and possible treatment strategies for ARID1A mutations in gastric cancer in this review.
Keywords: ARID1A mutation, Gastric cancer, Prognosis, Treatment, Biomarker
Introduction
Gastric cancer (GC) is a disease that affects individuals worldwide. In 2021, there were reportedly 1.1 million new cases, and diagnosing stomach carcinoma at an advanced stage resulted in high mortality, a high recurrence rate, and a bad prognosis [1]. Research has confirmed that various etiologies play a crucial part in the pathogenesis of GC, such as microbiota, smoking, alcohol consumption, family genetic factors, previous gastric surgery, and poor eating habits. With the rapid development of research in molecular biology and the ongoing maturation of sequencing technology, the connection between classical genetics and epigenetics and stomach cancer has received attention. The research directions mainly include gene mutation, gene deletion, DNA methylation, histone modification, and chromatin remodeling. AT-rich interactive domain 1 A (ARID1A) genes usually have inactive mutations. Clear cell carcinoma (40–57%), GC (8–27%), bladder cancer (about 20%), hepatocellular carcinoma (10–17%), melanoma (about 12%), colon cancer (about 9%), and lung cancer (about 8%) are a few varieties of cancer that show a high incidence of mutations that render ARID1A inactive with loss of expression [2–9]. Various cancer treatment options associated with ARID1A mutations are undergoing clinical trials; they have been listed in Table 1. The clinical importance and molecular mechanism of the ARID1A mutation in GC and its role in traditional targeted treatment and emerging immune checkpoint blockade (ICB) therapy have not been thoroughly investigated. From molecular mechanisms to potential clinical treatment strategies, this article comprehensively reviews the functionality of ARID1A mutation in GC, especially its possible correlation with other biological molecules in tumorigenesis and its predictive value in treatment selection and prognosis. Studying the function and role of ARID1A in GC carcinogenesis will help to improve patients’ prognoses and guide clinical practice.
Table 1.
Clinical trials for cancer therapies involving ARID1A
| Number | Study types | Status | Phase | Cancer types | Intervention | Primary endpoint |
|---|---|---|---|---|---|---|
| NCT04065269 | Interventional (Clinical Trial) | Recruiting | II | Gynecological Cancers | ATR inhibitor (AZD6738), PARP inhibitor (Olaparib) | ORR |
| NCT04957615 | Interventional (Clinical Trial) | Recruiting | II | Metastatic Malignant Solid Neoplasm, Unresectable Solid Neoplasm | Nivolumab | ORR, OS |
| NCT05523440 | Interventional (Clinical Trial) | Recruiting | II |
Recurrent Endometrial Carcinoma Recurrent Ovarian Carcinoma |
Bevacizumab, Niraparib | ORR |
| NCT05690035 | Interventional (Clinical Trial) | Not yet recruiting | II | Metastatic Colorectal Cancer | Tislelizumab & Fruquintinib | ORR |
| NCT04953104 | Interventional (Clinical Trial) | Not yet recruiting | II | Urologic Neoplasms | Nivolumab | ORR, OS |
| NCT03682289 | Interventional (Clinical Trial) | Recruiting | II | Solid Tumors | Ceralasertib, Olaparib, Durvalumab | ORR |
| NCT04042831 | Interventional (Clinical Trial) | Recruiting | II | Advanced Biliary Tract Cancer | Olaparib | ORR |
| NCT04284202 | Interventional (Clinical Trial) | Unknown | II | NSCLC Stage IV | PD-1 plus Dasatinib | PFS |
ATR: ataxia telangiectasia and rad3-related; PARP: poly (ADP-ribose) polymerase; PD-1: Programmed cell death protein 1; ORR: Objective Response Rate; OS: overall survival; PFS: Progress Free Survival
Chromatin remodeling and chromatin remodeling complexes
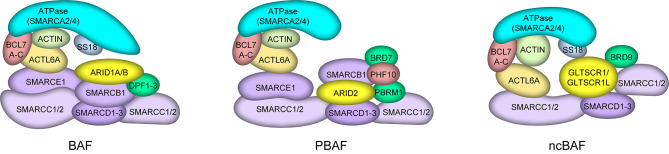
As the basic structural unit of chromatin, the nucleosome consists of an octamer of histones and double-stranded DNA surrounded by negative supercoils that are highly concentrated to produce a dense structure. Dynamic alterations such as histone insertion, expulsion, and nucleosome sliding cause changes in the spatial location of histones and DNA, making it simpler for protein regulators to approach double-stranded DNA to complete DNA replication, transcription, recombination, and other processes. This is known as chromatin remodeling [10]. The energy released by ATP hydrolysis is used by the ATP-dependent chromatin remodeling complex to drive chromatin conformational changes and control gene expression. The ATPase subunit, which belongs to the SF2 helicase family, is the most important protein component in the ATP-dependent chromatin remodeling complex. Based on their domains, ATPase subunits are divided into four subfamilies: SWI/SNF, INO80, ISWI, and CHD [11]. The switch/sucrose-non-fermentable (SWI/SNF) chromatin remodeling complex was first discovered in S. cerevisiae in the 1990s. It is a transcriptional regulatory complex composed of multiple genes encoding proteins. In mammalian cells, three types of SWI/SNF complexes have been thoroughly investigated: canonical BAF (CBAF) containing accessory subunits ARID1A/ARID1B and DPF1/2/3, polybromo-associated BAF (PBAF) with ARID2, PHF10, PBRM1, and BRD7 as marker subunits and non-canonical BAF (ncBAF) with BRD9 and GLTSCR1 or GLTSCR1L (GLTSCR1-like) as its distinct subunit. They all include the ATPase catalytic subunit SMARCA4 or SMARCA2, and they all also have individual subunits that make different complexes unique [12–16] (Fig. 1).
Fig. 1.
The structure of three types of mammalian switch/sucrose-non-fermentable complexes (BAF, PBAF, and ncBAF).
Structure and function of ARID1A
Structure of ARID1A
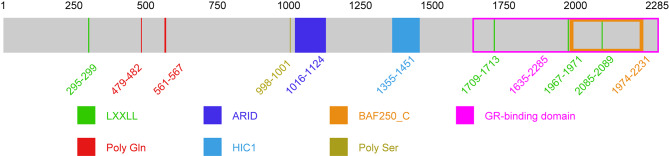
ARID1A, also named as OSA1, P270, hOSA1, BAF250, C1orf4, BAF250a, and SMARCF1, is a SWI/SNF complex component. The ARID1A gene, which codes for a protein with 2285 amino acid residues and a relative molecular mass of 240 KD, is found on chromosome 1p36.11. ARID1A, typically located in the nucleus, is strongly expressed in various body tissues [17]. ARID1A protein mainly contains a conserved domain (aa 1016–1124) that binds to adenine (A) and thymine (T)-rich DNA sequences, a HIC1-binding domain (aa 1355–1451), a glucocorticoid receptor (GR) binding domain (aa 1635–2285), and four LXXLL motif structures [18, 19] (Fig. 2). ARID1B has a conserved domain that is highly similar to ARID1A. Their expression in development and cell regulation differs, resulting in different biological functions in vivo [20]. In recent years, ARID1A and ARID1B co-mutations have been found in various cancers, but at least one functional allele has survived. A recent study found that their double deletion may lead to the redistribution of the cBAF complex after division and may affect the oligomerization of PBAF [21]. In ARID1A mutant tumor cells, the loss of ARID1B leads to the loss of enhancer structure and changes in chromatin accessibility, making it challenging for cancer cells to survive. Similar to the interdependence of SMARCA4 or SMARCA2, the interplay between ARID1A and ARID1B synthetic lethality in oncology needs to be further studied [22–25].
Fig. 2.
The structure of AT-rich interactive domain 1 A
Biological function of ARID1A
ARID1A and cell stemness
ARID1A plays an outstanding role in regulating the differentiation of a wide assortment of stem cells, including cardiac progenitor cells, neural stem/progenitor cells, as well as embryonic stem cells. In the mouse model, by altering the accessibility of chromatin, ARID1A controls the expression of crucial genes during myocardial development, promoting the differentiation of cardiac progenitor cells into normal cardiomyocytes [26]. ARID1A gene knockout inhibits the self-renewal characteristics of embryonic stem cells, and mesoderm differentiation is negatively impacted, seriously impeding embryonic development [27]. Liu et al. used Cre/loxP to construct ARID1A neural stem cell conditional knockout mice, demonstrating that the absence of ARID1A function impairs radial glial cell proliferation and leads to the dysregulation of genes related to neural stem progenitor cell differentiation, for instance, Fezf2, Rgs6, Ptk2b, and Lpar1 [28]. According to Wang et al., the lack of ARID1A causes the destruction of the structure and function of the SWI/SNF complex, which in turn leads to an imbalance in the expression of genes involved in cell stemness, cell differentiation, and liver function, which promotes the growth of liver cancer [29]. Meanwhile, research has demonstrated that ARID1A is crucial for preserving the functionality of pancreatic acinar cells and re-proliferation following damage. The differentiation of ARID1A-deficient pancreatic tumors is blocked and they are endowed with high mobility, invasiveness, and stem-like properties [30].
ARID1A and DNA damage repair
DNA is vulnerable to various endogenous and exogenous factors, resulting in changes in genetic information carriers, leading to major diseases such as cancer. The DNA damage response (DDR) allows organisms to sense DNA damage signals, slow down or block cell cycle progression, and activate different DNA repair mechanisms or apoptosis mechanisms. DNA double-strand break (DSB) is a common form of DNA damage. In mammalian cells, DSB has two common repair pathways: non-homologous end joining (NHEJ), which occurs mainly in the S phase of the cell, and homologous recombination (HR), which occurs primarily in the G1 and G2 phases. The balanced development of the two repair pathways keeps the genome stable. Recent findings have pointed out that ARID1A is essential in the two DNA damage repair pathways. The recruitment of the SWI/SNF complex ATPase subunit to the DNA damage site depends on the presence of ARID1A. Inhibition of ARID1A reduces the accumulation of NHEJ pathway initiator KU70/KU80 in DNA DSB, leading to the inactivation of the NHEJ pathway [31]. Ataxia telangiectasia mutant gene (ATM) and ataxia telangiectasia andrad3-related (ATR), both members of the PI3/PI4 kinase family, are essential for the HR-mediated DSB response. ARID1A is recruited to DNA DSBs and interacts with the vital kinase ATR. As a result, the cell cycle is stopped, and broken DNA repair is facilitated [32].
ARID1A and tumor cell proliferation, invasion, metastasis, and apoptosis
ARID1A is generally considered a tumor suppressor gene that can inhibit the biological behavior of malignant tumors and regulate the cell cycle to promote apoptosis to exert anticancer effects. He et al. [33] discovered that human hepatocellular carcinoma (HCC) cell lines Huh-7 and MHCC-97 H express ARID1A differently. The former was relatively high, and the latter was deficient. ShRNA-mediated ARID1A knockdown significantly promoted the migration and invasion of Huh-7 cells. On the contrary, overexpressing ARID1A markedly inhibited the ability of MHCC-97 H cells to invade and migrate. In addition, in vivo experiments of mouse xenograft tumors showed that lung metastasis occurred in HCC cells knocking down ARID1A in 50% (3/6) of mice, demonstrating that a decrease in ARID1A expression was related to HCC metastasis.
Angiogenesis is an essential process of tumor growth, invasion, and metastasis. Yoodee et al. [34] found that after knocking down ARID1A in colorectal cancer cell line Caco-2, the secretion level of the angiogenic factor VEGF was significantly increased by ELISA. A previous study by his team also showed that the human endothelial cells’ down-regulation of ARID1A promotes Ang2 secretion and endothelial cell activity, which induces angiogenesis [35].
Apoptosis is the term used to describe spontaneous and planned cell death, which is regulated by genes to preserve the stability of the internal environment. Some researchers have shown that the knockdown of ARID1A in leukemia cell lines can resist FAS-mediated apoptosis [36]. Zhang et al. [37] found that siRNA knockdown of ARID1A significantly increased the expression of cyclin D1, Bcl-2, and Akt phosphorylation and inhibited paclitaxel-induced apoptosis. Xie et al. [38] found that the depletion of ARID1A promoted the proliferation of colorectal cancer cell lines and inhibited 5-fluorouracil-induced apoptosis. In recent years, researchers have shown that impaired ARID1A expression in GC cells may resist Harakiri-mediated apoptosis and lead to disease [39]. These findings demonstrate the role of ARID1A in apoptosis.
It has been found that the absence of ARID1A may lead to changes in EMT markers. Tomihara et al. [40] used pancreatic ductal adenocarcinoma cell lines to study the interaction between ARID1A and EMT regulatory proteins. After ARID1A knockout, the levels of epithelial markers cytokeratin-19 and E-cadherin decreased, and the level of cytoplasmic marker vimentin increased. Somsuan et al. [41] found that siARID1A transfection increased the levels of renal interstitial markers (fibronectin and vimentin) and decreased the levels of epithelial markers (E-cadherin and ZO-1) when they examined the effect of ARID1A expression reduction on the characteristics of non-malignant renal cell carcinogenesis. This is likely to be related to the TGF-b1/SNAI1 signaling pathway. The same results were obtained in the malignant renal cell carcinoma cell line 786-O. According to these studies, a lack of ARID1A could trigger EMT and promote tumor cell metastasis.
Association between GC and ARID1A mutation or ARID1A protein expression loss
Relationship between GC subtypes and ARID1A mutation
GuanBin’s analysis of 257 cases of somatic ARID1A mutation data showed that ARID1A mutations were mostly frame-shift mutations or nonsense mutations. Nonsense-mediated RNA decay (NMD) or incorrect protein degradation leads to abnormal expression of ARID1A protein, ultimately leading to tumor inhibition loss [42]. Despite the fact that ARID1A mutation is linked to loss of expression, the gene mutation is not the only cause of the loss of protein expression. Recently, it was discovered that ubiquitination, followed by proteasomal degradation, was the cause of the ARID1A protein’s disappearance in GC cells [43]. ARID1A, the second-largest mutant gene after TP53 in GC, can detect mutations in 8–27% of GC cases [4–9, 44].
Interestingly, the frequency of ARID1A mutations differs significantly in different subtypes of GC. The Cancer Genome Atlas (TCGA) conducted complex statistics and informatics analysis on 295 cases of GC tissue and blood samples and formally divided GC into four molecular subtypes, including Epstein-Barr virus (EBV)-positive, microsatellite instability (MSI), chromosome instability (CIN), and genomic stability (GS). Frequent ARID1A mutations were found in GC that was EBV-positive [45]. In 2015, GC was reclassified into four subtypes by the Asian Cancer Research Group (ACRG) to better direct treatment and prognosis: MSI, MSS / EMT, MSS / TP53 +, and MSS / TP53-. The mutation rates of ARID1A were 19/43 (44.2%), 5/36 (13.9%), 11/59 (18.6%), and 5/85 (5.9%), respectively [46].
Mechanistically, the deletion of ARID1A significantly increased the efficiency of EBV infection in gastric epithelial cells and it was challenging to recruit mismatch repair proteins, which initiated the occurrence of EBV subtype and MSI subtype GC [47, 48]. Setia et al. used immunohistochemistry and in situ hybridization to classify GC more easily: EBV-positive, MSI-H, aberrant expression of E-cadherin, aberrant expression of P53, and normal expression of P53. It was found that EBV-positive and MSI-H (high microsatellite instability) gastric cancer had a better prognosis [49]. Subsequently, according to the above classification method, some researchers found that the loss of ARID1A protein expression in GC was significantly related to the positive expression of the MSI-H subtype and PD-L1 [50]. Since gastric cancer with the MSI-H subtype and PD-L1 positive expression more effectively responds to immune checkpoint inhibitors (ICIs), the expression of ARID1A may become a biomarker for GC immunotherapy [51].
Clinical and prognostic relevance of ARID1A mutation or protein loss in GC
A study that divided up GC tissue samples in accordance with the degree of ARID1A expression revealed that either total or partial loss of ARID1A expression was linked to a shorter progression-free survival (PFS) and overall survival (OS) [52]. Another study found a significant correlation between the deletion of ARID1A and tumor differentiation (P = 0.009), metastasis to lymph nodes (P = 0.030), and tumor size (P = 0.022) [9]. Zhou et al. [53] discovered that the expression level of ARID1A protein in GC tissues was significantly lower than in the normal tissues adjacent to the GC tissues. The expression level of ARID1A was interrelated with the depth of tumor invasion (P = 0.040). In line with this, Wang et al. [51] ascertained that ARID1A protein deletion is an independent risk factor for the poor prognosis of GC after analyzing 272 primary GC samples by Immunohistochemistry (IHC) and quantitative reverse transcription PCR (qRT-PCR). These findings demonstrate, from several angles, that the absence of ARID1A protein expression is related to a poor prognosis for GC.
However, not all studies support this view. For example, Ibarrola-Villava et al. [54] found that compared to patients with ARID1A positive expression, the OS of patients with ARID1A negative expression was considerably higher (P = 0.03). In a cohort study using tissue microarray technology (n = 173), There was no discernible link between OS and loss of ARID1A expression [55]. Although there is some controversy, a meta-analysis related to GC and ARID1A support that the decrease in ARID1A expression is connected to adverse clinical outcomes [56].
The following factors may be responsible for the disagreement regarding the relationship between the expression level of ARID1A and the prognosis of GC: (1) The individual heterogeneity of GC is due to the pathological features of GC being affected by a variety of genetic and environmental factors, and thus the molecular and morphological heterogeneity of GC is formed. (2) The limitation of the number of patients may lead to limited experimental results. (3) In the process of immunohistochemical staining, due to the lack of sensitivity of the detection method, a negative reaction is caused, or the differences in criteria for the results of immunohistochemical staining and the different reagents lead to differing experimental results. These elements might be the leading causes of the differing findings among many studies. At the same time, it is worth noting that ARID1A is not the only factor affecting the prognosis of GC. Many other factors may lead to changes in the clinical prognosis of GC, such as different GC subtypes. In short, ARID1A mutation or loss of expression may result in a worsening of the biological behavior of GC, indicating that ARID1A expression level may be an essential determinant in judging the prognosis of GC. However, many experiments are still needed to confirm this.
Interaction between ARID1A mutation or protein deletion and other genes and gene pathways in GC
It is generally known that the TP53 gene acts as a tumor suppressor gene. The P53 protein translated by the TP53 gene can block cell cycle progression, repair damaged DNA, and promote apoptosis. It has been found that the expression of two downstream target genes of P53 decreased after ARID1A silencing in GC cells, indicating that ARID1A and P53 may synergistically activate the transcription of target genes and inhibit tumor growth [57], which confirmed the previous view of Guan in ovarian cancer research [58]. Further studies have found that ARID1A mutation or absence of ARID1A protein expression in GC negatively correlates with TP53 mutation [59]. Recently, Loe et al. [60] established a clinically applicable gastric tumor model with ARID1A loss of heterozygosity. After in-depth analysis and in vivo verification, they found that the lack of ARID1A heterozygosity in GC will lead to the extensive loss of H3K27ac modification in the enhancer region of genes related to TP53 tumor suppressor pathway and apoptosis pathway, resulting in the inhibition of p53 apoptosis-related gene expression, thereby promoting tumor progression. In addition, the team further found that activation of the TP53 signal pathway may possess a therapeutic impact on AIRD1A heterozygous GC.
The PIK3CA gene, which codes for 1068 amino acids, is found on chromosome 3q26.3. It is a catalytic subunit of the IA-type PI3Ks family. PIK3CA gene mutation activates the PI3K/AKT signaling pathway, leading to tumorigenesis. Zhang et al. [57] found that knockout of ARID1A in GC cell lines directly targets PDK1 and PIK3CA transcription in the PIK3/AKT pathway, resulting in phosphorylation changes in the main components of the PIK3/AKT signaling path, including AKT, mammalian target of rapamycin (mTOR),and glycogen synthase kinase 3α/β (GSK3α/β), among others. In this process, the HIC1 binding domain in ARID1A may play a crucial part. A similar in vitro study also confirmed that in ARID1A-deficient GC cells, the PI3K/AKT pathway was activated, which induced the proliferation of GC cells, and ARID1A-deficient GC cells were more sensitive to PI3K and AKT inhibitors [61]. Despite the low number of clinical studies, it seems that the loss of ARID1A expression can be used as a biomarker for AKT pathway activation and predict the effect of AKT inhibitors in patients with GC. Yang et al. [62] found that miR-233-3P can stimulate GC cell growth and migration by specifically targeting ARID1A, and the NF-κB/miR-223-3p/ARID1A pathway is an essential pathway for HP-mediated chronic gastritis to GC transformation.
Potential therapeutic strategies for ARID1A and GC
ARID1A and immunotherapy of GC
GC is traditionally treated with surgery, radiotherapy, and chemotherapy. With the advancement of medical technology, targeted therapy, immunotherapy, angiogenesis therapy, and other new treatment methods for GC have emerged as research hotspots. It is a widely held belief that ICIs are effective in treating GC, and the clinical trials of PD-1/PD-L1 inhibitors have garnered much interest recently.
ATTRACTION-2 is a phase III multicenter trial that evaluates nivolumab versus placebo in treating patients with advanced GC who progressed after ≥ 2 lines of chemotherapy. Regarding OS and PFS, the results showed that nivolumab significantly outperformed the placebo [63]. Subsequently, as a result of the findings of the CheckMate-649 study, the FDA granted approval for the combination of nivolumab and chemotherapy in 2021, making it the first immunotherapy treatment to be authorized as a first-line treatment for GC anywhere in the world [64].
Despite the fact that ICB therapy has changed the treatment strategy for malignant tumors, a significant number of GC patients do not respond well to immunotherapy. Therefore, it is urgent to further screen out relevant biomarkers to choose patients who could gain from ICBs.
The KETNOTE-059 test showed that pembrolizumab had a more favorable therapeutic impact on gastric or esophagogastric junction (EGJ) adenocarcinoma with a PD-L1 combined positive score (CPS) ≥ 1 [65]. CHECKMATE-649 also showed that in advanced GC (AGC) and EGJ cancers, nivolumab combined with chemotherapy resulted in better OS than chemotherapy alone, especially when PD-L1 is combined with CPS ≥ 5 [64]. These critical findings highlight the importance of PD-L1 expression levels in determining how ICIs should be used.
The tumor mutation burden (TMB) can be used to assess the extent and capacity of a tumor to produce new antigens and predict how well immunotherapy will work for various tumor types. Wang et al. [66] found that TMB was connected to significant survival benefits of AGC using whole exome sequencing. The OS of the high TMB group was significantly better than that of the low TMB group (14.6 vs. 4.0 months, HR = 0.48, P = 0.038). It was confirmed that the TMB level might be a predictive biomarker for screening the survival benefit population of ICI toripalimab in treating AGC.
Different mismatch repairs (dMMRs) are usually caused by mutations in the gene encoding mismatch repair proteins. If there is a problem with the mismatch repair system, the length of the short tandem repeats changes to form microsatellite instability (MSI-H). Patients with MSI-H/dMMR have severe defects in the tumor DNA repair mechanism, but numerous studies, including KEYNOTE-016, 164, 012, 028, and 158, have demonstrated that patients with MSI-H / dMMR tumors have better immunotherapy effects. The FDA currently approves Pembrolizumab to treat patients with metastatic or unresectable solid tumors carrying dMMR or MSI-H biomarkers [67].
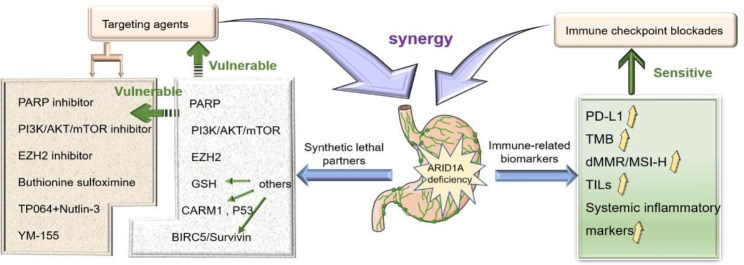
Many studies have confirmed that the degree of tumor-infiltrating Lymphocytes (TILs) can serve as a biomarker to forecast the favorable outcome of PD-1/PD-L1 immunosuppressive therapy by detecting the subgroup, number, and regional distribution of TILs [68, 69]. Based on three systemic inflammatory markers, Formica et al. established the Gastric Inflammation Prognostic Index (GIPI) to evaluate the prognosis of patients with a metastatic gastro-esophageal junction (mGOJ)/GC (GC) after ICI treatment [70]. Surprisingly, the expression of ARID1A in GC is intimately connected to these biomarkers that affect immune blockade therapy. The positive expression of PD-L1 in tumors is closely correlated with the loss of ARID1A in GC. In terms of the mechanism, ARID1A deficiency up-regulates PD-L1 expression by activating the PI3K/AKT/mTOR pathway [71]. In addition, researchers used bioinformatics methods to find that the increased immune activity of gastrointestinal cancer with ARID1A mutation is attributed to higher TMB levels and has better advantages in immunotherapy [72]. It has been reported that ARID1A deletion cannot recruit mismatch repair (MMR) protein MSH2 during DNA replication, resulting in increased tumor MSI subtypes and TMB levels [48]. Wang et al. [51] found that the loss of ARID1A protein expression in GC is related to dMMR status, systemic inflammatory markers, and increased PD-L1 expression levels. A study found that the expression level of ARID1A protein in early-onset GC tissues and normal mucosal tissues around the tumor was associated with tumor T-cell infiltration [73].
A recent study discovered that the SWI/SNF complex gene (especially the ARID1A gene) has a high mutation rate in various cancers and is associated with high TMB status and MSI subtypes. After ICI treatment for malignant tumors, the three significant tumor treatment endpoint indicators (progression-free survival [PFS], overall response rate [ORR], and disease control rate [DCR]) were significantly prolonged [74]. The discovery of a link between ARID1A inactivation and PD-L1, TMB, MMR, TILs, and systemic inflammatory markers in GC raises the possibility that ARID1A deletion might serve as a predictive biomarker for ICBs therapy for GC. More research is needed to understand the underlying mechanisms of the correlation between immunotherapy biomarkers so that patients with malignant tumors have a better chance of survival after receiving ICBs treatment.
Other treatments for ARID1A-deficient GC
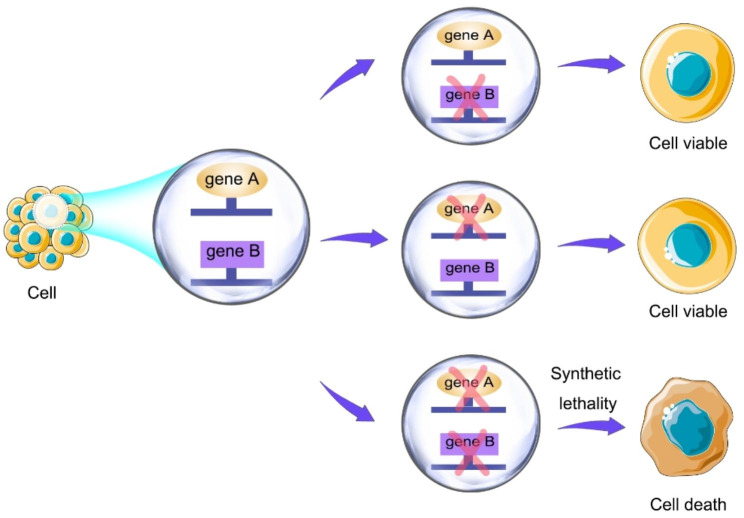
Synthetic lethality refers to the simultaneous mutation of two genes leading to cell death (Fig. 3); based on the concept of ‘synthetic lethality,’ oncologists have continuously explored cancer-targeted therapy strategies to provide innovative ideas for developing tumor drugs. Due to the diversity of the SWI/SNF complex structure, combination, and function, SWI/SNF mutations in cancer can use the synthetic lethal interaction mode to disturb other SWI/SNF subunits or some molecules related to function, highlighting their potential as drug development target molecules [75]. To date, targeted molecular inhibitors for GC, such as PARP inhibitors, PI3K / AKT / mTOR inhibitors, EZH2 inhibitors, and others, have been investigated extensively based on ARID1A gene mutation or defect combined with the concept of synthetic lethality.
Fig. 3.
Basic principles of synthetic lethal therapy. When gene A and gene B exist at the same time, the cells survive. If one of gene A and gene B is absent, the cells can survive. If gene A and gene B lose expression at the same time, cell death will occur
PARP inhibitors
Inhibitors of PARP were the first synthetic lethal concept–based anticancer drugs that gained approval for clinical use. In breast cancer and ovarian cancer, tumor suppressor gene BRCA1/2 defects or mutations while inhibiting the activity of PARP can target tumor cells, leading to synthetic death [76]. In addition to BRCA1/2, ARID1A-deficient tumors also show sensitivity to PARP inhibitors [32]. However, PARP inhibitor monotherapy has minimal effect on cancers lacking ARID1A, and it often needs to work in combination with other drugs. The mechanism may be that the HR pathway in ARID1A-deficient tumors is not impaired and can repair DSB induced by replication fork stagnation and PARP [77]. In addition, it has been found that the combination of PARP inhibitor olaparib and PI3K inhibitor BKM120 may be an emerging treatment for ARID1A-deficient GC [78]. In a phase 1 clinical trial (NCT03842228), individuals with advanced solid cancers were chosen based on more than 20 gene mutations, including ARID1A, MSH2, PTEN, BARD1, BRCA1, and BRCA2. PARP inhibitors, PI3K inhibitors, and ICIs were used to evaluate treatment efficacy, but an accurate conclusion has yet to be reached.
Targeted PI3k/AKT/mTOR inhibitors
The PI3K/AKT/mTOR pathway regulates GC cells’ function through various mechanisms, including promoting cell invasion, metastasis, epithelial-mesenchymal transition (EMT), angiogenesis, and activating tumor chemotherapy resistance. Pan-PI3K inhibitors such as BKM120, BAY80-6946, and PI3K subunit selective inhibitors such as BYL719 and TAK117 showed sound anti-tumor effects in in vitro experiments of GC [78–83]. Still, they did not produce promising results in clinical studies. The pan-AKT inhibitor AZD5363 has been used in phase I and II clinical studies of GC with good drug resistance and safety. In addition, MK-2206, which is currently in the early clinical trial stage, is a potent and highly selective pan-AKT inhibitor [84, 85]. Traditional mTOR inhibitors that block the PI3K/AKT/mTOR signaling pathway through mTORC1, including rapamycin and its analog everolimus, have not achieved satisfactory clinical results in a phase III clinical study of everolimus as a second or third-line treatment for advanced GC [86]. The clinical effects of second- and third-generation mTOR inhibitors still require accurate reporting of extensive sample clinical data.
Although a variety of targeted molecular inhibitors can exert potential anti-tumor effects through the PI3K/AKT/mTOR pathway, to date, the clinical efficacy of these inhibitors as a monotherapy has shown meager response rates. It is possible that these failures are the result of a requirement for appropriate patient selection based on dependable biomarkers. Lee et al. [61] found that patients selected based on ARID1A expression in GC tissues had increased sensitivity to drugs that block the activity of AKT and greatly improved clinical results. A combination therapy consisting of the AKT inhibitor GSK690693 and standard chemotherapy enhances the efficacy of ARID1A knockdown GC cells and has potential in future research. A recent study showed that ARID1A could be used to screen GC patients who profit from mTOR inhibitor therapy. In terms of the mechanism, mTOR inhibitors can effectively target the activated pS6 and SOX9 in ARID1A-deficient GC [87]. In addition, Hanahan et al. [88] found that PI3K-AKT-mTOR inhibitors additionally have a significant function in regulating the tumor immune microenvironment. It has been demonstrated that inhibiting the PI3K/AKT pathway increases the sensitivity to tumor-specific CD8 + T cell-mediated cytotoxicity [89, 90]. According to other research findings, combining PI3K/AKT/mTOR inhibitors with ICIs (which include PD-1 inhibitors in parallel with CTLA-4 inhibitors) or other anti-tumor immunotherapy may enable patients to obtain the best efficacy [91].
EZH2 inhibitors
Recent research has revealed that numerous cancer cells exhibit EZH2 overexpression and aberrant regulation. EZH2-targeted inhibitors based on synthetic lethal effects play an essential role in ARID1A mutant cancers. The researchers found that in ovarian cancer cells with ARID1A mutation, PIK3IP1, a direct target gene of ARID1A and EZH2, was up-regulated after EZH2 was inhibited, leading to cell death by inhibiting the PI3K-AKT signaling pathway. Moreover, EZH2 inhibitor GSK126 can lead to the decline of ovarian cancer carrying ARID1A mutation in vivo [92]. Another study confirmed that EZH2 inhibitors increased the selective sensitivity of ARID1A-deficient GC [93]. In addition, inhibition of EZH2 by Tazestat or GSK126 can lead to the synthetic lethality of SMARCA4, SMARCB1, PBRM1, and other SWI/SNF chromatin remodeling complex subunit-deficient cancers [94–97]. As a popular epigenetic target, there is mounting evidence demonstrating that EZH2 can control a wide range of tumor-infiltrating lymphocytes, form an immunosuppressive microenvironment within the tumor, and allow tumor cells to escape the recognition and destruction of the immune system. Inhibition of EZH2 as an attractive therapeutic strategy can strengthen existing immunotherapy. Studies have shown that the use of EZH2 inhibitors in conjunction with ICB results in a significant degree of synergy in the treatment of specific tumors [98, 99]. Still, it must be considered that this combination therapy may lead to excessive immune system activation and increase the potential risk of autoimmune diseases. There needs to be more research on how EZH2 inhibitors and ICB work together to treat ARID1A mutant cancer.
Other
Other targeted drugs with an anti-tumor ability for ARID1A-deficient GC include glutathione (GSH) inhibitors, YM-155, and Nutlin-3 + TP064. Reduced solute carrier family seven-member 11(SLC7A11) recombinant protein expression decreases GSH synthesis, making ARID1A-deficient stomach cancer cells vulnerable to GSH inhibition [100]. It has been discovered that the catalytic subunit of the rate-limiting enzyme in GSH synthesis, the Glutamate-Cysteine Ligase Catalytic Subunit (GCLC), is a promising therapeutic target for tumors lacking ARID1A [101]. Therefore, developing drugs targeting the glutathione metabolic pathway may be a prospective treatment strategy for ARID1A-deficient tumors. A novel inhibitor called YM-155, which has been shown to inhibit apoptosis in vitro, targets the survivin protein. Lo et al. [102] discovered that survivin (BIRC5) might be a lethal partner for ARID1A synthesis in a genetically engineered human gastric organoid model, particularly in the early stages of ARID1A-deficient GC. Additional clinical studies are still required to understand better the connection between YM155 and malignant tumors that diminish the ARID1A gene. At the same time, the combination therapy of CARM1 inhibitor TP064, a vital regulator of the BAF complex, and P53 agonist Nutlin-3 also provides a potentially effective treatment option for GC patients with ARID1A mutation [44].
Conclusions and prospects
To summarize, due to its unique advantages, the ARID1A mutation plays an essential role in the clinical practice of GC. ARID1A can be a screening biomarker for individuals whose cancer is responsive to targeted therapy and immunotherapy. Combining the two therapies may significantly increase the likelihood of successfully treating GC (Fig. 4). Studies, both preclinical and clinical, have demonstrated that the role of genetics and epigenetics in malignant tumors will be explored further, and mutation of the chromatin remodeling complex subunit, ARID1A, may become a promising tool for individualized treatment targets in patients with GC. However, some challenges need to be overcome. First, since the majority of current clinical findings were derived from a retrospective analysis of a relatively tiny sample, selection bias and confounding factors may interfere with the result. Secondly, based on the defects of single therapy resistance and combined treatment of adverse reactions, it is necessary to strictly control the dose and usage of ARID1A-mutant GC targeted therapy drugs. Third, the role of ARID1A mutation in tumorigenesis, tumor development, prediction of therapeutic response, and potential biological mechanisms needs to be further examined. Prospective clinical studies conducted across multiple centers and on a large scale are required to draw more reliable results regarding the predictive and prognostic value of ARID1A mutations in GC. On the basis of this review, we hope that the subset of patients suffering from GC with ARID1A mutation will have improved clinical outcomes.
Fig. 4.
ARID1A-deficient gastric cancer immunotherapy synergistic targeted therapy strategy. ARID1A, the AT-rich interaction domain 1 A; PD-L1, programmed cell death ligand 1; TMB, tumor mutation burden; dMMR, different Mismatch Repair; MSI-H, microsatellite instability-high; TILs, Tumor Infiltrating Lymphocytes; PARP, poly (ADP-ribose) polymerase; EZH2, enhancer of zeste homolog 2; GSH, glutathione
Authors’ contributions
Shan Lu participated in the literature search and data analysis, and drafted and revised the article’s content;Ruifeng Duan, Liang Cong, and Ying Song participated in drafting and revising the key content of the article and made key contributions to the intellectual content of the article.
Funding
This work was supported by the 13th Five-year Science and Technology Project of Jilin Provincial Education Department [NO. JJKH20201088KJ].
Data Availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
All the authors have no conflict of interests to disclose.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Wang TL, Shih IM, et al. Frequent mutations of chromatin remodeling gene ARID1A in Ovarian Clear Cell Carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. New Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, Li M, Parsons DW, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several Tumor types. Hum Mutat. 2012;33:100–3. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–23. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 6.Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 7.Rokutan H, Hosoda F, Hama N, et al. Comprehensive mutation profiling of mucinous gastric carcinoma. J Pathol. 2016;240:137–48. doi: 10.1002/path.4761. [DOI] [PubMed] [Google Scholar]
- 8.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu YP, Sheng LL, Wu J, et al. Loss of ARID1A expression is associated with poor prognosis in patients with gastric cancer. Hum Pathol. 2018;78:28–35. doi: 10.1016/j.humpath.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Oike T, Ogiwara H, Nakano T, et al. Inactivating mutations in SWI/SNF chromatin remodeling genes in human cancer. Jap J Clin Oncol. 2013;43:849–55. doi: 10.1093/jjco/hyt101. [DOI] [PubMed] [Google Scholar]
- 11.Clapier CR, Iwasa J, Cairns BR, et al. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–22. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Côté J, Xue Y, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–82. doi: 10.1002/j.1460-2075.1996.tb00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–6. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mashtalir N, D’Avino AR, Michel BC, et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175:1272–88e20. doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alpsoy A, Dykhuizen EC. Glioma Tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol Chem. 2018;293:3892–903. doi: 10.1074/jbc.RA117.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reisman DN, Sciarrotta J, Bouldin TW, et al. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl Immunohistochem Mol Morphol. 2005;13:66–74. doi: 10.1097/00129039-200503000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Wu JN, Roberts CWM. ARID1A mutations in cancer: another epigenetic Tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Rechem C, Boulay G, Leprince D. HIC1 interacts with a specific subunit of SWI/SNF complexes, ARID1A/BAF250A. Biochem Biophys Res Commun. 2009;385:586–90. doi: 10.1016/j.bbrc.2009.05.115. [DOI] [PubMed] [Google Scholar]
- 19.Nie Z, Xue Y, Yang D, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–88. doi: 10.1128/MCB.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato E, Nakayama K, Razia S, et al. ARID1B as a potential therapeutic target for ARID1A-mutant ovarian clear cell carcinoma. Int J Mol Sci. 2018;19:9. doi: 10.3390/ijms19061710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Chen K, Jia Y, et al. Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptive redistribution of BAF complexes. Nat Cancer. 2020;1:909–22. doi: 10.1038/s43018-020-00109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helming KC, Wang X, Wilson BG, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20:251–4. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathur R, Alver BH, San Roman AK, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon Cancer in mice. Nat Genet. 2017;49:296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelso TWR, Porter DK, Amaral ML et al. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. ELife. 2017;6. [DOI] [PMC free article] [PubMed]
- 25.Watanabe R, Ui A, Kanno S-I, et al. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014;74:2465–75. doi: 10.1158/0008-5472.CAN-13-3608. [DOI] [PubMed] [Google Scholar]
- 26.Lei IL, Gao XL, Sham MH, et al. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J Biol Chem. 2012;287:24255–62. doi: 10.1074/jbc.M112.365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X, Tate P, Hu P, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–61. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Dai SK, Liu PP, et al. Arid1a regulates neural stem/progenitor cell proliferation and differentiation during cortical development. Cell Prolif. 2021;54:e13124. doi: 10.1111/cpr.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Deng CH, Luo Q, et al. Inhibition of Arid1a increases stem/progenitor cell-like properties of Liver cancer. Cancer Lett. 2022;546:215869. doi: 10.1016/j.canlet.2022.215869. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Friedland SC, Guo B, et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut. 2019;68:1245–58. doi: 10.1136/gutjnl-2017-315541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe R, Ui A, Kanno S, et al. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014;74:2465–75. doi: 10.1158/0008-5472.CAN-13-3608. [DOI] [PubMed] [Google Scholar]
- 32.Shen J, Peng Y, Wei L, et al. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 2015;5:752–67. doi: 10.1158/2159-8290.CD-14-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He F, Li J, Xu J, et al. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. J Exp Clin Cancer Res. 2015;34:47. doi: 10.1186/s13046-015-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peerapen P, Sueksakit K, Boonmark W, et al. ARID1A knockdown enhances carcinogenesis features and aggressiveness of Caco-2 colon Cancer cells: an in vitro cellular mechanism study. J Cancer. 2022;13:373–84. doi: 10.7150/jca.65511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoodee S, Peerapen P, Plumworasawat S, et al. ARID1A knockdown in human endothelial cells directly induces angiogenesis by regulating angiopoietin-2 secretion and endothelial cell activity. Int J Biol Macromol. 2021;180:1–13. doi: 10.1016/j.ijbiomac.2021.02.218. [DOI] [PubMed] [Google Scholar]
- 36.Luo B, Cheung HW, Subramanian A, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Xu X, Zhang M, et al. ARID1A is downregulated in non-small cell Lung cancer and regulates cell proliferation and apoptosis. Tumour Biol. 2014;35:5701–7. doi: 10.1007/s13277-014-1755-x. [DOI] [PubMed] [Google Scholar]
- 38.Xie C, Fu L, Han Y, et al. Decreased ARID1A expression facilitates cell proliferation and inhibits 5-fluorouracil-induced apoptosis in colorectal carcinoma. Tumour Biol. 2014;35:7921–7. doi: 10.1007/s13277-014-2074-y. [DOI] [PubMed] [Google Scholar]
- 39.Sakuratani T, Takeuchi T, Yasufuku I, et al. Downregulation of ARID1A in gastric cancer cells: a putative protective molecular mechanism against the Harakiri-mediated apoptosis pathway. Virchows Arch. 2021;478:401–11. doi: 10.1007/s00428-020-02899-1. [DOI] [PubMed] [Google Scholar]
- 40.Tomihara H, Carbone F, Perelli L, et al. Loss of ARID1A promotes epithelial-mesenchymal transition and sensitizes pancreatic tumors to proteotoxic stress. Cancer Res. 2021;81:332–43. doi: 10.1158/0008-5472.CAN-19-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somsuan K, Peerapen P, Boonmark W, et al. ARID1A knockdown triggers epithelial-mesenchymal transition and carcinogenesis features of renal cells: role in renal cell carcinoma. FASEB J. 2019;33:12226–39. doi: 10.1096/fj.201802720RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan B, Gao M, Wu CH, et al. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its Tumor suppressor functions. Neoplasia. 2012;14:986–93. doi: 10.1593/neo.121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang ZH, Peng T, Qian HL, et al. DNA damage-induced activation of ATM promotes β-TRCP-mediated ARID1A ubiquitination and destruction in gastric cancer cells. Cancer Cell Int. 2019;19:162. doi: 10.1186/s12935-019-0878-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loe AKH, Francis R, Seo J et al. Uncovering the dosage-dependent roles of Arid1a in gastric tumorigenesis for combinatorial drug therapy. J Exp Med. 2021;218. [DOI] [PMC free article] [PubMed]
- 45.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 47.Abe H, Kunita A, Otake Y, et al. Virus-host interactions in carcinogenesis of Epstein-Barr virus-associated gastric carcinoma: potential roles of lost ARID1A expression in its early stage. PLoS ONE. 2021;16:e0256440. doi: 10.1371/journal.pone.0256440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen J, Ju Z, Zhao W, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–62. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Setia N, Agoston AT, Han HS, et al. A protein and mRNA expression-based classification of gastric cancer. Mod Pathol. 2016;29:772–84. doi: 10.1038/modpathol.2016.55. [DOI] [PubMed] [Google Scholar]
- 50.Mehrvarz Sarshekeh A, Alshenaifi J, Roszik J, et al. ARID1A mutation may define an immunologically active subgroup in patients with microsatellite stable Colorectal cancer. Clin Cancer Res. 2021;27:1663–70. doi: 10.1158/1078-0432.CCR-20-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Che K, Shi T, et al. Loss of ARID1A expression is associated with systemic inflammation markers and has important prognostic significance in gastric cancer. J Cancer Res Clin Oncol. 2022;148:1583–95. doi: 10.1007/s00432-022-03971-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim YB, Ham IH, Hur H, et al. Various ARID1A expression patterns and their clinical significance in gastric cancers. Hum Pathol. 2016;49:61–70. doi: 10.1016/j.humpath.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 53.Zhou H, Tan S, Li H, et al. Expression and significance of EBV, ARID1A and PIK3CA in gastric carcinoma. Mol Med Rep. 2019;19:2125–36. doi: 10.3892/mmr.2019.9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ibarrola-Villava M, Llorca-Cardeñosa MJ, Tarazona N, et al. Deregulation of ARID1A, CDH1, cMET and PIK3CA and target-related microRNA expression in gastric cancer. Oncotarget. 2015;6:26935–45. doi: 10.18632/oncotarget.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiegand KC, Sy K, Kalloger SE, et al. ARID1A/BAF250a as a prognostic marker for gastric carcinoma: a study of 2 cohorts. Hum Pathol. 2014;45:1258–68. doi: 10.1016/j.humpath.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Kim YS, Jeong H, Choi JW, et al. Unique characteristics of ARID1A mutation and protein level in gastric and Colorectal cancer: a meta-analysis. Saudi J Gastroenterol. 2017;23:268–74. doi: 10.4103/sjg.SJG_184_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, Yan HB, Wang J, et al. Chromatin remodeling gene AT-rich interactive domain-containing protein 1A suppresses gastric cancer cell proliferation by targeting PIK3CA and PDK1. Oncotarget. 2016;7:46127–41. doi: 10.18632/oncotarget.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a Tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–27. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosse T, ter Haar NT, Seeber LM, et al. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod Pathol. 2013;26:1525–35. doi: 10.1038/modpathol.2013.96. [DOI] [PubMed] [Google Scholar]
- 60.Loe AKH, Francis R, Seo J, et al. Uncovering the dosage-dependent roles of Arid1a in gastric tumorigenesis for combinatorial drug therapy. J Exp Med. 2021;218:25. doi: 10.1084/jem.20200219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee D, Yu EJ, Ham IH, et al. AKT inhibition is an effective treatment strategy in ARID1A-deficient gastric cancer cells. Onco Targets Ther. 2017;10:4153–9. doi: 10.2147/OTT.S139664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang F, Xu Y, Liu C, et al. NF-κB/miR-223-3p/ARID1A axis is involved in Helicobacter pylori CagA-induced gastric carcinogenesis and progression. Cell Death Dis. 2018;9:12. doi: 10.1038/s41419-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–71. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 64.Janjigian YY, Shitara K, Moehler M, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet. 2021;398:27–40. doi: 10.1016/S0140-6736(21)00797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuchs CS, Doi T, Jang RW, et al. Safety and Efficacy of Pembrolizumab Monotherapy in patients with previously treated Advanced gastric and gastroesophageal Junction Cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 2018;4:e180013. doi: 10.1001/jamaoncol.2018.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang F, Wei XL, Wang FH, et al. Safety, efficacy and Tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann Oncol. 2019;30:1479–86. doi: 10.1093/annonc/mdz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemery S, Keegan P, Pazdur R. First FDA approval Agnostic of cancer site - when a biomarker defines the indication. N Engl J Med. 2017;377:1409–12. doi: 10.1056/NEJMp1709968. [DOI] [PubMed] [Google Scholar]
- 68.Dong ZY, Wu SP, Liao RQ, et al. Potential biomarker for checkpoint blockade immunotherapy and treatment strategy. Tumour Biol. 2016;37:4251–61. doi: 10.1007/s13277-016-4812-9. [DOI] [PubMed] [Google Scholar]
- 69.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–91. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 70.Formica V, Morelli C, Patrikidou A, et al. Gastric inflammatory Prognostic Index (GIPI) in patients with Metastatic Gastro-Esophageal Junction/Gastric Cancer treated with PD-1/PD-L1 Immune Checkpoint inhibitors. Target Oncol. 2020;15:327–36. doi: 10.1007/s11523-020-00723-z. [DOI] [PubMed] [Google Scholar]
- 71.Kim YB, Ahn JM, Bae WJ, et al. Functional loss of ARID1A is tightly associated with high PD-L1 expression in gastric cancer. Int J Cancer. 2019;145:916–26. doi: 10.1002/ijc.32140. [DOI] [PubMed] [Google Scholar]
- 72.Li L, Li M, Jiang Z et al. ARID1A mutations are associated with increased immune activity in gastrointestinal cancer. Cells. 2019;8. [DOI] [PMC free article] [PubMed]
- 73.Zou J, Qin W, Yang L, et al. Genetic alterations and expression characteristics of ARID1A impact Tumor immune contexture and survival in early-onset gastric cancer. Am J Cancer Res. 2020;10:3947–72. [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Yang X, Zhu W, et al. SWI/SNF complex gene variations are associated with a higher Tumor mutational burden and a better response to immune checkpoint inhibitor treatment: a pan-cancer analysis of next-generation sequencing data corresponding to 4591 cases. Cancer Cell Int. 2022;22:347. doi: 10.1186/s12935-022-02757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wanior M, Krämer A, Knapp S, et al. Exploiting vulnerabilities of SWI/SNF chromatin remodelling complexes for cancer therapy. Oncogene. 2021;40:3637–54. doi: 10.1038/s41388-021-01781-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park Y, Chui MH, Suryo Rahmanto Y, et al. Loss of ARID1A in Tumor cells renders selective vulnerability to combined ionizing radiation and PARP inhibitor therapy. Clin Cancer Res. 2019;25:5584–94. doi: 10.1158/1078-0432.CCR-18-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang L, Yang G, Ding Y, et al. Combined treatment with PI3K inhibitor BKM120 and PARP inhibitor olaparib is effective in inhibiting the gastric cancer cells with ARID1A deficiency. Oncol Rep. 2018;40:479–87. doi: 10.3892/or.2018.6445. [DOI] [PubMed] [Google Scholar]
- 79.Mueller A, Bachmann E, Linnig M, et al. Selective PI3K inhibition by BKM120 and BEZ235 alone or in combination with chemotherapy in wild-type and mutated human gastrointestinal cancer cell lines. Cancer Chemother Pharmacol. 2012;69:1601–15. doi: 10.1007/s00280-012-1869-z. [DOI] [PubMed] [Google Scholar]
- 80.Giraud J, Bouriez D, Seeneevassen L et al. Orthotopic patient-derived xenografts of gastric Cancer to Decipher Drugs effects on Cancer Stem cells and metastatic dissemination. Cancers (Basel). 2019;11. [DOI] [PMC free article] [PubMed]
- 81.Liu N, Rowley BR, Bull CO, et al. BAY 80-6946 is a highly selective intravenous PI3K inhibitor with potent p110α and p110δ activities in Tumor cell lines and xenograft models. Mol Cancer Ther. 2013;12:2319–30. doi: 10.1158/1535-7163.MCT-12-0993-T. [DOI] [PubMed] [Google Scholar]
- 82.Garrett JT, Sutton CR, Kurupi R, et al. Combination of antibody that inhibits ligand-independent HER3 dimerization and a p110α inhibitor potently blocks PI3K signaling and growth of HER2 + breast cancers. Cancer Res. 2013;73:6013–23. doi: 10.1158/0008-5472.CAN-13-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramanathan RK, McDonough SL, Kennecke HF, et al. Phase 2 study of MK-2206, an allosteric inhibitor of AKT, as second-line therapy for advanced gastric and gastroesophageal junction cancer: a SWOG cooperative group trial (S1005) Cancer. 2015;121:2193–7. doi: 10.1002/cncr.29363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Almhanna K, Strosberg J, Malafa M. Targeting AKT protein kinase in gastric cancer. Anticancer Res. 2011;31:4387–92. [PubMed] [Google Scholar]
- 86.Takahari D. Second-line chemotherapy for patients with advanced gastric cancer. Gastric Cancer. 2017;20:395–406. doi: 10.1007/s10120-017-0707-8. [DOI] [PubMed] [Google Scholar]
- 87.Dong X, Song S, Li Y, et al. Loss of ARID1A activates mTOR signaling and SOX9 in gastric adenocarcinoma-rationale for targeting ARID1A deficiency. Gut. 2022;71:467–78. doi: 10.1136/gutjnl-2020-322660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 89.Noh KH, Kang TH, Kim JH, et al. Activation of akt as a mechanism for Tumor immune evasion. Mol Ther. 2009;17:439–47. doi: 10.1038/mt.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Donnell JS, Massi D, Teng MWL, et al. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin Cancer Biol. 2018;48:91–103. doi: 10.1016/j.semcancer.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 91.Smyth MJ, Ngiow SF, Ribas A, et al. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13:143–58. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 92.Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–8. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamada L, Saito M, Thar Min AK, et al. Selective sensitivity of EZH2 inhibitors based on synthetic lethality in ARID1A-deficient gastric cancer. Gastric Cancer. 2021;24:60–71. doi: 10.1007/s10120-020-01094-0. [DOI] [PubMed] [Google Scholar]
- 94.Wang Y, Chen SY, Karnezis AN, et al. The histone methyltransferase EZH2 is a therapeutic target in small cell carcinoma of the ovary, hypercalcaemic type. J Pathol. 2017;242:371–83. doi: 10.1002/path.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gounder MM, Zhu G, Roshal L, et al. Immunologic correlates of the Abscopal Effect in a SMARCB1/INI1-negative poorly differentiated Chordoma after EZH2 Inhibition and Radiotherapy. Clin Cancer Res. 2019;25:2064–71. doi: 10.1158/1078-0432.CCR-18-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang K, Sun R, Chen J, et al. A novel EZH2 inhibitor induces synthetic lethality and apoptosis in PBRM1-deficient cancer cells. Cell Cycle. 2020;19:758–71. doi: 10.1080/15384101.2020.1729450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hong YK, Li Y, Pandit H, et al. Epigenetic modulation enhances immunotherapy for hepatocellular carcinoma. Cell Immunol. 2019;336:66–74. doi: 10.1016/j.cellimm.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 99.Zhou L, Mudianto T, Ma X, et al. Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents Anti-PD-1 resistance in Head and Neck cancer. Clin Cancer Res. 2020;26:290–300. doi: 10.1158/1078-0432.CCR-19-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sasaki M, Chiwaki F, Kuroda T, et al. Efficacy of glutathione inhibitors for the treatment of ARID1A-deficient diffuse-type gastric cancers. Biochem Biophys Res Commun. 2020;522:342–7. doi: 10.1016/j.bbrc.2019.11.078. [DOI] [PubMed] [Google Scholar]
- 101.Ogiwara H, Takahashi K, Sasaki M, et al. Targeting the vulnerability of glutathione metabolism in ARID1A-deficient cancers. Cancer Cell. 2019;35:177–90e8. doi: 10.1016/j.ccell.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 102.Lo YH, Kolahi KS, Du Y, et al. A CRISPR/Cas9-engineered ARID1A-deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov. 2021;11:1562–81. doi: 10.1158/2159-8290.CD-20-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.