Abstract
As an emerging optical imaging modality, stimulated Raman scattering (SRS) microscopy provides invaluable opportunities for chemical biology studies using its rich chemical information. Through rapid progress over the last decade, the development of Raman probes harnessing the chemical biology toolbox has proven to play a key role in advancing SRS microscopy and expanding biological SRS applications. In this perspective, we first discuss the development of biorthogonal SRS imaging using small tagging of triple bonds or isotopes and highlight their unique advantages for metabolic pathway analysis and microbiology investigations. The potential opportunities for chemical biology studies by integrating small tagging with SRS imaging are also proposed. We next summarize the current designs of highly sensitive and super-multiplexed SRS probes, as well as provide future directions and considerations for next-generation functional probe design. These rationally designed SRS probes are envisioned to bridge the gap between SRS microscopy and chemical biology research and should benefit their mutual development and applications.
Graphical Abstract
Introduction
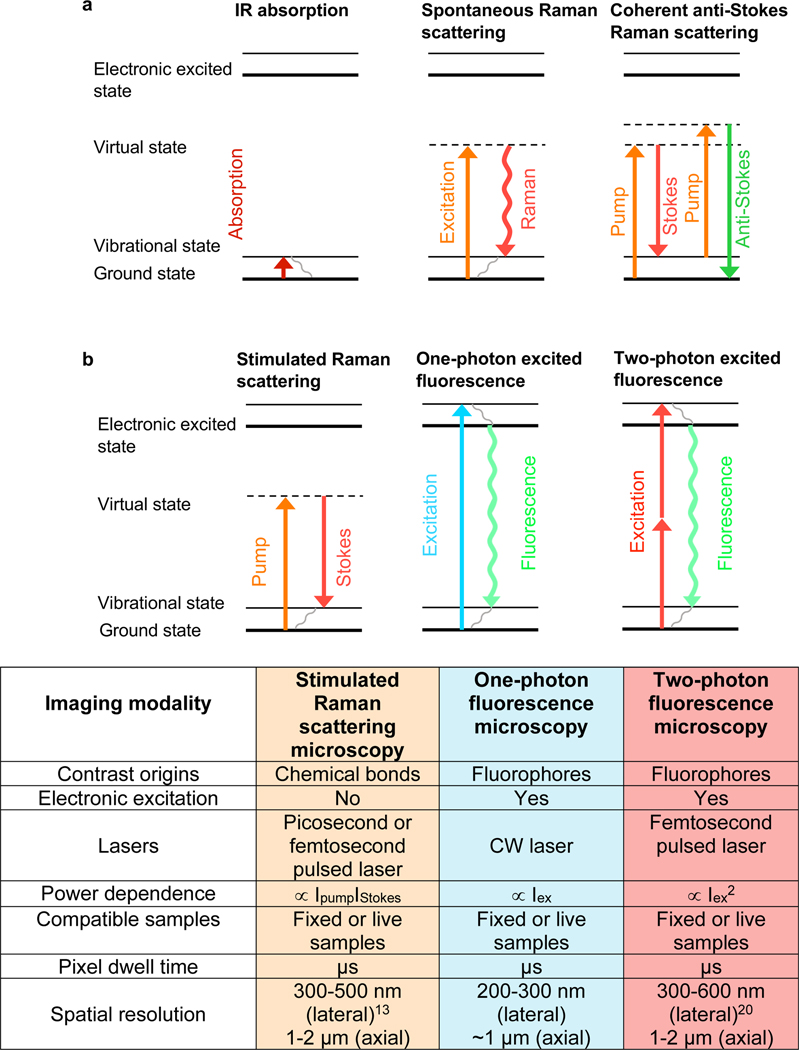
Vibrational spectroscopy, including infrared (IR) absorption and Raman scattering (Figure 1a), reveals molecular information through probing the inherent vibrations of chemical bonds. It has been extensively used for characterizing molecules1, deciphering reaction mechanisms2,3, and interpreting molecule-environment interactions4,5. Raman spectroscopy was first discovered by the physicist Sir C.V. Raman in 1928, where the inelastic Raman scattering shifts a very small portion of photons to lower frequency upon interacting with molecules6. The changes of the photon energy reflect the vibrational energy levels of the chemical bonds within the molecules and thus carry rich chemical information. Although both IR and Raman spectroscopy contain vibrational information of the molecules, they have different selection rules. For example, water has very high IR absorption but weak Raman scattering. In addition, Raman typically utilizes UV to near-infrared light (200 – 1100 nm) while IR relies on mid-infrared light (2500 – 50000 nm), thus the spatial resolution (scaling inversely with the light wavelength) is much higher for Raman (Figure 1a). Both the subcellular spatial-resolution and the minimal water background make Raman scattering as a better-suited technique for the biological Raman applications.
Figure 1. Energy diagram of different vibrational micro-spectroscopy and fluorescence microscopy.
(a) Energy diagram of IR absorption, spontaneous Raman scattering and CARS spectroscopy. (b) Energy diagram and key parameters of SRS microscopy, one- and two-photon excited fluorescence microscopy.
However, the utility of spontaneous Raman is largely limited by its feeble signals (~ 1010 smaller than fluorescence), which requires a long acquisition time and is easily overwhelmed by the auto-fluorescence background from the biological samples. To address both issues, the coherent anti-Stokes Raman Scattering (CARS) microscopy (Figure 1a) was invented. CARS microscopy significantly boosted the imaging sensitivity and speed and pushed Raman spectro-microscopy to be more compatible with biological imaging7. However, CARS signals suffer from severe nonresonant background and have non-linear concentration dependence, which sacrifices imaging quality and adds complexity for imaging annotation8. Around 2008, stimulated Raman scattering (SRS) microscopy was introduced, providing even better sensitivity than CARS and tackling the above issues present with CARS9–11. SRS utilizes two spatially and temporally overlapped laser pulse trains (pump and Stokes, Figure 1b), which enhances the otherwise weak Raman transitions by up to 108-fold through stimulated emission quantum amplification. This two-photon excitation feature also yields SRS intrinsic 3D optical sectioning capability for deep tissue imaging. In addition, the technical implementation of the high-frequency modulation transfer scheme also pushes the detection sensitivity of SRS close to the theoretical limit and removes the potential fluorescence and other interfering background. The readers are encouraged to check more technical details of SRS in other reviews12,13.
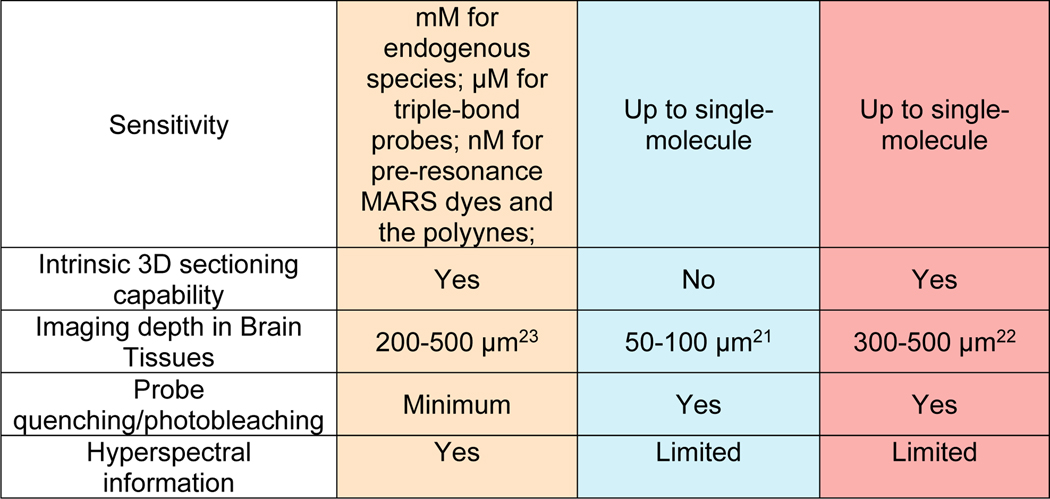
It is now widely recognized that SRS is the most suitable far-field Raman imaging modality for live biological studies with provided image quality comparable to fluorescence microscopy. Prominently, SRS also achieved a high imaging speed up to video-rate (110 frames/s) 14–16 and a spatial resolution within 100 nm with recent instrumentation and sample-expansion strategies17–19. It would be informative to compare the key technical and application features of SRS microscopy with the popular one- and two-photon fluorescence microscopy. Below, we provide the energy diagram and a table summarizing key imaging parameters for the three techniques (Figure 1b). Compared to fluorescence microscopy, the absence of electronic excitation provides SRS with general imaging capability of all types of molecules with minimum photobleaching or environmental quenching. In addition, SRS typically uses near-infrared excitation lasers (800–1100 nm) and thus offers less phototoxicity compared to one-photon fluorescence with visible laser excitation. This near-infrared excitation together with the intrinsic 3D sectioning makes the penetration depth of SRS into thick tissues similar to that obtained by two-photon fluorescence. Moreover, the common utilization of pico-second lasers in SRS renders a much smaller laser peak power compared to that from femto-second lasers for two-photon fluorescence, resulting in less nonlinear photodamage. However, the μM-mM detection sensitivity is still the major challenge in SRS that limits its application to detect molecules with low abundance in live biological samples. Recent developments in probe engineering such as the creation of highly sensitive MARS and polyynes dyes has largely improved the sensitivity down to nM and broken the traditional color barrier in fluorescence microscopy with super-multiplexed imaging.
Label-free vibrational imaging
Since Raman signals originate from chemical bonds instead of conjugated fluorophores, SRS offers general imaging applicability for versatile molecules. Since its inception, SRS microscopy has established itself as a powerful label-free bioimaging modality. Targeting the fingerprint region (500 – 1700 cm−1) or high wavenumber carbon-hydrogen (C-H) stretching region (2800 – 3100 cm−1), nucleic acids, proteins, lipids, carbohydrates, neuron-transmitters and other endogenous biomolecules carrying O-P-O, C=O, C=C, C-H2, C-H3 bonds are readily imaged with subcellular resolution (Figure 2a). Additionally, hyperspectral SRS24,25 adds another layer of information for subcellular spectral analysis. With these capabilities, label-free SRS paves the way for various applications, including sensing environmental cues26, studying lipid metabolism and identifying druggable targets27–31, tracking drug delivery and distribution10,32–34, multicolor cell sorting35,36, fast diagnosis of tumors37,38, and investigations of amyloid plaques in neurodegenerative diseases39,40. The current imaging speed, throughput and detection sensitivity are still being continuously improved with rapid instrumental innovations41–43. In parallel, emerging data processing approaches, particularly the machine learning algorithms, further upgrade image quality and enable data mining with rich chemical information18,29,43–45.
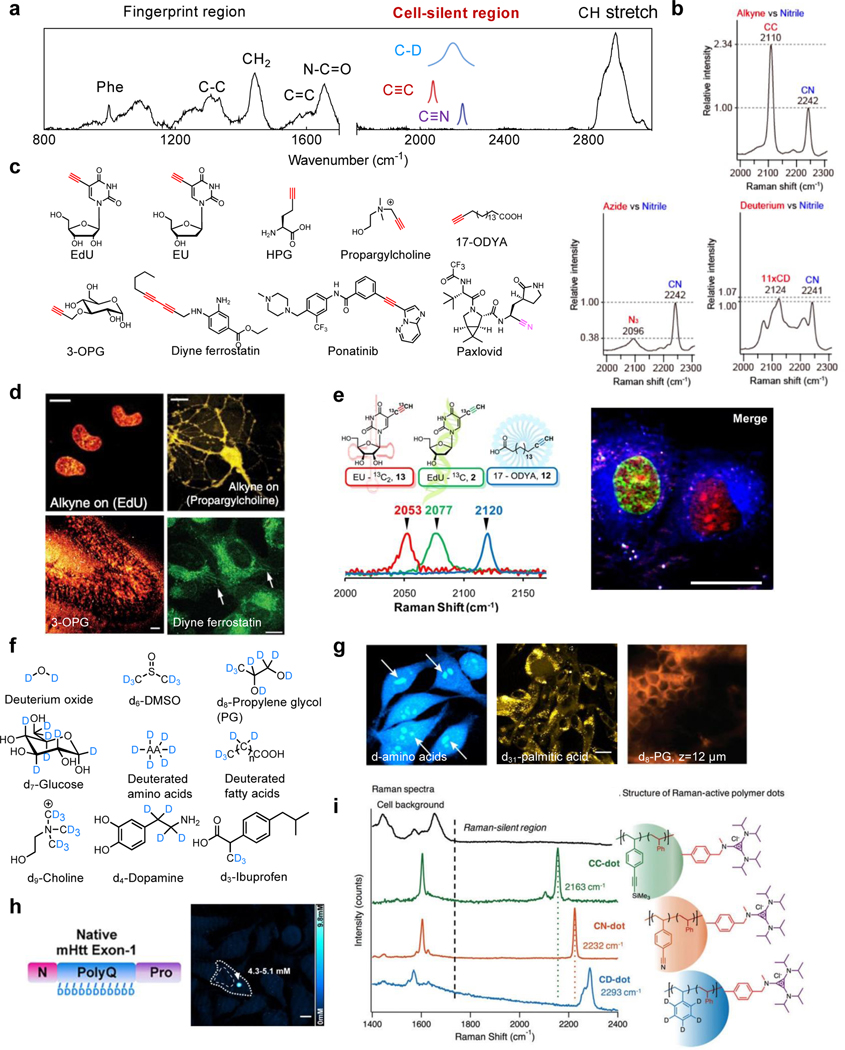
Figure 2. Small biorthogonal Raman tagging with C≡C, C≡N and C-D.
(a) A typical Raman spectrum of a mammalian cell (on glass slides) depicting the crowded fingerprint region, the C-H stretch region, and the cell-silent region (pink). (b) Relative Raman peak intensities of common silent-region Raman tags (i.e. C≡C, C≡N and C-D). Adapted in part with permission from ref 64. Copyright 2010 American Chemical Society. (c) Representative triple-bond tagged small metabolites and drugs. (d) SRS Imaging targeting the alkyne vibrations for incorporation of thymidine-analogue EdU in live HeLa cells, choline-analogue propargylcholine in live neurons, glucose-analogue 3-OPG in cultured mouse brain tissue slices (Scale bar: 40 μm) and alkyne-tagged ferrostatin in live HT-1080 cells. Other scale bars: 10 μm. Adapted in part with permission from ref32, Copyright 2014 Nature publishing group. Adapted in part with permission from ref65, Copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. Adapted in part with permission from ref59. Copyright 2018 American Chemical Society. (e) 13C isotope editing enables three-color imaging of EdU, EU and 17-ODYA with alkyne probes. Scale bar, 25 μm. Adapted in part with permission from ref61. Copyright 2014 American Chemical Society. (f) Representative small molecules with deuterium tagging. (g) SRS imaging targeting the C-D vibration for the incorporation of deuterated amino acids in live HeLa cells, d31-palmitic acid in melanoma cells (Scale bar: 20 μm) and d8-PG in the mice stratum corneum at the depth of 12 μm after 124 min of treatment. Adapted in part with permission from ref66, Copyright 2013 United States National Academy of Sciences. Adapted in part with permission from ref29, Copyright 2020 Nature publishing group. Adapted in part with permission from ref67. Copyright 2014 American Chemical Society. (h) SRS imaging of d5-Gln-labeled mHtt-97Q aggregates in live HeLa cells. Quantifications for the local polyQ concentrations within the aggregates are also shown. Scale bar: 10 μm. Adapted in part with permission from ref68. Copyright 2020 American Chemical Society. (i) C-D and triple-bond enriched polymer dots with amplified Raman signals. Adapted in part with permission from ref69. Copyright 2017 The Royal Society of Chemistry.
Despite the success of label-free SRS imaging, there are several fundamental limitations. First, the specificity of targets is often compromised since endogenous biomolecules tend to share multiple chemical bonds and therefore overlapping spectra. Second, the detection limit of SRS is still relatively low compared with fluorescence - at the scale of millimolar for most biomolecules. Thus, label-free SRS is more suited for investigating relatively abundant molecules including proteins, lipids, nucleic acids and carbohydrates. Third, label-free imaging is not capable of tracking many dynamic processes such as uptake, synthesis, catabolism, and intracellular-to-extracellular interactions. These limitations largely restrict the applications of SRS imaging but can be greatly diminished through the implementation of Raman probes.
Labeling with bioorthogonal probes
Driven by the need of higher specificity, sensitivity, and functionality, which are fundamentally limited in the label-free approaches, Raman labels have been introduced to shift SRS imaging from the label-free to the labeling paradigm46. Fortunately, the cell-silent spectral region (1800–2800 cm−1), where there are no endogenous Raman signals from cells, leaves spacious spectral room for background-free Raman labeling and imaging (Figure 2a). Bioorthogonal chemical bonds, including alkynes (C≡C), nitriles (C≡N) and carbon-deuterium bonds (C-D) are small, nontoxic and Raman active in this clean region, making them well suitable for tagging with low perturbation to the biological systems46. As such, biorthogonal SRS is especially beneficial for live-cell interrogations of small molecules including metabolites and drugs whose label-free vibrational signatures are overwhelmed by cellular background and whose physiological functions are perturbed by conventional fluorophore labeling.
Furthermore, by harnessing the narrow linewidth of Raman peaks (50–100 times narrower than fluorescence peaks), the development of highly sensitive Raman probe palettes followed. The matching dye palettes enable super-multiplexed (more than 20 channels) optical imaging for organelles or protein profiling with sensitivity down to 250 nM, bridging the optical imaging’s subcellular spatial resolution with system biology’s high information throughput47–49. Moreover, chemically activatable and photochromic SRS probes have also been developed lately for intracellular sensing and multiplexed tracking, empowering the functional SRS imaging50–54.
All these recently established Raman probes have greatly expanded the application boundary of vibrational imaging. Efforts in the past decade have proven that probe development plays a central role in driving the next frontiers of SRS microscopy. The growing chemical biology toolbox inspires the development of new SRS imaging functionalities. In turn, the SRS platform also finds a unique niche for chemical biology studies. Exploring new opportunities for merging SRS imaging with chemical biology is worth brainstorming. In this perspective, we first review recent notable advances in the development of Raman probes and their biological applications. On this basis, we further provide an outlook to further expand multiplexing, enhance Raman signals and utilize SRS imaging to decipher new biology.
Small-molecule Raman tagging
Triple bonds (e.g. C≡C, C≡N) and stable-isotope-substituted chemical bonds (e.g. C-D, N-D, O-D) vibrate in the cell-silent region (Figure 2a). Among these chemical bonds, C≡C has the highest Raman signals (Figure 2b). One representative molecule tagged by C≡C is 5-ethynyl-2′-deoxyuridine (EdU), the well-adopted thymidine analogue with an SRS detection limit of 200 μM32, and is now frequently used as a benchmark for Raman intensity quantification. Pioneered by the click chemistry field, chemists have developed a suite of alkyne-tagged molecular handles for bio-labeling, many of which can now be directly detected by Raman without the subsequent click reactions. With these available and newly developed Raman-tailored alkyne probes (Figure 2c), the dynamic metabolic processes including DNA synthesis, choline and glucose uptake can be readily visualized in live cells and tissues (Figure 2d with the corresponding analog structures shown in Figure 2c)32,55–59. Additionally, drugs bearing native alkynes (e.g. ponatinib) or nitriles (e.g. paxlovid) or upon proper alkyne derivatization such as diyne-ferrostatin (Figure 2c&d) can be quantitatively imaged for intracellular distribution with well-maintained pharmacokinetics32,59,60. Inspired by the colorful fluorescent protein palette, alkyne “vibrational colors” are also tunable through an isotope-editing strategy based on the dependence of Raman frequency on the bond mass. A set of 13Cedited probes were developed that enabled multicolor SRS imaging of DNA, RNA and lipids in the same set of live cells61 (Figure 2e).
Compared to C≡C, C≡N has lower (about 40%) Raman cross-sections (Figure 2b). However, as the peak frequencies of C≡N are sensitive to the physical environment (particularly the electrostatic interactions4,62,63), they provide additional functions as vibrational sensors. Additionally, the C≡N vibration occupies the higher frequency region (2200 – 2300 cm−1), which separates well from that of C≡C (2100 – 2200 cm−1). Therefore, nitriles with similar isotope-editing for frequency shifting could be combined with alkynes for expanded imaging multiplexing.
As the stable isotope of hydrogen, deuterium-labeled chemical bonds (exemplified by the C-D) have unmatched advantages. Most importantly, since stable isotopes have almost the same physicochemical properties as their counterparts, the labeled molecules could be processed by cells’ natural machineries with minimal perturbation to the native biological functions. In addition, compared to the label-free imaging of C-H, C-D yields an improved SRS detection limit due to the absence of interfering cellular background. Although the Raman cross section of C-D is smaller than that of the triple bonds (Figure 2b), what is lacking in cross section could be compensated by the large labeling number. For example, palmitic acid, the most common long-chain saturated fatty acid in mammalian cells, can have up to 31 deuteriums per molecule as d31palmitic acid. Owing to these features, deuterium plays a significant role in Raman labeling and has been applied to interrogating a wide range of uptake and metabolic dynamics for targets including amino acids66, glucose70, fatty acids71, choline72, cholesterol73, water74, solvents33 and drugs67 (Figure 2f). For instance, the employment of deuterated amino acids allows imaging of complex protein metabolism, including synthesis, degradation, and analysis of temporally defined populations66 (Figure 2g). Similarly, d31palmitic acid enabled quantitative SRS visualization of fatty acid uptake and their metabolic incorporation29; and the deuteration of propylene glycol (PG) allowed the capture of real-time 3D penetration for this common pharmaceutical cosolvent/excipient across the mice stratum corneum67 (Figure 2g).
Deuterium labeling can also obtain protein-specific imaging in live cells for certain targets. For example, the aggregation-prone mutant Huntington (mHtt) protein harboring polyglutamine (polyQ) expansions has been shown to be specifically labeled by deuterated glutamine for their enrichment in the polyglutamine expansions68 (Figure 2h). This approach enabled the first quantitative analysis of mHtt and non-mHtt proteins inside the same protein aggregates in live cells without the need of any fluorescent labels. To expand the library of labeled targets apart from proteins, similar selective labeling strategies may be developed, such as the selective labeling of glycogen by deuterated glucose in live cancer cells75. The concept of harnessing such repeating units toward higher sensitivity is also adopted for C-D and triple-bond containing polymers, which could amplify the Raman signals by up to 105 fold69,76–79 (Figure 2i). The versatile applications of Raman-tagged cellular imaging are still expanding, and would benefit from easy synthetic accessibility to new vibrationally-tagged molecules. Toward this front, we envision that the recent synthetic advances in late-stage functionalization80–82 could provide convenient synthetic routes to deuterium or triple-bond tagged molecular targets beyond currently available pools.
Metabolic pathway analysis with deuterium labeling: from cell metabolism to microbiology
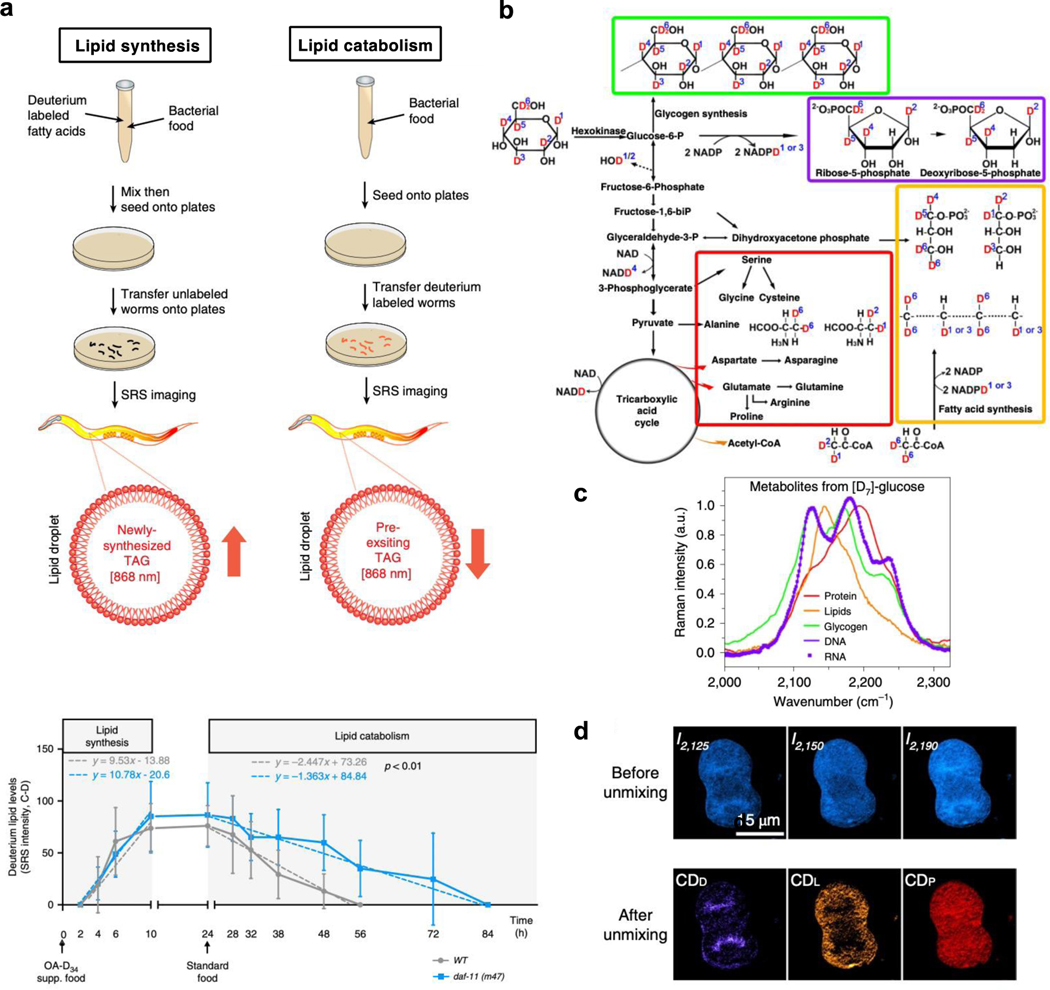
Various small Raman labels offer a wide range of applicability especially for investigating different aspects of cellular metabolism. To assay the uptake of metabolites (e.g. glucose), triple bonds are usually the top choice for their higher detectability57,65. However, even triple-bond tagged metabolic analogs usually stop at the early catabolic steps in the metabolic pathway. In this case, stable isotope tagged metabolites are superior for tracing transformations into the downstream metabolic products from live cells to organisms with minimum toxicity13. For example, with deuterated fatty acid labeling, lipid synthesis and mobilization could be non-invasively probed in live Caenorhabditis elegans (C. elegans) by SRS with high throughput83,84 (Figure 3a). C. elegans with daf-11 mutants were discovered to have no changes in the rate of lipid synthesis, but have a significant reduction in the rate of lipid catabolism84.
Figure 3. Metabolic pathway analysis with deuterium labeling.
(a) Utilizing SRS imaging assays to determine the rate of lipid synthesis (left) and lipid mobilization (right) with deuterium-labeled fatty acids in C. elegans. Signals derived from deuterium-labeled lipids in intestinal lipid droplets were quantified (bottom). daf-11 mutants have similar rates of lipid synthesis, but with reduced rates of lipid catabolism. Adapted in part with permission from ref83,84, Copyright 2017, 2020 Nature publishing group. (b) The biological pathway from d7-glucose incorporation for spectral tracing of deuterium isotopes (STRIDE) of various metabolic products. Adapted in part with permission from ref85, Copyright 2019 Nature publishing group. (c) Normalized C–D Raman spectra of five d7-glucose-derived biomolecules, including proteins, lipids, glycogen, DNA and RNA. Adapted in part with permission from ref85, Copyright 2019 Nature publishing group. (d) Images of a d7-glucose metabolically labeled mitotic HeLa cell before and after spectral unmixing. Adapted in part with permission from ref85, Copyright 2019 Nature publishing group.
Comprehensive cellular metabolism beyond metabolite uptake or distribution can be probed with suitable deuterium labeled probes. Glucose is the primary energy source for mammalian cells as well as an important precursor of downstream metabolites including amino acids, lipids, nucleic acids, glycogen and adenine dinucleotide phosphate (NADPH)86. While 3-OPG (i.e. alkyne-tagged glucose, Fig. 2d) is able to capture the glucose uptake in live cells and tissues, it stops at the phosphorylation step when going into the glucose metabolism pathway61. Recently, with deuterated glucose (i.e. d7-glucose) labeling, diverse downstream products, such as DNA/RNA, proteins, lipids and glycogen, have been shown to be sparsely labeled with deuterium through each corresponding metabolic pathway85 (Figure 3b). these sparsely labeled downstream products are spectrally separatable with varied features due to the different chemical environments surrounding the deuterium (Figure 3b&c)70,75,85. A linear combination algorithm can then be utilized to quantitatively retrieve the relative C-D enrichment maps in each identified species75,85 (Figure 3d). Alternatively, site-specific deuterated glucose instead of d7-glucose allows for tracing specific metabolic pathways. For example, 3-D-glucose ([3-D]Glc) was shown to monitor NADPH-mediated lipid synthesis through oxidative pentose phosphate pathway (oxPPP) by targeting lipid droplets88.
Metabolic reprogramming serves as a unique hallmark for cancer and neurodegenerative diseases. This Raman-based imaging platform for complex glucose metabolism hence forms a live-cell spatially resolved assay with subcellular resolution. Indeed, cancers cells have been shown to exhibit different levels of glucose uptake rate versus metabolism rate87. Subcellular glycogen accumulation through d7-glucose labeling was also discovered in cancer cells as a potential indicator for their resistance to glucose deficiency75. A similar spectral tracing strategy can be applied to cost-effective heavy water labeling (DO-SRS)74. Since water is the most abundant molecule in biological systems, the incorporation of deuterium from D2O to C-D in macromolecules is highly efficient even at low and biologically-safe D2O concentrations (e.g. 20% D2O). The resulting distinct spectral signatures of C-D enable visualizing both lipid and protein metabolism in animals with long-term incubation (26 days)74.
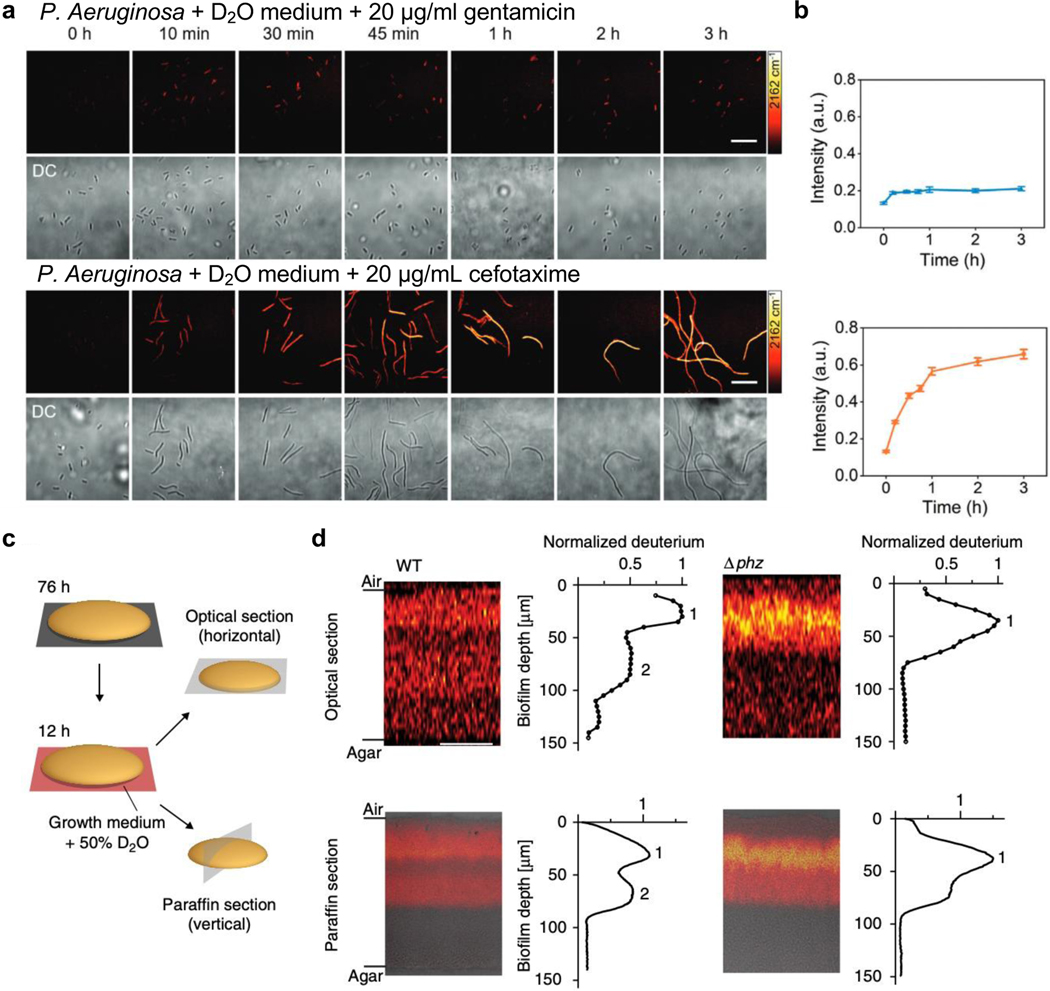
Assaying microbial metabolism is another application field that can be empowered by the metabolic Raman platform. The role of microbiota is increasingly recognized in human health. One of the most important problems associated with microbes is antibiotic resistance. Current standard antimicrobial susceptibility testing (AST) requires 16 – 24 h for multiple cell cycle growth. By culturing the microbes in 70% D2O medium and tracing the metabolic incorporation of deuterium into the biomass, varied metabolic responses to antibiotics can be probed in as short as 10 min, the fastest method to date89. The C-D signals indicated that Pseudomonas aeruginosa (P. aeruginosa), a common cause of hospital-acquired infection, exhibits distinct metabolic rate under different common antibiotics (gentamicin and cefotaxime) treatment (Figure 4a&b). As such, SRS imaging provides a rapid and cost-effective AST assay. In a more clinical-relevant P. aeruginosa biofilm system, 3D metabolic activity deep inside the film was visualized by SRS with 50% D2O medium labeling90. SRS’s inherent optical sectioning capability provides a convenient way to visualize 3D metabolic activity without the need of traditional paraffin embedding and sectioning (Figure 4c). The active metabolism in the hypoxic deep region was revealed to be supported by the small redox metabolite phenazine (phz) in the optical sections90 (Figure 4d top, WT vs Δphz, peak 2). This finding was validated by paraffin sections ((Figure 4d bottom, WT vs Δphz, peak 2). In different Staphylococcus aureus (S. aureus) biofilms, tagging antibiotic vancomycin with alkynes uncovered a non-uniform and limited penetration of antibiotic into biofilm with preferential affinity of the antibiotic to the cells instead of the extracellular polymeric matrix (EPM)91.
Figure 4. Assaying microbial metabolism with the biorthogonal SRS platform.
(a) Time-lapse SRS (targeting the C-D vibration, 2162 cm−1) and the corresponding transmission imaging of P. aeruginosa after culturing in D2O-containing medium with the addition of 20 μg mL−1 gentamicin (top) or cefotaxime (bottom). Scale bars: 20 μm. Adapted in part with permission from ref89, Copyright 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) Corresponding average C–D intensity plot (designating labeled biomass) over time for P. aeruginosa after culturing in D2O-containing medium with gentamicin (top blue) or cefotaxime (bottom orange) treatment. Number of cells N ≥ 10 per group. Error bars: SEM. Adapted in part with permission from ref89, Copyright 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (c) Schematic of two methods to visualize metabolic activity in colony biofilms. Optical sectioning uses the inherent 3D sectioning capability of SRS to acquire images in z-direction in 5 μm steps without sample preparation procedure. The paraffin section requires paraffin embedding and sectioning to provide 10-μm-thin slices for imaging. Adapted in part with permission from ref90, Copyright 2019 Nature publishing group. (d) SRS images and the corresponding plots of deuterium signals per biofilm depth for both optical sections and paraffin sections of colony P. aeruginosa biofilms after a 12-h incubation in D2O-containing medium. Data plots show the mean deuterium signal per biofilm depth. One replicate each of WT and Δphz is shown and is representative of at least five biological replicates. Scale bars: 50 μm. Adapted in part with permission from ref90, Copyright 2019 Nature publishing group.
Proposition of SRS imaging with small tagging
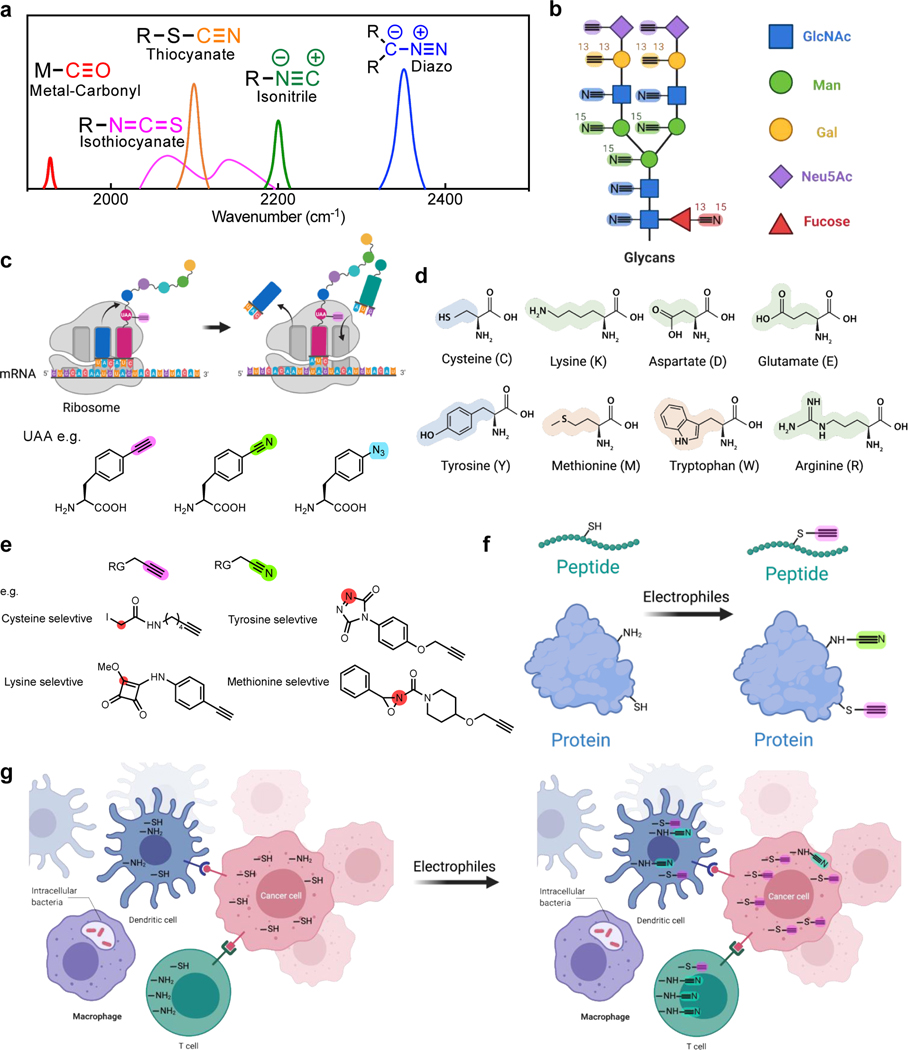
The coupling of SRS with small vibrational tags has established itself as a powerful live-cell imaging platform. Going beyond what has been achieved, below we discuss some of our thoughts for further improvements of molecular probes and potential opportunities of the platform in chemical biology. First, the wide cell-silent region remains spacious with room for spectral multiplexing (Figure 2a). Other triple bonds (Figure 5a), including metal-carbonyl (M-C≡O)92, isothiocyanate (-N=C=S), diazo (-N≡N), isonitrile (-N≡C) and thiocyanate (-S-C≡N), with strong vibrations in this region are worth in-depth investigations and engineering for their biological utilities. If these additional tags are available, the multiplexing of Raman-based profiling strategies could be largely expanded. New applications, such as imaging-based glycans profiling of cells, could then be envisioned. Glycans are oligosaccharides attached to biomacromolecules including proteins. They are regarded as post-translational modifications for modulating cell functions, but are still less understood due to the lack of studying methods93. A typical way to image glycan directly from cells is through metabolic incorporation of unnatural monosaccharides with small chemical reporters (e.g. azides or alkynes), which undergo sequential labeling via biorthogonal chemistry94. However, vastly different kinetics, selectivity and the scarce availability of the biorthogonal reactions limit the applicability of multiplexed monosaccharide labeling. With SRS imaging, we envision that spatial glycan profiling of monosaccharides covering N-acetylglucosamine (GlcNAc), mannose (Man), galactose (Gal), sialic acid (Neu5Ac), fucose in live systems will be possible with proper isotope-edited triple-bond tagging (Figure 5b) together with the expandable tag repertoire (Figure 5a).
Figure 5. Proposed applications for SRS imaging with small tagging.
(a) Additional triple bonds for SRS imaging which vibrate in the cell-silent region (The spectral features of these additional triple bonds are based on the reference compounds including Mn(tpm)(CO)3)Cl (metal-carbonyl), benzyl isothiocyanate (isothiocyanate), 4-bromobenzenediazonium tetrafluoroborate (diazo), benzyl isocyanide (isonitrile) and benzyl thiocyanate (thiocyanate).). (b) Spatial glycan profiling with triple bonds labeled monosaccharides. (c) Genetically-encoded protein labeling using triple-bond tagged unnatural amino acids (UAA). (d) Representative nucleophilic amino acid residues. (e) Representative triple-bond containing electrophiles that target specific amino acid residues. RG: reaction group. Red circles indicate the initial site of electrophilic reactivity. (f) Site-specific peptides and proteins labeling with amino-acid selective electrophiles (exemplified in e). (g) Potential spatial mapping for the reactivity of specific amino acids in complex biological systems with selective triple bond containing electrophiles (exemplified in e).
In addition to metabolic labeling, site-specific labeling through genetic encoding, such as utilizing unnatural amino acids (UAA) with genetic code expansion (Figure 5c), is another direction that is worth exploring for protein-selective Raman imaging. Although numerous UAAs carrying alkynes or nitriles have been developed (Figure 5c)62,95, SRS signals from these single-UAA labeled proteins are not sufficient96. While technical innovations are in urgent needs of sensitivity improvement42, new chemical or biological labeling strategies to incorporate an increased number (> 10) of triple bonds in one protein would also be a breakthrough for obtaining satisfying SRS signals for general protein imaging applications. Towards higher sensitivity, aqueous Glaser-Hay bioconjugation97, which may extend the terminal alkynes from UAAs into polyynes in situ, could be an alternative option to reduce the required labeling number of UAA by serving as a sequential signal-amplification method.
Compared to fluorophores, the above-shown small and multiplexed triple-bond Raman tags allow wider accessibility due to much smaller tag sizes and require no sequential labeling (e.g. click reaction) for highly-multiplexed applications. We hence expect that SRS imaging will contribute to solving certain important chemical biology questions, such as spatially resolving the proteome reactivity in live cells, in which the signal level is not a problem. The side chain reactivity of canonical amino acids together with the local protein microenvironment builds up “hotspots” in the proteome98. The electron-rich side chains of various amino acids including cysteine, lysine, aspartate, glutamate, tyrosine, and methionine are naturally the targets for electrophiles (Figure 5d). These functional amino acids are always catalytic residues or sites of post-translational modifications, and thus they are the keys in modulating cellular functions. The side-chain reactivity of the amino acids can be quantitatively analyzed by isotopic tandem orthogonal proteolysis activity based protein profiling (isoTOP-ABPP) platform98,99. This mass spectrometry method has high throughput and unmatched protein resolvability, but it lacks spatial information and cannot track dynamic processes with intra- and intercellular interaction in complex biological systems.
Recently, researchers comprehensively profiled the proteome-wide reactivity for a library of 54 alkyne-bearing electrophiles100. They identified highly selective probes specific to a total of 9 amino acids plus the N-terminus with different reactivities (several representative probes are listed in Figure 5e). These alkyne-tagged electrophiles provide an effective site-selective labeling tool for proteins or peptides (Figure 5f). The alkyne handles across these electrophile probes (Figure 5e) could be substituted with color-resolvable isotope-labeled triple bonds from the Raman probe repertoire (Figure 5a&5b). With this design, it is possible to generate maps for amino acid reactivities in a complex biological system such as cancer immunology and microbe-host environment. By supplying the probes into the systems, the subsequent multiplexed SRS imaging would allow single-cell profiling en masse (Figure 5g). Enough signals should be expected with this strategy since similar proteome-wide new protein synthesis using alkyne-tagged methionine (HPG, Figure 2c) labeling was already demonstrated by SRS imaging in live cells32. This application would be highly challenging for direct fluorescent labeling as the active amino acid residues are likely less accessible to bulkier fluorophores without the sequential labeling step. Together with isoTOP-ABPP and protein targeting labeling, proteome-wide SRS screening may contribute to annotating specific proteins and elucidating their spatially-resolved dynamic activity changes.
Next-generation Raman probe palettes for highly sensitive and super-multiplexed imaging
Even with the signal amplification from SRS and the introduced Raman tags, there is still a sensitivity gap between the SRS and the fluorescence microscopy. The SRS detection limit of EdU is 200 μM in live cells while the most sensitive fluorophores offer single-molecule sensitivity (< 10 nM). One central drive in SRS imaging is to narrow this sensitivity gap. A promising solution that has been proven successful is the development of highly sensitive Raman probes. As the Raman bands are inherently narrow (peak width about 10 cm−1, ~ 50–100 times narrower than that for fluorescent peaks), these Raman probes would enable high-throughput super-multiplexed (> 20 channels) imaging once the requirement for sensitivity is met. Therefore, Raman imaging holds the promise of breaking the color barrier of fluorescence imaging and should greatly benefit systematic biology investigations.
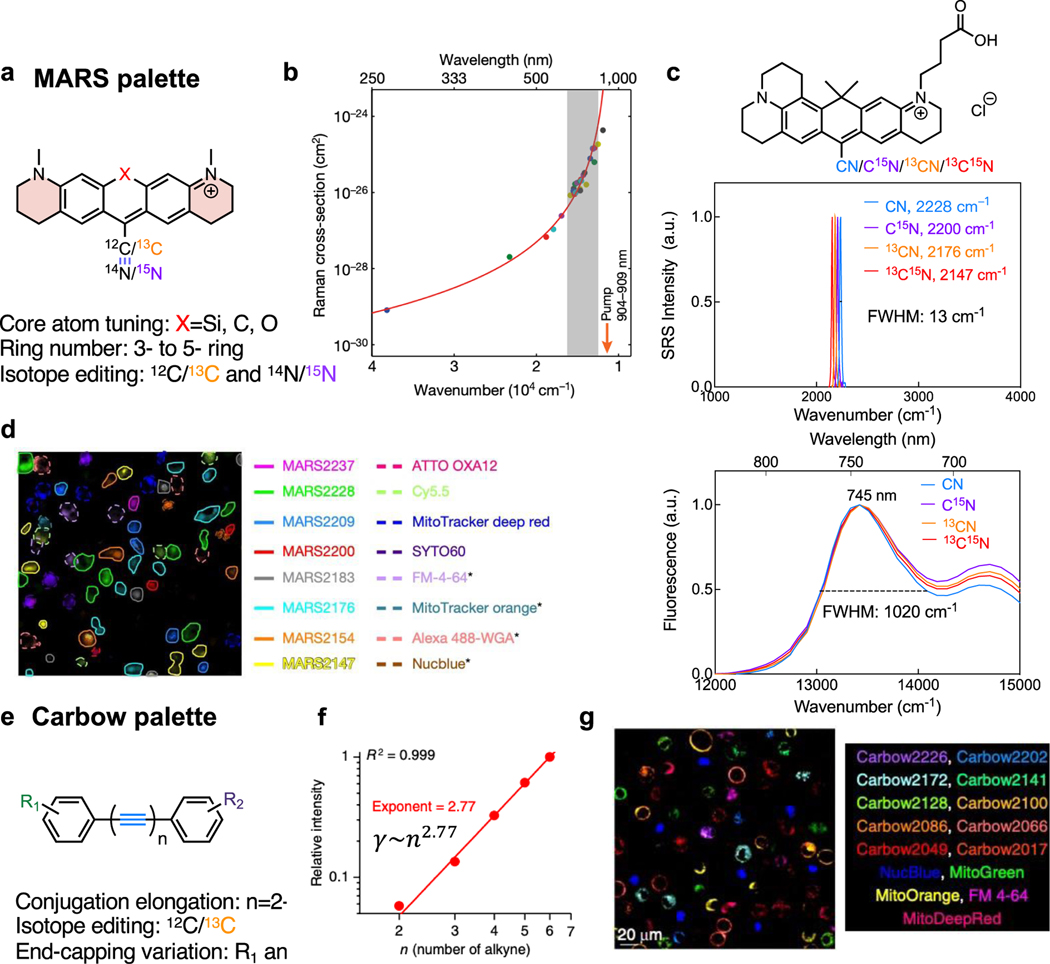
Two sets of highly sensitive and multiplexed Raman palettes have been developed for SRS imaging over the past five years. The first is xanthene-based electronic pre-resonance enhanced Manhattan Raman scattering (MARS) dyes47,48 (Figure 6a). By carefully tuning the absorption of the dyes (650 – 760 nm) to moderately close to the laser wavelength (~ 900 nm), SRS intensities of the nitrile vibration from these dyes could be pre-resonantly enhanced by up to 104 (detection limit up to 250 nM) with a well-maintained high signal-to-background ratio (Figure 6b&c). Taking advantage of the much narrower (78 times, 13 vs 1020 cm−1 of FWHM) SRS peak compared with the fluorescence absorption peak and the multiplexing from isotope labeling as illustrated in the example of MARS2228 series dyes (Figure 6c), MARS dyes have the inherent capability for super-multiplexed imaging. With central atom (position 10) replacement, ring expansion and isotope editing, an SRS dye palette with up to 24 plex was created for super-multiplexed imaging47 (Figure 6d). Through investigations in our lab, we later found that different scaffolds could present vastly different pre-resonance SRS signals even with the same absorption wavelength, possibly due to the complicated electronic–vibrational coupling strength. This indicates that finding the right chemical scaffold is as important as physically modulating the absorption of the electronic structures. To facilitate the development of new palettes, theoretical models and computational tools are also urgently needed101. While the construction of near-infrared chromophores is more challenging, the suitable electronic pre-resonance palettes could be largely expanded with freely tunable laser sources into the visible range102,103.
Figure 6. Physical principles for current strong and super-multiplexed Raman probes.
(a) Construction of 24-color MARS xanthene palette. (b) The dramatic increase of SRS cross sections by electronic pre-resonance enhancing effects. Adapted in part with permission from ref47, Copyright 2017 Nature publishing group. (c) The nitrile SRS peak (top) and the fluorescence absorption spectra (bottom) of a series of four isotope labeled MARS dyes (structure drawn at the top). The full width half maximum (FWHM) of the nitrile SRS peaks and the dominant absorption peak is listed. (d) Live-cell 16-color imaging with MARS and commercial dyes. Adapted in part with permission from ref47, Copyright 2017 Nature publishing group. (e) Construction of 20-color Carbow polyyne palette. (f) The super-linear (an exponent of 2.77) SRS-signal growth with the number of conjugation alkynes. Adapted in part with permission from ref104, Copyright 2018 Nature publishing group. (g) Live-cell 15-color imaging with Carbow and commercial dyes. Adapted in part with permission from ref104, Copyright 2018 Nature publishing group.
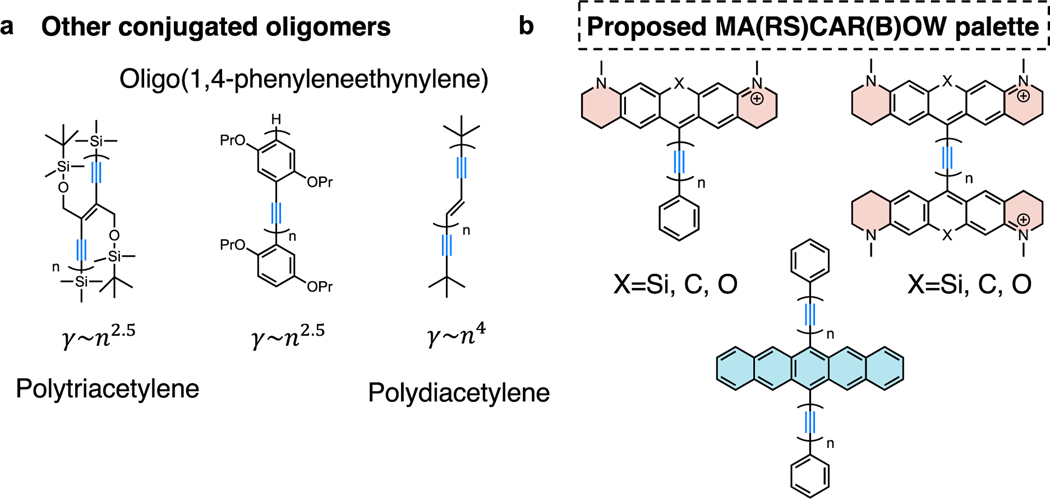
The second established set of highly-sensitive Raman probes is the Carbow series, which is a set of linear conjugated alkynes with aromatic capping104 (Figure 6e). Different from the electronic enhancement mechanism, the strong SRS signals of polyynes originate from the amplified second-order hyperpolarizability () from conjugated alkynes. When the number of conjugated alkynes increases from 2 to 6, the Raman intensity grows super-linearly with an exponent of 2.77 (i.e. 2.77, Figure 6f), offering a desirable detectability down to 630 nM for 4-yne. The increase of conjugation length is also accompanied by a desirable Raman peak shift, offering more spectral resolvability for multiplexed applications. Further combined with end-capping substitution and isotope editing, another 20-color CARBOW palette was created (Figure 6g). Without the involvement of electronic excitation, polyynes are free of photobleaching or environmental quenching. Although slightly smaller in Raman cross section compared to the MARS palette, the Carbow palette has neutral scaffolds and is more suitable for live-cell targeted imaging with lower non-specific background. Theoretically, the longest conjugation length of polyynes could be over 6 with higher sensitivity than MARS, but at the risk of decreased stability. Going beyond the polyyne structures, other oligomers such as polytriacetylenes, oligo(1,4-phenyleneethynylene)s105, polydiacetylenes77 have also shown exponential power-law relationship between the second-order hyperpolarizability ( and repeating units (n) with slightly increased sizes (Figure 7a) 106, thus are promising candidates for next-generation strong Raman probes.
Figure 7. Possible oligomers and proposed MA(RS)CAR(B)OW palette as next-generation strong Raman probes.
(a) Other alkyne-conjugated oligomers with a similar exponential power-law relationship between second-order hyperpolarizability () and repeating units (n). (b) Molecule designs for building stronger Raman probes by simultaneously utilizing the electronic pre-resonance and hyperpolarizability enhancing effects.
The ultimate goal of improving the signals of Raman probes is to achieve single-molecule SRS imaging, meaning that even for the most sensitive MARS probes, there still needs a sensitivity improvement of ~ 30–50 folds. Towards this goal, a possible design direction for Raman probes is to combine MARS and Carbow scaffolds in the same molecules for cooperative enhancement from electronic pre-resonance and amplified (Figure 7b). That is to replace the single alkyne in the current alkyne-containing MARS probes to be conjugated polyynes (Figure 7b). In the case that the pre-resonance effect remains, the Raman signal is also expected to undergo exponential increase with the number of conjugated alkynes, thus the resulting MA(RS)CAR(B)OW palette might have another dozens of times enhancement compared with the MARS palette to meet the desired single-molecule detectability.
Functional Raman Imaging Probes
Probing biological systems through chemical probes is a central topic for chemical biology study. Leveraging the live-cell compatibility, strict linear-concentration dependence and non-quenching nature of Raman signals, SRS probes are preferred for quantitative analysis. Although small vibrational probes have been extensively utilized for spectroscopic analysis of the targeted cellular environment, such as electrostatic interactions, electrical currents, and temperature4,62,63, the development of highly sensitive Raman imaging probes for environmental sensing is still in its infancy. Benefiting from the unique super-multiplexed Raman features, functional SRS imaging probes should open the door for comprehensive investigations to elucidate the intricate intracellular and cell-to-cell interactions. Over the past four years, we witnessed the rapid growth of the development of such highly sensitive SRS sensors. Based on their Raman spectroscopic features, we categorized these Raman sensors into four classes: sensing by peak shifts, peaks enhancement, peak generation, and peak switching. Through the following discussions, we aim to summarize systematic guidelines for designing new probes with tailored functions.
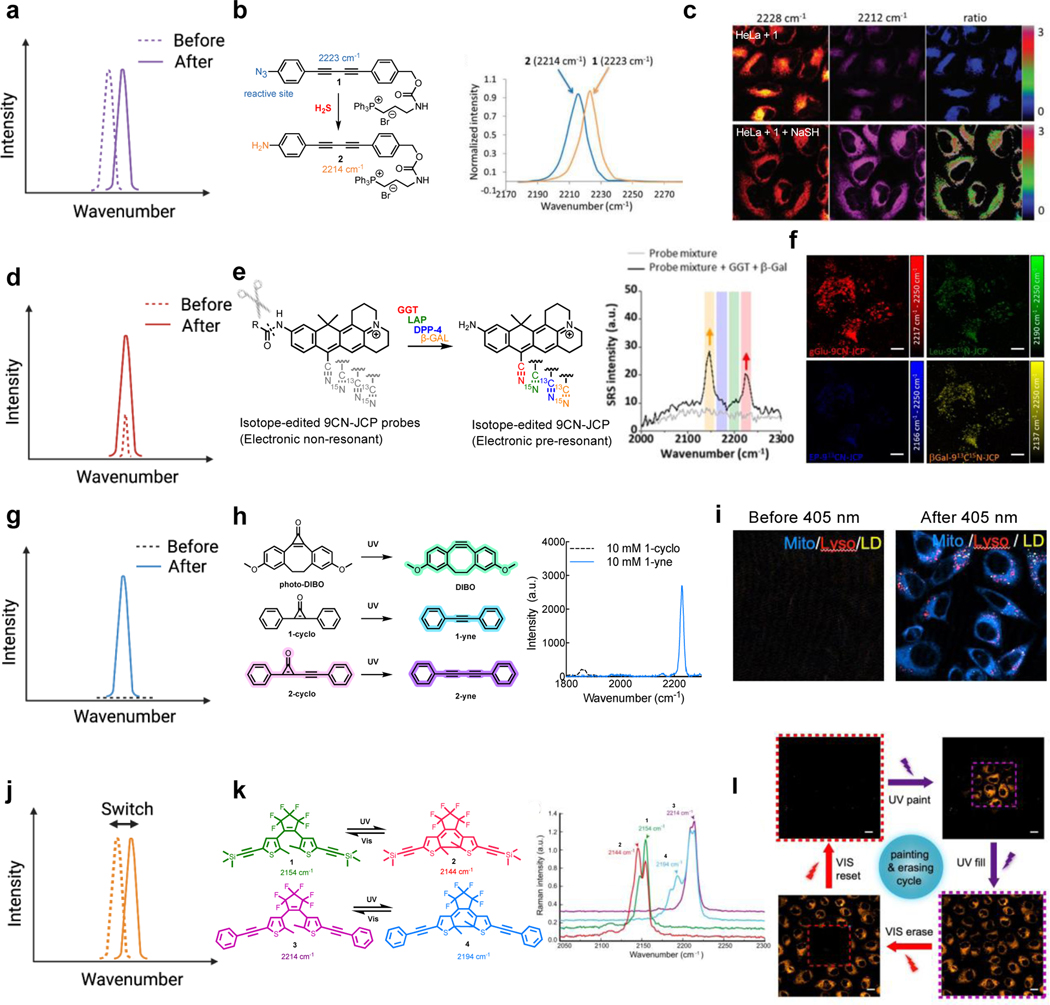
Most current Raman sensors are designed for sensing the targeted chemical environment through Raman peak shifts (Figure 8a). The chemical reactions triggered by environmental stimuli are designed to change the chemical structures adjacent to the Raman reporters (e.g. alkynes or nitriles), therefore resulting in distinct Raman peak shifts. For example, in a 2-yne scaffold, when the electron-withdrawing azide group on the end-phenyl cap is reduced to the electron donating amine in the presence of the reductive hydrogen sulfide species (such as H2S and NaSH), the alkyne Raman peak shows a 9 cm−1 red shift107 (Figure 8b), which is significant enough to be distinguished by SRS imaging (the typical full-width-half-maximum (FWHM) for alkyne peak is 15 cm−1 and the typical spectral width of SRS lasers is ~ 12–15 cm−1). The demonstrated SRS ratiometric imaging indeed showed a strong response to NaSH level changes in the mitochondria of live cells (Figure 8c). With similar targeted reactions, triple-bond Raman probes with the peak-shift principles have also been rationally designed and developed for sensing pH108, fluoride109 and metal ions110. Recently, the isotope exchange reactions (especially the H/D exchange) have been harnessed on terminal alkynes for both two-color imaging and cellular environmental sensing applications111,112, taking advantage of the dramatic alkyne Raman peak shift (> 130 cm−1) due to both the large mass difference between D and H and the quantum coupling between the alkyne and the adjacent C-D. In addition to chemical reactions, Raman peak frequency can also be tuned by the surrounding physical environment. A well-known example is that the hydrogen-bounding and electrostatics can shift the peak frequencies of triple bonds especially nitriles, an effect known as vibrational solvatochromism. By specifically mapping the peak frequency of nitrile-bearing MARS Raman dyes in live cells, the bound-water percentage in cytoplasm was revealed to be around 60% while that in nucleus was about 30%113,114. Recently, the SRS peak of voltage-sensitive rhodopsin has also been shown to shift upon voltage changes115.
Figure 8. SRS probes for functional imaging with different spectroscopic signatures and their chemical designs.
(a) SRS sensors based on peak shifts. (b) The conversion of -N3 to –NH2 induces a Raman peak shift of 9 cm−1 on the core of 2-yne. Adapted in part with permission from ref107. Copyright 2018 The Royal Society of Chemistry.(c) Ratiometric imaging for NaSH sensing from the peak shift of the probe 1. Adapted in part with permission from ref107. Copyright 2018 The Royal Society of Chemistry. (d) SRS sensors based on peak enhancements. (e) The electronic non-resonant 9CN-JCP probes undergo a large enzyme-induced red-shift in absorption to the electronic pre-resonant region with dramatic enhancement of SRS signals. Adapted in part with permission from ref50. Copyright 2020 American Chemical Society. (f) The enhanced SRS signals enable multiplexed imaging for live-cell enzymes (GGT, LAP, DPP-4, and β-Gal) sensing. Scale bars: 10 μm. Adapted in part with permission from ref50. Copyright 2020 American Chemical Society. (g) SRS sensors based on the peak generation. (h) The photoreactive conversion of cyclopropenones into alkynes generates strong SRS contrast with a sharp Raman peak. Adapted in part with permission from ref54. Copyright 2022 American Chemical Society. (i) Engineered cyclopropenone probes enable three-color organelle-target photoactivatable SRS imaging in live HeLa cells (Mito: mitochondria channel; Lyso: lysosome channel; LD: lipid droplet channel). Adapted in part with permission from ref54. Copyright 2022 American Chemical Society. (j) SRS sensors based on switchable peaks. (k) The Raman peak shifts reversibly upon photoisomerization of the diarylethene when irradiated by ultraviolet (UV) or visible light. Adapted in part with permission from ref51, Copyright 2021 Nature publishing group. (l) This photo-switchable peak shift enables SRS painting/erasing of cells with labelled alkyne-diarylethene. Scale bars: 20 μm. Adapted in part with permission from ref51, Copyright 2021 Nature publishing group.
Intensity enhancement represents another less explored category of design principle for SRS sensors (Figure 8d). Recently, modulation of electronic pre-resonance SRS effect was implemented for intracellular enzyme activity sensing. The nitrile xanthene scaffold, once caged by amides structures (9CN-JCP probes), will absorb in the visible region (506 nm, electronic non-resonance) with almost invisible SRS signals. However, the enzymatic conversion of amide structures to amine structures (9CN-JCPs) will dramatically boost SRS signals by shifting the absorption to the near-infrared region (630 nm, electronic pre-resonance) (Figure 8e). With isotope editing on the nitrile to generate multiple colors, this elegant intensity-modulation principle was successfully exploited to create a 4-color enzymatic sensing probe palette50 (Figure 8e). In this case, simultaneously detecting the activities of four distinct enzymes was demonstrated for effective profiling of different cancer cell phenotypes (Figure 8f). With the super-multiplexed MARS dye palette, this enzyme-activatable sensor design holds high promises for profiling more than ten enzymes simultaneously.
Peak generation allows background-free imaging and is highly beneficial for sensitive and multiplexed detection (Figure 8g). The Raman intensity activation does not solely come from the cellular or the chemical environment, but it can also originate from the photons. In a recent design, the first photoactivatable SRS imaging probes were developed based on the photocaged alkynes, the cyclopropenones (Figure 8h)54. Such rationally designed cyclopropenone structures were optimized with live-cell compatibility and have been proven to be well-suitable for multiplexed live-cell imaging and tracking54 (Figure 8i). With the high precision spatial-temporal control offered by photoactivation, this series of isotope editing probes was demonstrated for multiplexed tracking from the subcellular to the single-cell level. Upon further improvement of multiplexing, these new Raman sensors may illuminate complex cell-to-cell interactions and facilitate massive-parallel cell profiling. For example, combining photoactivatable SRS probes and single-cell RNA sequencing (scRNA-seq) could likely enable spatially-resolved transcriptomics profiling116.
Raman peaks could also be reversibly switchable with light manipulations (Figure 8j). Such features are crucial to a variety of biological investigations including tracking protein dynamics, subcellular environment sensing and super-resolution imaging. In 2021, three groups independently reported photophysical or photochemical approaches to achieve photoswitchable SRS imaging51–53. To name one example, the alkyne-tagged diarylethene showed impressive photo-switchable property in the cell-silent spectral window51 (Figure 8k). The UV induced photoisomerization converts diarylethene from the open-ring state to the closed-ring state while visible light would induce the reverse conversion, accompanied by the switching of SRS peak intensities. These alkynes-tagged diarylethenes were demonstrated for photo-rewritable patterning and mitochondria tracking (Figure 8l). It is noteworthy that the SRS readout lasers would induce the off-switch pathway, competing with the UV induced photo-cyclization, underscoring the careful choice of molecular absorption when designing the new probes.
Conclusion
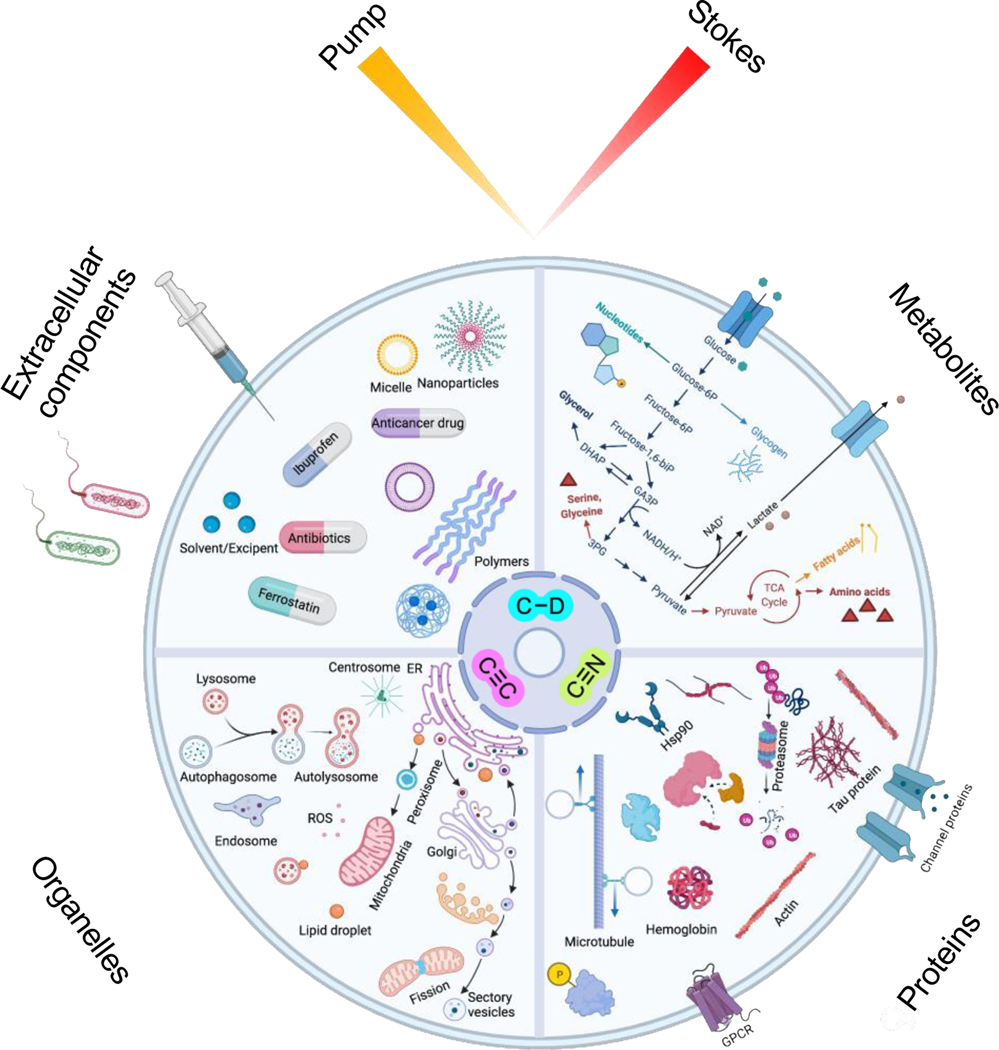
Through the past decade we have seen that the SRS microscopy has transited from the initial technical demonstration into a powerful method with the potential to answer many biological questions in a way no other methods can (Figure 9). Such transformation is impossible without appropriate Raman probes. With small triple-bond or isotope tagging, SRS microscopy enables the visualization and analysis of dynamic metabolism pathways in live cells and animals. From chemistry innovations, the construction of highly sensitive MARS and Carbow palettes realized the full potential of Raman for super-multiplexed imaging. The recently developed functional Raman sensors further expanded the applications for multiplexed cellular environment sensing and high precision spatial-temporal tracking. We also envision the design with further improved multiplexing and sensitivity of the Raman probes as a next step towards interrogating more complex biology.
Figure 9. Representative and envisioned applications of SRS imaging, particularly with the development of Raman-tailored bioorthogonal chemical tags of C≡C, C≡N and C-D.
The dynamic processes of targets across different molecular scales, from small-molecule metabolites to proteins, organelles, and extracellular components, can be visualized with high specificity and low perturbation.
As the development of Raman probes continue to be a central topic for the SRS community, we also hope to invoke brainstorming to borrow the wisdom from the chemical biology field and to promote vibrational imaging to solve biological questions. In retrospect, much of the recent progress of SRS probes was inspired by other fields: deuterium and other isotope labeling are prevalent in mass spectrometry; alkynes are the most important biorthogonal chemical group handles; MARS dyes originate from the design of commercially available fluorophores; diarylethene is a class of well-established photoswitchable chromophores, etc. The knowledge across fields should accelerate the advances of SRS probes into higher selectivity, sensitivity, photostability, biocompatibility, and multiplexing capability. Together with the developments of instrumentation, biorthogonal chemistry, molecular delivery and labeling methods, data analysis and more, SRS microscopy will further develop into an indispensable tool for chemical biology studies.
Acknowledgement:
L. Wei acknowledges the startup funds from California Institute of Technology and Grant No. DP2 GM140919 from National Institute of Health. We thank A.Colazo for helpful discussion.
Reference:
- (1).Kneipp K; Kneipp H; Itzkan I; Dasari RR; Feld MS Ultrasensitive Chemical Analysis by Raman Spectroscopy. Chem. Rev. 1999, 99 (10), 2957–2975. 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- (2).Fang C; Frontiera RR; Tran R; Mathies RA Mapping GFP Structure Evolution during Proton Transfer with Femtosecond Raman Spectroscopy. Nature 2009, 462 (7270), 200–204. 10.1038/nature08527. [DOI] [PubMed] [Google Scholar]
- (3).Kukura P; McCamant DW; Mathies RA Femtosecond Stimulated Raman Spectroscopy. Annu. Rev. Phys. Chem. 2007, 58, 461–488. 10.1146/annurev.physchem.58.032806.104456. [DOI] [PubMed] [Google Scholar]
- (4).Chattopadhyay A; Boxer SG Vibrational Stark Effect Spectroscopy. J. Am. Chem. Soc. 1995, 117 (4), 1449–1450. 10.1021/ja00109a038. [DOI] [Google Scholar]
- (5).Thomas GJ Raman Spectroscopy of Protein and Nucleic Acid Assemblies. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 1–27. 10.1146/annurev.biophys.28.1.1. [DOI] [PubMed] [Google Scholar]
- (6).Raman CV; Krishnan KS A New Type of Secondary Radiation [11]. Nature 1928, 121 (3048), 501–502. 10.1038/121501c0. [DOI] [Google Scholar]
- (7).Zumbusch A; Holtom GR; Xie XS Three-Dimensional Vibrational Imaging by Coherent Anti-Stokes Raman Scattering. Phys. Rev. Lett. 1999, 82 (20), 4142–4145. 10.1103/PhysRevLett.82.4142. [DOI] [Google Scholar]
- (8).Min W; Freudiger CW; Lu S; Xie XS Coherent Nonlinear Optical Imaging: Beyond Fluorescence Microscopy. Annu. Rev. Phys. Chem. 2011, 62, 507–530. 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ploetz E; Laimgruber S; Berner S; Zinth W; Gilch P Femtosecond Stimulated Raman Microscopy. Appl. Phys. B Lasers Opt. 2007, 87 (3), 389–393. 10.1007/s00340-007-2630-x. [DOI] [Google Scholar]
- (10).Freudiger CW; Min W; Saar BG; Lu S; Holtom GR; He C; Tsai JC; Kang JX; Xie S Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science 2008, 322 (19), 1857–1861. 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ozeki Y; Dake F; Kajiyama S; Fukui K; Itoh K Analysis and Experimental Assessment of the Sensitivity of Stimulated Raman Scattering Microscopy. Opt. Express 2009, 17 (5), 3651. 10.1364/oe.17.003651. [DOI] [PubMed] [Google Scholar]
- (12).Cheng JX; Xie XS Vibrational Spectroscopic Imaging of Living Systems: An Emerging Platform for Biology and Medicine. Science 2015, 350 (6264). 10.1126/science.aaa8870. [DOI] [PubMed] [Google Scholar]
- (13).Hu F; Shi L; Min W Biological Imaging of Chemical Bonds by Stimulated Raman Scattering Microscopy. Nat. Methods 2019, 16 (9), 830–842. 10.1038/s41592-019-0538-0. [DOI] [PubMed] [Google Scholar]
- (14).Saar BG; Freudiger CW; Reichman J; Stanley CM; Holtom GR; Xie XS Video-Rate Molecular Imaging in Vivo with Stimulated Raman Scattering. Science 2010, 330 (6009), 1368–1370. 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Camp CH; Lee YJ; Heddleston JM; Hartshorn CM; Walker ARH; Rich JN; Lathia JD; Cicerone MT High-Speed Coherent Raman Fingerprint Imaging of Biological Tissues. Nat. Photonics 2014, 8 (8), 627–634. 10.1038/nphoton.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wakisaka Y; Suzuki Y; Iwata O; Nakashima A; Ito T; Hirose M; Domon R; Sugawara M; Tsumura N; Watarai H; et al. Probing the Metabolic Heterogeneity of Live Euglena Gracilis with Stimulated Raman Scattering Microscopy. Nat. Microbiol. 2016, 1 (16124). 10.1038/nmicrobiol.2016.124. [DOI] [PubMed] [Google Scholar]
- (17).Bi Y; Yang C; Chen Y; Yan S; Yang G; Wu Y; Zhang G; Wang P Near-Resonance Enhanced Label-Free Stimulated Raman Scattering Microscopy with Spatial Resolution near 130 Nm. Light Sci. Appl. 2018, 7 (1), 1–10. 10.1038/s41377-018-0082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Qian C; Miao K; Lin LE; Chen X; Du J; Wei L Super-Resolution Label-Free Volumetric Vibrational Imaging. Nat. Commun. 2021, 12 (1), 1–10. 10.1038/s41467-021-23951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shi L; Klimas A; Gallagher B; Cheng Z; Fu F; Wijesekara P; Miao Y; Ren X; Zhao Y; Min W Super-Resolution Vibrational Imaging Using Expansion Stimulated Raman Scattering Microscopy. bioRxiv 2021, 2021.12.22.473713. 10.1101/2021.12.22.473713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zipfel WR; Williams RM; Webb WW Nonlinear Magic: Multiphoton Microscopy in the Biosciences. Nat. Biotechnol. 2003, 21 (11), 1369–1377. 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- (21).Jonkman J; Brown CM; Wright GD; Anderson KI; North AJ Tutorial: Guidance for Quantitative Confocal Microscopy. Nat. Protoc. 2020, 15 (5), 1585–1611. 10.1038/s41596-020-0313-9. [DOI] [PubMed] [Google Scholar]
- (22).Miller DR; Jarrett JW; Hassan AM; Dunn AK Deep Tissue Imaging with Multiphoton Fluorescence Microscopy. Curr. Opin. Biomed. Eng. 2017, 4, 32–39. 10.1016/j.cobme.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hill AH; Manifold B; Fu D Tissue Imaging Depth Limit of Stimulated Raman Scattering Microscopy. Biomed. Opt. Express 2020, 11 (2), 762. 10.1364/boe.382396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fu D; Holtom G; Freudiger C; Zhang X; Xie XS Hyperspectral Imaging with Stimulated Raman Scattering by Chirped Femtosecond Lasers. J. Phys. Chem. B 2013, 117 (16), 4634–4640. 10.1021/jp308938t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhang D; Wang P; Slipchenko MN; Ben-Amotz D; Weiner AM; Cheng JX Quantitative Vibrational Imaging by Hyperspectral Stimulated Raman Scattering Microscopy and Multivariate Curve Resolution Analysis. Anal. Chem. 2013, 85 (1), 98–106. 10.1021/ac3019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Figueroa B; Hu R; Rayner SG; Zheng Y; Fu D Real-Time Microscale Temperature Imaging by Stimulated Raman Scattering. J. Phys. Chem. Lett. 2020, 11 (17), 7083–7089. 10.1021/acs.jpclett.0c02029. [DOI] [PubMed] [Google Scholar]
- (27).Yue S; Li J; Lee S-Y; Lee HJ; Shao T; Song B; Cheng L; Masterson TA; Liu X; Ratliff TL; et al. Cholesteryl Ester Accumulation Induced by PTEN Loss and PI3KAKT Activation Underlies Human Prostate Cancer Aggressiveness. Cell Metab. 2014, 19, 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li J; Condello S; Thomes-Pepin J; Ma X; Xia Y; Hurley TD; Matei D; Cheng JX Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 2017, 20 (3), 303–314.e5. 10.1016/j.stem.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Du J; Su Y; Qian C; Yuan D; Miao K; Lee D; Ng AHC; Wijker RS; Ribas A; Levine RD; et al. Raman-Guided Subcellular Pharmaco-Metabolomics for Metastatic Melanoma Cells. Nat. Commun. 2020, 11 (1), 4830. 10.1038/s41467-020-18376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chen T; Yavuz A; Wang MC Dissecting Lipid Droplet Biology with Coherent Raman Scattering Microscopy. J. Cell Sci. 2022, 135 (5). 10.1242/jcs.252353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Shen Y; Zhao Z; Zhang L; Shi L; Shahriar S; Chan RB; Di Paolo G; Min W Metabolic Activity Induces Membrane Phase Separation in Endoplasmic Reticulum. Proc. Natl. Acad. Sci. 2017, 114 (51), 13394–13399. 10.1073/pnas.1712555114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wei L; Hu F; Shen Y; Chen Z; Yu Y; Lin CC; Wang MC; Min W Live-Cell Imaging of Alkyne-Tagged Small Biomolecules by Stimulated Raman Scattering. Nat. Methods 2014, 11 (4), 410–412. 10.1038/nmeth.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chiu WS; Belsey NA; Garrett NL; Moger J; Delgado-Charro MB; Guy RH Molecular Diffusion in the Human Nail Measured by Stimulated Raman Scattering Microscopy. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (25), 7725–7730. 10.1073/pnas.1503791112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Fu D; Zhou J; Zhu WS; Manley PW; Wang YK; Hood T; Wylie A; Xie XS Imaging the Intracellular Distribution of Tyrosine Kinase Inhibitors in Living Cells with Quantitative Hyperspectral Stimulated Raman Scattering. Nat. Chem. 2014, 6 (7), 614–622. 10.1038/nchem.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Huang KC; Li J; Zhang C; Tan Y; Cheng JX Multiplex Stimulated Raman Scattering Imaging Cytometry Reveals Lipid-Rich Protrusions in Cancer Cells under Stress Condition. iScience 2020, 23 (3), 100953. 10.1016/j.isci.2020.100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Nitta N; Iino T; Isozaki A; Yamagishi M; Kitahama Y; Sakuma S; Suzuki Y; Tezuka H; Oikawa M; Arai F; et al. Raman Image-Activated Cell Sorting. Nat. Commun. 2020, 11 (1), 1–16. 10.1038/s41467-020-17285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ji M; Orringer DA; Freudiger CW; Ramkissoon S; Liu X; Lau D; Golby AJ; Norton I; Hayashi M; Agar NYR; et al. Rapid, Label-Free Detection of Brain Tumors with Stimulated Raman Scattering Microscopy. Sci. Transl. Med. 2013, 5 (201). 10.1126/scitranslmed.3005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hollon TC; Pandian B; Adapa AR; Urias E; Save AV; Khalsa SSS; Eichberg DG; D’Amico RS; Farooq ZU; Lewis S; et al. Near Real-Time Intraoperative Brain Tumor Diagnosis Using Stimulated Raman Histology and Deep Neural Networks. Nat. Med. 2020, 26 (1), 52–58. 10.1038/s41591-019-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ji M; Arbel M; Zhang L; Freudiger CW; Hou SS; Lin D; Yang X; Bacskai BJ; Sunney Xie X Label-Free Imaging of Amyloid Plaques in Alzheimer’s Disease with Stimulated Raman Scattering Microscopy. Sci. Adv. 2018, 4 (11), 1–9. 10.1126/sciadv.aat7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lin LE; Miao K; Qian C; Wei L High Spatial-Resolution Imaging of Label-Free in Vivo Protein Aggregates by VISTA. Analyst 2021, 146 (13), 4135–4145. 10.1039/d1an00060h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Berto P; Andresen ER; Rigneault H Background-Free Stimulated Raman Spectroscopy and Microscopy. Phys. Rev. Lett. 2014, 112 (5), 1–5. 10.1103/PhysRevLett.112.053905. [DOI] [PubMed] [Google Scholar]
- (42).Casacio CA; Madsen LS; Terrasson A; Waleed M; Barnscheidt K; Hage B; Taylor MA; Bowen WP Quantum-Enhanced Nonlinear Microscopy. Nature 2021, 594 (7862), 201–206. 10.1038/s41586-021-03528-w. [DOI] [PubMed] [Google Scholar]
- (43).Lin H; Lee HJ; Tague N; Lugagne JB; Zong C; Deng F; Shin J; Tian L; Wong W; Dunlop MJ; et al. Microsecond Fingerprint Stimulated Raman Spectroscopic Imaging by Ultrafast Tuning and Spatial-Spectral Learning. Nat. Commun. 2021, 12 (1), 1–12. 10.1038/s41467-021-23202-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kobayashi-Kirschvink KJ; Gaddam S; James-Sorenson T; Grody E; Ounadjela JR; Ge B; Zhang K; Kang JW; Xavier R; So PTC; et al. Raman2RNA: Live-Cell Label-Free Prediction of Single-Cell RNA Expression Profiles by Raman Microscopy. bioRxiv 2021, 2021.11.30.470655. 10.1101/2021.11.30.470655. [DOI] [Google Scholar]
- (45).Manifold B; Men S; Hu R; Fu D A Versatile Deep Learning Architecture for Classification and Label-Free Prediction of Hyperspectral Images. Nat. Mach. Intell. 2021, 3 (4), 306–315. 10.1038/s42256-021-00309-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wei L; Hu F; Chen Z; Shen Y; Zhang L; Min W Live-Cell Bioorthogonal Chemical Imaging: Stimulated Raman Scattering Microscopy of Vibrational Probes. Acc. Chem. Res. 2016, 49 (8), 1494–1502. 10.1021/acs.accounts.6b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wei L; Chen Z; Shi L; Long R; Anzalone AV; Zhang L; Hu F; Yuste R; Cornish VW; Min W Super-Multiplex Vibrational Imaging. Nature 2017. 10.1038/nature22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Miao Y; Qian N; Shi L; Hu F; Min W 9-Cyanopyronin Probe Palette for Super-Multiplexed Vibrational Imaging. Nat. Commun. 2021, 12, 4518. 10.1038/s41467-021-24855-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Shi L; Wei M; Miao Y; Qian N; Shi L; Singer RA; Benninger RKP; Min W Highly-Multiplexed Volumetric Mapping with Raman Dye Imaging and Tissue Clearing. Nat. Biotechnol. 2021. 10.1038/s41587-021-01041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Fujioka H; Shou J; Kojima R; Urano Y; Ozeki Y; Kamiya M Multicolor Activatable Raman Probes for Simultaneous Detection of Plural Enzyme Activities. J. Am. Chem. Soc. 2020, 142 (49), 20701–20707. 10.1021/jacs.0c09200. [DOI] [PubMed] [Google Scholar]
- (51).Ao J; Fang X; Miao X; Ling J; Kang H; Park S; Wu C; Ji M Switchable Stimulated Raman Scattering Microscopy with Photochromic Vibrational Probes. Nat. Commun. 2021, 12 (1), 1–8. 10.1038/s41467-021-23407-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Shou J; Ozeki Y Photoswitchable Stimulated Raman Scattering Spectroscopy and Microscopy. Opt. Lett. 2021, 46 (9), 2176. 10.1364/ol.418240. [DOI] [PubMed] [Google Scholar]
- (53).Lee D; Qian C; Wang H; Li L; Miao K; Du J; Shcherbakova DM; Verkhusha VV; Wang LV; Wei L Toward Photoswitchable Electronic Pre-Resonance Stimulated Raman Probes. J. Chem. Phys. 2021, 154 (135102), 1–10. 10.1063/5.0043791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Du J; Wei L Multicolor Photoactivatable Raman Probes for Subcellular Imaging and Tracking by Cyclopropenone Caging. J. Am. Chem. Soc. 2021. 10.1021/jacs.1c09689. [DOI] [PubMed] [Google Scholar]
- (55).Hong S; Chen T; Zhu Y; Li A; Huang Y; Chen X Live-Cell Stimulated Raman Scattering Imaging of Alkyne-Tagged Biomolecules. Angew. Chemie - Int. Ed. 2014, 53 (23), 5827–5831. 10.1002/anie.201400328. [DOI] [PubMed] [Google Scholar]
- (56).Lee HJ; Zhang W; Zhang D; Yang Y; Liu B; Barker EL; Buhman KK; Slipchenko LV; Dai M; Cheng JX Assessing Cholesterol Storage in Live Cells and C. Elegans by Stimulated Raman Scattering Imaging of Phenyl-Diyne Cholesterol. Sci. Rep. 2015, 5, 1–10. 10.1038/srep07930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).de Moliner F; Knox K; Gordon D; Lee M; Tipping WJ; Geddis A; Reinders A; Ward JM; Oparka K; Vendrell M A Palette of Minimally Tagged Sucrose Analogues for Real-Time Raman Imaging of Intracellular Plant Metabolism. Angew. Chemie - Int. Ed. 2021, 60 (14), 7637–7642. 10.1002/anie.202016802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Seidel J; Miao Y; Porterfield W; Cai W; Zhu X; Kim SJ; Hu F; Bhattarai-Kline S; Min W; Zhang W Structure-Activity-Distribution Relationship Study of Anti-Cancer Antimycin-Type Depsipeptides. Chem. Commun. 2019, 55 (63), 9379–9382. 10.1039/c9cc03051d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Gaschler MM; Hu F; Feng H; Linkermann A; Min W; Stockwell BR Determination of the Subcellular Localization and Mechanism of Action of Ferrostatins in Suppressing Ferroptosis. ACS Chem. Biol. 2018, 13 (4), 1013–1020. 10.1021/acschembio.8b00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Koike K; Bando K; Ando J; Yamakoshi H; Terayama N; Dodo K; Smith NI; Sodeoka M; Fujita K Quantitative Drug Dynamics Visualized by Alkyne-Tagged Plasmonic-Enhanced Raman Microscopy. ACS Nano 2020, 14 (11), 15032–15041. 10.1021/acsnano.0c05010. [DOI] [PubMed] [Google Scholar]
- (61).Chen Z; Paley DW; Wei L; Weisman AL; Friesner RA; Nuckolls C; Min W Multicolor Live-Cell Chemical Imaging by Isotopically Edited Alkyne Vibrational Palette. J. Am. Chem. Soc. 2014, 136 (22), 8027–8033. 10.1021/ja502706q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Waegele MM; Culik RM; Gai F Site-Specific Spectroscopic Reporters of the Local Electric Field, Hydration, Structure, and Dynamics of Biomolecules. J. Phys. Chem. Lett. 2011, 2 (20), 2598–2609. 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Bakthavatsalam S; Dodo K; Sodeoka M A Decade of Alkyne-Tag Raman Imaging (ATRI): Applications in Biological Systems. RSC Chem. Biol. 2021, 2, 1415–1429. 10.1039/d1cb00116g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Yamakoshi H; Dodo K; Palonpon A; Ando J; Fujita K; Kawata S; Sodeoka M Alkyne-Tag Raman Imaging for Visualization of Mobile Small Molecules in Live Cells. J. Am. Chem. Soc. 2012, 134 (51), 20681–20689. 10.1021/ja308529n. [DOI] [PubMed] [Google Scholar]
- (65).Hu F; Chen Z; Zhang L; Shen Y; Wei L; Min W Vibrational Imaging of Glucose Uptake Activity in Live Cells and Tissues by Stimulated Raman Scattering. Angew. Chemie - Int. Ed. 2015, 54 (34), 9821–9825. 10.1002/anie.201502543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Wei L; Yu Y; Shen Y; Wang MC; Min W Vibrational Imaging of Newly Synthesized Proteins in Live Cells by Stimulated Raman Scattering Microscopy. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (28), 11226–11231. 10.1073/pnas.1303768110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Saar BG; Contreras-Rojas LR; Xie SX; Guy RH Imaging Drug Delivery to Skin with Coherent Raman Scattering Microscopy. Mol. Pharm. 2014, 8, 969–975. 10.1007/978-3-642-32109-2_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Miao K; Wei L Live-Cell Imaging and Quantification of PolyQ Aggregates by Stimulated Raman Scattering of Selective Deuterium Labeling. ACS Cent. Sci. 2020, 6 (4), 478–486. 10.1021/acscentsci.9b01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Hu F; Brucks SD; Lambert TH; Campos LM; Min W Stimulated Raman Scattering of Polymer Nanoparticles for Multiplexed Live-Cell Imaging. Chem. Commun. 2017, 53 (46), 6187–6190. 10.1039/c7cc01860f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Li J; Cheng JX Direct Visualization of de Novo Lipogenesis in Single Living Cells. Sci. Rep. 2014, 4, 1–8. 10.1038/srep06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Zhang D; Slipchenko MN; Cheng JX Highly Sensitive Vibrational Imaging by Femtosecond Pulse Stimulated Raman Loss. J. Phys. Chem. Lett. 2011, 2 (11), 1248–1253. 10.1021/jz200516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Hu F; Wei L; Zheng C; Shen Y; Min W Live-Cell Vibrational Imaging of Choline Metabolites by Stimulated Raman Scattering Coupled with Isotope-Based Metabolic Labeling. Analyst 2014, 139 (10), 2312–2317. 10.1039/c3an02281a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Alfonso-García A; Pfisterer SG; Riezman H; Ikonen E; Potma EO D38-Cholesterol as a Raman Active Probe for Imaging Intracellular Cholesterol Storage. J. Biomed. Opt. 2015, 21 (6), 061003. 10.1117/1.jbo.21.6.061003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Shi L; Zheng C; Shen Y; Chen Z; Silveira ES; Zhang L; Wei M; Liu C; de Sena-Tomas C; Targoff K; et al. Optical Imaging of Metabolic Dynamics in Animals. Nat. Commun. 2018, 9 (1). 10.1038/s41467-018-05401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Lee D; Du J; Yu R; Su Y; Heath JR; Wei L Visualizing Subcellular Enrichment of Glycogen in Live Cancer Cells by Stimulated Raman Scattering. Anal. Chem. 2020, 92 (19), 13182–13191. 10.1021/acs.analchem.0c02348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Zhu W; Cai EL; Li HZ; Wang P; Shen AG; Popp J; Hu JM Precise Encoding of Triple-Bond Raman Scattering of Single Polymer Nanoparticles for Multiplexed Imaging Application. Angew. Chemie - Int. Ed. 2021, 60 (40), 21846–21852. 10.1002/anie.202106136. [DOI] [PubMed] [Google Scholar]
- (77).Tian S; Li H; Li Z; Tang H; Yin M; Chen Y; Wang S; Gao Y; Yang X; Meng F; et al. Polydiacetylene-Based Ultrastrong Bioorthogonal Raman Probes for Targeted Live-Cell Raman Imaging. Nat. Commun. 2020, 11 (1), 1–9. 10.1038/s41467-019-13784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Zhao Z; Chen C; Wei S; Xiong H; Hu F; Miao Y; Jin T; Min W Ultra-Bright Raman Dots for Multiplexed Optical Imaging. Nat. Commun. 2021, 12, 1305. 10.1038/s41467-021-21570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Jin QQ; Fan X; Chen C; Huang L; Wang J; Tang X Multicolor Raman Beads for Multiplexed Tumor Cell and Tissue Imaging and in Vivo Tumor Spectral Detection. Anal. Chem. 2019, 91 (6), 3784–3789. 10.1021/acs.analchem.9b00028. [DOI] [PubMed] [Google Scholar]
- (80).Farizyan M; Mondal A; Mal S; Deufel F; Van Gemmeren M Palladium-Catalyzed Nondirected Late-Stage C-H Deuteration of Arenes. J. Am. Chem. Soc. 2021, 143 (40), 16370–16376. 10.1021/jacs.1c08233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zhao D; Petzold R; Yan J; Muri D; Ritter T Tritiation of Aryl Thianthrenium Salts with a Molecular Palladium Catalyst. Nature 2021, 600 (7889), 444–449. 10.1038/s41586-021-04007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Porey S; Zhang X; Bhowmick S; Kumar Singh V; Guin S; Paton RS; Maiti D Alkyne Linchpin Strategy for Drug:Pharmacophore Conjugation: Experimental and Computational Realization of a Meta-Selective Inverse Sonogashira Coupling. J. Am. Chem. Soc. 2020, 142 (8), 3762–3774. 10.1021/jacs.9b10646. [DOI] [PubMed] [Google Scholar]
- (83).Yu Y; Mutlu AS; Liu H; Wang MC High-Throughput Screens Using Photo-Highlighting Discover BMP Signaling in Mitochondrial Lipid Oxidation. Nat. Commun. 2017, 8 (1), 1–11. 10.1038/s41467-017-00944-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Mutlu AS; Gao SM; Zhang H; Wang MC Olfactory Specificity Regulates Lipid Metabolism through Neuroendocrine Signaling in Caenorhabditis Elegans. Nat. Commun. 2020, 11 (1), 1–15. 10.1038/s41467-020-15296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Zhang L; Shi L; Shen Y; Miao Y; Wei M; Qian N; Liu Y; Min W Spectral Tracing of Deuterium for Imaging Glucose Metabolism. Nat. Biomed. Eng. 2019, 3 (5), 402–413. 10.1038/s41551-019-0393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Deberardinis RJ; Thompson CB Cellular Metabolism and Disease: What Do Metabolic Outliers Teach Us? Cell 2012, 148 (6), 1132–1144. 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Long R; Zhang L; Shi L; Shen Y; Hu F; Zeng C; Min W Two-Color Vibrational Imaging of Glucose Metabolism Using Stimulated Raman Scattering. Chem. Commun. 2017, 54 (2), 152–155. 10.1039/c7cc08217g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Hong S; Chen T; Liu L; Cao C; Lv F; Rabinowitz JD; Huang Y; Chen X Live-Cell Imaging of NADPH Production from Specific Pathways. CCS Chem. 2021, 3 (6), 1642–1648. 10.31635/ccschem.020.202000346. [DOI] [Google Scholar]
- (89).Zhang M; Hong W; Abutaleb NS; Li J; Dong PT; Zong C; Wang P; Seleem MN; Cheng JX Rapid Determination of Antimicrobial Susceptibility by Stimulated Raman Scattering Imaging of D2O Metabolic Incorporation in a Single Bacterium. Adv. Sci. 2020, 7 (19), 1–14. 10.1002/advs.202001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Schiessl KT; Hu F; Jo J; Nazia SZ; Wang B; Price-Whelan A; Min W; Dietrich LEP Phenazine Production Promotes Antibiotic Tolerance and Metabolic Heterogeneity in Pseudomonas Aeruginosa Biofilms. Nat. Commun. 2019, 10 (1), 1–10. 10.1038/s41467-019-08733-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Bae K; Zheng W; Ma Y; Huang Z Real-Time Monitoring of Pharmacokinetics of Antibiotics in Biofilms with Raman-Tagged Hyperspectral Stimulated Raman Scattering Microscopy. Theranostics 2019, 9 (5), 1348–1357. 10.7150/thno.32043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Meister K; Niesel J; Schatzschneider U; Metzler-Nolte N; Schmidt DA; Havenith M Label-Free Imaging of Metal-Carbonyl Complexes in Live Cells by Raman Microspectroscopy. Angew. Chemie - Int. Ed. 2010, 49 (19), 3310–3312. 10.1002/anie.201000097. [DOI] [PubMed] [Google Scholar]
- (93).Bird RE; Lemmel SA; Yu X; Zhou QA Bioorthogonal Chemistry and Its Applications. Bioconjug. Chem. 2021, 32 (12), 2457–2479. 10.1021/acs.bioconjchem.1c00461. [DOI] [PubMed] [Google Scholar]
- (94).Scinto SL; Bilodeau DA; Hincapie R; Lee W; Nguyen SS; Xu M; am Ende CW; Finn MG; Lang K; Lin Q; et al. Bioorthogonal Chemistry. Nat. Rev. Methods Prim. 2021, 1 (1), 30. 10.1038/s43586-021-00028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Saleh AM; Wilding KM; Calve S; Bundy BC; Kinzer-Ursem TL Non-Canonical Amino Acid Labeling in Proteomics and Biotechnology. J. Biol. Eng. 2019, 13 (1), 1–14. 10.1186/s13036-019-0166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Zhang J; Yan S; He Z; Ding C; Zhai T; Chen Y; Li H; Yang G; Zhou X; Wang P Small Unnatural Amino Acid Carried Raman Tag for Molecular Imaging of Genetically Targeted Proteins. J. Phys. Chem. Lett. 2018, 9 (16), 4679–4685. 10.1021/acs.jpclett.8b01991. [DOI] [PubMed] [Google Scholar]
- (97).Lampkowski JS; Villa JK; Young TS; Young DD Development and Optimization of Glaser-Hay Bioconjugations. Angew. Chemie - Int. Ed. 2015, 54 (32), 9343–9346. 10.1002/anie.201502676. [DOI] [PubMed] [Google Scholar]
- (98).Weerapana E; Wang C; Simon GM; Richter F; Khare S; Dillon MBD; Bachovchin DA; Mowen K; Baker D; Cravatt BF Quantitative Reactivity Profiling Predicts Functional Cysteines in Proteomes. Nature 2010, 468 (7325), 790–797. 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Backus KM; Correia BE; Lum KM; Forli S; Horning BD; González-Páez GE; Chatterjee S; Lanning BR; Teijaro JR; Olson AJ; et al. Proteome-Wide Covalent Ligand Discovery in Native Biological Systems. Nature 2016, 534 (7608), 570–574. 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Zanon PRA; Yu F; Musacchio PZ; Lewald L; Zollo M; Krauskopf K; Mrdović D; Raunft P; Maher TE; Cigler M; et al. Profiling the Proteome-Wide Selectivity of Diverse Electrophiles. ChemRxiv 2021, 1, 1–10. [Google Scholar]
- (101).Miao Y; Shi L; Hu F; Min W Probe Design for Super-Multiplexed Vibrational Imaging. Phys. Biol. 2019, 16 (4), 41003. 10.1088/1478-3975/ab0fcd. [DOI] [PubMed] [Google Scholar]
- (102).Zhuge M; Huang KC; Lee HJ; Jiang Y; Tan Y; Lin H; Dong PT; Zhao G; Matei D; Yang Q; et al. Ultrasensitive Vibrational Imaging of Retinoids by Visible Preresonance Stimulated Raman Scattering Microscopy. Adv. Sci. 2021, 8 (9), 1–11. 10.1002/advs.202003136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Xiong H; Qian N; Miao Y; Zhao Z; Min W Stimulated Raman Excited Fluorescence Spectroscopy of Visible Dyes. J. Phys. Chem. Lett. 2019, 10 (13), 3563–3570. 10.1021/acs.jpclett.9b01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Hu F; Zeng C; Long R; Miao Y; Wei L; Xu Q; Min W Supermultiplexed Optical Imaging and Barcoding with Engineered Polyynes. Nat. Methods 2018, 15 (3), 194–200. 10.1038/nmeth.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Su X; Liu R; Li Y; Han T; Zhang Z; Niu N; Kang M; Fu S; Wang D; Wang D; et al. Aggregation-Induced Emission-Active Poly(Phenyleneethynylene)s for Fluorescence and Raman Dual-Modal Imaging and Drug-Resistant Bacteria Killing. Adv. Healthc. Mater. 2021, 10 (24). 10.1002/adhm.202101167. [DOI] [PubMed] [Google Scholar]
- (106).Lucotti A; Tommasini M; Fazzi D; Del Zoppo M; Chalifoux WA; Tykwinski RR; Zerbi G Absolute Raman Intensity Measurements and Determination of the Vibrational Second Hyperpolarizability of Adamantyl Endcapped Polyynes. J. Raman Spectrosc. 2012, 43 (9), 1293–1298. 10.1002/jrs.3166. [DOI] [Google Scholar]
- (107).Zeng C; Hu F; Long R; Min W A Ratiometric Raman Probe for Live-Cell Imaging of Hydrogen Sulfide in Mitochondria by Stimulated Raman Scattering. Analyst 2018, 143 (20), 4844–4848. 10.1039/c8an00910d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Wilson LT; Tipping WJ; Jamieson LE; Wetherill C; Henley Z; Faulds K; Graham D; MacKay SP; Tomkinson NC O. A New Class of Ratiometric Small Molecule Intracellular PH Sensors for Raman Microscopy. Analyst 2020, 145 (15), 5289–5298. 10.1039/d0an00865f. [DOI] [PubMed] [Google Scholar]
- (109).Tipping WJ; Wilson LT; Blaseio SK; Tomkinson NCO; Faulds K; Graham D Ratiometric Sensing of Fluoride Ions Using Raman Spectroscopy. Chem. Commun. 2020, 56 (92), 14463–14466. 10.1039/d0cc05939k. [DOI] [PubMed] [Google Scholar]
- (110).Takemura S; Watanabe H; Nishihara T; Okamoto A; Tanabe K Monitoring Intracellular Metal Ion Complexation with an Acetylene-Tagged Ligand by Raman Spectroscopy. RSC Adv. 2020, 10 (59), 36119–36123. 10.1039/d0ra06329k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Egoshi S; Dodo K; Ohgane K; Sodeoka M Deuteration of Terminal Alkynes Realizes Simultaneous Live Cell Raman Imaging of Similar Alkyne-Tagged Biomolecules. Org. Biomol. Chem. 2021, 19 (38), 8232–8236. 10.1039/d1ob01479j. [DOI] [PubMed] [Google Scholar]
- (112).Bi X; Miao K; Wei L Alkyne-Tagged Raman Probes for Local Environmental Sensing by Hydrogen − Deuterium Exchange. J. Am. Chem. Soc. 2022, 144, 8504–8514. 10.1021/jacs.2c01991. [DOI] [PubMed] [Google Scholar]
- (113).Shi L; Hu F; Min W Optical Mapping of Biological Water in Single Live Cells by Stimulated Raman Excited Fluorescence Microscopy. Nat. Commun. 2019, 10 (1), 1–8. 10.1038/s41467-019-12708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Lang X; Welsher K Mapping Solvation Heterogeneity in Live Cells by Hyperspectral Stimulated Raman Scattering Microscopy. J. Chem. Phys. 2020, 152 (17). 10.1063/1.5141422. [DOI] [PubMed] [Google Scholar]
- (115).Lee HJ; Huang KC; Mei G; Zong C; Mamaeva N; Degrip WJ; Rothschild KJ; Cheng JX Electronic Preresonance Stimulated Raman Scattering Imaging of Red-Shifted Proteorhodopsins: Toward Quantitation of the Membrane Potential. J. Phys. Chem. Lett. 2019, 10 (15), 4374–4381. 10.1021/acs.jpclett.9b01337. [DOI] [PubMed] [Google Scholar]
- (116).Tang Q; Liu L; Guo Y; Zhang X; Zhang S; Jia Y; Du Y; Cheng B; Yang L; Huang Y; et al. Optical Cell Tagging for Spatially Resolved Single-Cell RNA Sequencing. Angew. Chemie Int. Ed. 2021, 100730. 10.1002/anie.202113929. [DOI] [PubMed] [Google Scholar]