Abstract
Among nematodes, the free-living model organism Caenorhabditis elegans boasts the most advanced portfolio of high-quality omics data. The resources available for parasitic nematodes, including Strongyloides spp., however, are lagging behind. While C. elegans remains the most tractable nematode and has significantly advanced our understanding of many facets of nematode biology, C. elegans is not suitable as a surrogate system for the study of parasitism and it is important that we improve the omics resources available for parasitic nematode species. Here, we review the omics data available for Strongyloides spp. and compare the available resources to those for C. elegans and other parasitic nematodes. The advancements in C. elegans omics offer a blueprint for improving omics-led research in Strongyloides. We suggest areas of priority for future research that will pave the way for expansions in omics resources and technologies.
This article is part of the Theo Murphy meeting issue ‘Strongyloides: omics to worm-free populations’.
Keywords: Strongyloides, genomics, transcriptomics, proteomics, small RNA, transposable elements
1. Application of omics, bioinformatics pipelines, functional genomics
The advent of high-throughput, sensitive and cost-effective omics technologies has revolutionized biological research. ‘Omics’ encompasses a range of data types including genomics, transcriptomics, proteomics, peptidomics and metabolomics, which facilitate unbiased, largescale analyses of all of the molecules in a cell, organism or population. In addition, omics also incorporates advanced functional genomics tools, such as RNAi, transgenesis and CRISPR, which can enable a more focused analysis of the function of one, or a few, specific genes of interest. Indeed, for many higher organisms, multi-omics approaches that incorporate multiple omics technologies, tools and data types have become integrated into well-developed platforms that enhance our capability to unravel complex biological questions.
Among nematodes, the free-living model organism Caenorhabditis elegans boasts the most advanced portfolio of high-quality omics data that, in tandem with a robust functional omics toolkit, has become one of the most valuable and widely exploited research platforms in biological and biomedical research [1]. While C. elegans remains the most tractable nematode and has significantly advanced our understanding of many facets of nematode biology, including parasite systems, the sole use of C. elegans as a surrogate system for the study of parasitism and infection biology should be approached cautiously. For example, several parasite-specific gene families are absent or functionally divergent in C. elegans [2] hindering the translation of C. elegans-derived functional data to parasites. Despite this, the C. elegans platform should provide a blueprint for the translation of omics tools and technologies to other nematodes that will pave the way for expansions in omics resources and technologies in therapeutically relevant and experimentally tractable parasitic nematodes [3].
The availability of high-quality species and life cycle stage-specific omics datasets forms the foundation of robust omics platforms. The accessibility and quality of parasitic nematode omics data are continually improving, providing large-scale, rich datasets ripe for exploitation. Indeed, WormBase ParaSite now provides a centralized repository for 177 nematode genomes (WBPS18, April 2023; https://parasite.wormbase.org/index.html) and a range of transcriptome resources, consistently updating and improving assemblies and annotations where possible [4]. In addition to the provision of high-quality omics data, standardized bioinformatics pipelines are also essential to foster a consistent approach to omics analysis that will, in turn, seed downstream functional omics studies in tractable model systems; we are beginning to see the parasitology community recognizing this through the provision of gold standard experimental workflows for helminths [5].
The translation and application of omics approaches to parasitic species continue to be challenged by the nature of the parasitic lifestyles where the inaccessibility of parasite material and difficulty in maintaining life cycle stages in vitro hinder progress. Regardless, several species, including Ascaris suum, Brugia malayi and Strongyloides spp., possess traits that have driven their emergence as forerunners in the translation and application of omics technologies to nematode parasites [3,6]. Among these, Strongyloides spp. are rapidly becoming the most advanced parasitic nematode model system available, boasting an omics toolkit (including RNAi, transgenesis and CRISPR/Cas9-mediated targeted mutagenesis capabilities) that is currently unrivalled by any other nematode parasite (reviewed extensively elsewhere; see [6]). Further, recent advances in the quality Strongyloides omics data also present new opportunities to integrate multiple omics levels (multi-omics) and rapidly progress understanding of parasite biology by connecting genotype and phenotype data in a therapeutically relevant parasitic nematode.
Sustained investment in the development of parasite omics platforms and multi-omics analyses for Strongyloides and other nematodes causing Neglected Tropical Diseases will revolutionise the ability to perform comparative omics, evolution, drug resistance and molecular interaction studies that will drive the discovery of new targets for therapeutic intervention, vaccine development and improved diagnostics. Here, we provide an overview of the omics technologies and datasets currently available for Strongyloides species, and discuss approaches to data acquisition, data analysis, challenges and future directions.
2. Strongyloides genome assemblies and annotation
Published genome assemblies are available for four Strongyloides species—two parasites of rats, S. ratti (44 Mb) and S. venezulensis (44 Mb), the livestock parasite S. papillosus (61 Mb) and the human parasite S. stercoralis (43 Mb). There are also genome assemblies available for two closely related species—a parasite of possums, Parastrongyloides trichosuri (43 Mb), and a free-living species, Rhabditiophanes ditinus (45 Mb) [7]. While originally the genomes of all six species were sequenced using short-read technologies, four of them: S. ratti, S. stercoralis, S. venezulensis and R. ditinus, have since been improved using Oxford Nanopore Technologies (ONT) long-read sequencing [8] (NCBI GCA_029582065.1). S. ratti, S. venezulensis and R. ditinus genome assemblies also incorporated Hi-C maps that map long-range interactions between regions of the genome in 3D space, which can be used to further improve the assemblies as chromosomes tend to have more contacts with parts of the genome that are physically close in the genome [9]. Contiguity of the genome assemblies varies widely; S. venezulensis and R. ditinus both have chromosome-length scaffolds, and S. ratti has two autosomal scaffolds and the X chromosome is in two scaffolds. The assembly of S. stercoralis has two autosomes at chromosome-level scaffolds and the X chromosome is in six scaffolds [8] (NCBI GCA_029582065.1). The genome assemblies for S. papillosus and P. trichosuri are more fragmented due to only being based on short-read Illumina sequencing, and contain 4703 and 1810 scaffolds, respectively, which, in contrast to the other species, lack assignment to chromosomes (table 1). A custom pipeline for Strongyloides gene model annotation was built around AUGUSTUS [19] and MAKER [20] training tools using manual annotations of between 197 and 423 genes (depending on species), RNA-seq data and genes from closely related species. This resulted in the annotation of 12 474, 16 904, 18 456 and 13 123 genes in S. ratti, S. venezulensis, S. papillosus and S. stercoralis, respectively [7].
Table 1.
Summary of genome assemblies for Strongyloides spp., the closely related species P. trichosuri and R. ditinus, C. elegans and a selection of parasitic nematodes with high-quality genome assemblies.
| species | clade | genome size (Mbp) | no. of chromosomesa | N50 (Mbp) | no. of scaffolds | no. of annotated protein-coding genes (%) | BUSCO assembly score | data used in assembly | reference |
|---|---|---|---|---|---|---|---|---|---|
| Strongyloides species | |||||||||
| S. ratti | IV | 43.9 | 3 (1) | 12.1 | 4 | 87.0 | Illumina, ONT, Hi-C | Kounosu et al. [8] | |
| S. ratti | IV | 43.17 | 3 (1) | 11.7 | 136 | 12 464 | 78.1 | genetic map, Illumina, Sanger, 454 pyrosequencing | Hunt et al. [7]; Nemetschke et al. [10] |
| S. stercoralis | IV | 42.7 | 3 (1) | 11.7 | 8 | 86.6 | ONT | NCBI accession: GCA_029582065.1 | |
| S. stercoralis | IV | 42.7 | 3 (1) | 0.4 | 800 | 13 123 | 78.5 | Illumina | Hunt et al. [7] |
| S. venezulensis | IV | 44.4 | 2 (0) | 31.3 | 2 | 85.5 | Illumina, ONT, Hi-C | Kounosu et al. [8] | |
| S. venezulensis | IV | 52.2 | 2 (0) | 0.7 | 587 | 16 904 | 76.7 | genetic map, Illumina | Hunt et al. [7] |
| S. papulosis | IV | 60.5 | 2 (0) | 0.1 | 4703 | 18 456 | 77.2 | Illumina | Hunt et al. [7] |
| closely related species and C. elegans | |||||||||
| P. trichosuri | IV | 42.5 | 3 (1) | 0.8 | 1810 | 15 010 | 78.1 | Illumina | Hunt et al. [7] |
| R. ditinus | IV | 44.9 | 6 (1) | 7.8 | 6 | 86.6 | Illumina, ONT, Hi-C | Kounosu et al. [8] | |
| R. ditinus | IV | 47.3 | 5 (0) | 0.5 | 471 | 13 496 | 74.1 | Illumina | Hunt et al. [7] |
| C. elegans | V | 100.3 | 6 (1) | 17.5 | 6 | 19 981 | 98.6 | genetic map, Sanger | the C. elegans Sequencing Consortium [11] |
| other parasitic nematodes | |||||||||
| A. suum | III | 278.6 | 24 (5) | 6.4 | 108 | 16 778 | 93.8 | PacBio, Hi-C | Wang et al. [12] |
| B. malayi | III | 88.2 | 5 (2) | 14.2 | 205 | 10 878 | 96.8 | optical map, Illumina, Sanger, 454 pyrosequencing, PacBio | Tracey et al. [13] |
| B. xylophilus | IV | 78.3 | 6 (0) | 12.8 | 11 | 15 884 | 77.7 | Illumina, ONT, Hi-C | Dayi et al. [14] |
| H. contortus | V | 283.4 | 6 (1) | 47.4 | 7 | 19 623 | 86.6 | optical map, genetic map, Illumina, 454 pyrosequencing, PacBio, 10× linked long-range | Doyle et al. [15] |
| H. glycines | IV | 158 | 9 (0) | 17.9 | 2121 | 22 465 | 55.2 | Illumina, PacBio, Hi-C | Masonbrink et al. [16] |
| O. volvulus | III | 96.4 | 5 (2) | 25.5 | 1006 | 12 109 | 97.6 | optical map, Illumina | Cotton et al. [17] |
| T. muris | I | 111.8 | 4 (2) | 25.5 | 803 | 14 995 | 69.9 | Illumina, 454 pyrosequencing | Foth et al. [18] |
aThe numbers of chromosomes that are sex chromosomes are indicated in brackets.
(a) . Comparison with Caenorhabditis elegans
Caenorhabditis elegans was the first multicellular organism to have its genome assembled. This was achieved using a genetic map built during the 1980s and Sanger capillary sequencing with targeting of regions without a known sequence, resulting in an essentially complete assembly, with only a few known gaps, that has been used for most subsequent research [11]. Comparative analysis with a long read-based assembly suggests this is 99.98% identical to current C. elegans genomes, having missed only 1.8 Mb of repetitive regions [21]. Compared to Strongyloides spp., the C. elegans genome is 100.29 Mb, more than double the 43.9 Mb S. ratti genome, and is comprised of six chromosomes, compared to the three chromosomes for S. ratti and S. stercoralis and two chromosomes for S. papillosus and S. venezuelensis. The C. elegans genome also has a higher number of annotated genes—19 981 (WormBase Release 289)—compared to S. ratti's 12 474. The N50 size is also larger (17.5 Mb for C. elegans versus 12.1 Mb for S. ratti); however, this is likely due to the larger size of the C. elegans genome, which has longer chromosomes (size range 12–20 Mb versus 12–18 Mb for S. ratti) (table 1).
(b) . Comparison with other parasitic nematodes
S. ratti has had one of the most contiguous genome assemblies among parasitic nematodes since the assembly became available in 2016 [22]. However, in recent years, new assemblies with near chromosome-length contigs have been created for parasitic nematodes such as A. suum [12], Haemonchus contortus [15], Bursaphelenchus xylophilus [14], B. malayi [13] and Heterodera glycines [16]. Like the most recent 2023 Strongyloides genome assemblies, these assemblies have taken advantage of long-read sequencing (e.g. ONT or PacBio HiFi) and Hi-C contact data (table 1). Annotation methods vary between genome assemblies and have mostly been generated using in-house pipelines incorporating programs such as AUGUSTUS [23] and MAKER [20]. Gene model annotations vary between 10 878 and 22 465 annotated protein-coding genes in the genome (table 1).
(c) . Future areas for priority
(i) Improvement of existing reference genome assemblies of S. papillosus and P. trichosuri using long-read sequencing and Hi-C contact maps to scaffold is a priority. This will generate assemblies with chromosome length contigs that would open opportunities for comparative genomic analysis such as evaluating the role of gene organization in infection. (ii) Genome sequencing of more Strongyloides species, particularly those that are medically important such as the human parasite S. fuelleborni, to improve genome-level understanding of therapeutically relevant parasitic nematode species. (iii) Current genome assemblies are limited to laboratory-based cultures. S. ratti and S. stercoralis have been maintained in the laboratory since 1960 (M Viney 2023, personal communication) and 1984 [24], respectively. High-quality sequencing and assembly of wild isolate genomes is important for understanding the genomics of Strongyloides in real-world infections. Combining these data with clinical information or information about infectivity is also important to determine the genetic basis of Strongyloides parasitism and addressing questions around the zoonotic characteristics of S. stercoralis.
3. Strongyloides phylogenetics, phylogenomics and population genetics and genomics
Although not strictly an omics application, phylogenetics and population genetics are useful tools for exploring evolutionary histories and relatedness across Strongyloides samples within a population (population genetics) and between species (phylogenetics). Phylogenomics and population genomics, i.e. where information from genome sequences are used instead of information for a single or a small selection of genes or sequences, is fast becoming a popular method to better study understand relatedness and differentiation within and between populations and species. Here, we summarize the research on Strongyloides using both genetic and genomic-based approaches. Most phylogenetics datasets for Strongyloides focus on the epidemiology of S. stercoralis or S. fuelleborni in two key areas: (i) infection surveillance to track transmission in areas where Strongyloides is endemic and (ii) zoonotic transmission, to identify shared haplotypes of parasites infecting both humans and other animals (particularly non-human primates and dogs; table 2). Such studies are largely based on reconstructing phylogenies or categorizing haplotypes using mitochondrial DNA sequences, including the 18S rRNA subunit or the cox1 (cytochrome c oxidase subunit 1) gene [49]. The hypervariable regions of the 18S small subunit [50] have species-specific arrangements and facilitate differentiation between Strongyloides species.
Table 2.
Summary of phylogenetics, population genetics, phylogenomics and population genomics studies on Strongyloides species. SSU, small subunit; LSU, large subunit.
| species | host | genetic marker(s) | reference |
|---|---|---|---|
| marker gene studies | |||
| S. ratti | Rattus norvegicus | Actin, BSP-8, CM-2 | Fisher & Viney [25] |
| S. ratti | Rattus norvegicus | Actin, BSP-8 | Paterson et al. [26] |
| mtDNA studies | |||
| S. papillosus, unknown Strongyloides of cows (suggested name S. vituli) | Ovis aries | 18S SSU | Eberhardt et al. [27] |
| Bos taurus | |||
| unknown Strongyloides infections of humans, S. stercoralis, S. fuelleborni, S. planiceps | Homo sapiens | 18S SSU, cox1 | Hasegawa et al. [28] |
| Canis familiaris | |||
| primate (various) | |||
| S. fuelleborni | Papio sp. | 18S SSU | Anderson et al. [29] |
| S. stercoralis | Homo sapiens | 18S SSU | Schär et al. [30] |
| S. stercoralis | Canis familiaris | 18S SSU, 28S SSU, cox1, MSP gene | Nagayasu et al. [31] |
| Homo sapiens | |||
| S. stercoralis, S. fuelleborni | Papio papio | 18S SSU | Barratt et al. [32] |
| Canis familiaris | |||
| Homo sapiens | |||
| S. stercoralis | Canis familiaris | 18S SSU, cox1 | Basso et al. [33] |
| S. stercoralis | Canis familiaris | 18S SSU, cox1 | Beknazarova et al. [34] |
| Homo sapiens | |||
| S. stercoralis | Canis familiaris | 18S SSU, cox1 | Sanpool et al. [35] |
| Homo sapiens | |||
| S. stercoralis | n.a. | cox1, retrieved from published data repositories | Spotin et al. [36] |
| S. fuelleborni | Macaca fascicularis | 18S SSU, cox1 | Thanchomnang et al. [37] |
| S. stercoralis, S. tunefaciens | Felis catus | cox1 | Wulcan et al. [38] |
| S. stercoralis | n.a. | 18S SSU and cox1 retrieved from published data repositories | Barret & Sapp [39] |
| S. fuelleborni | |||
| S. fuelleborni | Macaca nemestrina, | Am | Janwan et al. [40] |
| Homo sapiens | |||
| S. stercoralis, S. procyonis, S. planiceps, unknown Strongyloides | Procyon lotor | 18S SSU, 28S LSU, cox1 | Ko et al. [41] |
| Meles anakuma | |||
| Nyctereutes procyonoides Paguma larvata Mustela sibirica | |||
| Felis catus | |||
| S. stercoralis | Homo sapiens | cox1 | Repetto et al. [42] |
| S. fuelleborni | Macaca mulatta | 18S SSU, cox1 | Ko et al. [43] |
| also found: S. cebus, S. vituli | |||
| S. fuelleborni | Macaca mulatta | 18S SSU, cox1, complete mitochondrial genome | Ko et al. [43] |
| S. vituli | |||
| S. cebus | Macaca fuscata | ||
| Trachypithecus francoisi | |||
| Pygathrix nemaeus | |||
| Pongo pygmaeus | |||
| Symphalangus syndactylus | |||
| Saimiri boliviensis | |||
| whole genome shotgun studies | |||
| S. stercoralis | Homo sapiens | whole genome shotgun | Kikuchi et al. [44] |
| S. stercoralis | Canis familiaris | 18S SSU, cox1, whole genome shotgun (subset) | Jaleta et al. [45] |
| Homo sapiens | |||
| S. stercoralis | Homo sapiens | 18S SSU, cox1 | Zhou et al. [46] |
| whole genome shotgun (subset) | |||
| S. stercoralis | Homo sapiens | 18S SSU, cox1, whole genome shotgun (subset) | Aupalee et al. [47] |
| S. ratti | Rattus norvegicus | whole genome shotgun | Cole et al. [48] |
More recently, there has been an increase in the use of phylogenomics and population genomics, using shotgun genome sequencing and evaluation of SNPs to try to resolve more detailed phylogenies and thereby improve understanding of how Strongyloides species evolve, and identify differences in genomic variation within and between populations and species. Population genomics has not been explored deeply in Strongyloides spp. Two studies have performed whole genome shotgun sequencing and examined genetic diversity in S. stercoralis isolates from infected patients [44,46]. Both studies found geographical clustering of the samples based on their genomes and little evidence of recombination, suggesting that the sexually reproducing free-living stage is rare in the wild populations. A population genomics study on S. ratti from wild rat faeces collected in England and Wales found that a few long-lived lines infect many rats across the three distinct sampling sites studies and there was little variation between the sites [48]. There is a strong positive selection pressure, with a high number of non-synonymous SNPs, on gene families previously associated with Strongyloides parasitism in S. ratti (astacin-like metalloendopeptidases, CAP-domain containing proteins and acetylcholinesterases) [48]. For all of these studies, only samples from a single geographical area (East Asia and the UK, respectively) are used so they are unlikely to be capturing all of the genetic diversity of either Strongyloides species.
(a) . Comparison with Caenorhabditis elegans
Recently, C. elegans population genomics has been used to (i) discover novel genotypes and unravel the genetic underpinnings of phenotypes [51]; (ii) investigate the evolution of hermaphrodism and the mating system [52]; (iii) assess genetic effects on the starvation response [53]; and (iv) assess genetic diversity and adaption to different environments [54]. There have also been studies on C. elegans laboratory strains to, for example, investigate experimental evolution [55] and study evolutionary responses to bacteria [56]. Most Strongyloides research in this area is solely concerned with transmission or species specificity; therefore, these C. elegans applications are much more diverse in comparison. There is scope to expand the types of questions and analyses carried out in Strongyloides studies, for example by focusing on other features of their life history or ecology and evolution.
(b) . Comparison with other parasitic nematodes
Phylogenetics studies have long been carried out on parasitic nematodes using marker genes, and more recently this has extended to phylogenomics and population genomics studies from whole genome sequencing data [57]. These studies span nematodes that infect plants [57], wild animals [58] and humans [59]; they are more diverse than those conducted in Strongyloides, addressing a wide range of topics including identifying a genetic locus associated with anthelminthic drug resistance in multiple distinct populations of H. contortus [60], and understanding zoonotic reservoirs and distribution expansion of Trichuris trichiura [61].
(c) . Future areas for priority
A priority is to perform population genomics studies for a wider range of Strongyloides species and over a wider geographical distribution of sampling sites. This would provide information about which regions of the genome are under more evolutionary pressure and how the populations in endemic areas are linked or associations of genetic signatures with anthelminthic drug resistance. This knowledge could be useful when selecting vaccine or drug targets [62], as well as providing insights into the evolution of parasitism. Additionally, this would allow identification and monitoring of antihelminth resistance variants in populations, which is an important goal in the World Health Organisation's Roadmap [63]. Improving our understanding of the distribution of Strongyloides infection genotypes, and matching this with information on infection phenotypes, e.g. severity of infection or symptoms associated with infection, should be a key focus for this research area. This information could facilitate the prediction of important information associated with infection including transmission dynamics, disease severity and treatment susceptibility. These insights would provide both epidemiological and clinical benefits, resulting in improved modelling of transmission and development of genomic tests to identify more appropriate treatments. We also need to diversify the range of Strongyloides species studied as the majority of published studies focus on human-infecting species. Generating data from a broader range of Strongyloides species, such as S. papillosus, would aid the creation of similar models for livestock treatment. In species such as S. ratti, whole genome population data would additionally facilitate complementary laboratory-based experiments e.g. to test hypothesizes about genotype–phenotype associations.
4. Strongyloides transcriptomics
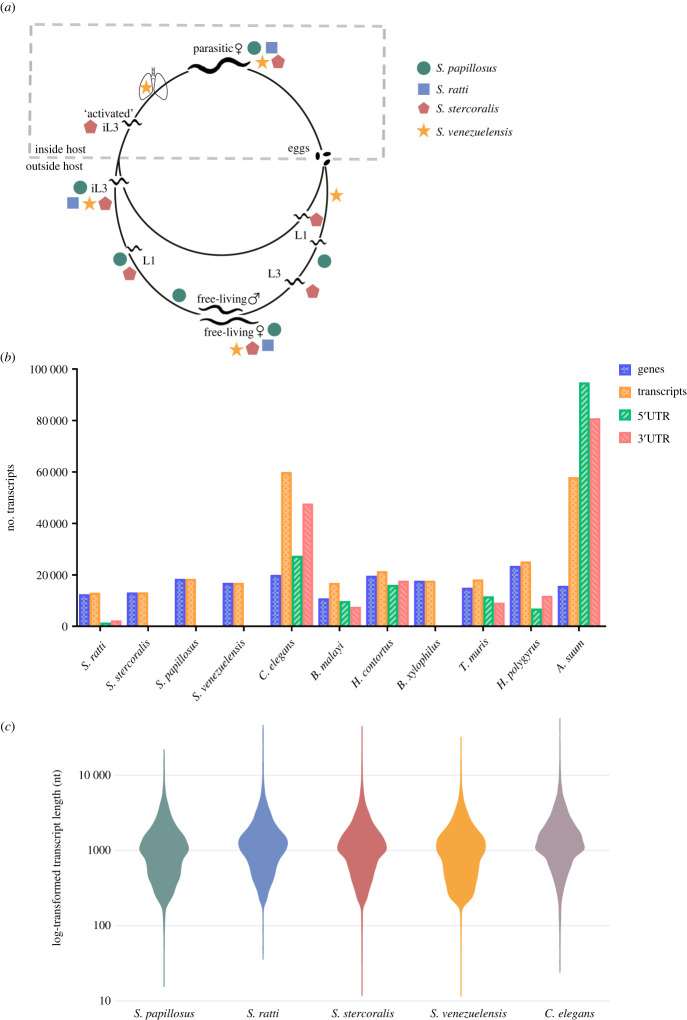
The transcriptome represents a portion of the genome transcribed into RNA molecules (expressed) in a given tissue, life cycle or organism [64] and by comparing transcriptomes we are able to identify the genes that are up- or downregulated in different life cycle stages or conditions. Transcripts are composed of introns, exons, 3′ and 5′ untranslated regions (UTRs), a 5′ cap and a 3′ poly A tail. Using the 454/Roche pyrosequencing technology [65] the transcriptomes of four developmental stages of S. venezuelensis were sequenced [66]. The majority of differentially expressed genes were in parasitic adults (226 genes) compared to eggs/L1, third stage infective larvae (iL3s) and lung-stage larvae (33, 68 and 37 genes, respectively; figure 1a). Genes coding for nicotinamide phosphoribosyltransferase, tuberin and ferrochelatase were identified as putative parasitism-related genes. Strongyloides spp. exhibit a unique life cycle in which genetically identical parasitic adult female (pF) and free-living adult female (flF) generations exist. These life cycle stages can be directly compared to investigate the genes underpinning parasitism; for example, genes significantly upregulated in the pFs versus flFs can identify genes putatively involved in parasitism or other features associated with the parasitic life stage [7]. RNAseq data were analysed using this comparative approach for S. stercoralis, S. ratti, S. papillosus and S. venezuelensis [7,67–69] (figure 1a). In total, 10–18% of expressed genes were identified as differentially expressed between flFs and pFs in the four Strongyloides species. Across Strongyloides spp., genes from 29 families are upregulated in pFs and are thought to have a putative role in parasitism; including genes that encode acetylcholinesterase, astacin-like metalloendopeptidase and CAP-domain containing proteins (also referred to as SCP/TAPs) [68,69].
Figure 1.
Transcriptome information for the Strongyloides species. (a) Availability of transcriptome data divided by life cycle stage for S. papillosus (green circle), S. ratti (blue square), S. stercoralis (red hexagon) and S. venezuelensis (yellow star) [7,66–69]. (b). Number of transcripts, 5′UTR and 3′UTR annotations for 11 nematode species including four Strongyloides spp. Note that multiple UTR annotations are sometimes reported for a single transcript. (c) S. papillosus, S. ratti, S. stercoralis, S. venezuelensis and C. elegans comparison of log-transformed transcript length. Transcript length is significantly different between all pairwise comparisons apart from S. ratti and S. stercoralis, and S. venezuelensis and S. papillosus (ANOVA, F = 485.7, p < 0.0001; Games–Howell multiple comparison test: Sr–Ss md = 65.72, p = 0.0013; Sr–Sp md = 221.9, p < 0.0001; Sr–Sv md = 234.4, p < 0.0001; Sr–Ce md = −238.2, p < 0.0001; Ss–Sp md = 156.2, p < 0.0001; Ss–Sv md = 168.6, p < 0.0001; Ss–Ce md = −303.9, p < 0.0001; Sp–Sv md = 12.42, p = 0.8606; Sp–Ce md = −460.1, p < 0.0001; Sv–Ce md = −472.5, p < 0.0001). (Online version in colour.)
Of the 1188 genes in S. stercoralis and 909 in S. ratti which were specifically upregulated in pFs compared with flFs, 18–19% code for astacin-like metalloproteases or CAP-domain containing proteins [69]. The types of gene families upregulated in the pF (cf. flF) of S. stercoralis, S. ratti, S. papillosus and S. venezuelensis differs between species. Specifically, 25–305 gene families were uniquely upregulated in pFs of one species but not the pFs of the other three Strongyloides species. S. venezuelensis is the most divergent and 305 gene families containing 327 genes upregulated in the pF (compared to the flF) are unique to this species [68], suggesting different species may use distinct molecular toolboxes to infect their host. Interestingly, the upregulated parasitism-associated gene families are expanded in Strongyloides spp. coinciding with the evolution of parasitism within the Strongyloides clade. Also, genes upregulated in the S. ratti pF, compared with the flF or iL3 stage, are physically clustered together on regions of chromosome II, which could be important for the co-regulation of transcription of parasitism-associated genes [69].
The transcriptome of the iL3 life cycle stage has provided further insight into the genetic basis of Strongyloides parasitism. The iL3 has both a free-living stage where it is seeking a host, and a parasitic stage where it penetrates and migrates through host tissue en route to the small intestine. iL3s have been recovered from host lungs and the lung is believed to be a common route of infection for iL3s migrating to the small intestine. After the intestine, the lungs are the most common organ to find Strongyloides iL3s located during a disseminated infection [70]. The iL3 lung stage in S. venezuelensis has been associated with an upregulation of six astacin-like metalloproteases and two glycoprotein coding genes, predicted to code for proteins in excretory/secretory products [66]. In general, much less is known about the genes involved during migration or disseminated infection in the lungs compared with other life cycle stages. The transcriptome varies between the parasitic and free-living larval stages in both S. venezuelensis and S. stercoralis and based on their transcriptome, larval stages appear to be separated by life cycle stage rather than species, despite S. venezuelensis and S. stercoralis infecting different hosts (rats and humans, respectively) [71].
RNAseq transcriptomics of seven life cycle stages of S. stercoralis [67] was used for comparative analysis of developmental larval forms of Strongyloides and C. elegans, to investigate similarities of molecular regulation of developmental arrest between the C. elegans' dauer larvae and parasitic iL3s [71,72]. Genes encoding components in the cGMP pathway (which controls dauer arrest in C. elegans) were upregulated in Strongyloides iL3s, compared to other larval stages. In addition, gene homologues of C. elegans dauer arrest and activation insulin-like signalling, and TGFβ signalling, are also upregulated in S. stercoralis iL3s, suggesting a role in iL3 development. In S. papillosus, transcript expression analysis of 10 RNAseq datasets across six life cycle stages indicates a high degree of conservation in developmental regulation across Strongyloides species [69]. The majority of S. papillosus genes are developmentally regulated with 73% of genes differentially expressed across life cycle stages; 10% of S. papillosus genes were differentially expressed between pFs and flFs, 21% were differentially expressed between free-living males (flM) and flFs and 42–45% were differentially expressed between flF/pF and iL3s. In a comparative transcriptomic analysis, 55% of orthologues from S. venezuelensis and S. stercoralis were assigned to different developmental stages including eggs, larvae and adult stages [71].
Investigation of genes that are differentially expressed when Strongyloides spp. are exposed to different host conditions contributes to our understanding of Strongyloides survival and adaptation to their host [66,73–75]. In pF S. ratti, genes involved in collagen regulation, muscle function and repair, are upregulated in immunized versus naive mice, indicating that the S. ratti transcriptome can be altered to facilitate protection against damage and expulsion from the host [73]. However, the mouse is not a natural host of S. ratti and this must be taking into consideration when interpreting these results. A comparison of gene expression in S. ratti pF derived from the natural rat host compared with pFs derived from a permissive gerbil host, demonstrated an increase in astacin-like metalloproteases and acetylcholinesterase genes in the gerbil-derived parasites further highlighting a putative link between astacin-like metalloprotease expression and parasitism. Even though fewer iL3s successfully establish infection in gerbils, gerbil-derived S. ratti parasitic adults produce more male larvae and survive/reproduce for longer, thus highlighting differences in S. ratti survival and reproduction when exposed to different hosts [74].
There is a lack of information about how Strongyloides spp. respond at the transcriptome level to drug treatments. When comparing human patients' faecal samples cultured on nutrient agar plates that are either untreated or treated with the corticosteroid dexamethasone (DXM), free-living adult stages of S. stercoralis display differential expression of 199 and 263 genes upregulated in flFs and flMs, respectively, that are involved in developmental processes, multicellular organismal processes and embryonic morphogenesis [75]. This raises questions about how clinical steroids and other medication can affect parasite development and survival in endemic strongyloidiasis. As we continue to generate and collate Strongyloides transcriptome data, data repositories and software such as the Strongyloides RNA-seq Browser [76] will be useful in combining existing data from Strongyloides species and will facilitate easier comparative gene expression and enrichment analyses.
(a) . Comparison with Caenorhabditis elegans
The C. elegans transcriptome (PRJNA13758) contains 31 989 transcript variants for 19 981 protein-coding genes with further 28 138 transcripts for other RNA sequences such as pseudogenes and non-coding RNAs. In Strongyloides spp. the total number of transcripts annotated is similar to the total number of genes (figure 1b) (WormBase ParaSite Version WBPS18) [4] because the transcript variants for Strongyloides genes are not well annotated. Furthermore, while most transcripts have an average read length of 1202–1237 bp across all four Strongyloides species sequenced, similar to C. elegans, there are an increased number of shorter length transcripts in Strongyloides spp. especially in S. papillosus and S. venezuelensis (figure 1c). This may represent genuinely shorter transcripts in Strongyloides spp. but is likely to be indicative of annotations that require improvement. Information on the UTR regions of transcripts is also lacking for Strongyloides spp. C elegans remains at the forefront of nematode transcriptome data availability and annotation quality and is continuously improving through sustained contributions from a large scientific community. Current C. elegans annotations contain 27 371 5′UTR and 47 676 3′UTRs (n.b. some transcript variants have multiple UTR annotations). In C. elegans, long-read cDNA and direct RNA sequencing has further improved transcriptome annotations. Using ONT long-read sequencing 23 865 isoforms have been confirmed from 14 611 genes, out of which 3452 were previously undiscovered, with 16 342 isoforms in the 3′UTR region and polyA identification and annotation in all the major life cycle stages, excluding embryos and the arrested dauer stage [77]. ONT sequencing has also been applied to C. elegans to further improve the transcript annotations, identify trans-splicing of mRNAs and confirm splice leaders 1 and 2 (SL1, SL2) [78].
(b) . Comparison with other parasitic nematodes
With the improved availability and affordability of RNAseq, transcriptome data is available for many parasitic nematodes, especially those from the nematode clades III–V [79–82]. The availability and quality of stage-specific expression varies between parasitic nematode species, with adult life cycle stages more likely to be sequenced and egg stages the least likely [79]. While many parasitic UTRomes and alternative splicing annotations remain unexplored, with the advance of RNAseq there has been a recent increase in UTR and splicing annotations. Brugia malayi (PRJNA10729), Heligmosomoides polygyrus (PRJEB15396) and Trichuris muris (PRJEB126) have transcript variant data available, and 27–64% of transcripts contain UTR annotations (WormBase ParaSite Version WBPS18). In A. suum, over 60 000 transcripts have been annotated and validated from RNAseq data, defining alternatively spliced and trans-spliced transcripts and allowing for an extended annotation of UTR regions of previously annotated transcripts [83]. Using PacBio Iso-seq, transcriptome annotations and isoform classification have been improved for H. contrortus [15], however, most parasitic nematode species do not have long-read transcriptome data available. In H. contrortus 67.8% of transcripts were annotated with UTR regions following long-read sequencing and manual curation, many of which were previously misannotated as coding exons [15,22], further highlighting the importance of long-read sequencing and high-quality annotations.
(c) . Priorities for future work
There is limited egg and early stage host-migrating larvae transcript information available for Strongyloides spp. and other parasitic nematodes. In addition, time point comparisons within life cycle stages have demonstrated differential expression [73] but these data are lacking for most life cycle stages and species. Improved annotation of transcripts including identifying transcript variants is essential to improve gene expression and regulation studies, and for functional genomic applications. Because conventional RNAseq methods use short RNA fragments (approx. 50–150 nt) and are mainly optimized for sequencing protein-coding regions of genes, there is a lack of precision in defining ends of genes and information on alternative splicing and UTR regions [84]. While UTR sequencing (3P-Seq) and pipelines for UTR prediction from RNAseq data are available, these methods are technically challenging, require separate library preparations, mainly focus on 3′UTR regions and can be difficult outside of model organisms [85]. Additionally, short-read sequencing can cause mapping errors of exons and UTRs which fails to recognize alternatively spliced isoforms of transcripts and UTRs. Data on 5′UTR and 3′UTRs, which play an importance in gene regulation, is missing entirely from S. papillosus, S. venezuelensis and S. stercoralis (figure 1b) and partially from S. ratti for which we have 5′UTR and 3′UTR annotation for only 11% and 17% of the transcriptome (figure 1b). Improved UTR data will enable us to bioinformatically more accurately predict target sites for small RNAs (sRNAs) and to improve transcript comparisons between life cycle stages, e.g. are different transcript variants of the same gene used in different life cycle stages? Long-read sequencing technologies such as ONT and PacBio are important tools for improving these annotations in the future. Long-read sequencing enables us to sequence full-length transcripts, accurately characterizing the transcripts from 5′UTR to 3′UTR region, including alternatively splicing of exons. Long-read sequences also reduce the GC and amplification bias of RNAseq, improving the quantification of transcripts [86]. Sequencing data from single cell and spatial transcriptomics to map localization of gene expression are also not available either for Strongyloides species or for most other parasitic nematodes, but would offer further insights into genes that are important for parasitism and development. The increased availability of sequencing data for more Strongyloides species and strains, along with further research into their life cycle stages—including time points within these stages and data on gene expression for wild isolates—will aid our understanding of Strongyloides species’ adaptation to their environment and their gene regulation response to environmental change.
5. Strongyloides proteomics
Proteomics is an important tool for understanding the molecular basis of parasitism, where the application of proteomics tools has the potential to characterize protein fingerprints associated with parasite infection, development and parasite–host–microbe interactions. With the advent of tandem liquid-chromatography–mass spectrometry (LC–MS/MS) and improvements in nematode genomic and transcriptomic resources, research into nematode proteomics has expanded in recent years. Most of the current nematode proteomics research can be divided into three main categories based on the source material: (i) somatic (whole-worm) lysate, (ii) excretory–secretory products (ESPs) and (iii) extracellular vesicles (EVs).
Nematode somatic proteomes are typically derived from whole-worm lysates whereby nematode tissue has been mechanically disrupted, resuspended in a lysis buffer and subjected to protein extraction. Most of the somatic proteomics research in Strongyloides spp. has focused on the iL3 stage to better understand strategies used to establish infection [7,87–89] (table 3). The first major proteomics study in Strongyloides spp. was conducted with S. stercoralis, followed by studies in S. ratti [87] and S. venezuelensis [88]. The seminal S. stercoralis proteomics study used iL3s obtained from infected patients and identified 26 proteins including myosin, actin, elongation factors, ATP synthases, galectin and stress response proteins [90]. However, one limitation of this work was that a large portion of mass-spectrometry (MS) spectra remained unassigned following LC–MS/MS data analysis; this may be explained by the unavailability of genomic information and expressed-sequence tag (EST) datasets at the time of the study [90]. The release of the S. stercoralis draft genome in 2016 [7] enabled Dishnica and colleagues [89] to build upon the S. stercoralis iL3 proteome to identify 430 proteins (3.3% of the predicted protein dataset) in S. stercoralis iL3 whole-worm lysates [89] including 3 of the 4 major antioxidant families previously identified in the LC/MS analysis of S. ratti [87] and S. venezuelensis [88] ESP (table 3). Many of the protein categories proposed to be involved in iL3 parasitism [7] were also identified, for example astacin-like metalloproteases that have been reported to be fundamental to the initial phases of S. stercoralis host tissue penetration, parasite development and host immune evasion [96]. Overall, this LC–MS/MS analysis supports the predictions from other Strongyloides spp. transcriptomic, genomic and proteomic analyses by confirming the presence of proteins that were predicted to be associated with Strongyloides spp. parasitism. Collectively the data derived from the above Strongyloides spp. studies highlight the benefits of a multi-omics approach whereby the combination of in silico approaches (genomic and transcriptomic) with experimental proteomics (LC–MS/MS) has the potential to identify proteins associated with parasitism and reveal complex host–parasite interactions, reviewed here and also elsewhere in this issue [97].
Table 3.
Summary of Strongyloides proteomics studies. ESP, excretory/secretory products; iL3, infective L3 larvae; fLF, free living female; pF, parasitic female.
| species | life cycle stage | sample type | reference |
|---|---|---|---|
| S. stercoralis | iL3 | somatic extract—whole worm lysate | Marcilla et al. [90] |
| S. stercoralis | iL3 | somatic extract—whole worm lysate | Rodpai et al. [91] |
| S. stercoralis | iL3 | somatic extract—whole worm lysate | Dishnica et al. [89] |
| S. stercoralis | L3 | somatic extract—whole worm lysate | Rodpai et al. [92] |
| S. ratti | iL3, flF, pF | ESP | Soblik et al. [87] |
| S. ratti | pF | ESP | Younis et al. [93] |
| S. ratti | flF, pF | somatic extract—whole worm lysate | Hunt et al. [7] |
| S. venezuelensis | iL3s, pF | ESP | Maeda et al. [88] |
| S. venezuelensis | iL3 | somatic extract—whole worm lysate | Fonseca et al. [94] |
| S. venezuelensis | iL3 | somatic extract—whole worm lysate | Corral et al. [95] |
There is a demand for the identification of new biomarkers for novel Strongyloides diagnostics [98]. Proteomics has the potential to identify proteins with potential immunogenic properties that may serve as biomarkers for novel diagnostic tests. Previous somatic proteomic analysis of S. venezuelensis iL3s identified 877 proteins that included numerous proteins with biomarker potential such as antioxidants, astacin-like metallopeptidases, proteases and galectins [94] (table 3). Study [94] also highlighted the need for characterization of CAP domain-containing proteins, which are thought to be immunogenic [99]; indeed, nine proteins, including CAP domain-containing proteins, were also detected in S. stercoralis somatic extracts [89]. Interestingly, CAP domain-containing proteins are thought to be associated with parasitism due to their expansion in Strongyloides and Parastrongyloides spp. but their absence from the closely related free-living species Rhabditophanes sp.. Comparison of CAP domain-containing protein detection in other parasitic nematode species in combination with functional characterization may shed light on their potential as strongyloidiasis-specific diagnostic markers [7]. Both studies provide a catalogue of potential diagnostic biomarkers for human and animal strongyloidiasis; however, further functional work is required to characterize their immunogenic properties and promise as novel biomarkers.
An emerging area of research is the use of reverse in silico approaches to predict putative immunogenic proteins for diagnostic/vaccine development [100]. A similar approach was recently applied to predict 34 immunogenic proteins from S. stercoralis proteomics data that hold potential as novel vaccines or diagnostic biomarker candidates [101]. None of the 34 proteins identified in S. stercoralis were identified in a previous S. venezuelensis LC–MS/MS analysis [94]; this may reflect species differences and/or differences in experimental approaches.
Excretory–secretory products (ESPs), released by helminths into the host environment, are considered an important facet of host–parasite–microbe interactions [102]. As such, ESP characterization will drive an improved understanding of the complexity of the host–parasite–microbe interface [103]. To date Strongyloides spp. ESP proteomics analysis has been limited to S. ratti and S. venezuelensis species. A large scale comparative analysis, performed on S. ratti iL3, pF and fLF ESP, revealed a total of 586 proteins across all three life stages with 450, 335 and 219 proteins identified in iL3s, pFs and fLFs, respectively [87] (table 3). Of these 586 proteins, 140 were identified across all three life stages and included proteins such as antioxidants, heat shock proteins, carbohydrate-binding proteins, enolases, galectin, transthyretins and CAP domain-containing proteins [87]. Similarly, in S. venezuelensis the highest number of proteins were identified in iL3 ESP (436 in iL3 versus 196 in pF), with CAP domain-containing proteins, galectins and enolases among the most abundant proteins detected [88] (table 3). Notably, the astacin-like metalloproteases and CAP-domain containing proteins were identified in both S. ratti and S. venezuelensis iL3 ESP and are upregulated (at both the gene and protein levels) in S. ratti pFs [7,87,88,104]. Interestingly, CAP domain-containing proteins were also identified in Ancylostoma caninum iL3 ESP, where they are associated with the free-living to parasitic lifestyle transition, host invasion and host immune system cross-talk [105]; comparison of other Strongyloides spp. iL3 ESP proteomes will shed light on the importance of these proteins in iL3 biology. Further life stage-specific ESP comparisons identified both conserved and species-specific proteins in S. venezuelensis and S. ratti [7,87,88]. In S. ratti, 196, 79 and 35 proteins were unique to iL3s, pFs and fLFs, respectively [87] while in S. venezuelensis, 350 proteins were specific to iL3s and 94 proteins specific to pFs [88]. The detection of unique ESP proteins in different life cycle stages highlights the need for this analysis to be translated to other Strongyloides and Parastronyloides spp. to aid the characterization of proteins that may be key to parasitism or parasite development.
EVs are membrane-bound vesicles (40–1000 nm) [106] that are released into the extracellular space within an organism and also, in the case of parasitic nematodes, into the host environment [107]. EV cargo includes microRNAs, proteins/peptides, lipids and signalling molecules [108] that, if released into the host, may modulate host innate and adaptive host immune responses and/or interact with the host environment [107,108]. EV proteomics tools have not yet been exploited in Strongyloides spp.; however, translation of established parasitic nematode EV pipelines [109–111] provides an opportunity to characterize Strongyloides spp. EV cargo to reveal novel insights into parasitism, host–parasite communication and identify new diagnostic biomarkers.
(a) . Comparison with Caenorhabditis elegans
By exploiting LC–MS/MS and shotgun proteomics, Merrihew et al. [112] identified approximately 7000 proteins in C. elegans tissue [112]. Following this, Schrimpf et al. [113] detected approximately 11 000 proteins representing over half of the predicted proteins present in C. elegans [113]. Since then, proteomics studies have contributed to the understanding of C. elegans biology, including reproduction and development [114], ageing and longevity [115,116] and innate immunity mechanisms against pathogens [117–119]. One hundred and eighty-four proteins have been identified in C. elegans ESP, including transthyretin-like proteins, C-type lectins, proteases and fatty acid binding proteins [120]. Interestingly, while some ESP proteins are common to parasitic and free-living species, others appear to be parasite-specific [120]. A number of proteins (enolase, aldolase, peroxiredoxin, hydroperoxide reductase, thiol-specific antioxidants) are absent in C. elegans ESP but present in parasitic nematode ESP, suggesting a common functional role in parasitism [120,121]. Of note, Strongyloides spp. pFs appear to secrete unique trypsin-inhibitor like (TIL) domain-containing proteins [87,88,120,122] which have been linked to Stongyloides spp. parasitism [7]. Like parasitic nematodes, proteomics data on C. elegans EV cargo is lagging behind the somatic and ESP proteome analysis; however, 224 proteins have been identified in C. elegans EVs and demonstrated that C. elegans EV protein composition differs considerably from whole-worm lysate [123].
(b) . Comparison with other parasitic nematodes
Over the past 10–15 years significant progress has been made in proteomics platforms for identification of nematode somatic proteins that hold potential as biomarkers or therapeutic targets [124]. Life stage-specific somatic proteomics data exist for several parasitic nematode species including Trichinella spiralis, H. contortus, H. polygyrus and Nippostrongylus brasiliensis that enable species and life stage comparative analyses [125–130]. Indeed, similar to Strongyloides spp., there appear to be a core set of proteins (ca. 20% in T. spiralis and ca. 30% in H. contortus) that are conserved between parasite life stages, indicating that the majority of proteins detected appear to be life stage-specific [125,131]. Parasitic nematode ESP proteomics has received a lot of attention. Beyond Strongyloides spp., there are ESP proteomic profiles for 20 parasitic nematode species including A. caninum, Necator americanus, H. contortus and H. polygyrus [132–136]. Comparative analyses of ESP proteomes across parasitic species from different clades and lifestyles reveal conservation of protein families. For example, CAP domain-containing proteins are some of the most abundant proteins detected across multiple parasitic nematode ESPs [134,135,137–139] and they have been implicated in host invasion and parasite development [137]. Other highly conserved ESP proteins include metallopeptidases, transthyretin-like family proteins, astacin-like metalloproteases and enolases [120]. While progress has been slow for Strongyloides spp., numerous studies have characterized EV protein cargo in other parasitic nematode spp. [109,110,140–142]. Similar to the somatic and ESP proteomics data, EV proteomics studies have identified proteins conserved across parasitic nematode species [109,140,142]; however, the reproducibility of EV proteomics datasets can be problematic. For example, two independent analyses of T. muris EV proteomes yielded different data [142,143] and this highlights the need for a standardized pipeline for the characterization of nematode derived EVs, as outlined elsewhere [5]. Continued proteome comparisons across relevant parasitic nematodes and life stages will aid the identification of proteins of interest that will seed functional characterization and reveal novel biomarkers and vaccine targets [120,124].
(c) . Priorities for future work
Expansion and improvement of genome and predicted protein datasets will facilitate the growth and enhancement of nematode proteomic data derived from somatic (whole-worm) lysate, ESP and EV sources [7]. The continued advancement in proteomics tools, technologies and analysis pipelines in addition to sustained efforts to translate these to parasitic nematodes will also drive a better understanding of Strongyloides spp. biology. Indeed, more generally, the standardization of proteomics methods is also critical to the delivery of experimentally robust datasets that will inform nematode biology. Primarily, the priority for Strongyloides spp. should be the generation of equivalent proteomics data for somatic, ESP and EVs across Strongyloides spp. and life stages to facilitate robust comparative analyses and drive the discovery of new biomarkers and control targets for strongyloidiasis. These data would also enable interrogation of the Strongyloides host–parasite interface and the influence of abiotic/biotic environmental factors on the Strongyloides ESP and EV proteomes.
6. Strongyloides peptidomics
Peptidomics is classed as a subset of proteomics and is defined as the peptides (approx. less than 10 kDa) produced in a cell, tissue or organism at a specific timepoint [144]. Relative to proteomics, peptidomics is in its infancy; peptidomics studies in nematode parasites first emerged in the literature around the early 2000s [144–146] and have not yet advanced to include Strongyloides spp.
(a) . Comparison with Caenorhabditis elegans
The Schoofs laboratory has developed a high-throughput LC–MS/MS peptidomics approach adapted from Drosophila melanogaster for application in C. elegans. This has resulted in the identification of a raft of C. elegans neuropeptides [147–149] that have also been detected in parasitic nematodes [150,151]. Advances in C. elegans peptidomics highlights the need for expanded LC–MS/MS studies in parasitic nematodes that exploit in silico analyses [152,153] and interrogate somatic, ESP and EV extracts. In addition, as C. elegans peptidomics data parallel advances in LC–MS/MS technology, it will become possible to map specific cell/neuron peptide expression and dynamics to provide novel insights into nematode peptide biology that will also aid our understanding of parasite systems.
(b) . Comparison with other parasitic nematodes
Much of the initial parasitic nematode peptidomics research was focused on the pig parasite A. suum [154–157]. Since the advent of high-throughput peptidomics only H. contortus has been studied in addition to A. suum, and the focus has primarily been the identification of neuropeptide and antimicrobial peptide profiles in somatic extracts [150,151,158]. Integrating in silico genomics and transcriptomics with LC–MS/MS has further advanced proteomic and peptidomics research [159], and these approaches are beginning to emerge within parasitic nematode literature [160].
(c) . Priorities for future work
The advancement of Strongyloides spp. peptidomics studies will rely on the continued expansion, improvement and integration of in silico datasets, peptidomics tools, LC–MS/MS technologies and optimized analysis pipelines. Specifically, future research in this area should prioritize the translation of the peptidomics pipelines used for H. contortus [150,151] and C. elegans [148] to probe Strongyloides spp., life cycle stage, somatic, ESP and EV peptidomes. These analyses will facilitate downstream functional analyses to inform aspects of Strongyloides peptide biology, including host–parasite–microbiome interactions.
7. Strongyloides small RNAs
sRNAs are non-coding RNA molecules of typically 18–30 nt in length that regulate genes at the post transcriptional level. The regulation of genes by sRNAs is an important mechanism required for the survival and reproduction of parasitic nematodes in the host and environment [161]. The role of sRNAs has been extensively studied in gene silencing, nematode development, transposon silencing and chromatin regulation [162]. sRNAs can be divided into three main classes: microRNAs (miRNAs), short-interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs). Microarrays, reverse transcription-quantitative PCR (RT-qPCR) and deep sRNA sequencing (sRNAseq) are commonly used for sRNA identification. Although both microarrays and RT-qPCR are valuable tools, these methods can only detect known and predefined sRNAs, and sRNAseq is the preferred method for identifying novel sRNAs.
sRNA sequencing has confirmed that sRNAs are expressed in S. ratti, S. papillosus, S. stercoralis and the close relative P. trichosuri [163–166]. The first study characterizing sRNA expression in Strongyloides was carried out in S. ratti iL3s and free-living adults [163]. Using a modified miRDeep2 pipeline [167], 106 mature and precursor S. ratti miRNA sequences were annotated, of which 18 were identified in mixed life cycle stages, eight in iL3s and 80 in both stages. Two miRNAs (mir-34 and mir-71) were highlighted as potentially important in regulating stress and ageing. Further analysis on the miRNA seed sequence (the 2–8 nt region associated with regulating gene expression) revealed that 37 of the 106 miRNAs contain conserved miRNA seed families shared with C. elegans and Pristionchus pacificus, among 24 miRNA seed families conserved between the three species investigated [163]. sRNA expression in S. ratti, S. papillosus and P. trichosuri iL3s and free-living adults (mixed sex) was further compared using two different library preparations to capture sRNAs that have either a 5′ monophosphate or 5′ polyphosphate modification [164]. Annotation of S. ratti miRNAs identified 33 new miRNAs in addition to the 106 previously reported, bringing the total number of characterized miRNAs in S. ratti to 139 [164]. A total of 140 and 163 miRNAs have been annotated for S. papillosus and P. trichosuri, respectively, and miRNA sequences are largely conserved across Strongyloides spp. and P. trichosuri. miRNAs 22–23nt in length represent 79%, 55% and 80% of all genome-mapped sRNA reads in the 5′ monophosphate-enriched library for S. ratti, S. papillosus and P. trichosuri, respectively, and have a propensity for a 5′ uracil (U) in all three species [164]. Among all three species (S. ratti, S. papillosus and P. trichosuri), 65, 73 and 99 miRNAs, respectively, exhibited a differential level of expression between iL3s and free-living adult life cycle stages. Strongyloides stercoralis miRNA expression has been investigated in L1 and iL3s isolated from human stool [165]. A total of 385 and 208, mature and precursor miRNAs were identified in S. stercoralis, substantially more than the number of miRNAs identified in other Strongyloides spp. Upon comparison, 169 novel miRNAs showed no sequence similarity with miRNAs from S. ratti, S. papillosus or C. elegans, suggesting that there are many species-specific miRNAs in S. stercoralis or that miRNAs associated with the L1 stage in other Strongyloides spp. are yet to be discovered. Interestingly, the miRNAs expressed in iL3s were more transcriptionally active than those derived from L1s and were predicted to target and regulate the expression of parasitism-associated genes including astacin-like metalloproteases [165] (table 4).
Table 4.
Summary of small RNA studies in Strongyloides species.
| species | sRNA type | length of sRNA and 5′ nt | life cycle stage | modification | reference |
|---|---|---|---|---|---|
| S. ratti | miRNA | ≥18 nt | iL3 | 5′ monophosphate | Ahmed et al. [163] |
| free-living mixed stage | |||||
| S. ratti | miRNA | 22–23U | iL3 | 5′ monophosphate | Holz & Streit [164] |
| S. papillosus | free-living mixed stage | ||||
| P. trichosuri | |||||
| S. stercoralis | miRNA | 21–23U | L1 | 5′ monophosphate | Pomari et al. [165] |
| iL3 | |||||
| S. ratti | miRNA | 21–23U | pF | 5′ monophosphate | Suleiman et al. [166] |
| free-living female | |||||
| S. ratti | siRNA | 27GA | iL3 | 5′ all-phosphate | Holz & Streit [164] |
| S. papillosus | free-living mixed stage | ||||
| P. trichosuri | |||||
| S. ratti | siRNA | 27GA | pF | 5′ polyphosphate | Suleiman et al. [166] |
| free-living female | |||||
| S. ratti | piRNA-like sRNA | 21–22U | pF | 5′ monophosphate | Suleiman et al. [166] |
Non-miRNA classes of sRNAs including siRNAs and piRNAs have also been investigated in Strongyloides. Strongyloides spp. express sRNAs that originate from tRNAs, rRNAs, transposable elements (TEs) and intergenic sequences in the genome, hypothesized to be siRNAs [164]. These predicted siRNAs are more abundant in the 5′ modification-independent libraries, indicating that they have a 5′ modification such as a polyphosphate 5′ end, similar to the secondary siRNAs reported in C. elegans. The siRNAs expressed in S. ratti, S. papilllosus and P. trichosuri have a length of 27nt with an equal bias towards a 5′ guanine (G) and adenine (A) (27GAs). The targets of 27GAs based on sequence complementarity were predicted to be TEs in S. ratti pFs, free-living adults and iL3s, S. papillosus and P. trichosuri free-living adults and iL3s [164]. In S. ratti pF and flF, the 27GA siRNAs are predicted to target and regulate the expression of TEs predominantly located on the X-chromosome [166], but this information is not known for other species. A notable difference between 27GAs expressed in pF and flF life cycle stages is that the pF 27GAs mostly target DNA transposons, in comparison to the flF 27GAs that target retrotransposons [166]. Although sRNA data are available [165], siRNA expression in S. stercoralis has not yet been investigated.
The piRNA class of sRNAs has been lost in nematodes outside of clade V nematodes, including Strongyloides [68,161,164] evident from the absence of the Argonaute proteins from the PIWI family that interacts with and facilitates biogenesis of piRNAs. sRNA sequencing has not detected piRNA sequences in S. ratti, S. papillosus and P. trichosuri iL3 and free-living adults. However, the pF stage of S. ratti expresses a piRNA-like class of sRNAs with a length of 21–22 nt and propensity towards a 5′ uracil (21–22U) [166]. The piRNA-like sRNAs show striking resemblance to piRNAs of C. elegans and D. melanogaster, including their length, 5′ uracil, 5′ monophosphate modification, clustering and overlapping of sequences in the genome and an AT rich downstream sequence. However, these piRNA-like sRNAs did not have an upstream Ruby motif, as found for C. elegans piRNAs. The 21–22U piRNA-like sRNAs are specifically highly expressed in the pF compared with flF, indicating that they may be directly related to parasitism or features associated with the parasitic life cycle [166].
(a) . Comparison with Caenorhabditis elegans
sRNAs were originally identified in C. elegans and this species has become an important model organism for the study sRNA biology [168,169]. C. elegans expresses 253 precursors and 437 mature miRNAs [170]. These miRNAs have similar features to Strongyloides miRNAs, including a 5′ monophosphate modification, with a length of 21–22 nt and bias towards 5′ U or 5′ A. Comparison of Strongyloides miRNAs to those found in C. elegans has revealed that 31 seed families are shared between S. ratti and C. elegans, while 30 are shared between S. papillosus and C. elegans [164]. Among the three species, 29 seed families were shared that include miRNAs such as let-7, lin-4, mir-34 and mir-37, indicating that the conservation of these miRNAs is essential in nematodes. In comparison to Strongyloides, C. elegans siRNAs and the sRNA pathways they belong to are well characterized, with distinct biological functions depending on their biogenesis. C. elegans expresses primary 26G siRNAs containing a 5′ monophosphate modification, which is important in the regulation of gene expression during spermatogenesis and zygotic development [171]. The 26G siRNAs, alongside piRNAs, can initiate the activation of secondary siRNAs, termed 22G siRNAs. The 22Gs contain a 5′ polyphosphate modification and have a role in regulating the expression of genes, pseudogenes and transposons, predominantly in the germline [172]. As there are no 26G and 22G siRNAs in Strongyloides, it has been suggested that 27GA siRNAs are equivalent to 22Gs in C. elegans [164,166]. As discussed above, unlike nematodes from clades I–IV—including Strongyloides spp.—C. elegans is known to produce and express piRNAs, also known as 21U sRNAs, that are important in regulating the activity of TEs [173]. Strongyloides have evolved an alternative class of piRNA-like sRNAs, the 21–22Us, which are predicted to be important in regulating TEs in the absence of piRNAs [166].
(b) . Comparison with other parasitic nematodess
Nematodes in clades I–V express conserved miRNAs [162]. However, similar to Strongyloides, piRNAs in clades I–IV nematode including A. suum (clade III) [174] and B. pahangi [175] are lost. Some of the species from clades III and IV have diverged and compensated for the loss of the PIWI pathway through higher expression of the secondary siRNA 22Gs that target and defend the germline against TEs, similar to the C. elegans piRNAs [161,174,176] and the piRNA-like sRNAs observed in S. ratti [166]. In addition to a role in regulating TEs and endogenous transcripts, sRNAs also have a role in parasite–host interactions. Parasitic nematodes secrete ELVs containing sRNAs that are internalized by host cells and alter host gene expression. The secretion of ELVs containing sRNAs was first identified in the parasitic nematode H. polygyrus [177]. These ELVs containing sRNAs suppress genes associated with innate immunity and are important for establishing parasitism in the host [178]. miRNAs in H. polygyrus ELVs shared sequence similarity with the host miRNAs. Further analysis of sRNAs using the 5′ phosphate-independent library revealed a prevalence of siRNAs derived from repetitive elements and intergenic regions [179].
(c) . Priority areas for future research
(i) sRNA expression across a range of Strongyloides species. To gain a deeper understanding of the role of sRNAs in parasitism, it is crucial to conduct research on a range of life cycle stages, such as the pF and free-living adult stages of S. stercoralis, and species, such as S. fuelleborni and S. venezuelensis. (ii) Secretion of parasite-derived EVs containing sRNAs taken up by host cells. Further research is needed to understand the specific functions of sRNAs in these interactions, including how parasites use sRNAs to manipulate the host environment. This knowledge could not only enhance our understanding of host–parasite relationships, but also provide valuable diagnostic tools. ELV-bound sRNAs have not been investigated in Strongyloides. (iii) sRNAs as biomarkers. sRNAs have potential to be useful biomarkers for disease diagnosis and prognosis in humans and animals. As there is currently no gold standard method for detecting Strongyloides infections in the host, sRNAs present an opportunity to be developed as a diagnostic tool. For instance, the research carried out on sRNAs in S. stercoralis from human infections [165] provides insight into which sRNAs could be detected and exploited as putative biomarkers for the diagnostics of Strongyloides in humans. (iv) Validation of sRNAs and their targets. Most sRNA research to date has used sequencing followed by computational analysis, predicting targets through sequence complimentary. This work needs to be validated using high-throughput approaches such as Argonaute cross-linking immunoprecipitation (CLIP) [180] that identifies endogenous in vivo sRNA–target interactions, allowing us to better understand the role that sRNAs play in parasitism and host–parasite interactions. Understanding the regulatory networks of sRNAs could provide insight into their roles in the control of gene expression and other biological processes.
8. Transposable elements in Strongyloides
TEs are mobile genetic elements occupying the genomes of organisms across all branches of life. TEs can be a major driving force behind genetic variation but can also disrupt regular gene activity via different routes of mutagenesis [181]. The persistence of TEs in populations is maintained through vertical inheritance from one generation to the next. Although the study of TEs is not traditionally classified within the ‘omics’ field, unlike the large-scale study of genes, proteins and metabolites, recent advancements in sequencing technologies and downstream bioinformatic analyses have enabled researchers to explore TEs experimentally on a broader scale, akin to more conventional omics methodologies. Furthermore, omics methodologies such as transcriptomics and genomics enable comprehensive research into the TE biology of a species or at a population level, and can provide a deeper understanding of the effects of TEs on gene expression, genome organization and evolution. Eukaryotes have adopted post-transcriptional silencing via RNAi to counteract the harmful effects of TE insertions. The two known classes of sRNAs involved in these pathways in Strongyloides spp. are described above [164,166]. TEs are tightly controlled in the gonads of most animals because transposition into, and the potential disruption of sexual development genes can be lethal to the host organism [182]. In S. ratti, TE sequences are distributed throughout the genome but TE-associated sRNAs and their predicted TE targets are clustered on the X-chromosome of both S. ratti female adult stages [166].
Mutations induced by strong negative selection pressure have left TE sequences deteriorated and repetitive in nature, which makes their annotation notoriously difficult, especially in assemblies produced from short-read sequences. While several bioinformatics tools with de novo and homology-based algorithms have been developed to identify and annotate TEs, often a substantial amount of manual curation is still required to acquire a reliable TE consensus sequence library [183]. TE sequence annotation of the S. ratti genome has been carried out by four groups employing different methods of annotation: (i) Suleiman et al. [166] constructed a repeat library with RepeatModeler2 [184] and RepeatMasker [185] to annotate repeats via sequence comparison against the Dfam library [186]. As LTR retrotransposons seemed overrepresented in the S. ratti genome, they were further annotated by using the LTR specific tools LTRharvest [187] and LTRdigest [188]; (ii) WormBase ParaSite have recently announced a new repeat annotation feature, where RepeartModeler2 [184] was used to generate custom repeat models for all available genome assemblies including S. ratti, S. venezuelensis, S. stercoralis and S. papillosus, as well as close relatives P. trichosuri and R. ditinus. To annotate the repeat features, a pipeline including RepeatMasker [185], DustMasker [189] and TRF [190] was employed; (iii) Szitenberg et al. [191] have assembled TE libraries for several nematode species, including S. ratti, S. venezuelensis, S. papillosus and P. trichosuri. This curation used a homology-based method that based TE sequence searches on a nematode-specific de novo constructed library, rather than the more general Dfam or RepBase libraries [191,192]. After repeat sequence identification and compilation using RepeatModeler and RepeatMasker, a non-redundant library was pooled together using USEARCH [193] and One Code to Find Them All [194], while further consensus sequence classification was done with CENSOR [195] and LTRHarvest [187]. (iv) An in silico study conducted by the International Helminth Genomes Consortium constructed repeat libraries for the genomes of 56 parasitic and free-living nematodes. For each species, repeat libraries were constructed by combining libraries generated from RepeatModeler [184], TransposonPSI and LTRharvest [187]. The library was then used to mask genomic repeats using RepeatMasker [196].
The curation of TE annotations for Strongyloides spp. by Suleiman et al. [166] and WormBase ParaSite have run RepeatMasker [185] with its native Dfam and Repbase libraries that consist of eukaryotic repeat sequences predominantly belonging to model organisms. Hence, repeat sequences and TEs belonging exclusively to non-model organisms would not be as accurately represented. Conversely, the Szitenberg and International Helminth Genomes Consortium consensus libraries for S. ratti, S. venezuelensis, S. papillosus and P. trichosuri [191] have been built by running RepeatMasker with custom de novo nematode libraries based on repeat sequences generated through running RepeatModeler in addition to other tools mentioned above. This technique would offer a more specific annotation through the identification of species-specific repeats that would not be as represented by using the more universal Dfam and RepBase libraries on their own. The Szitenberg S. ratti TE library comprises 657 TE families occupying 11.2% of the genome, while the International Helminth Genomes Consortium library consists of 3 TE families covering 1.8% of the genome. The Suleiman library contains 5526 TE families that comprise 8.45% of the genome, and the WormBase ParaSite library contains 111 TE families that cover 5.64% of the genome. In addition to mapping to variable proportions of the S. ratti genome, the four TE consensus libraries also vary in the annotation and quantification of different TE families (figure 2). An unknown TE class occupies much of the genome according to the WormBase ParaSite and Suleiman libraries. The DNA element Merlin is also shown to occupy approximately 0.25% of the genome by both libraries. An LTR belonging to an unknown family is shown to have the highest level of expansion in the S. ratti genome by the Szitenberg and the International Helminth Genomes Consortium annotations. Interestingly, this expansion has not been highlighted in the WormBase ParaSite and Suleiman annotations, and instead an unknown TE has shown increased genomic proliferation. The unknown TE family emphasized in the WormBase ParaSite and Suleiman libraries could in fact be the unclassified LTR family highlighted in the Szitenberg and the International Helminth Genomes Consortium libraries. This element could have been highlighted in only two of the available S. ratti repeat annotations due to the use of de novo libraries during the analysis or as a result of different cut-offs and LTR characterization techniques. The DNA transposon TcMar-Mariner and the LTR Gypsy elements are highlighted in the genome by all libraries at different amounts apart from the International Helminth Genomes Consortium library. The International Helminth Genomes Consortium annotation covers the lowest proportion of the S. ratti genome as their annotation only represents specific TE families. RC-Helitron DNA elements are annotated in the Szitenberg and Suleiman libraries, where it is shown at a higher abundance by the Suleiman library. SINEs have only been annotated and detected by the Szitenberg TE annotation.
Figure 2.
Comparison of TE consensus libraries for S. ratti for the four currently available TE libraries constructed by the International Helminth Genomics Consortium 2019 [179], WormBaseParaSite 2023 (v.18), Suleiman et al. [166] and Szitenberg et al. [191]. The percentage of the genome occupied by each TE family is illustrated on the x-axis and annotation of different classes of TE are highlighted by colour. (Online version in colour.)
(a) . Comparison with Caenorhabditis elegans
Despite being the first animal to have its genome sequenced and assembled to a complete level, little is known about TE dynamics in the model organism C. elegans. Various methods have been used to annotate the repetitive sequences, including TEs, in the C. elegans genome [197–200] and consistently conclude that TEs occupy approximately 12% of the genome. Similar to Meloidogyne spp. genomes, DNA transposons make up the majority of expanded TEs. Furthermore, repetitive sequences of the free-living nematode P. pacificus were recently annotated via a custom ‘sliding window’ method, where 1 kbp sequence ‘windows’ are extracted and analysed for repeats using 11 different de novo and library-based tools illustrating that 24% of the P. pacificus genome consists of TEs [201]. Similar to the genome of S. ratti, retrotransposons are the most abundant class of TEs in the P. pacificus genome, accounting for 50% of masked repeats.
(b) . Comparison with other parasitic nematodes
The International Helminth Genomes Consortium TE annotation of 56 helminth genomes was analysed via a multiple regression model to reveal that variations in genome size within the nematode phylum were primarily influenced by variations in genome repeat content, ranging from 3.8% to 54.5%. Notably, the significant contributors to this variation were LTRs, DNA transposons and simple repeats [196]. The aforementioned libraries were constructed from genome assemblies produced by short-read sequences, which are limited in their ability to fully represent TE content due to the repetitive and divergent nature of these elements' sequences [202]. Combining long-read and short-read sequences can increase assembly accuracy and aid in improving the annotation of repetitive regions and TE boundaries [203]. The genome of the plant parasite Meloidogyne enterolobii was recently sequenced and assembled using PacBio and Illumina reads [204]. A TE consensus library was built by utilizing the Tedenovo pipeline [205] for TE predictions. The clustering of similar sequences was carried out using Recon [206], where a multi-sequence alignment of similar clusters was then completed to deduce a consensus sequence using MAP [56] and PASTEClassifier [207]. It was found that 47.62% of the M. enterolobii genome is composed of repetitive elements. However, only 8.7% of the repeats were annotated as TEs, where DNA elements seem to have expanded at a greater rate (5.83%) than retrotransposons (2.87%). A consensus TE library was deduced for the closely related species M. incognita using the same workflow described by Koutsovoulos et al. [204]. The repeatome was shown to span 26.38% of the genome, 4.67% of which is subjugated by retrotransposons (0.9%) and DNA elements (3.77%) [208]. Meloidogyne spp. have a lower proportion of TEs in their genomes compared to Strongyloides spp. despite them leading similar parasitic lifestyles. Furthermore, DNA transposons have experienced a much larger expansion in the genomes of Meloidogyne spp. in comparison to S. ratti, whose genome seems to include a large number of LTRs. The repeatome of the pig parasite A. suum was also identified using a combination of homology and de novo-based tools [176]. The Ascaris genome was assembled using long PacBio reads, revealing that approximately 41% of the genome comprises repetitive sequences. However, the specific classification of these repeats and the familial identities of Ascaris TEs have not been reported [171].
(c) . Areas for future priority
TEs contribute to gene regulation by acting as regulatory elements that manipulate transcription and epigenetic states [209]. By studying TE activity in parasitic and free-living stages of Strongyloides spp., researchers have the opportunity to explore potential variations in transposition and TE repression patterns between free-living and parasitic stages. This exploration may elucidate the molecular mechanisms responsible for variances in gene regulation and epigenetic patterns resulting from TE activity, particularly within germline cells [210] and how these mechanisms relate to different lifestyles, i.e. parasitic versus free-living, parthenogenic versus sexual, long versus short lifespans (pF versus flM/flF, respectively). To better understand Strongyloides TEs, a library consisting of consensus sequences corresponding to the different TE families present in the genomes of Strongyloides species must be curated. Utilizing long and short sequencing in a de novo pipeline as described by Koutsovoulos et al. [204] could be used for Strongyloides spp. to construct consensus libraries. However, as some previously uncharacterized TE families could be Strongyloides-specific, some manual curation will be required to compile an accurate consensus library, as described by Goubert et al. [183]. The established consensus library can be employed in transcriptomic analyses to examine TE dynamics across life cycle stages. Additionally, exploring different histone marks in the context of masked TEs within the Strongyloides genome will allow for a deeper investigation into the role of TEs in genome architecture and their expression patterns. On a broader scale, accurately annotated TE libraries can facilitate comparative studies on TE load across different species and populations. These results can shed light on whether factors such as environment, lifestyle and stress influence the abundance and diversity of an organism's TE dynamics, and to help elucidate what regulatory roles differences in composition, load and activity may play. Furthermore, investigating the TE content of diverse Strongyloides species and strains inhabiting different environments can shed light on the impact of the environment on genome organization, as shown previously in the parasitic flatworms Schistosoma mansoni and S. japonicum [211].
9. Conclusion
Recent advances in technologies and omics analysis pipelines will continue to reduce the gap between omics data availability for Strongyloides spp. and C. elegans. Moving forward, the priority in Strongyloides omics includes (i) investigating more species, including those important to human and livestock health; (ii) investigating samples from wild and clinically important settings as well as laboratory-based populations to understand variation and diversity; and (iii) the application of the latest technologies e.g. long-read sequencing, to improve genome and transcriptome assemblies. There are also areas of omics that have received little or no attention, such as peptidomics, metabolomics, long non-coding RNA, chromatin organization and histone modifications. Numerous projects are now underway within the Strongyloides scientific community to address these areas, which will further close the gap between C. elegans and Strongyloides spp. and improve our understating of the omics biology underpinning parasitism. Advances in omics technologies and data availability will aid identification and development of new ways to detect and control Strongyloides infections and treat the disease that they cause.
Contributor Information
Louise Atkinson, Email: l.atkinson@qub.ac.uk.
Vicky Hunt, Email: bs1vlh@bath.ac.uk.
Data accessibility
This article has no additional data.
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors' contributions
R.A.-J: writing—original draft, writing—review and editing; D.L.: writing—original draft, writing—review and editing; D.M.: writing—original draft, writing—review and editing; K.R.: writing—original draft, writing—review and editing; M.S.: writing—original draft, writing—review and editing; A.M.: conceptualization, supervision, writing—original draft, writing—review and editing; L.A.: conceptualization, supervision, writing—original draft, writing—review and editing; V.H.: conceptualization, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported by Biotechnology and Biological Sciences Research Council-funded South West Biosciences Doctoral Training Partnership (grant no. BB/T008741/1) awarded to R.A.-J., the Department of Agriculture, Environment and Rural Affairs (DAERA) studentship awarded to D.M., GW4 BIOMED MRC DTP (MR/W006308/1) awarded to K.R., University of Bath Research Studentship Award to M.S., Academy of Medical Sciences Springboard Award (grant no. SBF004\1018) to L.A., Biotechnology and Biological Sciences Research Council (grant no. BB/T016396/1) award to A.M. and a Wellcome Trust Sir Henry Dale Fellowship (211227/Z/18/Z) awarded to V.H.
References
- 1.Salinas G, Risi G. 2018. Caenorhabditis elegans: nature and nurture gift to nematode parasitologists. Parasitology 145, 979-987. ( 10.1017/S0031182017002165) [DOI] [PubMed] [Google Scholar]
- 2.Hahnel SR, Dilks CM, Heisler I, Andersen EC, Kulke D. 2020. Caenorhabditis elegans in anthelmintic research – Old model, new perspectives. Int. J. Parasitol. Drugs Drug Resist. 14, 237-248. ( 10.1016/j.ijpddr.2020.09.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward JD. 2015. Rendering the intractable more tractable: tools from Caenorhabditis elegans ripe for import into parasitic nematodes. Genetics 201, 1279-1294. ( 10.1534/genetics.115.182717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. 2017. WormBase ParaSite − a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 215, 2-10. ( 10.1016/j.molbiopara.2016.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White R, et al. 2023. Special considerations for studies of extracellular vesicles from parasitic helminths: a community-led roadmap to increase rigour and reproducibility. J. Extracell. Vesicles 12, e12298. ( 10.1002/jev2.12298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castelletto ML, Gang SS, Hallem EA. 2020. Recent advances in functional genomics for parasitic nematodes of mammals. J. Exp. Biol. 223, jeb206482. ( 10.1242/jeb.206482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt VL, et al. 2016. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 48, 299-307. ( 10.1038/ng.3495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kounosu A, Sun S, Maeda Y, Dayi M, Yoshida A, Maruyama H, Hunt V, Sugimoto A, Kikuchi T. 2023. Syntenic relationship of chromosomes in Strongyloides species and Rhabditophanes diutinus based on the chromosome-level genome assemblies. Phil. Trans. R. Soc. B 379, 20220446. ( 10.1098/rstb.2022.0446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi K, Kadota M, Nishimura O, Ohishi Y, Naito Y, Kuraku S. 2021. Technical considerations in Hi-C scaffolding and evaluation of chromosome-scale genome assemblies. Mol. Ecol. 30, 5923-5934. ( 10.1111/mec.16146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemetschke L, Eberhardt AG, Viney ME, Streit A. 2010. A genetic map of the animal-parasitic nematode Strongyloides ratti. Mol. Biochem. Parasitol. 169, 124-127. ( 10.1016/j.molbiopara.2009.10.008) [DOI] [PubMed] [Google Scholar]
- 11.The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012-2018. ( 10.1126/science.282.5396.2012) [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Veronezi GMB, Kang Y, Zagoskin M, O'Toole ET, Davis RE. 2020. Comprehensive chromosome end remodeling during programmed DNA elimination. Curr. Biol. 30, 3397-3413.e4. ( 10.1016/j.cub.2020.06.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tracey A, et al. 2020. Nearly complete genome sequence of Brugia malayi strain FR3. Microbiol. Resour. Announc. 9, e00154-20. ( 10.1128/MRA.00154-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dayi M, Sun S, Maeda Y, Tanaka R, Yoshida A, Tsai IJ, Kikuchi T. 2020. Nearly complete genome assembly of the pinewood nematode Bursaphelenchus xylophilus strain Ka4C1. Microbiol. Resour. Announc. 9, 10-128. ( 10.1128/MRA.01002-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle SR, et al. 2020. Genomic and transcriptomic variation defines the chromosome-scale assembly of Haemonchus contortus, a model gastrointestinal worm. Commun. Biol. 3, 656. ( 10.1038/s42003-020-01377-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masonbrink RE, Maier TR, Hudson M, Severin A, Baum T. 2021. A chromosomal assembly of the soybean cyst nematode genome. Mol. Ecol. Resour. 21, 2407-2422. ( 10.1111/1755-0998.13432) [DOI] [PubMed] [Google Scholar]
- 17.Cotton JA, et al. 2016. The genome of Onchocerca volvulus, agent of river blindness. Nat. Microbiol. 2, Article 2. ( 10.1038/nmicrobiol.2016.216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foth BJ, et al. 2014. Whipworm genome and dual-species transcriptome analyses provide molecular insights into an intimate host-parasite interaction. Nat. Genet. 46, Article 7. ( 10.1038/ng.3010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33(Web Server issue), W465-W467. ( 10.1093/nar/gki458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinf. 12, 491. ( 10.1186/1471-2105-12-491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshimura J, et al. 2019. Recompleting the Caenorhabditis elegans genome. Genome Res. 29, 1009-1022. ( 10.1101/gr.244830.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle SR. 2022. Improving helminth genome resources in the post-genomic era. Trends Parasitol. 38, 831-840. ( 10.1016/j.pt.2022.06.002) [DOI] [PubMed] [Google Scholar]
- 23.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32(Web Server issue), W309-W312. ( 10.1093/nar/gkh379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schad GA, Hellman ME, Muncey DW. 1984. Strongyloides stercoralis: hyperinfection in immunosuppressed dogs. Exp. Parasitol. 57, 287-296. ( 10.1016/0014-4894(84)90103-6) [DOI] [PubMed] [Google Scholar]
- 25.Fisher MC, Viney ME. 1998. The population genetic structure of the facultatively sexual parasitic nematode Strongyloides ratti in wild rats. Proc. R. Soc. Lond. B 265, 703-709. ( 10.1098/rspb.1998.0350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterson S, Fisher MC, Viney ME. 2000. Inferring infection processes of a parasitic nematode using population genetics. Parasitology 120, 185-194. ( 10.1017/S0031182099005417) [DOI] [PubMed] [Google Scholar]
- 27.Eberhardt AG, Mayer WE, Bonfoh B, Streit A. 2008. The Strongyloides (Nematoda) of sheep and the predominant Strongyloides of cattle form at least two different, genetically isolated populations. Vet. Parasitol. 157, 89-99. ( 10.1016/j.vetpar.2008.07.019) [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa H, et al. 2010. Molecular identification of the causative agent of human strongyloidiasis acquired in Tanzania: dispersal and diversity of Strongyloides spp. and their hosts. Parasitol. Int. 59, 407-413. ( 10.1016/j.parint.2010.05.007) [DOI] [PubMed] [Google Scholar]
- 29.Anderson J, Upadhayay R, Sudimack D, Nair S, Leland M, Williams JT, Anderson TJC. 2012. Trichuris sp. and Strongyloides sp. infections in a free-ranging baboon colony. J. Parasitol. 98, 205-208. ( 10.1645/GE-2493.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schär F, Guo L, Streit A, Khieu V, Muth S, Marti H, Odermatt P. 2014. Strongyloides stercoralis genotypes in humans in Cambodia. Parasitol. Int. 63, 533-536. ( 10.1016/j.parint.2014.01.010) [DOI] [PubMed] [Google Scholar]
- 31.Nagayasu E, et al. 2017. A possible origin population of pathogenic intestinal nematodes, Strongyloides stercoralis, unveiled by molecular phylogeny. Sci. Rep. 7, 4844. ( 10.1038/s41598-017-05049-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barratt JLN, et al. 2019. A global genotyping survey of Strongyloides stercoralis and Strongyloides fuelleborni using deep amplicon sequencing. PLoS Negl. Trop. Dis. 13, e0007609. ( 10.1371/journal.pntd.0007609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basso W, Grandt L-M, Magnenat A-L, Gottstein B, Campos M. 2019. Strongyloides stercoralis infection in imported and local dogs in Switzerland: from clinics to molecular genetics. Parasitol. Res. 118, 255-266. ( 10.1007/s00436-018-6173-3) [DOI] [PubMed] [Google Scholar]
- 34.Beknazarova M, Barratt JLN, Bradbury RS, Lane M, Whiley H, Ross K. 2019. Detection of classic and cryptic Strongyloides genotypes by deep amplicon sequencing: a preliminary survey of dog and human specimens collected from remote Australian communities. PLoS Negl. Trop. Dis. 13, e0007241. ( 10.1371/journal.pntd.0007241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanpool O, Intapan PM, Rodpai R, Laoraksawong P, Sadaow L, Tourtip S, Piratae S, Maleewong W, Thanchomnang T. 2019. Dogs are reservoir hosts for possible transmission of human strongyloidiasis in Thailand: molecular identification and genetic diversity of causative parasite species. J. Helminthol. 94, e110. ( 10.1017/S0022149X1900107X) [DOI] [PubMed] [Google Scholar]
- 36.Spotin A, Mahami-Oskouei M, Nami S. 2019. Assessment of the global paradigms of genetic variability in Strongyloides stercoralis infrapopulations determined by mitochondrial DNA sequences. Comp. Immunol. Microbiol. Infect. Dis. 67, 101354. ( 10.1016/j.cimid.2019.101354) [DOI] [PubMed] [Google Scholar]
- 37.Thanchomnang T, et al. 2019. First molecular identification of Strongyloides fuelleborni in long-tailed macaques in Thailand and Lao People's Democratic Republic reveals considerable genetic diversity. J. Helminthol. 93, 608-615. ( 10.1017/S0022149X18000512) [DOI] [PubMed] [Google Scholar]
- 38.Wulcan JM, Dennis MM, Ketzis JK, Bevelock TJ, Verocai GG. 2019. Strongyloides spp. in cats: a review of the literature and the first report of zoonotic Strongyloides stercoralis in colonic epithelial nodular hyperplasia in cats. Parasites Vectors 12, 349. ( 10.1186/s13071-019-3592-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barratt JLN, Sapp SGH. 2020. Machine learning-based analyses support the existence of species complexes for Strongyloides fuelleborni and Strongyloides stercoralis. Parasitology 147, 1184-1195. ( 10.1017/S0031182020000979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janwan P, Rodpai R, Intapan PM, Sanpool O, Tourtip S, Maleewong W, Thanchomnang T. 2020. Possible transmission of Strongyloides fuelleborni between working Southern pig-tailed macaques (Macaca nemestrina) and their owners in Southern Thailand: molecular identification and diversity. Infect. Genet. Evol. 85, 104516. ( 10.1016/j.meegid.2020.104516) [DOI] [PubMed] [Google Scholar]
- 41.Ko PP, et al. 2020. Phylogenetic relationships of Strongyloides species in carnivore hosts. Parasitol. Int. 78, 102151. ( 10.1016/j.parint.2020.102151) [DOI] [PubMed] [Google Scholar]
- 42.Repetto SA, et al. 2021. Molecular typing of Strongyloides stercoralis in Latin America, the clinical connection. Parasitology 149, 24-34. ( 10.1017/S0031182021001517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ko PP, et al. 2022. Population genetics study of Strongyloides fuelleborni and phylogenetic considerations on primate-infecting species of Strongyloides based on their mitochondrial genome sequences. Parasitol. Int. 92, 102663. ( 10.1016/j.parint.2022.102663) [DOI] [PubMed] [Google Scholar]
- 44.Kikuchi T, et al. 2016. Genome-wide analyses of individual Strongyloides stercoralis (Nematoda: Rhabditoidea) provide insights into population structure and reproductive life cycles. PLoS Negl. Trop. Dis. 10, e0005253. ( 10.1371/journal.pntd.0005253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaleta TG, Zhou S, Bemm FM, Schär F, Khieu V, Muth S, Odermatt P, Lok JB, Streit A. 2017. Different but overlapping populations of Strongyloides stercoralis in dogs and humans—dogs as a possible source for zoonotic strongyloidiasis. PLoS Negl. Trop. Dis. 11, e0005752. ( 10.1371/journal.pntd.0005752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou S, et al. 2019. Characterization of a non-sexual population of Strongyloides stercoralis with hybrid 18S rDNA haplotypes in Guangxi, Southern China. PLoS Negl. Trop. Dis. 13, e0007396. ( 10.1371/journal.pntd.0007396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aupalee K, Wijit A, Singphai K, Rödelsperger C, Zhou S, Saeung A, Streit A. 2020. Genomic studies on Strongyloides stercoralis in northern and western Thailand. Parasites Vectors 13, 250. ( 10.1186/s13071-020-04115-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole R, Holroyd N, Tracey A, Berriman M, Viney M. 2022. The parasitic nematode Strongyloides ratti exists as populations of long-lived asexual lineages. Nat. Commun. 14, 6427. (https://www.nature.com/articles/s41467-023-42250-1.epdf) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradbury RS, Pafčo B, Nosková E, Hasegawa H. 2021. Strongyloides genotyping: a review of methods and application in public health and population genetics. Int. J. Parasitol. 51, 1153-1166. ( 10.1016/j.ijpara.2021.10.001) [DOI] [PubMed] [Google Scholar]
- 50.Hasegawa H, Hayashida S, Ikeda Y, Sato H. 2009. Hyper-variable regions in 18S rDNA of Strongyloides spp. as markers for species-specific diagnosis. Parasitol. Res. 104, 869-874. ( 10.1007/s00436-008-1269-9) [DOI] [PubMed] [Google Scholar]
- 51.Andersen EC, Rockman MV. 2022. Natural genetic variation as a tool for discovery in Caenorhabditis nematodes. Genetics 220, iyab156. ( 10.1093/genetics/iyab156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L. 2008. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature 454, 1019-1022. ( 10.1038/nature07171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webster AK, et al. 2022. Using population selection and sequencing to characterize natural variation of starvation resistance in Caenorhabditis elegans. Elife 11, e80204. ( 10.7554/eLife.80204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee D, et al. 2021. Balancing selection maintains hyper-divergent haplotypes in Caenorhabditis elegans. Nat. Ecol. Evol. 5, 794-807. ( 10.1038/s41559-021-01435-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teotónio H, Estes S, Phillips PC, Baer CF. 2017. Experimental evolution with Caenorhabditis nematodes. Genetics 206, 691-716. ( 10.1534/genetics.115.186288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y, Kammenga JE. 2020. Genetic variation in Caenorhabditis elegans responses to pathogenic microbiota. Microorganisms 8, 618. ( 10.3390/microorganisms8040618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montarry J, Mimee B, Danchin EGJ, Koutsovoulos GD, Ste-Croix DT, Grenier E. 2021. Recent advances in population genomics of plant-parasitic nematodes. Phytopathology 111, 40-48. ( 10.1094/PHYTO-09-20-0418-RVW) [DOI] [PubMed] [Google Scholar]
- 58.Cole R, Viney M. 2018. The population genetics of parasitic nematodes of wild animals. Parasites Vectors 11, 590. ( 10.1186/s13071-018-3137-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Small ST, Tisch DJ, Zimmerman PA. 2014. Molecular epidemiology, phylogeny and evolution of the filarial nematode Wuchereria bancrofti. Infect. Genet. Evol. 28, 33-43. ( 10.1016/j.meegid.2014.08.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doyle SR, et al. 2019. Population genomic and evolutionary modelling analyses reveal a single major QTL for ivermectin drug resistance in the pathogenic nematode, Haemonchus contortus. BMC Genom. 20, 218. ( 10.1186/s12864-019-5592-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doyle SR, et al. 2022. Population genomics of ancient and modern Trichuris trichiura. Nat. Commun. 13, 3888. ( 10.1038/s41467-022-31487-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su XZ, Lane KD, Xia L, Sá JM, Wellems TE. 2019. Plasmodium genomics and genetics: new insights into malaria pathogenesis, drug resistance, epidemiology, and evolution. Clin. Microbiol. Rev. 32, 10-128. ( 10.1128/cmr.00019-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ntuli MM (ed.) 2021. Ending the neglect to attain the Sustainable Development Goals: A road map for neglected tropical diseases 2021–2030.
- 64.Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. 2017. Transcriptomics technologies. PLoS Comput. Biol. 13, e1005457. ( 10.1371/journal.pcbi.1005457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376-380. ( 10.1038/nature03959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagayasu E, Ogura Y, Itoh T, Yoshida A, Chakraborty G, Hayashi T, Maruyama H. 2013. Transcriptomic analysis of four developmental stages of Strongyloides venezuelensis. Parasitol. Int. 62, 57-65. ( 10.1016/j.parint.2012.09.006) [DOI] [PubMed] [Google Scholar]
- 67.Stoltzfus JD, Minot S, Berriman M, Nolan TJ, Lok JB. 2012. RNAseq analysis of the parasitic nematode Strongyloides stercoralis reveals divergent regulation of canonical dauer pathways. PLoS Negl. Trop. Dis. 6, e1854. ( 10.1371/journal.pntd.0001854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hunt VL, Hino A, Yoshida A, Kikuchi T. 2018. Comparative transcriptomics gives insights into the evolution of parasitism in Strongyloides nematodes at the genus, subclade and species level. Sci. Rep. 8, 5192. ( 10.1038/s41598-018-23514-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baskaran P, Jaleta TG, Streit A, Rödelsperger C. 2017. Duplications and positive selection drive the evolution of parasitism-associated gene families in the nematode Strongyloides papillosus. Genome Biol. Evol. 9, 790-801. ( 10.1093/gbe/evx040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benhur Junior A, Serufo JC, Lambertucci JR. 2004. Pulmonary strongyloidiasis. Rev. Soc. Bras. Med. Trop. 37, 359-360. ( 10.1590/S0037-86822004000400015) [DOI] [PubMed] [Google Scholar]
- 71.Lu MR, Lai CK, Liao BY, Tsai IJ. 2020. Comparative transcriptomics across nematode life cycles reveal gene expression conservation and correlated evolution in adjacent developmental stages. Genome Biol. Evol. 12, 1019-1030. ( 10.1093/gbe/evaa110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viney ME. 2009. How did parasitic worms evolve? Bioessays 31, 496-499. ( 10.1002/bies.200900010) [DOI] [PubMed] [Google Scholar]
- 73.O'Meara H, Barber R, Mello LV, Sangaralingam A, Viney ME, Paterson S. 2010. Response of the Strongyloides ratti transcriptome to host immunological environment. Int. J. Parasitol. 40, 1609-1617. ( 10.1016/j.ijpara.2010.06.005) [DOI] [PubMed] [Google Scholar]
- 74.Jaleta TG, Rödelsperger C, Streit A. 2017. Parasitological and transcriptomic comparison of Strongyloides ratti infections in natural and in suboptimal permissive hosts. Exp. Parasitol. 180, 112-118. ( 10.1016/j.exppara.2016.12.003) [DOI] [PubMed] [Google Scholar]
- 75.Rodpai R, et al. 2021. Exposure to dexamethasone modifies transcriptomic responses of free-living stages of Strongyloides stercoralis. PLoS ONE 16, e0253701. ( 10.1371/journal.pone.0253701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bryant AS, DeMarco SF, Hallem EA. 2021. Strongyloides RNA-seq Browser: a web-based software platform for on-demand bioinformatics analyses of Strongyloides species. G3 (Bethesda) 11, jkab104. ( 10.1093/g3journal/jkab104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roach NP, Sadowski N, Alessi AF, Timp W, Taylor J, Kim JK. 2020. The full-length transcriptome of C. elegans using direct RNA sequencing. Genome Res. 30, 299-312. ( 10.1101/gr.251314.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernard F, Dargère D, Rechavi O, Dupuy D. 2023. Quantitative analysis of C. elegans transcripts by nanopore direct-cDNA sequencing reveals terminal hairpins in non trans-spliced mRNAs. Nat. Commun. 14, 1229. ( 10.1038/s41467-023-36915-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jex AR, Gasser RB, Schwarz EM. 2019. Transcriptomic resources for parasitic nematodes of veterinary importance. Trends Parasitol. 35, 72-84. ( 10.1016/j.pt.2018.09.010) [DOI] [PubMed] [Google Scholar]
- 80.Choi YJ, Ghedin E, Berriman M, McQuillan J, Holroyd N, Mayhew GF, Christensen BM, Michalski ML. 2011. A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi. PLoS Negl. Trop. Dis. 5, e1409. ( 10.1371/journal.pntd.0001409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laing R, et al. 2013. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 14, R88. ( 10.1186/gb-2013-14-8-r88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanaka SE, et al. 2019. Stage-specific transcriptome of Bursaphelenchus xylophilus reveals temporal regulation of effector genes and roles of the dauer-like stages in the lifecycle. Sci. Rep. 9, 6080. ( 10.1038/s41598-019-42570-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J, et al. 2017. Comparative genome analysis of programmed DNA elimination in nematodes. Genome Res. 27, 2001-2014. ( 10.1101/gr.225730.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schurch NJ, et al. 2014. Improved annotation of 3' untranslated regions and complex loci by combination of strand-specific direct RNA sequencing, RNA-Seq and ESTs. PLoS ONE 9, e94270. ( 10.1371/journal.pone.0094270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corley SM, Troy NM, Bosco A, Wilkins MR. 2019. QuantSeq. 3' sequencing combined with Salmon provides a fast, reliable approach for high throughput RNA expression analysis. Sci. Rep. 9, 18895. ( 10.1038/s41598-019-55434-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bayega A, Oikonomopoulos S, Gregoriou ME, Tsoumani KT, Giakountis A, Wang YC, Mathiopoulos KD, Ragoussis J. 2021. Nanopore long-read RNA-seq and absolute quantification delineate transcription dynamics in early embryo development of an insect pest. Sci. Rep. 11, 7878. ( 10.1038/s41598-021-86753-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soblik H, Younis AE, Mitreva M, Renard BY, Kirchner M, Geisinger F, Steen H, Brattig NW. 2011. Life cycle stage-resolved proteomic analysis of the excretome/secretome from Strongyloides ratti--identification of stage-specific proteases. Mol. Cell. Proteom. 10, M111 010157. ( 10.1074/mcp.M111.010157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maeda Y, Palomares-Rius JE, Hino A, Afrin T, Mondal SI, Nakatake A, Maruyama H, Kikuchi T. 2019. Secretome analysis of Strongyloides venezuelensis parasitic stages reveals that soluble and insoluble proteins are involved in its parasitism. Parasites Vectors 12, 21. ( 10.1186/s13071-018-3266-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dishnica K, Piubelli C, Manfredi M, Kondaveeti RT, Longoni SS, Degani M, Buonfrate D, Giorgetti A, Tiberti N. 2023. Novel insights into the somatic proteome of Strongyloides stercoralis infective third-stage larvae. Parasites Vectors 16, 45. ( 10.1186/s13071-023-05675-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marcilla A, et al. 2010. Proteomic analysis of Strongyloides stercoralis L3 larvae. Parasitology 137, 1577-1583. ( 10.1017/S0031182010000314) [DOI] [PubMed] [Google Scholar]
- 91.Rodpai R, Intapan PM, Thanchomnang T, Sanpool O, Janwan P, Laummaunwai P, Wongkham C, Insawang T, Maleewong W. 2016. Strongyloides stercoralis diagnostic polypeptides for human strongyloidiasis and their proteomic analysis. Parasitol. Res. 115, 4007-4012. ( 10.1007/s00436-016-5170-7) [DOI] [PubMed] [Google Scholar]
- 92.Rodpai R, Intapan PM, Thanchomnang T, Sanpool O, Janwan P, Laummaunwai P, Wongkham C, Insawang T, Maleewong W. 2017. Identification of antigenic proteins in Strongyloides stercoralis by proteomic analysis. Parasitol. Res. 116, 1687-1693. ( 10.1007/s00436-017-5443-9) [DOI] [PubMed] [Google Scholar]
- 93.Younis AE, et al. 2011. Stage-specific excretory-secretory small heat shock proteins from the parasitic nematode Strongyloides ratti—putative links to host's intestinal mucosal defense system. FEBS J. 278, 3319-3336. ( 10.1111/j.1742-4658.2011.08248.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fonseca PDM, et al. 2020. Shotgun proteomics of Strongyloides venezuelensis infective third stage larvae: insights into host–parasite interaction and novel targets for diagnostics. Mol. Biochem. Parasitol. 235, 111249. ( 10.1016/j.molbiopara.2019.111249) [DOI] [PubMed] [Google Scholar]
- 95.Corral MA, et al. 2017. Potential immunological markers for diagnosis of human strongyloidiasis using heterologous antigens. Parasitology 144, 124-130. ( 10.1017/S0031182016001645) [DOI] [PubMed] [Google Scholar]
- 96.Varatharajalu R, Parandaman V, Ndao M, Andersen JF, Neva FA. 2011. Strongyloides stercoralis excretory/secretory protein strongylastacin specifically recognized by IgE antibodies in infected human sera. Microbiol. Immunol. 55, 115-122. ( 10.1111/j.1348-0421.2010.00289.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tiberti N, Manfredi M, Piubelli C, Buonfrate D. 2023. Progresses and challenges in Strongyloides spp. proteomics. Phil. Trans. R. Soc. B 379, 20220447. ( 10.1098/rstb.2022.0447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Levenhagen MA, Costa-Cruz JM. 2014. Update on immunologic and molecular diagnosis of human strongyloidiasis. Acta Trop. 135, 33-43. ( 10.1016/j.actatropica.2014.03.015) [DOI] [PubMed] [Google Scholar]
- 99.Cantacessi C, et al. 2009. A portrait of the "SCP/TAPS" proteins of eukaryotes — developing a framework for fundamental research and biotechnological outcomes. Biotechnol. Adv. 27, 376-388. ( 10.1016/j.biotechadv.2009.02.005) [DOI] [PubMed] [Google Scholar]
- 100.Evangelista FMD, van Vliet AHM, Lawton SP, Betson M. 2022. A reverse vaccinology approach identifies putative vaccination targets in the zoonotic nematode Ascaris. Front. Vet. Sci. 9, 1014198. ( 10.3389/fvets.2022.1014198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Culma MF. 2021. Strongyloides stercoralis proteome: a reverse approach to the identification of potential immunogenic candidates. Microb. Pathog. 152, 104545. ( 10.1016/j.micpath.2020.104545) [DOI] [PubMed] [Google Scholar]
- 102.Ranganathan S, Garg G. 2009. Secretome: clues into pathogen infection and clinical applications. Genome Med. 1, 113. ( 10.1186/gm113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rooney J, Northcote HM, Williams TL, Cortes A, Cantacessi C, Morphew RM. 2022. Parasitic helminths and the host microbiome – a missing 'extracellular vesicle-sized' link? Trends Parasitol. 38, 737-747. ( 10.1016/j.pt.2022.06.003) [DOI] [PubMed] [Google Scholar]
- 104.Hunt VL, Tsai IJ, Selkirk ME, Viney M. 2017. The genome of Strongyloides spp. gives insights into protein families with a putative role in nematode parasitism. Parasitology 144, 343-358. ( 10.1017/S0031182016001554) [DOI] [PubMed] [Google Scholar]
- 105.Hawdon JM, Narasimhan S, Hotez PJ. 1999. Ancylostoma secreted protein 2: cloning and characterisation of a second member of nematode secreted proteins from Ancylostoma caninum. Mol. Biochem. Parasitol. 99, 149-165. ( 10.1016/S0166-6851(99)00011-0) [DOI] [PubMed] [Google Scholar]
- 106.Drurey C, Maizels RM. 2021. Helminth extracellular vesicles: interactions with the host immune system. Mol. Immunol. 137, 124-133. ( 10.1016/j.molimm.2021.06.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Doyle LM, Wang MZ. 2019. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8, 727. ( 10.3390/cells8070727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maas SLN, Breakefield XO, Weaver AM. 2017. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 27, 172-188. ( 10.1016/j.tcb.2016.11.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hansen EP, et al. 2019. Exploration of extracellular vesicles from Ascaris suum provides evidence of parasite–host cross talk. J. Extracell. Vesicles 8, 1578116. ( 10.1080/20013078.2019.1578116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tzelos T, et al. 2016. A preliminary proteomic characterisation of extracellular vesicles released by the ovine parasitic nematode, Teladorsagia circumcincta. Vet. Parasitol. 221, 84-92. ( 10.1016/j.vetpar.2016.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gao X, Yang Y, Liu X, Xu F, Wang Y, Liu L, Yang Y, Liu M, Bai X. 2022. Extracellular vesicles from Trichinella spiralis: proteomic analysis and protective immunity. PLoS Negl. Trop. Dis. 16, e0010528. ( 10.1371/journal.pntd.0010528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Merrihew GE, et al. 2008. Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations. Genome Res. 18, 1660-1669. ( 10.1101/gr.077644.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schrimpf SP, et al. 2009. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7, e48. ( 10.1371/journal.pbio.1000048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zimmerman SM, Hinkson IV, Elias JE, Kim SK. 2015. Reproductive aging drives protein accumulation in the uterus and limits lifespan in C. elegans. PLoS Genet. 11, e1005725. ( 10.1371/journal.pgen.1005725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reis-Rodrigues P, et al. 2012. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell 11, 120-127. ( 10.1111/j.1474-9726.2011.00765.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liang Z, Schmerberg CM, Li L. 2015. Mass spectrometric measurement of neuropeptide secretion in the crab, Cancer borealis, by in vivo microdialysis. Analyst 140, 3803-3813. ( 10.1039/C4AN02016B) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mir DA, Balamurugan K. 2019. Global proteomic response of Caenorhabditis elegans against PemK(Sa) toxin. Front. Cell Infect. Microbiol. 9, 172. ( 10.3389/fcimb.2019.00172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu Y, Sellegounder D, Sun J. 2016. Neuronal GPCR OCTR-1 regulates innate immunity by controlling protein synthesis in Caenorhabditis elegans. Sci. Rep. 6, 36832. ( 10.1038/srep36832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang W, Dierking K, Esser D, Tholey A, Leippe M, Rosenstiel P, Schulenburg H. 2015. Overlapping and unique signatures in the proteomic and transcriptomic responses of the nematode Caenorhabditis elegans toward pathogenic Bacillus thuringiensis. Dev. Comp. Immunol. 51, 1-9. ( 10.1016/j.dci.2015.02.010) [DOI] [PubMed] [Google Scholar]
- 120.Tritten L, Ballesteros C, Beech R, Geary TG, Moreno Y. 2021. Mining nematode protein secretomes to explain lifestyle and host specificity. PLoS Negl. Trop. Dis. 15, e0009828. ( 10.1371/journal.pntd.0009828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Duguet TB, Soichot J, Kuzyakiv R, Malmstrom L, Tritten L. 2020. Extracellular vesicle-contained microRNA of C. elegans as a tool to decipher the molecular basis of nematode parasitism. Front. Cell. Infect. Microbiol. 10, 217. ( 10.3389/fcimb.2020.00217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Younis AE, Brattig NW. 2018. Identification, isolation, and cloning of the full-length sequence of a gene encoding trypsin inhibitor-like protein (TIL) secreted by the intestinal parasitic nematode Strongyloides ratti. J. Basic Appl. Zool. 79, 1-9. ( 10.1186/s41936-018-0023-9) [DOI] [Google Scholar]
- 123.Russell JC, Kim TK, Noori A, Merrihew GE, Robbins JE, Golubeva A, Wang K, Maccoss MJ, Kaeberlein M. 2020. Composition of Caenorhabditis elegans extracellular vesicles suggests roles in metabolism, immunity, and aging. Geroscience 42, 1133-1145. ( 10.1007/s11357-020-00204-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang T, Gasser RB. 2021. Prospects of using high-throughput proteomics to underpin the discovery of animal host–nematode interactions. Pathogens 10, 825. ( 10.3390/pathogens10070825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang T, et al. 2019. Somatic proteome of Haemonchus contortus. Int. J. Parasitol. 49, 311-320. ( 10.1016/j.ijpara.2018.12.003) [DOI] [PubMed] [Google Scholar]
- 126.Zaragoza-Vera M, et al. 2022. Identification of somatic proteins in Haemonchus contortus infective larvae (L3) and adults. Helminthologia 59, 143-151. ( 10.2478/helm-2022-0017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Maizels RM, et al. 2012. Immune modulation and modulators in Heligmosomoides polygyrus infection. Exp. Parasitol. 132, 76-89. ( 10.1016/j.exppara.2011.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang J, Pan W, Sun X, Zhao X, Yuan G, Sun Q, Huang J, Zhu X. 2015. Immunoproteomic profile of Trichinella spiralis adult worm proteins recognized by early infection sera. Parasites Vectors 8, 20. ( 10.1186/s13071-015-0641-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ren HN, Liu RD, Song YY, Zhuo TX, Guo KX, Zhang Y, Jiang P, Wang ZQ, Cui J. 2019. Label-free quantitative proteomic analysis of molting-related proteins of Trichinella spiralis intestinal infective larvae. Vet. Res. 50, 70. ( 10.1186/s13567-019-0689-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sotillo J, et al. 2014. Secreted proteomes of different developmental stages of the gastrointestinal nematode Nippostrongylus brasiliensis. Mol. Cell. Proteom. 13, 2736-2751. ( 10.1074/mcp.M114.038950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu JY, et al. 2016. Proteomic analysis of differentially expressed proteins in the three developmental stages of Trichinella spiralis. Vet. Parasitol. 231, 32-38. ( 10.1016/j.vetpar.2016.06.021) [DOI] [PubMed] [Google Scholar]
- 132.Mulvenna J, Hamilton B, Nagaraj SH, Smyth D, Loukas A, Gorman JJ. 2009. Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol. Cell. Proteom. 8, 109-121. ( 10.1074/mcp.M800206-MCP200) [DOI] [PubMed] [Google Scholar]
- 133.Morante T, Shepherd C, Constantinoiu C, Loukas A, Sotillo J. 2017. Revisiting the Ancylostoma caninum secretome provides new information on hookworm–host interactions. Proteomics 17, 1700186. ( 10.1002/pmic.201700186) [DOI] [PubMed] [Google Scholar]
- 134.Logan J, et al. 2020. Comprehensive analysis of the secreted proteome of adult Necator americanus hookworms. PLoS Negl. Trop. Dis. 14, e0008237. ( 10.1371/journal.pntd.0008237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang T, Ma G, Ang CS, Korhonen PK, Koehler AV, Young ND, Nie S, Williamson NA, Gasser RB. 2019. High throughput LC-MS/MS-based proteomic analysis of excretory-secretory products from short-term in vitro culture of Haemonchus contortus. J. Proteom. 204, 103375. ( 10.1016/j.jprot.2019.05.003) [DOI] [PubMed] [Google Scholar]
- 136.Moreno Y, Gros PP, Tam M, Segura M, Valanparambil R, Geary TG, Stevenson MM. 2011. Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with next-generation sequencing transcriptomic information. PLoS Negl. Trop. Dis. 5, e1370. ( 10.1371/journal.pntd.0001370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wilbers RHP, Schneiter R, Holterman MHM, Drurey C, Smant G, Asojo OA, Maizels RM, Lozano-Torres JL. 2018. Secreted venom allergen-like proteins of helminths: conserved modulators of host responses in animals and plants. PLoS Pathog. 14, e1007300. ( 10.1371/journal.ppat.1007300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang T, Van Steendam K, Dhaenens M, Vlaminck J, Deforce D, Jex AR, Gasser RB, Geldhof P. 2013. Proteomic analysis of the excretory-secretory products from larval stages of Ascaris suum reveals high abundance of glycosyl hydrolases. PLoS Negl. Trop. Dis. 7, e2467. ( 10.1371/journal.pntd.0002467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maruszewska-Cheruiyot M, Szewczak L, Krawczak-Wojcik K, Glaczynska M, Donskow-Lysoniewska K. 2021. The production of excretory-secretory molecules from Heligmosomoides polygyrus bakeri fourth stage larvae varies between mixed and single sex cultures. Parasites Vectors 14, 106. ( 10.1186/s13071-021-04613-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harischandra H, Yuan W, Loghry HJ, Zamanian M, Kimber MJ. 2018. Profiling extracellular vesicle release by the filarial nematode Brugia malayi reveals sex-specific differences in cargo and a sensitivity to ivermectin. PLoS Negl. Trop. Dis. 12, e0006438. ( 10.1371/journal.pntd.0006438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sheng ZA, et al. 2023. Proteomic analysis of exosome-like vesicles from Fasciola gigantica adult worm provides support for new vaccine targets against fascioliasis. Parasites Vectors 16, 62. ( 10.1186/s13071-023-05659-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eichenberger RM, Talukder MH, Field MA, Wangchuk P, Giacomin P, Loukas A, Sotillo J. 2018. Characterization of Trichuris muris secreted proteins and extracellular vesicles provides new insights into host–parasite communication. J. Extracell. Vesicles 7, 1428004. ( 10.1080/20013078.2018.1428004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tritten L, Tam M, Vargas M, Jardim A, Stevenson MM, Keiser J, Geary TG. 2017. Excretory/secretory products from the gastrointestinal nematode Trichuris muris. Exp. Parasitol. 178, 30-36. ( 10.1016/j.exppara.2017.05.003) [DOI] [PubMed] [Google Scholar]
- 144.Baggerman G, Cerstiaens A, De Loof A, Schoofs L. 2002. Peptidomics of the larval Drosophila melanogaster central nervous system. J. Biol. Chem. 277, 40 368-40 374. ( 10.1074/jbc.M206257200) [DOI] [PubMed] [Google Scholar]
- 145.Clynen E, et al. 2001. Peptidomics of the pars intercerebralis-corpus.pdf. Eur. J. Biochem. 268, 1929-1939. ( 10.1046/j.1432-1327.2001.02067.x) [DOI] [PubMed] [Google Scholar]
- 146.Schrader M, Schulz-Knappe P. 2001. Peptidomics technologies for human body fluids. Trends Biotechnol. 19, 55-60. ( 10.1016/S0167-7799(01)00010-5) [DOI] [PubMed] [Google Scholar]
- 147.Schoofs L, Baggerman G. 2003. Peptidomics in Drosphilia melanogaster. Brief. Funct. Genom. 2, 114-120. ( 10.1093/bfgp/2.2.114) [DOI] [PubMed] [Google Scholar]
- 148.Van Bael S, Zels S, Boonen K, Beets I, Schoofs L, Temmerman L. 2018. A Caenorhabditis elegans mass spectrometric resource for neuropeptidomics. J. Am. Soc. Mass Spectrom. 29, 879-889. ( 10.1007/s13361-017-1856-z) [DOI] [PubMed] [Google Scholar]
- 149.Husson SJ, Landuyt B, Nys T, Baggerman G, Boonen K, Clynen E, Lindemans M, Janssen T, Schoofs L. 2009. Comparative peptidomics of Caenorhabditis elegans versus C. briggsae by LC-MALDI-TOF MS. Peptides 30, 449-457. ( 10.1016/j.peptides.2008.07.021) [DOI] [PubMed] [Google Scholar]
- 150.Atkinson LE, et al. 2021. Ascaris suum informs extrasynaptic volume transmission in nematodes. ACS Chem. Neurosci. 12, 3176-3188. ( 10.1021/acschemneuro.1c00281) [DOI] [PubMed] [Google Scholar]
- 151.Buzy A, Allain C, Harrington J, Lesuisse D, Mikol V, Bruhn DF, Maule AG, Guillemot J-C. 2021. Peptidomics of Haemonchus contortus. ACS Omega 6, 10 288-10 305. ( 10.1021/acsomega.1c00650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McCoy CJ, Atkinson LE, Zamanian M, McVeigh P, Day TA, Kimber MJ, Marks NJ, Maule AG, Mousley A. 2014. New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteom. 3, 262-272. ( 10.1016/j.euprot.2014.04.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McKay FM, McCoy CJ, Crooks B, Marks NJ, Maule AG, Atkinson LE, Mousley A. 2022. In silico analyses of neuropeptide-like protein (NLP) profiles in parasitic nematodes. Int. J. Parasitol. 52, 77-85. ( 10.1016/j.ijpara.2021.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cowden C, Stretton AO, Davies RE. 1989. AF1, a sequenced bioactive neuropeptide isolated from the nematode Ascaris suum. Neuron 2, 1465-1473. ( 10.1016/0896-6273(89)90192-X) [DOI] [PubMed] [Google Scholar]
- 155.Cowden C, Stretton AO. 1993. AF2, an Ascaris neuropeptide: isolation, sequence, and bioactivity. Peptides 14, 423-430. ( 10.1016/0196-9781(93)90127-3) [DOI] [PubMed] [Google Scholar]
- 156.Cowden C, Stretton AO. 1995. Eight novel FMRFamide-like neuropeptides isolated from the nematode Ascaris suum. Peptides 16, 491-500. ( 10.1016/0196-9781(94)00211-N) [DOI] [PubMed] [Google Scholar]
- 157.Yew JY, Kutz KK, Dikler S, Messinger L, Li L, Stretton AO. 2005. Mass spectrometric map of neuropeptide expression in Ascaris suum. J. Comp. Neurol. 488, 396-413. ( 10.1002/cne.20587) [DOI] [PubMed] [Google Scholar]
- 158.Midha A, Janek K, Niewienda A, Henklein P, Guenther S, Serra DO, Schlosser J, Hengge R, Hartmann S. 2018. The intestinal roundworm Ascaris suum releases antimicrobial factors which interfere with bacterial growth and biofilm formation. Front. Cell. Infect. Microbiol. 8, 271. ( 10.3389/fcimb.2018.00271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Romanova EV, Sweedler JV. 2015. Peptidomics for the discovery and characterization of neuropeptides and hormones. Trends Pharmacol. Sci. 36, 579-586. ( 10.1016/j.tips.2015.05.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Irvine A, McKenzie D, McCoy CJ, Graham RLJ, Graham C, Huws SA, Atkinson LE, Mousley A. 2023. Novel integrated computational AMP discovery approaches highlight diversity in the helminth AMP repertoire. PLoS Pathog. 19, e1011508. ( 10.1371/journal.ppat.1011508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sarkies P, et al. 2015. Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS Biol. 13, e1002061. ( 10.1371/journal.pbio.1002061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Britton C, Laing R, Devaney E. 2020. Small RNAs in parasitic nematodes – forms and functions. Parasitology 147, 855-864. ( 10.1017/S0031182019001689) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ahmed R, Chang Z, Younis AE, Langnick C, Li N, Chen W, Brattig N, Dieterich C. et al. 2013. Conserved miRNAs are candidate post-transcriptional regulators of developmental arrest in free-living and parasitic nematodes. Genome Biol. Evol. 5, 1246-1260. ( 10.1093/gbe/evt086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Holz A, Streit A. 2017. Gain and loss of small RNA classes—characterization of small RNAs in the parasitic nematode family Strongyloididae. Genome Biol. Evol. 9, 2826-2843. ( 10.1093/gbe/evx197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pomari E, et al. 2022. Identification of miRNAs of Strongyloides stercoralis L1 and iL3 larvae isolated from human stool. Sci. Rep. 12, 9957. ( 10.1038/s41598-022-14185-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Suleiman M, et al. 2022. piRNA-like small RNAs target transposable elements in a Clade IV parasitic nematode. Sci. Rep. 12, 10156. ( 10.1038/s41598-022-14247-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37-52. ( 10.1093/nar/gkr688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854. ( 10.1016/0092-8674(93)90529-Y) [DOI] [PubMed] [Google Scholar]
- 169.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901-906. ( 10.1038/35002607) [DOI] [PubMed] [Google Scholar]
- 170.Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47(D1), D155-D162. ( 10.1093/nar/gky1141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 2009. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 106, 18 674-18 679. ( 10.1073/pnas.0906378106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ketting RF, Cochella L. 2021. Concepts and functions of small RNA pathways in C. elegans. Curr. Top. Dev. Biol. 144, 45-89. ( 10.1016/bs.ctdb.2020.08.002) [DOI] [PubMed] [Google Scholar]
- 173.Batista PJ, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 31, 67-78. ( 10.1016/j.molcel.2008.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, Davis RE. 2011. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 21, 1462-1477. ( 10.1101/gr.121426.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Winter AD, Weir W, Hunt M, Berriman M, Gilleard JS, Devaney E, Britton C. 2012. Diversity in parasitic nematode genomes: the microRNAs of Brugia pahangi and Haemonchus contortus are largely novel. BMC Genom. 13, 1-9. ( 10.1186/1471-2164-13-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zagoskin MV, Wang J, Neff AT, Veronezi GMB, Davis RE. 2022. Small RNA pathways in the nematode Ascaris in the absence of piRNAs. Nat. Commun. 13, 837. ( 10.1038/s41467-022-28482-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Buck AH, et al. 2014. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 5, 5488. ( 10.1038/ncomms6488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Coakley G, et al. 2017. Extracellular vesicles from a helminth parasite suppress macrophage activation and constitute an effective vaccine for protective immunity. Cell Rep. 19, 1545-1557. ( 10.1016/j.celrep.2017.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.White R, Kumar S, Chow FW, Robertson E, Hayes KS, Grencis RK, Duque-Correa MA, Buck AH. 2020. Extracellular vesicles from Heligmosomoides bakeri and Trichuris muris contain distinct microRNA families and small RNAs that could underpin different functions in the host. Int. J. Parasitol. 50, 719-729. ( 10.1016/j.ijpara.2020.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.König J, Zarnack K, Luscombe NM, Ule J. 2012. Protein–RNA interactions: new genomic technologies and perspectives. Nat. Rev. Genet. 13, 77-83. ( 10.1038/nrg3141) [DOI] [PubMed] [Google Scholar]
- 181.Bourque G, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19, 199. ( 10.1186/s13059-018-1577-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dechaud C, Volff JN, Schartl M, Naville M. 2019. Sex and the TEs: transposable elements in sexual development and function in animals. Mob. DNA 10, 42. ( 10.1186/s13100-019-0185-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Goubert C, Craig RJ, Bilat AF, Peona V, Vogan AA, Protasio AV. 2022. A beginner's guide to manual curation of transposable elements. Mob. DNA 13, 7. ( 10.1186/s13100-021-00259-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl Acad. Sci. USA 117, 9451-9457. ( 10.1073/pnas.1921046117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Smit AF, Hubley R, Green P. 2015. Repeatmasker Open 4.0. See www.repeatmasker.org.
- 186.Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF. 2021. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob. DNA 12, 2. ( 10.1186/s13100-020-00230-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinf. 9, 18. ( 10.1186/1471-2105-9-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Steinbiss S, Willhoeft U, Gremme G, Kurtz S. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37, 7002-7013. ( 10.1093/nar/gkp759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. 2006. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J. Comput. Biol. 13, 1028-1040. ( 10.1089/cmb.2006.13.1028) [DOI] [PubMed] [Google Scholar]
- 190.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573-580. ( 10.1093/nar/27.2.573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Szitenberg A, Cha S, Opperman CH, Bird DM, Blaxter ML, Lunt DH. 2016. Genetic drift, not life history or RNAi, determine long-term evolution of transposable elements. Genome Biol. Evol. 8, 2964-2978. ( 10.1093/gbe/evw208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11. ( 10.1186/s13100-015-0041-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460-2461. ( 10.1093/bioinformatics/btq461) [DOI] [PubMed] [Google Scholar]
- 194.Bailly-Bechet M, Haudry A, Lerat E. 2014. ‘One code to find them all’: a perl tool to conveniently parse RepeatMasker output files. Mob. DNA 5, 13. ( 10.1186/1759-8753-5-13) [DOI] [Google Scholar]
- 195.Jurka J, Klonowski P, Dagman V, Pelton P. 1996. CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem. 20, 119-121. ( 10.1016/S0097-8485(96)80013-1) [DOI] [PubMed] [Google Scholar]
- 196.International Helminth Genomes Consortium. 2019. Comparative genomics of the major parasitic worms. Nat. Genet. 51, 163-174. ( 10.1038/s41588-018-0262-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Laricchia KM, Zdraljevic S, Cook DE, Andersen EC. 2017. Natural variation in the distribution and abundance of transposable elements across the Caenorhabditis elegans species. Mol. Biol. Evol. 34, 2187-2202. ( 10.1093/molbev/msx155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kozlowski DKL, Hassanaly-Goulamhoussen R, Da Rocha M, Koutsovoulos GD, Bailly-Bechet M, Danchin EGJ. 2021. Movements of transposable elements contribute to the genomic plasticity and species diversification in an asexually reproducing nematode pest. Evol. Appl. 14, 1844-1866. ( 10.1111/eva.13246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ansaloni F, Scarpato M, Di Schiavi E, Gustincich S, Sanges R. 2019. Exploratory analysis of transposable elements expression in the C. elegans early embryo. BMC Bioinf. 20(Suppl 9), 484. ( 10.1186/s12859-019-3088-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bessereau J-L. 2006. Transposons in C. elegans. In WormBook: The Online Review of C. elegans Biology (ed. C. elegans research community). ( 10.1895/wormbook.1.70.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Athanasouli M, Rödelsperger C. 2022. Analysis of repeat elements in the Pristionchus pacificus genome reveals an ancient invasion by horizontally transferred transposons. BMC Genom. 23, 523. ( 10.1186/s12864-022-08731-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193-1207. ( 10.1016/j.cell.2006.10.040) [DOI] [PubMed] [Google Scholar]
- 203.Shahid S, Slotkin RK. 2020. The current revolution in transposable element biology enabled by long reads. Curr. Opin. Plant Biol. 54, 49-56. ( 10.1016/j.pbi.2019.12.012) [DOI] [PubMed] [Google Scholar]
- 204.Koutsovoulos GD, et al. 2020. Genome assembly and annotation of Meloidogyne enterolobii, an emerging parthenogenetic root-knot nematode. Sci. Data 7, 324. ( 10.1038/s41597-020-00666-0) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 205.Rodriguez M, Makałowski W. 2022. Software evaluation for de novo detection of transposons. Mob. DNA 13, 14. ( 10.1186/s13100-022-00266-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bao Z, Eddy SR. 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12, 1269-1276. ( 10.1101/gr.88502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hoede C, Arnoux S, Moisset M, Chaumier T, Inizan O, Jamilloux V, Quesneville H. 2014. PASTEC: an automatic transposable element classification tool. PLoS ONE 9, e91929. ( 10.1371/journal.pone.0091929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Koutsovoulos GD, et al. 2020. Population genomics supports clonal reproduction and multiple independent gains and losses of parasitic abilities in the most devastating nematode pest. Evol. Appl. 13, 442-457. ( 10.1111/eva.12881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272-285. ( 10.1038/nrg2072) [DOI] [PubMed] [Google Scholar]
- 210.Senft AD, Macfarlan TS. 2021. Transposable elements shape the evolution of mammalian development. Nat. Rev. Genet. 22, 691-711. ( 10.1038/s41576-021-00385-1) [DOI] [PubMed] [Google Scholar]
- 211.Venancio TM, Wilson RA, Verjovski-Almeida S, DeMarco R. 2010. Bursts of transposition from non-long terminal repeat retrotransposon families of the RTE clade in Schistosoma mansoni. Int. J. Parasitol. 40, 743-749. ( 10.1016/j.ijpara.2009.11.013) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.