Abstract
The diversity of dinitrogenase reductase gene (nifH) fragments in Paenibacillus azotofixans strains was investigated by using molecular methods. The partial nifH gene sequences of eight P. azotofixans strains, as well as one strain each of the close relatives Paenibacillus durum, Paenibacillus polymyxa, and Paenibacillus macerans, were amplified by PCR by using degenerate primers and were characterized by DNA sequencing. We found that there are two nifH sequence clusters, designated clusters I and II, in P. azotofixans. The data further indicated that there was sequence divergence among the nifH genes of P. azotofixans strains at the DNA level. However, the gene products were more conserved at the protein level. Phylogenetic analysis showed that all nifH cluster II sequences were similar to the alternative (anf) nitrogenase sequence. A nested PCR assay for the detection of nifH (cluster I) of P. azotofixans was developed by using the degenerate primers as outer primers and two specific primers, designed on the basis of the sequence information obtained, as inner primers. The specificity of the inner primers was tested with several diazotrophic bacteria, and PCR revealed that these primers are specific for the P. azotofixans nifH gene. A GC clamp was attached to one inner primer, and a denaturing gradient gel electrophoresis (DGGE) protocol was developed to study the genetic diversity of this region of nifH in P. azotofixans strains, as well as in soil and rhizosphere samples. The results revealed sequence heterogeneity among different nifH genes. Moreover, nifH is probably a multicopy gene in P. azotofixans. Both similarities and differences were detected in the P. azotofixans nifH DGGE profiles generated with soil and rhizosphere DNAs. The DGGE assay developed here is reproducible and provides a rapid way to assess the intraspecific genetic diversity of an important functional gene in pure cultures, as well as in environmental samples.
The genus Paenibacillus (1) represents a new phylum that encompasses several species described as nitrogen-fixing bacilli, including Paenibacillus polymyxa, Paenibacillus macerans, and Paenibacillus azotofixans (1). P. azotofixans (formerly Bacillus azotofixans) is a nitrogen-fixing bacterium that is often found in the rhizospheres of important crop plants in tropical regions (32). This species is a potential plant growth-promoting rhizobacterium since it produces antimicrobial substances, solubilizes organic phosphates (unpublished data), and is stimulated in plant rhizospheres (28). Moreover, P. azotofixans has a capacity to fix atmospheric nitrogen that is greater than the capacities of all other Paenibacillus species. In contrast to other nitrogen-fixing microorganisms, expression of the nitrogenase genes in P. azotofixans is not affected by the presence of nitrate (32). Hence, fixation of atmospheric nitrogen is feasible in soils with input nitrate.
The nitrogenase enzyme complex consists of the following two conserved proteins: the MoFe protein, composed of subunits encoded by the nifD and nifK genes; and the Fe protein, encoded by the nifH gene (5, 43). The nitrogenase iron protein gene, nifH, is one of the oldest existing functional genes in the history of gene evolution, and the relationships among bacteria based on sequence divergences of this gene have been reported to be in agreement with the phylogeny inferred from 16S rRNA gene sequences (3, 13, 21, 23, 38, 43, 45). This feature of the nifH gene has provided a strategy for a molecular approach to address the diversity of nitrogen fixation genes in bacteria of interest, as well as for characterizing such genes in natural soil microbial communities (2, 23, 32, 45).
Although such studies provided new insights into the genetic diversity of nitrogen-fixing bacteria, the cloning and sequencing strategies used were rather cumbersome and time-consuming. Hence, they are not suitable for monitoring large numbers of samples (e.g., for monitoring shifts in microbial communities during different stages of plant growth).
A new approach to the study of the diversity of natural microbial communities is analysis of PCR products generated with primers homologous to relatively conserved regions in the genome (11) via denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel electrophoresis (14, 18, 19). These approaches allow separation of DNA molecules that differ by single bases (20) and hence have the potential to provide information about variations in target genes in bacterial populations in natural systems. Moreover, by adjusting the primers used for amplification, both major and minor constituents of microbial communities can be characterized (14). Temperature gradient gel electrophoresis performed with 16S ribosomal DNA (rDNA) amplicons has already been used to study the genetic diversity of microbial communities in environments such as the potato rhizosphere (14) and also to detect sequence heterogeneities in single genomes (22). PCR-DGGE has also been used similarly in a range of different environments (7, 8, 17–19, 26). Despite the great potential for detecting sequence variations, the use of such techniques for studying functional genes has been limited to the [NiFe] hydrogenase gene of Desulfovibrio spp. in experimental bioreactors (42). In studies of functional gene diversity, the major obstacle has often been the lack of sufficient sequence information. Ideally, several genes should be sequenced and aligned to obtain consensus sequences in order to design oligonucleotide primers which anneal to regions conserved in a family of similar but not identical genes (42).
The objective of this study was to obtain information about the diversity of Paenibacillus sp. nifH gene sequences in pure culture and in soil. When this study was started, only one partial Paenibacillus nifH sequence had been deposited in a database (45). To provide material for diversity assessments, degenerate primers (44) were used to obtain nifH amplicons of diverse strains of P. azotofixans and other Paenibacillus spp., as well as to generate sequence information in order to design specific primers for P. azotofixans nifH genes. The usefulness of DGGE separation for analyzing the diversity of nifH gene fragments of different P. azotofixans strains, as well as populations from bulk and rhizosphere soils, was then assessed.
MATERIALS AND METHODS
Bacterial strains.
The strains used and their sources are listed in Table 1. Strains were kept (for short periods of time) at room temperature on GB agar slants (31) supplemented with 1% (wt/vol) CaCO3 to maintain the pH at 7.4. Long-term storage was at −80°C in 20% (vol/vol) glycerol. To propagate cultures, the TBN medium described by Seldin and Penido (33) was employed. Several nitrogen-fixing bacterial strains (Table 1) or their genomic DNAs were kindly provided by A. D. L. Akkermans and H. Ramirez (Wageningen Agricultural University, Wageningen, The Netherlands).
TABLE 1.
Bacterial strains and specificity of the PCR based on the nifH gene of P. azotofixans
| Species | Strain | PCR producta | Origin or referenceb |
|---|---|---|---|
| P. azotofixans | ATCC 35681 | + | 32 |
| F102 | + | 32 | |
| 2RC1 | + | 32 | |
| SD20 | + | 32 | |
| C3L4 | + | 32 | |
| RCPG7 | + | 32 | |
| P3E20 | + | 32 | |
| RBN4 | + | 32 | |
| BE1 | + | 32 | |
| SD17 | + | ||
| P. durum | DSMZ 1735 | + | DSMZ |
| P. polymyxa | DSMZ 356 | − | DSMZ |
| LMAU B58 | − | Akkermans | |
| Loutit | − | CIM | |
| P. macerans | LMD 24.3 | − | CIM |
| LMAU B57 | − | DSMZ | |
| Frankia sp. | Br | − | Ramirez |
| AgKg4 | − | Ramirez | |
| Hrl1 | − | Ramirez | |
| Arl3 | − | Ramirez | |
| Azospirillum brasilense | LMAU A150 | − | Akkermans |
| Azospirillum lipoferum | LMAU A146 | − | Akkermans |
| Azotobacter beijerinckii | LMAU A34 | − | Akkermans |
| Azotobacter chroococcum | LMAU A95 | − | Akkermans |
| Azotobacter vinelandii | LMAU A66 | − | Akkermans |
| Beijerinckia sp. | LMAU B50 | − | Akkermans |
| Bradyrhizobium japonicum | LMAU R87 | − | Akkermans |
| Rhizobium meliloti | LMAU R13 | − | Akkermans |
| Rhizobium phaseoli | LMAU R19 | − | Akkermans |
The PCR was performed with primers NHA1 and NHA2 specific to the nifH gene of P. azotofixans. +, present; −, absent.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; Akkermans, A. D. L. Akkermans, Microbiology Department, Wageningen Agricultural University, Wageningen, The Netherlands; CIM, Collection of Instituto de Microbiologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil; Ramirez, H. Ramirez, Microbiology Department, Wageningen Agricultural University, Wageningen, The Netherlands.
Rhizosphere and soil samples.
Maize rhizosphere samples were obtained from plants growing in two different Brazilian soils, Cerrado and Várzea, in the state of Minas Gerais, Brazil. These rhizosphere soil samples were taken after 10, 30, and 60 days of plant growth. They were kindly provided by Edilson Paiva (Empresa Brasileira de Pesquisa Agropecuária, Sete Lagoas, Brazil). Samples of soils from a Brazilian grassland region with sugarcane plantations (Guaíra soils P04 and P33) were provided by Heitor Coutinho (Centro Nacional de Pesquisa do Meio Ambiente [CNPMA], Jaguaruna, SP, Brazil). Two Dutch soils, Flevo silt loam and Ede loamy sand (39), were also used in the nifH diversity assessments.
Extraction of DNA from pure cultures and from bulk and rhizosphere soil samples.
Total genomic DNA was isolated from pure cultures of bacterial strains as previously described (27). For extraction of DNA from soil, 2-g samples of bulk or rhizosphere soil were subjected to direct extraction by using a modification of the protocol of Smalla et al. (36). The modification consisted of a final purification step based on Wizard resin spin columns (Promega, Madison, Wis.) instead of the standard glassmilk purification (40). Following purification, the soil DNA was visualized on 0.8% (wt/vol) agarose gels (30) to assess its purity and molecular size. DNA concentrations were determined with a GeneQuant apparatus (Pharmacia, Uppsala, Sweden). The final DNA extracts obtained from the soils were not colored, indicating that they did not contain substantial amounts of humic compounds, had large molecular sizes (>10 kb), and could be amplified by PCR.
PCR amplification of nifH gene fragments.
Two sets of oligonucleotide primers (Table 2) were used for PCR amplification of nifH gene fragments. First, a set of degenerate primers described by Zehr and McReynolds (44) was used to amplify the nifH gene of Paenibacillus species for the cloning-sequencing experiments. The second primer set (primers NHA1 and NHA2) was based on specific regions of the P. azotofixans nifH gene identified in this study.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Reference(s) |
|---|---|---|
| Forward | 5′-TG YGA YCC NAA RGC NGA-3′ | 44, 45 |
| Reverse | 5′-AD NGC CAT CAT YTC NCC-3′ | 44, 45 |
| NHA1 | 5′-TCC ACT CGT CTG ATC CTG-3′ | This study |
| NHA2 | 5′-CTC GCG GAT TGG CAT TGCG-3′ | This study |
| GC clampb | 5′-CGCCCGCCGCGCCCCGCGCCCGT CCCGCCGCCCCCGCCCG-3′ | 19 |
Y = T or C; N = A, C, G, or T; R = A or G; and D = A, G, or T.
The GC clamp was attached to the 5′ end of the primer NHA1.
A nested PCR approach (12) was used with soil and rhizosphere samples in order to obtain P. azotofixans-specific nifH amplicons with a high degree of specificity. The first PCR was performed with the degenerate primers by using 10 thermal cycles consisting of 93°C for 1.2 min, 50°C for 1 min, and 70°C for 1.5 min. Then, 1 μl of the reaction mixture was used for a second PCR with primers NHA1 and NHA2. PCR amplifications with the specific primers were performed in mixtures (final volume, 50 μl) containing target DNA, 5 μl of 10× PCR buffer (10 mM Tris-HCl [pH 8.3], 10 mM KCl), 10 μl of a mixture containing each deoxynucleoside triphosphate at a concentration of 200 μM, 3.75 mM MgCl2, 20 pmol of each primer, 1% (vol/vol) formamide, 0.25 μg of T4 gene 32 protein (Boehringer, Mannheim, Germany), 5 U of the Taq DNA polymerase Stoffel fragment (Perkin-Elmer, Nieuwerkerk, The Netherlands). T4 gene 32 protein was added since it improved the efficiency of amplification of targets in soil DNA (40). The reaction mixtures were overlaid with 2 drops of mineral oil (Sigma, Zwijndrecht, The Netherlands). A hot-start procedure (5 min, 94°C) was used before enzyme was added to avoid initial mispriming. In addition, to enhance the specificity, a touchdown PCR (19) was performed as follows. The annealing temperature was initially set at 67°C and then was decreased by 1°C every second cycle until it was 57°C; then 10 additional cycles were carried out at 57°C. Strand separation was carried out at 94°C for 1 min, primer annealing was performed (by using the scheme described above) for 1 min, and primer extension was performed at 72°C for 2 min. The final extension step consisted of 10 min at 72°C, after which the reaction mixtures were cooled to 4°C. The amplification products were analyzed by electrophoresis in 1.4% (wt/vol) agarose gels in 0.5× TBE buffer (30) and were stored at −20°C.
Cloning and nucleotide sequencing of nifH-specific amplicons.
The PCR products obtained with the degenerate primers were gel purified with a GeneClean II kit (Bio 101, La Jolla, Calif.) and were cloned into cloning vector pCRII, after which competent Escherichia coli Inv-alfa cells were transformed by using the protocol of the manufacturer (TA cloning kit; Invitrogen, Leek, The Netherlands). Small-scale preparations of plasmid DNA were obtained as described by Sambrook et al. (30) from randomly picked recombinant clones. The plasmid DNA was purified by using Wizard resin spin columns (Promega) and was used as a template in sequencing reactions with a Thermo sequenase fluorescently labelled primer cycle sequencing kit, 7-deaza-dGTP (Amersham Nederland BV, ’s Hertogenbosch, The Netherlands), and an automatic sequence analyzer (model ALF DNA sequencer; Pharmacia). Both strands of the amplification products were sequenced for each clone. The DNA sequence data were analyzed by using the sequence analysis software package developed by the University of Wisconsin Genetics Computer Group (10) at the CAOS/CAMM center (University of Nijmegen, Nijmegen, The Netherlands). The database nifH sequences used for comparison in this study were retrieved from the GenBank and EMBL databases. Sequences were translated and aligned by using Genetics Computer Group software (10) and then were corrected manually. Phylogenetic trees were inferred from the deduced amino acid sequences by both parsimony and distance matrix (neighbor-joining program, PHYLIP package) methods (6).
Analysis of PCR products by DGGE.
DGGE was performed with the phorU2 system (Ingeny, Leiden, The Netherlands). PCR products (15 to 20 μl) were applied directly onto 6% (wt/vol) polyacrylamide gels in 0.5× TAE buffer (20 mM Tris-acetate [pH 7.4], 10 mM sodium acetate, 0.5 mM disodium EDTA) containing a linear denaturing gradient (in general, the concentration of the denaturant ranged from 35 to 65%). The gradients were formed with 6% (wt/vol) acrylamide stock solutions (19) that contained no denaturant and 100% denaturant (the 100% denaturant solution contained 7 M urea and 40% [vol/vol] formamide deionized with AG501-X8 mixed-bed resin [Bio-Rad, Veenendaal, The Netherlands]). The gels were electrophoresed for 4 to 5 h at 60°C and 200 V.
After electrophoresis, the gels were stained for 30 min with SYBR Green I nucleic acid gel stain (Molecular Probes Europe, Leiden, The Netherlands) and were photographed under UV light by using a SYBR Green gel stain photographic filter (Molecular Probes) and a Docugel V system apparatus (Biozym, Landgraaf, The Netherlands).
Restriction enzyme cleavage.
Chromosomal DNA was digested with 10 to 20 U of EcoRI per μg of DNA for 20 h at 37°C by using the protocol supplied by Gibco-BRL (Breda, The Netherlands). Agarose (0.8%, wt/vol) gel electrophoresis of restricted DNA samples was performed in TBE buffer (30) at 2 V cm−1 for 16 h at room temperature. The gels were stained with ethidium bromide, photographed under UV illumination, and used, after blotting, in hybridization experiments.
Blotting and hybridization analysis.
The nifH PCR product generated with P. azotofixans ATCC 35681 (the type strain), which was obtained with the P. azotofixans nifH-specific primers NHA1 and NHA2, was gel purified by using a GeneClean II kit (Bio 101) and was used as a probe to analyze the blotted gels. Probes were labelled with a digoxigenin labelling kit by using the protocol of the manufacturer (Boehringer).
Agarose and DGGE gels were Southern blotted (30) onto nylon membranes (Boehringer), and the membranes were hybridized with specific probes at 42°C (high-stringency conditions; >90% probe-target homology) as described by Fulthorpe et al. (9). For DGGE gels, the denaturing and neutralization steps in the transfer procedure (30) were performed by applying the appropriate solutions for 15 to 30 min on top of the gels. After hybridization, the blots were subjected to sequential washing steps at increasing stringencies, as follows: two washes (5 min each) with 2× SSC–0.1% sodium dodecyl sulfate at room temperature (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate); and two washes (5 min each) with 0.1× SSC–0.1% sodium dodecyl sulfate at 50°C. After washing, a chemiluminescence detection kit (Boehringer) targeting the digoxigenin label was used for detection, and X-ray films were exposed for 15 to 30 min.
Nucleotide sequence accession numbers.
The sequence data determined in this study have been deposited in the EMBL nucleotide sequence database under accession no. AJ224418 through AJ224428.
RESULTS
Hybridization analysis of Paenibacillus sp. EcoRI-digested chromosomal DNA.
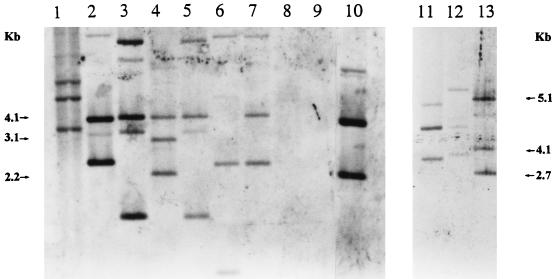
To determine the presence of sequences homologous to the P. azotofixans ATCC 35681 nifH sequence in the genomes of other P. azotofixans and Paenibacillus sp. strains, we performed a hybridization analysis of the EcoRI-digested genomic DNA by using the digoxigenin-labelled nifH PCR product as a probe. Figure 1 shows the results of this analysis. All strains of P. azotofixans, as well as Paenibacillus durum DSMZ 1735 (type strain) (recently proposed as a member of P. azotofixans [29]), showed homology to the nifH probe, whereas the closely related nitrogen-fixing species P. polymyxa and P. macerans did not. In all hybridization-positive strains, more than one hybridizing fragment was observed, and the hybridization patterns of these strains differed. Despite the fact that a series of factors can affect the signal strength distribution and the number of fragments observed, the results indicated that more than one copy of the nifH gene was present in several of the P. azotofixans strains examined.
FIG. 1.
Southern hybridization of EcoRI-restricted genomic DNA of P. azotofixans and Paenibacillus sp. strains with a nifH probe (PCR product from strain ATCC 35681 generated with primers NHA1 and NHA2). The figure is a composite of two blots. Lane 1, P. durum DSMZ 1735; lane 2, P. azotofixans SD20; lane 3, BE1; lane 4, P3E20; lane 5, RCPG7; lane 6, SD17; lane 7, 2RC1; lane 8, P. macerans LMD24.3; lane 9, P. polymyxa DSMZ 356; lane 10, ATCC 35681; lane 11, F102; lane 12, C3L4; lane 13, RBN4.
nifH gene sequence analysis.
Part of the nifH gene was amplified from a range of P. azotofixans and other Paenibacillus strains by using a set of degenerate primers (44), and the PCR products obtained were cloned and sequenced. An alignment of the amino acid sequences deduced from the nifH fragments amplified from the Paenibacillus sp. strains is shown in Fig. 2, together with corresponding nifH product sequences of selected eubacterial and archaeal strains. The similarities among the DNA and deduced amino acid sequences are presented in Table 3. The nifH sequences of P. azotofixans strains were 66 to 99% identical to each other at the DNA level (Table 3). Two main clusters of nifH sequences could be distinguished within the P. azotofixans strains. One cluster encompassed the sequences of strains BE1, RCPG7, 2RC1, ATCC 35681, F102, SD20, and C3L4. These DNA sequences exhibited 93 to 99% similarity among themselves (Table 3). The second cluster consisted of products obtained from P. azotofixans P3E20 and RBN4 and P. durum DSMZ 1735. The sequences in this cluster exhibited 97 to 98% similarity among themselves (Table 3). Although the sequences in each cluster revealed the presence of a characteristic and conserved initial region, some variations in nucleotide sequence were present in the overall sequences (data not shown). However, at the amino acid level, the nifH product sequences of P. azotofixans strains showed a high degree of conservation within each cluster but not between clusters (Fig. 2 and Table 3). Comparisons of the DNA sequences, as well as the amino acid sequences, of both nifH clusters with sequences from the GenBank and EMBL databases (10) revealed that the P. azotofixans nifH cluster I sequences were quite similar to corresponding sequences of the common dinitrogenase iron protein. Curiously, the DNA and amino acid sequences of nifH cluster II exhibited high levels of similarity with the gene sequence and, in particular, the amino acid sequence of the alternative nif (anf) system (Table 3).
FIG. 2.
Alignment of amino acid sequences deduced from the nifH sequence of Paenibacillus sp. and from the sequences of nifH genes of other organisms in the database. The sequences compared correspond to residues 48 to 155 of the Anabaena (Nostoc) sp. strain 7120 sequence (GenBank accession no. A00534). Abbreviations: av, Azotobacter vinelandii; kp, Klebsiella pneumoniae; C3l4, sd20, 2rc1, atcc, be1, rcpg7, and f102, P. azotofixans C3L4, SD20, 2RC1, ATCC 35681, BE1, RCPG7, and F102, respectively; rc, Rhodobacter capsulata; rm, Rhizobium meliloti; LL, Lyngbya lagerheimii; durum, P. durum; p3e20 and frbn4, P. azotofixans P3E20 and RBN4, respectively; cp3, C. pasteurianum nifH3 alternative (anf); cp1, C. pasteurianum nifH1; mtal, Methanococcus thermolithotrophicus alternative (anf); pol, P. polymyxa; mac, P. macerans. For database accession numbers see Fig. 3.
TABLE 3.
Levels of nucleotide and amino acid sequence identity for nifH genes of P. azotofixans and nifH sequences of other bacterial species
| Strain or sequence | % Nucleotide or amino acid identitya
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. azotofixans BE1 | P. azotofixans RCPG7 | P. azotofixans 2RC1 | P. azotofixans ATCC 35681 | P. azotofixans F102 | P. azotofixans SD20 | P. azotofixans C3L4 | Azotobacter vinelandii | Klebsiella pneumoniae | Rhodobacter capsulata | Lyngbya lagerheimii | Rhizobium meliloti | P. polymyxa | P. macerans | P. durum | P. azotofixans P3E20 | P. azotofixans RBN4 |
Clostridium pasteurianum nifH3 alternative (anf) |
Methanococcus thermolithotrophicus alternative (anf) |
|
| P. azotofixans BE1 | 99 | 97 | 96 | 96 | 95 | 93 | 76 | 76 | 76 | 72 | 73 | 72 | 71 | 67 | 65 | 66 | 59 | 57 | |
| P. azotofixans RCPG7 | 100 | 98 | 97 | 96 | 96 | 94 | 76 | 76 | 76 | 72 | 73 | 72 | 71 | 67 | 66 | 67 | 59 | 58 | |
| P. azotofixans 2RC1 | 99 | 99 | 98 | 97 | 95 | 94 | 76 | 77 | 76 | 73 | 73 | 72 | 72 | 66 | 65 | 66 | 59 | 57 | |
| P. azotofixans ATCC 35681 | 99 | 99 | 100 | 96 | 95 | 93 | 76 | 77 | 75 | 72 | 73 | 72 | 71 | 65 | 65 | 65 | 59 | 57 | |
| P. azotofixans F102 | 99 | 99 | 98 | 98 | 94 | 94 | 77 | 76 | 76 | 72 | 73 | 73 | 72 | 67 | 66 | 66 | 59 | 56 | |
| P. azotofixans SD20 | 99 | 99 | 98 | 98 | 98 | 95 | 76 | 76 | 76 | 72 | 73 | 73 | 72 | 67 | 66 | 66 | 60 | 59 | |
| P. azotofixans C3L4 | 99 | 99 | 98 | 98 | 98 | 100 | 76 | 75 | 75 | 72 | 72 | 73 | 71 | 67 | 66 | 66 | 59 | 56 | |
| Azotobacter vinelandii | 85 | 85 | 86 | 86 | 86 | 85 | 85 | 83 | 79 | 79 | 78 | 71 | 73 | 66 | 64 | 64 | 59 | 56 | |
| Klebsiella pneumoniae | 86 | 86 | 87 | 87 | 85 | 86 | 86 | 93 | 80 | 78 | 76 | 71 | 72 | 67 | 66 | 66 | 58 | 57 | |
| Rhodobacter capsulata | 88 | 88 | 87 | 87 | 89 | 88 | 88 | 86 | 86 | 76 | 76 | 70 | 72 | 64 | 63 | 64 | 57 | 56 | |
| Lyngbya lagerheimii | 84 | 84 | 85 | 85 | 84 | 84 | 84 | 87 | 84 | 83 | 74 | 70 | 67 | 63 | 62 | 62 | 56 | 55 | |
| Rhizobium meliloti | 85 | 85 | 86 | 86 | 86 | 85 | 85 | 84 | 83 | 91 | 82 | 70 | 71 | 63 | 63 | 63 | 58 | 57 | |
| P. polymyxa | 75 | 75 | 74 | 74 | 74 | 75 | 75 | 68 | 68 | 68 | 68 | 65 | 74 | 59 | 58 | 59 | 57 | 56 | |
| P. macerans | 48 | 48 | 48 | 48 | 49 | 49 | 49 | 44 | 42 | 46 | 46 | 44 | 49 | 60 | 59 | 60 | 53 | 54 | |
| P. durum | 74 | 74 | 73 | 73 | 75 | 74 | 74 | 78 | 76 | 76 | 74 | 72 | 59 | 41 | 98 | 97 | 72 | 66 | |
| P. azotofixans P3E20 | 73 | 73 | 72 | 72 | 74 | 73 | 73 | 77 | 75 | 75 | 73 | 72 | 58 | 41 | 99 | 97 | 71 | 65 | |
| P. azotofixans RBN4 | 73 | 73 | 72 | 72 | 74 | 73 | 73 | 77 | 75 | 75 | 73 | 72 | 58 | 41 | 97 | 98 | 73 | 66 | |
| Clostridium pasteurianum nifH3 alternative (anf) | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 74 | 76 | 73 | 72 | 71 | 57 | 38 | 86 | 86 | 86 | 68 | |
| Methanococcus thermolithotrophicus alternative (anf) | 68 | 68 | 68 | 68 | 67 | 68 | 68 | 72 | 72 | 69 | 69 | 67 | 54 | 35 | 80 | 79 | 79 | 79 | |
The values on the upper right are levels of DNA identity, and the values on the lower left are levels of amino acid identity. For database accession numbers see Fig. 3.
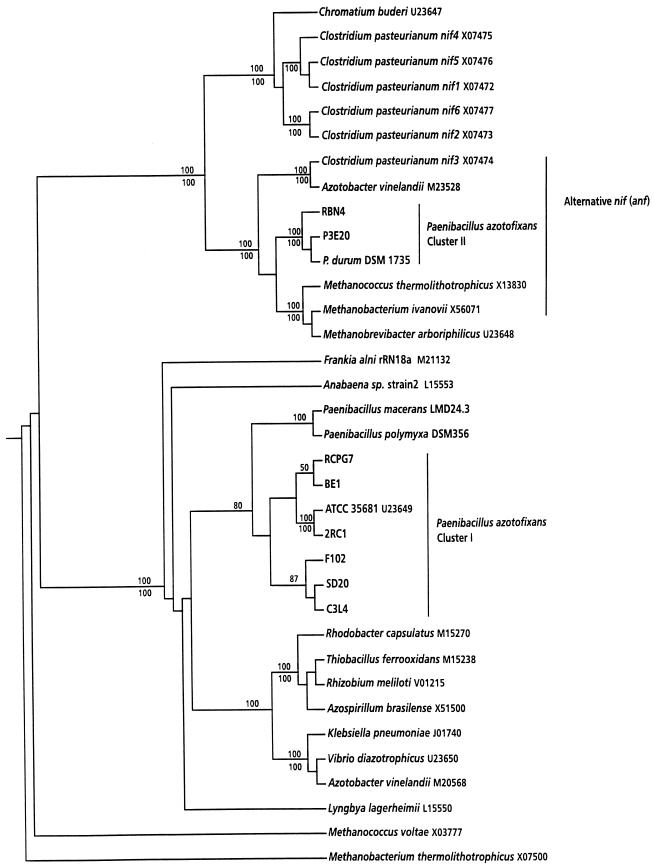
The nifH tree constructed in this study (Fig. 3) had a topology similar to that of previously published trees of nifH phylogeny (13, 23, 38, 43, 45) with respect to the database sequences and was consistent with several aspects of relatedness based on 16S rRNA phylogenies (25). As expected, the nifH cluster II sequences of P. azotofixans and P. durum clustered together and were closely related to sequences of alternative nifH genes of several organisms (Fig. 3). The P. azotofixans cluster I nifH sequences (seven strains) grouped together with the previously determined partial nifH sequence of P. azotofixans ATCC 35681 (45). The partial nifH sequences of the closest relatives of P. azotofixans, P. polymyxa and P. macerans, clustered together and were more closely related to P. azotofixans nifH (cluster I) sequences than to any other nifH sequence from another bacterium. These sequences were only distantly related to a cluster composed of various different eubacterial nifH sequences (Fig. 3).
FIG. 3.
Phylogenetic tree based on partial nifH product amino acid sequences, including the sequences of P. azotofixans strains and Paenibacillus spp. and other sequences from the database. The location of the nifH fragments used for the analysis corresponds to Anabaena (Nostoc) sp. strain 7120 residues 48 to 155 (GenBank accession no. A00534). The database accession numbers are indicated after the bacterial names. Bootstrap values (percentages) for parsimony (above the lines) and neighbor joining (below the lines) are shown for clusters supported by both analyses; only values greater than 50% are shown.
Design of PCR primers.
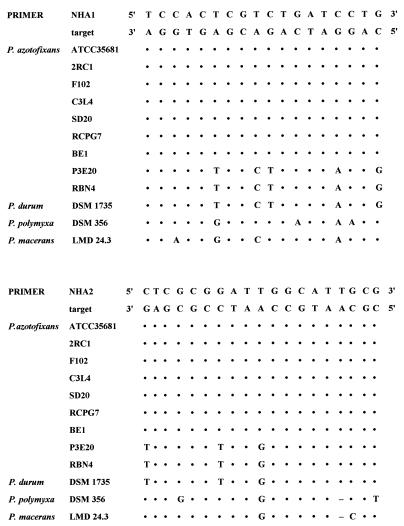
The partial nifH gene sequences of the different P. azotofixans and P. durum strains, as well as those of P. polymyxa and P. macerans, were aligned by using PILEUP (sequence analysis software package of the University of Wisconsin Genetics Computer Group [10]). From the alignment, two consensus sequences for P. azotofixans nifH cluster I were obtained (Fig. 4), and primers were designed on the basis of invariable stretches at both ends of these sequences. Table 2 shows the primer sequences (see also Fig. 4). These sequences were compared with sequences obtained from the EMBL database by using fastA homology searches. No similarities were found that would give rise to false-positive PCR products. The possibility that there was amplification of cluster II fragments was excluded on the basis of five and three differences in the sequences of the forward and reverse primers, respectively (Fig. 4). For DGGE analysis, a 40-bp GC clamp (19) was attached to the 5′ end of primer NHA1 (forward primer).
FIG. 4.
Sequence alignment of P. azotofixans-specific stretches of the nifH gene with closely related nifH sequences, showing target regions used to design specific primers. Most of the Paenibacillus sp. sequences were obtained in this study; the only exception was the sequence of P. azotofixans ATCC 35681 (45). DSM, DSMZ. Dots indicate bases identical to those of the target sequence.
To explore the specificity of the PCR assay developed, we amplified the nifH gene by using genomic DNA of several P. azotofixans strains, strains of closely related Paenibacillus species, and strains of a variety of nitrogen-fixing and non-nitrogen-fixing bacteria (Table 1). To reduce the formation of spurious by-products, a touchdown PCR protocol (19) was used. The results revealed that amplification products of the expected size, approximately 360 bp, were obtained with P. azotofixans strains and with P. durum DSMZ 1735 (Table 1), whereas no PCR products were obtained with DNA from any of the other bacterial strains used. This revealed that P. azotofixans P3E20 and RBN4, as well as P. durum DSMZ 1735, which had been found to contain cluster II nifH sequences, also contain copies of cluster I nifH sequences in their genomes (Fig. 2).
Despite the specificity and sensitivity of primers NHA1 and NHA2 for amplification of (cluster I) nifH gene sequences with genomic DNA from pure cultures, the yield of the PCR product was poor with soil DNA. To enhance sensitivity, we used a nested PCR protocol (12, 37) in which the primers of the degenerate primer set (Table 2) (44) were used as the external primers and primers NHA1 and NHA2 were used as internal primers (touchdown reaction). This nested PCR protocol resulted in efficient PCR amplification with genomic DNAs of all P. azotofixans strains and generated PCR products of the expected sizes (data not shown). Experiments were then performed to directly amplify the nifH genes of P. azotofixans populations in bulk and maize rhizosphere soil samples by using soil DNA. Positive results were obtained with DNA recovered from seven of the different soil and rhizosphere samples tested. All PCR products were the same size, 360 bp, which was identical to the size of the PCR product obtained when genomic DNA from pure cultures was the target.
PCR-DGGE analysis of nifH gene sequences with pure-culture DNA.
To analyze the melting behavior of the (cluster I) nifH amplicons, MELT95 (Ingeny), a modified version of the FORTRAN program MELT87, was used. The theoretical melting characteristics of the sequences from strains ATCC 35681 and 2RC1 were very similar. Three melting domains, one the result of the GC clamp and the other two intrinsic, were found and the main differences were in one melting domain. When analyzed by DGGE, the amplicons were predicted to comigrate until 50% urea-formamide, after which they would separate due to their different melting behaviors. On the basis of the melting maps, a 35 to 65% denaturant gradient was selected. A time travel experiment (19) performed with products obtained from several different strains established that the optimal electrophoresis time was 4 to 5 h at 200 V.
Cluster I nifH-specific DGGE was first used to separate the P. azotofixans nifH fragments amplified from pure cultures. The results (Fig. 5) confirmed the diversity of P. azotofixans nifH cluster I gene sequences that had been observed in hybridization and cloning experiments (Fig. 1 and 2). For all strains tested, patterns consisting of more than one band were found in the DGGE analysis (Fig. 5). The patterns were reproducible and characteristic for each strain tested, indicating that there was interstrain sequence divergence. For several strains, the DGGE band patterns contained two bands that migrated very close together (Fig. 5). This observation indicates that there were DNA molecules with slightly different melting behaviors, possibly caused by incomplete extension of the same template due to the GC clamp (22). Furthermore, for some strains (e.g., BE1 and RCPG7), similar band patterns were obtained. Strains F102, 2RC1, and P2E20 also produced similar band patterns, differing only in the two upper bands produced by strain 2RC1. Strains BE1, RCPG7, RBN4, and ATCC 35681 produced double bands that migrated in the same position, indicating that they may have similar copies of nifH.
FIG. 5.
DGGE analysis of PCR-amplified nifH gene fragments from P. azotofixans and P. durum strains: parallel DGGE separation patterns of gene fragments obtained with genomic DNA from strains DSMZ 1735 (lane 1), BE1 (lane 2), SD20 (lane 3), SD17 (lane 4), RCPG7 (lane 5), F102 (lane 6), 2RC1 (lane 7), C3L4 (lane 8), RBN4 (lane 9), P3E20 (lane 10), and ATCC 35681 (lane 11).
Hybridization of Southern blots of the DGGE gel with the nifH probe under high-stringency conditions (>90% homology [9]) revealed positive signals that confirmed the presence of homology with the nifH probe in all of the bands observed in DGGE gels (data not shown).
PCR-DGGE analysis of bulk and rhizosphere soil samples.
Different soil and maize rhizosphere samples were screened twice to determine the putative diversity of P. azotofixans cluster I nifH amplicons. Both data sets obtained are described below, and one is shown in Fig. 6. The DGGE analyses performed with directly extracted DNA (36) revealed different, low levels of complexity of nifH sequence types in the different soil and rhizosphere samples. Most strikingly, in Guaíra soil, in all Cerrado and Várzea maize rhizosphere soils, and possibly in Flevo silt loam soil, a single dominant band was always found (Fig. 6). This band was also generated with genomic DNA of P. azotofixans LV17 obtained from the Várzea samples, indicating that this strain and its partial nifH sequence might be representative of the dominant P. azotofixans cell and cluster I nifH gene populations in these soils. On the other hand, other strains also isolated from the Várzea samples produced diverse nifH amplicons, several of which were similar to the amplicon generated with P. azotofixans C3L4 (Fig. 6).
FIG. 6.
DGGE analysis of PCR-amplified nifH gene fragments generated with DNA from Dutch and Brazilian bulk and rhizosphere soil samples and individual P. azotofixans strains. Lane 1, Ede loamy sand bulk soil; lane 2, Flevo silt loam bulk soil; lane 3, Guaíra bulk soil; lane 4, Cerrado soil, maize rhizosphere, 30 days; lane 5, Cerrado soil, maize rhizosphere, 60 days; lane 6, Várzea soil, maize rhizosphere, 10 days; lane 7, Várzea soil, maize rhizosphere, 30 days; lane 8, Várzea soil, maize rhizosphere, 60 days; lane 9, Várzea soil isolate LV17; lane 10, Várzea soil isolate PV55; lane 11, Várzea soil isolate PV18; lane 12, Várzea soil isolate PV9; lane 13, strain C3L4; lanes M, markers (16S rDNA-based products of [from top to bottom] Listeria innocua ALM105, Arthrobacter sp., and Burkholderia cepacia P2).
Furthermore, the profiles obtained with maize rhizosphere samples taken over time in Cerrado and Várzea soils indicated that there were possible shifts in the dominant populations, since at 60 and 30 days, respectively, additional strong bands appeared which were absent in the initial samples.
Soils P04 and P33 exhibited the most complex nifH DGGE profiles, with three and four or five visible bands, respectively (data not shown). Both profiles contained similar bands that comigrated with the product generated with strain C3L4 and had homology to the nifH cluster I probe, as shown by hybridization. As expected, blotting of the DGGE gels and high-stringency (8) hybridization with the nifH cluster I-specific probe revealed the presence of homology to the probe in all lanes (data not shown).
DISCUSSION
In this paper, we describe a molecular approach to analyze the genetic diversity of P. azotofixans structural nitrogen fixation genes in pure cultures, soils, and rhizospheres. Although the nifH gene and gene product are quite conserved, mainly at the amino acid sequence level (43), several reports have also shown that there is sufficient variation at the DNA sequence level to distinguish similarity groups that may represent taxonomic groups. Thus, specific primers or probes can be generated that can be used to assess the affiliation and diversity of nifH genes found in soil (2, 35). Similarly, a set of Frankia-specific PCR primers and strain-specific probes have been designed to study the competitiveness of two Frankia strains in the nodulation of host plant roots (35). Ben-Porath and Zehr (2), using the same set of degenerate primers which we used, confirmed that the sequence of the 359-bp product is sufficiently variable to distinguish cyanobacterial nifH gene sequences from eubacterial and archaeal nifH gene sequences, as well as heterocystous nifH genes from nonheterocystous nifH genes.
Phylogenetic analysis of the partial nifH gene sequences of the Paenibacillus spp. demonstrated that there was sufficient variation at the DNA sequence level to distinguish the nifH genes of P. azotofixans from the nifH genes of related species (i.e., P. polymyxa and P. macerans). The nifH sequence obtained for P. durum DSMZ 1735 clustered together with the nifH sequence of P. azotofixans P3E20 and RBN4 (nifH cluster II). Furthermore, for these three strains amplicons were also generated with nifH cluster I-specific primers. These results indicated that these strains are closely affiliated, as judged by the presence of copies of nifH cluster I and cluster II genes. As P. durum (4) has been recently proposed as a member of P. azotofixans based on several criteria (29), its inclusion in this nifH-based group was not surprising. Moreover, the results of the nifH sequencing experiments provided several interesting leads. First, the finding that there are two nifH gene clusters suggested that in addition to the genes encoding the common nitrogenase, there are genes encoding the alternative nitrogenase system (anf) in the P. azotofixans genome. This possibility was supported by the fact that the cluster II sequences were related to known anf sequences found in strains belonging to other taxonomic groups (Fig. 3). Second, when the nifH DNA sequences of P. azotofixans strains were compared to each other, several sequence divergences which indicated microheterogeneities were found, which should permit diversity studies in which gradient gel electrophoresis is used. However, the differences were less evident at the amino acid sequence level (Table 3). The occurrence of multiple copies of nifH genes and/or alternative nitrogenase systems has been reported for several other bacterial species (2, 16, 41). For instance, Clostridium pasteurianum has six copies of the nifH gene, including one copy encoding an alternative nitrogenase (41). In this context, Nübel et al. (22) showed that P. polymyxa has several copies of the 16S rRNA gene, which differ from each other. Thus, the exploitation of 16S rDNA sequences for phylogenetic and ecological studies can meet with problems like interstrain (intraspecific) variability and the presence of multiple copies of the operon (15). Therefore, results obtained with nifH sequences retrieved from natural environments also should be analyzed with caution.
The divergence found in the P. azotofixans nifH gene sequences was considerable and cannot be explained by Taq polymerase errors. Furthermore, when P. azotofixans DNA was digested with EcoRI and probed with the nifH probe generated via PCR with P. azotofixans ATCC 35681, two to four bands were observed (Fig. 1). This result supports the presence of multiple nifH-homologous regions in P. azotofixans. In a previous study (34), DNA homology to Klebsiella pneumoniae nifH and nifD genes was found in 22 strains of P. azotofixans, and the hybridization patterns of the strains differed. For several strains, the patterns found with the K. pneumoniae probe (34) were similar to the patterns reported in this paper, supporting the localization of nifH in several genomic positions. The possibility of reiteration and rearrangement of this gene during P. azotofixans evolution has been suggested (34). On the other hand, in a similar study no evidence of either reiteration or rearrangement of nif genes was found in P. polymyxa and P. macerans (24).
Several aspects of the nifH phylogenetic tree proposed here were consistent with phylogenies based on the 16S rRNA gene sequence (25) derived by other workers (13, 23, 38, 43, 45); one exception is the separation into cluster I and cluster II (anf) sequences (45). Whereas the cluster II nifH sequences grouped together with other anf gene sequences in the database, the P. azotofixans cluster I nifH sequences grouped together and close to the nifH sequences of other Paenibacillus spp. There was only a distant relationship with the other nifH sequences, as shown previously for P. azotofixans ATCC 35681 (45). Moreover, the nifH sequences of P. azotofixans and C. pasteurianum did not cluster together, even though both species belong to the low-G+C-content gram-positive bacteria. This fact was in agreement with previous reports (23, 45).
Some recent reports have assessed nifH sequences that were directly amplified and isolated from natural environments (23, 38, 45). These studies suggested that natural bacterial communities can have considerable diversity in their nifH sequences and that communities of diazotrophs may consist of diverse nitrogen-fixing organisms, including as-yet-uncharacterized organisms. The diversity of nif genes in the underlying organisms may therefore be greater than previously thought. Since the diversity of the P. azotofixans nifH genes in the soil environment has been largely unexplored, we directly addressed this intraspecies diversity by performing specific PCR-DGGE with soil DNA. The PCR assay, based on conserved regions of P. azotofixans nifH cluster I, amplified this region from DNA of all of the P. azotofixans strains tested, as well as from P. durum DNA. The absence of PCR products from DNA of any other strain tested suggested that the primers were indeed specific for nifH cluster I of P. azotofixans.
In order to enhance the probability of specific amplification of P. azotofixans nifH DNA from soil, we used nested PCR (12). Nested PCR can enhance the sensitivity and specificity of amplification by allowing a first round of amplification with less stringent external primers, followed by a second round with internal primers designed to recognize specific regions within the initial amplicon. Nonspecific primer annealing is thus minimized (12), which is particularly useful when organisms or genes are detected in environmental samples in which the exact specificity or uniqueness of the primers is unknown (37). The nifH-specific nested PCR applied to soil DNA also effectively diluted putative PCR inhibitors to acceptable levels and thus allowed successful amplification.
The optimized PCR-DGGE protocol revealed the existence of considerable diversity, including multiple bands, in the nifH sequences of different strains of P. azotofixans. Thus, DGGE analysis confirmed the results of the cloning-sequencing and hybridization experiments, i.e., that P. azotofixans strains harbor multiple copies of nifH and that these copies exhibit sequence divergence. These results also supported the hypothesis that the nifH gene in P. azotofixans underwent rearrangement during evolution (34). Factors that might have contributed to sequence heterogeneity in the P. azotofixans nifH genes could range from gene conversion and/or selection to horizontal gene transfer (13, 22).
The putative functions of the multiple nifH-like sequences need to be investigated further. Possible explanations for C. pasteurianum have been discussed by Wang et al. (41). These authors suggested that some copies of nifH in this organism could play a role in the regulation of nitrogenase activity under certain ecological conditions (41). The identification of primers specific for cluster I nifH genes on the one hand and for cluster II (anf) genes on the other hand opens the possibility for in situ detection of the expression of either system via reverse transcription-PCR of mRNA.
The upper bands in DGGE profiles often represent heteroduplex molecules (8), and we cannot exclude this possibility for some of the profiles which we obtained (e.g., the strain 2RC1 and C3L4 profiles) (Fig. 5). Heteroduplexes may form during mixed-template PCR due to annealing between similar but not identical products (8). Despite this problem, the bands generated with the different strains revealed nifH sequence diversity, as well as multiple copies (Fig. 5). On the other hand, some strains (e.g., BE1, RCPG7, RBN4, and ATCC 35681) shared bands, which resulted in identical or similar profiles.
PCR-DGGE analysis of bulk and rhizosphere soils revealed possible similarities, as well as differences, among the different profiles (Fig. 6). First, the stable presence of one dominant band in the Guaíra, Cerrado, and Várzea soil profiles, in combination with the comigrating band generated with P. azotofixans LV17 isolated from Várzea soil, suggested that strain LV17 might be an example of the hosts of the nifH type identified in these soils. On the other hand, the differences and possible population shifts observed both over time (Cerrado and Várzea soils) and between soils indicated that other, diverse P. azotofixans nifH types also played a role in the soils and rhizospheres examined. One of these types might be exemplified by the C3L4 nifH type that was found in soils P04 and P33. Selection exerted by sugarcane might link these observations, as strain C3L4 was originally obtained from a sugarcane rhizosphere in Hawaii (32), and soils P04 and P33 have been used for sugarcane plantations. Selection by sugarcane was recently also suggested for other P. azotofixans strains (29a). Overall, these data might indicate that different P. azotofixans and nifH populations were selected by different soils or plants. However, since some P. azotofixans strains produced more than one band in the DGGE profiles of the nifH amplicons, numbers of bands cannot be strictly related to numbers of different strains. Moreover, identical band positions do not unequivocally demonstrate sequence identity as overlapping bands with different sequences can occur (19).
The PCR-DGGE approach taken here is a sensitive indicator of the diversity of nifH genes in P. azotofixans populations and can be used as an efficient way to perform further in-depth ecological studies in soil. The advantages of PCR-DGGE analysis over classical cloning and sequencing of amplicons are manifold, as the method is rapid, simple, and relatively inexpensive (16). Thus, the laborious screening necessary due to redundancy of clones in a library can be eliminated.
Finally, the occurrence of nifH genes in a soil system does not a priori indicate that nitrogenase activity is present (5). Also, it is not known whether all P. azotofixans nifH gene copies revealed by DGGE in our study are functional copies. Such questions can only be addressed by targeting mRNA in a direct reverse transcription-PCR-DGGE approach.
ACKNOWLEDGMENTS
This work was supported by the EU-IC Programme. A.S.R. and G.F.D. were recipients of fellowships awarded by the National Research Council of Brazil (CNPq). We thank A. D. L. Akkermans and H. Ramirez for providing bacterial strains and/or DNA and H. Coutinho and E. Paiva for supplying soil samples. The excellent technical assistance of A. C. Keijzer and L. Lankwarden is gratefully acknowledged.
REFERENCES
- 1.Ash C, Farrow J A E, Priest F G, Collins M D. Molecular identification of rRNA group 3 bacilli using a PCR probe test. Proposal for the creation of a genus Paenibacillus. Antonie Leeuwenhoek. 1993;64:253–260. doi: 10.1007/BF00873085. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Porath J, Zehr J P. Detection and characterization of cyanobacterial nifH genes. Appl Environ Microbiol. 1994;60:880–887. doi: 10.1128/aem.60.3.880-887.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 5.Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–834. [Google Scholar]
- 6.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 7.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulthorpe R R, McGowan C, Maltseva O V, Holben W E, Tiedje J M. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genetics Computer Group. Program manual for the Wisconsin package. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 11.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 12.Haqqi T M, Sarkar G, David C S, Sommer S S. Specific amplification with PCR of a refractory segment of genomic DNA. Nucleic Acids Res. 1988;16:11844. doi: 10.1093/nar/16.24.11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennecke H, Kaluza K, Thony B, Fuhrmann M, Ludwig W, Stackebrandt E. Concurrent evolution of nitrogenase genes and 16S rRNA in Rhizobium species and other nitrogen fixing bacteria. Arch Microbiol. 1985;142:342–348. [Google Scholar]
- 14.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities. In: van Elsas J D, Wellington E M H, Trevors J T, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 353–373. [Google Scholar]
- 15.Johansen T, Carlson C R, Kolsto A-B. Variable numbers of rRNA gene operons in Bacillus cereus strains. FEMS Microbiol Lett. 1996;136:325–328. doi: 10.1111/j.1574-6968.1996.tb08068.x. [DOI] [PubMed] [Google Scholar]
- 16.Kirshtein J D, Paerl H W, Zehr J. Amplification, cloning, and sequencing of a nifH segment from aquatic microorganisms and natural communities. Appl Environ Microbiol. 1991;57:2645–2650. doi: 10.1128/aem.57.9.2645-2650.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalchuk G A, Stephen J R, de Boer W, Prosser J L, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muyzer G, Hottentrager S, Teske A, Wawer C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities, section 3.4.4. In: Akkermans A D L, Van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–23. [Google Scholar]
- 20.Myers R M, Maniatis T, Lerman L S. Detection and localization of single base changes by denaturing gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- 21.Normand P, Simonet P, Bardin R. Conservation of nif sequences in Frankia. Mol Gen Genet. 1988;213:238–246. doi: 10.1007/BF00339587. [DOI] [PubMed] [Google Scholar]
- 22.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira S S, Seldin L, Bastos M C F. Identification of structural nitrogen-fixation genes in Bacillus polymyxa and Bacillus macerans. World J Microbiol Biotechnol. 1993;9:387–389. doi: 10.1007/BF00383088. [DOI] [PubMed] [Google Scholar]
- 25.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolleke S, Muyzer G, Wawer C, Wanner G, Lubitz W. Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1996;62:2059–2065. doi: 10.1128/aem.62.6.2059-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosado A S, Seldin L. Isolation and partial characterization of a new linear DNA plasmid isolated from Bacillus polymyxa SCE2. J Gen Microbiol. 1993;139:1277–1282. [Google Scholar]
- 28.Rosado A S, Seldin L, Wolters A C, van Elsas J D. Quantitative 16S rDNA targeted polymerase chain reaction and oligonucleotide hybridization for the detection of Paenibacillus azotofixans in soil and the wheat rhizosphere. FEMS Microbiol Ecol. 1996;19:153–164. [Google Scholar]
- 29.Rosado A S, van Elsas J D, Seldin L. Reclassification of Paenibacillus durum (formerly Clostridium durum Smith and Cato 1974) Collins et al. 1994 as a member of the species P. azotofixans (formerly Bacillus azotofixans Seldin et al. 1984) Ash et al. 1994. Int J Syst Bacteriol. 1997;47:569–572. doi: 10.1099/00207713-47-2-569. [DOI] [PubMed] [Google Scholar]
- 29a.Rosado A S, de Azevedo F S, da Cruz D W, van Elsas J D, Seldin L. Phenotypic and genetic diversity of Paenibacillus azotofixans strains isolated from the rhizoplane or rhizosphere soil of different grasses. J Appl Bacteriol. 1998;84:216–226. [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Seldin L, van Elsas J D, Penido E G C. Bacillus nitrogen fixers from Brazilian soils. Plant Soil. 1983;70:243–255. [Google Scholar]
- 32.Seldin L, van Elsas J D, Penido E G C. Bacillus azotofixans sp. nov., a nitrogen-fixing species from Brazilian soils and grass roots. Int J Syst Bacteriol. 1984;34:451–456. [Google Scholar]
- 33.Seldin L, Penido E G C. Identification of Bacillus azotofixans using API tests. Antonie Leeuwenhoek. 1986;52:403–409. doi: 10.1007/BF00393468. [DOI] [PubMed] [Google Scholar]
- 34.Seldin L, Bastos M C F, Penido E G C. Identification of Bacillus azotofixans nitrogen fixation genes using heterologous nif probes. In: Skinner F A, Boddey R M, Fendrik I, editors. Nitrogen fixation with non-legumes. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1989. pp. 179–187. [Google Scholar]
- 35.Simonet P, Normand P, Moiroud A, Bardin R. Identification of Frankia strains in nodules by hybridization of polymerase chain reaction products with strain-specific oligonucleotide probes. Arch Microbiol. 1990;153:235–240. doi: 10.1007/BF00249074. [DOI] [PubMed] [Google Scholar]
- 36.Smalla K, Creswell N, Mendonca-Hagler L C, Wolters A, Van Elsas J D. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol. 1993;74:78–85. [Google Scholar]
- 37.Steffan R J, Atlas R M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- 38.Ueda T, Suga Y, Yashiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Elsas J D, Dijkstra A F, Govaert J M, van Veen J A. Survival of Pseudomonas fluorescens and Bacillus subtilis introduced into two soils of different texture in field microplots. FEMS Microbiol Ecol. 1986;38:151–160. [Google Scholar]
- 40.Van Elsas J D, Mäntynen V, Wolters A C. Soil DNA extraction and assessment of the fate of Mycobacterium chlorophenolicum strain PCP-1 in different soils by 16S ribosomal RNA gene sequence based most-probable-number PCR and immunofluorescence. Biol Fertil Soils. 1997;24:188–195. [Google Scholar]
- 41.Wang S-Z, Chen J-S, Johnson J L. The presence of five nifH-like sequences in Clostridium pasteurianum: sequence divergence and transcription properties. Nucleic Acids Res. 1988;16:439–454. doi: 10.1093/nar/16.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wawer C, Muyzer G. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gel electrophoresis of [NiFe] hydrogenase gene fragments. Appl Environ Microbiol. 1995;61:2203–2210. doi: 10.1128/aem.61.6.2203-2210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 43–86. [Google Scholar]
- 44.Zehr J, McReynolds L. Use of degenerate oligonucleotide primers for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zehr J P, Mellon M, Braun S, Litaker W, Steppe T, Paerl H W. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]