Abstract
Several cutinase variants derived by molecular modelling and site-directed mutagenesis of a cutinase gene from Fusarium solani pisi are poorly secreted by Saccharomyces cerevisiae. The majority of these variants are successfully produced by the filamentous fungus Aspergillus awamori. However, the L51S and T179Y mutations caused reductions in the levels of extracellular production of two cutinase variants by A. awamori. Metabolic labelling studies were performed to analyze the bottleneck in enzyme production by the fungus in detail. These studies showed that because of the single L51S substitution, rapid extracellular degradation of cutinase occurred. The T179Y substitution did not result in enhanced sensitivity towards extracellular proteases. Presumably, the delay in the extracellular accumulation of this cutinase variant is caused by the enhanced hydrophobicity of the molecule. Overexpression of the A. awamori gene encoding the chaperone BiP in the cutinase-producing A. awamori strains had no significant effect on the secretion efficiency of the cutinases. A cutinase variant with the amino acid changes G28A, A85F, V184I, A185L, and L189F that was known to aggregate in the endoplasmic reticulum of S. cerevisiae, resulting in low extracellular protein levels, was successfully produced by A. awamori. An initial bottleneck in secretion occurred before or during translocation into the endoplasmic reticulum but was rapidly overcome by the fungus.
Cutinases are produced by several phytopathogenic fungi, including Fusarium solani pisi (29), Magnaportha grisea (30), and Colletotrichum gloeosporioides (7). These enzymes can hydrolyze ester bonds in the cutin polymer, an insoluble lipid-polyester matrix covering the surfaces of plant leaves (18). Cutinase belongs to a class of esterases that are able to hydrolyze fatty acid esters and emulsified triglycerides as efficiently as lipases, without showing enhancement of activity in the presence of a lipid-water interface (18). It has been found that cutinase has an “in-the-wash” effect, making this protein suitable for use in laundry detergents (31). The three-dimensional X-ray structure of the cutinase from F. solani pisi (21) was used to design variants in a protein-engineering study aimed at increasing knowledge of the structure-function relationship of cutinase and improving its performance in detergent formulations. The amino acid sequence was modified in such a way that the hydrophobicity at the surface of the enzyme was increased to form an enlarged lipid contact zone (5) and decreased sensitivity towards anionic surfactants present in detergents (6).
A synthetic copy of the cDNA encoding the cutinase from F. solani pisi has been expressed successfully in Saccharomyces cerevisiae and Aspergillus awamori (32). About 20% of cutinase variants expressed in S. cerevisiae could not be produced at the same extracellular levels as the wild-type cutinase is produced (unpublished results) (Table 1). In this study, we analyzed whether these variants could be produced by the filamentous fungus A. awamori.
TABLE 1.
Rationale for design of some cutinase variants produced poorly by S. cerevisiae
| Cutinase variant(s)a | Target | Rationale | % Production by yeastb |
|---|---|---|---|
| Wild type | 100 | ||
| L51S | Anionic compatibility | Distortion of hydrophobic patch | 10 |
| E201K | Performance, adsorption | Altered hydrophobicity | 71 |
| G82A, A85F, V184I, A185L, L189F | Performance | Increased hydrophobicity of the binding site | 22 |
| A29−, S30− | Anionic compatibility | Stabilization | 24 |
| T179Y | Structure-function | Increased hydrophobicity | 42 |
| W69Y | Structure-function | Trp replaced by Tyr | 12 |
Amino acid numbers start at the propeptide of the natural cutinase.
Production levels were determined as described by Sagt et al. (26).
Several studies have described the overproduction of both a molecular chaperone and a heterologous protein with the aim of increasing the yield of the heterologous protein (19, 25). Overexpression of the BiP-encoding gene in S. cerevisiae resulted in an increase in heterologous protein production (12). This suggests that overproduction of foreign proteins can result in saturation of the protein-folding machinery caused by limited availability of BiP in the endoplasmic reticulum of eukaryotes. A similar situation may occur in filamentous fungi. Therefore, the gene encoding the BiP protein from A. awamori, bipA, was cloned (34) and subsequently overexpressed in the cutinase-producing strains.
MATERIALS AND METHODS
Strains, media, and transformations.
Standard molecular biology procedures were performed as described by Sambrook et al. (27). Escherichia coli JM109 (37) and 1046 (F− met hsdS supE supF recA56) were used as hosts for molecular cloning.
AW4-20, a pyrG (orotidine-5′-phosphate decarboxylase)-deficient derivative of A. awamori CBS 115.52 that contains a defined mutation at the BglII site of the pyrG gene (9), was used as a recipient strain for transformation experiments carried out as described by Punt and Van den Hondel (23). Transformants of the pyrG-deficient strain with cutinase vectors were selected on the basis of uridine prototrophy. Transformants with additional copies of the A. awamori bipA gene were obtained by selection on medium containing 100 μg of hygromycin per ml. All strains were cultivated in Aspergillus minimal medium (3) supplemented with 0.5 or 0.1% (wt/vol) yeast extract (Difco Laboratories, Detroit, Mich.).
Construction of plasmids.
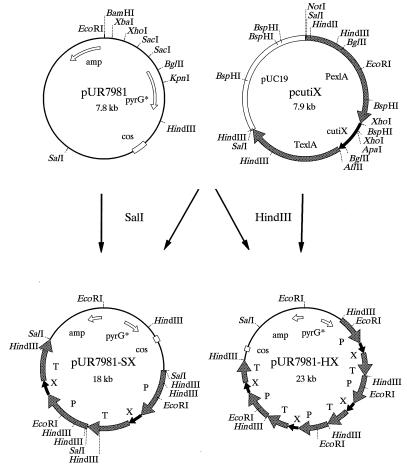
Mutations in the cutinase gene were obtained by site-directed mutagenesis via PCR (13). The expression signals from the endoxylanase II (exlA) gene (10) were used for inducible overexpression of the cutinase alleles and bipA genes in A. awamori strains. The mature cutinase genes were fused to the exlA presequence, and the bipA gene was expressed with its natural presequence. The coding regions were adapted for cloning into the general fungal expression vector pAW14B12 (33). A BspHI site was introduced at the start codon and an AflII site was introduced at the stop codon of the genes, which were subsequently cloned into the BbsI and AflII sites of pAW14B12. Studies in one of our laboratories have shown that the HindIII fragment containing the exlA expression signals is sufficient for high levels of gene expression in A. awamori (unpublished results). Vectors containing four to eight HindIII fragments of cutinase cassettes in tandem at the HindIII site of pUR7981 (Fig. 1), cosmid pJB8 (14) containing a pyrG gene with a defined mutation at the SalI site (9), were obtained via packaging by using a Gigapack Gold kit from Stratagene (La Jolla, Calif.). Because homologous integration of these large vectors at the pyrG locus of A. awamori could not be accomplished in 30 separate transformation experiments, cotransformations were performed with plasmid pAW4-1 (9). This plasmid contains the intact pyrG gene, and cotransformation with pAW4-1 resulted in several random multicopy transformants of cosmid pUR7981-HX (Fig. 1). To obtain homologous integration at the pyrG locus of AW4-20, two SalI cutinase cassettes were cloned into the SalI site of pUR7981, resulting in plasmid pUR7981-SX (Fig. 1). The A. awamori bipA gene, derived from plasmid pAWBiP (34), was adapted and cloned into pAW14B12, resulting in pUR7987 (35). For selection, pAWBiP and pUR7987 were provided with the hygromycin resistance expression cassette derived from pAW15-7 (10), resulting in pUR7381 and pUR7988, respectively (35).
FIG. 1.
Schematic diagram of the construction of cutinase variant gene (cutiX) integration vectors. pcutiX is a model expression vector, in which cutiX is regulated by the exlA (endoxylanase II) promoter (P) and terminator (T). pUR7981 is a derivative of pJB8 containing the pyrG gene with a defined SalI mutation, pyrG*. For random integration, more than four cutiX expression cassettes were cloned into the HindIII site of pUR7981, resulting in pUR7981-HX. Targeted integration at the pyrG locus was performed with pUR7981-SX, with pUR7981 containing two cutiX expression cassettes at the SalI site.
Protein analysis.
Screening of cutinase variant transformants for cutinase activity was carried out with BYPO plates containing an olive oil-arabic gum emulsion (8), with previously described adjustments (33). The formation of clear zones surrounding the colonies was used as an indication of active enzyme production. Shake flask induction experiments were performed as described by Gouka et al. (10) by using 5% d-xylose as an inducer. In these analyses, the single-copy cutinase strain AW85 (33) containing a wild-type cutinase expression cassette at the pyrG locus was used as a control strain. The amount of cutinase produced extracellularly 42 h after induction was measured spectrophotometrically by using p-nitrophenyl butyrate (PNPB) (Sigma) as the substrate as described by Van Gemeren et al. (32). In the PNPB measurements, standard amounts of all of the purified variant cutinases except L51S produced similar increases in absorbance, indicating that the specific activities with this substrate were similar. L51S was not produced in sufficient quantities to be tested in the purified form in the PNPB assay, but the activities of crude preparations were comparable to those of the other enzymes.
Intracellular cutinase was analyzed after washing of the mycelium with Triton X-100, disruption, and sodium dodecyl sulfate (SDS) treatment of the mycelium as described by Van Gemeren et al. (33). The samples were subjected to Western blot analysis (27) by using a cutinase-specific polyclonal antiserum raised in rabbits. The amounts of BiP protein present in the different strains were determined by a Western blot analysis of mycelial extracts by using an Aspergillus niger BiP-specific antiserum raised in rabbits (35).
DNA and RNA procedures.
Mycelium samples were taken 15 or 22 h after induction with d-xylose in shake flask experiments. A. awamori chromosomal DNA and total RNA were isolated from mycelial powder as described by Kolar et al. (17). A Northern blot analysis with a Hybond membrane (Amersham International, Little Chalfont, Buckinghamshire, England) was performed by using standard procedures (27). In the slot blot analysis of DNA and RNA, twofold dilutions were blotted onto a GeneScreen Plus membrane (Dupont NEN Products, Boston, Mass.) by using a Milliblot-S apparatus (Millipore Corp., Bedford, Mass.). The blots were hybridized with the ApaI-AflII cutinase fragment or the XhoI bipA fragment (34). The coding region of the glyceraldehyde-3-phosphate dehydrogenase (gpdA) gene of A. niger (1.4-kb HindIII fragment from pAB5-2 [22]) was used as an internal control for the amount of nucleic acid blotted. The multiprime DNA labelling system (Amersham International) was used to label the DNA probes with 32P. Hybridization signals were quantified with an InstantImager (Packard, Canberra, Australia). Transformants containing the bipA expression vectors were identified by colony hybridization (16) by using the internal XhoI bipA fragment as a probe.
Metabolic labelling and immunoprecipitation.
Cutinase-producing strains were cultured in shake flasks, and 5-ml samples were taken 22 h after induction with 5% d-xylose. Each mycelium sample was filtered over Miracloth (Calbiochem, La Jolla, Calif.), washed with 5 ml of minimal medium without yeast extract (MM-Y), and induced with 5 ml of this medium containing 2.5% d-xylose. The mycelium was shaken for 1 h at 30°C and 125 rpm, and then 20 to 30 μCi of 35S-labelled methionine-cysteine (1,000 Ci/mmol; Pro-mix; Amersham) per ml was added. After 20 min, the mycelium was washed again with MM-Y, suspended in MM-Y containing xylose, and 25 mM methionine and 25 mM cysteine were added. One-milliliter samples were taken at intervals and kept on ice during further treatments. The medium was removed by centrifugation, and the mycelium was washed with ice-cold phosphate buffer (50 mM Na2HPO4/NaH2PO4 [pH 7.0], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The mycelium samples were disrupted in 1 ml of solubilization buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% SDS, 0.5% Triton X-100, 1 mM EDTA, 1 mM PMSF) by adding 0.8 g of acid-washed glass beads and vortexing three times for 30 s in glass vials. The disrupted mycelium samples were incubated for 5 min at 94°C in 2-ml Eppendorf tubes and centrifuged for 10 min at 14,000 × g and 4°C. The supernatant collected was designated the soluble intracellular protein fraction.
Immunoprecipitation of the labelled cutinase was performed with a solution containing 200 μl of the soluble intracellular protein fraction, 200 μl of immunoprecipitation buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Tween 20, 1 mM EDTA, 1 mM PMSF), and 25 μl of cutinase-specific polyclonal antiserum. The samples were rotated at room temperature for 2 h, 30 μl of protein A-Sepharose CL-4B (0.14 g/ml; Sigma) was added, and the mixture was incubated for an additional 2 to 16 h. Subsequently, the samples were centrifuged for 10 min at 14,000 × g and 4°C. The immunoprecipitates were washed with immunoprecipitation buffer, and 10 μl of SDS sample buffer was added. Extracellular proteins were precipitated by adding 15 μl of 50% (vol/vol) trichloroacetic acid to 200 μl of extracellular medium, incubating the preparation on ice for 30 min, and centrifuging it for 10 min at 14,000 × g and 4°C. The precipitates were washed with 200 μl of ice-cold acetone and resuspended in 10 μl of sample buffer. Prior to SDS-polyacrylamide gel electrophoresis, the samples were heated at 94°C for 5 min and centrifuged for 1 min. After Coomassie blue staining and destaining, the gels were treated for 15 min with Amplify (Amersham), dried, and exposed to X-ray film for 1 to 2 weeks.
RESULTS
Analysis of the production of cutinase variants by A. awamori.
Six cutinase variants that were poorly secreted by S. cerevisiae were studied further with A. awamori (Table 1). The expression of the genes was controlled by the inducible exlA promoter (10). In an earlier study, twofold more cutinase was produced by strains containing a cassette with the mature cutinase region fused directly to the exlA presequence than by strains containing constructs with the cutinase prosequence in addition to the exlA presequence (33). Similar cassettes expressing cutinase variants were cloned in a tandem array into cosmids and were randomly integrated into the genome of A. awamori. As a result, the locus was unknown, but the cassettes were integrated in a defined head-to-tail manner.
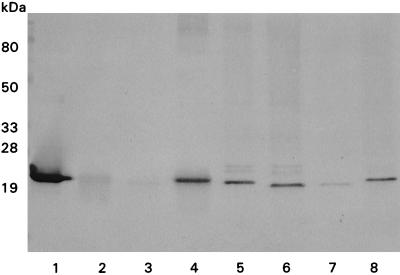
Transformants were prescreened for cutinase activity with a plate assay and with shakeflask induction experiments. The transformants with the highest cutinase activities were selected for determinations of the numbers of expression cassettes and the cutinase-specific mRNA levels (Table 2). The extracellular cutinase protein levels determined by hydrolysis of PNPB were in agreement with results obtained by Western blotting (Fig. 2, lanes 1 through 4), indicating similar immunogenic reactivities.
TABLE 2.
Expression and production of cutinase variants by A. awamori
| Strain | Mutation(s) | Cutinase gene copy no. | Level of cutinase mRNAa | Cutinase concn (mg/liter)b |
|---|---|---|---|---|
| AW85 | 1 | 1 | 14 ± 3 | |
| AW91 | 2 | 2–5 | 74 ± 19 | |
| AW23 | L51S | 2 | 2–5 | 6 ± 2 |
| AW91-16 | 10–15 | 5–10 | 55 ± 3 | |
| AW23-16 | L51S | 2–5 | 2–5 | 1 ± 1 |
| AW20 | E201K | 5–10 | 5–10 | 51 ± 6 |
| AW28 | G82A, A85F, V184I, A185L, L189F | 5–10 | 2–5 | 66 ± 10 |
| AW34 | A29−, S30− | 2–5 | 2–5 | 62 ± 18 |
| AW48-15 | T179Y | 5–10 | 2–5 | 25 ± 11 |
| AW48-04 | T179Y | 15–30 | 2–5 | 6 ± 2 |
| AW55 | W69Y | 10–15 | 5–10 | 50 ± 14 |
mRNA level relative to the mRNA level in single-copy strain AW85, which was defined as 1.
Mean ± standard deviation from two or more independent shake flask induction experiments. Enzyme levels were obtained by relating the PNPB activities of samples to the activity of a purified wild-type cutinase sample having a known concentration.
FIG. 2.
Western blot analysis of cutinase variant production by A. awamori after 42 h of induction with 5% d-xylose. For analysis of extracellular cutinase production a fraction of the culture medium was used (lanes 1 through 4), and intracellular fractions were obtained after SDS treatment of pulverized mycelia (lanes 5 through 8). The following transformants were analyzed: AW91 (lanes 1 and 5), AW23 (lanes 2 and 6), AW23-16 (lanes 3 and 7), and AW48-15 (lanes 4 and 8).
The production of four of the cutinase variants by A. awamori transformants AW20-02/09 (E201K), AW28 (G82A, A85F, V184I, A185L, and L189F), AW34 (A29- and S30-), and AW55-01/05 (W69Y) was comparable to the production of the wild-type cutinase by AW91-06. The protein levels of cutinase variant T179Y produced by transformants AW48-04 and AW48-15, however, were considerably lower than the wild-type protein level. Moreover, there was almost no extracellular production of cutinase variant L51S by transformant AW23-16. To facilitate a precise comparison of the production of this cutinase variant with wild-type cutinase production, two cassettes containing the wild-type gene or the L51S cutinase mutant alleles were cloned into a cosmid and integrated at the pyrG locus. The resulting defined transformants, AW91 and AW23, exhibited similar levels of transcription of the cutinase cDNA (Table 2). The level of extracellular production of L51S cutinase by AW23 was more than 10-fold lower than the level of wild-type cutinase production by AW91. This result is comparable to the results obtained with the randomly transformed counterparts AW91-6 and AW23-16 (Table 2). Western blot analysis of the culture media from these strains revealed patterns similar to the activity measurement patterns (Fig. 2, lanes 1 through 4). The amounts of intracellular cutinase produced by AW91, AW23, AW23-16, and AW48-15 were determined by Western blot analysis of SDS-treated, pulverized mycelia (Fig. 2, lanes 5 through 8). Assuming that there were no differences in translational efficiency, this analysis revealed that there is little intracellular accumulation of the wild-type and variant cutinases in A. awamori.
Effect of overexpression of bipA.
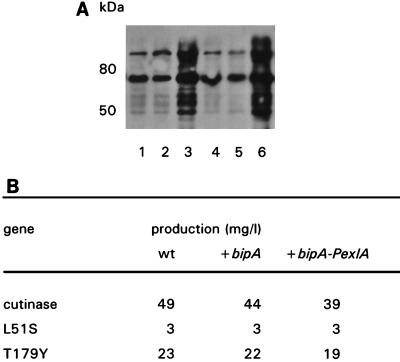
To study whether an increase in the level of the chaperone protein BiP could improve the secretion of the cutinase variants, the A. awamori bipA gene (34) controlled either by its own promoter or by the exlA expression signals was introduced into the wild-type or cutinase variant-producing transformants AW91, AW23, and AW48-15. Northern blot analysis revealed a significant increase in the A. awamori bipA mRNA in transformants of AW91, AW23, and AW48-15 containing multiple copies of the bipA gene (results not shown). The A. awamori BiP protein level at 38 h after induction, however, was increased only in transformants of AW91 and AW23 containing extra bipA copies controlled by the exlA promoter (Fig. 3A, lanes 3 and 6, respectively). The 75-kDa band corresponds to the intact protein, which could have formed a dimer under nonreducing conditions via one S-S bridge, resulting in the more slowly migrating band. The more rapidly migrating bands presumably represent degradation products of BiPA (12). Strains containing extra copies of the A. awamori bipA gene with its own promoter did not contain increased BiPA levels (Fig. 3A, lanes 2 and 5), presumably because of negative feedback regulation on the transcription-translation level (35). The elevated A. awamori BiP levels did not result in higher cutinase production levels. Production of the wild-type cutinase even decreased slightly (Fig. 3B).
FIG. 3.
Effect of overexpression of bipA in A. awamori. (A) Western blot analysis of BiP protein levels by using BiP-specific antiserum of mycelial extracts from the following transformants: AW91 (lane 1), AW91 containing bipA (under control of the natural promoter) (lane 2), AW91 containing bipA-PexlA (under control of the exlA promoter) (lane 3), AW23 (lane 4), AW23 containing bipA (lane 5), and AW23 containing bipA-PexlA (lane 6). (B) Effect of overexpression of bipA on extracellular cutinase production. Averages of the values from three independent shake flask induction experiments are shown. wt, wild type.
Metabolic labelling analysis of cutinase production.
Secretion of wild-type and variant cutinases by A. awamori was analyzed by metabolic labelling experiments performed with [35S] methionine-cysteine. The mycelial wet weights and protein patterns on SDS gels were identical for all strains at specific time points, indicating that the cultures grew equally and induction of protein synthesis was the same in all cultures.
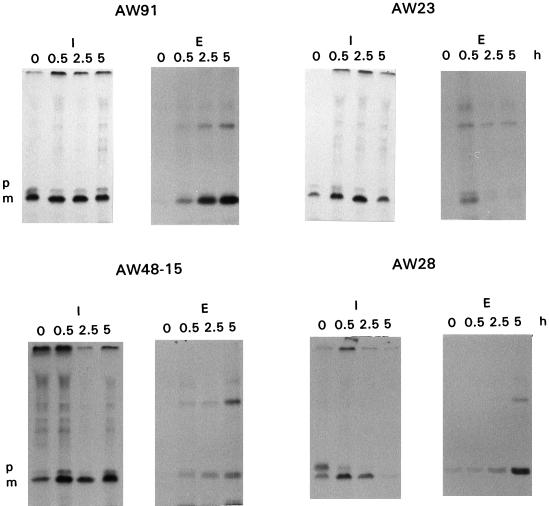
Analysis of cutinase production by AW91 expressing the wild-type cutinase showed that the increase in extracellular cutinase levels over time was not reflected by a similar decrease in intracellular cutinase levels (Fig. 4). This suggests that labelled cutinase was synthesized even after removal of the radiolabelled substrate, probably by ongoing translation of cutinase-specific mRNA with labelled methionine and cysteine which had formed an intracellular storage pool. Cycloheximide was not added to the cultures to arrest translation because this treatment could also have blocked the production of necessary chaperones.
FIG. 4.
Pulse-labelling analysis of strains producing cutinase variants. Samples were taken at several time points during the chase. These samples were subjected to immunoprecipitation with cutinase-specific antiserum and separated by SDS-polyacrylamide gel electrophoresis and autoradiography was performed. The following transformants were analyzed: AW91, AW23, AW48-15, and AW28. Representative results from one of three analyses are shown. Abbreviations: I, intracellular fractions; E, extracellular fractions; p, precutinase; m, mature cutinase.
Transformant AW23 expressing cutinase variant L51S exhibited reduced intra- and extracellular cutinase levels compared to AW91 (Fig. 4). The extracellular cutinase level of AW23 increased slightly for 0.5 h and then declined rapidly (Fig. 4). This result is consistent with extracellular proteolytic degradation of the L51S variant. To analyze the possibility that production of L51S induced synthesis of extracellular proteases, purified wild-type cutinase was added to AW23 culture medium. The cutinase was not degraded after 24 h of incubation at 30°C, suggesting that there was no increased induction of extracellular protease production and that this variant has increased proteolytic susceptibility compared with that of the wild-type cutinase.
The cutinase variant T179Y production pattern of AW48-15 resembled that of wild-type strain AW91 (Fig. 4). Nevertheless, the extracellular cutinase level was significantly lower than the extracellular cutinase level in AW91. The possibility that extracellular proteolytic breakdown occurred was excluded, as purified T179Y was stable for 24 h at 30°C in the culture medium of wild-type A. awamori or of AW48-15. This suggests that there was an intracellular bottleneck in the production of variant T179Y.
A surprising result was obtained with transformant AW28, which contained a gene encoding a cutinase variant with multiple mutations. Although this variant was produced at wild-type levels after 42 h (Table 2), the pattern of intracellular cutinase production differed from that of the other cutinase variants (Fig. 4). The intracellular fractions of AW91 and AW48 contained two forms of cutinase, a 22.2-kDa precursor with a prepeptide and the mature 20.6-kDa form. In most cases, the precursor form was present at a low level (10%) compared to the mature cutinase and stayed visible over the entire 5-h experiment. In the intracellular fraction of AW28, 66% of the cutinase was present in the preform at zero time (Fig. 4). After 0.5 and 2.5 h of chase, 11 and 1% of the intracellular cutinase contained a prepeptide, respectively. In addition, the level of the intracellular mature cutinase form decreased more rapidly in AW28 than in the other strains (Fig. 4).
DISCUSSION
Previous studies have shown that the cutinase cDNA from F. solani pisi is efficiently expressed by the industrial hosts S. cerevisiae and A. awamori (32, 33). Protein-engineering studies have been performed to obtain a more stable enzyme with greater performance in detergents. In these studies, about 20% of the cutinase variants were not produced efficiently by S. cerevisiae (unpublished results) (Table 1). In this report, we describe the production of several of these variants by A. awamori.
Most of the cutinase variants were produced efficiently by A. awamori. However, two variants, L51S and T179Y, were produced at low levels, and the bottleneck was shown to be located after transcription. The possibility that there was less efficient translation of the transcripts encoding these two variants cannot be excluded. However, according to the general codon usage bias of aspergilli (20; unpublished results), the alterations of L51S (TCT) and T179Y (TAC) do not require the use of rare tRNAs. The drastic effect of the substitution of one amino acid on protein production has been described earlier for bacterial, yeast, and mammalian cells (15, 24, 36). When A. awamori glucoamylase was expressed in yeast, amino acid deletions resulted in both intracellular accumulation and higher susceptibility to proteolytic degradation of this protein (2). Protein aggregation and intracellular accumulation often are observed during heterologous protein production in S. cerevisiae (26, 28). In general, filamentous fungi do not exhibit pronounced accumulation (11, 33), and this was also the case with the cutinase variants.
The extracellular level of variant L51S produced by A. awamori AW23 declined rapidly. The L51S mutation was designed to obtain more compatibility with anionic surfactants by distortion of the hydrophobic patch near the active site. A serine located at the surface of a molecule frequently introduces flexibility, resulting in a more open structure. This could have rendered the cutinase molecule more susceptible to extracellular proteases.
The slow extracellular accumulation of variant T179Y by strain AW48-15 was not due to extracellular proteolytic breakdown. The T179Y substitution probably causes increased sensitivity to intracellular proteolytic enzymes. As the rationale for generating this cutinase variant was to increase hydrophobicity, it is conceivable that the intracellular form of the protein aggregates transiently, is recognized as foreign, and subsequently is rapidly degraded. The variant protein showed a slight improvement in performance on lipid substrates and enhanced stability with anionic detergents (unpublished results). Disruption of protease-encoding genes, which has been shown to be a useful approach for optimization of protein production by aspergilli (1, 4), could improve the production of cutinase variants L51S and T179Y.
In contrast to S. cerevisiae (26), A. awamori AW28 produced the cutinase variant with five amino acid substitutions at a level comparable to that of the wild type. The hydrophobicity of the surface around the active site has been increased in this variant, resulting in greater stability of the enzyme and better washing performance (26). On the other hand, this increased hydrophobicity could have caused reduced efficiency in the translocation process across the endoplasmic reticulum membrane. This problem appears to be solved by the fungus, possibly with the aid of chaperones. However, overexpression and overproduction of the gene encoding the BiP protein from A. awamori (34) did not stimulate production of the wild-type cutinase or of variants L51S and T179Y, indicating that the level of this chaperone is not a major limiting factor in the secretion of these proteins. Further studies will focus on the effect of the BiP concentration on the production of other secretory proteins by Aspergillus strains.
ACKNOWLEDGMENTS
C. Hjort (Novo Nordisk, Bagsværd, Denmark) is acknowledged for providing the preliminary methods used for metabolic labelling. C. Visser and L. van Schie (Unilever Research, Vlaardingen, The Netherlands) are thanked for providing cutinase PCR fragments. J. W. Kalhorn is thanked for construction of the strains with extra bipA gene copies.
This project was supported by SENTER, a program of the Dutch Ministry of Economical Affairs.
REFERENCES
- 1.Archer D B, Mackenzie D A, Jeenes D J, Roberts I N. Proteolytic degradation of heterologous proteins expressed in Aspergillus niger. Biotechnol Lett. 1992;14:357–362. [Google Scholar]
- 2.Baker Libby C, Cornett C A G, Reilly P J, Ford C. Effect of amino acid deletions in the O-glycosylated region of Aspergillus awamori glucoamylase. Protein Eng. 1994;7:1109–1114. doi: 10.1093/protein/7.9.1109. [DOI] [PubMed] [Google Scholar]
- 3.Bennet J W, Lasure L L. More gene manipulations in fungi. San Diego, Calif: Academic Press; 1991. Growth media; p. 445. [Google Scholar]
- 4.Broekhuijsen M P, Mattern I E, Contreras R, Kinghorn J R, Van den Hondel C A M J J. Secretion of heterologous proteins by Aspergillus niger: production of active human interleukin-6 in a protease deficient mutant by KEX2-like processing of a glucoamylase-HIL6 fusion protein. J Biotechnol. 1993;31:135–145. doi: 10.1016/0168-1656(93)90156-h. [DOI] [PubMed] [Google Scholar]
- 5.Egmond, M. R., H. T. Van der Hijden, W. Musters, H. Peters, C. T. Verrips, and J. De Vlieg. July 1994. Eukaryotic cutinase variant with increased lipolytic activity. Unilever patent WO 9414963.
- 6.Egmond, M. R., H. T. Van der Hijden, W. Musters, H. Peters, C. T. Verrips, and J. De Vlieg. July 1994. Eukaryotic cutinase variant with improved lipolytic activity. Unilever patent WO 9414964.
- 7.Ettinger W F, Thukral S K, Kolattukudy P E. Structure of cutinase gene, cDNA, and the derived amino acid sequence from phytopathogenic fungi. Biochemistry. 1987;26:7883–7892. [Google Scholar]
- 8.Frenken L G J, Egmond M R, Batenburg A M, Bos J W, Visser C, Verrips C T. Cloning of the Pseudomonas glumae lipase gene and determination of the active site residues. Appl Environ Microbiol. 1992;58:3787–3791. doi: 10.1128/aem.58.12.3787-3791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouka R J, Hessing J G M, Stam H, Musters W, Van den Hondel C A M J J. A novel strategy for the isolation of defined pyrG mutants and the development of a site-specific integration system for Aspergillus awamori. Curr Genet. 1995;27:536–540. doi: 10.1007/BF00314444. [DOI] [PubMed] [Google Scholar]
- 10.Gouka R J, Hessing J G M, Punt P J, Stam H, Musters W, Van den Hondel C A M J J. An expression system based on the promoter region of the Aspergillus awamori 1,4-β-endoxylanase A gene. Appl Microbiol Biotechnol. 1996;46:28–35. doi: 10.1007/s002530050779. [DOI] [PubMed] [Google Scholar]
- 11.Gouka R J, Punt P J, Hessing J G M, Van den Hondel C A M J J. Analysis of heterologous protein production in defined recombinant Aspergillus awamori strains. Appl Environ Microbiol. 1996;62:1951–1957. doi: 10.1128/aem.62.6.1951-1957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmsen M M, Bruyne M I, Raué H A, Maat J. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosine but not plant thaumatin in yeast. Appl Microbiol Biotechnol. 1996;46:365–370. doi: 10.1007/BF00166231. [DOI] [PubMed] [Google Scholar]
- 13.Hedstrom L, Graf L, Steward C-B, Rutter W J, Phillips M A. Modulation of enzyme specificity by site directed mutagenesis. Methods Enzymol. 1991;202:671–687. doi: 10.1016/0076-6879(91)02031-4. [DOI] [PubMed] [Google Scholar]
- 14.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahn M J, Kieber-Emmons T, Vilaire G, Murali R, Poncz M, Bennett J S. Effect of mutagenesis of GPIIb amino acid 273 on the expression and conformation of the platelet integrin GPIIb-IIIa. Biochemistry. 1996;35:14304–14311. doi: 10.1021/bi961702x. [DOI] [PubMed] [Google Scholar]
- 16.Kinsey J A. A simple colony blot procedure for Neurospora. Fungal Genet Newsl. 1989;36:45–46. [Google Scholar]
- 17.Kolar M, Punt P J, Van den Hondel C A M J J, Schwab H. Transformation of Penicillium chrysochenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene. 1988;62:127–134. doi: 10.1016/0378-1119(88)90586-0. [DOI] [PubMed] [Google Scholar]
- 18.Kolattukudy P E. Cutinases from fungi and pollen. In: Borgström B, Brockman H, editors. Lipases. Amsterdam, The Netherlands: Elsevier; 1984. pp. 471–504. [Google Scholar]
- 19.Lee S C, Olins P O. Effect of overproduction of heat shock chaperones GroESL and DnaK on human procollagenase production by Escherichia coli. J Biol Chem. 1992;267:2849–2852. [PubMed] [Google Scholar]
- 20.Lloyd A T, Sharp P M. Codon usage in Aspergillus nidulans. Mol Gen Genet. 1991;230:288–294. doi: 10.1007/BF00290679. [DOI] [PubMed] [Google Scholar]
- 21.Martinez C, de Geus P, Lauwereys M, Matthysens G, Cambillau C. Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature. 1992;356:615–618. doi: 10.1038/356615a0. [DOI] [PubMed] [Google Scholar]
- 22.Punt P J, Dingemanse M A, Jacobs-Meijsing B J M, Pouwels P H, Van den Hondel C A M J J. Isolation and characterisation of the glyceraldehyde-3-phosphate dehydrogenase gene of Aspergillus nidulans. Gene. 1988;69:49–57. doi: 10.1016/0378-1119(88)90377-0. [DOI] [PubMed] [Google Scholar]
- 23.Punt P J, Van den Hondel C A M J J. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 1992;216:447–457. doi: 10.1016/0076-6879(92)16041-h. [DOI] [PubMed] [Google Scholar]
- 24.Rad M R, Katz H. Retention of a co-translational translocated mutant protein of carboxypeptidase Y of Saccharomyces cerevisiae in endoplasmic reticulum. FEMS Microbiol Lett. 1993;111:165–170. doi: 10.1111/j.1574-6968.1993.tb06380.x. [DOI] [PubMed] [Google Scholar]
- 25.Robinson A S, Hines V, Wittrup K D. Protein disulphide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Bio/Technology. 1994;12:381–384. doi: 10.1038/nbt0494-381. [DOI] [PubMed] [Google Scholar]
- 26.Sagt C M J, Müller W H, Boonstra J, Verkleij A J, Verrips C T. Impaired secretion of a hydrophobic cutinase by Saccharomyces cerevisiae correlates with an increased association with BiP. Appl Environ Microbiol. 1998;64:316–324. doi: 10.1128/aem.64.1.316-324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Smith R A, Duncan M J, Moir D T. Heterologous protein secretion from yeast. Science. 1985;229:1219–1224. doi: 10.1126/science.3939723. [DOI] [PubMed] [Google Scholar]
- 29.Soliday C L, Flurkey W H, Okita T W, Kolattukudy P E. Cloning and structure determination of cDNA for cutinase, an enzyme involved in fungal penetration of plants. Proc Natl Acad Sci USA. 1984;82:3939–3943. doi: 10.1073/pnas.81.13.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweigard J A, Chumley F G, Valent B. Cloning and analysis of CUT1, a cutinase gene from Magnaporta grisea. Mol Gen Genet. 1992;232:174–182. doi: 10.1007/BF00279994. [DOI] [PubMed] [Google Scholar]
- 31.Van der Hijden, H. T., J. D. Marugg, J. F. Warr, J. Klugkist, W. Musters, and D. H. A. Hondmann. February 1994. Enzyme-containing surfactants compositions. Unilever patent WO 9403578.
- 32.Van Gemeren I A, Musters W, Van den Hondel C A M J J, Verrips C T. Construction and heterologous expression of a synthetic copy of the cutinase cDNA from Fusarium solani pisi. J Biotechnol. 1995;40:155–162. doi: 10.1016/0168-1656(95)00041-n. [DOI] [PubMed] [Google Scholar]
- 33.Van Gemeren I A, Beijersbergen A, Musters W, Gouka R J, Van den Hondel C A M J J, Verrips C T. The effect of pre- and pro-sequences and multi-copy integration on heterologous expression of the Fusarium solani pisi cutinase gene in Aspergillus awamori. Appl Microbiol Biotechnol. 1996;45:755–763. doi: 10.1007/s002530050759. [DOI] [PubMed] [Google Scholar]
- 34.Van Gemeren I A, Punt P J, Drint-Kuijvenhoven A, Broekhuijsen M P, Van’t Hoog A, Beijersbergen A, Verrips C T, Van den Hondel C A M J J. The ER chaperone encoding bipA gene of black Aspergilli is induced by heat shock and unfolded proteins. Gene. 1997;198:43–52. doi: 10.1016/s0378-1119(97)00290-4. [DOI] [PubMed] [Google Scholar]
- 35.Van Gemeren, I. A., P. J. Punt, A. Drint-Kuijvenhoven, J. G. M. Hessing, M. Van Muijlwijk, A. Beijersbergen, C. T. Verrips, and C. A. M. J. J. Van den Hondel. Analysis of the role of the major ER-chaperone encoding gene bipA in the secretion of homologous and heterologous proteins in black Aspergilli. Submitted for publication. [DOI] [PubMed]
- 36.Wetzel R. Mutations and off-pathway aggregation of proteins. TIBTECH. 1994;12:193–198. doi: 10.1016/0167-7799(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 37.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]