Abstract
Rhodococcus sp. strain AD45 was isolated from an enrichment culture on isoprene (2-methyl-1,3-butadiene). Isoprene-grown cells of strain AD45 oxidized isoprene to 3,4-epoxy-3-methyl-1-butene, cis-1,2-dichloroethene to cis-1,2-dichloroepoxyethane, and trans-1,2-dichloroethene to trans-1,2-dichloroepoxyethane. Isoprene-grown cells also degraded cis-1,2-dichloroepoxyethane and trans-1,2-dichloroepoxyethane. All organic chlorine was liberated as chloride during degradation of cis-1,2-dichloroepoxyethane. A glutathione (GSH)-dependent activity towards 3,4-epoxy-3-methyl-1-butene, epoxypropane, cis-1,2-dichloroepoxyethane, and trans-1,2-dichloroepoxyethane was detected in cell extracts of cultures grown on isoprene and 3,4-epoxy-3-methyl-1-butene. The epoxide-degrading activity of strain AD45 was irreversibly lost upon incubation of cells with 1,2-epoxyhexane. A conjugate of GSH and 1,2-epoxyhexane was detected in cell extracts of cells exposed to 1,2-epoxyhexane, indicating that GSH is the physiological cofactor of the epoxide-transforming activity. The results indicate that a GSH S-transferase is involved in the metabolism of isoprene and that the enzyme can detoxify reactive epoxides produced by monooxygenation of chlorinated ethenes.
Perchloroethene and trichloroethene (TCE) have been widely used as solvents and degreasing agents, and improper disposal and spillage of these compounds have frequently resulted in contamination of groundwater. Dechlorination reactions occurring in situ under anaerobic conditions may result in the accumulation of cis-1,2-dichloroethene (cis-1,2-DCE) and vinyl chloride at contaminated locations (34). There is great interest in biological methods for treatment of sites that are contaminated with these compounds. Under aerobic conditions, vinyl chloride has been shown to serve as a growth substrate (14), whereas for TCE and dichloroethenes only cometabolic degradation has been reported. Conversion of chlorinated ethenes has been described for organisms that produce dioxygenases or monooxygenases with broad substrate ranges (3, 7, 12, 20, 23, 36, 37).
Oxidation of chlorinated ethenes by monooxygenases results in the formation of epoxides (12, 16, 32). These electrophilic compounds are unstable in aqueous solutions. The reactivities of the epoxides and their degradation products often result in covalent modification of cellular components, causing toxic effects (12, 23, 33). Consequently, the amount of chlorinated ethene that can be converted by cells is limited, and continuous-treatment systems may be unstable (11, 22). Since the toxicity that is associated with oxidative cometabolic degradation of chlorinated ethenes is the main limiting factor for the application of monooxygenase-expressing organisms, it is desirable to find ways to biologically detoxify reactive transformation products.
Theoretically, enzymatic conversion of TCE epoxide or dichloroethene epoxides to nonreactive products may decrease the toxic effects, but information about the microbial conversion of these compounds is scarce. Degradation of vinyl chloride by Mycobacterium aurum L1 proceeds via chloroepoxyethane, but the enzyme(s) that converts this epoxide has not been characterized (14). Cells of Methylosinus trichosporium OB3b expressing soluble methane monooxygenase converted cis-1,2-dichloroepoxyethane, but rapid inactivation occurred during this transformation, indicating that even products that are more toxic were generated (32, 33).
Epoxides occur in the degradation pathways for many unsaturated aliphatic compounds (5, 6, 15, 38), and the epoxide-converting enzymes in organisms utilizing these compounds as growth substrates may also exhibit activity with chlorinated epoxyethanes. Indeed, the presence of epoxide-transforming enzymes has been proposed as an explanation for the decreased toxicity of trichloroethene for isoprene-utilizing bacteria (8).
Isoprene is emitted by bacteria, fungi, animals, and plants in large amounts (26). Rates of isoprene synthesis increase under thermal stress conditions (27), when isoprene may function as a stabilizing agent for biological membranes (26). Trees may emit about 2% of the carbon assimilated as isoprene. The global emission of isoprene is estimated to be about 3 × 1014 g year−1, which is roughly equal to the global methane emission (4). Isoprene is a reactive compound compared to other atmospheric hydrocarbons due to the presence of two unsaturated bonds, and it plays a role in ozone formation via a series of photochemical reactions (28). However, despite its important role in atmospheric chemistry, little is known about the microbial degradation of this compound (9, 31).
In this paper we report the isolation and characterization of an isoprene-utilizing organism that dechlorinates cis-1,2-dichloroepoxyethane. Furthermore, we show that a glutathione (GSH) S-transferase is involved in epoxide metabolism in this organism.
MATERIALS AND METHODS
Growth conditions.
In all batch experiments MMY medium was used (30). Stock solutions of carbon sources were sterilized with a 0.2-μm-pore-size filter. The organic solvents that were tested as carbon sources were found to be sterile. Batch cultures were grown at 30°C in 100-ml or 1- or 3-liter serum flasks filled to one-fourth their volume with medium, and the flasks were incubated with rotary shaking (200 rpm). Growth was monitored by measuring the turbidity at 450 nm with a Hitachi model 100-60 spectrophotometer.
Batch enrichments were carried out at 30°C as described previously (30). Pure cultures were obtained by repeated streaking onto MMY agar plates that were incubated in a desiccator with isoprene in the gas phase. The organisms were maintained on 0.8% nutrient broth agar plates.
In continuous culture, the organisms were grown at a dilution rate of 0.026 h−1 in MMY medium to which extra (NH4)SO4 (0.5 g liter−1), MgSO4 (0.2 g liter−1), and yeast extract (20 mg liter−1) were added. All components except phosphate buffer were sterilized separately to prevent the formation of precipitates. Cells were grown in a continuous culture at a dilution rate of 0.026 h−1. The pH was regulated continuously by titration with 1 N NaOH. The growth substrate was added by bubbling air (flow rate, 4.1 ml min−1) through a flask containing isoprene kept on ice. Other conditions were as follows: working volume, 2.35 liters; temperature, 30°C; impeller speed, 1,175 rpm; and airflow rate, 29.2 ml min−1. This resulted in a steady state in which the cell density was 2.1 mg ml−1, the growth yield was 0.54 g of cells g of isoprene−1, and the dissolved oxygen concentration was 7 to 10% of air saturation.
Identification of strain AD45.
Taxonomic identification was carried out by workers at the LMG Culture Collection (University of Ghent, Ghent, Belgium), who used fatty acid analysis and a metabolic fingerprint.
For analysis of the 16S rRNA gene, genomic DNA was isolated from a 2-ml overnight culture grown on nutrient broth. Ampicillin 200 (μg ml−1) and lysozyme 100 (μg ml−1) were added, and the culture was incubated for 2 h at 30°C. After centrifugation (10,000 × g, 10 min), 5 μl of 1 M Tris-chloride (pH 8.4), 1 μl of 0.5 M EDTA (pH 8.0), and 5 μl of 5 M NaCl were added. Lysozyme was added to a final concentration of 2 mg ml−1, and the cells were incubated at 37°C for 2 h. The cells were lysed by adding 80 μl of 10% sodium dodecyl sulfate, followed by overnight incubation at 65°C. After 60 μl of 3 M sodium acetate (pH 7.0) was added, the lysate was incubated at 65°C. After 60 μl of 3 M sodium acetate (pH 7.0) was added, the lysate was incubated at 65°C for another 2 h. The DNA was purified and isolated by standard phenol-chloroform extraction and ethanol precipitation methods (25). The 16S rRNA gene was analyzed by PCR amplification of a ca. 1,320-bp fragment of the 16S rRNA gene. Amplification, sequencing, and sequence analysis were carried out as described by Marchesi and coworkers (19) by using the data bank and analysis tools of the Ribosomal Database Project (18).
Degradation experiments with cell suspensions.
Experiments to examine degradation of all compounds except cis-1,2-dichloroepoxyethane were carried out with cells grown in batch cultures on 2 mM isoprene. Cells were centrifuged and resuspended to a density of 0.2 mg ml−1. Substrate was added, and degradation was monitored as described previously (30). The kinetics of degradation of isoprene and cis-1,2-DCE were determined by headspace analysis. In the case of cis-1,2-DCE, 1 mM sodium succinate was added as a reductant. Substrate depletion was monitored by analyzing seven headspace samples over a period of 15 min. The kinetic parameters were estimated by transforming the data by the method of Hanes Woolf as described by Oldenhuis et al. (23).
For cis-1,2-dichloroepoxyethane degradation experiments, strain AD45 cells grown in continuous culture were centrifuged (10,000 × g, 10 min) and resuspended to a density of 40 mg ml−1 in MMY medium. Inactivation with 1,2-epoxyhexane was carried out by incubating cells with 1 mM 1,2-epoxyhexane for 15 min at 30°C. The cells were washed twice with MMY medium to remove excess 1,2-epoxyhexane. Activities were measured by monitoring substrate concentrations by on-line gas chromatography essentially as described by Van Hylckama Vlieg et al. (32), with some minor modifications. A 38-ml incubation vessel which contained 7 ml of a vigorously stirred cell suspension was used. Gas was continuously withdrawn from the headspace, and after passage through a 35-μl sample loop it was reinjected into the headspace, since reinjection in the water phase resulted in excessive foam formation at the high cell densities that were used. At 1-min time intervals, the contents of the sample loop were injected into the gas chromatograph and analyzed isothermally at 90°C. Assays were started by adding cis-1,2-dichloroepoxyethane from a 50 mM stock solution in 10 mM sodium phosphate buffer (pH 7.0). Gas chromatography-mass spectrometry was performed as described by Van Hylckama Vlieg et al. (32). To determine chloride levels, parallel incubations were carried out, from which 300-μl samples were removed at different times. The samples were rapidly chilled on ice and centrifuged (15,000 × g, 1 min) to remove the cells. Each supernatant (200 μl) was lyophilized to remove excess cis-1,2-dichloroepoxyethane. Water (200 μl) was added, and chloride levels were determined by the method of Bergmann and Sanik (3a).
Preparation of cell extracts and enzyme assays.
The cells used to prepare cell extracts were harvested from late-exponential-phase batch cultures or from continuous cultures. After centrifugation (15 min, 10,000 × g), the cells were resuspended in 50 mM Tris-HCl buffer (pH 7.5) (Tris buffer). All subsequent steps were carried out at 0 to 4°C. The cells were washed twice with Tris buffer before they were resuspended in 3 volumes of 10 mM Tris buffer containing 1 mM β-mercaptoethanol and 1 mM EDTA (TEM buffer). The cells were disrupted by sonication (4 ml, 250-W output) 10 times for 10 s with 1-min intervals to cool the suspension on ice. A cell extract was obtained by centrifugation (60 min, 40,000 × g).
GSH S-transferase activities were assayed at 30°C in 50 mM Tris-Cl buffer (pH 8.5) (assay buffer) containing substrate at a concentration of 5 mM (all substrates except cis- and trans-1,2-dichloroepoxyethanes) or 1 mM (cis- and trans-1,2-dichloroepoxyethanes) and 5 mM of glutathione (GSH), which was added from a 0.5 M stock solution in assay buffer. Substrate depletion was monitored by on-line gas chromatography. A dimensionless Henry coefficient for cis-1,2-dichloroepoxyethane of 0.011 was used to calculate activities (32). The following dimensionless Henry coefficients for the other epoxides were determined as described previously (32): epoxypropane, 0.007; 1,2-epoxyhexane, 0.02; and 3,4-epoxy-3-methyl-1-butene, 0.02. Chloride liberation in assays performed with cis-1,2-dichloroepoxyethane was monitored by removing 300-μl samples from parallel incubation mixtures as described above. Samples were quenched with 1% H2O2 to avoid nonenzymatic reaction of GSH with cis-1,2-dichloroepoxyethane. Activities were expressed in units per milligram of protein. One unit was defined as the activity that catalyzed the conversion of 1 μmol of substrate per min.
Detection of GSH-epoxide conjugates in deproteinized cell extracts.
An isoprene-grown cell suspension (18 mg ml−1) harvested from a continuous culture was divided into two equal portions. One portion was incubated with 1 mM 1,2-epoxyhexane at room temperature for 15 min, and the other was used as a control. All subsequent steps were carried out at 0 to 4°C. The cells were centrifuged and washed twice before they were resuspended in 6 ml of 10 mM potassium phosphate buffer (pH 7.0). Lysozyme and EDTA were added to final concentrations of 0.17 mg ml−1 and 1 mM, respectively, and after 1 h the cells were disrupted by sonication. Deproteination of cell extracts was carried out essentially as described by Fahey et al. (10). The extracts were lyophilized and resuspended in 200 ml of water. Proteins were precipitated by adding HCl to a final concentration of 0.1 N, and after vortexing the precipitates were removed by centrifugation at 15,000 × g for 10 min. The supernatants were mixed with an equal volume of 4 M sodium methanesulfonate, and the mixtures were frozen in liquid nitrogen. After warming, the insoluble portions were removed by centrifugation at 15,000 × g.
The GSH-epoxyhexane conjugate that was used as a reference was synthesized with partially purified enzyme in a standard enzyme assay by using 5 mM 1,2-epoxyhexane and 10 mM GSH. After 90% of the 1,2-epoxyhexane was converted, the sample was lyophilized to remove the remaining epoxide.
The GSH-epoxide conjugates present in the supernatants were analyzed by reversed-phase high-performance liquid chromatography (HPLC) with a Merck Hitachi model L-6200A system equipped with a Lichrosorb 5C18 column (20 by 4.6 mm) and a Merck Hitachi model L4000 UV detector. For data acquisition a Merck Hitachi model D-2500 Chromato-Integrator was used. The buffer system consisted of 0.1% trifluoroacetic acid in water (buffer A) and 0.1% trifluoroacetic acid in acetonitrile (buffer B). The following elution protocol was used: 0 to 5 min, 0% isocratic buffer B; 5 to 75 min, 0 to 67% buffer B linear gradient; 75 to 80 min, 67 to 100% buffer B linear gradient (column regeneration). The elution profile was obtained by measuring the absorbance at 214 nm.
Chemicals.
Both cis-1,2-dichloroepoxyethane and trans-1,2-dichloroepoxyethane were synthesized with M. trichosporium OB3b as described previously (32). The concentrations in the stock solutions that were obtained were determined by overnight hydrolysis in 50 mM KOH at 80°C and subsequent determination of chloride levels. Other chemicals were obtained from AGA Gas B.V. (Amsterdam, The Netherlands), Acros Organics (’s-Hertogenbosch, The Netherlands), or Aldrich (Milwaukee, Wis.).
RESULTS
Degradation of halogenated ethenes by isoprene-utilizing cultures.
Four different pure cultures capable of growing with isoprene as the sole source of carbon and energy were isolated from freshwater sediment. The doubling times in liquid media containing isoprene as a growth substrate ranged from 3.5 to 16 h. All strains were gram positive and nonfermentative and differed in colony morphology and color during growth on nutrient broth agar plates. Suspensions of washed cells (0.2 mg ml−1) prepared from batch cultures grown on isoprene were tested for the ability to degrade TCE and trans-1,2-DCE. The culture with the highest growth rate on isoprene, designated strain AD45, showed the highest activity with chlorinated ethenes, as judged by gas chromatography and the amount of chloride released.
We also tested whether enrichment with other compounds having isoprene-like structures would result in the isolation of new cultures that are capable of cometabolic degradation of chlorinated ethenes. Four mixed cultures were isolated with 3-methyl-3-butene-1-ol, 2-methyl-3-butene-2-ol, 2-methyl-2-butene, and 2-methyl-2-pentene and were grown for 10 days on one of these substrates in the presence of 200 μM TCE or 200 μM trans-1,2-DCE. None of the cultures degraded TCE or trans-1,2-DCE, and no chloride release was detected.
All subsequent experiments were carried with strain AD45, since this culture showed the highest activity with halogenated ethenes. Strain AD45 was tentatively identified as a Rhodococcus sp. based on fatty acid analysis and a metabolic fingerprint. Analysis of the 16S rRNA gene sequence revealed that the closest organisms as determined by similarity rank (Sab, 0.850 to 0.942) were all Rhodococcus spp., and the highest score was obtained with Rhodococcus globerulus (18).
In batch cultures strain AD45 grew on isoprene, 3,4-epoxy-3-methyl-1-butene, phenol, 3-methyl-3-butene-1-ol, 3-methyl-1-butanol, glycerol, 1,2-propanediol, ethanol, and glucose. This organism did not grow on 2,3-dimethyl-2-butene, 3,3-dimethyl-1-butene, 2-methyl-2-pentene, 2-methyl-2-butene, benzene, toluene, 3-methyl-2-butene-1-ol, 2-methyl-3-butene-1-ol, 1-pentene, 1-hexene, epoxypropane, 1,2-epoxybutane, glycolate, glyoxylate, citrate, allylalcohol, or methanol.
Identification of the primary oxidation products of isoprene, cis-1,2-DCE, and trans-1,2-DCE.
A concentrated cell suspension (7.5 mg ml−1) harvested from a continuous culture was used to identify the primary oxidation product of isoprene. When a pulse of isoprene was added, which resulted in a concentration of isoprene in the liquid phase of 3 mM, accumulation of a product was observed. The retention time of this product during gas chromatography was identical to the retention time of 3,4-epoxy-3-methyl-1-butene. This compound was well separated from 3,4-epoxy-2-methyl-1-butene, which is the other epoxide that can be generated by the oxidation of isoprene. A mass spectrometry analysis of the primary oxidation product and commercially available 3,4-epoxy-3-methyl-1-butene revealed the presence of ions with m/z (relative intensity of the primary oxidation product, relative intensity of standard 3,4-epoxy-3-methyl-1-butene) 39 (100, 100), 55 (74, 74), 29 (68, 41), 43 (63, 74), 53 (50, 56), 41 (44, 29), 27 (40, 36), 56 (31, 17), 83 (23, 23), 54 (22, 30), 69 (22, 22), 50 (18, 18), 51 (15, 18), 26 (12, 9), 84 (molecular ion) (6, 6), and 38 (5, 8). Thus, the compound was identified as 3,4-epoxy-3-methyl-1-butene.
In analogous experiments with cis-1,2-DCE and trans-1,2-DCE products accumulated that were identified as the corresponding epoxides as described previously (32).
Kinetics of biodegradation.
The kinetics of degradation of isoprene and cis-1,2-DCE by cell suspensions of Rhodococcus sp. strain AD45 were determined. The Km for isoprene conversion was 0.8 μM, and the Vmax was 76 nmol min−1 mg of cells−1. The Km for cis-1,2-DCE was 63 μM, and the Vmax was 8 nmol min−1 mg of cells−1.
The cells also degraded toluene, styrene, and propylene. The rate of toluene oxidation was 10 nmol min−1 mg of cells−1 at a concentration of 73 μM in the medium. Propylene was converted to epoxypropane. Degradation of toluene and cis-1,2-DCE was inhibited by isoprene, indicating that these compounds compete for the same active site.
Conversion of cis-1,2-dichloroepoxyethane by cell suspensions.
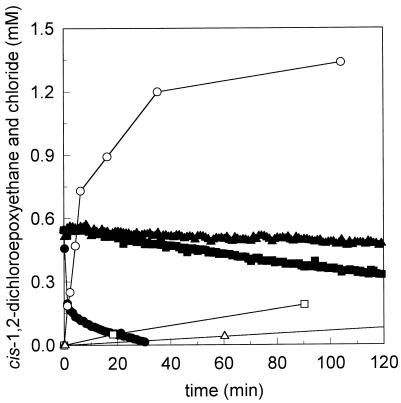
Cell suspensions harvested from a continuous culture were used to test whether Rhodococcus sp. strain AD45 can degrade cis-1,2-dichloroepoxyethane. All organic chlorine was liberated as chloride during degradation of cis-DCE epoxide (Fig. 1). The following two different stages of epoxide degradation were distinguished: a first stage (from zero time to 2 min), during which approximately 0.3 to 0.32 mM epoxide was rapidly degraded; and a second stage (after 2 min), during which degradation proceeded at a lower rate, 0.12 nmol min−1 mg of cells−1.
FIG. 1.
Conversion of cis-1,2-dichloroepoxyethane by cell suspensions of Rhodococcus sp. strain AD45 (40 mg [dry weight] ml−1). The depletion of cis-1,2-DCE (solid symbols) and the generation of chloride (open symbols) were monitored with time. Incubations were carried out with active cells (circles), 1,2-epoxyhexane-inactivated cells (squares), and heat-killed cells (triangles).
No degradation of cis-1,2-dichloroepoxyethane was observed with heat-killed cells. Cells that were treated with 1,2-epoxyhexane almost completely lost cis-1,2-dichloroepoxyethane-degrading activity.
Epoxide-degrading activity in cell extracts.
We prepared cell extracts to determine which type of enzymes was involved in epoxide conversion. A GSH-dependent specific activity of 5.4 U mg of protein−1 towards 3,4-epoxy-3-methyl-1-butene was observed in extracts of isoprene-grown cultures. GSH could not be replaced by other thiols, such as cysteine, lipoic acid, or coenzyme A. No activity was detected in assays for epoxide dehydrogenase (6), epoxide isomerase (15), or epoxide hydrolase (24). In the presence of GSH, other epoxides were also converted by the cell extracts. The specific activities were 2.5 U mg−1 with epoxypropane, 0.3 U mg−1 with cis-1,2-dichloroepoxyethane, and 0.5 U mg−1 with trans-1,2-dichloroepoxyethane.
The GSH-dependent activities in cell extracts of cultures grown on other carbon sources were also determined. A specific activity of 2.1 U mg−1 with 3,4-epoxy-3-methyl-1-butene was detected in an extract of a culture grown on 3,4-epoxy-3-methyl-1-butene. In an extract of glucose-grown cells the activities were 0.1 U mg−1 with 3,4-epoxy-3-methyl-1-butene, 0.1 U mg−1 with epoxypropane, and less than 0.05 U mg−1 with cis-1,2-dichloroepoxyethane. With ethanol-grown cells the specific activities were 0.3 U mg−1 with 3,4-epoxy-3-methyl-1-butene, 0.2 U mg−1 with epoxypropane, and less than 0.05 U mg−1 with cis-1,2-dichloroepoxyethane. No activity was detected with extracts from cells grown on 3-methyl-3-butene-1-ol or 2-methyl-butanol. The results indicated that an inducible GSH S-transferase is involved in epoxide conversion.
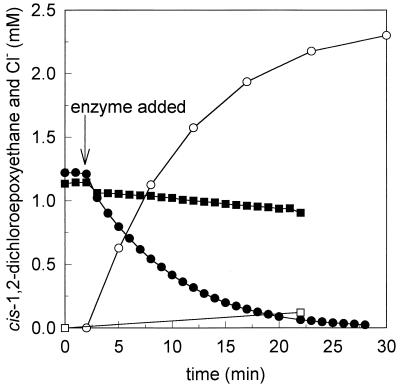
A sample of partially purified enzyme (33a) was used to study the conversion of cis-1,2-dichloroepoxyethane. All organic chlorine was released as chloride. This activity was absent with heat-killed enzyme (Fig. 2).
FIG. 2.
Conversion of cis-1,2-dichloroepoxyethane by partially purified GSH S-transferase from Rhodococcus sp. strain AD45. Depletion of cis-1,2-DCE (solid symbols) and generation of chloride (open symbols) were determined in incubations with active enzyme (0.08 mg ml−1) (circles) or heat-inactivated enzyme (squares).
Accumulation of GSH–1,2-epoxyhexane conjugate in 1,2-epoxyhexane-inactivated cells.
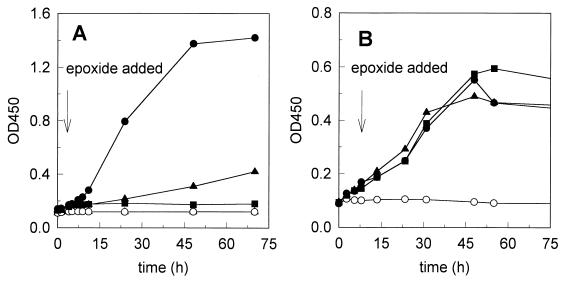
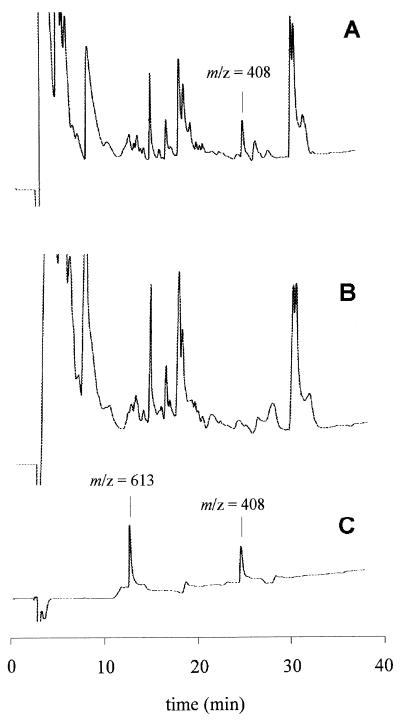
Epoxide degradation by cell extracts of Rhodococcus sp. strain AD45 was dependent on the addition of GSH. However, we could not exclude the possibility that in vivo another thiol may act as a cofactor, as has been observed with other epoxide-degrading organisms. For instance, GSH was not the physiological cofactor in the metabolism of epoxypropane by Xanthobacter sp. strain Py2 despite that fact that after GSH was added, activity could be detected (39). We also observed that addition of low concentrations of 1,2-epoxybutane and 1,2-epoxyhexane strongly inhibited growth on isoprene but not growth on ethanol (Fig. 3) and irreversibly inhibited the epoxide-degrading activity of cell suspensions (Fig. 1). Since the epoxide-transforming GSH-dependent enzyme exhibited activity with various epoxides, the inhibition observed may have been caused by the accumulation of nonmetabolizable conjugates of thiol and 1,2-epoxyhexane. Therefore, we analyzed cell extracts prepared from 1,2-epoxyhexane-inactivated cells for the presence of such conjugates. In HPLC traces obtained with deproteinized extracts of inactivated cells, a compound eluting at 26 min was detected which was absent in traces of extracts of active cells (Fig. 4). The retention time of this compound was identical to that of the GSH–1,2-epoxyhexane conjugate that was synthesized with partially purified GSH S-transferase. Analysis by HPLC-mass spectrometry showed that the two peaks represented a compound with a molecular mass (m/z) of 408, which is consistent with the theoretical value for the protonated molecular ion. Analysis of the sample prepared with partially purified enzyme also revealed the presence of a compound eluting at 13 min with m/z 613, which is in agreement with the theoretical value for the protonated molecular ion oxidized GSH (GSSG). This compound may be generated by autooxidation from excess GSH during sample preparation (1). From Fig. 4, traces A and C, we calculated that approximately 3 nmol of GSH was present per mg (dry weight) of cells, assuming that all intracellular GSH was converted to the conjugate.
FIG. 3.
Toxicity of 1,2-epoxybutane and 1,2-epoxyhexane for Rhodococcus sp. strain AD45 growing on 0.9 mM isoprene (A) or 4 mM ethanol (B). Symbols: •, no epoxide added; ▴, 0.1 mM 1,2-epoxybutane added; ■, 0.1 mM 1,2-epoxyhexane added; ○, no growth substrate. OD450, optical density at 450 nm.
FIG. 4.
Formation of the conjugate of GSH and 1,2-epoxyhexane in cell suspensions of strain AD45. (A and B) HPLC profiles recorded by measuring the absorbance at 214 nm of a deproteinized cell extract prepared from a cell suspension that was inactivated with 1,2-epoxyhexane (A) or from a suspension that was not inactivated (B). (C) Control (GSH–1,2-epoxyhexane conjugate synthesized with partially purified GSH S-transferase). The m/z values (408 and 613 Da) are in agreement with the theoretical values for the protonated molecular ions of the conjugate of GSH and 1,2-epoxyhexane and for oxidized GSH (GSSG), respectively.
DISCUSSION
We isolated a Rhodococcus sp. that utilizes the important environmental hydrocarbon isoprene as a sole source of carbon and energy. Previously, Van Ginkel et al. (30) reported the isolation of several isoprene-utilizing strains that were tentatively identified as Nocardia sp. strains. Suspensions of these organisms that were inactivated with 1,2-epoxybutane accumulated both 3,4-epoxy-3-methyl-1-butene and the diepoxide (1,2-3,4-diepoxy-butane). However, Rhodococcus sp. strain AD45 accumulated mainly 3,4-epoxy-3-methyl-1-butene, indicating that these strains have different metabolic features.
The results show that in strain AD45 isoprene degradation starts with the oxidation of the sterically most hindered double bond, resulting in the formation of 3,4-epoxy-3-methyl-1-butene. The metabolism of isoprene in strain AD45 is similar to the metabolism of isoprene in mammals. In liver microsomes of various rodent species, for instance, the methyl-substituted double bond rather than the unsubstituted bond is oxidized by cytochrome P-450 (13).
An inducible GSH-dependent activity towards 3,4-epoxy-3-methyl-1-butene was detected in cell extracts of Rhodococcus sp. strain AD45, suggesting that a GSH S-transferase is involved in the metabolism of isoprene. This activity was also detected with cis- and trans-1,2-dichloroepoxyethanes and epoxypropane. The accumulation of a GSH-epoxyhexane conjugate in cells poisoned with 1,2-epoxyhexane indicates that GSH is the physiological cofactor for the epoxide-transforming enzyme. The level of conjugate accumulation corresponds to a GSH concentration of approximately 3 nmol mg (dry weight) of cells−1 or a concentration of 2 mM in the cytoplasm. This concentration is very high for nocardioform actinomycetes, whereas similar values have been reported for Streptococcus, Enterococcus, and various gram-negative species (21). Previously, Ewers and Knackmuss (9) described a GSH-dependent activity towards 3,4-epoxy-3-methyl-1-butene in cell extracts of an isoprene-utilizing Rhodococcus sp., but the enzyme responsible for this activity has not been further characterized.
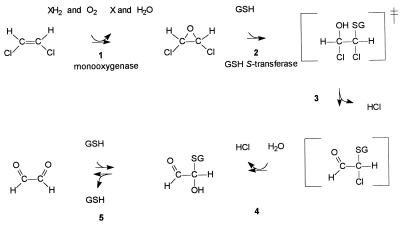
The monooxygenase involved in isoprene metabolism in strain AD45 also exhibits activity with chlorinated ethenes and toluene. Oxidation of cis-1,2-DCE and trans-1,2-DCE resulted in the formation of the corresponding epoxides, as has been found with other organisms that express monooxygenases (3, 7, 12, 20, 23, 36, 37). These epoxides were also converted by cell suspensions of strain AD45, but the transformation rates for cis-1,2-dichloroepoxyethane were approximately 70-fold lower than the transformation rates for cis-1,2-DCE. Both 1,2-dichloroepoxyethanes are substrates for the GSH S-transferase. Reaction of cis-1,2-dichloroepoxyethane with GSH resulted in complete liberation of organic chlorine as chloride, indicating that no toxic halogenated metabolites were generated. This suggests that there is a pathway in which, after nucleophilic attack of GSH, an unstable product is formed that nonbiologically decomposes to glyoxal (Fig. 5). Analysis of glyoxal by osazone formation with 2,4-dinitrophenylhydrazine failed due to the presence of GSH. HPLC analysis also did not reveal the generation of a stable GSH conjugate. The dechlorination in reaction step 4 (Fig. 5) is analogous to the dechlorination by nonenzymic hydrolysis of S-chloromethyl GSH. The latter compound is the product of nucleophilic displacement with dichloromethane, a reaction that is catalyzed by dichloromethane dehalogenases (17). In aqueous solution, 2-oxoaldehydes occur in nonhydrated, monohydrated, and even dihydrated forms (29). Nonhydrated 2-oxoaldehydes rapidly react with GSH to form a hemithioacetal. The best-studied compound in this respect is methylglyoxal, which is generated in vivo from glyceraldehyde 3-phosphate and dihydroxyacetone phosphate by phosphate elimination. Under physiological conditions in erythrocytes, for instance, only 0.04% of the methylglyoxal exists as the 2-oxoaldehyde and 41% exists as the hemithioacetal, whereas the rest is present in hydrated form. The two-stage degradation of cis-1,2-dichloroepoxyethane (Fig. 1) may be caused by decreased free GSH concentrations due to accumulation of the hemithioacetal of glyoxal.
FIG. 5.
Proposed pathway for the degradation of cis-1,2-DCE in Rhodococcus sp. strain AD45. Reactions 3 to 5 are nonbiological reactions postulated on the basis of the observation that all organic chlorine was liberated as chloride during degradation of cis-1,2-dichloroepoxyethane and on the basis of previously published data. The dechlorination in reaction 4 is analogous to the nonbiological dechlorination of S-chloromethyl GSH during dichloromethane degradation (17). In aqueous solutions, 2-oxoaldehydes, such as glyoxal, also occur in hydrated form. Nonhydrated glyoxal is chemically in equilibrium with GSH (29).
Bacterial conversion of epoxides is a topic that has drawn interest from people working on biocatalysis and biodegradation of organic compounds. A wide range of enzymes are involved in microbial metabolism of epoxides; these enzymes include epoxide isomerases, carboxylases, dehydrogenases, hydrolases, reductases, and lyases (5, 6, 15, 24, 38, 39). However, activity with chloroepoxyethanes has not been reported for any of these enzymes.
Apart from the GSH-dependent activity described here, the only bacterial activity with 1,2-dichloroepoxyethane that has been reported is the activity of M. trichosporium OB3b. The soluble methane monooxygenase of this organism has activity with cis-1,2-dichloroepoxyethane but not with trans-1,2-dichloroepoxyethane. However, this activity results in extreme toxicity since monooxygenase activity and the viability of cells were significantly inhibited (31, 32).
Data on bacterial GSH S-transferases have been reviewed recently (35). Some of these enzymes are associated with the metabolism of aromatic compounds. Other GSH S-transferases are involved in the reductive cleavage of ether bonds in lignin or in reductive or hydrolytic dehalogenation reactions. An enzyme homologous to extradiol dioxygenases catalyzing ring opening in epoxides by GSH is involved in resistance to the antibiotic fosfomycin. Divalent cations are needed for optimal activity of this enzyme (2), unlike the enzyme of strain AD45. Thus, in view of its substrate range, the GSH S-transferase of strain AD45 may be a novel type of GSH S-transferase. The biochemistry and genetics of isoprene degradation are currently being studied.
ACKNOWLEDGMENTS
The work of J.E.T.v.H.V. was financed by grant IOP91204 from the Dutch IOP Environmental Biotechnology Program.
Julian R. Marchesi (School of Pure and Applied Biology, University of Wales, Cardiff, United Kingdom) is acknowledged for performing the 16S rRNA gene sequence analysis, and Piet Wietzes is acknowledged for technical support. C. Margot Jeronimus-Stratingh and Andries P. Bruins (Department of Pharmacy, University of Groningen, Groningen, The Netherlands) are acknowledged for performing the mass spectrometry analysis.
REFERENCES
- 1.Akerboom T P M, Sies H. Assay for glutathione, glutathione disulfide, and glutathione mixed disulfides in biological samples. Methods Enzymol. 1981;77:373–382. doi: 10.1016/s0076-6879(81)77050-2. [DOI] [PubMed] [Google Scholar]
- 2.Arca P, Hardisson C, Suarez J E. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob Agents Chemother. 1990;34:844–848. doi: 10.1128/aac.34.5.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arciero D, Vanelli T, Logan T M, Hooper A B. Degradation of trichloroethylene by the ammonia oxidizing bacterium Nitrosomonas europaea. Biochem Biophys Res Commun. 1989;159:640–643. doi: 10.1016/0006-291x(89)90042-9. [DOI] [PubMed] [Google Scholar]
- 3a.Bergmann J G, Saniko J. Determination of trace amounts of chloride in naphtha. Anal Chem. 1957;29:241–243. [Google Scholar]
- 4.Brasseur G P, Chatfield R B. The fate of biogenic trace gasses in the atmosphere. In: Sharkey T D, Holland E A, Mooney H A, editors. Trace gas emission from plants. San Diego, Calif: Academic Press; 1991. pp. 1–27. [Google Scholar]
- 5.Chion C K, Leak D J. Purification and characterization of two components of an epoxypropane isomerase/carboxylase of Xanthobacter Py2. Biochem J. 1996;319:299–406. doi: 10.1042/bj3190499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Bont J A M, Harder W. Metabolism of ethylene by Mycobacterium E20. FEMS Microbiol Lett. 1980;3:89–93. [Google Scholar]
- 7.Ensign S A, Hyman M R, Arp D A. Cometabolic degradation of chlorinated alkanes by alkene monooxygenase in a propylene-grown Xanthobacter strain. Appl Environ Microbiol. 1992;58:3038–3046. doi: 10.1128/aem.58.9.3038-3046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewers J W C, Freier-Schröder D, Knackmuss H J. Selection of trichloroethylene (TCE) degrading bacteria that resist inactivation by TCE. Arch Microbiol. 1990;154:410–413. doi: 10.1007/BF00276540. [DOI] [PubMed] [Google Scholar]
- 9.Ewers J W C, Knackmuss H-J. Biodegradation of chloroethenes using isoprene as a co-substrate. In: Verachtert H, Verstraete W, editors. Proceedings of the International Symposium on Environmental Biotechnology Oostende. Oostende, Belgium: Royal Flemish Society of Engineers; 1991. pp. 77–83. [Google Scholar]
- 10.Fahey R C, Newton G L, Dorian R, Kosower E M. Analysis of biological thiols at the picomole level based upon derivatization with monobromobimames and separation by cation-exchange chromatography. Anal Biochem. 1981;111:357–365. doi: 10.1016/0003-2697(81)90573-x. [DOI] [PubMed] [Google Scholar]
- 11.Fitch M W, Weisman D, Phelps P, Georgiou G, Speitel G E., Jr Trichloroethylene degradation by Methylosinus trichosporium OB3b mutants in a sequencing biofilm reactor. Water Res. 1996;11:2655–2664. [Google Scholar]
- 12.Fox B G, Borneman J G, Wackett L P, Lipscomb J D. Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry. 1990;29:6419–6427. doi: 10.1021/bi00479a013. [DOI] [PubMed] [Google Scholar]
- 13.Gervasi P G, Longo V. Metabolism and mutagenicity of isoprene. Environ Health Perspect. 1990;86:85–87. doi: 10.1289/ehp.908685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartmans S, de Bont J A M. Aerobic vinyl chloride metabolism in Mycobacterium L1. Appl Environ Microbiol. 1992;58:1220–1226. doi: 10.1128/aem.58.4.1220-1226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmans S, Smits J P, van der Werf M J, Volkering F, De Bont J A M. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter 124X. Appl Environ Microbiol. 1989;55:2850–2855. doi: 10.1128/aem.55.11.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen D B, Grobben G, Hoekstra R, Oldenhuis R, Witholt B. Degradation of trans-1,2-dichloroethene by mixed cultures of methanotrophic bacteria. Appl Environ Biotechnol. 1988;29:392–399. [Google Scholar]
- 17.Leisinger T, Bader R, Hermann R, Schmid-Appert M, Vuilleumier S. Microbes, enzymes and genes involved in dichloromethane utilization. Biodegradation. 1994;5:237–248. doi: 10.1007/BF00696462. [DOI] [PubMed] [Google Scholar]
- 18.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchesi J R, Sago T, Weightman A J, Martin T A, Fry J C, Hiom S J, Wade W G. Design and evaluation of useful bacterium-specific primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson M J K, Montgomery S O, Mahaffery W R, Pritchard P H. Biodegradation of trichloroethylene and involvement of an aromatic pathway. Appl Environ Microbiol. 1987;53:949–954. doi: 10.1128/aem.53.5.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton G L, Arnold K, Price M S, Sherill C, Delcadayre S B, Aharonowitz Y, Cohen G, Davies J, Fahey R, Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1995;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldenhuis R, Janssen D B. Degradation of trichloroethylene by methanotrophic bacteria. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Andover, United Kingdom: Intercept Ltd.; 1993. pp. 121–133. [Google Scholar]
- 23.Oldenhuis R, Oedzes J Y, van der Waarde J J, Janssen D B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991;57:7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rink R, Fennema M, Smids M, Dehmel U, Janssen D B. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Sharkey T D. Isoprene synthesis by plants and animals. Endeavour (Cambridge) 1996;20:74–78. doi: 10.1016/0160-9327(96)10014-4. [DOI] [PubMed] [Google Scholar]
- 27.Sharkey T D, Singsaas E L. Why plants emit isoprene. Nature. 1995;374:769. [Google Scholar]
- 28.Thompson A M. The oxidizing capacity of the earth’s atmosphere; probable past and future changes. Science. 1992;256:1157–1165. doi: 10.1126/science.256.5060.1157. [DOI] [PubMed] [Google Scholar]
- 29.Thornally P J. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van den Wijngaard A J, Prins J, Smal A J A C, Janssen D B. Degradation of 2-chloroethylvinylether by Ancylobacter aquaticus AD25 and AD27. Appl Environ Microbiol. 1993;59:2777–2783. doi: 10.1128/aem.59.9.2777-2783.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Ginkel C G, De Jong E, Tilanus J W R, De Bont J A M. Microbial oxidation of isoprene, a biogenic foliage volatile, and of 1,3-butadiene, an anthropogenic gas. FEMS Microbiol Ecol. 1987;45:275–279. [Google Scholar]
- 32.Van Hylckama Vlieg J E T, de Koning W, Janssen D B. Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl Environ Microbiol. 1996;62:3304–3312. doi: 10.1128/aem.62.9.3304-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Hylckama Vlieg J E T, de Koning W, Janssen D B. Effect of chlorinated ethene conversion on viability and activity of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1997;63:4961–4964. doi: 10.1128/aem.63.12.4961-4964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Van Hylckama Vlieg, J. E. T., and D. B. Janssen. Unpublished data.
- 34.Vogel T M, McCarthy P L. Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions. Appl Environ Microbiol. 1985;49:1080–1083. doi: 10.1128/aem.49.5.1080-1083.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wackett L P, Brusseau G A, Householder S R, Hanson R S. Survey of microbial oxygenases: trichloroethylene degradation by propane-oxidizing bacteria. Appl Environ Microbiol. 1989;55:2960–2964. doi: 10.1128/aem.55.11.2960-2964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wackett L P, Gibson D T. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microbiol. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weijers C A G M, de Haan A, de Bont J A M. Microbial production and metabolism of epoxides. Microbiol Sci. 1988;5:156–159. [PubMed] [Google Scholar]
- 39.Weijers C A G M, Jongejan H, Franssen M C R, de Groot A, de Bont J A M. Dithiol- and NAD-dependent degradation of epoxyalkanes by Xanthobacter PY2. Appl Microbiol Biotechnol. 1995;42:775–781. [Google Scholar]