Abstract
During a bacterial survey of the Huon Estuary in southern Tasmania, Australia, we isolated a yellow-pigmented Pseudoalteromonas strain (class Proteobacteria, gamma subdivision), designated strain Y, that had potent algicidal effects on harmful algal bloom species. This organism was identified by 16S rRNA sequencing as a strain with close affinities to Pseudoalteromonas peptidysin. This bacterium caused rapid cell lysis and death (within 3 h) of gymnodinoids (including Gymnodinium catenatum) and raphidophytes (Chattonella marina and Heterosigma akashiwo). It caused ecdysis of armored dinoflagellates (e.g., Alexandrium catenella, Alexandrium minutum, and Prorocentrum mexicanum), but the algal cultures then recovered over the subsequent 24 h. Strain Y had no effect on a cryptomonad (Chroomonas sp.), a diatom (Skeletonema sp.), a cyanobacterium (Oscillatoria sp.), and two aplastidic protozoans. The algicidal principle of strain Y was excreted into the seawater medium and lost its efficacy after heating. Another common bacterial species, Pseudoalteromonas carrageenovora, was isolated at the same time and did not have these algicidal effects. The minimum concentrations of strain Y required to kill G. catenatum were higher than the mean concentrations found in nature under nonbloom conditions. However, the new bacterium showed a chemotactic, swarming behavior that resulted in localized high concentrations around target organisms. These observations imply that certain bacteria could play an important role in regulating the onset and development of harmful algal blooms.
Historically, the dynamics of marine bacterial and algal populations have been studied largely in isolation. Increasing evidence is now pointing toward a close spatial and temporal association between the two and recently attention has been focused on phagocytosis of bacteria by photosynthetic flagellates (21, 28, 30). In contrast, the importance of inhibitory or predatory bacteria in regulating populations of different algal species has received relatively little attention (9, 11). Some bacteria may selectively promote bloom formation by algal species (13), while other bacteria have algicidal effects and are involved in the termination and decomposition of algal blooms (12). The latter finding has raised the possibility of bacterial control of harmful algal blooms (19). There is little data on the occurrence of marine algicidal bacteria outside Japan, where toxic blooms are frequent events (20), and algicidal bacteria have been isolated during toxic blooms of naked dinoflagellates and raphidophytes (9).
Gymnodinium catenatum (a causative organism of paralytic shellfish poisoning) is thought to have been introduced into southern Tasmania via ballast water after 1973, and in some years it has a severe negative impact on the shellfish industry (16). Previous efforts to understand and predict the seasonal and interannual variability of harmful algal blooms have largely focused on the environmental factors that affect dinoflagellate growth in the water column, notably water temperature, rainfall, and water column stability (16). Rainfall and estuarine flow patterns also largely determine the allochthonous input of dissolved organic matter (DOM), which is a source of organic carbon for bacteria (27) and is possibly involved in micronutrient dynamics that promote G. catenatum growth (3, 6). As part of a study investigating DOM, bacteria, and algal interactions in the Huon Estuary (24), we isolated two bacterial strains that we tested for possible alga-bacterium interactions by using cultures of G. catenatum. Both bacteria appeared to be Pseudoalteromonas species, which are extremely common, slightly halophilic, gram-negative bacteria found in many marine ecosystems. Preliminary observations indicated that one of the strains was extremely toxic towards G. catenatum, while the other was more benign. The aims of this study were (i) to determine the taxonomic identity of the bacteria, (ii) to document by light microscopy the sequence of algal cell lysis after exposure to an algicidal Pseudoalteromonas strain and compare this lysis to the effect of the more benign Pseudoalteromonas species, (iii) to define the minimum bacterial concentrations required for algicidal effects and compare these concentrations to concentrations in natural water samples, and (iv) to investigate the range of potential target organisms for the bacterium.
MATERIALS AND METHODS
Sample collection and environmental variables.
Bacterial sampling was carried out in conjunction with a hydrological study performed by the CSIRO Division of Marine Research of the Huon Estuary, southern Tasmania, Australia. Samples for microbial analyses were transferred from 3-liter Niskin type bottles into 1-liter Nalgene sterile sample bottles which had been rinsed three times with sample water prior to filling. Samples were stored in the dark in an ice chest until processing in the laboratory within 4 h of collection. The culturable bacteria were assayed by filtering 5 ml of a sample water onto Sartorius cellulose nitrate 0.2-μm-pore-size filters (diameter, 47 mm; gridded) which were inverted onto ZoBell agar plates (36). The plates with the filters were incubated at 17°C, and colonies were counted after 2 days. Single-cell clones were obtained by progressive streaking onto ZoBell agar plates (36). The growth rates of the isolates were determined at 17°C with cultures growing in 10% ZoBell nutrient broth made with filtered (pore size, 0.2 μm) 28‰ seawater; cell counts were determined with a Neubauer grid at a magnification of ×1,000 by using a Carl Zeiss Photo Microscope. Growth rates were calculated as described by Stolp (33).
Natural populations of Huon Estuary bacteria were preserved with 1% (vol/vol) electron microscope grade glutaraldehyde and were stored in the dark at 4°C. The bacteria were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) for 30 min, filtered onto black 0.22-μm-pore-size membrane filters, mounted on slides, and counted with a Leitz fluorescent microscope at a magnification of ×1,000 (29) within 3 days. Chromophoric DOM (CDOM) (also known as gelbstoff) is a tracer of river water in coastal zones (5, 8). CDOM was measured by performing fluorescence emission scans with samples that had been filtered (pore size, 0.22 μm; Sartorius Minisart syringe filter) and stored in the dark at 2°C. The scans were made from 400 to 600 nm at 5-nm intervals (emission bandwidth, 7 nm) with a Perkin-Elmer model LS5 luminescence spectrophotometer set to an excitation wavelength of 348 nm (10-nm excitation bandpass) (22). A new 10-mm Starna PMMA disposable cuvette was used for each sample. The height of the CDOM fluorescence emission peak at 435 nm was divided by the area of the Raman scattering peak for water, as described by Determann et al. (5), which resulted in estimates of Raman-normalized fluorescent CDOM (FCDOM) (nanometer−1).
Bacterial identification.
The bacterial strains were identified by PCR amplification of the 16S rRNA gene (4), BLAST analysis (1), and comparison with sequences in the GenBank nucleotide database. Specifically, the 16S rRNA gene from strain Y was amplified by PCR by using primers and conditions previously described by Dobson and Franzmann (7). The amplicons were purified with a QiaQuick PCR purification kit (Qiagen, La Jolla, Calif.). The 16S rRNA sequences were then generated with PRISM dye terminator cycle sequencing ready reaction kits and a model A377 automated DNA sequencer (Applied Biosystems, Foster City, Calif.). The sequence data were manually aligned with Pseudoalteromonas 16S rRNA sequences. PHYLIP, version 3.57c (10), was used to further analyze the sequence data. DNADIST, performed with the maximum-likelihood option, was employed to determine sequence similarities, and NEIGHBOR was used to create a phylogenetic tree. Sequences of the following organisms were utilized in the phylogenetic analysis and were obtained directly from GenBank: Pseudoalteromonas antarctica (GenBank accession no. X98336), Pseudoalteromonas atlantica (X82134), Pseudoalteromonas aurantia (X82135), Pseudoalteromonas carrageenovora (X82136), Pseudoalteromonas citrea (X82137), Pseudoalteromonas denitrificans (X82138), Pseudoalteromonas espejiana (X82143), Pseudoalteromonas haloplanktis subsp. haloplanktis (X67024), Pseudoalteromonas haloplanktis subsp. tetraodonis (X82139), Pseudoalteromonas luteoviolacea (X82144), Pseudoalteromonas nigrefaciens (X82146), Pseudoalteromonas peptidysin (AF007286), Pseudoalteromonas piscicida (X82141), Pseudoalteromonas rubra (X82147), and Pseudoalteromonas undina (X82140).
Transmission electron microscopy of the bacteria was done by using negative staining with uranyl acetate of osmium tetroxide-fixed bacteria which had been grown on ZoBell agar. Grids were examined with a Phillips model CM 100 transmission electron microscope.
Algal cultures.
The biocidal effects of the bacteria were tested by using the algae and protozoans listed in Table 1. Protist cultures were maintained at 17°C with cycles consisting of 12 h of darkness and 12 h of cool white fluorescent light. The media used for bacteria, algae, and protozoans were made by using filtered seawater obtained off the Tasman Peninsula. The salinity of the seawater was adjusted to 28‰ by using Milli-Q-deionized filtered (pore size, 0.22 μm) water. Gymnodinium species were grown in GSe medium (3). Other cultures were grown in standard media listed in Table 1 (14, 15). All dinoflagellate and raphidophyte cultures and the Chroomonas culture were unialgal and had little bacterial contamination. The Oscillatoria sp. and amoebae were grown as a stable (for 4 months) two-species culture, and the Skeletonema, Bodo, and Mantoniella strains were grown as a stable (for 2 months) three-species culture.
TABLE 1.
Algae and protozoans tested for bacterial reactions
| Taxon | Class | Mediuma | Strainb | Strain source and history |
|---|---|---|---|---|
| Gymnodinium catenatum Graham | Dinophyceae | GSe | GCDE08 | S. Blackburn; Derwent, Tasmania, Australia |
| Gymnodinium sanguineum Hirasaki | Dinophyceae | GSe | CS 35 | F. T. Haxo; La Jolla, Calif. |
| Gyrodinium sp. strain 1 | Dinophyceae | GSe | GYPA01 | C. Bolch; Port Aurthur, Tasmania, Australia |
| Gyrodinium sp. strain 2 | Dinophyceae | GSe | GYDE02 | C. Bolch; Derwent, Tasmania, Australia |
| Gyrodinium cf. uncatenum | Dinophyceae | GSe | GyDE01 | C. Bolch; Derwent, Tasmania, Australia |
| Gyrodinium sp. strain 3 | Dinophyceae | GSe | CS 289 | CSIRO; Bathurst Harbour, Tasmania, Australia |
| Alexandrium minutum Halim | Dinophyceae | GSe | AMAD 06 | J. Cannon and S. Blackburn; Port River, South Australia, Australia |
| Alexandrium catenella (Whedon and Kofoid) Balech | Dinophyceae | GSe | ACCS01 | S. Norwood; Sydney Harbour, New South Wales, Australia |
| Prorocentrum mexicanum Tafall | Dinophyceae | GSe | PMAL01 | C. Bolch; Albury, New South Wales, Australia |
| Chattonella marina (Subrahmanyan) Hara & Chihara | Rhaphidophyceae | GSe | CMPtL01 | J. Marshall; Port Lincoln, South Australia, Australia |
| Heterosigma akashiwo (Hada) Hada | Rhaphidophyceae | GSe | CS169 | J. Stauber; West Lake, South Australia, Australia |
| Skeletonema costatum (Greville) Cleve | Bacillariophyceae | Fe20 | SkeH0A | C. Lovejoy; Huon Estuary; this study |
| Mantoniella sp. | Prasinophyceae | Fe20 | ManH0A | C. Lovejoy; Huon Estuary; this study |
| Chroomonas placoidea Butcher | Cryptophyceae | Fe2 | CS200 | Butcher; England; CCAP 978/8 |
| Oscillatoria sp. | Cyanobacteria | F20 | OscH0A | C. Lovejoy; Huon Estuary; this study |
| Bodo sp. | Kinetoplastidea | Fe20 | BodH0A | C. Lovejoy; Huon Estuary; this study |
| Amoebae | Amoebidae | F20 | AmoH0A | C. Lovejoy; Huon Estuary; this study |
Toxicity testing.
To test the bacterial effects on algae and protozoans, bacteria were maintained in 10% ZoBell nutrients in filtered seawater (28‰) broth. At the start of each experiment, 3-ml portions of the inoculating bacterial cultures were preserved in 1% (vol/vol) glutaraldehyde, and the initial concentrations were determined by direct counting at a magnification of ×1,000 by using a Neubauer grid and a Carl Zeiss Photo Microscope. All experiments were carried out in sterile 12- or 24-well tissue culture plates. The first experiment testing for differences between logarithmic-phase and stationary-phase bacterial cultures was carried out by using 12-well plates. One milliliter of a bacterial culture, a filtrate from a culture, or bacterium-free filtered sterile fresh medium was placed in each well and diluted with 2 ml of F2 algal culture medium (14, 15). Subsequently, 0.5 ml of a logarithmic-phase culture of G. catenatum was added. In the second experiment to test the effects of different concentrations of bacteria and bacterial filtrates we used 24-well plates. Filtered fresh GSe medium was placed into wells (0.5 ml in the first well, 0.9 ml in subsequent wells). Initially, 0.5 ml of either a bacterial culture or a filtrate (pore size, 0.22 μm) from a bacterial culture was put into the first well and then diluted by using an automatic pipette. Then 0.2 ml of G. catenatum culture was added. To test for heat liability, 3 ml of a filtrate from a bacterial culture was placed into a Schött bottle and microwaved for 20 s (95°C). The cooled filtrate was added to wells containing G. catenatum as described above. In all cases once the algal culture and test bacteria (or filtrate or medium control) were mixed, the plates were sealed with Parafilm, and reactions to the bacteria were monitored visually by using an inverted Carl Zeiss Axiovert 25 microscope at a magnification of ×100.
In experiments testing the bacterial effects on different algae and protozoans we used 24-well plates. Each well contained 1 ml of algal culture, to which 0.4 or 0.5 ml of a bacterial culture (or filtrate or medium control) was added. The plates were sealed with Parafilm and monitored as described above at a magnification of ×100 or ×400 depending on the size of the organism being tested. Different types of organisms reacted differently to the bacteria; the most severe effect (MSE) was noted along with the time when this effect was first seen in each test well. Several transects of each well were inspected, and the condition of the alga (or other protist) was noted. It took between 10 and 15 min to examine each 24-well plate, and the plates were inspected every 20 to 30 min for the first 6 h and then less frequently over the next 2 days. The MSE on the dinoflagellates in a well was noted and was categorized as follows: lysis (there was rapid leaking, and cell contents were destroyed); disintegrated, (the cell membrane was disrupted, and the internal contents were in disarray); rounded (flagella were not visible, and each cell resembled a temporary resting stage); or ecdysis (a theca was shed [thecate species]). The MSE for other groups are shown in Table 2. A crude test for the toxicity of the algae towards the bacteria was carried out by reinoculating agar plates after 3 days with bacteria from the test wells. A loopful from each well was streaked onto ZoBell agar and examined for growth of the inoculating bacteria. Videos and photographs of the interactions between bacteria and G. catenatum and were taken at magnifications of ×400 and ×1,000 by using a Carl Zeiss Photo Microscope 2 and a Umatic video system.
TABLE 2.
Bacterial effects on a wide range of algae and protozoans
| Test organism | Strain Ya
|
P. carrageenovorab
|
Control actived | ||
|---|---|---|---|---|---|
| MSE (time)c | Recovery | MSE (time)c | Recovery | ||
| Gymnodinium catenatum | Lysis (1 h 50 min) | No | Disintegrated (27 h 40 min) | No | No |
| Gymnodinium sanguineum | Lysis (1 h 30 min) | No | Disintegrated (3 h 40 min) | No | No |
| Prorocentrum mexicanum | Ecdysis (1 h 50 min) | Yes | None | NAe | Yes |
| Heterosigma akashiwo | Lysis (1 h 50 min) | No | None | NA | Yes |
| Chattonella marina | Lysis (1 h 50 min) | No | None | NA | Yes |
| Skeletonema costatum | None | NA | None | NA | Yes |
| Chroomonas placoidea | None | NA | None | NA | Yes |
| Mantoniella sp. | NS (6 h 55 min) | Yes | None | NA | Yes |
| Oscillatoria sp. | None | NA | None | NA | Yes |
| Bodo sp. | None | NA | None | NA | Yes |
| Amoebae | PC (1 h 10 min) | Yes | PC (1 h 30 min) | Yes | Yes |
The initial strain Y concentration was 0.78 × 107 cells per ml.
The initial P. carrageenovora concentration was 1.81 × 107 cells per ml.
NS, not swimming (most individuals had stopped swimming or moving); PC, pseudopodia contracted (pseudopodia were normally extended by unperturbed feeding cells). For an explanation of other MSE see Table 5, footnote c. The times are the times when the MSE were first observed.
The control was considered to have had no effect if the test organism was still active after 2 days.
NA, not applicable.
Nucleotide sequence accession number.
The 16S rRNA sequence of strain Y has been deposited in the GenBank database under accession no. AF030381.
RESULTS
Bacteria and FCDOM.
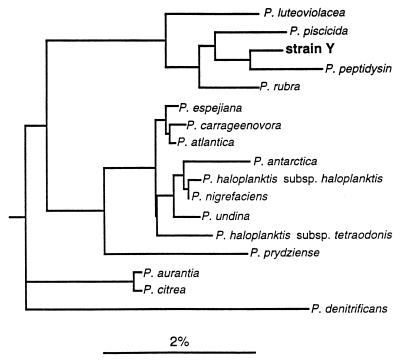
The total numbers of bacteria in different parts of the Huon Estuary (range, 0.4 × 105 to 4.5 × 105 cells ml−1) and observations on bacterial colonies recovered from water samples are shown in Table 3. Less than 1% of the total bacteria counted by using DAPI were culturable. The two bacteria described here were isolated as part of an effort to assay culturable bacteria in the Huon Estuary and to isolate bacterial species to be offered as food in studies on algal phagotrophy. During these studies, it was noted that one of the bacterial strains caused the toxic dinoflagellate G. catenatum to lyse within minutes after small volumes of the two cultures were mixed on a microscope slide. Three types of bacterial colonies from the Huon Estuary were culturable on ZoBell agar (Table 3). The first samples tested (taken on 28 February 1997) were collected from the upper to central portion of the Huon Estuary, where only two types of colonies were observed. The two colony types were isolated in monoclonal cultures and were identified by using 16S rRNA. The first strain closely resembled P. carrageenovora (99.7% similarity; only 2 of 510 bases differed). This bacterium formed light cream-colored colonies which tended to liquefy agar, and transmission electron micrographs showed that the cells were 1.6 by 0.5 μm and had one polar flagellum. The second strain, Pseudoalteromonas sp. strain Y, formed compact yellow colonies, and the cell dimensions as determined by transmission electron microscopy were 1.4 to 1.5 by 0.4 to 0.5 μm. This bacterium exhibited the highest level of 16S rRNA similarity to P. peptidysin (Fig. 1). Further testing is required to determine whether strain Y is a member of a separate species. Both organisms used for subsequent genetic characterization originated from the sample collection on 28 February 1997 at site L1 at a depth of 2 m. The Huon Estuary is a typical saltwedge estuary, and site L1 is located in the upper estuary and was density stratified at the time (24). The influence of freshwater input at this site is shown by the high FCDOM levels relative to other sites (Table 3). There was a positive relationship between the CFU of P. carrageenovora and FCDOM (analysis of variance on regression with r2 = 0.51; P < 0.05). A third bacterium, which formed pink colonies and was not subcultured or identified, was obtained later in the season. The yellow bacterium, strain Y, was not recovered on 15 April 1997 from samples taken from lower in the estuary, when P. carrageenovora and colonies of pinkish cells were present. On 31 May, yellow colonies of strain Y were present in the surface water obtained at site H3 but not at the other sites, while P. carrageenovora and the organism that formed pink colonies were present at all of the sites tested.
TABLE 3.
Results of bacterial surveys of Huon Estuary, Tasmania, Australia, on three dates in summer and autumn
| Date (mo/day/yr) | Sitea | Depth (m) | FCDOM (nm−1) | Concentration of:
|
|||
|---|---|---|---|---|---|---|---|
| DAPI-stained bacteria (cells ml−1)b | P. carrageenovora (CFU ml−1) | Strain Y (CFU ml−1) | Pink bacteria (CFU ml−1)c | ||||
| 2/28/97 | H2 | 2 | 0.28 | 4.52 × 105 | 8.4 | 0.2 | NDd |
| J1 | 2 | 0.10 | 3.98 × 105 | 4.6 | ND | ND | |
| L1 | 2 | 0.33 | 3.47 × 105 | 16.8 | 0.4 | ND | |
| 4/15/97 | H3 | 2 | 0.25 | 2.43 × 105 | 9.4 | ND | 0.4 |
| F3 | 2 | 0.22 | 3.91 × 105 | 8.8 | ND | 0.4 | |
| B1 | 3 | 0.25 | 2.68 × 105 | 2.2 | ND | 0.2 | |
| X3 | 3 | 0.23 | —e | 12.8 | ND | 0.4 | |
| 5/31/97 | H3 | 0 | 0.10 | 2.85 × 105 | 4.0 | 0.8 | 5.6 |
| H3 | 2 | 0.10 | 3.81 × 105 | 3.0 | ND | 3.0 | |
| F3 | 2 | 0.11 | 0.40 × 105 | 9.6 | ND | 0.2 | |
| B1 | 3 | 0.06 | 3.46 × 105 | 0.2 | ND | 2.6 | |
| X3 | 3 | 0.09 | 3.67 × 105 | 3.0 | ND | 2.8 | |
The sites occur up the estuary (from marine to riverine) in the following order: B, F, X, H, J, L. The numbers indicate positions from east to west across the mouth of the estuary, which is oriented predominantly north-south.
Total bacteria.
Unidentified bacteria that form pink colonies.
ND, not detected.
—, no sample.
FIG. 1.
Phylogenetic tree based on 16S rRNA sequences of Pseudoalteromonas species (class Proteobacteria, gamma subdivision), showing the location of strain Y. Bar = sequence dissimilarity of 2%.
Algicidal activity.
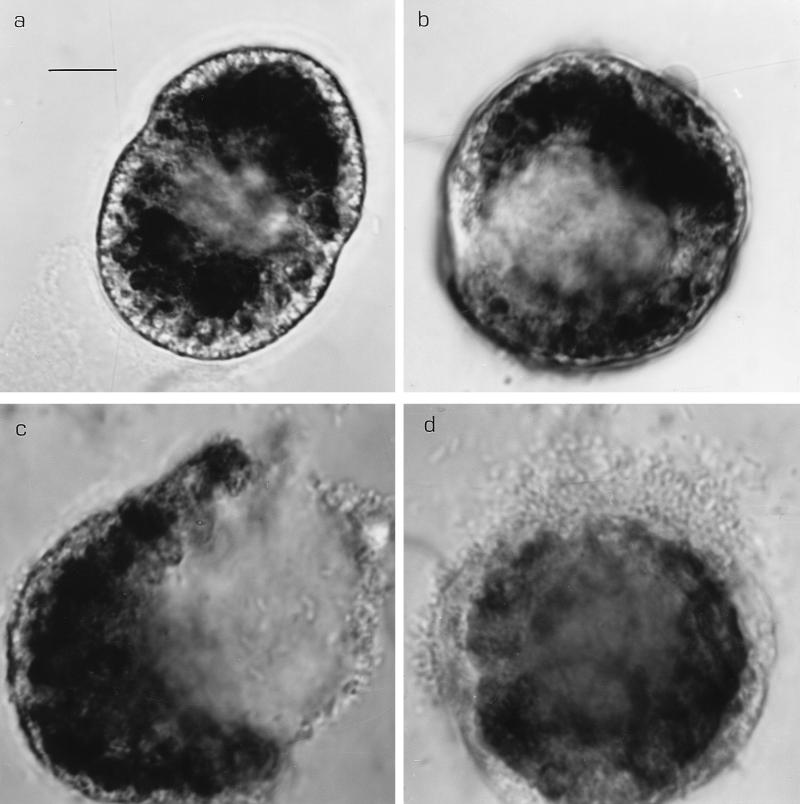
We confirmed the effect of the yellow bacterium on the dinoflagellate G. catenatum by testing logarithmic-phase (16-h-old) and stationary-phase (3-day-old) batch cultures of both Pseudoalteromonas strains. Strain Y logarithmic- and stationary-phase bacteria and filtrates first caused dinoflagellate chains to fall apart, then rounding up of the resulting single cells, swelling, and finally lysis of G. catenatum within 2 to 4 hours under the conditions in the well plates (Table 4). After the cell walls were breached, bacteria from outside the field of view swam towards the lysing cells and entered the cells (mobbing or swarming behavior). Figures 2a and b show ventral and apical views, respectively, of G. catenatum cells in the process of rounding up and swelling 30 to 60 min after the initial exposure to strain Y. Figure 2c shows the same cell as Fig. 2a less than 1 h later, just after lysis. Figure 2d shows the same cell as Fig. 2b following lysis and after the cell was subjected to bacterial swarming. The contents of wells containing filtrates from both logarithmic- and stationary-phase bacterial cultures also caused G. catenatum chains to disintegrate and the cells to swell and lyse. All G. catenatum cells in strain Y-treated wells were dead and most cells had lysed after 2 days. Stationary-phase P. carrageenovora cultures caused many single G. catenatum cells to round up and form temporary resting cysts (3) after 4 h. Some dinoflagellate cells were swarmed upon by large numbers of bacteria, but 24 h later (and also on day 2) most G. catenatum cells were still swimming and intact. There was no change in G. catenatum in wells containing P. carrageenovora filtrate or fresh sterile media (Table 4).
TABLE 4.
MSE on G. catenatum of log-phase and stationary-phase cultures and of filtrates from log-phase cultures over short time intervals
| Time (min) | MSEa
|
||||||
|---|---|---|---|---|---|---|---|
| Strain Yb
|
P. carrageenovorac
|
Control | |||||
| Log-phase culture | Stationary-phase culture | Filtrate | Log-phase culture | Stationary-phase culture | Filtrate | ||
| 10 | Rounded | Rounded | Few rounded | None | None | None | None |
| 90 | Swollen | Swollen | Swollen | None | None | None | None |
| 255 | Lysed | Lysed | Lysed | None | Rounded | None | None |
For an explanation of MSE see the text.
The strain Y log-phase and stationary-phase cultures contained 1.47 × 107 and 2.14 × 107 cells per ml, respectively.
The P. carrageenovora log-phase and stationary-phase cultures contained 3.91 × 107 and 4.85 × 107 cells per ml, respectively.
FIG. 2.
Lytic effect of strain Y on G. catenatum. Approximately 60 min after the bacterium was added, the cells exhibited swelling and separation of chloroplasts from the cell wall region. (a) Ventral view. (b) Apical view of a second cell at approximately the same time. Lysis occurred 30 min later. (c) Ventral view of the cell in panel a after cell lysis. (d) Apical view of the cell in panel b after lysis, showing the swarming of bacteria around the breached cell wall. Note how the cells continued to enlarge up to lysis. Light micrographs (oil immersion). Bar = 10 μm.
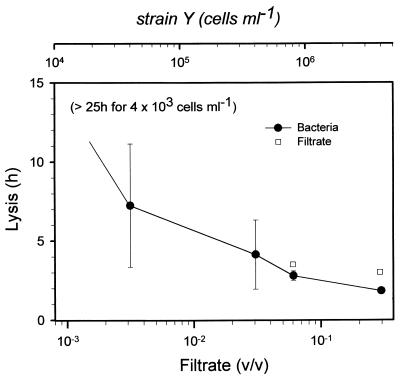
The minimum concentrations of strain Y required to cause lysis in G. catenatum were in the range 103 to 104 cells ml−1 (Fig. 3), assuming that no bacterial growth occurred. The growth rate of strain Y was 0.263 h−1 at 17°C (doubling time, ca. 2 h 40 min). If it is assumed that the bacteria continued to grow in the wells, the concentration would have been ca. 2.7 × 105 cells ml−1 after 7 h 15 min, which was the average time before cell lysis occurred during the 15 h when the experiment was closely monitored. At 25 h a few cells had also lysed in the most dilute wells (initial concentration, 4 × 103 bacteria ml−1). The filtrate was less effective than living bacteria in causing cells to lyse (Fig. 3). No dilution series was prepared with heated filtrate, but there was no response by G. catenatum to the presence of microwaved filtrate at the same concentration as the highest concentration of nonheated filtrate tested.
FIG. 3.
Mean times to lysis of G. catenatum cells in the presence of different bacterial concentrations (top scale). The error bars indicate standard deviations for duplicates of each treatment. The cultures were not monitored between 15 and 25 h after bacteria were added but were checked 48 h after the start of the experiment. All G. catenatum cultures in experimental wells containing bacteria showed mortality after 2 days. Filtrates had no effect on algae at concentrations lower than 0.003 ml of filtrate/ml of algal culture medium (bottom scale). Filtrate which had been microwaved (95°C) caused no mortality.
Species specificity of the algicidal effect.
Two tests of the effect of the bacteria on other protists were conducted. In the first experiment, we tested the bacteria with a number of unarmored Gymnodinium and Gyrodinium dinoflagellate species and two armored Alexandrium spp. (both strains tested are known to produce saxitoxins). The gymnodinoids were all adversely affected and quickly lysed in the strain Y-treated wells (Table 5). G. catenatum, Gymnodinium sanguineum, and two of the Gyrodinium spp. also appeared to be sensitive to P. carrageenovora. In these cases the dinoflagellate cells rounded up but did not exhibit swelling. Many G. sanguineum strains and two Gyrodinium spp. eventually disintegrated under the experimental conditions used, and there were no swimming cells left after 2 days. G. sanguineum and Gyrodinium species strain 2 also appeared to be dead after 2 days in the control wells but did not lyse. The armored dinoflagellates Alexandrium minutum and Alexandrium catenella reacted to strain Y, and many cells shed their thecae over the first few hours and rounded up, forming temporary resting stages. Two days later, however, the cultures appeared to recover, with many cells swimming normally. Some individual Alexandrium cells of both species stopped swimming in the P. carrageenovora-containing wells but recovered after 2 days. Alexandrium cells in control wells were not affected and remained healthy.
TABLE 5.
Results of the first screening experiment performed with log-phase cultures (20-h cultures in 0.1× ZoBell broth [see text])
| Test organism | Strain Ya
|
P. carrageenovorab
|
Control activee | ||
|---|---|---|---|---|---|
| MSE (time)c | Recoveryd | MSE (time)c | Recoveryd | ||
| Gymnodinium catenatum | Lysis (1 h 55 min) | No | Rounded (3 h 50 min) | No | Yes |
| Gymnodinium sanguineum | Lysis (1 h 30 min) | No | Disintegrated (15 min) | No | No |
| Gyrodinium sp. strain 1 | Lysis (3 h 50 min) | No | Rounded (1 h 30 min) | Yes | Yes |
| Gyrodinium cf. uncatenum | Lysis (1 h 55 min) | No | Rounded (3 h 50 min) | Yes | Yes |
| Gyrodinium sp. strain 2 | Lysis (1 h 55 min) | No | Disintegrated (5 h 10 min) | No | No |
| Gyrodinium sp. strain 3 | Lysis (1 h 30 min) | No | Rounded (30 min) | No | Yes |
| Alexandrium minutum | Ecdysis (50 min) | Yes | Rounded (50 min) | Yes | Yes |
| Alexandrium catenella | Ecdysis (50 min) | Yes | Rounded (1 h 30 min) | Yes | Yes |
The initial strain Y concentration was 0.91 × 107 cells per ml.
The initial P. carrageenovora concentration was 2.11 × 107 cells per ml.
Lysis, rapid leaking and cell contents destroyed; disintegrated, cell membrane disrupted and internal contents in disarray; rounded, flagella not visible and each cell resembled a temporary resting stage; ecdysis, theca shed (thecate species). The times are the times when the MSE were first observed.
Swimming cells were present in wells after 2 days.
The control was considered to have had no effect if the test organism was still active after 2 days.
In the second experiment (Table 2) a wide range of protists representing eight major groups (dinoflagellates, raphidophytes, diatoms, cryptomonads, prasinophytes, cyanobacteria, kinoplastidia, and amoebae) were tested. G. sanguineum and G. catenatum were tested again as positive controls and reacted in a fashion similar to that described above. In this experiment, G. catenatum also died after 2 days in the control wells. Both raphidophytes tested, Chattonella marina and Heterosigma akashiwo, reacted quickly to strain Y and lysed after swelling. The chloroplasts of H. akashiwo first leaked out of the lysed cells and then lysed themselves after about 30 min. No effect on raphidophytes was observed in the wells containing P. carrageenovora or in the control wells. Other protist groups did not appear to be adversely affected by the added bacteria, with the following exceptions in the strain Y-treated wells. The dinoflagellate Prorocentrum mexicanum reacted like the Alexandrium spp. in the previous experiment; i.e., they ecdysed but recovered. The small green flagellate Mantoniella sp. ceased swimming after 6 h but also recovered after 2 days. The amoebae tested usually occurred in a free-floating form with radiating pseudopodia, but these contracted soon after bacteria (both strains) were added and then recovered rapidly. After 2 days all pseudopodia were extended normally in both experimental and control wells. Amoebae and bodonids both appeared to capture and ingest the large bacteria. The cryptomonad, Chroomonas placoides, showed visible growth after 3 days in the wells. In both experiments, we recovered colonies of both bacterial types from the respective experimental wells after 3 days, suggesting that there were no antibacterial substances produced by any of the protists tested against the two Pseudoalteromonas spp.
DISCUSSION
Both pseudoalteromonads tested, strain Y and P. carrageenovora, were fatal to several of the gymnodinoid dinoflagellates. P. carrageenovora appeared to stress the gymnodinoids, and G. sanguineum and Gyrodinium species 2 were particularly sensitive. Experimental conditions in the 24-well plates were not optimal for the naked dinoflagellates since there was some mortality in the controls in some experiments after 2 days. However, the impact of strain Y was always rapid, and cells exhibited characteristic swelling prior to lysis (Fig. 2a and b). All Gymnodinium and Gyrodinium species tested were lysed within 4 h by strain Y. The experimental conditions in the well plates, including the small volume and the effect of frequent motion when the wells were inspected, did not adversely affect the raphidophytes; Chattonella and Heterosigma cells remained intact and swimming in both the P. carrageenovora-treated and control wells. However, they were severely affected by strain Y and lysed within 2 h of exposure (Table 2). The effects of strain Y on thecate dinoflagellates, shedding of their thecae and rounding up, may indicate that these organisms have a mechanism that protects them against the bacterial compound produced by Pseudoalteromonas sp. strain Y. The recovery of these organisms indicated that at the concentrations used, the algicidal action was not effective against members of the thecate groups. Strain Y was lethal only to the unarmored gymnodinoid dinoflagellates and the raphidophytes. The other groups, including species isolated from the Huon Estuary over the study period (Table 2), tolerated the added bacteria.
Bacterial interactions with harmful algal bloom species have been reviewed recently by Doucette et al. (9). Bacteria may both promote and regulate algal blooms (11). In general, bacteria that inhibit algal growth are effective through direct or indirect attack. An example of direct attack by marine bacteria is the attack by the gliding bacterium Cytophaga sp. strain J18/M01 (17). This strain effectively kills both diatoms and raphidophytes when it is added to algal cultures but not when filtrate alone is added. Indirect attacks are thought to be chemically mediated, and some seem to be species specific. Yoshinaga et al. (34) showed that Flavobacterium sp. strain C49 effectively inhibited H. akashiwo but did not affect the dinoflagellates or diatoms tested. Previously, Yoshinaga and coworkers had monitored a Gymnodinium mikimotoi bloom in Tanabe Bay, Japan, and had isolated bacteria over the course of the bloom. A total of 27 bacterial isolates had a negative effect on the growth of the target species, G. mikimotoi (35). In subsequent work, the growth responses of the other algal species to these inhibiting bacteria were investigated (34). Experiments were conducted over a 2-week period, and in-flask growth of algae in response to inoculated bacteria was examined. The initial bacterial concentrations were ca. 103 cells ml−1, and the final concentrations were 106 to 107 cells ml−1. After 2 weeks all G. mikimotoi cells and most H. akashiwo cells had died, while the four other algal species tested (A. catenella and the diatoms Thalassiosira sp., Ditylum brightwellii, and Skeletonema costatum) were not affected by the majority of the isolates. Two Vibrio strains were fatal to the Thalassiosira sp. tested, and another Vibrio strain killed A. catenella. In summary, the Gymnodinium strains and raphidophytes were sensitive to exposure to high concentrations of most bacterial strains tested (34, 35). In our screening procedure we did not document growth responses of algae but tested only for immediate effects (hours) and recovery from initial contact (adjustment), and direct comparisons between the two studies are difficult to make. It appears that the P. carrageenovora reaction is comparable to the reactions of the majority of the G. mikimotoi-inhibiting bacteria and would have impaired growth and eventually overwhelmed the gymnodinoid cultures. In contrast, strain Y affected G. catenatum quickly and at comparatively low bacterial concentrations (Fig. 3). Both pseudoalteromonads obtained from the Huon Estuary adversely affected naked dinoflagellates, but they acted via different mechanisms. Interestingly, species closely related to strain Y, such as P. rubra and P. luteoviolacea (Fig. 1), produce high-molecular-weight antibacterial substances (2).
The concentrations of the two pseudoalteromonads were very low, as determined by CFU measurements, compared to the concentrations of bacteria used in the experiments. However, given the relatively high growth rate of strain Y, it would take just 3 days to reach levels of 106 to 107 cells ml−1. Gymnodinium doubling times are typically around 3 days (3), and given favorable conditions, the bacteria could potentially reach effective concentrations in nature. We demonstrated that strain Y was present in the Huon Estuary under nonbloom conditions and that it can effectively kill gymnodinoid dinoflagellates and raphidophytes. This means that for a G. catenatum bloom to occur, conditions must be favorable for the algae to bloom but not for the bacteria at the same time. The phytoplankton species composition of the Huon Estuary was quite diverse throughout the 1997 summer-fall period; the most obvious dinoflagellates present were thecate species, such as Ceratium tripos, Ceratium furcus, and Dinophysis spp., along with diatoms and small flagellates. The concentrations of gymnodinoid dinoflagellates and raphidophytes (a small Heterosigma sp.) throughout the 1997 study period were also low (25). The frequency of encounters between bacteria and target algae would have been low (31). Bacterial persistence throughout autumn in the absence of any algal blooms indicates that the bacteria probably also utilize DOM and do not depend on algal predation in the natural environment. Culturable Pseudoalteromonas spp. were found in conjunction with higher FCDOM levels, and the number of CFU of P. carrageenovora correlated with the FCDOM concentration in the Huon Estuary (Table 3). Unfortunately, strain Y was relatively rare, and we did not have sufficient data to test the correlation further. Strain Y originally was isolated from samples with high FCDOM concentrations (Table 3) and subsequently was recovered from a surface sample with a lower FCDOM concentration. However, given that FCDOM breaks down under UV (solar) radiation (23), the available DOM concentrations in that surface sample may well have been underestimated.
Despite more than 30 years of investigations into the dynamics of phytoplankton species succession (26, 32), remarkably little progress has been made in predicting algal blooms at the species level. To a large extent, this may be due to the absence of information on conditions preceding bloom events, including the incidence of biological control by viruses and bacteria which could lose their effectiveness under specific environmental conditions. Fukami et al. (11) investigated the effect of bacterial assemblages during different stages of phytoplankton succession. These assemblages selectively inhibited dominant phytoplankton species, which suggested that there was strong bacterial control of phytoplankton succession. If inhibition by bacteria were a factor in suppressing algal bloom growth rates, hydrodynamic conditions, such as rainfall events, could affect inhibitory and algicidal bacteria in the system and create an opportunity for species to bloom. This proposed mechanism would not exclude the role of other variables, including endogenous timing of excystment of some species and a sudden input of a limiting micronutrient (6). In the present work, we documented how gymnodinoid dinoflagellates and raphidophytes are affected by two closely related Pseudoalteromonas bacteria, albeit by obviously different mechanisms. Both pseudoalteromonads occurred as incidental but persistent members of the bacterial flora of the Huon Estuary but did not adversely affect a range of other planktonic organisms isolated over the same period as the bacteria. Bacterial species composition may play a significant role in harmful algal bloom dynamics.
ACKNOWLEDGMENTS
This work was supported in part by Australian Research Council grants to J.P.B. and G.M.H. Fieldwork was undertaken as part of the Huon Estuary Study under the auspices of the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Division of Marine Research, Australia, and was funded in part by Fisheries Research & Development Corporation project 96/284.
Thanks are extended to E. Butler of CSIRO and Huon Estuary team leader Sue Blackburn, who also provided cultures from the CSIRO culture collection. Chris Bolch generously provided additional dinoflagellate cultures. Lesley Clementson, Pru Bonham, Alison Turnbull, and the boat drivers from Tassal Ltd. kindly aided C.L. in the field. We thank A. Davidson of the CSIRO Antarctic Division for use of a spectrofluorometer, J.-M. Leroy and J. Marshal for laboratory support, and J. Skerrett for discussions about strain Y. W. F. Vincent provided personal and intellectual support for C.L. while she was in Tasmania.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P, Gauthier M J, Baumann L. Genus Alteromonas Baumann, Baumann, Mandel and Allen 1972, 418.AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams and Wilkins Co.; 1984. pp. 343–352. [Google Scholar]
- 3.Blackburn S I, Hallegraeff G M, Bolch C J. Vegetative reproduction and sexual life cycle of the toxic dinoflagellate Gymnodinium catenatum from Tasmania, Australia. J Phycol. 1989;25:577–590. [Google Scholar]
- 4.Bowman J P, Austin J J, Cavanagh J, Sanderson K. Novel species of Psychrobacter from Antarctic ornithogenic soils. Int J Syst Bacteriol. 1996;46:841–848. doi: 10.1099/00207713-46-4-841. [DOI] [PubMed] [Google Scholar]
- 5.Determann S, Reuter R, Wagner P, Willkomn R. Fluorescent matter in the eastern Atlantic Ocean. Part 1. Method of measurement and near-surface distribution. Deep-Sea Res. 1994;41:659–675. [Google Scholar]
- 6.Doblin, M. A., S. I. Blackburn, and G. M. Hallegraeff. Growth of the toxic dinoflagellate Gymnodinium catenatum: interaction of humic substances with selenium and nutrients. In B. Reguera, J. Blanco, M. L. Fernandez, and T. Wyatt (ed.), Proceedings of the 8th International Conference on Toxic Algae, in press.
- 7.Dobson S J, Franzmann P D. Unification of the genera Deleya (Baumann et al. 1983), Halomonas (Vreeland et al. 1980), and Halovibrio (Fendrich 1988) and the species Paracoccus halodenitrificans (Robinson and Gibbons 1952) into a single genus, Halomonas, and placement of the genus Zymobacter into the family Halomonadaceae. Int J Syst Bacteriol. 1996;46:550–558. [Google Scholar]
- 8.Dorsch J E, Bidleman T F. Natural organics as fluorescent tracers of river-sea mixing. Estuarine Coastal Shelf Sci. 1982;15:701–707. [Google Scholar]
- 9.Doucette, G. J., M. Kodama, S. Franca, and S. Gallacher. Bacterial interactions with harmful algal bloom species: bloom ecology, toxigenesis and cytology. In D. A. Anderson, A. Cembella, and G. Hallegraeff (ed.), The physiological ecology of harmful algal blooms, in press. Springer Verlag, Berlin, Germany.
- 10.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5. Seattle: University of Washington; 1993. [Google Scholar]
- 11.Fukami K, Sakaguchi K, Kanou M, Nishijima T. Effect of bacterial assemblages on the succession of blooming phytoplankton from Skeletonema costatum to Heterosigma akashiwo. In: Yasumota T, Oshima Y, Fukuyo Y, editors. Harmful and toxic algal blooms. Paris, France: Intergovernmental Oceanographic Commission of UNESCO; 1996. pp. 335–338. [Google Scholar]
- 12.Fukami K, Yuzawa A, Nishijima T, Hata Y. Isolation and properties of a bacterium inhibiting the growth of Gymnodinium nagasakiense. Nippon Suisan Gakkaishi. 1991;58:1073–1077. [Google Scholar]
- 13.Furuki M, Kobayashi M. Interaction between Chattonella and bacteria and prevention of this red tide—EMECS ’90. Mar Pollut Bull. 1991;23:189–193. [Google Scholar]
- 14.Guillard R R L. Culture of phytoplankton for feeding marine invertebrates. In: Smith W L, Chanley M H, editors. Culture of marine invertebrate animals. New York, N.Y: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- 15.Guillard R R L, Ryther J H. Studies of marine plankton diatoms. I. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 16.Hallegraeff G M, McCausland M A, Brown R K. Early warning of toxic dinoflagellate blooms of Gymnodinium catenatum in southern Tasmanian waters. J Plankton Res. 1995;17:1163–1176. [Google Scholar]
- 17.Imai I, Ishida Y, Hata Y. Killing of marine phytoplankton by a gliding bacterium Cytophaga sp., isolated from the coastal sea of Japan. Mar Biol. 1993;116:527–532. [Google Scholar]
- 18.Imai I, Ishida Y, Sakaguchi K, Hata Y. Algicidal marine bacteria isolated from northern Hiroshima Bay, Japan. Fish Sci. 1995;61:628–636. [Google Scholar]
- 19.Ishida, Y., I. Yoshinaga, K. Mu-Chan, and A. Uchida. Possibility of bacterial control of harmful algal blooms. In Proceedings of the 7th International Symposium on Microbial Ecology, in press.
- 20.Iwasaki H. Recent progress of red tide studies in Japan: an overview. In: Okaichi T, Anderson D M, Nemoto T, editors. Red tides: biology, environmental science, and toxicology. New York, N.Y: Elsevier; 1989. pp. 3–9. [Google Scholar]
- 21.Jones H L J. A classification of mixotrophic protists based on their behavior. Freshwater Biol. 1997;37:35–43. [Google Scholar]
- 22.Laurion I, Vincent W F, Lean D S. Underwater ultraviolet radiation: development of spectral models for northern high latitude lakes. Photochem Photobiol. 1997;65:107–114. [Google Scholar]
- 23.Lindell M J, Granéli W, Tranvik L J. Enhanced bacterial growth response to photochemical transformations of dissolved organic matter. Limnol Oceanogr. 1995;40:195–199. [Google Scholar]
- 24.Lovejoy, C., and E. Butler (Commonwealth Scientific and Industrial Research Organisation). Unpublished data.
- 25.Lovejoy, C., S. Blackburn, and P. Bonham. Unpublished data.
- 26.Margalef R. Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanol Acta. 1978;1:493–509. [Google Scholar]
- 27.Meyer J L, Edwards R T, Risley R. Bacterial growth on dissolved organic carbon from a blackwater river. Microb Ecol. 1987;13:13–29. doi: 10.1007/BF02014960. [DOI] [PubMed] [Google Scholar]
- 28.Nygaard K, Tobiesen A. Bacterivory in algae: a survival strategy during nutrient limitation. Limnol Oceanogr. 1993;38:273–279. [Google Scholar]
- 29.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–947. [Google Scholar]
- 30.Rothhaupt K O. Laboratory experiments with a mixotrophic chrysophyte and obligately phagotrophic and phototrophic competitors. Ecology. 1996;77:716–724. [Google Scholar]
- 31.Shimeta J. Diffusional encounters of submicrometer particles and small cells by suspension feeders. Limnol Oceanogr. 1993;38:456–465. [Google Scholar]
- 32.Smayda T J. Phytoplankton species succession. In: Morris I, editor. The physiological ecology of phytoplankton. Berkeley: University of California Press; 1980. pp. 493–570. [Google Scholar]
- 33.Stolp H. Microbial ecology: organisms, habitats, activities. Cambridge, United Kingdom: Cambridge University Press; 1988. p. 21. [Google Scholar]
- 34.Yoshinaga I, Kawai T, Ishida Y. Analysis of algicidal ranges of the bacteria killing the marine dinoflagellate Gymnodinium mikimotoi isolated from Tanabe Bay, Wakayama Pref., Japan. Fish Sci. 1997;63:94–98. [Google Scholar]
- 35.Yoshinaga I, Kawai T, Takeuchi T, Ishida Y. Distribution and fluctuation of bacteria inhibiting the growth of a marine red tide phytoplankton Gymnodinium mikimotoi in Tanabe Bay (Wakayama Pref., Japan) Fish Sci. 1995;61:780–786. [Google Scholar]
- 36.ZoBell C E. Marine microbiology. Waltham, Mass: Chronica Botanica; 1946. [Google Scholar]