Abstract
Naturally occurring plasmids isolated from heterotrophic bacterial isolates originating from coastal California marine sediments were characterized by analyzing their incompatibility and replication properties. Previously, we reported on the lack of DNA homology between plasmids from the culturable bacterial population of marine sediments and the replicon probes specific for a number of well-characterized incompatibility and replication groups (P. A. Sobecky, T. J. Mincer, M. C. Chang, and D. R. Helinski, Appl. Environ. Microbiol. 63:888–895, 1997). In the present study we isolated 1.8- to 2.3-kb fragments that contain functional replication origins from one relatively large (30-kb) and three small (<10-kb) naturally occurring plasmids present in different marine isolates. 16S rRNA sequence analyses indicated that the four plasmid-bearing marine isolates belonged to the α and γ subclasses of the class Proteobacteria. Three of the marine sediment isolates are related to the γ-3 subclass organisms Vibrio splendidus and Vibrio fischeri, while the fourth isolate may be related to Roseobacter litoralis. Sequence analysis of the plasmid replication regions revealed the presence of features common to replication origins of well-characterized plasmids from clinical bacterial isolates, suggesting that there may be similar mechanisms for plasmid replication initiation in the indigenous plasmids of gram-negative marine sediment bacteria. In addition to replication in Escherichia coli DH5α and C2110, the host ranges of the plasmid replicons, designated repSD41, repSD121, repSD164, and repSD172, extended to marine species belonging to the genera Achromobacter, Pseudomonas, Serratia, and Vibrio. While sequence analysis of repSD41 and repSD121 revealed considerable stretches of homology between the two fragments, these regions do not display incompatibility properties against each other. The replication origin repSD41 was detected in 5% of the culturable plasmid-bearing marine sediment bacterial isolates, whereas the replication origins repSD164 and repSD172 were not detected in any plasmid-bearing bacteria other than the parental isolates. Microbial community DNA extracted from samples collected in November 1995 and June 1997 and amplified by PCR yielded positive signals when they were hybridized with probes specific for repSD41 and repSD172 replication sequences. In contrast, replication sequences specific for repSD164 were not detected in the DNA extracted from marine sediment microbial communities.
The maintenance and horizontal transfer of extrachromosomal elements provide one mechanism by which microbial communities can rapidly adapt to changes in environmental conditions. This adaptation can be in the form of plasmid rearrangements and duplications (18, 40), a change in the plasmid copy number (40, 54), or lateral or horizontal movement of plasmids within bacterial populations. An example demonstrating the importance of plasmid-mediated genetic adaptation in natural microbial communities, likely caused by lateral transfer, is the increased frequencies (2- to 10-fold) of catabolic plasmids reported in bacterial isolates obtained from polluted marine and freshwater environments compared to isolates from nonpolluted or less impacted ecosystems (8, 23, 43). Plasmids also play a major role in promoting the widespread distribution of antibiotic resistance genes attributed to the intense and increased use of antibiotics (42).
The ability of plasmids to self-transfer or to be mobilized by transfer-proficient plasmids and the ability to replicate in different bacterial hosts are key factors in the spread of plasmid-encoded genes within microbial communities. Plasmids which are considered to have broad host ranges in nature have the potential to significantly affect the microbial community structure and function due to their ability to replicate and be maintained in members of distantly related genera. Thus, to better understand gene flux in natural systems and hence the potential role of plasmids in promoting horizontal transfer within microbial communities, knowledge of the distribution, diversity, and host ranges of naturally occurring plasmids is necessary.
At present, most indigenous plasmids from marine and freshwater systems have been only partially characterized with respect to host range, replication mechanisms, incompatibility groups, and conjugal abilities. Plasmids containing similar or related replication systems are considered incompatible if they cannot coexist in a host cell (12, 41). This trait has facilitated the grouping of plasmids from gram-negative bacteria, mainly members of the family Enterobacteriaceae, into more than 30 different incompatibility groups (3). While molecularly based plasmid classification or replicon typing by using DNA sequences of replication origins and incompatibility loci of well-characterized plasmids has been useful in classifying plasmids from bacterial isolates of medical importance (9, 10, 14), plasmids from various marine microbial communities, including sediments, biofilms, bulk water, and the marine air-water interface, have been recently shown to contain incompatibility and replication regions unrelated to those currently defined (11, 53).
The present study was undertaken to characterize, at the molecular level, the replication and incompatibility loci of naturally occurring plasmids isolated from gram-negative marine heterotrophs for use as replicon probes to classify and type, at the molecular level, plasmids present in bacterial populations of marine sediments. Replication origins were obtained from plasmids ranging in size from 6 to 30 kb isolated from culturable bacteria of coastal California marine sediments (53). Phylogenetic analysis indicated that the plasmids were initially isolated from bacteria belonging to the α and γ-3 subclasses of the class Proteobacteria. Although a sequence and hybridization analysis of the replication origins from the marine plasmids confirmed the lack of homology with previously described plasmids, the replication regions contained features commonly found in previously characterized plasmid replication origins. The replication origins of the naturally occurring plasmids appear to have a broad host range, as indicated by their ability to replicate in members of diverse gram-negative marine genera. In addition to molecular characterization of the indigenous plasmids, the persistence of the replicons in marine sediment bacterial populations was determined by PCR amplification of microbial community DNA extracted on different dates and examined for the presence of homologous plasmid replication sequences.
MATERIALS AND METHODS
Isolation and identification of plasmid-bearing marine isolates.
The bacterial strains and plasmids used in this study are listed in Table 1. DAPI (4′,6-diamidino-2-phenylindole) direct bacterial counts were obtained by using 1-g samples of marine sediments serially diluted in artificial seawater by the method of Porter and Feig (43a). Bacteria were isolated from coastal marine sediments by serially diluting sediment samples (1 g) in artificial seawater, spreading the dilutions onto solid media (e.g., YTSS, 0.5× YTSS, and TSS [53]), and incubating the resulting plates for 1 to 14 days at 30°C (53). Colonies were picked from the plates and restreaked at least twice on the same medium to ensure purity (53). The presence of plasmids in the marine bacterial isolates was determined by a modification of the Kieser method (28, 53). Specifically, plasmid presence was determined by centrifuging (6,000 × g, 10 min) 5 to 10 ml of an overnight cell culture grown in the same medium in which the isolate was initially cultured, draining the cell pellet, resuspending it in 500 μl of solution A (2 mg of lysozyme per ml, 0.3 M sucrose, 25 mM Tris [pH 8.0], 25 mM EDTA [pH 8.0], 0.02% bromocresol green), and incubating the preparation at 37°C for 30 min; this was followed by adding 250 μl of solution B (0.3 M NaOH, 2% sodium dodecyl sulfate [SDS]), mixing the preparation by inverting it several times, and incubating it at 55°C for 30 min. Samples were allowed to cool to room temperature before 180 μl of solution C (5 g of phenol, 5 ml of chloroform, 1 ml of distilled water, 5 mg of 8-hydroxyquinoline) was added, and they were quickly vortexed to mix them and centrifuged (8,000 × g, 5 min). The supernatants were carefully removed and immediately loaded onto 0.6 to 0.8% horizontal agarose gels. The gels were electrophoresed at 5 V per cm, stained with ethidium bromide, destained in water, and photographed on a UV transilluminator.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| C2110 | polAI | 27 |
| HB101 | recA13 | 6 |
| DH5α | recA1 | 4 |
| Marine strains | ||
| Achromobacter sp. | Georgia salt marsh isolate, Rifr | 52a |
| Pseudomonas nautica ATCC 35188 | Rifr | ATCCa |
| Pseudomonas duodoroffii ATCC 27123 | Rifr | ATCC |
| Roseobacter sp. strain 164 | San Diego salt marsh isolate, source of repSD164 | This study |
| Serratia rubidaea ATCC 27164 | Rifr | ATCC |
| Vibrio sp. | Georgia salt marsh isolate, Rifr | 52 |
| Vibrio sp. strain 41 | San Diego salt marsh isolate, source of repSD41 | This study |
| Vibrio sp. strain 121 | San Diego salt marsh isolate, source of repSD121 | This study |
| Vibrio sp. strain 172 | San Diego salt marsh isolate, source of repSD172 | This study |
| Plasmids | ||
| pUC4K | Source of Tn903 npt gene | 59 |
| pRK2013 | ColE1 replicon, RK2 Tra+ | 16 |
| pFF1 | Mini-RK2 replicon, bla cat oriT | 17 |
| pBR325 | pBR322 + Cmr (ColE1) | 44 |
| pTM41 | pBR325 + repSD41 | This study |
| pTM121 | pBR325 + repSD121 | This study |
| pTM164 | pBR325 + repSD164 | This study |
| pTM172 | pBR325 + repSD172 | This study |
ATCC, American Type Culture Collection.
The identities of selected plasmid-bearing marine isolates were determined by 16S rRNA analysis as follows. Genomic DNA was purified from 1 ml of an overnight cell culture grown at 30°C in YTSS broth (52a) by using an anion-exchange column (Genomic-Tip 20) as recommended by the manufacturer (Qiagen, Chatsworth, Calif.). The entire 16S rRNA gene was amplified from approximately 0.1 to 0.5 μg of genomic DNA by using fD1 and rD1 as the primers (60). A total of 35 cycles were used under the following conditions: denaturation at 95°C for 1 min, primer annealing at 52°C for 1 min, and DNA extension at 72°C for 1 min, with initial incubation at 95°C for 2 min and at 60°C for 2 min. The amplified product was electrophoresed on 1.0% agarose and purified by electroelution (37), and partial insert sequences were obtained by using three primers corresponding to the following positions in the Escherichia coli sequence: primer 1, positions 519 to 536; primer 2, positions 907 to 926; and primer 3, positions 1392 to 1406 (32). The rRNA gene sequence of each of the marine isolates was compared to the Ribosomal Database Project (36) SSU_Prok data set (release 6.0). Nearest-neighbor sequences for the plasmid-bearing marine sediment isolates were identified by using the RDP SSU Prok data set (36), Suggest Tree, and Similarity Rank analysis. Aligned 16S rRNA sample sequences, supplemented with 16S rRNA sequences from GenBank (region spanning positions 228 to 1295 of the E. coli numbering system), were obtained by using the Genetics Computer Group Inc. package (19a). Phylogenetic trees were constructed and a bootstrap analysis (100 replicates) was performed with the PHYLIP package (18a) by using the evolutionary distances (Jukes-Cantor distances) and the neighbor-joining method.
Large-scale isolation of plasmid DNA from marine isolates.
A modification of the alkaline lysis method of Birnboim and Doly (5) was used to isolate supercoiled plasmid DNA from gram-negative marine bacteria. Each cell pellet obtained from centrifugation (6,000 × g, 10 min) of 1 liter of an overnight cell culture grown in either TSS or 0.5× YTSS (53) at 30°C was thoroughly drained and resuspended in 50 ml of solution 1 (10 mg of lysozyme per ml, 50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 100 μg of RNaseA per ml); this was followed by addition of 50 ml of solution 2 (200 mM NaOH, 1% SDS), mixing of the preparation by inverting it several times, and incubation for 5 min at room temperature and then by addition of 50 ml of solution 3 (3 M potassium acetate, pH 5.5), mixing of the preparation by inverting it several times, and incubation on ice for 30 min. Samples were centrifuged (20,000 × g, 30 min), each supernatant was extracted with 0.5 volume of phenol-chloroform, the phases were separated by centrifugation (10,000 × g, 10 min), and the aqueous layer was precipitated with 0.8 volume of isopropanol at room temperature and immediately centrifuged (20,000 × g, 30 min). Plasmid DNA was subsequently purified by cesium chloride gradient centrifugation (37).
Cloning and sequencing of plasmid replication origins from marine bacterial isolates.
Approximately 1 μg of plasmid DNA was partially digested with restriction endonuclease Sau3AI for 1, 10, and 30 min, and the terminal 5′ phosphates were removed with shrimp alkaline phosphatase as recommended by the manufacturer (United States Biochemical, Cleveland, Ohio). The partially digested plasmid DNA was ligated to the Tn903 npt gene isolated as a BamHI fragment from pUC4K (37, 59) with T4 DNA ligase as recommended by the manufacturer (Promega, Madison, Wis.). The ligation mixture was transformed into E. coli DH5α and C2110 and was grown on Lennox L agar (39) (Gibco Scientific, Grand Island, N.Y.) supplemented with 50 μg of kanamycin per ml at 30°C for 18 to 48 h. Plasmid DNA was obtained from 5 ml of an overnight cell culture grown in Lennox L broth at 30°C by alkaline lysis (37) and was digested with PstI restriction endonuclease to identify the transformant(s) containing the smallest replication-proficient fragment. Plasmids pTM41, pTM121, pTM164, and pTM172 were generated by inserting the replication-proficient fragments, isolated as PstI fragments, into the PstI site of pBR325. Plasmids were purified from 500 to 1,000 ml of cell culture grown in Lennox L broth at 30°C by cesium chloride density gradient centrifugation (37). The nucleotide sequences of the cloned replication origins from the marine plasmid-bearing isolates were determined by using the vector primer sequences 5′-ATTGTTGCCGGGAAGCTAGAGTAAGTAGTT-3′ and 5′-AATGAAGCCATACCAAACGACGAGCGTGAC-3′ and a model ABI 373 automated DNA sequencer (Perkin-Elmer Applied Biosystems). Both strands of repSD41, repSD121, repSD164, and repSD172 were sequenced from pTM41, pTM121, pTM164, and pTM172, respectively. DNA sequences were compiled and analyzed with Oligo version 5.0 (National Biosciences, Inc., Plymouth, Minn.). DNA sequence analysis and alignment were performed by the pairwise correlation method by using Macaw 2.0.5 (National Center for Biotechnology Information, Bethesda, Md.). Different alignments were examined to maximize sequence identity, which was calculated by using GeneDoc 2.1.
Curing of large native plasmid.
Electrocompetent cell stocks of Vibrio sp. strain 172 were prepared as recommended by the manufacturer (Invitrogen, San Diego, Calif.). Plasmid pTM172 (1 μg) was introduced into the Vibrio strain by electroporation by using the following conditions: pulse length, 7.5 ms; resistance, 150 Ω; voltage, 1.8 kV; cuvette gap, 0.1 cm. Electroporated cells were then incubated at 30°C in 1 ml of 0.5× YTSS broth (53) shaken at 200 rpm for 2 h, plated onto 0.5× YTSS plates supplemented with 40 μg of chloramphenicol per ml, and incubated overnight at 30°C. Colonies were restreaked onto fresh selective media, and single colonies were grown at 30°C overnight in selective 0.5× YTSS broth to screen for native plasmid loss by the modified Kieser method (53).
Southern hybridizations.
Plasmids pTM41, pTM121, pTM164, and pTM172 were digested with PstI restriction endonuclease, and the DNA fragment containing the replication regions from the plasmids was eluted with QIAquick gel extraction (Qiagen). The isolated DNA fragments which were to be used as probes were labeled by random priming by using [α-32P]dATP (6,000 Ci/mmol; NEN Dupont) and the Boehringer Mannheim (Indianapolis, Ind.) randomly primed DNA labeling system. Following electrophoresis of plasmid DNA on 0.8% LE agarose (FMC, Rockland, Maine), the gels were denatured, neutralized, and blotted onto nylon membranes (Schleicher & Schuell, Inc., Keene, N.H.) essentially as recommended by the manufacturer; however, the DNA was routinely allowed to transfer overnight to ensure complete transfer. Following transfer, the membranes were rinsed in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4 [pH 7.7], and 1 mM EDTA) (37), baked for 2 h at 80°C under a vacuum, and stored until hybridization. The membranes were then washed in prewarmed (42°C) 2× SSPE, placed in hybridization bottles (Hybaid Instruments, Holbrook, N.Y.), and prehybridized in 30 ml of hybridization solution consisting of 50% (vol/vol) deionized formamide, 6× SSPE, 5× Denhardt’s solution, 1% SDS, and 100 μg of salmon sperm DNA per ml at 37 to 42°C for 4 to 8 h at 7 rpm. Radiolabeled probes were added at a concentration of approximately 2 × 106 cpm per ml, and the preparations were incubated at 37 to 42°C for 16 h at 7 rpm. Unbound probe was removed by washing the membranes twice in 2× SSPE–0.1% SDS for 15 min at 65°C and twice in 1× SSPE–0.1% SDS at 65°C for 30 min in a HybAid oven at 10 rpm. The final wash step was in 0.1× SSPE for 10 min at 65°C, and the membranes were exposed to BioMax X-ray film (Kodak) at −70°C with an intensifying screen. When it was necessary to reprobe membranes with a different environmental plasmid replication probe, bound labeled probe was removed by using the recommendations of the manufacturer (Schleicher & Schuell).
DNA extraction and purification from marine sediments.
Total DNA (genomic DNA and plasmid DNA) was extracted from sediment samples (1 to 5 g) by the method of Tsai and Olson (56), with slight modifications. The modifications consisted of two phenol-chloroform extractions rather than one phenol extraction and one phenol-chloroform extraction. The crude DNA extract was purified to remove humic acids and other coextracted contaminants by the method of Tebbe and Vahjen (55) with ion-exchange columns (Qiagen-Tip 500).
PCR amplification.
The primers (18- to 21-mers) used for detection of the plasmid replication sequences were based on sequences obtained during this study. The 5′-to-3′ sequences of repSD41-1, repSD41-2, repSD164-1, repSD164-2, repSD172-1, and repSD172-2 were AGCAAAACACCCTCTCAG, GTATCAGCGAACTCAACAAA, GCAAGACCAAGCATCACGAAG, CAGCAATCACGCCCCAAT, CCCGTTAAATTGCTAATCAC, and AAGCCTTACAGCGAAAAAG, respectively. Primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). Amplification was performed as described by Saiki et. al (47) by using Taq 2000 polymerase (Stratagene, La Jolla, Calif.). Each PCR mixture contained 1× PCR amplification buffer (1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl [pH 8.8], 0.001% gelatin), deoxynucleoside triphosphates (each at a concentration of 200 μM), 1 μl of a sediment DNA extract (corresponding to the DNA recovered from approximately 5 to 10 mg of sediment), which was used as the DNA template, 2.5 U of Taq polymerase per 100 μl, and 2.5 μg of T4 gene 32 protein (Boehringer Mannheim) per 100 μl. The addition of T4 gene 32 protein, which binds and stabilizes single-stranded DNA, has been shown to improve PCR amplification (55). Amplification of the repSD41 and repSD172 sequences was performed for 35 PCR cycles under the following conditions: denaturation at 95°C for 1 min, primer annealing at 53°C for 1 min, and DNA extension at 72°C for 1 min, with initial incubation at 98°C for 5 min and at 68°C for 5 min. Amplification of the repSD164 sequence was performed for 35 PCR cycles under the following conditions: denaturation at 95°C for 1 min and primer annealing at 70°C for 2 min, with initial incubation at 95°C for 2 min and at 70°C for 2 min. Amplified products were detected on 1.0% agarose gels electrophoresed in TBE buffer, stained with ethidium bromide, and photographed on a UV transilluminator.
Determination of plasmid host range.
A modification of the method of Ditta et al. (16) was used to test for the ability of the plasmid origins to replicate in diverse hosts. One milliliter of an overnight cell culture grown in either Luria-Bertani broth containing the appropriate antibiotic or 0.5× YTSS (53) was centrifuged (10,000 × g, 30 s) and resuspended in 0.1 ml of fresh broth. Mating mixtures were prepared by mixing together 1 volume of an overnight liquid culture of the donor strain (E. coli DH5α containing either pFF1, pTM41, pTM121, pTM164, or pTM172), 1 volume of E. coli HB101(pRK2013) (a helper strain), and 5 volumes of the marine bacterial recipient (Table 1). Thirty to thirty-five microliters was spotted onto 0.5× YTSS agar and incubated at 30°C for 2 to 5 days before cells were resuspended and plated. Transconjugants were obtained on 0.5× YTSS containing 40 μg of chloramphenicol per ml and either 75 μg of rifampin per ml to select for the Achromobacter sp. or 200 μg of rifampin per ml to select for Pseudomonas duodoroffii, Pseudomonas nautica, Serratia rubidaea, and Vibrio sp. Putative transconjugants were restreaked several times, and plasmid presence was verified by isolation of plasmid DNA by either the alkaline lysis method of Birnboim and Doly (5) or the modified Kieser method (53). Plasmid integrity was determined by cleavage with PstI. Restriction enzyme-digested DNA was electrophoresed on a 0.7% agarose gel, stained with ethidium bromide, and photographed.
Nucleotide sequence accession numbers.
The nucleotide sequences of plasmid replication origins repSD164, repSD41, repSD172, and repSD121 have been deposited in the GenBank database under accession no. AF020624, AF020625, AF020626, and AF03157, respectively. The four 16S rRNA gene sequences obtained in this study have been deposited in the GenBank database under accession no. AF064556, AF064557, AF064558, and AF064559.
RESULTS
Analysis of plasmid replication origins from marine sediment bacterial isolates.
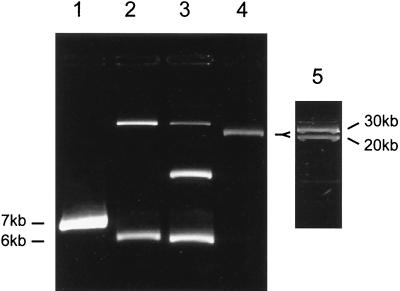
While numerous studies have reported on the incidence and frequency of plasmids in marine water column and sediment bacterial isolates (1, 2, 26, 31, 33, 51, 62), few studies have attempted to classify plasmids from the marine environment by characteristics such as maintenance requirements, host range, incompatibility group, and conjugal proficiency. Using a method to obtain replication-proficient fragments from indigenous plasmids for use as replicon probes, we isolated replication origins from plasmid-bearing marine sediment strains 41, 121, 164, and 172 (Table 1), which belong to the genera Vibrio and Roseobacter and are capable of replicating in E. coli C2110 and DH5α. The four replication origins were rescued from plasmids having approximate sizes of 6.0 and 7.0 kb (Fig. 1, lanes 1 through 3) and 30 kb (Fig. 1, lanes 4 and 5).
FIG. 1.
Supercoiled plasmid contents of bacterial isolates from coastal marine sediments obtained by ethidium bromide-cesium chloride gradient centrifugation and visualized by ethidium bromide staining of a 0.7% agarose gel. Lane 1, Vibrio sp. isolate 41 (7 kb); lane 2, Vibrio sp. isolate 121 (6 kb); lane 3, Roseobacter sp. isolate 164 (6.5 kb); lane 4, Vibrio sp. isolate 172 (with 0.7-V/cm gel running conditions only one plasmid band was evident); lane 5, Vibrio sp. isolate 172 (20- and 30-kb plasmid bands were visible when the gel running conditions were 0.1 V/cm). Approximate sizes in parentheses were obtained from restriction digests of the plasmids from which functional replication origins were obtained in E. coli. Note that isolates 121, 164, and 172 contain multiple plasmids.
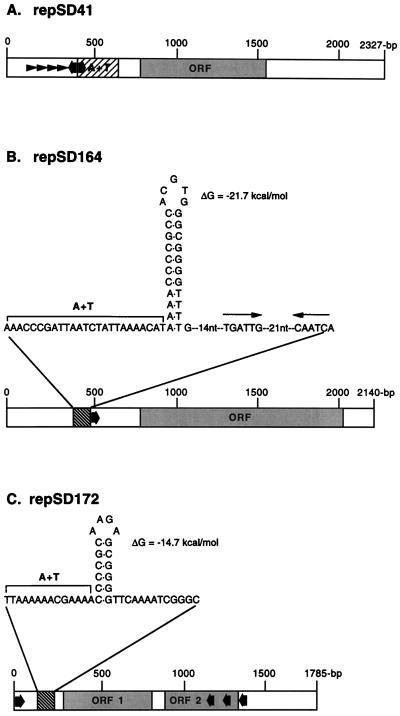
A sequence analysis was carried out for a replication-proficient 2.3-kb region derived from plasmid pTM41 (Table 1) that is present in marine Vibrio sp. strain 41 (Fig. 1 and Fig. 2A). This replicon, designated repSD41, exhibited the following features: (i) four conserved 21-bp direct repeats (TAAAAAGACAAATCTGGGAA[A/G]) which are present in pairs, starting at nucleotides 333 and 386, and which have an 11-nucleotide spacer region between the second and the third direct repeats; (ii) an AT-rich region of approximately 100 nucleotides starting at base position 459; (iii) a putative open reading frame (base positions 724 to 1540) with an ATG start codon and a TAA stop codon, which has the potential to encode a polypeptide of 29 kDa, a molecular mass similar to the molecular masses of most Rep proteins encoded by plasmid replicons (25); and (iv) two sequences in an indirect orientation corresponding to the DnaA box consensus sequence 5′-(T/C) (T/C) (A/T/C) T (A/C) C (A/G) (A/C/T) (A/C)-3′ (49), which are present between nucleotides 450 and 458 (5′-CTATCCACA-3′) (Fig. 2A).
FIG. 2.
Schematic diagram of the structural organization of the plasmid replication origins isolated from gram-negative marine sediment bacterial isolates capable of replicating in E. coli, as deduced from the nucleotide sequence. The directions of DnaA boxes are indicated by broad arrows, and the sequences of the plasmid replication origins are similar to the consensus sequence 5′-(T/C)(T/C)(A/T/C)T(A/C)C(A/G)(A/T/C)(A/C)-3′ (49). (A) The 2,327-bp fragment containing the repSD41 replication origin obtained from a 7-kb plasmid isolated from marine Vibrio sp. strain 41. ORF, open reading frame that may encode a replication initiation protein. Direct repeats are indicated by arrowheads. The location of the approximately 100-bp AT-rich region is indicated by the cross-hatched box. (B) The 2,140-bp fragment containing the repSD164 replication origin obtained from a 6.5-kb plasmid isolated from marine Roseobacter sp. strain 164 is diagrammed. The open reading frame (ORF) may encode a replication initiation protein. The locations of a region that potentially is able to form a hairpin structure is indicated by the cross-hatched box. Inverted repeats in close proximity to the repSD164 hairpin structure are indicated by thin arrows. nt, nucleotides. (C) The 1,785-bp fragment containing the repSD172 replication origin obtained from a 30-kb plasmid isolated from marine Vibrio sp. strain 172 is diagrammed. Open reading frames 1 and 2 (ORF 1 and ORF 2) may encode proteins involved in plasmid origin replication. The locations of a region that potentially is able to form a hairpin structure is indicated by the cross-hatched box.
Sequence analysis of the 2.1-kb repSD164 replication-proficient region of pTM164 (Table 1) isolated from an approximately 6.5-kb plasmid present in marine Roseobacter sp. isolate 164 revealed the following features (Fig. 1 and 2B); (i) a GC-rich region between nucleotides 393 and 420 capable of forming a cruciform structure, an AT-rich region adjacent to the putative hairpin structure, and a downstream inverted repeat that is 6 nucleotides long (Fig. 2B); (ii) a putative open reading frame (base positions 567 to 1932) with an ATG start codon and a TGA stop codon, which has the potential to encode a polypeptide of approximately 50 kDa; and (iii) a single sequence corresponding to the DnaA box consensus sequence (49) beginning at nucleotide 488 (5′-TCATCCAAA-3′).
Sequence analysis of repSD172 (Table 1), a 1.8-kb plasmid replication-proficient fragment obtained from an approximately 30-kb plasmid isolated from marine Vibrio sp. strain 172 (Fig. 1 and 2C), revealed the following features: (i) a GC-rich region capable of forming a potential cruciform or hairpin structure between nucleotides 130 and 145 (Fig. 2C); (ii) two putative open reading frames with ATG start codons at base positions 294 to 795 and 894 to 1338 which have the potential to encode polypeptides of 19 and 17 kDa, respectively; and (iii) four sequences corresponding to the DnaA box consensus sequence starting at nucleotide position 16 (5′-CTTTCCG-3′), nucleotide 1082 (5′-CCATCCATA-3′), nucleotide 1202 (5′-TCATACACA-3′), and nucleotide 1349 (5′-TCCTCCGCA-3′).
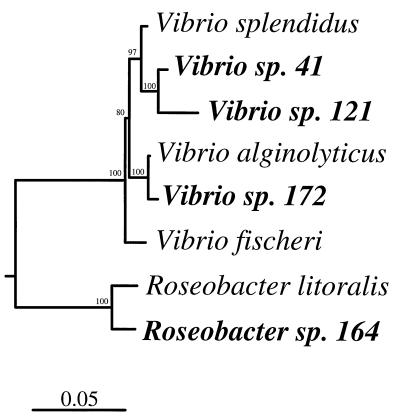
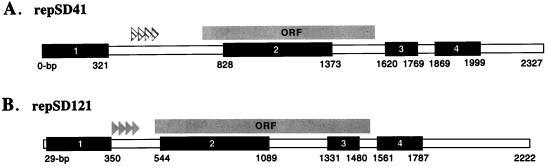
The replication-proficient fragment repSD121 was isolated from pTM121 (Table 1), a 6-kb plasmid found in a marine sediment bacterial isolate that was phylogenetically distinct from the isolate from which repSD41 was obtained (strain 41) (Fig. 3). The 7- and 6-kb parental plasmids, the sources of repSD41 and repSD121, respectively (Fig. 1, lanes 1 and 2), displayed different restriction endonuclease patterns (data not shown). However, analysis of the replication-proficient sequence of repSD121 (Fig. 4) revealed four regions with significant levels of similarity (77 to 94%, as determined by pairwise correlation) to repSD41. The similar regions of the two replication sequences, which on average were 434 nucleotides long, were found to include the site of a putative replication protein gene (Fig. 4). Interestingly, pTM41 and pTM121 were found to be able to coexist in the same E. coli host cell, indicating that these plasmids are compatible even though they exhibit significant DNA homology in the replication origin. However, this result is not surprising since repSD41 and repSD121 lack homology in the direct repeat region (Fig. 4), which is known to be one of the main determinants of incompatibility (25).
FIG. 3.
Phylogenetic relationships among plasmid-containing marine bacterial isolates from which plasmid replication fragments were isolated. The tree is unrooted and was constructed with 1,000 bases of aligned 16S rRNA gene sequences by using Bacillus subtilis as the outgroup. The bootstrap values (100 replicates) are indicated above the lines. The bar indicates Jukes-Cantor distances.
FIG. 4.
Comparison of the regions of sequence homology in repSD41 and repSD121. Four extended regions with significant levels of similarity (77 to 94%) were aligned on the basis of nucleotide sequence homology as determined by pairwise correlation, and these regions are indicated by solid boxes. The regions of similarity corresponding to base positions 0 to 321 and 29 to 350 had a level of similarity of 94%. The direct repeat (iteron) units of repSD41 and repSD121 are indicated by cross-hatched arrowheads and solid arrowheads, respectively. Base positions 828 to 1373 and 544 to 1089 of repSD41 and repSD121 have a level of similarity of 77%. Analysis of a third region and a fourth region corresponding to base positions 1620 to 1769 and 1869 to 1999 (repSD41) and to base positions 1331 to 1480 and 1561 to 1787 (repSD121) also revealed significant similarity between the repSD41 and repSD121 DNA sequences (levels of similarity, 79 to 81%). ORF, open reading frame.
The direct repeats (iterons) found at the origins of a large number of plasmids are essential for origin activity and serve as the binding sites for the plasmid-specified replication initiation protein. When inserted into a heterologous compatible plasmid, the iterons also display the incompatibility properties of the parent plasmid. Previously, Filutowicz et al. (19) reported the presence of a conserved hexanucleotide sequence (5′-TGAGPuG-3′) in the iterons of broad-host-range plasmids RK2 and RSF1010 and narrow-host-range plasmids, F, λ, R6K, and P1. Although the exact role of the hexanucleotide sequence in plasmid replication initiation in these plasmids is unclear, the direct repeats displayed incompatibility properties characteristic of the parental plasmid when they were cloned into a heterologous plasmid replicon. Analysis of the two replication origins containing direct repeats, repSD41 and repSD121, also revealed the presence of the hexanucleotide region in the iterons (Table 2), suggesting that these iterons are important in the replication activity and incompatibility properties of the plasmids. Direct repeats were not observed in either the repSD164 sequence or the repSD172 sequence.
TABLE 2.
Comparison of nucleotide sequence repeats (iterons) of replication origins of broad- and narrow-host-range plasmids and plasmids from marine bacterial isolates
| Plasmid | Length of iterons (nucleotides) | Direct repeat nucleotide sequencea |
|---|---|---|
| RK2 | 17 | 5′-TGACABWTGAGGGGCGG-3′ |
| RSF1010 | 20 | 5′-CGTGACAGTTATTGCAGGGG-3′ |
| R6K | 22 | 5′-ARMCATGAGDGBWTAGTWCGAX-3′ |
| Mini-F (repFIA) | 22 | 5′-TGAGGGYDRWYYRTCACAKWTK-3′ |
| pTM41 | 21 | 5′-TAAAAAGACAAATCTGGGAAA-3′ |
| pTM121 | 21 | 5′-TTAAAACTACAAATTGGGGMA-3′ |
B = not A; D = A, G, or T; K = G or T; M = A or C; R = A or G; W = A or T; X = any base; Y = C or T.
Due to the large size (30 kb) of the wild-type plasmid isolated from Vibrio sp. strain 172, an additional origin of replication could have been responsible for replication and maintenance in the marine host. To ensure that repSD172 was the fragment responsible for plasmid replication, curing studies were done in which pTM172 was introduced into the parent strain. Transformants arose that contained pTM172 when they were maintained under selective pressure. Southern analysis of total bacterial DNA revealed a loss of the wild-type 30-kb plasmid in Vibrio transformants containing pTM172 (data not shown) and confirmed that two different plasmids were present in the native Vibrio strain (Fig. 1, lanes 4 and 5).
Host range analysis of plasmid origins from marine isolates.
Plasmids capable of autonomous replication in distantly related host backgrounds have important implications for horizontal gene transfer in bacterial communities. While plasmids from nonmarine isolates belonging to incompatibility groups IncN, IncP, IncQ, and IncW have been demonstrated to be capable of replication in a large number of gram-negative bacteria, few plasmids isolated from gram-negative marine bacteria have been shown to have broad-host-range replication capabilities (48). Therefore, we determined the abilities of the various plasmid replicons isolated in this study to replicate in a diverse set of gram-negative marine bacteria belonging to the class Proteobacteria (29, 46). The fact that the four marine plasmid replicons (repSD41, repSD121, repSD164, and repSD172 [Table 3]) can become established in the polA1 E. coli C2110 host indicated that host DNA polymerase I is not required, unlike the ColE1 type family of plasmids, pCU1 (incompatibility group IncN), and gram-positive bacterial plasmid pAMβ1 (Table 3) (25). All four of the plasmid origins in plasmids pTM41, pTM121, pTM164, and pTM172 could become established after conjugal transfer from an E. coli host and, therefore, are able to replicate in a Vibrio sp. previously isolated from a Georgia salt marsh and in S. rubidaea ATCC 27614 (Table 3). However, the plasmids failed to replicate in the marine host P. nautica (Table 3). Plasmid pFF1 containing the replication origin from the well-characterized broad-host-range plasmid RK2 (IncPα), which is stably maintained in a wide range of gram-negative bacteria (50), can replicate in P. nautica and each of the other four marine isolates studied (Table 3). Although the repSD41 and repSD121 replicons appear to be related, they exhibited some differences in host ranges (Achromobacter sp. and P. duodoroffii) (Table 3), suggesting that there are differences in plasmid-host interactions (15). Since each of the four marine plasmid replicons is present as a cointegrate with plasmid pBR325, it was important to demonstrate that pBR325 cannot by itself become established in any of the strains tested (Table 3). In the absence of an environmental replicon (e.g., repSD41, repSD121, repSD164, or repSD172), pBR325 was not maintained in any of the strains tested (Table 3). Restriction enzyme analysis of the plasmid in each marine host after establishment of the plasmid replicons did not reveal any gross rearrangements (e.g., sequence amplification, deletion, or loss) of the established plasmid (data not shown).
TABLE 3.
Host ranges of plasmid replication origins isolated from marine sediment isolates
| Organism | Replication with plasmid:
|
|||||
|---|---|---|---|---|---|---|
| pBR325 | pFF1 | pTM41 | pTM121 | pTM164 | pTM172 | |
| E. coli C2110 | − | + | + | + | + | + |
| Achromobacter sp. | − | + | + | − | + | + |
| P. duodoroffii ATCC 27123 | − | + | + | + | + | − |
| P. nautica ATCC 35188 | − | + | − | − | − | − |
| S. rubidaea ATCC 27614 | − | + | + | + | + | + |
| Vibrio sp. | − | + | + | + | + | + |
Southern hybridization analysis of culturable bacterial populations.
To determine the specificity of the four broad-host-range marine plasmid replicons for typing and classifying plasmids from culturable marine bacteria, a Southern blot analysis of E. coli strains containing plasmids belonging to the known broad-host-range incompatibility groups (groups N, P, Q, and W) was conducted. There was a lack of homology between the four incompatibility groups and the DNA probes prepared from repSD41, repSD121, repSD164, and repSD172 (data not shown). While there was no cross-reactivity between the marine replicons repSD41, repSD164, and rep172, there was cross-reactivity between the repSD41 and repSD121 replicons when they were probed with each other (data not shown).
To determine the incidence of the various marine plasmids in the culturable microbial community from which they were initially isolated, DNA obtained by the modified Kieser method was blotted from agarose gels and hybridized to each of the three plasmid-specific replication probes (repSD41, repSD164, and repSD172). The DNA of as many as 150 marine isolates containing one or more plasmids collected in November 1995 did not hybridize to either repSD164 or repSD172. In contrast, approximately 6 of 111 culturable plasmid-bearing isolates exhibited homology to replication probe repSD41 (data not shown). Although cross-reactivity between repSD41 and repSD121 was detected, the profiles of isolates containing plasmids hybridizing to repSD41 (except the isolate that carried the plasmid from which repSD121 was obtained) were identical to the profile of the parental isolate from which the repSD41 probe was derived (data not shown). Lowering the stringency of hybridization (i.e., <75% homology) did not reveal any additional naturally occurring plasmids from culturable marine bacteria that had significant homology to any of the marine replication probes obtained in this study (data not shown).
Plasmid replication sequences in microbial community DNA.
The bacterial isolation method used in this study resulted in cultivation of 0.01% of the total bacterial population from marine sediments (based on DAPI direct counts) (data not shown) regardless of the sampling date. Using primers based on the sequences obtained for the replication origins of plasmids isolated from culturable plasmid-bearing marine sediment bacteria, we attempted to determine the presence of three plasmid replicons (repSD41, repSD164, and repSD172) in microbial community DNA extracted from coastal marine sediment samples collected on different dates.
Community DNA was screened by PCR amplification for plasmid replicons repSD41, repSD164, and repSD172 (Fig. 5A). We were able to detect a positive signal from microbial community DNA extracted from the sediment samples collected in November 1995 and June 1997 and amplified with repSD41 replicon-specific primers (Fig. 5A, lanes 3 and 4). The specificity of the PCR products obtained with the repSD41 primers was confirmed by Southern hybridization by using a 1,302-bp repSD41 probe (Fig. 5B, lanes 2 and 4). PCR amplification of the original plasmid from which repSD121 was derived yielded a PCR product which differed in size and was distinguishable from the 875-bp repSD41-specific PCR product (data not shown).
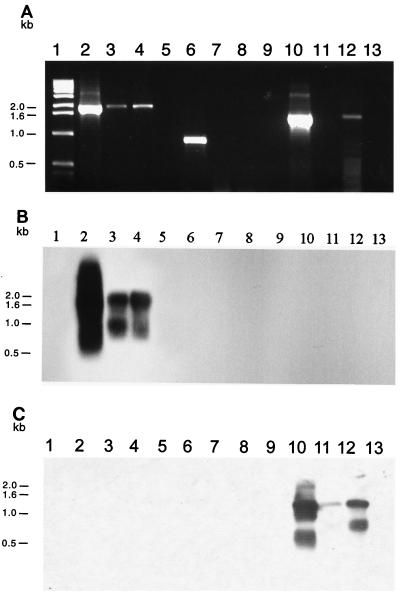
FIG. 5.
(A) Amplification of indigenous plasmid replication sequences from DNA extracted from marine sediment microbial communities: ethidium bromide-stained 0.8% agarose gel electrophoresis of PCR-amplified DNA obtained with either repSD41-specific primers (lanes 2 to 5), repSD164-specific primers (lanes 6 to 9), or repSD172-specific primers (lanes 10 to 13). Lane 1, 1-kb DNA size standard; lane 2, positive control (cesium chloride-purified plasmid pTM41 DNA); lane 3, total community DNA extracted from 1 g of sediment collected in November 1995; lane 4, total community DNA extracted from 1 g of sediment collected in June 1997; lane 5, negative control (sterilized sediment with no template DNA added); lane 6, positive control (cesium chloride-purified plasmid pTM164 DNA); lane 7, total community DNA extracted from 1 g of sediment collected in August 1995; lane 8, total community DNA extracted from 1 g of sediment collected in November 1995; lane 9, negative control (sterilized sediment with no template DNA added); lane 10, positive control (cesium chloride-purified plasmid pTM172 DNA); lane 11, total community DNA extracted from 1 g of sediment collected in November 1995; lane 12, total community DNA extracted from 1 g of sediment collected in June 1997; lane 13, negative control (sterilized sediment with no template DNA added). (B) Corresponding Southern blot analysis performed with the 1,302-bp repSD41 probe. (C) Corresponding Southern blot analysis performed with the 875-bp repSD172 probe. The band showing homology to the repSD172 probe below the predominant PCR product after 36 to 48 h of exposure to X-ray film represents smaller amplified replication products likely due to differential annealing of the PCR primers to the template DNA.
In contrast to the results obtained with primers for repSD41, we were unable to detect a positive signal when we probed with repSD164 replicon-specific primers from microbial community DNA extracted in August 1995 (data not shown), November 1995, and June 1997 (Fig. 5A, lanes 7 and 8). Attempts to detect a positive signal by Southern hybridization of the PCR products with a DNA probe internal to the repSD164 replicon sequence were also unsuccessful (data not shown). A positive signal was obtained with microbial community DNA extracted from the sediment samples collected in June 1997 and amplified with repSD172 replicon-specific primers (Fig. 5C, lane 12). Although we were unable to detect a positive signal with community DNA extracted in November 1995 (Fig. 5A, lane 11), we were able to confirm the presence of the repSD172 replicon by Southern hybridization (Fig. 5C, lane 11). Although typically we were able to visualize only one predominant PCR product for the repSD172 origin by eithidium bromide staining, subsequent hybridization with an 875-bp probe that was internal to the amplified PCR product revealed two hybridization signals (Fig. 5C, lanes 11 and 12). Two hybridization signals were detected with both community DNA and cesium-purified plasmid DNA from the parental plasmid-bearing marine isolate (Fig. 5C, lanes 12 and 10, respectively). Since additional PCR products obtained with replicon-specific primers for repSD41 and repSD164 were not detected with the repSD172 probe (data not shown), the additional hybridization signal was likely due to differential annealing of the PCR primers to the template DNA.
Since plasmid DNA is routinely isolated from laboratory-grown bacterial strains, it is imperative to control for PCR false positives caused by laboratory cross-contamination of community DNA extracted from marine sediments. To rule out such contamination, an autoclave-sterilized sediment sample was included as a negative control in each sediment extraction assay to monitor for contamination of the marine plasmid sequences. Laboratory-generated plasmid contamination of the sterilized sediment samples was never observed during these analyses (Fig. 5A through C, lanes 5, 9, and 13).
DISCUSSION
The typical clustering of genes essential for plasmid replication has facilitated isolation and characterization of replicons from plasmids isolated from bacteria of clinical and animal origins. Minimal replicons containing replication and incompatibility loci have been demonstrated to be valuable in molecular typing of plasmids from bacterial isolates of medical importance (9, 10). Replicon probes derived from plasmids found in members of the Enterobacteriaceae have not been suitable, however, for classifying plasmids isolated from either indigenous bacterial populations in coastal marine sediments (53), the marine air-water interface bulk water and biofilm communities (11), or the natural bacterial flora associated with the sugar beet phyllosphere (30). With the exception of recent studies performed with plasmids isolated from bacterial populations associated with terrestrial plant communities (24, 58), little, if any, information is available regarding the replication and incompatibility functions of bacterial plasmids occurring in natural microbial communities.
To date, only a limited number of plasmids from marine bacteria have been characterized at the molecular level (38, 57). Since molecular characterization (replicon typing) of plasmids from marine bacterial populations requires probes specific for a plasmid replication region, we isolated replication regions from several indigenous marine isolates for use as replicon probes. The same probes can be used eventually for assigning plasmids to specific incompatibility groups. Along with the four replicons used in this study, we recently isolated an additional replicon (data not shown) which can be stably propagated in E. coli hosts, indicating that a number of indigenous plasmids from marine bacteria have an extended replication host range. We failed to isolate replicons from six other plasmids, which may indicate either that the plasmids have a limited or narrow host range or that the necessary genetic information for replication is not tightly clustered and, therefore, is not be readily isolated by the method used in this study. The structural features of the repSD164 replicon suggest that pTM164 replicates via a rolling-circle mechanism, a widely dispersed strategy common in plasmids that are less than 10 kb long (25). The presence of features similar to the features of a number of plasmids that contain iterons as essential components of the origin and the presence of an open reading frame that potentially encodes a replication initiation protein suggest that the related replicons repSD41 and repSD121 use the theta mode of replication (25). Of course, much more work must be done to determine what elements in the replicons of plasmids pTM41, pTM121, pTM164, and pTM172 are absolutely essential for replicon activity before a specific replication control mechanism can be ascribed to each of these replicons. Sequence analysis revealed that the replication origins of four plasmids from gram-negative marine bacteria characterized in our study contained structural features and general organization patterns common to known replication origins (15, 25). However, sequence analysis of the replication origins of plasmids from a marine environment failed to reveal homology (e.g., there was no similarity to known rep proteins) to previously characterized replicons derived from clinical sources.
Plasmids that can replicate and be stably maintained in members of distantly related bacterial families are considered to have broad host ranges (13, 22). DNA fragments containing a functional replicon obtained from the indigenous plasmids of marine bacteria were propagated in distantly related members of the Proteobacteria, indicating that all necessary information for replication was contained on the isolated fragments. Early events in the initiation of plasmid replication independent of host factors and versatility in the interactions between plasmid-specific and host-encoded initiation proteins are two strategies thought to contribute to the ability of broad-host-range plasmids to become established in different hosts (25). The extent of plasmid host range has important implications for gene exchange in bacterial populations. Plasmids with broad host ranges are likely to mediate the dissemination of genes, presumably encoding advantageous traits, throughout microbial communities. While we did not determine the transfer abilities of the naturally occurring plasmids characterized in this study, our findings at least indicate that plasmids isolated from microbial communities of coastal marine sediments encode replication origins that have broad-host-range capabilities. Upon transfer, either through conjugation, transformation, or transduction events, such replicons should allow replication to proceed in both similar and unrelated hosts, thereby greatly increasing the potential spread of advantageous plasmid-encoded genes throughout the microbial population. It is interesting that two recent studies have suggested that plasmid-mediated gene exchange may occur infrequently between some indigenous populations of Rhizobium sp. and Bacillus sp. (61, 63).
Previously, Gotz et al. (21) used PCR-based methods to detect broad-host-range plasmids belonging to the classic incompatibility groups (groups IncN, IncP, IncQ, and IncW) in soil and manure slurries. We were able to amplify from total environmental DNA repSD41- and repSD172-specific replication sequences of marine sediment microbial community DNA obtained on sampling dates in November 1995 and June 1997. Although we have recently demonstrated that many plasmids present in culturable marine sediment bacterial populations do not carry readily assayed selective traits (53), it is likely that the metabolic burden of maintaining plasmids requires that some selective advantage be conferred to the host bacterium. Our findings therefore suggest that the repSD41 and repSD172 replicons have an established presence in the marine sediment microbial community, thus presumably conferring some advantageous trait(s) to the host cells. Previously, Lilley et al. (35) demonstrated the long-term persistence of distinct groups of large (>250-kb) self-transmissible plasmids in the natural bacterial populations of sugar beet crops sampled at the same site for 3 consecutive years. However, apart from conferring narrow-spectrum resistance to mercury, there was no obvious selective advantage encoded on these plasmids to account for their continued persistence in the sugar beet microbial community. It is of considerable interest to determine if plasmids in marine bacteria have evolved mechanisms similar to the partitioning and postsegregational killing systems found in a number of plasmids found in bacteria of clinical origin to provide for their stable maintenance (7, 20, 34, 45).
Although efforts in this study focused on characterizing plasmids from a limited number of bacteria from a culturable marine sediment population, the extent of replicon diversity and the potential for broad-host-range replication of plasmids occurring in marine microbial communities are striking. The fact that plasmid replication origins with broad host ranges can be readily isolated from marine bacterial isolates suggests that plasmid-mediated gene exchange between members of diverse bacterial genera is an important mechanism by which marine sediment microbial communities can evolve and adapt to changes or fluctuations in environmental conditions. The continued development of replicon probes specific for plasmids from environmental sources, such as marine sediments, should greatly enhance future studies of plasmid diversity and the distribution and flow of genes in natural microbial communities.
ACKNOWLEDGMENTS
This work was supported by Office of Naval Research grant N00014-95-1-0606-01.
We thank Igor Konieczny and Jed Fuhrman for helpful discussions and Alison Buchan for help with the phylogenetic analyses.
REFERENCES
- 1.Aviles M, Codina J C, Perez-Garcia A, Cazorla F, Romero P, de Vicente A. Occurrence of resistance to antibiotics and metals and of plasmids in bacterial strains isolated from marine environments. Water Sci Technol. 1993;27:475–478. [Google Scholar]
- 2.Belliveau B H, Starodub M E, Trevors J T. Occurrence of antibiotic and metal resistance and plasmids in Bacillus strains isolated from marine sediment. Can J Microbiol. 1991;37:513–520. doi: 10.1139/m91-087. [DOI] [PubMed] [Google Scholar]
- 3.Bergquist P L. Incompatibility. In: Hardy K G, editor. Plasmids—a practical approach. Oxford, United Kingdom: IRL Press; 1987. pp. 37–78. [Google Scholar]
- 4.Bethesda Research Laboratories. BRL pUC host: E. coli DH5αTM competent cells. Bethesda Res Lab Focus. 1986;8:9. [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1522. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Bravo A, Ortega S, de Torrontegui G, Diaz R. Killing of Escherichia coli cells modulated by components of the stability system ParD of plasmid R1. Mol Gen Genet. 1988;215:146–151. doi: 10.1007/BF00331316. [DOI] [PubMed] [Google Scholar]
- 8.Burton N F, Day M J, Bull A T. Distribution of bacterial plasmids in clean and polluted sites in a South Wales river. Appl Environ Microbiol. 1982;44:1026–1029. doi: 10.1128/aem.44.5.1026-1029.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaslus-Dancla E, Pohl P, Meurisse M, Marine M, Lafont J P. High genetic homology between plasmids of human and animal origins conferring resistance to the aminoglycosides gentamicin and apramycin. Antimicrob Agents Chemother. 1991;35:590–593. doi: 10.1128/aac.35.3.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couturier M F, Bex F, Bergquist P L, Maas W K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlberg C, Linberg C, Torsvik V L, Hermansson M. Conjugative plasmids isolated from bacteria in marine environments show various degrees of homology to each other and are not closely related to well-characterized plasmids. Appl Environ Microbiol. 1997;63:4692–4697. doi: 10.1128/aem.63.12.4692-4697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta N. Plasmid classification: incompatibility grouping. In: Timmis K N, Puhler A, editors. Plasmids of medical, environmental and commercial importance. Amsterdam, The Netherlands: Elsevier/North Holland Publishing Co.; 1979. pp. 3–12. [Google Scholar]
- 13.Datta N, Hedges R W. Host ranges of R factors. J Gen Microbiol. 1972;70:453–460. doi: 10.1099/00221287-70-3-453. [DOI] [PubMed] [Google Scholar]
- 14.Davey R B, Bird P I, Nikoletti S M, Prazkier J, Pittard J. The use of mini-gal plasmids for rapid incompatability grouping of conjugative R plasmids. Plasmid. 1984;11:234–242. doi: 10.1016/0147-619x(84)90029-5. [DOI] [PubMed] [Google Scholar]
- 15.del Solar G, Alonso J C, Espinsoa M, Diaz-Orejas R. Broad-host-range plasmid replication: an open question. Mol Microbiol. 1996;21:661–666. doi: 10.1046/j.1365-2958.1996.6611376.x. [DOI] [PubMed] [Google Scholar]
- 16.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durland R H, Toukdarian A, Fang F, Helinski D R. Mutations in the trfA replication gene of the broad-host-range plasmid RK2 result in elevated plasmid copy numbers. J Bacteriol. 1990;172:3859–3867. doi: 10.1128/jb.172.7.3859-3867.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberhard W G. Evolution in bacterial plasmids and levels of selection. Q Rev Biol. 1990;65:3–22. doi: 10.1086/416582. [DOI] [PubMed] [Google Scholar]
- 18a.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 19.Filutowicz M, McEachern M J, Greener A, Mukhopadhyay P, Uhlenhopp E, Durland R, Helinski D R. Role of the p initiation protein and direct nucleotide sequence repeats in the regulation of plasmid R6K replication. In: Helinski D R, Cohen S N, Clewell D B, Jackson D A, Hollaender A, editors. Plasmids in bacteria. New York, N.Y: Plenum Publishing Co.; 1986. pp. 125–140. [DOI] [PubMed] [Google Scholar]
- 19a.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 20.Gerdes K, Rasmussen P B, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cell. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotz A, Pukall R, Smit E, Tietze E, Prager R, Tschape H, van Elsas J D, Smalla K. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guiney D G. Broad-host-range conjugative and mobilizable plasmids in gram-negative bacteria. In: Clewell D, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 75–103. [Google Scholar]
- 23.Hada H S, Sizemore R K. Incidence of plasmids in marine Vibrio spp. isolated from an oil field in the northwestern Gulf of Mexico. Appl Environ Microbiol. 1981;41:199–202. doi: 10.1128/aem.41.1.199-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasnain S, Thomas C M. Two related rolling circle replication plasmids from salt-tolerant bacteria. Plasmid. 1996;36:191–199. doi: 10.1006/plas.1996.0046. [DOI] [PubMed] [Google Scholar]
- 25.Helinski D R, Toukdarian A E, Novick R P. Replication control and other stable maintenance mechanisms of plasmids. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 2295–2324. [Google Scholar]
- 26.Hermansson M, Jones G W, Kjelleberg S. Frequency of antibiotic and heavy metal resistance, pigmentation, and plasmids in bacteria of the marine air-water interface. Appl Environ Microbiol. 1987;53:2338–2342. doi: 10.1128/aem.53.10.2338-2342.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahn M, Ow D, Sauer R, Rabinowitz A, Calendar R. Genetic analysis of bacteriophage P4 using P4-plasmid ColE1 hybrids. Mol Gen Genet. 1980;177:399–412. doi: 10.1007/BF00271478. [DOI] [PubMed] [Google Scholar]
- 28.Kieser T. Factors affecting the isolation of ccc DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 29.Kita-Tsukamoto K, Oyaizu H, Nanba K, Simidu U. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int J Syst Bacteriol. 1993;43:8–19. doi: 10.1099/00207713-43-1-8. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi N, Bailey M J. Plasmids isolated from sugar beet phyllosphere show little or no homology to molecular probes currently available for plasmid typing. Microbiology. 1994;140:289–296. doi: 10.1099/13500872-140-2-289. [DOI] [PubMed] [Google Scholar]
- 31.Kobori H, Sullivan C W, Shizuya H. Bacterial plasmids in Antarctic natural assemblages. Appl Environ Microbiol. 1984;48:515–518. doi: 10.1128/aem.48.3.515-518.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leahy J G, Somerville C C, Cunningham K A, Adamantiades G A, Byrd J J, Colwell R R. Hydrocarbon mineralization in sediments and plasmid incidence in sediment bacteria from the Campeche Bank. Appl Environ Microbiol. 1990;56:1565–1570. doi: 10.1128/aem.56.6.1565-1570.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehnherr H, Yarmolinsky M B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilley A K, Bailey M J, Day M J, Fry J C. Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive years. FEMS Microbiol Ecol. 1996;20:211–227. [Google Scholar]
- 36.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 38.Matsunaga T, Mihashita H, Miyake M, Burgess J G. Nucleotide sequence of the replication region of the marine Rhodobacter plasmid pRD31. FEBS Lett. 1991;283:263–266. doi: 10.1016/0014-5793(91)80603-z. [DOI] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 40.Modi R I, Adams J. Coevolution in bacterial-plasmid populations. Evolution. 1991;45:656–667. doi: 10.1111/j.1558-5646.1991.tb04336.x. [DOI] [PubMed] [Google Scholar]
- 41.Novick R P. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Brien T F, Pla M P, Mayer K H, Kishi H, Gilleece E, Syvanen M, Hopkins J D. Intercontinental spread of a new antibiotic resistance gene on an epidemic plasmid. Science. 1985;230:87–88. doi: 10.1126/science.2994226. [DOI] [PubMed] [Google Scholar]
- 43.Ogunseitan O A, Tedford E T, Pacia D, Sirotkin K M, Sayler G S. Distribution of plasmids in groundwater bacteria. J Ind Microbiol. 1987;1:311–317. [Google Scholar]
- 43a.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 44.Prentki P, Karch F, Iida S, Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981;14:289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- 45.Roberts R C, Strom A R, Helinski D R. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J Mol Biol. 1994;237:35–51. doi: 10.1006/jmbi.1994.1207. [DOI] [PubMed] [Google Scholar]
- 46.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christen R. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int J Syst Bacteriol. 1994;44:416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 47.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of b-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 48.Sandaa R-A, Enger O. High frequency transfer of a broad host range plasmid present in an atypical strain of the fish pathogen Aeromonas salmonicida. Dis Aquat Org. 1996;24:71–75. [Google Scholar]
- 49.Schaeffer C, Messer W. DnaA protein/DNA interaction. Modulation of the recognition sequence. Mol Gen Genet. 1991;226:34–40. doi: 10.1007/BF00273584. [DOI] [PubMed] [Google Scholar]
- 50.Schmidhauser T J, Helinski D R. Regions of the broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacterial. J Bacteriol. 1985;164:446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sizemore R K, Colwell R R. Plasmids carried by antibiotic resistant marine bacteria. Antimicrob Agents Chemother. 1977;12:372–382. doi: 10.1128/aac.12.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobecky, P. A. Unpublished data.
- 52a.Sobecky P A, Schell M A, Moran M A, Hodson R E. Impact of a genetically engineered bacterium with enhanced alkaline phosphatase activity on marine phytoplankton communities. Appl Environ Microbiol. 1996;62:6–12. doi: 10.1128/aem.62.1.6-12.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobecky P A, Mincer T J, Chang M C, Helinski D R. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol. 1997;63:888–895. doi: 10.1128/aem.63.3.888-895.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeyama H, Burgess J G, Sodo H, Sode K, Matsunaga T. Salinity-dependent copy number increase of a marine cyanobacterial endogenous plasmid. FEMS Microbiol Lett. 1991;90:95–98. [Google Scholar]
- 55.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van der Plas J, Oosterhoff-Teertstra R, Borrias M, Weisbeek P. Identification of replication and stability functions in the complete nucleotide sequence of plasmid pHU24 from the cyanobacterium Synechococcus sp. PCC7942. Mol Microbiol. 1992;6:653–664. doi: 10.1111/j.1365-2958.1992.tb01513.x. [DOI] [PubMed] [Google Scholar]
- 58.Viegas C A, Lilley A K, Bruce K, Bailey M J. Description of a novel plasmid replicative origin from a genetically distinct family of conjugative plasmids associated with phytosphere microflora. FEMS Microbiol Lett. 1997;149:121–127. doi: 10.1111/j.1574-6968.1997.tb10318.x. [DOI] [PubMed] [Google Scholar]
- 59.Vieria J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 60.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wernegreen J J, Harding E E, Riley M A. Rhizobium gone native: unexpected plasmid stability of indigenous Rhizobium leguminosarum. Proc Natl Acad Sci USA. 1997;94:5483–5488. doi: 10.1073/pnas.94.10.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wortman A T, Colwell R R. Frequency and characteristics of plasmids in bacteria isolated from deep sea amphipods. Appl Environ Microbiol. 1988;54:1284–1288. doi: 10.1128/aem.54.5.1284-1288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zawadzaki P, Riley M A, Cohan F M. Homology among nearly all plasmids infecting three Bacillus species. J Bacteriol. 1996;178:191–198. doi: 10.1128/jb.178.1.191-198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]