Abstract
The most prevalent malignant illness of the gastrointestinal system, colorectal cancer, is the third most prevalent cancer in males and the second most prevalent cancer in women. Importin-11 is a protein that acts as a regulator of cancer cell proliferation in colorectal tumours by conveying -catenin to the cell nucleus. However, the IPO11 gene was found to encode a protein called Importin-11, which functions as a nucleus importer for the cell. As a result, preventing -catenin from entering the nucleus requires blocking Importin-11. As a result, we conducted a multi-omics investigation to assess IPO11 gene potential as a therapeutic biomarker for human colorectal cancer (CC). Oncomine, GEPIA2, immunohisto-chemistry, and UALCAN databases were used to analyses the mRNA expression profiles of IPO11 in CC. The investigation has yielded clear evidence of the increase of IPO11 expression in CC subtypes, as indicated by the data acquired. Analysing CC research from the cBioPortal database, the study discovered three new missense mutations in the importin-11 protein sequence at a frequency of 0.00–1.50% copy number changes. Additionally, the Kaplan-Meier plots demonstrated a strong connection concerning IPO11 downregulation and a poorer CC patient survival rate. The co-expressed gene profile of IPO11 was likewise associated with the onset of CC. IPO11 co-expressed gene profile was also linked to CC development. Moreover, the correlation analysis using bc-GenExMiner and the UCSC Xena server identified KIF2A as the most positively co-expressed gene. The study found that KIF2A and its co-expressed genes were involved in a wide variety of cancer progression pathways using the Enrichr database. Cumulatively, this result will not only provide new information about the expression of IPO11 associated with CC progression and patient survival, but could also serve as a therapeutic biomarker for treating CC in a significant and worthwhile manner.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13755-023-00259-2.
Keywords: IPO11, Colorectal cancer, Therapeutic target mutation in IPO11, Prognostic rate
Introduction
Cancer is a genetic or acquired disease in which a normal cell changes into a malevolent growth because of changes in the cell’s genetic material. It is the world’s second-most frequent cause of mortality, accounting for almost one in every six fatalities. In 2018, the World Health Organization ed that therewould be 18.1 million new cases of cancer and 9.6 million deaths globally [1]. Cancer of the colorectal area (CRC) is an infectious illness that begins in the gut’s epithelial cells and spreads via the Wnt signaling pathway. Human mutations can be passed down or picked up, and both kinds have been linked to stem cells in the crypts of the small intestine [2]. Still, CRC is an adenocarcinoma; by 2020, it is expected to cause about 10% of cancer cases. “Traditional therapeutic approaches” like surgery, radiation therapy, and chemotherapy cannot cure advanced-stage CRC, but they are good at treating early and late-stage disease [3]. The accumulation of gene changes is a major driver of oncogenesis and correlates favorably with cancer patients’ outcomes. Cancer diagnostic markers are used to find genes in which differential expression is linked to the survival of cancer patients, allowing for earlier detection of the disease. In addition, once the cause of the differences in the diagnostic biomarkers has been found, they could be used as medicines [4]. Gene mutations produce altered gene expression, a primary cause of colorectal cancer. However, the differentially expressed genes (DEGs) indicate colorectal cancer’s clinical features and patient survival. DEGs associated with a colorectal cancer diagnosis can be used as indicators for early identification and more effective therapy [5]. One must understand their important cancer progression pathways to validate DEGs as therapeutic targets. Importin 11 (IPO11) is a gene that codes for proteins [6]. The Gene Ontology (GO) annotations include binding and small GTPase binding. IPO8 is a significant paralog of this gene serving as a nuclear transport receptor to import nuclear proteins. IPO11 is a transport receptor in the importin family. It helps move proteins and RNAs from the nucleus to the cytoplasm. The protein importin-11 moves substances from the cytoplasm to the nucleus. Lower nuclear catenin protein levels and reduced catenin target gene activation in IPO11 cells suggest that IPO11 aids catenin nuclear import. The IPO11 deletion inhibited both the growth of patient-derived CRC organoids and the establishment of colony-forming units from CRC cell lines [7]. The susceptibility of cancer cells to anti-cancer medications can also be utilized to develop therapeutic targets and biomarkers [8]. Multi-omics studies can simplify the prediction of therapeutic targets and biomarkers for various cancer diseases by using genomics, epigenomics, proteomics, transcriptomics, and metabolomics data. Genomic data are more mature than other data, as established genetic types of disease and treatment can be used to predict a patient’s prognosis [9]. However, epigenomics influences genetic and environmental variables through reversible DNA methylation changes. It identified the evolution of diseases at specific periods [10]. On the other hand, the significance of transcriptomics in the physiological activities of living organisms is crucial. Long non-coding RNA was linked to numerous viral illnesses. Thus, the dysregulation of short and long RNA can be exploited as biomarkers and therapeutic targets for various illnesses [11]. Moreover, proteomics can show the abundance, changes, and interactions of proteins from the cell, which play key roles in signaling, control, activity, turnover, and transport [12]. Finally, metabolomics can predict genetic loci by regulating small biological molecules. It can be used as a biomarker or therapeutic target through the breakdown of amino acids, fatty acids, carbohydrates, and other cellular functions [13]. We can benefit from multi-omics because it allows visualization of the entire process, from the genetic, environmental, or developmental origins of disease through their functional outcomes and related interactions. This study conducted a comprehensive analysis of IPO11 expression and clinical outcomes in several tumors to evaluate its potential prognostic utility in managing colorectal malignancies. We used one of many available expression and patient survival datasets from reputable web sources. However, changes to the gene under study are linked to high IPO11 expression in colorectal cancer. In view of this, it has been determined the IPO11 expression can be examined as a biomarker for the prognosis of human colorectal cancer, which might eventually be helpful for colorectal cancer diagnosis and treatment.
Methods
IPO11 mRNA expression and human malignancies
Several human malignancies were confirmed to affect the mRNA expression level of IPO11 through the Oncomine database (https://www.oncomine.org) and constant default threshold values. The web-based data mining tool Oncomine has been utilized to find cancer-related genes from the analysis of multi-omics expression [14]. UALCAN is a public-accessible database used to characterize the differentially expressed genes (DEGs) in terms of multi-cancer progression based on the TCGA dataset [15].
IPO11 expression analysis in both CC and tumor malignancies
The mRNA expression analysis of IPO11 in colorectal cancer has been performed by using the Oncomine database, Gene Expression Profiling Interactive Analysis 2 (GEPIA2) tools and the UALCAN web server [16]. Threshold limitation was fixed for determining significant result with a standard p value of 0.05. The Oncomine study analyzes the expression of IPO11 in normal and malignant tissues. The Human Protein Atlas (HPA) database was also used to explore the variant expression of IPO11 in carcinogenic settings from the colorectal cancer cells by examining immunohistochemistry output [17]. The UALCAN database’s TCGA data were also custom to observe IPO11 mRNA manifestation as associated with clinicopathological characteristics of colorectal tumor prognosis.
Identified clinical characteristics of IPO11 promoter methylation level based on CC
In response to the methylation of the IPO11 promoter in colorectal cancer, the UALCAN database was used to generate the TCGA data. IPO11’s methylation status was examined using several clinicopathological features of colorectal cancer patients. The methylation analysis only considered statistically significant results [18].
Examined IPO11 protein sequence for the genomic mutation and copy number alteration (CNAs)
To identify the genomic mutations and copy number alterations (CNAs) in the IPO11 protein sequence, the study examined sixteen different colorectal cancer studies stored on the cBioPortal server (https://www.cbioportal.org/). These studies were retrieved from the cBioPortal website [19]. The mutation analysis not only provides an estimation of the location and frequency but also focuses on the kind of genetic alterations situated within the amino acid residues.
IPO11 expression in comparison to survival of colorectal cancer patient
Kaplan-Meier Plotter service was used to consider survival data and determine the experimental significance of IPO11 expression in colorectal malignancy development. The Kaplan-Meier plotter is a freely available online resource that uses multi-omics data to conduct a meta-analysis. The prognostic significance was evaluated by dividing the range of gene expression into the top and bottom quartiles. Colorectal cancer patients’ survival rates were shown to be correlated with their IPO11 mRNA expression levels [20].
Examine the co-expressed genes with IPO11
The popular database Oncomine database was used to determine the correlation of genes with IPO11. The function and similar expression ere compiled from the genes identified as having a positive correlation with IPO11. The KIF2A gene was confirmed through the maximum co-expressed gene with IPO11 by using the Oncomine database. As a result, both genes IPO11 and KIF2A were further visualized graphically based on co-expression correlation using the UCSC Xena web-based server [21].
Confirmed the signaling pathway and genes co-expresses with IPO11
The Oncomine server was used to identify and retrieve the co-expressed gene of IPO11. The signaling pathway was determined from the co-expressed genes related to IPO11. The popular webserver Enrichr was used to analyze related features. Diverse gene-specific experimentation was included in the Enrichr web server, where a large set of gene libraries were evaluated to interpret the signaling pathways [22].
Results
IPO11 mRNA expression in CC
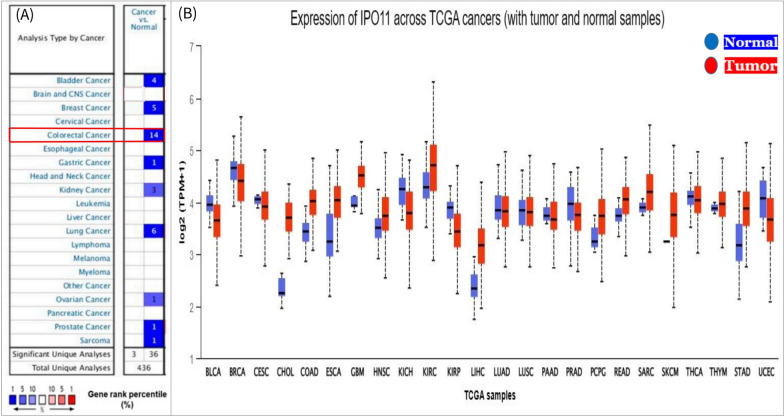
The pattern of IPO11 expression in many cancer studies was compared to the pattern of IPO11 expression in normal tissues. The aim was to see if any significant variation in IPO11 expression exists in conditions known to promote malignancy. However, the systematic review posed information on 15 different cancer types, including bile duct, colorectal, esophageal, Glioblastoma multiforme head and neck, kidney, lung, Skin Cutaneous Melanoma, and pancreatic, in which IPO11 is highly expressed in the majority of them. The highest number of meaningful individual analyses correlating with IPO11 expression was found in the colorectal cancer subtype, which is relevant to consider (Fig. 1A). Then, we analyzed the IPO11 expression levels across 24 separate cancer studies using the UALCAN database. Here, we found that IPO11 expression is high in eleven but low in thirteen cancers (Fig. 1B).
Fig. 1.
IPO11 mRNA expression in diverse cancer types: A mRNA levels going up (red) and down (blue) in different kinds of cancer. IPO11 is turned up in several types of cancer, including colorectal cancer, which is shown in blue. B Boxplots displayed the expression of IPO11 in several TCGA cancer data sets with tumor (red) and normal (blue) samples. The values between the upper and lower quartiles within the box and the dashed lines represented the upper and lower limits of average expression
Receptor expression of IPO11 in CC
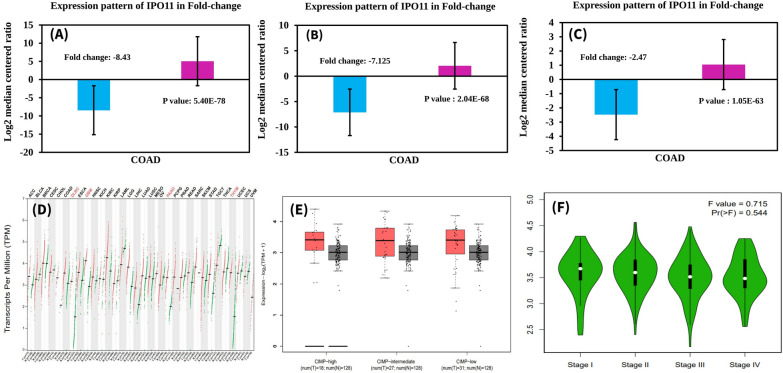
IPO11 was expressed in each type of colorectal cancer using the Oncomine database. However, we discriminated the IPO11 expression in colorectal cancer subtypes by comparing the level of IPO11 expression in the normal condition with the expression level found in malignant carcinoma. The colorectal cancer subtypes greatly overexpressed the IPO11 gene (Fig. 2A–C). We found similar results using the GEPIA2 database, indicating the elevation of IPO11 expression in colorectal cancer (Fig. 2D). Using the immunohistochemistry data that the Human Protein Atlas Project stored, we next investigated the characteristics of IPO11 protein expression in normal tissue (glandular cells) and tissue taken from colorectal cancer patients (tumor cells). In this case, we discovered that the staining signal in normal glandular cells was of moderate intensity, but the staining level of tumor cell expression was significantly greater (Fig. 2E). In addition, the intensity signal of glandular cells was found to be at a level in the middle. Nevertheless, in the case of cancer cells, we discovered a high-intensity signal in each of the many stages of the tumor (Fig. 2F).
Fig. 2.
The expression pattern of IPO11 was associated with tumor and normal tissue. A–C Fold-change of IPO11 in three forms of tumors was found from data analyses, shown as a box plot. The box plot on the right is a result of an analysis of the ONCOMINE database, and it compares the levels of IPO11 expression in normal tissue with those in cancerous tissue (D). GEPIA collected IPO11 expression data from the TCGA database (E). Box plots displaying the IPO11 mRNA expression in the primary tumour (red plot) and normal tissues (black plot) using data from the TCGA database retrieved via UALCAN. A stage plot depicting the expression of the IPO11 gene in different cancer stages (F)
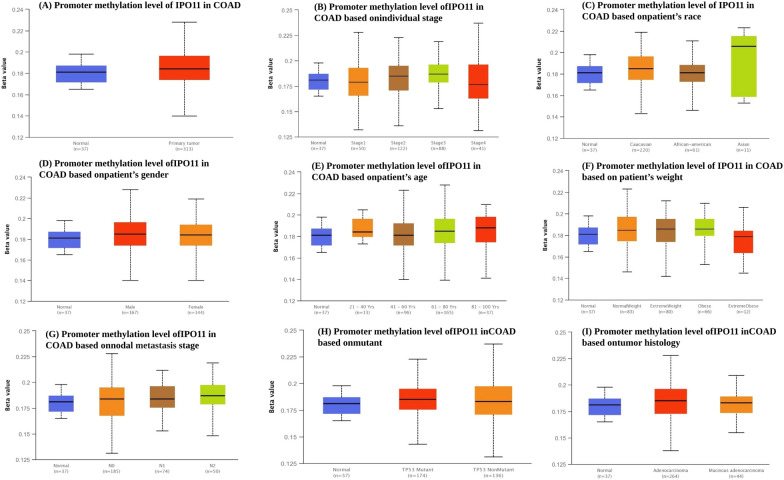
Analysis of IPO11 promoter methylation in CC based on multiple clinicopathological factors
Cliniopathological characteristics were used to analyse the different types of promoter methylation levels depending on IPO11 expression. The Beta number reflects the degree of DNA methylation, from 0 (unmethylated) to 1 (fully methylated). Beta values between 0.7 and 0.5 have been recognised as the threshold for hyper-methylation and hypo-methylation, respectively. However, compared to normal tissues, the study revealed that IPO11 promoter methylation has increased in primary cancers (Fig. 3A). The study revealed that, in terms of particular cancer stages, the Beta value of methylation was higher in tumour tissue than in normal tissue. IPO11 was found to have the least amount of methylation at stage one, compared to stages three and four (Fig. 3B). Moreover, in comparison to all other races, Asian patients had the highest level of IPO11 methylation. In contrast, African Americans had the lowest level (Fig. 3C). Increased IPO11 promoter methylation was observed more frequently in female patients than male patients (Fig. 3D). Patients between the ages of 21 and 40 had the highest amount of promoter methylation when compared to other age groups. On the other hand, we found that the level of IPO11 promoter methylation was most downregulated in people aged 41 to 60 (Fig. 3E). Compared to all other races, people with normal weights had the highest level of IPO11 methylation. In contrast, people who were very overweight had the lowest level (Fig. 3F). The stage of nodal metastasis was observed in each of the three groups where the high methylation is more prevalent in the third group of patients than in previous groups (Fig. 3G). Cancer Genome Atlas (TCGA) entire exome sequencing data are used to determine TP53 mutation status. The statistical significance between the normal and TP53 mutant groups was 2.597200E−01 (Fig. 3H). We observed that adenocarcinoma had the highest promoter methylation level of all histological subtypes (Fig. 3I).
Fig. 3.
The TCGA dataset was used to look at the level of methylation of the IPO11 promoter in different clinic pathological conditions. The methylation level of IPO11 was compared to A sample type, B stages, C, race, D, gender, E age, F cancer subclass, G metastasis stage, H, mutant, and I tumour histology
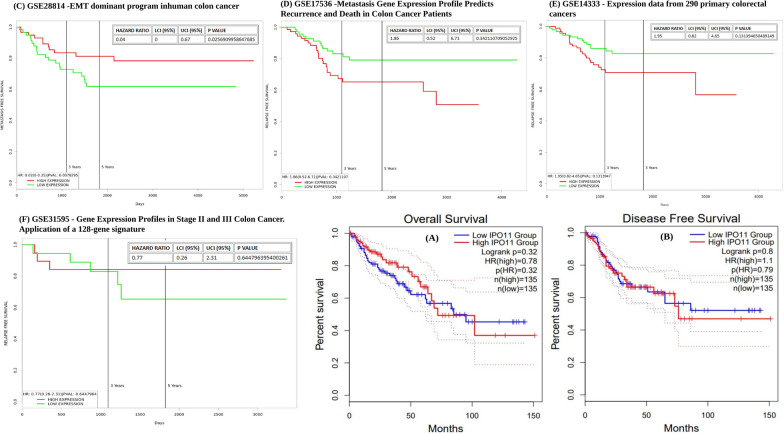
Analysis of the relationship between IPO11 expression and CC cancer patient survival
Despite the established involvement of IPO11, its clinical significance in determining the outcome of colorectal cancer remains unclear. Patients with colorectal cancer fared better when IPO11 levels were low. We also found that the expression of IPO11 was the opposite of how people with colorectal cancer were likely to fare in the future, although the computed p value for overall survival was not statistically significant. Based on the P HR value of 0.32, all of the findings were plotted in a forest plot (Fig. 4A). There was no significant difference in disease-free survival between the lower and higher IPO11 groups (Fig. 4A, B). Moreover, epithelial to mesenchymal transition (EMT) is linked to invasive or metastatic phenotypes in colorectal cancer (CRC) (Fig. 4C). It showed high expression until three years of death, where p value was 0.025. Patients with colon cancer had an insufficiently predicted metastatic risk based on staging. We employed a gene expression profile constructed from aggressive, metastatic colon cancer cells produced from the graph where hazard radio was 1.86 (Fig. 4D). Surgically resected tissues from 290 individuals with colorectal cancer were studied. The high expression of IPO11 gene was observed until 30 days, whereas low expression of IPO11 gene was expressed at the end of 4000 days (Fig. 4E). Patients with stages II and III colorectal tumors may benefit from testing a 128-gene signature designed to predict survival. The current investigation aimed to verify the 128-gene signature in new, unrelated data. The 128-gene signature was reliable in predicting stage III colon cancer recurrence but not stage II. Unsatisfactory individual patient predictions were found in a separate Danish source (Fig. 4F).
Fig. 4.
Survival predictions for colorectal cancer patients were associated with IPO11 expression. Based on IPO11 expression, relapse-free and overall survival probabilities were assessed for several colorectal cancer subtypes (A, B). Red indicates increased expression and black indicates decreased expression in the survival curves. In human colon cancer, epithelial-mesenchymal transition (EMT) is the driving force (C). Colon cancer patients’ metastatic gene expression profile predicts recurrence and mortality (D). Expression data were derived from 290 primary colorectal tumours (E). Gene expression profile in stage 11 and 111 colon cancer application of a 128-gene signature (F)
IPO11 mutation and CNA analysis associated with CC cancer development
Colorectal cancer and alterations in the IPO11 protein’s DNA sequence were investigated. Here, we report the discovery of three missense mutations in the human IPO11 gene (Table 1). The whole protein sequence of IPO11 has a mutation, which was identified (Fig. 5A). Next, we noticed several sorts of copy number modifications with a frequency ranging from 0.0% to 1.5%. Several investigations of colorectal cancer were used to construct the bar graphs, with the MBC project demonstrating the highest frequency rate (1.50%) of alterations. TCGA data on colorectal cancer revealed a greater mutation prevalence (1.5%). Amplification was determined to be the most prevalent change in these investigations (Fig. 5B). We noticed that shallow deletion-type CNAs predominated over all other types of mutations. Nevertheless, the greatest expression of IPO11 was seen for the shallow deletion type variant (Fig. 5C).
Table 1.
List of mutations identified in the IPO11 protein sequence from multiple colorectal cancer studies
| Cancer dataset | Sample size | Protein change | Mutation type | Sample ID |
|---|---|---|---|---|
| Colorectal Adenocarcinoma (DFCI, Cell Reports 2016) | 619 | R875Q | Missense | coadreadfci016 |
| Colorectal Adenocarcinoma (Genentech, Nature 2012) | 951 | V570A | Missense | coadreadenentech |
| Colorectal Adenocarcinoma (TCGA, Firehose Legacy) | 3504 | K268N | Missense | coadreadcga |
Fig. 5.
Genomic mutation determination of IPO11 in colorectal cancer (A). Lollypop plots show missense type mutation in IPO11 (B). The alteration frequency of CNAs in IPO11 is presented as a bar chart where the green blocks indicate mutation and the blue blocks indicate deep deletion type CNAs (C). Analysis of the mRNA expression of variant types of CNAs in IPO11 based on TCGA databases
Prediction of protein–protein interaction and cross-cancer analysis of IPO11 mutations and copy number alterations
The protein–protein interaction is not only collected co-expression, interaction, pathways, and prediction but also shared domains of protein through the GeneMANIA and STRING web. However, IPO11 is shows the interaction and analyzed the co-occurrence of functional proteins pattern (Fig. 6A). Changes in frequency were shown in blue (mutations), violet (no mutations), and red (not profiled) (Fig. 6B). The predicted protein partners of IPO11 were: SH3GL2, CACYBP (calcyclin binding protein), ADORA3 (adenosine A3 receptor), CSE1L (chromosome segregation 1 like), HUWE1 (HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1), IPO7 (importin 7), IPO8 (importin 8), PRDX2 (peroxiredoxin 2), TNPO1 (transportin 1), XPO7 (exportin 7), IPO9 (importin 9), TNPO2 (transportin 2), RANBP 17 (RAN binding protein 17), UBE1E2 (ubiquitin conjugating enzyme E2 E1), UBE2E3 (ubiquitin conjugating enzyme E2 E3), PLCD4 (phospholipase C delta 4), XPO1 (exportin 1), RPL12 (ribosomal protein L12), RAN (RAN, member RAS oncogene family) and DDX5 (DEAD-box helicase 5) (Fig. 6C). Hence, these anticipated interacting protein partners of IPO11 may be implicated in regulating cancer progression and prognosis mediated by IPO11. Using cBioPortal’s ONCOPRINT function, we analysed the mutations and CNAs of IPO11’s 30 predicted interacting protein partners. Finally, queried genes were altered in 4384 queried patients from the 4260 samples.
Fig. 6.
Functional protein interaction and co-occurrence partners of IPO11. A Essential structural proteins for IPO11 function predicted using GeneMANIA and STRING web. Nodes are represented on the screen by circles Co-expression, co-localization, genetic relationships, pathways, physical interactions, and shared protein domains suggest functional partners. B The highest reported rates of mutation (>10%) were found in studies of squamous cell cancer. Blue denoted mutations, violet non-mutations, and not profiled were all part of the frequency distribution of alterations (red). C Using cBioPortal online, the alteration frequency of an 11-gene signature from the GeneMANIA database was determined. Datasets with > 100 samples per cancer type and >20% modification frequency are displayed
Identification of IPO11 and others co-expressed gene-signaling pathways in CC
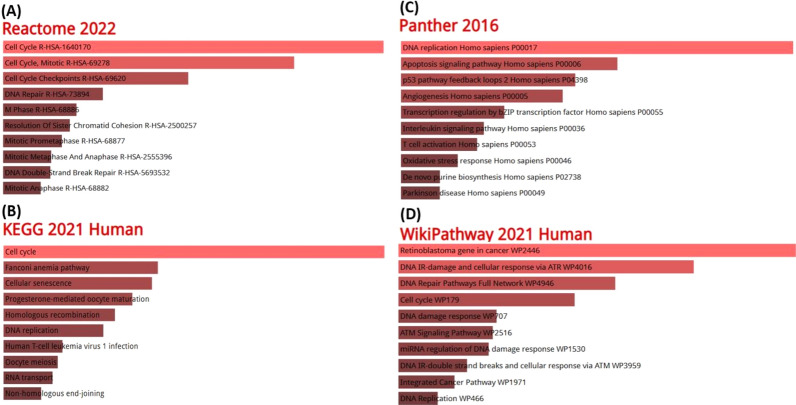
The IPO11 and related co-expressed genes contributed significantly to colorectal cancer-promoting pathways. Cell cycle, mitotic cell cycle, DNA repair, M phase, mitotic prophase, mitotic metaphase, and anaphase, and DNA double strand bank repair were among the top most represented reactome pathways, indicating IPO11 and other positively co-altered genes for incorporation with multidisciplinary pathways associated with cancer development (Fig. 7A). However, cell cycle, cellular senescence, progesterone-mediated oocyte maturation, homologous recombination, meiosis in oocytes, and RNA transport were some of the most common signalling pathways investigated by the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Fig. 7B). Additionally, Based on topmost panther pathways, we found that IPO11 and co-expressed genes were mostly related to DNA replication, Apoptosis signaling pathway, p53 signaling pathway, Angiogenesis homo sapiens, Transcription regulation, Interleukin signaling pathway, T cell activation Homo sapiens, and Oxidative stress response (Fig. 7C). Finally, We searched wikipathway that Retinoblastoma gene in cancer, DNA IR-damage and cellular response via WP4016, DNA repair pathways full network, cell cycle, DNA damage, ATM signaling pathway, Intregated cancer pathway and DNA replication.
Fig. 7.
The role of co-expressed genes in the IPO11 signalling pathway. The bar length represents the significance of that term and brighter color represents more significance A Reactome Pathway, B KEGG 2019 pathway, C Panther 2016 pathway, D WikiPathway 2021 human
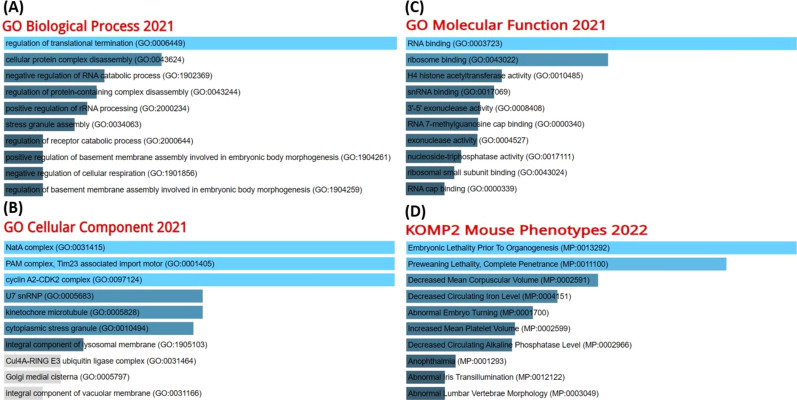
Finally, we used the list of IPO11’s co-expressed genes to investigate the molecular mechanism underlying prognosis in colorectal cancer. Go biological process pathways indicated IPO11 and other favorably co-altered genes for incorporation into interdisciplinary cancer development pathways such as regulation of transitional termination, cellular protein complex disassembly, negative regulation of RNA catabolic process, negative regulation of cellular respiration and regulation of basement membrane assembly involved in embryonic body morphogenesis. (Fig. 8A). The cellular component pathway signified the NatA complex, PAM complex, and integral component of lysosomal membrane (Fig. 8B) Go molecular function shows RNA binding and Ribosome binding (Fig. 8C) and Embryonic lethality before organogenesis preweaning lethality and decreased circulating iron level (Fig. 8D).
Fig. 8.
A GO Biological Process 2021, B GO Cellular Component 2021, C GO Molecular Function 2021 and D KOMP2 Mouse Phenotypes 2022
Discussion
Colorectal cancer affects both men and women. It is the third most common cancer in terms of new cases and the second most common cancer in terms of deaths [23]. There are approximately two hundred different types of cancer, each characterized by the unchecked growth and spread of cells in a specific organ or tissue. Lung, breast, prostate, and colon cancer are the four most frequent types of the disease. In terms of mortality, colorectal cancer (CRC) ranks second overall and third among both sexes. Around 500,000 people each year are killed by the disease, accounting for 10% of all male and 9.2% of all female cancer cases worldwide [24]. It is projected that there will be 147,950 new cases of CRC in the United States, with an associated mortality rate of roughly 53,200. Colon cancer is estimated to be fatal for 1 in 32 German men and 1 in 39 German women throughout a lifetime [25].
Molecular genetics studies have shown three distinct molecular subtypes of colorectal cancer. The “chromosomal unstable” category, characterized by a buildup of mutations in specific oncogenes and tumor suppressor genes, is by far the most common [26]. The most prevalent kind of genomic instability in CRC is chromosomal instability. Multiple copy numbers and structural alterations to chromosomes are hallmarks of this condition. The physical loss of a wild-type copy of a tumor suppressor gene, such as APC, P53, or SMAD4, can initiate a cascade of events that disrupt the normal functions of these genes [27]. When more genes were found to be impacted by the process, it became clear that some groups of genes had consistently enhanced methylation in certain malignancies; this led to the identification of the CpG Island Methylation phenotype (CIMP) [28].
Consequently, we conducted a full multi-omics investigation of the IPO11 expression pattern in colorectal cancer using a wide range of free bioinformatics tools. Intriguingly, we found that all of the cases of colorectal cancer we looked at had their IPO11 expression turned down. Since then, we’ve been looking at how IPO11 is expressed in different kinds of colorectal cancer. Subsequently, we compared colorectal cancer and healthy tissue using immunohistochemistry. The tumor cells in this study significantly differed from normal glandular cells in terms of staining, intensity, and quality. Finally, we used clinicopathological factors to assess the extent to which IPO11 was expressed and its promoter was methylated. Colorectal cancer has been linked to a decrease in IPO11 mRNA expression and an increase in promoter methylation levels across the board. Our investigation into the functional characterization of the IPO11 protein was motivated by a desire to understand better the factors that led to these shifts in gene expression. The 975-amino-acid human importin-11 protein was shown to have a frequency of genetic variations between 0.00 and 4.00%, with three missense kinds of mutation and many additional copy number modifications being discovered. These alterations in the gene sequence may be useful as biomarkers for IPO11 downregulation in colorectal cancer. This led us to investigate whether or not IPO11 expression was associated with a better prognosis for patients with colorectal cancer. This study found a statistically significant link between IPO11 expression and the likelihood of survival for patients with colorectal cancer.
In order to gain a better understanding of the role that IPO11 plays in the development of colorectal cancer, we focused our attention on the genes that co-expressed IPO11 at the highest levels. This study identified KIF2A as the most strongly related gene among 878 substantially co-expressed genes. A proper mitotic progression necessitates the protein produced by this gene, which is a plus end-directed motor. The encoded protein is essential for appropriate spindle activity during mitosis. MMany transcript variants have been identified for this gene, each of which encodes a unique isoform. An M-type nonmotile microtubule depolymerase, kinesin superfamily member 2A (KIF2A), has been studied for its potential role in carcinogenesis and prognostic significance in various cancers. In recent research, KIF2A expression was found to predict clinical outcomes in patients with colorectal cancer (CRC). KIF2A has been proposed as a novel predictive and prognostic marker for breast cancer and it may play a significant role in the development of the disease. According to a recent study, KIF2A overexpression has been linked to more advanced T and TNM stages in colorectal cancer patients. Moreover, The PANTHER pathway indicated the most probable interconnection of IPO11 is with the apoptosis signaling pathway.
The study revealed that, KIF2A regulates caspase activation and apoptosis by stabilizing microtubules, which can activate the pro-apoptotic protein Bax. Although IPO11 (Importin 11) is a nuclear transport receptor that plays a role in nuclear import and export. Recent studies have shown that IPO11 regulates apoptosis by interacting with various pro-apoptotic and anti-apoptotic proteins, including Bcl-2, Bak, and Bax [29]. One study found that KIF2A, along with IPO11, interacts with the pro-apoptotic protein Bax, which triggers the intrinsic pathway of apoptosis. Another showed that KIF2A and IPO11 bind to Bax and promote its mitochondrial localization, leading to the release of cytochrome c, activation of caspase-9, and, ultimately, apoptosis. The co-expression of KIF2A and IPO11 promoted cell proliferation and inhibited apoptosis in cancer cells. In this case, KAF2A acts as the apoptosis signalling pathway’s suppressor by inhibiting other complexes’ expression. Overall, our studies suggest that KIF2A and IPO11 may regulate apoptosis signaling pathways, leading to colorectal cancer progression.
Finally, we identified the important pathways that IPO11 and its most abundant co-expressed genes activated. Overall, we saw that IPO11 and the genes that work with it are linked to oncogenic signaling pathways. We used the KEGG pathway to find that IPO11 and its positively co-expressed genes were linked to cancer-related pathways. This pathway study provided substantial evidence linking IPO11 to carcinogenesis. The connection between IPO11 and its co-expressed genes and the cell cycle became apparent through the Reactome pathways. Dysfunctions in the cell cycle, a key biological system, have been related to cancer formation through fueling malignant growth. Mutagenic downregulation of the tumor suppressive IPO11 gene and its co-expressed genes may induce cell cycle disruption. According to the PANTHER pathway, IPO11 is most likely linked to the Fanconi anemia pathway. New evidence suggests that the FA pathway helps maintain genomic integrity by stabilizing replication forks, reducing replication stress, and controlling cytokinesis as part of a larger tumor-suppressor network. We have since turned to Gene Ontology (GO) analysis to learn more about the major roles played by IPO11 and its co-expressed genes at the cellular, molecular, and organismic levels. We discovered that IPO11 is most closely associated with the cellular response to cytokine stimulation. Cytokines can be produced by breast cells and used either to suppress tumor development or promote it. Because of the involvement of IPO11 in the cellular response to the Nata complex, manipulating NATs in cancer cells can trigger cell-cycle arrest, apoptosis, or autophagy. Gene Ontology (GO) molecular function analysis of colorectal cancer genes showed that IPO11 and its related co-altered genes are mostly involved in kinase binding [30]. Mutations in protein tyrosine kinases, which are oncogenic, are thought to be the tightly controlled catalyst in colorectal cancer [31]. As a result of its interaction with kinase binding, IPO11 may be involved in colorectal cancer. As a result, these studies support the idea that IPo11 functions as a biomarker for detecting colorectal cancer.
Conclusion
Multi-omics has contributed to understanding biological processes, including molecular functions, interactions, and cellular fate. In the era of precision medicine, multi-omics can facilitate the discovery of predictive or prognostic biomarkers and potentially repurposed and innovative therapeutic targets. However, IPO11 could be used as a therapeutic biomarker for colorectal cancer, confirmed through the different bioinformatics tools. In this extensive multi-omics study, we investigated the known contributors to the development and progression of colorectal cancer, including their expression, methylation, immunohistochemistry, functional status through mutation and copy number analysis, prognostic significance, and effects on various signalling pathways. The current findings investigate IPO11 as a possible tumor suppressor whose expression is considerably elevated in colorectal cancer growing tissues. This downregulation of IPO11 expression may result from elevated methylation levels or other genomic modifications, including mutations or copy number abnormalities. The ultimate outcomes of this research approach will likely determine the efficiency of IPO11 as a therapeutic biomarker for colorectal cancer.
Supplementary Information
Acknowledgements
The authors gratefully acknowledge technical and financial support provided by the Ministry of Education and King Abdulaziz University, DSR, Jeddah. Our appreciations are extended to the editor for handling the process and to the reviewers for their review and valuable comments.
Author contributions
MHRM and MOA: Conceptualization, MHRM, and MOA: methodology, formal analysis, and writing original draft preparation, MHRM and MOA: supervision, writing review and editing and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was funded by Institutional Fund Projects under Grant no. (IFPIP: 38-130-1443) King Abdulaziz University-Institutional Funding Program for Research and Development, Ministry of Education, Kingdom of Saudi Arabia.
Data availibility
Additional materials related with the study are available upon request to the authors.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Zhao H, Ming T, Tang S, Ren S, Yang H, Liu M, et al. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer. 2022;21:1–34. doi: 10.1186/s12943-022-01616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dovizio M, Ballerini P, Fullone R, Tacconelli S, Contursi A, Patrignani P. Multifaceted functions of platelets in cancer: from tumorigenesis to liquid biopsy tool and drug delivery system. Int J Mol Sci. 2020;21:1–31. doi: 10.3390/ijms21249585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith JC, Sheltzer JM. Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. elife. 2018;7:e39217. doi: 10.7554/eLife.39217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song D, Zhang D, Chen S, Wu J, Hao Q, Zhao L, et al. Identification and validation of prognosis-associated DNA repair gene signatures in colorectal cancer. Sci Rep. 2023;12(1):6946. doi: 10.1038/s41598-022-10561-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long T, Liu Z, Zhou X, Yu S, Tian H, Bao Y. Identification of differentially expressed genes and enriched pathways in lung cancer using bioinformatics analysis. Mol Med Rep. 2019;19(3):2029–2040. doi: 10.3892/mmr.2019.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plafker SM, Macara IG. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 2000;19:5502. doi: 10.1093/emboj/19.20.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mokhtari RB, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das B, et al. Combination therapy in combating cancer. Oncotarget. 2017;8:38022. doi: 10.18632/oncotarget.16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raufaste-Cazavieille V, Santiago R, Droit A. Multi-omics analysis: paving the path toward achieving precision medicine in cancer treatment and immuno-oncology. Front Mol Biosci. 2022;9:962743. doi: 10.3389/fmolb.2022.962743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumon MAA, Molla MHR, Hakeem IJ, Ahammad F, Amran RH, Jamal MT, et al. Epigenetics and probiotics application toward the modulation of fish reproductive performance. Fishes. 2022;7:189. doi: 10.3390/fishes7040189. [DOI] [Google Scholar]
- 11.Aljahdali MO, Molla MHR, Filfilan WM. Whole genome sequence of the newly prescribed subspecies Oreochromis spilurus saudii: a valuable genetic resource for aquaculture in saudi arabia. J Mar Sci Eng. 2021;9:506. doi: 10.3390/jmse9050506. [DOI] [Google Scholar]
- 12.Samad A, Haque F, Nain Z, Alam R, Al Noman MA, Rahman Molla MH, et al. Computational assessment of MCM2 transcriptional expression and identification of the prognostic biomarker for human breast cancer. Heliyon. 2020;6:e05087. doi: 10.1016/j.heliyon.2020.e05087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aljahdali MO, Molla MHR, Ahammad F. Compounds identified from marine mangrove plant (Avicennia alba) as potential antiviral drug candidates against WDSV, an in-silico approach. Mar Drugs. 2021;19:253. doi: 10.3390/md19050253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X, et al. Expression patterns and prognostic value of m6A-related genes in colorectal cancer. Am J Transl Res. 2019;11:3972–91. [PMC free article] [PubMed] [Google Scholar]
- 18.Coppedè F, Migheli F, Lopomo A, Failli A, Legitimo A, Consolini R, et al. Gene promoter methylation in colorectal cancer and healthy adjacent mucosa specimens: correlation with physiological and pathological characteristics, and with biomarkers of one-carbon metabolism. Epigenetics. 2014;9:621. doi: 10.4161/epi.27956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira DM, Santamaria G, Laudanna C, Migliozzi S, Zoppoli P, Quist M, et al. Identification of copy number alterations in colon cancer from analysis of amplicon-based next generation sequencing data. Oncotarget. 2018;9:20409. doi: 10.18632/oncotarget.24912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Romero J, Bueno-Fortes S, Martín-Merino M, Ramirez De Molina A, De Las Rivas J. Survival marker genes of colorectal cancer derived from consistent transcriptomic profiling. BMC Genomics 2018;19(18):45–60. [DOI] [PMC free article] [PubMed]
- 21.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:1–14. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawicki T, Ruszkowska M, Danielewicz A, Niedźwiedzka E, Arłukowicz T, Przybyłowicz KE. A review of colorectal cancer in terms of epidemiology, risk factors, development, symptoms and diagnosis. Cancers. 2025;2021:13. doi: 10.3390/cancers13092025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Gastroenterol Rev. 2019;14:89. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel RL, Miller KD, Sauer AG, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 26.Breuhahn K, Gores G, Schirmacher P. Strategies for hepatocellular carcinoma therapy and diagnostics: lessons learned from high throughput and profiling approaches. Hepatology. 2011;53:2112–21. doi: 10.1002/hep.24313. [DOI] [PubMed] [Google Scholar]
- 27.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mojarad EN, Kuppen PJK, Aghdaei HA, Zali MR. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol Hepatol. 2013;6:120. [PMC free article] [PubMed] [Google Scholar]
- 29.Molla MHR, Aljahdali MO, Sumon MAA, Asseri AH, Altayb HN, Islam MS, et al. Integrative ligand-based pharmacophore modeling, virtual screening, and molecular docking simulation approaches identified potential lead compounds against pancreatic cancer by targeting FAK1. Pharmaceuticals. 2023;16:120. doi: 10.3390/ph16010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Bao Y, Ma M, Yang W. Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int J Mol Sci. 2017;18(4):722. doi: 10.3390/ijms18040722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drake JM, Lee JK, Witte ON. Clinical targeting of mutated and wild-type protein tyrosine kinases in cancer. Mol Cell Biol. 2014;34:1722. doi: 10.1128/MCB.01592-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional materials related with the study are available upon request to the authors.