Abstract
Recent studies have shown that archaea which were always thought to live under strict anoxic or extreme environmental conditions are also present in cold, oxygenated seawater, soils, the digestive tract of a holothurian deep-sea-deposit feeder, and a marine sponge. In this study, we show, by using PCR-mediated screening in other marine eukaryotes, that marine archaea are also present in the digestive tracts of flounder and grey mullet, two fish species common in the North Sea, in fecal samples of flounder, and in suspended particulate matter of the North Sea water column. No marine archaea could be detected in the digestive tracts of mussels or the fecal pellets of a copepod species. The archaeal 16S ribosomal DNA clone libraries of feces of flounder and the contents of the digestive tracts of grey mullet and flounder were dominated by group II marine archaea. The marine archaeal clones derived from flounder and grey mullet digestive tracts and feces formed a distinct cluster within the group II marine archaea, with 76.7 to 89.8% similarity to previously described group II clones. Fingerprinting of the archaeal community of flounder digestive tract contents and feces by terminal restriction fragment length polymorphism of archaeal 16S rRNA genes after restriction with HhaI showed a dominant fragment at 249 bp, which is likely to be derived from group II marine archaea. Clones of marine archaea that were closely related to the fish-associated marine archaea clones were obtained from suspended particulate matter of the water column at two stations in the North Sea. Terminal restriction fragment length polymorphism fingerprinting of the archaeal community present in suspended particulate matter showed the same fragment pattern as was found for the archaeal community of the flounder digestive tract contents and feces. These data demonstrate that marine archaea are present in the digestive tracts and feces of very common marine fish. It is possible that the marine archaea associated with the digestive tracts of marine fish are liberated into the water column through the feces and subsequently contribute to the marine archaeal community of suspended particulate matter.
Until a few years ago, the domain of the Archaea was considered to consist of only methanogens that live under strict anoxic conditions and extremophiles that inhabit inhospitable environments (24, 28). However, with the discovery of 16S ribosomal DNA (rDNA) sequences of archaea in cold, oxygenated ocean water (3, 4, 8, 9), it became clear that archaea might be more widely distributed. In coastal waters of the Atlantic and Pacific oceans, marine archaea constitute between 2 and 8% of the prokaryotic community (3, 17). Occasionally they can be very abundant and contribute up to 34% of the prokaryotic biomass as was found for Antarctic waters (4). Archaea present in ocean water are designated marine archaea and can be divided into three phylogenetic lineages (3, 7, 8). The first lineage constitutes the group I marine archaea belonging to the subdomain of the Crenarchaeota, which includes extreme thermophilic species. The second lineage is the group II marine archaea and is part of the subdomain Euryarchaota, which includes thermophiles, sulfur-metabolizing microorganisms, and all known methanogens. The third lineage of marine archaea comprises clones that have been obtained from deep-water samples (7); closely related archaeal sequences have been retrieved from coastal (19) and continental shelf sediments (27). A recent study on water samples from the Santa Barbara Channel showed that the group I and II marine archaea have different vertical distributions (16). Group II is dominant in the surface layer, while group I becomes abundant at depths of 100 m (16), thus suggesting that representatives of the two groups have different ecological traits. Not-yet-cultivated archaea have also been found in other habitats. Currently, crenarchaeotal 16S rRNA gene sequences have been detected in agricultural and forest soils (2, 12, 25), freshwater and coastal sediments (15, 19, 23), and deep-sea sediment (13).
The difference in membrane lipid composition between bacteria and archaea (10) has been used to specifically look for archaeal lipids that cannot be assigned to known cultivated members of the Archaea. An unknown C40-ether-bound lipid, which was assigned to a planktonic marine archaeon, was detected in particulate organic matter of the Cariaco Trench and the Black Sea water column (11). Compound-specific isotope analysis of the carbon skeleton of this lipid suggested that this marine archaeon utilizes an isotopically heavy carbon source, such as algal carbohydrates or dissolved bicarbonate (11). Recently, the identification of specific lipids associated with group I marine archaea has been reported by DeLong et al. (5). The only other studies on marine archaea showed that members of the group I marine archaea were found in the gut contents of a deep-sea-deposit feeder (18) and in a marine sponge (21).
To extend knowledge on the distribution of marine archaea in marine animals, we screened digestive tract and fecal samples of two marine fish species, common mussels, and a copepod species for the presence and diversity of marine archaea. Here we report on the association of group I and II marine archaea with the digestive tracts of flounder and grey mullet, two fish species which are commonly found in the North Sea. It was also shown that feces of flounder and suspended particulate matter of the North Sea water column contain group I and II marine archaea. These clones were closely related to the ones found in the digestive tract contents. The data suggest that certain marine fish species host marine archaea in their digestive tracts and that the feces of these fish could be a source of particle-associated marine archaea.
MATERIALS AND METHODS
Community DNA samples and DNA extraction.
Large volume (100-liter) samples of the North Sea water column were taken in May 1996 from stations 10 and 235 miles northwest of the coast of Terschelling, The Netherlands (Fig. 1). These samples were designated TS 10 and TS 235, respectively. The water samples were stored at 4°C after sampling, and within several days the water was filtered over glass fiber filters (Whatman GF/C) with a low-pressure filtration setup. The filters were stored at −80°C until further use. Freshly caught species of grey mullet (Mugil cephalus) and flounder (Platichthys flesus), which are abundant in the North Sea, were purchased at the local fish market in Groningen, The Netherlands. The fish were dissected immediately after purchase. The contents of the digestive tract were carefully removed and stored at −80°C. Feces from flounder were collected from the bottom of an aquarium at the Department of Marine Biology, University of Groningen, and stored at −80°C until further use. Fecal pellets of a copepod species (Pseudocalamus sp.) were obtained from the Department of Biological Oceanography of The Netherlands Institute for Sea Research in Texel, The Netherlands. The copepods were kept in a large aquarium and fed with a mixture of three different algae. Mussels (Mytilus edilus) were collected from small rocks that can be found along the North Sea coast of The Netherlands. The digestive tract was dissected and, after being washed with sterile water, was stored at −80°C until further use.
FIG. 1.
Map of The Netherlands and part of the North Sea showing the locations of stations TS10 and TS235, from which samples in this study were taken.
DNA was extracted from one-fourth of the GF/C filter after it was cut into small pieces with a sterile surgical blade or from small subsamples of the fish or mussel digestive tract contents or feces by incubation in 1.8 ml of buffer containing 5% (wt/vol) sucrose, 50 mM EDTA, 5 mM Tris–HCl (pH 8), 1 M guanidinium thiocyanate, and 0.4 mg of lysozyme for 30 min on ice. After the addition of 67 μl of 25% sodium dodecyl sulfate solution, the mixture was incubated for 30 min at 37°C. Proteins were removed by incubation with 1 mg of proteinase K at 55°C for 3 to 4 h. The last step in the extraction procedure consisted of boiling the mixture for 1 to 2 min. Nucleic acids were extracted with phenol-chloroform-isoamylalcohol (25:24:1) and precipitated with 7.5 M ammonium acetate and 96% ethanol as described in Sambrook et al. (22). The precipitated nucleic acids were resuspended in a small volume of TE buffer containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA (pH 8.0). RNA was removed from the precipitated nucleic acids (50 to 100 μl) by incubation with 0.5 μg of DNase-free RNase for 1 h at 37°C. Extracted DNA was purified over a Wizard DNA purification column (Promega, Madison, Wis.).
PCR amplification, cloning of PCR products, and sequencing.
Archaeal 16S rRNA genes were amplified with two archaea-specific primers, S-D-Arch-0002-a-S-20 (Arch21F) and S-D-Arch-0940-a-A-20 (Arch958R), described by DeLong (3). The PCR mixture (50 μl) contained 25 to 50 ng of DNA, 50 μM (each) deoxynucleoside triphosphate, 0.2 μM (each) primer, 5 μl of 10× Taq DNA polymerase buffer, and 2.5 μg of bovine serum albumin (Boehringer, Mannheim, Germany). After an initial denaturation step of 5 min at 95°C, the temperature of the PCR mixture was lowered to 80°C and 1 to 2 U of Taq DNA polymerase (Pharmacia Biotech, Uppsala, Sweden) was added. The PCR conditions were as follows: 30 cycles of 1.5 min of denaturation at 95°C, 1.5 min of annealing at 55°C, and extension at 74°C for 1.5 min. The final step consisted of 5 min at 74°C and storage at 4°C. PCRs were done on a Progene thermal cycler (Techne, Cambridge, United Kingdom). The PCR products were analyzed by electrophoresis in 1% (wt/vol) agarose gels. Positive controls consisted of DNA from the methanogen Methanococcoides methylutens and the thermophile Sulfolobus acidocaldarius; negative controls consisted of DNA from Escherichia coli or no DNA addition.
PCR products were ligated into the p-GEM-T vector (Promega). To obtain the highest ligation efficiency under the conditions used, the PCR products were not purified and a vector/insert ratio of 1:30 was used instead of a prescribed ratio of 1:3. Ligation products were cloned into E. coli DH5α cells, which had been treated with 100 mM ice-cold CaCl2 (21). A number of transformants were randomly picked, and the plasmid was extracted with the plasmid purification kit (Qiagen Inc., Chatsworth, Calif.). The extracted plasmid was digested with either RsaI or DdeI (Pharmacia Biotech) to confirm the presence of an insert and to study the diversity of the inserts. In a final volume of 30 μl, these reaction mixtures contained 1 μl of extracted plasmid DNA, 3 μl of 10× One-Phor-All (Pharmacia BioTech) reaction buffer, and 3 to 5 U of enzyme. The restriction mixture was incubated for 1 to 2 h at 37°C, separated on a 2% low-melting-point agarose, and visualized by staining with ethidium bromide. A selected number of plasmids were sequenced on an ABI 310 automated DNA sequencer (Perkin-Elmer, Foster City, Calif.) with universal primers and the dye-terminator cycle sequencing reaction mixture according to the manufacturer’s guidelines.
Hybridization experiments.
The distribution of group I and II marine archaea among the different clonal libraries was determined by slot blot hybridization with chemiluminescently labelled oligonucleotide probes, which have been described previously by Massana et al. (16). One hundred microliters of plasmid DNA diluted 1,000 times was denatured by boiling for 5 min and spotted on a Hybond N+ membrane (Boehringer) with a slot blot setup and a vacuum pump (Pharmacia Biotech). After the membrane was dried at 50°C for approximately 15 min, the membrane was prehybridized for 1 to 3 h at 51°C with a solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% (wt/vol) blocking reagent (Boehringer), 0.1% (wt/vol) lauroylsarcosine, and 0.02% (wt/vol) sodium dodecyl sulfate followed by hybridization overnight with 10 pmol of digoxigenin-labelled probe in prehybridization solution. The probe was labelled with digoxigenin according to the manufacturer’s manual. After several washing steps and binding of a digoxygenin antibody to which alkaline phosphatase had been attached, bound probe was detected through the conversion of the substrate CSPD (Boehringer) with alkaline phosphatase and capture of the light signal on X-ray film.
Fingerprinting by terminal restriction fragment length polymorphism.
Archaeal community composition was analyzed by terminal restriction fragment length polymorphism as described by Liu et al. (14). The 16S rDNA of the archaeal community was amplified with the same primers as those described above for the amplification and cloning experiments. However, the Arch958R was labelled at the 5′ end with a 6-carboxyfluorescein (FAM; Perkin-Elmer). After amplification under the same conditions as those described above, the PCR products were purified with Wizard PCR purification columns (Promega). Twenty microliters of the purified PCR product was digested with 10 U of HhaI restriction enzyme (Promega) for 3 h at 37°C. The restricted DNA was precipitated with 0.1 volume of sodium acetate (3 M, pH 5.2) and 2 volumes of 96% ice-cold ethanol. After centrifugation at 14,000 rpm in an Eppendorf microcentrifuge, a washing with 80% ice-cold ethanol, and the drying of the pellet, the DNA was dissolved in 4 μl of sterile MilliQ water. To the concentrated DNA was added 5.0 μl of deionized formamide and 1.0 μl of DNA fragment length standard (TAMRA 2500; Perkin-Elmer). Prior to electrophoresis, the mixture was denatured at 95°C for 3 min and immediately put on ice. Electrophoresis of the restriction fragments was performed on an ABI 310 automated DNA sequencer in the gene scan mode with the POP4 gel matrix and a capillary column (47 cm by 50 μm). The exact lengths of the labelled fragments were determined by comparison with the internal TAMRA 2500 standard with ABI GeneScan software.
Phylogenetic analysis.
Preliminary determination of the phylogenetic affiliation of the clones consisted of a BLAST analysis (1) with the National Center for Biotechnology Information database. A number of sequences from the BLAST similarity ranking list were chosen for detailed phylogenetic analysis. The clone sequences were aligned with those from the database with the Dedicated Comparative Sequence Editor software program of de Rijk and de Wachter (6). Phylogenetic trees were generated and bootstrap analysis (100 replicates) was performed with the TREECON software package (26) by the algorithm described by Kimura and the neighbor joining method.
Nucleotide sequence accession numbers.
The sequences discussed in this study have been deposited in GenBank under accession no. AF052943 to AF052954.
RESULTS AND DISCUSSION
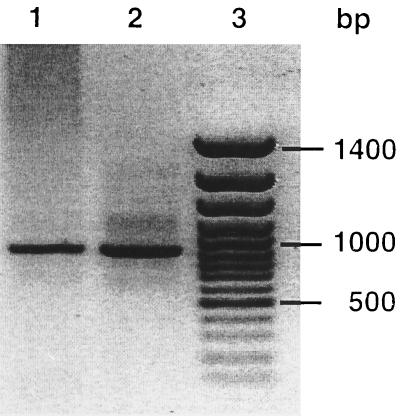
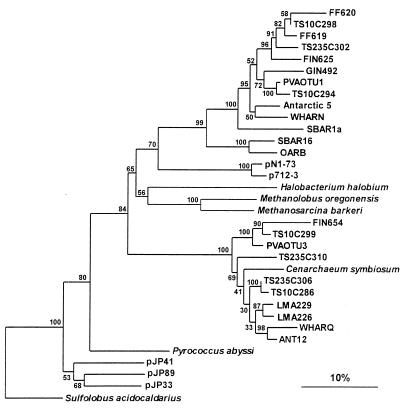
Marine archaeal 16S rDNA sequences have been found in the gut of a deep-sea-deposit feeder (18) and associated with a marine sponge (21). To investigate whether marine archaea are also present in other marine animals, digestive tract contents and feces were collected from a number of eukaryotic animals which are common in the North Sea. After extraction of DNA from these samples, PCRs with two archaea-specific 16S rDNA primers (3) were performed. Amplification products with the expected size of 950 bp were obtained with DNA extracted from the feces of flounder and the digestive tract contents of flounder and grey mullet (Fig. 2). After these amplification products were cloned into the p-GEM-T vector and transformed into competent E. coli cells, the plasmids of up to 30 randomly chosen white colonies were isolated for further analysis. The partial 16S rRNA gene insert of three clones, one from the digestive tract of flounder (FIN625), one from grey mullet digestive tract contents (GIN492), and two from flounder feces (FF619 and FF620), were sequenced. Phylogenetic analysis of these sequences showed that they clustered within the group II marine archaea (Fig. 3). The clones derived from flounder digestive tract and feces form a separate group within the lineage of the group II marine archaea and had only 76.7 to 89.8% similarity to the previously described group II marine archaea. The closest related sequences are clone PVAOTU1 (90.0 to 93.2% similarity), from a hydrothermal vent microbial community, Antarctic 5 (89.0 to 89.8% similarity), from Antarctic surface waters, and WHARN (87.7 to 88.5% similarity), from the coastal waters of the Atlantic Ocean near Woods Hole, Mass. (3). From the data presented in this study, it cannot be concluded exclusively whether the marine archaea found in the digestive tract and fecal samples are symbiotic members of the fish intestines or whether they originate from the seawater in which the fish lives and are passaged through the fish as it feeds. No amplification products were obtained from copepod fecal pellets or from the digestive tract contents of mussels with archaeal primers, whereas a PCR product was obtained with the universal primers S-*-Univ-50-a-S-19 and S-*-Univ-1492-a-A-22. Therefore, it appears that no archaeal DNA was recovered from the copepod and mussel samples. No amplification product was found when DNA of E. coli was used with the same archaea-specific primers and PCR conditions, indicating that the primers and the PCR conditions were specific for archaea and not bacteria.
FIG. 2.
PCR products obtained with two archaea-specific PCR primers, Arch2F and Arch958R. Lane 1, community DNA from flounder feces; lane 2, community DNA from flounder digestive-tract contents; lane 3, 100-bp ladder. PCR products were analyzed in 1% agarose and stained with ethidium bromide.
FIG. 3.
Phylogenetic positions of the archael clones derived from flounder feces (FF620 and FF619) or digestive tract (FIN625), grey mullet digestive tract (GIN492), and suspended particulate matter from stations TS10 and TS235 in the North Sea (TS10C286, TS10C294, TS10C298, TS10C299, TS235C302, TS235C306, and TS235C310). The dendrogram was constructed with the DCSE alignment program and the TREECON for Windows software package. The distance was estimated with the two-parameter model of Kimura, and the tree was constructed by the neighbor joining method. The numbers indicate absolute bootstrap values per 100 bootstraps performed. The bar represents 10% estimated nucleotide difference.
Phylogenetic analysis of four clones from the fish digestive-tract or fecal DNA showed that they are group II marine archaea. To determine the affiliation of other clones, hybridization experiments were carried out with chemiluminescently labelled group I and II probes. All 29 clones of the flounder feces 16S rDNA library and all 23 clones of the grey mullet 16S rDNA library hybridized with the group II probe (Table 1). However, of the 29 clones of the flounder digestive tract 16S rDNA library, 22 hybridized with the group II probe and 7 hybridized with the group I probe (Table 1). Plasmid DNA with a group I or group II insert was used as controls for the hybridization experiments. The control plasmids were obtained from subtropical Atlantic Ocean water samples and have been sequenced by us recently (26a). The exact affiliation of one of the group I clones of the flounder digestive tract was determined after sequencing. Clone FIN654 was found to cluster with the group I marine archaea and was most closely related to clone PVAOTU3 (91.3% similarity), which was derived from a hydrothermal vent community.
TABLE 1.
Analysis of the clone libraries of flounder and grey mullet digestive tracts and flounder feces by hybridization with chemiluminescently labelled marine archaea group I and II probes and by digestion with restriction enzymes
| Community | No. of clones | No. of clones hybridizing with:
|
No. of restriction groups with:
|
||
|---|---|---|---|---|---|
| Group I | Group II | DdeI | RsaI | ||
| Flounder feces | 29 | 0 | 29 | 4 | 4 |
| Flounder digestive tract | 29 | 7 | 22 | 5 | NDa |
| Grey mullet digestive tract | 23 | 0 | 23 | 3 | 4 |
ND, not determined.
Restriction fragment length polymorphism analysis of clones in the three libraries was performed by digestion of each clone with DdeI or RsaI. The restriction analysis of the grey mullet library showed three different restriction patterns with DdeI and four patterns with RsaI. Four different patterns were found for the library of the flounder feces, while five different patterns could be distinguished for the library of the flounder digestive tract (Table 1). These results indicate that a moderate degree of diversity was present among the inserts in the clone libraries and that more than one archaeal 16S rDNA was present in the original samples.
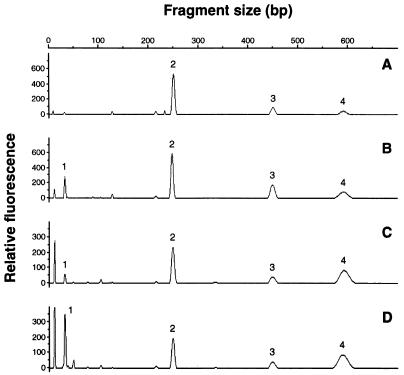
To study the diversity of archaea present in the digestive tract contents and feces of flounder in more detail, a molecular fingerprint of the archaeal community was made by terminal restriction fragment length polymorphism analysis as described by Liu et al. (14). In short, this method is based on the detection of fluorescently labelled 5′ terminal restriction fragments after digestion of the amplified 16S rDNA with a restriction enzyme. The archaeal 16S rRNA genes of the flounder digestive tract and feces were amplified with a nonlabelled Arch2F primer and a fluorescently labelled Arch958R primer, followed by digestion with HhaI and analysis of the labelled fragments with an automated sequencer. The profiles of the archaeal community present in the digestive tract and the feces of flounder were similar and showed four different fragments (Fig. 4A and B). To learn more about the potential origin of the prominent 249-bp fragment (no. 2), a simulated restriction analysis of 16S rRNA gene sequences in the database was performed (14). This method calculates the expected size of a labelled fragment for 16S rDNA sequences when the fluorescently labelled Arch958R primer and the restriction enzyme HhaI are used. A total of 35 archaeal 16S rDNA sequences, including all the sequences found in this study, were used for simulated restriction analysis. Of the 10 group II marine archaea sequences, 6 yielded a simulated restriction product of 248 bp. One sequence (WHARN) had a simulated product of 247 bp, two sequences of 249 bp (TS10C294 and TS235C302), and one sequence of 250 bp (TS10C298). The other 23 archaeal sequences, including 13 sequences of group I marine archaea, 6 sequences of methanogenic archaea, and 4 sequences of halo- or thermophilic archaea, all had simulated HhaI digests of different sizes. Only the sequence of the hyperthermophilic archaeon ES1, isolated from a hydrothermal vent, and that of Methanobacterium bryantii, a methanogen isolated from a bovine rumen, had a simulated HhaI restriction product of 249 bp. Based on the results of the simulated restriction analysis, it is concluded that the 249-bp peak of the digestive tract and fecal samples is likely derived from group II marine archaea. The possibility that the 249-bp fragment originated from an unknown fish digestive tract methanogen very closely related to M. bryantii cannot be excluded but seems unlikely because methanogenesis has been demonstrated only with the digestive tract contents of Dover sole and black cod (20), and no isolates of methanogenic archaea from marine fish digestive tract have been described.
FIG. 4.
Terminal restriction fragment length polymorphism analysis of 16S rRNA genes derived from flounder feces (A), digestive tract (B), and suspended particulate matter from stations TS 10 (C) and TS235 (D) with the PCR primers Arch2F and fluorescently labelled Arch 958R, followed by restriction with HhaI.
It is possible that the marine archaea present in the digestive tracts of marine fish are liberated into the water column and subsequently contribute to the marine archaeal community of suspended particulate matter. To investigate this, we isolated DNA from suspended particulate matter of the North Sea, which was retained on a GF/C filter, and used it for PCR detection, because until now all the studies on marine archaea have analyzed the fraction of the water that passes through a GF/C filter (3, 4, 8, 16, 17). From suspended particulate matter of two stations in the North Sea (see Materials and Methods for the exact locations), a PCR product of 950 bp was obtained. After the cloning of this amplification product, a number of clones were randomly selected and the sequences of the partial 16S rDNA inserts of these clones were determined. Four clones from station TS10, designated TS10C286, TS10C294, TS10C298, and TS10C299, and three from station TS235, designated TS235C302, TS235C306, and TS235C310, were found to cluster with the group I and II marine archaea (Fig. 1) and were related to the clones obtained from the fish digestive tract or fecal samples. The terminal restriction fragment length polymorphism analysis of the archaeal community of the suspended particulate matter of the two stations showed the same fragment pattern, similar to those obtained for the archaeal community for the flounder digestive tract and feces (Fig. 4C and D), indicating a high similarity between the archaeal communities of fish digestive tracts, feces, and suspended particulate matter. What the physiological characteristics of the fish-associated marine archaea are and whether or not they are actively present in suspended particulate matter cannot be concluded on the basis of the 16S rDNA sequences. The exact nature of fish-associated marine archaea remains unknown as long as no stable enrichments or pure cultures are available.
ACKNOWLEDGMENTS
This research was supported by the Beijerinck-Popping Foundation (grant 97-05).
We thank Jos de Wiljes for the fecal samples of the flounder, Wim Klein Breteler for the copepod fecal pellets, Jack van de Vossenberg for the DNA of S. acidocaldarius, and the Rijksinstituut voor Kust en Zee for collecting the North Sea samples. We also thank Marleen Otzen and Carien Booijink for their help with this research, Eijse Stamhuis and Theo Hansen for stimulating discussions, and Ine van Kuijk for critically reading the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLong E F, Wu K Y, Prézelin B B, Jovine R V M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 5.DeLong E F, King L L, Massana R, Cittone H, Murray A, Schleper C, Wakeham S G. Dibiphytanyl ether lipids in nonthermophilic crenarchaeotes. Appl Environ Microbiol. 1998;64:1133–1138. doi: 10.1128/aem.64.3.1133-1138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Rijk P, de Wachter R. DCSE v2.54, an interactive tool for sequence alignment and secondary structure research. Comput Appl Biosci. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrman J A, Davis A A. Widespread Archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- 8.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and the Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambacorta A, Tricone A, Niclaus B, Lama L, DeRosa M. Unique features of lipids of Archaea. Syst Appl Microbiol. 1994;16:518–527. [Google Scholar]
- 11.Hoefs M J L, Schouten S, de Leeuw J W, King L L, Wakeham S G, Sinninghe Damsté J S. Ether lipids of planktonic archaea in the marine water column. Appl Environ Microbiol. 1997;63:3090–3095. doi: 10.1128/aem.63.8.3090-3095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurgens G, Lindstrom K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato C, Li L, Tamaoka J, Horikoshi K. Molecular analysis of the sediment of the 11000-m deep Mariana Trench. Extremophiles. 1997;1:117–123. doi: 10.1007/s007920050024. [DOI] [PubMed] [Google Scholar]
- 14.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphism of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacGregor B J, Moser D P, Wheeler Alm E, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McInerney J O, Mullarkey M, Wernecke M E, Powell R. Phylogenetic analysis of group I marine archaeal rRNA sequences emphasizes the hidden diversity within the primary group Archaea. Proc R Soc Lond Ser B. 1997;264:1663–1669. [Google Scholar]
- 18.McInerney J O, Wilkinson M, Patching J W, Embley T M, Powell R. Recovery and phylogenetic analysis of novel archaeal rRNA sequences from a deep-sea deposit feeder. Appl Environ Microbiol. 1995;61:1646–1648. doi: 10.1128/aem.61.4.1646-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oremland R S. Methanogenic activity in plankton samples and fish digestive tracts: a mechanism for in situ methanogenesis in ocean surface waters. Limnol Oceanogr. 1979;24:1136–1141. [Google Scholar]
- 21.Preston C M, Ying Wu K, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov. sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Manniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schleper C, Holben W, Klenk H-P. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tindal B J. The archaebacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 677–808. [Google Scholar]
- 25.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 26.van de Peer Y, de Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 26a.van der Maarel, M. J. E. C. Unpublished data.
- 27.Vetriani C, Reysenbach A-L, Doré J. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol Lett. 1998;161:83–88. doi: 10.1111/j.1574-6968.1998.tb12932.x. [DOI] [PubMed] [Google Scholar]
- 28.Woese C R, Kandler O, Wheelis L M. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]