Abstract
A facultatively methylotrophic bacterium, strain IMB-1, that has been isolated from agricultural soil grows on methyl bromide (MeBr), methyl iodide, methyl chloride, and methylated amines, as well as on glucose, pyruvate, or acetate. Phylogenetic analysis of its 16S rRNA gene sequence indicates that strain IMB-1 classes in the alpha subgroup of the class Proteobacteria and is closely related to members of the genus Rhizobium. The ability of strain IMB-1 to oxidize MeBr to CO2 is constitutive in cells regardless of the growth substrate. Addition of cell suspensions of strain IMB-1 to soils greatly accelerates the oxidation of MeBr, as does pretreatment of soils with low concentrations of methyl iodide. These results suggest that soil treatment strategies can be devised whereby bacteria can effectively consume MeBr during field fumigations, which would diminish or eliminate the outward flux of MeBr to the atmosphere.
Methyl bromide (MeBr) is a fumigant used in the cultivation of selected fruits, vegetables, and flowers and in the preservation of stored grains and structures. Use of MeBr as a pesticide increases the yield and quality of crops without leaving behind toxic residues characteristic of more complex organopesticides. However, because bromine released from MeBr destroys stratospheric ozone (18, 22, 29, 33), its use will be eliminated in the United States and elsewhere under the auspices of the Clean Air Act and the Montreal Protocol unless effective mechanisms which prevent its escape to the atmosphere can be found (36). Currently, much uncertainty exists with regard to the tropospheric residence time (τ) of MeBr, a factor which is used to calculate its ozone degradation potential (2). Estimates of τ range from ∼1.7 years when only oxidation by tropospheric OH radicals is considered (22) to less than 1.2 years when oceanic sinks are factored in (20). The discovery that soil bacteria oxidize MeBr from the atmosphere, when quantified and combined with the two preceding sinks, lowers τ to ∼0.8 years (32). Chemical destruction of MeBr occurs by hydrolysis, exchange with other halides, and reaction with organic matter (8, 9, 12), but its destruction by microorganisms has been noted in soils and aquatic environments (3, 16, 17a, 19, 23, 27, 28, 32). In aerobic environments, MeBr is oxidized to CO2 and Br− (3, 16, 23, 27).
Bacterial oxidation of MeBr in soils has been reported both at very low (∼5 to 15 parts per trillion) ambient atmospheric mixing ratios (17a) and at the very high concentrations employed for field fumigation (23). The relative contributions that chemical reactions and bacterial oxidation make to the destruction of MeBr during agricultural fumigation are not yet known, but their combined effect will constrain the emissions of MeBr from soils. Reported destruction of MeBr within the soil matrix, as evidenced by the accumulation of Br−, can be substantial and account for as much as 39 to 70% of the applied MeBr in some cases (39, 40). Physical manipulations (e.g., soil compaction and deeper injection of MeBr) have been proposed to increase the retention time of MeBr within the soil matrix, thereby allowing for its more extensive degradation and subsequent decrease in its outward flux to the atmosphere (13). In addition, use of thicker, impermeable covering tarps has been proposed to reduce losses (14, 37), as has the substitution of methyl iodide for MeBr (11, 25). However, enhancement of microbial degradation of MeBr while it is present in the soil matrix may also be a means to eliminate emissions. This could be achieved by exploiting the ability of certain soil bacteria that use MeBr as a carbon and energy source (23). Here, we report further details on the characteristics of such an isolate (23), which we designate strain IMB-1. We demonstrate how the properties of IMB-1 can be used to greatly accelerate the oxidation of MeBr in fumigated soils. Because agricultural field fumigation represents the largest source of anthropogenic emissions of MeBr to the atmosphere, it is at least possible in theory that the overall goal of eliminating most human-derived emission of MeBr could be achieved by in situ biodegradation of this substance.
MATERIALS AND METHODS
Growth and cell suspension experiments with strain IMB-1.
The mineral salts medium described by Doronina et al. (6), as modified by Miller et al. (23), was employed to cultivate IMB-1. Cells were grown in crimp-seal Balch tubes filled with 10 ml of medium and sealed with a 15-ml-air headspace. Substrates were added (concentrations given in text) by syringe injections, and those tested for growth included methyl bromide, methyl iodide, methyl chloride, methyl fluoride, methane, sodium formate, methanol, monomethylamine, dimethylamine, trimethylamine, sodium acetate, glucose, sodium pyruvate, sodium citrate, sodium malate, and succinic acid. The pH was adjusted to 7.2, and after autoclaving, tubes were inoculated and incubated at 30°C with constant reciprocal shaking. Molar growth yield values were obtained by dividing the amount of substrate consumed into the final cell density achieved, assuming that the cell carbon content was 3.4 × 10−11 mg/cell for the IMB-1 isolate (1). The effect of chloropicrin (CCl3NO2; 0.5 to 500 μmol added per tube) on the growth of strain IMB-1 grown with MeBr or glucose as the source of carbon and energy was also investigated.
The MeBr oxidation assay was conducted on washed cell suspensions after cells were taken through two successive transfers on the substrate indicated. Ten milliliters of cells from the growth tubes was centrifuged (10,000 × g for 15 min at 7°C) and washed twice with mineral salts medium. The final pellets were resuspended in 5 ml of mineral salts medium, placed in 13-ml serum bottles, and sealed with crimped butyl rubber stoppers. [14C]MeBr (1.0 to 2.0 μCi/bottle; specific activity, 29.7 mCi/mmol; purity, 100%; New England Nuclear, Boston, Mass.) was injected, and cells were incubated statically for 4 to 6 h, at which time 0.25 ml of 6 N HCl was injected to stop the reaction and liberate 14CO2 into the gas phase. The tubes were vigorously hand shaken for 5 min before the gas phase was sampled for analysis. In another series of experiments, various trace levels of MeI were added to cells growing on glucose, methylamine, or acetate to determine if preexposure to MeI increased the ability of harvested cell suspensions to oxidize [14C]MeBr.
Determination of 16S rRNA gene sequences.
Chromosomal DNA was obtained from IMB-1 cells on agarose plates after washing them from the surface with 1.5 ml of a mixture of 50 mM Tris EDTA plus 150 mM NaCl. The collected liquid was centrifuged for 5 min at 10,000 × g, the pellet was resuspended in 1.4 ml of the above salts mixture plus 4 mg of lysozyme per ml, and the suspension was incubated overnight at 37°C, after which 0.05 ml of 20% sodium dodecyl sulfate was added and the tubes were incubated for 30 min at 45 to 50°C. DNA was then extracted with phenol and precipitated with ethanol as outlined by Sambrook et al. (31). PCRs were carried out in a Perkin-Elmer Gene Amp PCR System 9600 thermal cycler. Thirty cycles of 92, 60, and 72°C (1 min each) were performed with the 30-μl sample, followed by a final extension at 72°C for 5 min. The bacterial 16S rRNA gene was amplified with the bacterium-specific primers f27 and 1492r as detailed by Giovannoni (15). The PCR product was ligated into the pCRII vector of the T/A Cloning kit (Invitrogen, San Diego, Calif.). DNA sequencing from both strands was done with an Applied Biosystems automated sequencer. The phylogenetic classification was done by parsimony analysis of this sequence, together with similar sequences of the Ribosomal Database Project (21), by using the most parsimonious tree generated by PAUP branch and bound unweighed searching (34).
Soil experiments.
Loam soil of low organic content (0.4%) was employed. This soil was taken from a strawberry field located near Irvine, Calif., which has a past history of several previous MeBr fumigations. Details of this soil’s characteristics, storage, and handling are given elsewhere (23). Soil (5 g) was placed in serum vials (27 ml), sealed under air with butyl rubber stoppers, and injected with 0.05 ml of MeBr. In one experiment, soil received 0.5 ml of washed cell suspensions of either MeBr- or glucose-grown strain IMB-1. Live soil without added cells was incubated either with or without 0.5 ml of the mineral salts medium. One soil sample was autoclaved to serve as a killed control. Another set of killed controls consisted of autoclaving three soil samples after they were inoculated with 0.5 ml of MeBr-grown washed cells. In a second experiment, conditions were as described above, except that some soil was pretreated by receiving an injection of 75 μl of a 10% solution of MeI or of MeI plus 100 μl of 5 mM trimethylamine. After a pretreatment period lasting a few days (during which time the gas phase was analyzed for MeI), stoppers were removed and samples were flushed with a stream of air for ∼10 min to remove any residual MeI. Samples were resealed and injected with 0.2 ml of MeBr. All samples were incubated statically in the dark at ∼20°C.
Analytical.
Methyl halides in the headspace were analyzed by flame ionization gas chromatography, and the amount in the liquid phase was calculated from solubility coefficients applied to Henry’s Law as described by in Miller et al. (23). For MeI, a KH value of 0.2245 was used (24), resulting in a partitioning of 19% into the gas phase of the Balch tubes, with the remainder in the liquid phase. The amount of acetate was determined by high-performance liquid chromatography (HPLC) (5), and the amount of glucose was measured by a spectrophotometric kit assay (Sigma Diagnostics [procedure no. 315]). The amount of 14CO2 was determined by gas chromatography in series with gas proportional counting (4). Cell growth was quantified by acridine orange direct counts (17) and by turbidity (A680). The amount of iodide was determined by HPLC (26), and the amount of iodate was determined indirectly by its chemical reduction to iodide, followed by HPLC analysis and subtraction of the initial values for iodide. For reduction of iodate, sample aliquots (2 ml) were given 50 μl of 0.1 M ascorbic acid plus 55 μl of 6 N HCl (final pH, 1.5 to 2.0), and, after being stirred for 1 min, the pH was raised to >10 with NaOH (10).
Nucleotide sequence accession number.
The complete sequence of the 16S rRNA gene from IMB-1 has been deposited in the GenBank database under accession no. AF034798.
RESULTS
Morphology and phylogeny.
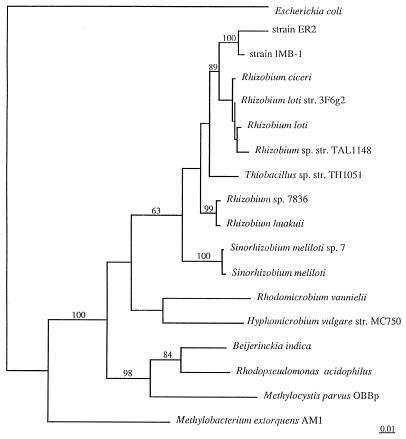
Strain IMB-1 is a motile, gram-negative rod (dimensions, ∼1.3 × 0.6 μm). A phylogenetic tree generated from comparisons of the 16S rRNA gene sequences classifies strain IMB-1 in the alpha subgroup of the class Proteobacteria. It is not closely related to recognized strains of methanotrophs or of methanol utilizers (Fig. 1) but rather to soil nitrogen-fixing bacteria of the genus Rhizobium. It is most closely related to strain ER2, a methylotroph which degrades methylcarbamate insecticides (38).
FIG. 1.
Phylogenetic analysis of the 16S rRNA gene from IMB-1. Bootstrap values are shown near the clades, and only values of 50% or higher are shown. The bar insert represents 1% sequence divergence as determined by measuring lengths of the horizontal lines connecting any two species.
Growth and cell suspension experiments.
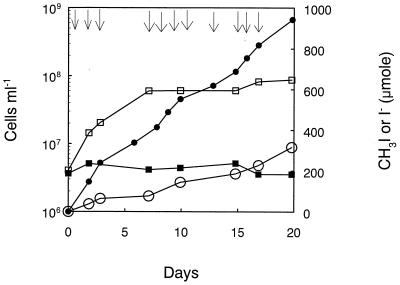
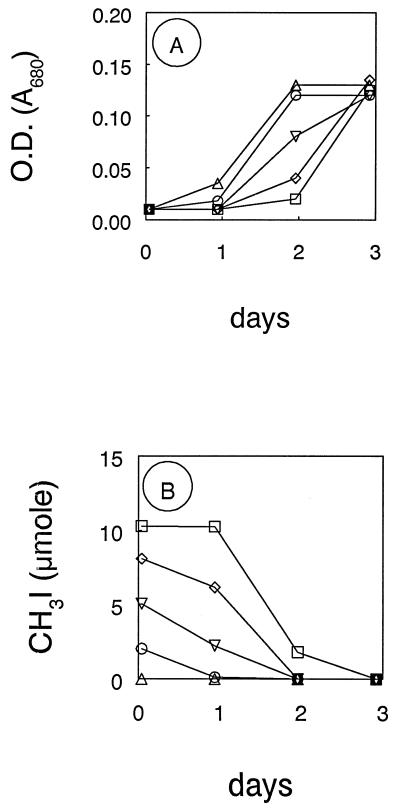
Strain IMB-1 was previously shown to grow with MeBr as the sole source of carbon and energy (23). Growth was also obtained when methyl iodide served as the electron donor and carbon source, and iodide accumulated in the medium as a consequence of this growth (Fig. 2). However, only about one-third of the methyl iodide consumed was recovered as iodide, possibly due to its oxidation to iodate, which is the most prevalent form of iodine in natural waters (30). However, we did not detect any additional iodide in these after we subjected them to chemical reduction with ascorbate. For example, the value of accumulated iodide was 316 μmol at the end of the incubation (Fig. 2), while after reduction the value was 290 μmol.
FIG. 2.
Growth of strain IMB-1 on methyl iodide. Arrows indicate additions of methyl iodide injected into the cultures. •, MeI consumed; □, cell counts in medium with MeI additions; ○, iodide; and ■, cell counts in medium without MeI additions.
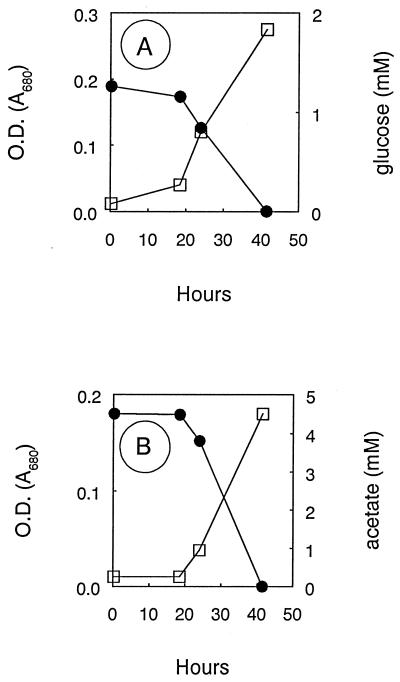
Strain IMB-1 also grew with glucose (Fig. 3A) or acetate (Fig. 3B) as electron donors. One-carbon compounds which supported growth included mono-, di-, and trimethylamine, but no growth occurred with methanol or formate (Table 1). Pyruvate supported growth, but not succinate, fumarate, or citrate, while weak growth was obtained on malate (Table 1). In addition to MeBr and methyl iodide, growth was also obtained on methyl chloride, but no growth occurred on methyl fluoride or methane (Table 2). Methyl fluoride (2 to 22 μmol/tube added) did not affect uptake of MeBr or growth of IMB-1 on MeBr (data not shown). Growth on glucose (Fig. 3A), acetate (Fig. 3B), and methylamines (Table 2) was much more rapid than that on the methyl halides and also achieved higher cell densities. Strain IMB-1 was unable to grow without the provision of ammonium salts in the medium (data not shown).
FIG. 3.
Growth of strain IMB-1 on glucose (A) and acetate (B). •, glucose or acetate; □, optical density (OD).
TABLE 1.
Growth of strain IMB-1 with various carbon and energy sources
| Substrate (concn [mM]) | A680a |
|---|---|
| Trimethylamine (5) | 0.230 |
| Dimethylamine (5) | 0.190 |
| Monomethylamine (5) | 0.095 |
| Pyruvate (5) | 0.230 |
| Malate (5) | 0.050 |
| Succinate (5) | 0.005 |
| Citrate (5) | 0.000 |
| Fumarate (5) | 0.010 |
| Formate (5) | 0.010 |
| Methanol (0.1)b | 0.005 |
| None | 0.000 |
Incubation period, 66 h.
No growth was obtained at higher concentrations of methanol.
TABLE 2.
Specific growth rates, molar growth yields, and MeBr oxidation activities in strain IMB-1
| Substrate (concn [mmol/liter])a | μ (h−1)b | YM (g mol−1)c | Amt of MeBr oxidized (pmol/106 cells/h) |
|---|---|---|---|
| MeBr (0.8) | 0.03 | 4.2 | 1.4 |
| MeCl (0.8) | 0.03 | 3.4 | 1.7 |
| MeI (0.3) | 0.07 | 2.7 | 2.4 |
| MeF (0.4) | 0.00 | 0.0 | 0.0 |
| Methane (3.7) | 0.00 | 0.0 | 0.0 |
| MMA (5.0) | 0.17 | NDd | 1.0 |
| DMA (4.0) | 0.19 | ND | 1.1 |
| TMA (4.0) | 0.16 | ND | 0.8 |
| Glucose (2.0) | 0.24 | 30.0 | 0.6 |
| Acetate (5.0) | 0.24 | 5.1 | 0.8 |
MeCl, MeI, and MeF, methyl chloride, methyl iodide, and methyl fluoride, respectively; MMA, DMA, and TMA, mono-, di-, and trimethylamine, respectively.
μ, specific growth rate, i.e., (ln Xt − ln X0)/(t − t0), where X is the cell biomass at times t and t0.
YM, molar growth yield.
ND, not determined.
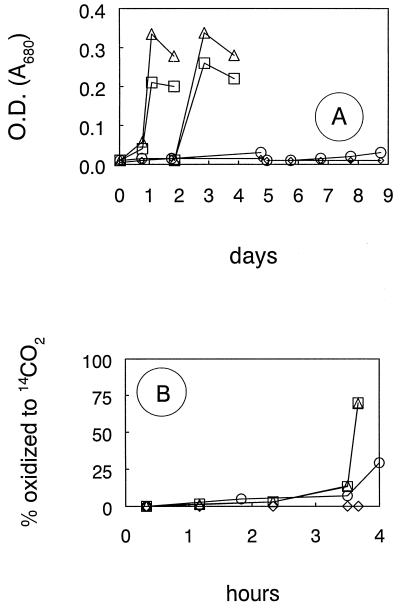
Cell suspensions readily oxidized [14C]MeBr to 14CO2 after two consecutive transfers in medium in which the growth substrate was not a methyl halide (Fig. 4). Thus, the ability of strain IMB-1 to oxidize MeBr was present regardless of the substrate that was utilized for growth (Table 2). However, MeBr oxidation rates in methyl halide-grown cells were significantly higher than those in cells grown on methylated amines, glucose, or acetate. Addition of methyl iodide to cells grown on methylamine initially retarded growth, resulting in a lag (Fig. 5A) during which methyl iodide was consumed (Fig. 5B). Cell suspensions harvested from these treatments all had equivalent capacities to oxidize [14C]MeBr regardless of whether they were exposed to methyl iodide. When normalized for cell densities, the rates of MeBr oxidation (in picomoles/106 cells/hour) were 1.2, 1.4, 1.1, 1.1, and 1.4 for cultures incubated with 0, 2, 5, 8, and 10 μmol of MeI, respectively. Similar results were obtained when acetate or glucose was used as the electron donor instead of methylamine (data not shown).
FIG. 4.
(A) Sequential growth of strain IMB-1 with two transfers on glucose (▵), acetate (□), or MeBr (○) or without substrate (◊). (B) Oxidation of [14C]MeBr by washed cell suspensions taken after growth of the second transfer. Symbols are the same as those for panel A. OD, optical density.
FIG. 5.
(A) Growth of IMB-1 on monomethylamine in the presence of 0 (▵), 2 (○), 5 (▿), 8 (◊), and 10 (□) μmol of methyl iodide per tube; (B) consumption of methyl iodide. OD, optical density.
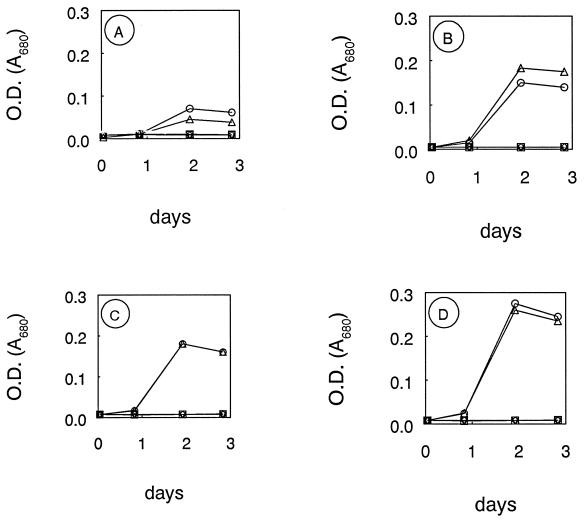
Chloropicrin had a pronounced inhibitory effect upon growth when applied at ≥0.05 μmol/tube, regardless of what growth substrate was present (Fig. 6). However, little or no inhibition was observed at the lowest chloropicrin application (0.005 μmol/tube). High concentrations of chloropicrin also caused substantial but not complete inhibition of [14C]MeBr oxidation by washed cell suspensions (Table 3).
FIG. 6.
Effect of chloropicrin on growth of strain IMB-1 on MeBr (A) monomethylamine (B), acetate (C), and glucose (D). ▵, ○, ▿, ◊, and □, 0, 0.005, 0.05, 0.5, and 5.0 μmol of chloramphenicol per tube, respectively. OD, optical density.
TABLE 3.
Effect of chloropicrin on the oxidation of [14C]MeBr to 14CO2 by cell suspensions of methylamine-grown IMB-1a
| Amt of chloropicrinb | Amt of 14CO2 formed (nCi) | % Inhibition |
|---|---|---|
| None | 0.65 | 0 |
| 0.01 | 0.40 | 38 |
| 0.05 | 0.16 | 76 |
| 0.50 | 0.08 | 88 |
| 5.00c | 0.09 | 86 |
Cells were incubated for 2 h with 1.55 nCi of [14C]MeBr and chloropicrin before being acidified.
Values are micromoles added per tube.
Equivalent to 0.5 mM.
Soil experiments.
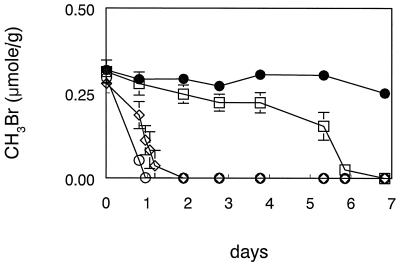
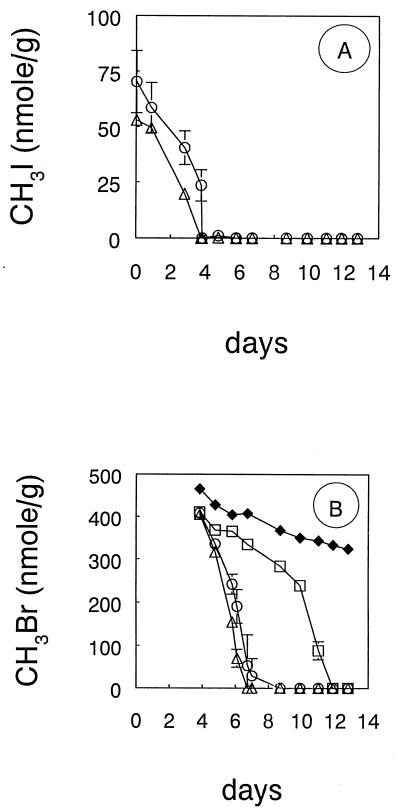
Addition of live cell suspensions of IMB-1 to agricultural soil resulted in a rapid removal of MeBr. All of the MeBr was consumed within 1 to 2 days, depending on whether cells were precultured on MeBr or on glucose (Fig. 7). In contrast, bacteria in the uninoculated soil required nearly 1 week to oxidize the MeBr. In the case of the uninoculated soil, addition of 0.5 ml of the mineral salts medium did not affect the pattern of MeBr consumption (not shown). No consumption of MeBr occurred in controls in which the soil was autoclaved after being inoculated with cell suspensions (Fig. 7) or in an uninoculated, autoclaved soil (not shown). Live soil degraded low concentrations of methyl iodide after several days of pretreatment incubation, while killed controls had only a minor amount of methyl iodide loss (Fig. 8A). When this pretreated soil was exposed to MeBr, there was a rapid degradation of the MeBr relative to the live soil which did not receive pretreatment (Fig. 8B). Soil preincubated with trimethylamine as well as methyl iodide exhibited slightly more rapid rates of MeBr degradation.
FIG. 7.
Consumption of unlabeled MeBr by agricultural soil. Symbols represent the means of three soil samples, and bars indicate ± 1 standard deviation. Absence of bars indicates that the error was smaller than the symbols. ○, soil incubated with 0.5 ml (3.0 × 108 cells) of a suspension of MeBr-grown IMB-1; ◊, soil incubated with 0.5 ml (6.6 × 108 cells) of a suspension of glucose-grown IMB-1; □, soil incubated with 0.5 ml of sterile medium; •, killed controls consisting of soil which had been autoclaved after receiving 0.5 ml of MeBr-grown cells.
FIG. 8.
Consumption of unlabeled MeBr in soil pretreated with MeI. (A) MeI levels in samples injected with MeI (○) and MeI plus trimethylamine (▵). (B) Levels of MeBr in untreated soil (□), and autoclaved soil (⧫). Symbols represent the means of three live soil samples, and bars indicate ± 1 standard deviation. The absence of bars from live samples indicates that the error was smaller than the symbols. Autoclaved control represents a single sample.
DISCUSSION
MeBr can be oxidized by methane-oxidizing bacteria (27) as well as by ammonia-oxidizing nitrifiers (28) via the monooxygenases of these organisms. However, neither methanotrophs nor nitrifiers can use MeBr as a substrate to support growth. Thus, the ability of strain IMB-1 to achieve growth on MeBr is unique (23). Previous results with methyl fluoride suggested that a nonmethanotrophic component of the flora of methane-oxidizing soils oxidized MeBr in the presence of this inhibitor (27). Since methyl fluoride is not metabolized by strain IMB-1 (Table 2) and has no effect on its ability to grow on or oxidize MeBr (see Results), it seems that organisms such as IMB-1 were responsible for the consumption of MeBr in soils which was not linked to methanotrophs or nitrifiers. Vannelli et al. (36a) recently reported that oxidation of methyl halides by Methylobacterium sp. strain CM4 are likely to proceed via a methyltransferase coupled with a dehydrogenase reaction sequence rather than by a monooxygenase. If such a methyltransferase-dehydrogenase system is not susceptible to inhibition by methyl fluoride, it could also serve as a model for MeBr oxidation by strain IMB-1.
Several facultative methylotrophs, including strains of Hyphomicrobium sp. and Methylobacterium extorquens, have been isolated which can grow on methyl chloride (6, 7) and oxidize MeBr (36a), but they are not phylogenetically related to strain IMB-1 (Fig. 1). Since strain IMB-1 does not grow on methane but does grow on other methyl halides (with the notable exception of methyl fluoride), methylamines, glucose, acetate, and pyruvate, it is clearly a facultative methylotroph (Fig. 2 and 3; Tables 1 and 2). In this respect, it shares some superficial substrate affinities with the facultative methylotrophs isolated from Russian soils (6, 7), as well as with strain ER2, a facultative methylotroph which degrades N-methyl carbamates (35). In this case, however, the two strains are closely related phylogenetically (Fig. 1). Both strain ER2 and IMB-1 are classed in the Rhizobium clade of the alpha subgroup of the Proteobacteria, which consist of aerobes noted for their abilities to fix atmospheric nitrogen either independently or when in symbiosis with plants. Although strain IMB-1 was unable to grow without combined nitrogen under an air atmosphere, this result does not totally eliminate the possibility that it is capable of fixing nitrogen under other physiological conditions. The presence of nif genes in IMB-1 is currently being investigated to answer this question.
The ability of cells to oxidize MeBr was constitutive in strain IMB-1, regardless of whether it was grown on methyl halides or on glucose, acetate, or methylamines (Fig. 4; Table 2). Therefore, it should be possible to mass culture strain IMB-1 on a conventional substrate, and it would still be able to degrade MeBr, a fact which eliminates the problem of having to employ a hazardous toxicant like MeBr as a substrate. When normalized for cell densities, however, cells grown on methyl halides had MeBr oxidation activities higher than those which were grown on other substrates (Table 2). We grew cells on conventional substrates in the presence of trace levels of methyl iodide in an attempt to see if this would induce higher MeBr oxidation activity in cell suspensions, but this did not occur (Fig. 5). Although cells were able to oxidize the methyl iodide, they did not achieve any greater capacity to oxidize MeBr after they were grown out on methylamine, glucose, or acetate.
Chloropicrin (i.e., tear gas) usually comprises about one-third of the MeBr fumigation mixture injected into soils and is used to enhance the overall biocidal effects of the mixture and to act as a warning agent to workers (38). We observed an inhibitory effect of chloropicrin on the capacity of agricultural soils to oxidize MeBr (23). Since chloropicrin inhibits the growth of strain IMB-1 (Fig. 6) as well as the ability of cell suspensions to oxidize MeBr (Table 2), it is likely that our soil observations were caused by the direct effects of chloropicrin on organisms such as strain IMB-1 which were present in the soil flora. Therefore, any attempts to enhance the biodegradation of MeBr during field fumigation operations must take into account the amount of chloropicrin employed in the fumigant mixture. Lower levels of chloropicrin in the fumigant mixtures could result in enhanced MeBr biodegradation without compromising its role as a warning agent.
The addition of live cells to soil greatly speeded its ability to consume MeBr (Fig. 7). Because no consumption of MeBr occurred in controls in which both the soil and the cells were heat killed, the observed consumption could not be attributed to chemical binding of the methyl group of MeBr to any of the organic material provided by the dead cells. Rather, it was clearly due to the biochemical oxidation of MeBr by strain IMB-1. This soil has been previously shown to oxidize MeBr to CO2 and Br− (23). We have obtained results identical to those given above with a low organic content loamy sand soil taken near Watsonville, Calif. (3a). These observations suggest that seeding soils with live cells of mass-cultured IMB-1 may be a viable option for enhancing the biodegradation of MeBr during fumigation of agricultural fields. Tarped periods of fumigation usually last for several days (23, 39), but in contrast the IMB-1 enhanced oxidation of fumigation levels of MeBr was so rapid (1 to 2 days) as to raise concern that insufficient levels of fumigant would be present over the course of the tarping period to effectively eliminate target pests. In practical terms, such a scenario might be avoided by seeding only the surface soils (e.g., upper 5 cm) with bacteria just prior to their being covered by tarps. This would create a zone of intense bacterial MeBr oxidation at the surface of the soil which would intercept the upward flux of MeBr from its deeper injection depth.
Another approach would be to pretreat fields with methyl iodide, a substance which has been proposed as an alternative ozone-safe fumigant in the event that MeBr use is eliminated by a worldwide ban (11, 25). Because IMB-1 also grows on methyl iodide (Fig. 2; Table 2), such a scenario would also increase the cell population of these organisms in the soil and speed the overall rate of MeBr biodegradation during fumigation operations. Experimental results with soil indicate that such an approach is feasible (Fig. 8).
It is clear that use of MeBr as a fumigant to enhance crop yield and to prevent destruction of grain stores by pests has considerable benefit to an expanding human population. Balanced against this stands the contribution that MeBr makes to the destruction of stratospheric ozone, with the largest component of anthropogenic emission coming from field fumigation. Although the global budget of sources and sinks of MeBr is not accurately known, it is generally believed that all anthropogenic emissions are outweighed by natural sources (2). If we extrapolate our laboratory results, it appears at least theoretically possible to use MeBr as an agricultural fumigant while employing naturally occurring soil bacteria to severely constrain its release to the atmosphere. However, to make this approach viable, clear success must also be achieved under complex field conditions and with soils of differing properties.
ACKNOWLEDGMENTS
The DNA extractions and sequencing were done by A. Costello and M. Lidstrom with the assistance of the AIDS Research Sequencing Facility at the University of Washington. The remainder of the research was conducted at the USGS. This work was supported by the USGS New Technologies Program and by NASA Earth Sciences Division Upper Atmosphere Research Program grant no. IAG-W-18267.
We thank P. Crill, B. F. Taylor, and three unidentified referees for their helpful reviews of the manuscript and A. Farrenkopf for discussions of iodine speciation.
REFERENCES
- 1.Bratbak G. Bacterial biovolume and biomass estimations. Appl Environ Microbiol. 1985;49:1488–1493. doi: 10.1128/aem.49.6.1488-1493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler J H, Rodriguez J M. Methyl bromide in the atmosphere. In: Bell C H, Price N, Chakrabarti B, editors. The methyl bromide issue. J. New York, N.Y: Wiley & Sons; 1996. pp. 28–90. [Google Scholar]
- 3.Connell T L, Joye S B, Miller L G, Oremland R S. Bacterial oxidation of methyl bromide in Mono Lake, California. Environ Sci Technol. 1997;31:1489–1495. [Google Scholar]
- 3a.Connell Hancock, T. L., L. G. Miller, and R. S. Oremland. Unpublished data.
- 4.Culbertson C W, Zehnder A J B, Oremland R S. Anaerobic oxidation of acetylene by estuarine sediments and enrichment cultures. Appl Environ Microbiol. 1981;41:396–403. doi: 10.1128/aem.41.2.396-403.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culbertson C W, Strohmaier F E, Oremland R S. Acetylene as a substrate in the development of primordial bacterial communities. Origins Life Evol Biosph. 1988;18:397–407. doi: 10.1007/BF01808218. [DOI] [PubMed] [Google Scholar]
- 6.Doronina N V, Sokolov A P, Trotsenko Y A. Isolation and initial characterization of aerobic chloromethane-utilizing bacteria. FEMS Microbiol Lett. 1996;142:179–183. [Google Scholar]
- 7.Doronina N V, Trotsenko Y A. Isolation and characterization of aerobic degraders of methyl chloride. Microbiology. 1997;66:57–64. [Google Scholar]
- 8.Elliott S, Rowland F S. Nucleophilic substitution rates and solubilities for methyl halides in seawater. Geophys Res Lett. 1993;20:1043–1046. [Google Scholar]
- 9.Elliott S, Rowland F S. Methyl halide hydrolysis rates in natural waters. J Atmos Chem. 1995;20:229–236. [Google Scholar]
- 10.Farrenkopf A M. Biologically catalysed redox cycling: productivity and denitrification reflected iodine speciation in the ocean. Ph.D. Dissertation. Lewes, Del: University of Delaware; 1998. [Google Scholar]
- 11.Gan J, Yates S R. Degradation and phase partition of methyl iodide in soil. J Agric Food Chem. 1996;44:4001–4008. [Google Scholar]
- 12.Gan J, Yates S R, Anderson M A, Spencer W F, Ernst F F, Yates M V. Effect of soil properties on degradation and sorption of methyl bromide in soil. Chemosphere. 1994;29:2685–2700. [Google Scholar]
- 13.Gan J, Yates S R, Wang D, Spencer W F. Effect of soil factors on methyl bromide volatilization after soil application. Environ Sci Technol. 1996;30:1629–1636. [Google Scholar]
- 14.Gan J, Yates S R, Spencer W F, Yates M V, Jury W A. Laboratory-scale measurements and simulations of effect of application methods on soil methyl bromide emission. J Environ Qual. 1997;26:310–317. [Google Scholar]
- 15.Giovannoni S J. Polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 177–204. [Google Scholar]
- 16.Goodwin K D, Lidstrom M E, Oremland R S. Marine bacterial degradation of brominated methanes. Environ Sci Technol. 1997;31:3188–3192. [Google Scholar]
- 17.Hobbie J E, Daley R L, Jaspar S. Use of nuclepore filters for counting bacteria for fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Hynes M E, Crill P M, Varner R K, Talbot R W, Shorter J H, Kolb C E, Harriss R C. Rapid consumption of low concentrations of methyl bromide by soil bacteria. Appl Environ Microbiol. 1998;64:1864–1870. doi: 10.1128/aem.64.5.1864-1870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalil M A K, Rasmussen R A, Gunawardena R. Atmospheric methyl bromide: trends and global mass balance. J Geophys Res. 1993;98:2887–2896. [Google Scholar]
- 19.King D B, Saltzman E S. Removal of methyl bromide in coastal water: chemical and biological rates. J Geophys Res. 1997;102:18715–18721. [Google Scholar]
- 20.Lobert J M, Butler J H, Montzka S A, Geller L S, Meyers R C, Elkins J W. A net sink for atmospheric CH3Br in the East Pacific Ocean. Science. 1995;267:1002–1005. doi: 10.1126/science.267.5200.1002. [DOI] [PubMed] [Google Scholar]
- 21.Maidak B L, Olsen G J, Larsen N, Overbeck R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellouki A, Talukar R K, Schmoltner A-M, Gierzak T, Mills M J, Solomon S, Ravishankara A R. Atmospheric lifetimes and ozone depletion potentials of methyl bromide (CH3Br) and dibromomethane (CH2Br2) Geophys Res Lett. 1992;19:2059–2062. [Google Scholar]
- 23.Miller L G, Connell T L, Guidetti J R, Oremland R S. Bacterial oxidation of methyl bromide in fumigated agricultural soils. Appl Environ Microbiol. 1997;63:4346–4354. doi: 10.1128/aem.63.11.4346-4354.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore R M, Geen C E, Tait V K. Determination of Henry’s Law constants for a suite of naturally occurring halogenated methanes in seawater. Chemosphere. 1995;30:1183–1191. [Google Scholar]
- 25.Ohr H D, Sims J J, Grech N M, Ole Becker J, McGiffen M E., Jr Methyl iodide, an ozone-safe alternative to methyl bromide as a soil fumigant. Plant Dis. 1996;80:731–735. [Google Scholar]
- 26.Oremland R S, Culbertson C W. Evaluation of methyl fluoride and dimethyl ether as inhibitors of aerobic methane oxidation. Appl Environ Microbiol. 1992;58:2983–2992. doi: 10.1128/aem.58.9.2983-2992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oremland R S, Miller L G, Culbertson C W, Connell T L, Jahnke L. Degradation of methyl bromide by methanotrophic bacteria in cell suspensions and soils. Appl Environ Microbiol. 1994;60:3640–3646. doi: 10.1128/aem.60.10.3640-3646.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou L-T, Joy P J, Thomas J E, Hornsby A G. Stimulation of degradation of methyl bromide in soil during oxidation of an ammonia fertilizer by nitrifiers. Environ Sci Technol. 1997;31:717–722. [Google Scholar]
- 29.Prather M J. Timescales in atmospheric chemistry: CH3Br, the ocean, and ozone depletion potentials. Global Biogeochem Cycles. 1997;11:393–400. [Google Scholar]
- 30.Riley J P M, Chester R. Introduction to marine chemistry. New York, N.Y: Academic Press; 1971. [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 32.Shorter J H, Kolb C E, Crill P M, Kerwin R A, Talbot R W, Hines M E, Harris R C. Rapid degradation of atmospheric methyl bromide in soils. Nature. 1995;377:717–719. [Google Scholar]
- 33.Singh H B, Kanakidou M. An investigation of the atmospheric sources and sinks of methyl bromide. Geophys Res Lett. 1993;20:133–136. [Google Scholar]
- 34.Swofford D L. PAUP: phylogenetic analysis using parsimony. Champaign, Ill: Illinois Natural History Survey; 1991. [Google Scholar]
- 35.Topp E, Hanson R S, Ringelberg D B, White D C, Wheatcroft R. Isolation and characterization of an N-methylcarbamate insecticide-degrading methylotrophic bacterium. Appl Environ Microbiol. 1993;59:3339–3349. doi: 10.1128/aem.59.10.3339-3349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.United Nations Environment Programme. Report of the methylbromide technical options committee. Nairobi, Kenya: United Nations Environment Programme; 1994. [Google Scholar]
- 36a.Vannelli T, Studer A, Kertesz M, Leisinger T. Chloromethane metabolism by Methylobacterium sp. strain CM4. Appl Environ Microbiol. 1998;64:1933–1936. doi: 10.1128/aem.64.5.1933-1936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Yates S R, Ennst F F, Gan J, Jury W A. Reducing methyl bromide emission with a high barrier plastic film and reduced dosage. Environ Sci Technol. 1997;31:3686–3691. [Google Scholar]
- 38.Wilhelm S N, Shepler K, Lawrence L J, Lee H. Environmental fate of chloropicrin. In: Seiber J N, et al., editors. Fumigants: environmental fate, exposure, and analysis. Washington, D.C: American Chemical Society; 1996. pp. 79–93. [Google Scholar]
- 39.Yagi K, Williams J, Yang N-Y, Cicerone R J. Atmospheric methyl bromide (CH3Br) from agricultural soil fumigations. Science. 1995;267:1979–1981. doi: 10.1126/science.267.5206.1979. [DOI] [PubMed] [Google Scholar]
- 40.Yates S R, Gan J, Ernst F F, Yates M V. Methyl bromide emissions from a covered field. I. Experimental conditions and degradation in soil. J Environ Qual. 1996;25:184–192. [Google Scholar]