Abstract
A β-N-acetylglucosaminidase gene (nagA) of Streptomyces thermoviolaceus OPC-520 was cloned in Streptomyces lividans 66. The nucleotide sequence of the gene, which encodes NagA, revealed an open reading frame of 1,896 bp, encoding a protein with an Mr of 66,329. The deduced primary structure of NagA was confirmed by comparison with the N-terminal amino acid sequence of the cloned β-N-acetylglucosaminidase expressed by S. lividans. The enzyme shares no sequence similarity with the classical β-N-acetylglucosaminidases belonging to family 20. However, NagA, which showed no detectable β-glucosidase activity, revealed homology with microbial β-glucosidases belonging to family 3; in particular, striking homology with the active-site regions of β-glucosidases was observed. Thus, the above-mentioned results indicate that NagA from S. thermoviolaceus OPC-520 is classified as a family 3 glycosyl hydrolase. The enzyme activity was optimal at 60°C and pH 5.0, and the apparent Km and Vmax values for p-nitrophenyl-β-N-acetylglucosamine were 425.7 μM and 24.8 μmol min−1 mg of protein−1, respectively.
Streptomycetes are gram-positive, mycelial soil bacteria with a high G+C content. In addition to having the ability to synthesize a wide variety of antibiotics and chemotherapeutic agents, they produce extracellular hydrolytic enzymes to obtain nutrients and energy by solubilizing polymeric compounds in soil. These enzymes include proteases, nucleases, lipases, and a variety of enzymes that hydrolyze different types of polysaccharides such as cellulose, chitin, and xylan (13). This last class of enzymes has received considerable attention not only from the standpoint of the utilization of renewable resources but also from that of basic research. Among actinomycetes, Streptomyces spp. make up one group regarded as particularly efficient in the breakdown of chitin (10). Following cellulose, chitin is the second most abundant polymer (β-1,4-linked polymer of N-acetylglucosamine) in nature. Efficient degradation of chitin by microorganisms is achieved by the concerted action of chitinase (EC 3.2.1.14) and β-N-acetylglucosaminidase (EC 3.2.1.30) (1, 19, 20).
We have been studying the chitinolytic system of Streptomyces thermoviolaceus OPC-520 to clarify the roles of individual enzymes involved in chitin degradation, the relationship between structure and function, and the regulation of gene expression. When S. thermoviolaceus OPC-520 is cultivated in the presence of chitin, this strain secretes three different chitinases and only one β-N-acetylglucosaminidase and the production is repressed by glucose (unpublished data). Previously, we purified and characterized a major chitinase (Chi40) produced by the strain, which shows a high optimum temperature (70 to 80°C), high optimum pH (pH 8.0 to 10.0), and heat stability (22), and recently reported the cloning and expression of the Chi40 gene (23).
While a number of chitinase genes have been isolated from a wide variety of organisms, including bacteria, fungi, insects, plants, and animals, examples of cloning of the β-N-acetylglucosaminidase gene involved in a chitinolytic system are few. To understand the role of β-N-acetylglucosaminidase in chitin degradation by strain OPC-520, its relationship to similar proteins isolated from other sources, and the regulatory system involved in the induction of the enzyme, we have isolated and expressed the gene encoding β-N-acetylglucosaminidase. Here we report the molecular cloning and biochemical characterization of a β-N-acetylglucosaminidase, designated NagA, from S. thermoviolaceus OPC-520. This novel enzyme, which is clearly different from the N-acetylglucosaminidases so far reported, is assigned to family 3 of the glycosyl hydrolases on the basis of sequence comparison. This is the first report of a β-N-acetylglucosaminidase gene isolated from the genus Streptomyces.
MATERIALS AND METHODS
Strains and plasmids.
S. thermoviolaceus OPC-520 was isolated from decayed wood in Osakasayama City, Osaka, Japan (22), and was used as the source of chromosomal DNA. Streptomyces lividans 66 was obtained from M. Sugiyama (Hiroshima University, Hiroshima, Japan) and used as a host for constructing gene libraries of S. thermoviolaceus OPC-520 in plasmid pIJ702. The vectors used for subcloning were pUC18 and pUC19, as well as pUWL219, an Escherichia coli-Streptomyces shuttle vector obtained from U. F. Wehmeier (Bergische Universität, Wuppertal, Germany). E. coli JM109 was used as the recipient for subcloning.
Media and culture conditions.
S. thermoviolaceus OPC-520 was grown at 50°C in a medium containing (in grams per liter) glucose, 10.0; yeast extract (Difco), 5.0; proteose peptone (Difco), 5.0; K2HPO4, 1.0; and MgSO4 · 7H2O, 0.2 (pH 7.0). E. coli was grown at 37°C in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml). For plates, LB medium was solidified with 1.5% (wt/vol) agar. For preparation of protoplasts and DNA isolation, S. lividans was cultivated at 27°C in GMP medium (1.0% glucose, 0.2% meat extract, 0.4% polypeptone, 0.2% yeast extract, 0.5% NaCl, 0.025% MgSO4 · 7H2O [pH 7.0]). For protoplast regeneration, R2YE agar plates were used (9). When the recombinant strain was grown, thiostrepton (Sigma) was added to solid and liquid media at a concentration of 50 μg/ml.
General DNA techniques.
Recombinant-DNA techniques with Streptomyces spp. were performed as described by Hopwood et al. (9). To clone a β-N-acetylglucosaminidase-encoding gene (nagA) from S. thermoviolaceus, chromosomal DNA was partially digested with BglII and 5- to 10-kb fragments were ligated to the multicopy plasmid pIJ702, which was cleaved at the unique BglII site within the mel gene. The ligation mixture was used to transform S. lividans 66. Thiostrepton-resistant Mel− transformants were grown for 72 h on screening plates (1% glycol chitin, 0.2% polypeptone, 0.1% yeast extract, 0.1% meat extract, 0.03% tyrosine, 1 ml of trace-element solution [9], 2.0% agar [pH 7.0]). For the screening of β-N-acetylglucosaminidase-producing clones, transformants grown on plates were sprayed with a 0.01 M solution of p-nitrophenyl-β-N-acetylglucosaminide (Seikagaku Kogyo, Tokyo, Japan) in 0.1 M sodium phosphate buffer pH 7.5 (28). Next, the plates were incubated at 50°C for 30 min. By this method, colonies producing a bright yellow color are putative clones containing hybrid plasmids with genomic inserts coding for β-N-acetylglucosaminidase activity. General DNA techniques with E. coli were carried out as described by Sambrook et al. (17). Restriction endonucleases and T4 DNA ligase were purchased from Toyobo (Tokyo, Japan) and were used according to the manufacturer’s specifications. The nucleotide sequence was determined by the dideoxy chain termination method (18) with a Thermo Sequenase fluorescence-labelled primer cycle sequencing kit (Amersham International plc) as specified by the manufacturer. DNA fragments were analyzed on a DNA sequencer (SQ3000; Hitachi).
Purification of recombinant β-N-acetylglucosaminidase.
A β-N-acetylglucosaminidase-positive clone of S. lividans, designated pNAG207, was grown in minimal medium (NMMP) (9) supplemented with 1% (wt/vol) chitin. Cultures were grown at 27°C with agitation at 200 rpm on a rotary shaker for 5 days. After filtration of the culture with Toyoroshi no. 2 filter paper (Toyoroshi Co., Ltd., Tokyo, Japan), the filtrate was used as crude β-N-acetylglucosaminidase. All purification steps were carried out at 4°C unless otherwise mentioned. The crude enzyme (500 ml) was dialyzed overnight against 50 mM Tris-HCl buffer, pH 7.5. The dialyzed enzyme solution was applied to a DEAE-Toyopearl 650 M column (1.9 by 45 cm; Tosoh, Tokyo, Japan) equilibrated with the same buffer. The column was washed first with buffer (300 ml) and then with a linear gradient of NaCl (0 to 1.0 M) at a flow rate of 36 ml/h. The enzyme was eluted at about 0.35 M NaCl. The pooled active fractions (32 ml) were concentrated by ultrafiltration with NanoSpin Plus (Gelman Sciences, Ann Arbor, Mich.). The concentrated sample was applied to a Sephadex G-100 column (1.9 by 90 cm; Pharmacia Biotech Inc.) equilibrated with buffer containing 0.1 M NaCl. The pooled active fraction was chromatographed by using a fast-performance liquid chromatography Q2 anion-exchange column (0.7 by 5.2 cm; Bio-Rad) equilibrated with buffer. The column was washed with buffer, and then the enzyme was eluted with a linear gradient of 0 to 0.5 M NaCl. It was eluted as a symmetrical peak at a concentration of about 0.15 M NaCl.
Assays for enzyme activity.
β-N-Acetylglucosaminidase was assayed by mixing a 0.1-ml aliquot of appropriately diluted enzyme with 0.2 ml of 2.5 mM p-nitrophenyl-β-N-acetylglucosaminide (PNP-β-GlcNAc) in 50 mM acetate buffer, pH 5.0. After incubation at 60°C for 10 min, the reaction was terminated by adding 2 ml of 0.2 M Na2CO3 and p-nitrophenol was measured at 420 nm. The other p-nitrophenyl derivatives were also used at the concentration of 2.5 mM. One unit of β-N-acetylglucosaminidase was defined as the amount of enzyme that liberated 1 μmol of p-nitrophenol in 1 min under the conditions described above. The assay system for chitin oligosaccharides from dimer to hexamer consisted of 0.1 ml each of enzyme solution, 50 mM acetate buffer (pH 5.0), and 5 mM substrate. After incubation at 60°C for 30 min, N-acetylglucosamine produced was measured by the method of Reissig et al. (15). The enzyme activity was measured at pH values from 4 to 10 under standard conditions with PNP-β-GlcNAc as a substrate. Buffers used were 50 mM acetate buffer (pH 4 to 6), 50 mM Tris-HCl buffer (pH 7 to 8), and 50 mM glycine-NaOH buffer (pH 9 to 10). The enzyme activities were also assayed at temperatures from 30 to 80°C at pH 5.0.
Other procedures.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and amino-terminal amino acid sequencing were performed as described before (24). Protein was assayed by the method of Bradford (3) with bovine serum albumin as a standard.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession number AB008771.
RESULTS
Gene isolation and sequence analysis.
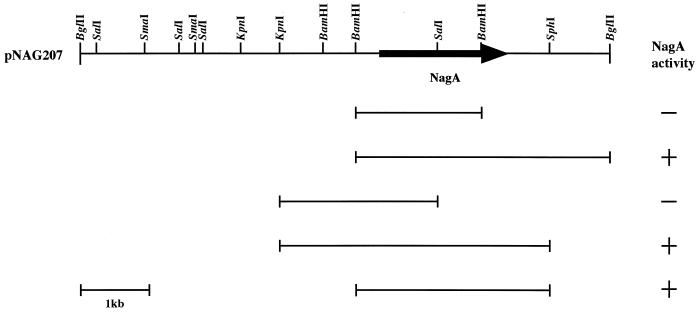
The gene encoding a β-N-acetylglucosaminidase was isolated by expression screening of a plasmid library on thiostrepton-containing plates. Positive clones were indicated by the production of a bright yellow color. Three clones expressing enzyme activity were isolated. The clones were analyzed by restriction endonuclease digestion and found to contain a common 8.0-kb insert (Fig. 1). To determine the location of the β-N-acetylglucosaminidase-encoding gene (nagA), a restriction map was constructed and various subclones were prepared. The results of subcloning showed that the 2.8-kb BamHI-SphI fragment was the region necessary for the expression of the enzyme activity (Fig. 1). The 2.8-kb DNA fragment isolated from plasmid pNAG207 was cloned in pUC18 and pUC19, and both strands were sequenced. A single 1,896-bp typical Streptomyces open reading frame (ORF) was found, which is in agreement with the high G+C content and the preferential codon usage in Streptomyces (29). This ORF starts with a GTG codon at position 400 and ends with a TAG translational stop codon at position 2298. This ORF could encode a protein of 632 amino acids with a calculated molecular weight of 66,329. The N-terminal sequence of the cloned β-N-acetylglucosaminidase from S. lividans was determined and coincides precisely with the sequence starting from His61 of the deduced amino acid sequence encoded by the gene. Cleavage of the signal peptide would yield a mature protein of 572 amino acids with a molecular weight of 60,380, which is in reasonable agreement with results obtained by SDS-PAGE (Fig. 2). However, the signal peptide cleavage site is not compatible with the −3, −1 rule of von Heijne (26). A putative ribosome binding site (Shine-Dalgarno sequence) with good complementarity to the 3′ end of 16S rRNA of S. lividans (2) was found 7 bp upstream from the start codon. The putative −10 and −35 regions, which showed homology with the consensus sequence of Streptomyces promoters (21), was found upstream of the Shine-Dalgarno sequence. A computer search of the promoter region pointed out operator-like sequences described as a complex array of tandem- or inverted-repeat sequences (6).
FIG. 1.
Restriction map of the recombinant plasmid containing nagA. The arrow indicates the ORF and the direction of transcription. Transformants grown on screening plates were sprayed with a 0.01 M solution of PNP-β-GlcNAc. β-N-Acetylglucosaminidase activity was determined by production of a bright yellow color. +, production of the color; −, no production of the color.
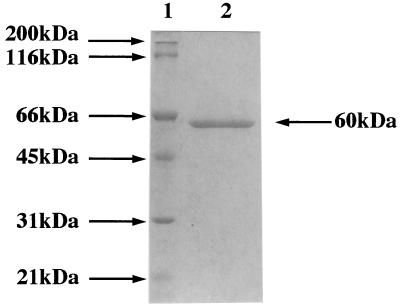
FIG. 2.
SDS-PAGE of purified NagA. Lanes: 1, marker proteins; 2, NagA.
Purification of the cloned β-N-acetylglucosaminidase.
To purify and characterize the cloned enzyme, S. lividans carrying shuttle plasmid pUWL219 containing the 2.8-kb BamHI-SphI fragment was grown in NMMP supplemented with 1% chitin for 5 days. A high level of β-N-acetylglucosaminidase activity against PNP-β-GlcNAc was observed in the culture filtrate. The enzyme was purified in three steps from the filtrate as shown in Table 1. In this procedure, the enzyme was purified 8.7-fold and total recovery was 68.7%. The purified enzyme was detected as a single band on SDS-PAGE (Fig. 2). The molecular masses of the enzyme were 60 and 59 kDa as determined by SDS-PAGE and analytical size exclusion fast-performance liquid chromatography (Superdex 200; Pharmacia), respectively. These results indicate that the enzyme is a monomeric protein.
TABLE 1.
Purification of cloned NagA
| Fraction | Total protein (mg) | Total activity (U) | Sp act (U/mg of protein) | Purification factor | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 53.6 | 412.3 | 7.2 | 1.0 | 100.0 |
| DEAE-Toyopearl 650M | 9.6 | 384.1 | 40.2 | 5.6 | 93.2 |
| Sephadex G-100 | 6.7 | 357.6 | 53.1 | 7.4 | 86.7 |
| Bio-scale Q2 | 4.4 | 283.3 | 64.4 | 8.7 | 68.7 |
Substrate specificity.
We investigated the substrate specificity of the enzyme by using various substrates (Table 2). Among the chromogenic substrates PNP-β-GlcNAc, PNP-α-GlcNAc, PNP-β-N-acetylgalactosamine (PNP-β-GalNAc), PNP-β-galactosamine (PNP-β-Gal), PNP-β-glucose (PNP-β-Glu), PNP-β-xylose (PNP-β-Xyl), and PNP-β-cellobioside (PNP-β-Cel), maximum activity was obtained with PNP-β-GlcNAc. The enzyme showed a trace of hydrolytic activity on PNP-β-GalNAc; however, it showed no activity on the other substrates tested. To clarify the role of the enzyme in the chitinolytic system of this strain, assays with chitin oligosaccharides of various lengths, from (GlcNAc)2 to (GlcNAc)6, were performed. Among the substrates tested, (GlcNAc)5 was the best, and the lowest activity was observed with (GlcNAc)2.
TABLE 2.
Substrate specificity of NagA
| Substrate | Relative rate (%)a |
|---|---|
| PNP-β-GlcNAc | 100.0 |
| PNP-α-GlcNAc | 0.0 |
| PNP-β-GalNAc | 2.6 |
| PNP-β-Gal | 0.0 |
| PNP-β-Glu | 0.0 |
| PNP-β-Xyl | 0.0 |
| PNP-β-Cel | 0.0 |
| Di-N-acetylchitobiose | 13.0 |
| Tri-N-acetylchitotriose | 72.0 |
| Tetra-N-acetylchitotetraose | 69.0 |
| Penta-N-acetylchitopentaose | 100.0 |
| Hexa-N-acetylchitohexaose | 31.0 |
Rate relative to activity with PNP-β-GlcNAc or penta-N-acetylchitopentaose as a substrate, which was taken as 100%. The enzyme activity was 12 mU.
Effect of pH, temperature, and metal ions on the activity.
The pH-activity profile obtained with PNP-β-GlcNAc showed a maximum at pH 5.0. At pH 6 and 7 the enzyme showed relatively high activity, >80% of the maximum activity, while a rapid decline was observed at pHs of <4. The temperature optimum for the hydrolysis of PNP-β-GlcNAc was 60°C, and even at 70°C the enzyme showed relatively high activity (>40%). For studying the effects of pH and temperature on enzyme stability, the enzyme solution was incubated for 30 min under various conditions. The enzyme was stable in the pH range from 5 to 7 up to 50°C. Enzyme activity was examined in the presence of metal ions at a 1 mM concentration. Zn2+ and Cu2+ inhibited the activity about 98%, while Ca2+, Mn2+, Co2+, and Mg2+ had practically no effect.
Kinetic characterization.
The effect of substrate (PNP-β-GlcNAc) concentration on enzyme activity was analyzed by hyperbolic regression analysis of the initial velocity and substrate concentration data. The apparent Km and Vmax values for PNP-β-GlcNAc were 425.7 μM and 24.8 μmol/min · mg of protein, respectively, at 60°C.
Comparison of the amino acid sequence of the cloned β-N-acetylglucosaminidase with those of other related enzymes.
The amino acid sequence of NagA was compared with available protein sequences from the GenBank and EMBL databases as well as those in the literature. A search for homology with proteins in databases by using FASTA demonstrated that the amino acid sequence of the enzyme deduced from the nucleotide sequence showed no significant homologies with other β-N-acetylglucosaminidases belonging to family 20, as described by Henrissat and Bairoch (8). However, the enzyme showed a similarity to two β-N-acetylglucosaminidases which have unique primary structures and have not yet been classified into a group of glycosyl hydrolases based on amino acid sequence similarities. One was a β-N-acetylglucosaminidase (Cht60, 27.3% identity) from the marine bacterium Alteromonas sp. strain O-7 in our laboratory (25), and the other was a cytoplasmic β-N-acetylglucosaminidase (ExoII, 16.8% identity) from Vibrio furnissii (5). Despite this similarity, there are significant differences in substrate specificity. Cht60 is a chitobiase, that is, it rapidly hydrolyzes (GlcNAc)2 and shows almost no activity with higher chitin oligosaccharides. On the other hand, ExoII hydrolyzes only PNP-β-GlcNAc and has no activity on chitin oligosaccharides.
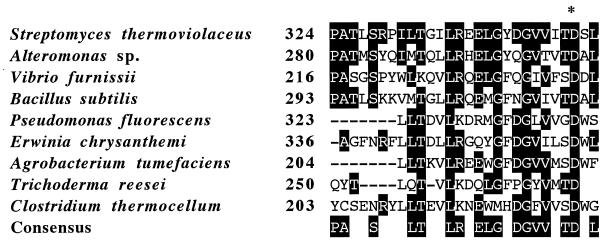
While the enzyme was different from any β-N-acetylglucosaminidase ever reported, the results of computer analysis also revealed sequence homology to microbial β-glucosidases from Bacillus subtilis (32.2% identity) (14), Pseudomonas fluorescens (20.8% identity) (16), Erwinia chrysanthemi (19.9% identity) (27), Agrobacterium tumefaciens (16.6% identity) (4), Trichoderma reesei (15.5% identity) (12), and Clostridium thermocellum (15.3% identity) (7). In particular, sequence comparison of these enzymes revealed high sequence homology with the active-site regions of the β-glucosidases (11), as shown in Fig. 3. This region contained an aspartate catalytic residue (D349 of NagA) well conserved in family 3 β-glucosidases, indicating that aspartic acid and the proximal residues could constitute the active site of NagA from S. thermoviolaceus. Thus, the above-mentioned results indicate that three β-N-acetylglucosaminidases, from S. thermoviolaceus OPC-520, Alteromonas sp. strain O-7, and V. furnissii, are family 3 glycosyl hydrolases.
FIG. 3.
Comparison of the amino acid sequence of NagA with those of the active-site regions of β-glucosidases. Numbers on the left are the residue numbers of the first amino acid in each line. Residues that are identical are indicated by white letters on a black background. The putative active-site aspartic acid residue is marked by an asterisk.
DISCUSSION
During growth on chitin as a carbon source, S. thermoviolaceus OPC-520 secretes a 60-kDa β-N-acetylglucosaminidase (NagA), which hydrolyzes chitin oligosaccharides to N-acetylglucosamine. The final goal of this research is to understand the relationship between the structure and function of enzymes and regulation of genes from the strain involved in chitin degradation. Here we report the isolation, expression, and characterization of a recombinant β-N-acetylglucosaminidase.
The β-N-acetylglucosaminidase gene (nagA) of S. thermoviolaceus OPC-520 was cloned in S. lividans 66 as a BglII fragment of 8.0 kb. The deletion experiments localized the coding region of the gene to a 2.8-kb BamHI-SphI segment. The 2.8-kb fragment contained only a single ORF, which encodes a β-N-acetylglucosaminidase with a predicted molecular mass of 66,329 Da. It is surprising that the mature NagA shares no sequence similarity with the available β-N-acetylglucosaminidase and hexosaminidase sequences except Cht60 (25) and ExoII (5). Although the amino acid sequence of NagA showed homology with those of Cht60 and ExoII, the enzyme clearly differs in important respects such as substrate specificity, kinetic parameters, and susceptibility to inhibitors. The more surprising finding was that NagA showed significant homology with microbial β-glucosidases belonging to glycosyl hydrolase family 3 (8) and that in particular, striking homology was observed in the active-site regions of the β-glucosidases. However, NagA showed no detectable β-glucosidase activity, despite prolonged incubation with excessive quantities of the enzyme. Chitlaru and Roseman reported that arbutin (p-hydroxyphenyl-β-glucoside) was an effective inhibitor of ExoII, because the enzyme showed high similarity to the active site of β-glucosidases (5). Unlike ExoII, arbutin had no inhibitory effect on the activity of NagA. These results indicate that the enzyme is a novel protein which is different from the β-N-acetylglucosaminidases, hexosaminidases, and β-glucosidases previously described. The enzyme hydrolyzed (GlcNAc)5 fastest, and its actions on (GlcNAc)3, (GlcNAc)4, and (GlcNAc)6 were relatively slow. (GlcNAc)2, unlike Cht60, showed the lowest relative hydrolysis rate among the oligomers tested. Thus, it is presumed that the role of the enzyme in the chitin degradation system of S. thermoviolaceus OPC-520 is to hydrolyze chitin oligosaccharides, which are the degradation products from chitin elicited by chitinases. Furthermore, judging from the result that NagA showed low activity on (GlcNAc)2, we might be able to infer that there is a secreted or cytoplasmic β-N-acetylglucosaminidase (chitobiase) that hydrolyzes (GlcNAc)2 escaping digestion by the enzyme to monosaccharide. Expression of the gene was induced by chitin or N-acetylglucosamine in both S. thermoviolaceus and S. lividans with cloned NagA genes and was repressed by glucose (data not shown). Experiments to investigate whether the 5′ and 3′ flanking regions of nagA are involved in the regulation of NagA production in S. lividans are currently in progress.
ACKNOWLEDGMENTS
We thank T. Watanabe and B. Henrissat for valuable discussion on the classification of β-N-acetylglucosaminidase from S. thermoviolaceus OPC-520.
REFERENCES
- 1.Bassler B L, Yu C, Lee Y C, Roseman S. Chitin utilization by marine bacteria: degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24276–24286. [PubMed] [Google Scholar]
- 2.Bibb M J, Cohen S N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187:265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Castle L A, Smith K D, Morris R O. Cloning and sequencing of an Agrobacterium tumefaciens β-glucosidase gene involved in modifying a vir-inducing plant signal molecule. J Bacteriol. 1992;174:1478–1486. doi: 10.1128/jb.174.5.1478-1486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chitlaru E, Roseman S. Molecular cloning and characterization of a novel β-N-acetyl-d-glucosaminidase from Vibrio furnissii. J Biol Chem. 1996;271:33433–33439. doi: 10.1074/jbc.271.52.33433. [DOI] [PubMed] [Google Scholar]
- 6.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gräbnitz F, Rücknagel K P, Seiß M, Staudenbauer W L. Nucleotide sequence of the Clostridium thermocellum bglB gene encoding thermostable β-glucosidase B: homology to fungal β-glucosidases. Mol Gen Genet. 1989;217:70–76. doi: 10.1007/BF00330944. [DOI] [PubMed] [Google Scholar]
- 8.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopwood D A, Bibb M J, Chater K F, Kieser T, Burton C J, Kieser H M, Lydiate D J, Smith C, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 10.Kutzner H J. The family Streptomycetaceae. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Berlin, Germany: Springer-Verlag; 1981. pp. 2028–2090. [Google Scholar]
- 11.Legler G, Roeser K-R, Illig H-K. Reaction of β-d-glucosidase A3 from Aspergillus wentii with d-glucal. Eur J Biochem. 1979;101:85–92. doi: 10.1111/j.1432-1033.1979.tb04219.x. [DOI] [PubMed] [Google Scholar]
- 12.Mach, R. L. 1994. GenBank accession no. U09580.
- 13.McCarthy A J, Williams S T. Actinomycetes as agents of biodegradation in the environment—a review. Gene. 1992;115:189–192. doi: 10.1016/0378-1119(92)90558-7. [DOI] [PubMed] [Google Scholar]
- 14.Quirk, P. G., A. A. Guffanti, S. Clejan, J. Cheng, and T. A. Krulwich. 1995. GenBank accession no. L19954.
- 15.Reissig J L, Strominger J L, Leloir L F. A modified colorimetric method for the estimation of N-acetylamino sugars. J Biol Chem. 1955;217:959–966. [PubMed] [Google Scholar]
- 16.Rixon J E, Ferreira L M A, Durrant A J, Laurie J I, Hazlewood G P, Gilbert H J. Characterization of the gene celD and its encoded product 1,4-β-d-glucan glucohydrolase D from Pseudomonas fluorescens subsp. cellulosa. Biochem J. 1992;285:947–955. doi: 10.1042/bj2850947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaikh S A, Deshpande M V. Chitinolytic enzymes: their contribution to basic and applied research. World J Microbiol Biotechnol. 1993;9:468–475. doi: 10.1007/BF00328035. [DOI] [PubMed] [Google Scholar]
- 20.Soto-Gil R W, Zyskind J W. N,N′-Diacetylchitobiase of Vibrio harveyi. J Biol Chem. 1989;264:14778–14783. [PubMed] [Google Scholar]
- 21.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsujibo H, Minoura K, Miyamoto K, Endo H, Moriwaki M, Inamori Y. Purification and properties of a thermostable chitinase from Streptomyces thermoviolaceus OPC-520. Appl Environ Microbiol. 1993;59:620–622. doi: 10.1128/aem.59.2.620-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsujibo H, Endo H, Minoura K, Miyamoto K, Inamori Y. Cloning and sequence analysis of the gene encoding a thermostable chitinase from Streptomyces thermoviolaceus OPC-520. Gene. 1993;134:113–117. doi: 10.1016/0378-1119(93)90183-4. [DOI] [PubMed] [Google Scholar]
- 24.Tsujibo H, Orikoshi H, Tanno H, Fujimoto H, Miyamoto K, Imada C, Okami Y, Inamori Y. Cloning, sequence, and expression of a chitinase gene from a marine bacterium, Alteromonas sp. strain O-7. J Bacteriol. 1993;175:176–181. doi: 10.1128/jb.175.1.176-181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsujibo H, Fujimoto K, Tanno H, Miyamoto K, Imada C, Okami Y, Inamori Y. Gene sequence, purification and characterization of N-acetyl-β-glucosaminidase from a marine bacterium, Alteromonas sp. strain O-7. Gene. 1994;146:111–115. doi: 10.1016/0378-1119(94)90843-5. [DOI] [PubMed] [Google Scholar]
- 26.von Heijne G. Patterns of amino acids near signal sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 27.Vroemen S, Heldens J, Boyd C, Henrissat B, Keen N T. Cloning and characterization of the bgxA gene from Erwinia chrysanthemi D1 which encodes a β-glucosidase/xylosidase enzyme. Mol Gen Genet. 1995;246:465–477. doi: 10.1007/BF00290450. [DOI] [PubMed] [Google Scholar]
- 28.Wortman A T, Somerville C C, Colwell R R. Chitinase determinants of Vibrio vulnificus: gene cloning and applications of a chitinase probe. Appl Environ Microbiol. 1986;52:142–145. doi: 10.1128/aem.52.1.142-145.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright F, Bibb M J. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]