Abstract
The widespread public health problem of esophageal squamous cell carcinoma (ESCC) is the cause of an increasing number of deaths each year due to delayed diagnosis. Therefore, we require specific and sensitive new biomarkers to manage ESCC better. The detection of diseases, such as cancer, can now be achieved through non-invasive circulating blood-based methods. Blood-based circulating non-coding RNAs, such as miRNA and lncRNA, have been extensively used as valuable markers for lung, esophageal, and breast cancer diagnostic purposes, as quoted in our previous research. Herein, we investigated the role of novel long non-coding RNA EWSAT1 as a blood-based liquid biopsy biomarker for the ESCC. Our findings indicate that EWSAT1 lncRNA has an increased tumor suppressive activity in ESCC, as it reduces by ∼2.59-fold relative to healthy controls. Moreover, we established that EWSAT1 expression can significantly distinguish between clinicopathological characteristics, including age, gender, and lifestyle choices such as smoking, alcohol consumption, and drinking hot beverages among patients with ESCC and healthy individuals. In addition, the expression levels of lncRNA EWSAT1 could distinguish between individuals with more advanced ESCC cancer and those without it, as illustrated by the ROC curve (AUC = 0.7174, 95 % confidence intervals = 0.5901 to 0.8448, p-value = 0.001). Our in-silico prediction methods demonstrated that miR-873-5p is the direct target of EWSAT1, which competes with the tumor suppressor candidate 3 (TUSC3) and EGL-9 family hypoxia-inducible factor 3 (EGLN3) mRNAs through a sponging mechanism, creating the EWSAT1/miR-873-5p/mRNA axis. We have analyzed the role of EWSAT1 in various cellular processes and signaling pathways, including mTOR, Wnt, and MAPK signaling pathways. Circulating EWSAT1 can be used as a liquid biopsy marker for diagnosis of ESCC and has the potential to serve as an effective therapeutic biomarker, according to this pilot study.
Keywords: Circulating long non-coding RNA, Ewing sarcoma associated transcript 1 (EWSAT1), Esophageal squamous cell carcinoma, Liquid biopsy, Diagnosis
1. Introduction
According to the most recent GLOBOCAN data, esophageal cancer (EC) is the eighth most frequent cancer and the sixth leading cause of death worldwide, with a survival rate of less than 18 % [1]. Additionally, according to our prior hospital-based study on the North Indian population, esophageal cancer ranked fourth among all malignancies [2]. Esophageal cancer mainly comprises of two histological subtypes: esophageal squamous cell carcinoma (ESCC) (90 %) and adenocarcinoma (AC) (10 %). The global incidence of ESCC is increasing, highlighting the urgent need to discover alternate and better treatments for ESCC [3]. Currently, multimodal treatment strategies, such as surgery, chemotherapy, and radiotherapy, have been employed for ESCC; however, tumor recurrence is expected due to late-stage diagnosis and ineffective drug molecules [4]. Currently, endoscopic ultrasonography, computed tomography (CT) scan, and tissue biopsy are normally used to diagnose patients; however, some limitations are high cost, late detection, and invasive painful procedures [5,6]. This emphasizes the need for blood-based minimally invasive methods along with reliable diagnostic and therapeutic biomarkers. Liquid biopsies have substantially changed the field of clinical oncology by making tumor collection easier, allowing for continuous monitoring through repeated sampling, screening for therapeutic resistance, and developing customized therapy regimens [7]. Liquid biopsies involve the removal of tumor-derived substances from cancer patients’ bodily fluids, such as blood-based circulating tumor cells, circulating tumor DNA, cell‐free long non‐coding RNAs (cf‐lncRNAs), circulating miRNA, tumor extracellular vesicles, and the subsequent analysis of the genomic, epigenomic and proteomic information they contain [8,9].
According to our previously published articles, circulating lncRNAs have recently undergone extensive research and have shown great promise as biomarkers for the diagnosis and prognosis of various cancers, such as thyroid, pancreatic, esophageal, and ovarian cancer [[10], [11], [12], [13]]. Recently, transcriptomic studies identified several cancer-associated lncRNAs, such as HOTAIR, MALAT1, and H19. Consequently, PCA3 lncRNA has been approved by the FDA for clinical use in prostate cancer [14]. Moreover, we also studied a panel of long-coding RNAs as liquid biopsy biomarkers for cervical and breast cancer [15]. Furthermore, our previous pilot study suggested that circulating lncRNA LOC100507053 and LINC00324 could be employed as liquid biopsy markers of ESCC [16]. As a result, discovering new lncRNA-based liquid biopsy biomarkers for early identification of ESCC has significant clinical implications. Thus, Ewing Sarcoma Associated Transcript 1 (EWSAT1) was identified to be dysregulated in ESCC patients compared to healthy controls, extending our prior investigation to look for potential circulating lncRNA biomarkers in blood samples of ESCC patients versus healthy people.
In various studies, EWSAT1 has already been known to play a significant role in the development and progression of multiple cancers [17]. EWSAT1 (NCBI Gene ID:283673) is a highly conserved lncRNA also known as LINC00277 or TMEM84 and is located on chromosome 15q23, with a length of 24,445 bases, and is found in both the nucleus and cytoplasm [18,19]. EWSAT1 has been reported to be dysregulated in many cancers, including Ewing sarcoma [20], osteosarcoma [21], glioma [22], ovarian cancer [23], and colorectal cancer [24], among others. It is well established that EWSAT1 exerts its oncogenic function through various mechanisms, including regulating gene expression, modulation of signaling pathways, and promoting tumor growth and metastasis [17]. Additionally, EWSAT1 has been suggested as a potential diagnostic and prognostic biomarker in several cancers and is a promising target for cancer therapy [17]. To date, no study has shown the role of EWSAT1 in diagnosing ESCC using a non-invasive strategy based on a circulating lncRNA biomarker.
Herein, we investigated the lncRNA EWSAT1 expression and its role as a liquid biopsy-based diagnostic biomarker for the timely screening of ESCC patients. Further, we also explored the association of EWSAT1 expression with the clinicopathological characters and life status of ESCC patients. Moreover, we also predicted the possible miRNA targets of EWSAT1 through which it regulated the expression of certain proteins and pathways. Using these targets, we also tried to find the possible biological, molecular, cellular processes, and KEGG pathways in which EWSAT1 has a direct or indirect role. Finally, we established a potential EWSAT1/miRNA/mRNA axis through an in-silico approach to be validated hereafter.
2. Methodology
2.1. Clinical sample collection and cell culture
The current study was planned following the Declaration of Helsinki ethical standards. It was approved by the Baba Farid University of Health Sciences, Faridkot (ERB/UCER/2019/4/17), and the Central University of Punjab, Bathinda, India (CUPB/IEC/2023/16), Ethics Review Boards. Blood samples were taken from ESCC patients and healthy individuals as controls, and all participants involved in the study signed informed written consent. To ensure the anonymity of their information, each participant was allocated a unique code that was only used for research purposes. The Tempus Blood RNA Tube (Cat. No. 4342792; Applied Biosystems, CA, United States of America) was used by a phlebotomist to collect samples of peripheral blood (3 ml) from 28 newly diagnosed ESCC patients (Aged 18 years and above) and 32 healthy individuals (Age 18 years and above). This study only included patients who had not yet begun treatment. The hospital's outpatient department followed up with all enrolled patients, either by phone or in person. Furthermore, we obtained KYSE-70 cells (CVCL_1356) (Catalog number: 94072012; Derived from a poorly differentiated invasive esophageal squamous cell carcinoma) from Sigma-Aldrich's European Collection of Authenticated Cell Culture (ECACC). KYSE-70 cells were grown at 37 °C in a humidified incubator with 5 % CO2 in RPMI-1640 media supplemented with 2 mM glutamine (Catalog number: 11875-093; Gibco, Thermo Fisher Scientific, Inc.), 10 % FBS, 1 % penicillin, and 1 % streptomycin. For control, we used surgically removed non-cancerous esophageal tissues that were washed three times with 1X PBS (Catalog number: 10010023; Gibco, Thermo Fisher Scientific, Inc.) and immediately processed for RNA isolation.
2.2. Characteristics of participants
Pathologists identified ESCC patients through histological examination of tissue biopsies in accordance with the eighth edition of the American Joint Committee on Cancer (AJCC) TNM staging system. The following inclusion criteria were used to select ESCC patients for the study: (i) patients who were solely diagnosed with ESCC before beginning treatment; (ii) histopathological stage confirmation as early or metastatic squamous cell carcinoma; (iii) patients with tumor grades ranging from IA to IV; (iv) patients aged 18 years or older, and their gender details; (v) lifestyle history; current or former smoker non‐smoker/passive smoker, and whether or not they consume alcohol and hot beverages. All ESCC patients are excluded: (i) if they have a history of or are currently suffering from any disease; (ii) if they are taking medications; (iii) if Patients had prior chemotherapy or radiotherapy.
Similarly, healthy persons with no history of cancer or other disorders were enlisted in the study as controls. The following criteria were used to choose healthy individuals: (i) individuals aged 18 years or older, as well as gender information; (ii) lifestyle history; non-smoker, nonalcoholic, and nontobacco chewer. Exclusion criteria for such people include (i) a history of cancer or another condition and (ii) the use of drugs currently. The distribution of patients on the basis of clinical features of the patient, including age, gender, tumor grade, TNM stages, and lifestyle habits such as consumption of alcohol, hot beverages, and smoking, are tabulated in Table 1.
Table 1.
Patients divided based on clinicopathological characters and lifestyle choices.
| Characteristics | Patients | |
|---|---|---|
| Age | 18 ≤ 50 | 14 |
| >50 | 14 | |
| Gender | Male | 14 |
| Female | 14 | |
| Tobacco smoking | Yes | 15 |
| No | 13 | |
| Hot beverages | Yes | 15 |
| No | 13 | |
| Alcohol consumption | Yes | 16 |
| No | 12 | |
| Tumor grading | Well-differentiated | 10 |
| Moderate differentiation | 4 | |
| Poor differentiation | 4 | |
| Unknown | 10 | |
| TNM stage | Stage (I + II) | 21 |
| Stage (III + IV) | 7 | |
2.3. Circulating lncRNA isolation and cDNA synthesis
Circulating lncRNAs were extracted from 3 mL of blood samples using the Qiamp RNA Blood mini kit (Cat. No.: 52304; Qiagen, Inc., Valencia, CA, U.S.A.) as previously described by our group [16]. Moreover, total RNA was extracted from normal esophageal tissue and the KYSE-70 cell line using the Trizol LS reagent (Catalog number: 10296010; Thermo Fisher Scientific, Inc). Next, isolated lncRNAs were quantified using a NanoDrop 2000 UV–Vis Spectrophotometer (Thermo Fisher Scientific). Further, using the RT2 First Strand Kit (Cat. No.: 330404, Qiagen, Inc., Valencia, CA, U.S.A.) and 200 ng of extracted lncRNAs as a template, cDNA library was synthesized in a reaction volume of 20 μL each. We prepared a 1:1 mixture of reverse transcriptase and genomic DNA elimination mix and incubated at 37 °C for 60 min, and the reaction was terminated by incubating at 95 °C for 5 min. The cDNA was diluted using 91 μL of nuclease-free water and stored at −20 °C until further use.
2.4. Quantitative real-time PCR (qRT-PCR)
Validation of the EWSAT1 endogenous expression was done using RT2 SYBR Green Mastermix (Qiagen), EWSAT1 Primer (Qiagen), and template (cDNA) according to the manufacturer's protocol. The PCR reaction was conducted at 95 °C for 10 min (holding stage), 95 °C for 5 s (denaturation), 60 °C for 1 min (annealing), 65 °C for 2 min (melt curve), and increments of 2 °C up to 95 °C using a QIAquant 96 5plex real-time PCR system (S/N: 3107V-3324269; Qiagen). All the reactions were performed in triplicates. The 2−ΔΔCT livak method was used to quantify relative gene expression, with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) serving as an internal reference to standardize the Ct values.
2.5. In-silico target prediction of lncRNA/miRNA/mRNA axis
To further explore the in-depth mechanism of EWSAT1 associated with the ESCC progression, we analyzed the subcellular distribution of EWSAT1 using the lncLocator 2.0 web tool (http://www.csbio.sjtu.edu.cn/cgi-bin/lncLocator.py). The subcellular location of EWSAT1 was predicted mostly in the cytoplasm. Afterward, we predicted its miRNA targets through online available computational software such as RNAInter (http://www.rnainter.org/search/), RAID v2.0 (https://www.rna-society.org/raid2/), and overlapped these miRNA targets with miRNAs found differentially expressed in our NGS data previously. Our research has previously demonstrated that by acting as "decoys" for miRNAs, cytoplasmic lncRNAs can affect mRNA stability or translation and influence signaling cascades. Afterward, we used RNA22 (https://cm.jefferson.edu/rna22/) to confirm the binding sites of the potential miRNA targets of EWSAT1. Finally, to complete the EWSAT1/miRNA/mRNA axis, miRNA-mRNA targets were predicted using miRDB (https://mirdb.org/), RNAInter, TargetScan, and RAID v2.0 web-based in-silico target prediction tools.
2.6. Gene ontology (GO) and KEGG pathway analysis
We further investigated the role of EWSAT1 and miRNA targets in different KEGG pathways and GO cellular, molecular, and biological processes. We performed gene set enrichment analysis in the GSEA MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/human/annotate.jsp) and starbase (https://starbase.sysu.edu.cn/) databases. Finally, we plotted the top 20 results based on the −log2(FDR) values.
2.7. Statistical analysis
For data analysis, we utilized GraphPad Prism v8.0 from La Jolla, California, in the U.S.A. To compare the expression levels of lncRNAs between ESCC patients and healthy individuals (control) blood samples. We used the Mann-Whitney test (a non-parametric test) and presented the data as mean ± standard error mean. Additionally, we used the unpaired Mann-Whitney test and paired t-test to examine the association between EWSAT1' expression and clinicopathological characteristics. To denote statistical significance, **** indicates a p-value of less than 0.0001, *** indicates a p-value of less than 0.001, ** indicates a p-value of less than 0.01, and * indicates a p-value of less than 0.05. Meanwhile, ns represents non-significant differences.
3. Results
3.1. EWSAT1 dysregulation in ESCC blood samples
To explore the role of EWSAT1 in ESCC, we first detected the expression of EWSAT1 in 28 ESCC blood samples. Results showed EWSAT1 downregulation in ESCC patients’ (Fold change ∼ 2.59; p-value = 0.0030) compared to healthy controls (Fig. 1A). In validation, we found that EWSAT1 was also downregulated (Fold change ∼ 2.5; p-value > 0.0042) in the ESCC cell line (KYSE-70) compared to non-cancerous esophageal tissue (Fig. 1B). These results confirm that EWSAT1 plays the role of tumor suppressor in ESCC patients. Next, to check the diagnostic potential of EWSAT1 in clinics, we plotted the ROC curve, as shown in Fig. 1C. The AUC value of the ROC curve was calculated to be 0.7174 [95 % CI = 0.5901 to 0.8448; p-value = 0.0034], which confirmed that EWSAT possesses a significant diagnostic value. Hence, EWSAT1 is highly sensitive lncRNA, which can significantly discriminate ESCC patients from healthy controls.
Fig. 1.
(A). Normalized expression (Ct) of EWSAT1 lncRNA was observed in blood samples from ESCC patients' when compared with healthy controls. (B). Relative expression of EWSAT1 in KYSE-70 cells compared to tumor-adjacent normal cells. (C). ROC curve confirming the diagnostic capability of lncRNA EWSAT1. A minimum of three separate experiments were carried out. Data are represented as the mean ± SEM. ΔCt values are calculated to show the normalized expression of EWSAT1 using ΔCt = mean Ct value of EWSAT1 – mean Ct value of GAPDH. A lower Ct value indicates higher expression, while a higher Ct value indicates lower expression. The data are presented as mean ± SEM, where * denotes p-values 0.05, ** denotes p-values 0.01, *** denotes p-values 0.001, and **** represents p-values 0.0001, determined using Mann-Whitney test.
3.2. EWSAT1 expression association with clinicopathological features and lifestyle choices
Additionally, we looked at the relationships between the expression of EWSAT1 and the clinicopathological traits and dietary habits of ESCC patients. Results demonstrated that in ESCC patients aged 18 ≤ 50 years (n = 14; p-value = 0.0028), EWSAT1 was ∼5.79-fold downregulated. There was no significant change in EWSAT1 expression between ESCC patients with ages greater than 50 years (n = 14; p-value = 0.1871) and healthy individuals. EWSAT1 expression did not differ significantly between the two age groups of ESCC patients (p-value = 0.3420) (Fig. 2A). Similarly, no significant change in EWSAT1 expression was seen between the two gender groups of ESCC patients (p-value = 0.9006). Female ESCC patients, on the other hand, had a ∼4.64-fold downregulation of EWSAT1 expression (n = 14; p-value = 0.0161) compared to female healthy individuals, whereas male ESCC patients had no significant association of EWSAT1 expression (n = 14; p-value = 0.1201) (Fig. 2B).
Fig. 2.
EWSAT1 expression association with ESCC patients'' lifestyle choices and clinicopathological traits compared to healthy people. (A). ESCC patient's age. (B). ESCC patients' gender. (C). ESCC patients with smoking habits. (D). ESCC patients with alcohol use status. (E). Hot beverage consumption status. (F). ESCC patients' histopathological grading. (G). ESCC patients' TNM staging. In comparison to healthy individuals, bar graphs show the normalized expression of EWSAT1 in ESCC patients. The data are presented as mean ± SEM, where * denotes p-values 0.05, ** denotes p-values 0.01, *** denotes p-values 0.001, and **** represents p-values 0.0001, determined using unpaired (Mann-Whitney test) and paired t-tests. [Tumor node metastasis (TNM)].
Following that, EWSAT1 expression levels in ESCC patients with a history of tobacco smoking did not alter significantly. In contrast, non-smoker ESCC patients had ∼4.934-fold lower levels of EWSAT1 (n = 13; p-value = 0.0032) than healthy individuals (Fig. 2C). There was no significant difference in EWSAT1 expression between smoking and non-smoking cancer patients. In addition, patients with a history of drinking hot beverages had a ∼4.038-fold downregulation (n = 15; p-value = 0.0161), whereas patients with no history of drinking hot beverages had a ∼3.631-fold downregulation (n = 13; p-value = 0.0321). At the same time, there was no significant difference between the two groups of patients (p-value = 0.9183) (Fig. 2D). Furthermore, ESCC patients with a history of alcohol consumption had a ∼4.63-fold downregulation (n = 16; p-value = 0.0066), whereas patients without a history of alcohol consumption had no statistically significant alteration in EWSAT1 expression (n = 12; p-value = 0.0768) when compared to healthy individuals. There was no significant difference in EWSAT1 expression levels across the two groups of ESCC patients (p-value = 0.4707) (Fig. 2E).
After that, we tested whether EWSAT1 expression can help differentiate various tumor grades in ESCC patients. However, no significant differences were found between patients with different tumor grades (well differentiated, moderate differentiated, poor differentiation) (p-values = 0.3477, 0.5720, 0.2327) (Fig. 2F). Furthermore, we tested whether EWSAT1 expression could distinguish between ESCC patients based on TNM stages. However, there was no significant difference between TNM stages (I + II) and (III + IV) of ESCC patients (p-value = 0.0815) (Fig. 2G).
3.3. EWSAT1 acts as ceRNA by sponging miR-873-5p and modulates (protein) expression
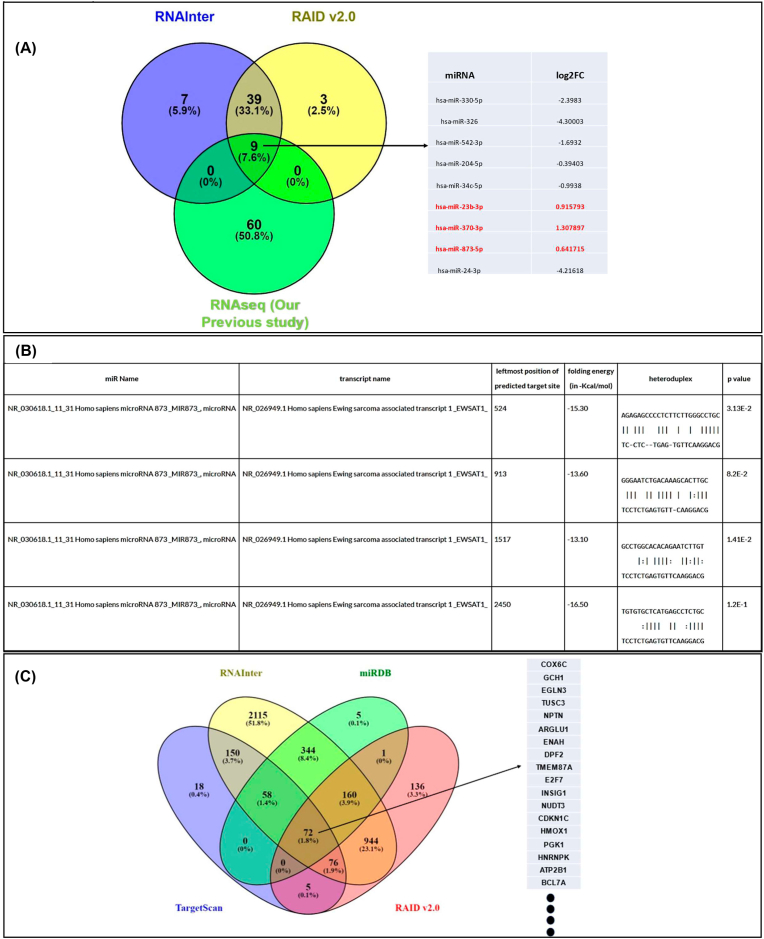
Using the lncLocator 2.0 in-silico tool, we found that lncRNA EWSAT1 is highly enriched in the cytoplasm. It has already been established that cytoplasmic lncRNAs function as a ceRNA and sponge miRNA, rendering them unable to bind to their mRNA target [25]. Thus, we predicted the miRNA targets of lncRNA EWSAT1 using in-silico tools, i.e., RNAInter, RAID v2.0, and RNAseq data of 4 ESCC patients’ blood samples from our previous study. Results showed nine common targets from which three potential miRNA targets were finalized, viz. miR-23b-3p, miR-370-3p, and miR-873-5p (Fig. 3A). Next, we used the RNA22 computational database to confirm the binding sites of these miRNAs with EWSAT1, and the results demonstrated miR-873-5p as the best-suited target with four potential binding sites and having significant folding energy (Fig. 3B). Further, to complete the EWSAT1/miR-873-5p/mRNA axis, we used in-silico miRNA target prediction web tools such as TargetScan, RNAInter, miRDB, and RAID v2.0. Results demonstrated seventy-two common mRNA targets, out of which we found two tumor suppressor mRNAs complementing EWSAT1 expression(Fig. 3C). Thus, we considered tumor suppressor candidate 3 (TUSC3) and Egl-9 family hypoxia-inducible factor 3 (EGLN3) as potential mRNA targets, which are already reported as tumor suppressors in ESCC. Next, we plan to do a luciferase assay to validate our in-silico predictions.
Fig. 3.
Using online databases and our earlier NGS data, in-silico target prediction of EWSAT1. (A). Three miRNAs identified in RNAInter, RAID v2.0, and our NGS data are expected to be EWSAT1 targets: hsa-miR-23b-3p, hsa-miR-370-3p, and hsa-miR-873-5p. (B). The hsa-miR-873-5p with the greatest number of putative binding sites was selected for further investigation. (C). The RNAInter, miRDB, RAID v2.0, and TargetScan databases all were having 72 common mRNA targets for hsa-miR-873-5p.
3.4. EWSAT1 targets play roles in various biological and cellular pathways
To understand the role of EWSAT1 in various KEGG pathways and GO biological, cellular, and molecular processes, we performed a Gene set enrichment analysis of the EWSAT1 targets. Results showed that EWSAT1 target genes are mainly involved in various biological processes, such as the negative regulation of cell cycle arrest, formation of a translation initiation complex, regulation of RNA binding, regulation of histone ubiquitination, actin polymerization and depolymerization, etc. (Fig. 4A). Also, they actively participate in various molecular processes such as PIP3 binding, CDK activity, GTPase inhibitor activity, RNA, enzyme, cytoskeletal protein binding, etc. (Fig. 4B). Further, they participate in many cellular processes involving transcription elongation factor complex, CDK holoenzyme complex, CDK positive transcription elongation factor complex, translation initiation factor 4f complex, etc. (Fig. 4C). Moreover, the KEGG pathways involving EWSAT1 targets are regulation of the actin cytoskeleton, citrate cycle TCA cycle, pyruvate metabolism, and mTOR signaling pathway (Fig. 4D).
Fig. 4.
Gene Set enrichment analysis of EWSAT1 target genes in various signaling pathways. (A). Significantly enriched activated and suppressed Gene Ontology (GO) biological processes. (B). Significantly enriched activated and suppressed GO molecular processes. (C). Significantly enriched activated and suppressed GO cellular processes. (D). Significantly enriched activated and suppressed KEGG pathway. The vertical items are the names of GO and KEGG terms, and the length of the horizontal lines represents the gene numbers and FDR value represented by respective colors in the index.
Next, we performed gene set enrichment analysis of the hsa-miR-873-5p targets to explore KEGG pathways and GO processes involving the EWSAT1/miR-873-5p axis. Results show that miR-873-5p targets are enriched in various biological processes such as negative regulation of the biosynthetic process, regulation of cell death, regulation of intracellular signal transduction, protein phosphorylation, apoptotic processes, cell cycle, intracellular transport, etc. (Fig. 5A). Furthermore, miR-873-5p targets are also involved in various molecular processes such as transcription factor binding, signaling receptor binding, chromatin binding, protein kinase activity, cell adhesion molecule binding, sequence-specific DNA binding, RNA binding, etc. (Fig. 5B). Many GO cellular processes that involve miR-873-5p targets include vesicle membrane, transferase complex, mitochondrion, plasma membrane region, chromatin, anchoring junction, endosome, chromosome, endoplasmic reticulum, Golgi apparatus, etc. (Fig. 5C). Moreover, KEGG pathways involving these targets are the Wnt signaling pathway, MAPK signaling pathway, and many other cancer-related pathways (Fig. 5D). These in-silico results revealed that the EWSAT1/miR-873-5p/mRNA axis has an active role in various biological processes and cancer-associated signaling pathways. Thus, we are exploring more about this axis using wet lab experimentation approaches to validate their molecular mechanism in ESCC etiology.
Fig. 5.
Gene Set enrichment analysis of miR-873-5p target genes in various signaling pathways. (A). Significantly enriched activated and suppressed Gene Ontology (GO) biological processes. (B). Significantly enriched activated and suppressed GO molecular processes. (C). Significantly enriched activated and suppressed GO cellular processes. (D). Significantly enriched activated and suppressed KEGG pathway. The vertical items are the names of GO and KEGG terms, and the length of the horizontal lines represents the gene numbers and FDR value represented by respective colors in the index.
4. Discussion
Currently, invasive endoscopy and painful histopathology biopsy are used to diagnose ESCC [5]; hence, a blood-based circulating biomarker for better detection and management of ESCC is necessary. Notably, as high-throughput transcriptome research became more prevalent, numerous lncRNAs were discovered to have an important role in cancer etiology. More study reveals that lncRNA can operate as a novel biomarker or target for cancer early detection and therapy due to their critical roles in disease formation, growth, progression, and metastasis [26]. Furthermore, when combined with various clinicopathological criteria, the status of each individual patient can be identified with higher precision, allowing for the formulation of more precise and individualized treatments [27].
In this regard, our prior study used high throughput RNAseq on blood samples from four ESCC patients and age-matched healthy controls. As a consequence, 296 differentially expressed lncRNAs were identified [16]. In this investigation, we chose one of the potential lncRNA "EWSAT1″ that was revealed to be downregulated in the previous study's RNAseq data. Using EWSAT1-specific primers, we confirmed its expression in blood samples from 28 ESCC patients and 34 healthy controls. The qRT-PCR results strongly corroborated the downregulation of EWSAT1 expression in ESCC patients, and we demonstrated the good diagnostic capability of lncRNA EWSAT1 in ESCC patients using the ROC curve with AUC value = 0.7174 (Fig. 1). Previous research has linked EWSAT1 dysregulation to a variety of malignancies. Upregulation of EWSAT1 has been linked to a variety of malignancies, including ovarian cancer [23], colorectal cancer [24], osteosarcoma [28], glioma [22], and hepatocellular carcinoma [29]. In contrast, a pan-cancer analysis based on the TCGA reveals that EWSAT1 is considerably downregulated in Testicular Germ Cell Tumors (TGCT) and Brain Lower Grade Glioma (LGG) [17]. Various factors could account for this kind of dual behavior of EWSAT1, including tissue-specific functions, cell type, and/or genetic and epigenetic differences in patients. Until now, no study has demonstrated EWSAT1 downregulation in ESCC, particularly with the benefit of liquid biopsy detection. Many lifestyle factors, such as cigarette use, diet (fried meals, hot and spicy foods, etc.), malnutrition, alcohol usage, and poor oral hygiene, are well-established to have a key role in ESCC pathogenesis [30]. As a result, we investigated the relationship between EWSAT1 expression and clinicopathological characteristics as well as ESCC patients' life status. Interestingly, the downregulation of EWSAT1 expression was found to be substantially linked with ESCC patients'' age group (18–50), female gender, hot beverage use, and alcohol consumption. While no significant connection was detected among ESCC patients of any age group (>50), male gender, cigarette smoking status, tumor grades, or TNM stages (Fig. 2).
Several signaling pathways and miRNAs have been found to interact with EWSAT1, implying that they are important molecules that influence its expression [24,31,32]. Cytoplasmic lncRNAs are well known for their role as ceRNAs that compete with other mRNAs for miRNA binding, forming an axis known as the lncRNA/miRNA/mRNA axis [25]. These ceRNA networks have already been explored in several cancers and are important in tumor etiology pathophysiology [33]. Thus, we identified miR-873-5p as a candidate target for EWSAT1 because it has four binding sites, and EWSAT1 downregulation and miR-873-5p overexpression have been verified in our prior RNAseq data. MiR-873-5p has already been thoroughly researched and found to be dysregulated in a number of malignancies, where it plays diverse roles [34]. In reality, miR-873-5p functions as an oncogene in certain malignancies, including non-small cell lung cancer (NSCLC) [35], Merkel cell carcinoma (MCC) [36], and lung adenocarcinoma (LUAD) [37]. It serves as a tumor suppressor in pancreatic cancer [38], cervical cancer [39], nasopharyngeal carcinoma [40], breast cancer [41], glioblastoma [42], and other cancers. Furthermore, multiple studies have identified miR-873-5p as a major diagnostic and prognostic biomarker for various malignancies [34]. Furthermore, miR-873-5p target genes are involved in various biological activities at the molecular level, including transcription regulation, signaling receptor binding, RNA binding, and so on. MiR-873-5p has also been identified to alter signaling pathways related to cancer growth, such as Wnt/-Catenin, MAPK, and others [34].
In this study, we used target prediction techniques to sort seventy-two candidate miR-873-5p targets and finally chose TUSC3 and EGLN3 as the best fit for the creation of the EWSAT1/miR-873-5p/mRNA axis because both targets had considerable binding affinity. TUSC3 has previously been identified as a tumor suppressor gene involved in ESCC prognosis [43]. EGLN3 has also been identified as a tumor suppressor gene in ESCC, as well as numerous other malignancies [44]. As a result, each of these targets can be employed to finish the EWSAT1 ceRNA axis. Various studies have established ceRNA interactions to regulate tumor cell proliferation, migration, invasion, apoptosis, or chemoresistance by regulating various signaling pathways. Likewise, EWSAT1/miR-873-5p/miRNA axis and its associated biological, molecular, and cellular processes, as well as KEGG pathways, demonstrated that the axis is vital in essential cellular functions and numerous signaling pathways, mainly linked to cancer hallmarks (Fig. 4, Fig. 5). Moreover, many ncRNAs facilitate antitumor effects by regulating the expression of immune and immunotherapy markers mostly through sponging miRNAs [45]. Unfortunately, we have not found any such function associated with lncRNA EWSAT1. Consequently, with high sensitivity and specificity (AUC value > 0.7174) along with advantage of blood-based detection of EWSAT1 confirms that EWSAT1 has significance over other diagnostic biomarkers and could be employed as a diagnostic biomarker for ESCC (Fig. 6).
Fig. 6.
Demonstration for using lncRNA EWSAT1 as a potential blood-based diagnostic biomarker for ESCC patients.
5. Conclusion
Overall, our findings point to a new blood-based circulating biomarker called "EWSAT1″ that can be used to diagnose ESCC. When compared to healthy controls, the LncRNA EWSAT1 was shown to be considerably downregulated in 28 ESCC patients. This confirmed that EWSAT1 is a tumor suppressor lncRNA in ESCC and that its dysregulation contributes to the disease's etiology. Furthermore, EWSAT1 expression was observed to be linked with clinicopathological variables such as age (18–50), female gender, and lifestyle choices such as cigarette smoking, alcohol drinking, and hot beverage intake. With these results, associations of dysregulated EWSAT1 with lifestyle choices demonstrate its significance and role in the etiology of ESCC. Furthermore, we estimated miR-873-5p as a direct target of EWSAT1, which might serve as a ceRNA and sponge miR-873-5p, leaving it impossible to bind its putative mRNA targets TUSC3 and EGLN3, both of which have a significant binding affinity. Finally, we discovered EWSAT1/miR-873-5p/mRNA axis participation in several GO biological, molecular, and cellular processes and KEGG pathways using in-silico techniques. Furthermore, because our current study has a limited cohort size, it can be converted into clinical trials with more significant cohorts. Additionally, as the EWSAT1/miR-873-5p/mRNA axis possesses a tumor suppressor role, it could be investigated for its therapeutic efficacy by employing various in-vitro and in-vivo techniques in the future.
Ethics approval and consent to participate
The present study was conducted in accordance with the declaration of Helsinki and approved by the Ethics Review Board of Baba Farid University of Health Sciences, Faridkot (ERB/UCER/2019/4/17), and the Institutional Ethics Committee of Central University of Punjab, Bathinda (CUPB/IEC/2023/16). All patients provided written informed consent.
CRediT authorship contribution statement
Vivek Uttam: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. Manjit Kaur Rana: Data curation, Investigation, Resources. Uttam Sharma: Methodology, Validation, Visualization. Karuna Singh: Data curation, Resources. Aklank Jain: Conceptualization, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
All the authors declare no conflict of interest.
Acknowledgments
Funding: The Central University of Punjab, Department of Zoology, supported the research by providing adequate research facilities and funds to Prof. Aklank Jain. University Grants Commission (U.G.C.), New Delhi, supported in the form of Junior Research Fellowship (J.R.F.) (U.G.C. ref. no.; 191620043900) under the National Fellowship for Schedule Caste (NFSC). This is the name of the grant sponsored by Ministry of Tribal Affairs, Government of Inida, scheme to Vivek Uttam.
Contributor Information
Vivek Uttam, Email: vivekuttam52@gmail.com.
Manjit Kaur Rana, Email: drmrsmannjitkaur@gmail.com.
Uttam Sharma, Email: uttamsharma1994@gmail.com.
Karuna Singh, Email: karuna.mamc@gmail.com.
Aklank Jain, Email: aklankjain@gmail.com.
References
- 1.Morgan E., Soerjomataram I., Rumgay H., Coleman H.G., Thrift A.P., Vignat J., et al. The global landscape of esophageal squamous cell carcinoma and esophageal adenocarcinoma incidence and mortality in 2020 and projections to 2040: new estimates from GLOBOCAN 2020. Gastroenterology. 2022;163(3):649–658 e2. doi: 10.1053/j.gastro.2022.05.054. [DOI] [PubMed] [Google Scholar]
- 2.Rana M.K., Barwal T.S., Sharma U., Bansal R., Singh K., Rana A.P.S., et al. Current trends of carcinoma: experience of a tertiary care cancer center in North India. Cureus. 2021;13(6) doi: 10.7759/cureus.15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q.L., Xie S.H., Wahlin K., Lagergren J. Global time trends in the incidence of esophageal squamous cell carcinoma. Clin. Epidemiol. 2018;10:717–728. doi: 10.2147/CLEP.S166078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J., Zhu Z., Liu Y., Jin X., Xu Z., Yu Q., et al. Diagnostic value of multiple tumor markers for patients with esophageal carcinoma. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0116951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allemani C., Matsuda T., Di Carlo V., Harewood R., Matz M., Niksic M., et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong F., Li N., Feng Z., Zheng Y., Zhu C., Zhang F. Exosomal microRNAs as novel diagnostic biomarkers in breast cancer: a systematic evaluation and meta-analysis. Asian J. Surg. 2023 doi: 10.1016/j.asjsur.2023.05.115. [DOI] [PubMed] [Google Scholar]
- 8.Alix-Panabieres C., Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021;11(4):858–873. doi: 10.1158/2159-8290.CD-20-1311. [DOI] [PubMed] [Google Scholar]
- 9.Roy R.K., Yadav R., Sharma U., Wasson M.K., Sharma A., Tanwar P., et al. Impact of non-coding RNAs on cancer-directed immune therapies: now then and forever. Int. J. Cancer. 2022;151(7):981–992. doi: 10.1002/ijc.34060. [DOI] [PubMed] [Google Scholar]
- 10.Barwal T.S., Sharma U., Rana M.K., Bazala S., Singh I., Murmu M., et al. A diagnostic and prognostic value of blood-based circulating long non-coding RNAs in thyroid, pancreatic and ovarian cancer. Crit. Rev. Oncol. Hematol. 2022;171 doi: 10.1016/j.critrevonc.2022.103598. [DOI] [PubMed] [Google Scholar]
- 11.Sanya D.R.A., Onesime D. Roles of non-coding RNAs in the metabolism and pathogenesis of bladder cancer. Hum. Cell. 2023;36(4):1343–1372. doi: 10.1007/s13577-023-00915-5. [DOI] [PubMed] [Google Scholar]
- 12.Sharma U., Barwal T.S., Murmu M., Acharya V., Pant N., Dey D., et al. Clinical potential of long non-coding RNA LINC01133 as a promising biomarker and therapeutic target in cancers. Biomarkers Med. 2022;16(5):349–369. doi: 10.2217/bmm-2021-0682. [DOI] [PubMed] [Google Scholar]
- 13.Sharma U., Barwal T.S., Malhotra A., Pant N., Vivek, Dey D., et al. Long non-coding RNA TINCR as a potential biomarker and therapeutic target for cancer. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Y., Shi L., Luo Z. Long non-coding RNAs in cancer: implications for diagnosis, prognosis, and therapy. Front. Med. 2020;7 doi: 10.3389/fmed.2020.612393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barwal T.S., Sharma U., Vasquez K.M., Prakash H., Jain A. A panel of circulating long non-coding RNAs as liquid biopsy biomarkers for breast and cervical cancers. Biochimie. 2020;176:62–70. doi: 10.1016/j.biochi.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Sharma U., Barwal T.S., Khandelwal A., Rana M.K., Rana A.P.S., Singh K., et al. Circulating long non-coding RNAs LINC00324 and LOC100507053 as potential liquid biopsy markers for esophageal squamous cell carcinoma: a pilot study. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.823953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen J., Li H., Li D., Dong X. Clinicopathological and prognostic significance of long non-coding RNA EWSAT1 in human cancers: a review and meta analysis. PLoS One. 2022;17(3) doi: 10.1371/journal.pone.0265264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barshir R., Fishilevich S., Iny-Stein T., Zelig O., Mazor Y., Guan-Golan Y., et al. GeneCaRNA: a comprehensive gene-centric database of human non-coding RNAs in the GeneCards suite. J. Mol. Biol. 2021;433(11) doi: 10.1016/j.jmb.2021.166913. [DOI] [PubMed] [Google Scholar]
- 20.Marques Howarth M., Simpson D., Ngok S.P., Nieves B., Chen R., Siprashvili Z., et al. Long non-coding RNA EWSAT1-mediated gene repression facilitates Ewing sarcoma oncogenesis. J. Clin. Invest. 2014;124(12):5275–5290. doi: 10.1172/JCI72124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G.Y., Zhang J.F., Hu X.M., Luo Z.P., Ma Y.Z. Clinical significance of long non-coding RNA EWSAT1 as a novel prognostic biomarker in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2017;21(23):5337–5341. doi: 10.26355/eurrev_201712_13918. [DOI] [PubMed] [Google Scholar]
- 22.Yang H., Chen W., Jiang G., Yang J., Wang W., Li H. Long non-coding RNA EWSAT1 contributes to the proliferation and invasion of glioma by sponging miR-152-3p. Oncol. Lett. 2020;20(2):1846–1854. doi: 10.3892/ol.2020.11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu X., Zhang L., Dan L., Wang K., Xu Y. LncRNA EWSAT1 promotes ovarian cancer progression through targeting miR-330-5p expression. Am J Transl Res. 2017;9(9):4094–4103. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Huang S., Liao X., Chen Z., Li L., Yu L., et al. LncRNA EWSAT1 promotes colorectal cancer progression through sponging miR-326 to modulate FBXL20 expression. OncoTargets Ther. 2021;14:367–378. doi: 10.2147/OTT.S272895. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L., Li C., Su X. Emerging impact of the long non-coding RNA MIR22HG on proliferation and apoptosis in multiple human cancers. J. Exp. Clin. Cancer Res. 2020;39(1):271. doi: 10.1186/s13046-020-01784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bashraheel S.S., Domling A., Goda S.K. Update on targeted cancer therapies, single or in combination, and their fine tuning for precision medicine. Biomed. Pharmacother. 2020;125 doi: 10.1016/j.biopha.2020.110009. [DOI] [PubMed] [Google Scholar]
- 28.Shen D., Liu Y., Liu Y., Wang T., Yuan L., Huang X., et al. Long non-coding RNA EWSAT1 promoted metastasis and actin cytoskeleton changes via miR-24-3p sponging in osteosarcoma. J. Cell Mol. Med. 2021;25(2):716–728. doi: 10.1111/jcmm.16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X., Chen J., Zhou J., Mao A., Xu W., Zhu H., et al. LncRNA-EWSAT1 promotes hepatocellular carcinoma metastasis via activation of the Src-YAP signaling axis. Faseb. J. 2022;36(12) doi: 10.1096/fj.202200825R. [DOI] [PubMed] [Google Scholar]
- 30.Uhlenhopp D.J., Then E.O., Sunkara T., Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13(6):1010–1021. doi: 10.1007/s12328-020-01237-x. [DOI] [PubMed] [Google Scholar]
- 31.Cui S., Yang C.L., Chen D.Y. LncRNA EWSAT1 regulates the tumorigenesis of NSCLC as a ceRNA by modulating miR-330-5p/ITGA5 Axis. Biochem. Genet. 2021;59(6):1441–1456. doi: 10.1007/s10528-021-10069-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Q., Xie Y., Wang L., Xu T., Gao Y. LncRNA EWSAT1 upregulates CPEB4 via miR-330-5p to promote cervical cancer development. Mol. Cell. Biochem. 2020;471(1–2):177–188. doi: 10.1007/s11010-020-03778-8. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Urrutia E., Bustamante Montes L.P., Ladron de Guevara Cervantes D., Perez-Plasencia C., Campos-Parra A.D. Crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: deciphering molecular mechanisms of master regulators in cancer. Front. Oncol. 2019;9:669. doi: 10.3389/fonc.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou Y., Zhong C., Hu Z., Duan S. MiR-873-5p: a potential molecular marker for cancer diagnosis and prognosis. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.743701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin S., He J., Li J., Guo R., Shu Y., Liu P. MiR-873 inhibition enhances gefitinib resistance in non-small cell lung cancer cells by targeting glioma-associated oncogene homolog 1. Thorac Cancer. 2018;9(10):1262–1270. doi: 10.1111/1759-7714.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ning M.S., Kim A.S., Prasad N., Levy S.E., Zhang H., Andl T. Characterization of the Merkel cell carcinoma miRNome. J Skin Cancer. 2014;2014 doi: 10.1155/2014/289548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y., Xue Q., Wang D., Du M., Zhang Y., Gao S. miR-873 induces lung adenocarcinoma cell proliferation and migration by targeting SRCIN1. Am J Transl Res. 2015;7(11):2519–2526. [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X.L., Ma Y.S., Liu Y.S., Jiang X.H., Ding H., Shi Y., et al. microRNA-873 inhibits self-renewal and proliferation of pancreatic cancer stem cells through pleckstrin-2-dependent PI3K/AKT pathway. Cell. Signal. 2021;84 doi: 10.1016/j.cellsig.2021.110025. [DOI] [PubMed] [Google Scholar]
- 39.Liang H.X., Li Y.H. MiR-873, as a suppressor in cervical cancer, inhibits cells proliferation, invasion and migration via negatively regulating ULBP2. Genes Genomics. 2020;42(4):371–382. doi: 10.1007/s13258-019-00905-8. [DOI] [PubMed] [Google Scholar]
- 40.Lv B., Li F., Liu X., Lin L. The tumor-suppressive role of microRNA-873 in nasopharyngeal carcinoma correlates with downregulation of ZIC2 and inhibition of AKT signaling pathway. Cancer Gene Ther. 2021;28(1–2):74–88. doi: 10.1038/s41417-020-0185-8. [DOI] [PubMed] [Google Scholar]
- 41.Tang L., Chen Y., Tang X., Wei D., Xu X., Yan F. Long non-coding RNA DCST1-AS1 promotes cell proliferation and metastasis in triple-negative breast cancer by forming a positive regulatory loop with miR-873-5p and MYC. J. Cancer. 2020;11(2):311–323. doi: 10.7150/jca.33982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang R.J., Li J.W., Bao B.H., Wu H.C., Du Z.H., Su J.L., et al. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J. Biol. Chem. 2015;290(14):8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X., Zhang J., Zhong H., Liu F., Liang N., Wang Y., et al. Decreased tumor suppressor candidate 3 predicts poor prognosis of patients' with esophageal squamous cell carcinoma. Int. J. Med. Sci. 2016;13(12):963–969. doi: 10.7150/ijms.16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fong L.Y., Taccioli C., Palamarchuk A., Tagliazucchi G.M., Jing R., Smalley K.J., et al. Abrogation of esophageal carcinoma development in miR-31 knockout rats. Proc. Natl. Acad. Sci. U. S. A. 2020;117(11):6075–6085. doi: 10.1073/pnas.1920333117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy R.K., Yadav R., Sharma U., Wasson M.K., Sharma A., Tanwar P., Jain A., Prakash H. Impact of noncoding RNAs on cancer directed immune therapies: now then and forever. Int. J. Cancer. 2022;151(7):981–992. doi: 10.1002/ijc.34060. [DOI] [PubMed] [Google Scholar]