Abstract
The gram-negative bacterium Pseudomonas cichorii 170, isolated from soil that was repeatedly treated with the nematocide 1,3-dichloropropene, could utilize low concentrations of 1,3-dichloropropene as a sole carbon and energy source. Strain 170 was also able to grow on 3-chloroallyl alcohol, 3-chloroacrylic acid, and several 1-halo-n-alkanes. This organism produced at least three different dehalogenases: a hydrolytic haloalkane dehalogenase specific for haloalkanes and two 3-chloroacrylic acid dehalogenases, one specific for cis-3-chloroacrylic acid and the other specific for trans-3-chloroacrylic acid. The haloalkane dehalogenase and the trans-3-chloroacrylic acid dehalogenase were expressed constitutively, whereas the cis-3-chloroacrylic acid dehalogenase was inducible. The presence of these enzymes indicates that 1,3-dichloropropene is hydrolyzed to 3-chloroallyl alcohol, which is oxidized in two steps to 3-chloroacrylic acid. The latter compound is then dehalogenated, probably forming malonic acid semialdehyde. The haloalkane dehalogenase gene, which is involved in the conversion of 1,3-dichloropropene to 3-chloroallyl alcohol, was cloned and sequenced, and this gene turned out to be identical to the previously studied dhaA gene of the gram-positive bacterium Rhodococcus rhodochrous NCIMB13064. Mutants resistant to the suicide substrate 1,2-dibromoethane lacked haloalkane dehalogenase activity and therefore could not utilize haloalkanes for growth. PCR analysis showed that these mutants had lost at least part of the dhaA gene.
1,3-Dichloropropene (γ-chloroallylchloride or 1,3-dichloropropylene) is a synthetic compound that is not known to be formed naturally. The industrial production of this compound started in the 1950s as the major and active ingredient of Shell D-D and Telone II. These commercial products are mixtures of cis-1,3-dichloropropene, trans-1,3-dichloropropene, and 1,2-dichloropropane and have been used worldwide in agriculture as preplant soil fumigants for control of plant-parasitic nematodes.
1,3-Dichloropropene is applied in The Netherlands to combat potato cyst nematodes on more than 30,000 ha/year. The mixture is usually applied by injection into the soil at a maximum dose of 170 kg/ha (23), which means that very large amounts (>5,000 tons/year) are used on potato fields. Although the soil is sealed by rolling, a large amount (50%) of the injected 1,3-dichloropropene evaporates into the atmosphere (23) and has a significant effect on the total air pollution caused by chlorinated hydrocarbons in The Netherlands. Fumigants such as Shell D-D and Telone II also represent an important class of carcinogenic water pollutants because their components are resistant to biological degradation and can easily permeate through soils into groundwater supplies (4, 23). The use of large amounts of these compounds in agriculture and the risk of undesirable side effects have led to several investigations into the fate and persistence of 1,3-dichloropropene and its degradation products.
Degradation of 1,3-dichloropropene in soil under laboratory and field conditions has been studied previously. Roberts and Stoydin (13) showed that both isomers of 1,3-dichloropropene were converted to the corresponding 3-chloroallyl alcohols and 3-chloroacrylic acids. Castro and Belser (3) proposed a degradation pathway for 1,3-dichloropropene and showed that the first step, chemical hydrolysis of 1,3-dichloropropene to 3-chloroallyl alcohol, does indeed take place in soil. Van Dijk (21) measured a much lower rate of disappearance of chloroallyl alcohols in sterilized soils than in nonsterilized soils, suggesting that the degradation in soil is mainly biological. These results suggest that environmental degradation of both 1,3-dichloropropene isomers is a result of microbial action, with the exception of the initial hydrolysis of 1,3-dichloropropene to 3-chloroallyl alcohol.
Bacterial degradation of 3-chloroallyl alcohol and 3-chloroacrylic acid by pure cultures has also been demonstrated (1, 6, 22), but little is known about the complete microbial degradation of 1,3-dichloropropene. The first report concerning enrichment and isolation of 1,3-dichloropropene-degrading organisms was published recently (24). In this report, Verhagen and coworkers demonstrated that repeated treatment of soils with 1,3-dichloropropene resulted in accelerated microbial degradation of this compound. Fifteen bacterial strains with 1,3-dichloropropene-degrading capacity were isolated from such adapted soils. One strain was characterized and identified as Pseudomonas cichorii 170, and this strain was thought to possess a dehalogenase gene homologous to the dhlA gene encoding haloalkane dehalogenase of Xanthobacter autotrophicus GJ10 (9).
Here we describe complete microbial degradation of 1,3-dichloropropene by P. cichorii 170 and propose a degradation pathway. The haloalkane dehalogenase gene involved in the conversion of 1,3-dichloropropene to 3-chloroallyl alcohol was cloned and sequenced, and this gene turned out to be identical to the dhaA gene encoding haloalkane dehalogenase of Rhodococcus rhodochrous NCIMB13064 (11), which exhibits some sequence similarity to the dehalogenase gene of X. autotrophicus GJ10. The presence and role of the dhaA gene in P. cichorii 170 were confirmed by isolation and characterization of dehalogenase-negative mutants.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The 1,3-dichloropropene-utilizing bacterium P. cichorii 170 was described previously by Verhagen et al. (24). Mutants of strain 170 resistant to the toxic compound 1,2-dibromoethane were isolated on MMY plates (15) containing 5 mM cis-3-chloroacrylic acid as a carbon source and 20 μl of 1,2-dibromoethane in the lid of each petri dish. Spontaneous mutants were observed after incubation for 2 weeks at 30°C. Four independently isolated mutants were purified and stored on Luria-Bertani (LB) medium (15). A representative mutant, strain 170M4, was chosen for further study.
Escherichia coli BL21(DE3), a strain expressing the RNA polymerase of bacteriophage T7 (19), and the vector pGEF+ (12) were used for the cloning experiments and for expression of the recombinant enzyme. E. coli JM101 (Promega) was used for isolation of single-stranded DNA from pGEF+ derivatives.
Media and growth conditions.
Cells of strains 170 and 170M4 were grown aerobically at 30°C in MMY (15) or LB medium (15). When required, Difco agar (15 g/liter) was added to the medium. To prevent evaporation of volatile substrates, cultivation was carried out in closed flasks filled to one-fifth of their volume with medium.
E. coli strains were grown at 30°C in LB medium with rotary shaking or on solid LB medium (15). Ampicillin (100 μg/ml) was used for detection of recombinant plasmids. LBi medium, which was used for pH indicator plates, was solid LB medium supplemented with 80 mg of bromothymol blue (Merck) per liter and adjusted to pH 8.0.
Gas chromatography.
Amounts of 1,3-dichloropropene and 3-chloroallyl alcohol were determined by capillary gas chromatography. Samples (1 ml) were extracted with 1 ml of diethyl ether containing 0.05 mM 1-bromohexane as an internal standard. Extracts were analyzed by split injection of 2- or 4-μl samples into a type HP-5 column (model HP 19091J-413; Hewlett-Packard) by using nitrogen as the carrier gas. The column was installed in a model 6890 gas chromatograph (Hewlett-Packard) equipped with a flame ionization detector. The oven was temperature programmed as follows: 3 min (isothermal) at 40°C, followed by an increase at a rate of 10°C/min to 90°C and then an increase at a rate of 30°C/min to 140°C for 1,3-dichloropropene; and 3 min (isothermal) at 60°C, followed by an increase at a rate of 10°C/min to 180°C for 3-chloroallyl alcohol. The different isomers of 1,3-dichloropropene and 3-chloroallyl alcohol were clearly separated from each other. Typical elution times for cis-1,3-dichloropropene, trans-1,3-dichloropropene, cis-3-chloroallyl alcohol, and trans-3-chloroallyl alcohol were 5.4, 4.9, 9.3, and 10.3 min, respectively.
Preparation of crude extracts.
Cells of strains 170 and 170M4 were harvested in the exponential growth phase by centrifugation, washed with 1 volume of TEMAG buffer (10 mM Tris-sulfate [pH 8.2], 1 mM EDTA, 1 mM β-mercaptoethanol, 0.02% sodium azide, 10% glycerol), and disrupted in an appropriate amount of this buffer by sonication. A crude extract was obtained by centrifugation (30 min at 50,000 rpm in a type 70 Ti rotor [Beckman]).
The recombinant dehalogenase was expressed in E. coli BL21(DE3) and a crude extract was prepared as described previously (17).
Enzyme purification.
For isolation and purification of the haloalkane dehalogenase of strain 170, cells were grown in LB medium or MMY–1% citrate. The cells were cultivated at 30°C until the early stationary growth phase. The cells were harvested by centrifugation, washed with 1 volume of TEMAG buffer, and disrupted in an appropriate amount of this buffer by sonication. Unbroken cells and debris were removed by centrifugation for 1 h at 50,000 rpm in a type 70 Ti rotor (Beckman). The crude extract was applied to a DEAE-cellulose column which was equilibrated with TEMAG buffer. The column was washed with 1 column volume of TEMAG buffer, and the proteins were eluted with a linear gradient of 0 to 1 M ammonium sulfate in TEMAG buffer. Fractions that showed dehalogenase activity with 1,2-dibromoethane were pooled and dialyzed overnight against PEMAG buffer (5 mM potassium phosphate [pH 6.5], 1 mM EDTA, 1 mM β-mercaptoethanol, 0.02% sodium azide, 10% glycerol). The dialysate was loaded onto a hydroxylapatite column which was equilibrated with PEMAG buffer. The column was washed with 1 column volume of PEMAG buffer, and the enzyme was eluted with a linear gradient of 0 to 100 mM potassium phosphate in PEMAG buffer. Fractions with the highest haloalkane dehalogenase activity were pooled and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Enzyme assays.
Haloalkane and 3-chloroacrylic acid dehalogenase activities were measured by incubating an appropriate amount of enzyme or cell extract with 3 ml of 5 mM substrate in 50 mM Tris-sulfate buffer (pH 8.2) at 30°C. Halide liberation was monitored colorimetrically as described previously (2, 10). All dehalogenase activities are expressed as units per milligram; 1 U was defined as 1 μmol of halide produced per min per mg of protein. Most enzyme assays were carried out twice, and the differences in specific activity were less than 10%.
Protein concentrations were estimated with Coomassie brilliant blue by using bovine serum albumin as the standard.
Biochemical characterization.
The molecular masses of denatured dehalogenases were determined by SDS-PAGE on gels containing 12.5% polyacrylamide. Phosphorylase b (molecular mass, 94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), soybean trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa), all obtained from Pharmacia, were used as reference proteins. The gels were stained with Coomassie brilliant blue.
To determine the amino-terminal amino acid sequence of the purified haloalkane dehalogenase (DhaA), 0.5 μg of protein was electrophoresed on a 12.5% polyacrylamide SDS-PAGE gel, transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore) by electroblotting, and stained with Coomassie brilliant blue. After the gel was washed with distilled water, the DhaA band was excised and immediately subjected to automated Edman degradation (Eurosequence BV, Groningen, The Netherlands).
DNA isolation.
To isolate total DNA, cells of strains 170 and 170M4 were grown in 50 ml of LB medium at 30°C until the early stationary growth phase. Ampicillin (200 μg/ml) and lysozyme (100 μg/ml) were added, and the culture was incubated at 30°C for another 1 h. Cells were harvested by centrifugation and resuspended in 10 ml of 10 mM Tris-buffer (pH 8.5) containing 1 mM EDTA and 50 mM NaCl. Then 10 mg of lysozyme was added immediately, and the mixture was incubated at 37°C for 2 h. After 1.6 ml of 10% SDS was added, the mixture was incubated at 65°C until lysis was complete. Finally, 1.2 ml of 3 M sodium acetate (pH 7.0) was added, and the mixture was incubated at 65°C for another 2 h. The preparation was extracted twice with an equal volume of phenol, then with phenol-chloroform (1/1, vol/vol), and finally with chloroform-isoamyl alcohol (24/1, vol/vol). Total DNA was precipitated by adding 2 volumes of cold ethanol (96%) and was collected with a glass rod. After washing with 70% ethanol, the DNA was resuspended in 1 ml of 10 mM Tris buffer (pH 7.4) containing 1 mM EDTA.
Cloning of the dhaA gene.
The general procedures used for cloning and DNA manipulation were essentially the procedures described previously (15). To clone dhaA, total DNA from strain 170 was directly used for PCR amplification by the standard protocol described by Innis and Gelfand (7). Total DNA and synthetic oligonucleotide primers were each used at a concentration of 100 ng per 100 μl of total PCR mixture. The reaction was performed with GoldStar DNA polymerase by using denaturation, annealing, and extension temperatures of 94, 58, and 72°C, respectively. The DNA oligonucleotides used as primers were designed on the basis of the N- and C-terminal DNA sequences of the dhaA gene of R. rhodochrous NCIMB13064 (GSDB accession no. L49435) and had the following nucleotide sequences: 5′-AAAATCGCCATGGCAGAAATCGGTA-3′ (the start codon is in boldface type, and the NcoI site is underlined) and 5′-TGGACATCGGACCATGGCGTGAACC-3′ (the C of the stop codon is in boldface type, and the NcoI site is underlined). After 30 cycles of amplification, formation of a DNA product was checked by agarose gel electrophoresis (15). The PCR product was purified with a QIAquick PCR purification kit (Qiagen), digested with NcoI, and ligated into the NcoI site of the T7 expression vector pGEF+. The ligation mixture was used to transform electrocompetent cells of E. coli BL21(DE3) by electroporation. Transformants were plated onto LBi medium plates containing 100 μg of ampicillin per ml. Resistant colonies were screened for the presence of dehalogenase activity as described previously (17). Plasmid DNA was isolated from a colony showing dehalogenase activity and was checked by restriction analysis. The recombinant plasmid pGEF(dhaA) was transformed into E. coli JM101 for isolation of single-stranded DNA (15), which was used as the template for DNA sequencing of the inserted gene by the dideoxy chain termination method of Sanger et al. (16).
Chemicals and enzymes.
Restriction enzymes, T4 DNA ligase, and molecular weight marker X were obtained from Boehringer (Mannheim, Germany). GoldStar DNA polymerase was purchased from Eurogentec (Seraing, Belgium). DEAE-cellulose was obtained from Whatman Ltd., Kent, England; and hydroxylapatite was obtained from Bio-Rad Laboratories, Richmond, Calif. All halogenated compounds, including the separate isomers of 1,3-dichloropropene, 3-chloroallyl alcohol, and 3-chloroacrylic acid, were supplied by Janssen Chimica (Beerse, Belgium) and were at least 97% pure according to the manufacturer. The DNA oligonucleotides used as primers were synthesized by Eurosequence BV.
RESULTS
Characterization of P. cichorii 170.
The 1,3-dichloropropene-degrading organism was identified by using the BIOLOG identification system as P. cichorii 170 (24). Strain 170 was able to utilize the following organic compounds as growth substrates: citrate, glucose, ethanol, 1-propanol, 1-butanol, 1-pentanol, and crotonic acid. No growth occurred with methanol, allyl alcohol, acrylic acid, ethylene glycol, or toluene.
Utilization of halogenated compounds.
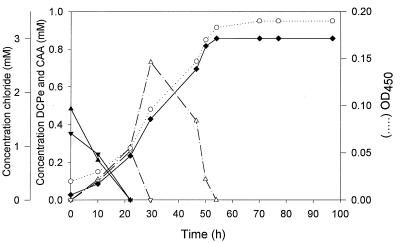
Growth inhibition experiments performed under standard conditions showed that when cis- or trans-1,3-dichloropropene was added at a concentration of more than 0.75 mmol/liter, it was very toxic for strain 170 and completely inhibited growth on citrate. This inhibition was caused by the toxic effects of 1,3-dichloropropene itself, because the corresponding 3-chloroallyl alcohols were not toxic and could even serve as growth substrates at concentrations up to 5 mM. However, strain 170 could efficiently utilize cis- and trans-1,3-dichloropropene when each of them was added at an amount of 0.075 mmol to 3-liter flasks with a high air/medium ratio (Fig. 1). Growth resulted in disappearance of the substrate and simultaneous formation of biomass and inorganic chloride. The final chloride concentration exceeded the low initial liquid phase concentration of dichloropropenes by more than twofold since the chloroalkenes were distributed in the gas and liquid phases. Both cis- and trans-3-chloroallyl alcohol transiently accumulated in the medium, indicating that they are the intermediates formed during conversion of cis- and trans-1,3-dichloropropenes, respectively.
FIG. 1.
Growth of strain 170 on a mixture of cis- and trans-1,3-dichloropropene. A 0.15-mmol portion of 1,3-dichloropropene (ratio of cis-1,3-dichloropropene to trans-1,3-dichloropropene, 1/1 [mol/mol]) was added to 100 ml of MMY in a 3-liter flask, which resulted in a low initial liquid phase concentration. Symbols: ▴, cis-1,3-dichloropropene concentration; ▾, trans-1,3-dichloropropene concentration; ▵, cis-3-chloroallyl alcohol concentration; ▿, trans-3-chloroallyl alcohol concentration; ○, optical density at 450 nm (OD450); ⧫, chloride concentration. Abbreviations: CAA, 3-chloroallyl alcohol; DCPe, 1,3-dichloropropene.
We determined whether strain 170 could utilize halogenated compounds that are structurally related to 1,3-dichloropropene and its possible degradation products. Growth tests were done under standard conditions in liquid medium supplemented with different carbon sources at a concentration of 2 mM (Table 1). P. cichorii 170 was capable of growth with 1,3-dichloropropane and with 1-halo-n-alkanes containing up to at least 10 carbon atoms. The environmentally important compounds 1,2-dichloroethane, 1,2-dibromoethane, 1,2-dichloropropane, and 1,2,3-trichloropropane were not growth substrates for strain 170. The lack of growth with these compounds was probably not due to toxicity, as observed with 1,3-dichloropropene, since these compounds lack the reactive allylic halogen, which can lead to alkylation of nucleophilic groups. Strain 170 was also able to grow on cis- or trans-3-chloroallyl alcohol and cis- or trans-3-chloroacrylic acid, which are possible degradation products of the corresponding 1,3-dichloropropenes. No growth was observed with other chloroallyl alcohols or halogenated acids.
TABLE 1.
Utilization of halogenated compounds by P. cichorii 170
| Compound | Utilization | Compound | Utilization | |
|---|---|---|---|---|
| Halogenated alkanes | Halogenated alcohols | |||
| 1,2-Dichloroethane | −a | 2-Chloroallyl alcohol | − | |
| 1,2-Dibromoethane | − | cis-3-Chloroallyl alcohol | + | |
| 1-Chloropropane | + | trans-3-Chloroallyl alcohol | + | |
| 1-Bromopropane | + | 3,3-Dichloroallyl alcohol | − | |
| 2-Chloropropane | − | 2,3,3-Trichloroallyl alcohol | − | |
| 1,2-Dichloropropane | − | |||
| 1,2-Dibromopropane | − | Halogenated acids | ||
| 1,3-Dichloropropane | + | Bromoacetic acid | − | |
| 1,3-Dibromopropane | − | Chloroacetic acid | − | |
| 1-Bromo-3-chloropropane | − | 2-Chloropropionic acid | − | |
| 1,2,3-Trichloropropane | − | 3-Chloropropionic acid | − | |
| 1,2,3-Tribromopropane | − | 3-Chlorocrotonic acid | − | |
| 1-Chlorobutane | + | cis-3-Chloroacrylic acid | + | |
| 1-Bromobutane | + | trans-3-Chloroacrylic acid | + | |
| 1-Chloropentane | + | |||
| 1-Bromopentane | + | |||
| 1-Chlorohexane | + | |||
| 1-Bromohexane | + | |||
| 1-Chlorooctane | − | |||
| 1-Chlorononane | − | |||
| 1-Bromononane | + | |||
| 1-Bromodecane | + |
−, no growth; +, visible growth after 1 week of cultivation in liquid MMY at room temperature. In all cases, growth was accompanied by halide release. Each carbon source was added at a concentration of 2 mM.
Metabolism of 1,3-dichloropropene.
Activities of enzymes that may be involved in 1,3-dichloropropene metabolism were tested with crude extracts prepared from cells grown on citrate, 1-chlorobutane, cis-3-chloroacrylic acid, and trans-3-chloroacrylic acid (Table 2). It appeared that both isomers of 1,3-dichloropropene were converted to the corresponding 3-chloroallyl alcohols, which indicates that dehalogenation of 1,3-dichloropropene is a hydrolytic reaction in this organism. The constitutively expressed haloalkane dehalogenase had a broad substrate range (Table 3) and did not require any cofactors or metal ions for activity. The highest level of dehalogenase activity was observed with 1,2-dibromoethane, while no activity was observed with the analog 1,2-dichloroethane.
TABLE 2.
Dehalogenase activities in crude extracts prepared from cells grown on different carbon sourcesa
| Enzyme substrate | Dehalogenase sp act (mU/mg of protein) with the following carbon sources:
|
|||
|---|---|---|---|---|
| Citrate | 1-Chloro- butane | cis-3-Chloro- acrylic acid | trans-3-Chloro- acrylic acid | |
| cis-1,3-Dichloropropene | 440 | 280 | 280 | 256 |
| trans-1,3-Dichloropropene | 238 | 203 | 167 | 147 |
| cis-3-Chloroallyl alcohol | <10 | <10 | <10 | <10 |
| trans-3-Chloroallyl alcohol | <10 | <10 | <10 | <10 |
| cis-3-Chloroacrylic acid | <10 | <10 | 161 | <10 |
| trans-3-Chloroacrylic acid | 230 | 63 | 134 | 177 |
Specific activities with various substrates (5 mM) were determined with extracts prepared from cells grown on different carbon sources.
TABLE 3.
Dehalogenating activities of a crude extract prepared from citrate-grown cells with various substrates
| Substrate | Sp act (mU/mg)a |
|---|---|
| 1,2-Dichloroethane | <10 |
| 1,2-Dibromoethane | 696 |
| 1-Chloropropane | 48 |
| 1-Bromopropane | 57 |
| 1,2-Dichloropropane | <10 |
| 1,2-Dibromopropane | 189 |
| 1,3-Dichloropropane | 119 |
| 1,3-Dibromopropane | 154 |
| 1-Bromo-3-chloropropane | 233 |
| 1,2,3-Trichloropropane | 10 |
| 1-Chlorobutane | 79 |
| 1-Bromobutane | 44 |
| 1-Chlorohexaneb | 88 |
| 1-Chlorononaneb | 28 |
| 1-Bromononaneb | 42 |
| 1-Bromodecaneb | 38 |
Halide production at 30°C with different substrates (5 mM) was determined at pH 8.2.
Assays were performed with substrate at a concentration of 3 mM.
Conversion of cis- and trans-3-chloroallyl alcohol is likely to proceed via 3-chloroacrolein to the corresponding 3-chloroacrylic acid isomers (3, 13). The extracts of cells grown on different substrates did not show chloride release upon incubation with cis- or trans-3-chloroallyl alcohol (Table 2), indicating that direct dechlorination of the 3-chloroallyl alcohol isomers indeed does not occur.
The initial step in 3-chloroacrylic acid metabolism in strain 170 was studied by incubating crude extracts separately with the 3-chloroacrylic acid isomers. The extracts of cells grown on different substrates showed chloride formation when trans-3-chloroacrylic acid was added. Dechlorination of cis-3-chloroacrylic acid was observed only with crude extracts prepared from cells grown on cis-3-chloroacrylic acid. These results indicate that two different enzymes are involved in the dechlorination of the 3-chloroacrylic acid isomers, a trans-specific dehalogenase that is constitutively expressed and an inducible cis-specific dehalogenase.
Purification of the haloalkane dehalogenase.
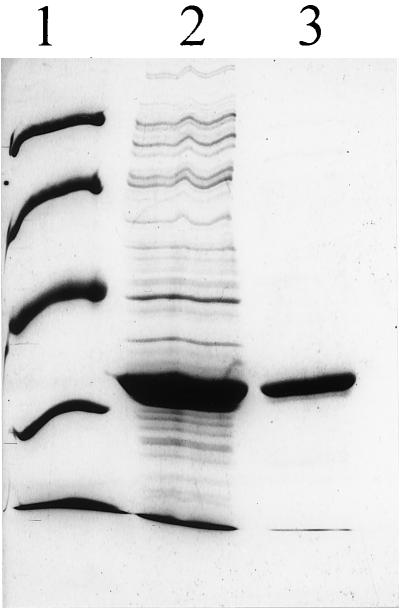
The haloalkane dehalogenase of strain 170 was purified 16-fold, indicating that this dehalogenase was present at a concentration equivalent to 6 to 7% of the total soluble cellular protein. After SDS-PAGE, only one protein band at approximately 33 kDa was observed (Fig. 2). The purified enzyme could be stored in TEMAG buffer at 4°C with no loss of activity. During purification, similar increases in specific activity were observed for cis- and trans-1,3-dichloropropene dehalogenase activities, indicating that both 1,3-dichloropropene isomers were converted by the same enzyme (Table 4). The purified haloalkane dehalogenase did not catalyze conversion of cis- or trans-3-chloroacrylic acid, indicating that other dehalogenases with activities for these compounds must be present in strain 170.
FIG. 2.
SDS-PAGE of crude extract of E. coli BL21(DE3)/pGEF(dhaA) (lane 2) and purified haloalkane dehalogenase from P. cichorii (lane 3). Lane 1 contained protein markers with molecular masses of 94, 67, 43, 30, and 20 kDa.
TABLE 4.
Dehalogenating activities of crude extracts and purified dehalogenase
| Enzyme substrate | Dehalogenase sp act (mU/mg of protein)a
|
||
|---|---|---|---|
| Cell extract of strain 170 | Cell extract of BL21(DE3)/ pGEF(dhaA) | Purified de- halogenase | |
| cis-1,3-Dichloropropene | 440 | 4,204 | 7,150 |
| trans-1,3-Dichloropropene | 238 | 2,140 | 3,549 |
| 1-Chloropropane | 48 | 449 | 744 |
| 1-Chlorobutane | 79 | 711 | 1,232 |
| cis-3-Chloroacrylic acid | NDb | <10 | <10 |
| trans-3-Chloroacrylic acid | 230 | <10 | <10 |
Halide production at 30°C with different substrates (5 mM) was determined at pH 8.2.
ND, not determined, because the enzyme is not expressed under the growth conditions used.
The amino-terminal amino acid sequence of the purified protein of the gram-negative organism P. cichorii was determined to be M-S-E-I-G-T-G-F-P-F-D-P-H-Y-V-E-V, which is identical to the amino-terminal sequences of the dehalogenases of the gram-positive strains R. rhodochrous NCIMB13064 (11), Rhodococcus erythropolis Y2 (14), and Arthrobacter sp. strain HA1 (18).
Cloning of the haloalkane dehalogenase gene dhaA.
Earlier studies (24) suggested that a plasmid-located dhlA-like gene may be involved in 1,3-dichloropropene degradation. To determine whether a haloalkane dehalogenase gene that was homologous to the dhlA gene of X. autotrophicus GJ10 (9) was present in strain 170, Southern hybridizations were performed with the Xanthobacter gene as a probe. These hybridizations with total DNA of strain 170 were done under nonstringent conditions, but no positive signal was detected.
The biochemical characteristics and amino-terminal sequence of the haloalkane dehalogenase that we purified from P. cichorii 170 suggested that the enzyme closely resembled the haloalkane dehalogenases present in a number of gram-positive strains (5, 8, 14, 18, 25). The DNA sequence of the haloalkane dehalogenase gene of one of these gram-positive strains, R. rhodochrous NCIMB13064, has been published recently (11). To determine whether the haloalkane dehalogenase gene of strain 170 was identical to the dhaA gene of R. rhodochrous, the putative dhaA gene of strain 170 was amplified by PCR with primers based on the N- and C-terminal dehalogenase sequences of R. rhodochrous. Electrophoresis on an agarose gel revealed that when total DNA of strain 170 was the template, a 0.9-kb DNA product was formed by PCR. The PCR fragment was purified and cloned in the expression vector pGEF+, which resulted in production of an active dehalogenase in E. coli BL21(DE3). SDS-PAGE of the purified haloalkane dehalogenase of strain 170 and the dhaA gene product overexpressed in E. coli BL21(DE3) showed that the enzymes had the same electrophoretic mobility and the same molecular mass, 33 kDa (Fig. 2). The ratio of trans-1,3-dichloropropene dehalogenase activity to cis-1,3-dichloropropene dehalogenase activity was 0.5 for the purified dehalogenase and also for the overexpressed dhaA gene product in E. coli (Table 4). Thus, the cloned gene indeed encodes the purified dehalogenase of strain 170. DNA sequencing of the cloned PCR-amplified dehalogenase gene revealed a sequence identical to the sequence of the dhaA gene of R. rhodochrous. Thus, the haloalkane dehalogenase gene of the gram-negative organism P. cichorii 170 is identical to the haloalkane dehalogenase gene of the gram-positive organism R. rhodochrous NCIMB13064.
Mutants affected in haloalkane utilization.
Strain 170 could not utilize 1,2-dibromoethane, although its dehalogenase was able to catalyze conversion of 1,2-dibromoethane to 2-bromoethanol and hydrolysis of the latter to ethylene glycol. 1,2-Dibromoethane was not used for growth since ethylene glycol did not support growth and because the intermediate 2-bromoethanol was oxidatively converted to a toxic product, presumably 2-bromoacetaldehyde. Therefore, we could use 1,2-dibromoethane as a suicide substrate to select for resistant mutants that were expected to have lost their dehalogenase activity.
Strain 170M4 was isolated by selecting for 1,2-dibromoethane resistance of strain 170 on MMY plates containing 5 mM cis-3-chloroacrylic acid and 20 μl of 1,2-dibromoethane in the lid of each petri dish. The mutant was not able to utilize 1,3-dichloropropene, 1-chloropropane, 1-chlorobutane, or 1-chloropentane as a sole carbon source. Growth on 3-chloroallyl alcohol and 3-chloroacrylic acid was not affected compared with wild-type growth.
Crude extracts prepared from 170M4 cells did not exhibit dehalogenase activity when 1,3-dichloropropene, 1,2-dibromoethane, 1-chloropropane, or 1-chlorobutane was the substrate, but dehalogenase activity with cis- and trans-3-chloroacrylic acids was present. A crude extract of strain 170 grown under the same conditions clearly exhibited haloalkane dehalogenase activity. Thus, strain 170M4 is defective in haloalkane dehalogenase activity and therefore is not able to utilize haloalkanes for growth.
To determine whether the structural dehalogenase gene in mutant strain 170M4 was deleted, a PCR was performed with the same primers that were used to amplify the dhaA gene from strain 170. Electrophoresis on an agarose gel revealed that with total DNA of strain 170M4 no DNA product was formed by PCR. The absence of the dhaA gene in strain 170M4 was confirmed by a Southern blot analysis performed with a probe based on the cloned dhaA gene of strain 170.
DISCUSSION
The fate of halogenated nematocidic soil fumigants, such as 1,3-dichloropropene, is largely dependent on the ability of microorganisms to initiate degradation through dehalogenation reactions. Although biodegradation of 1,3-dichloropropene by soil bacteria has been observed (3, 4, 21), little is known about the intermediates in the process and the dehalogenating enzymes involved in biodegradation. In this work, we describe the route of 1,3-dichloropropene metabolism in P. cichorii 170, an organism isolated by Verhagen and coworkers from soil that exhibited accelerated biodegradation of 1,3-dichloropropene (24).
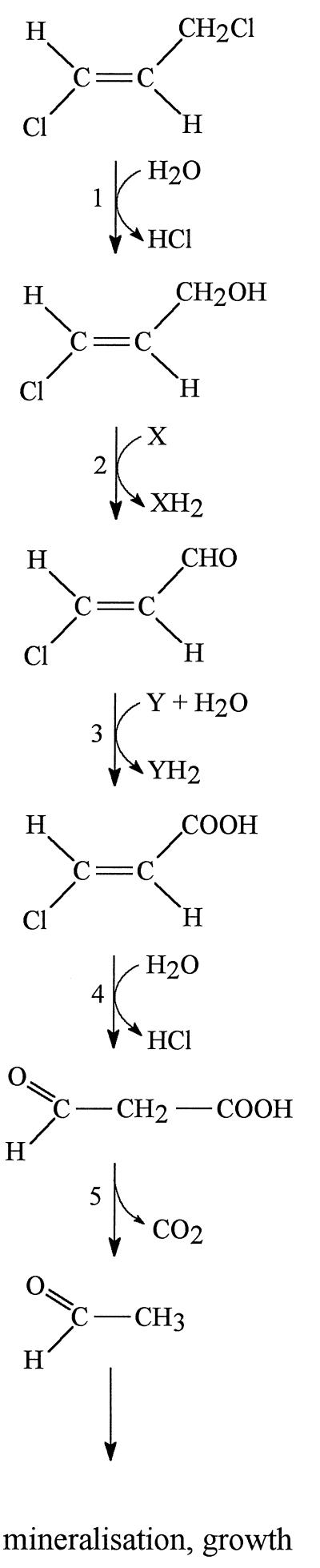
The first step in 1,3-dichloropropene metabolism in strain 170 was catalyzed by a hydrolytic haloalkane dehalogenase with broad substrate specificity (Fig. 3). This enzyme is different from the cis- and trans-3-chloroacrylic acid dehalogenases, as shown by substrate assays performed with purified haloalkane dehalogenase. Its involvement in the metabolism of several halogenated compounds was evident from the absence of the enzyme in a mutant that was impaired in utilization of haloalkanes and haloalkenes. PCR amplification of the haloalkane dehalogenase gene from strain 170 by using primers based on the expected similarity to the sequence of the dhaA gene of R. rhodochrous NCIMB13064 (5, 11), followed by DNA sequencing, indeed revealed a sequence identical to that of the dhaA gene of R. rhodochrous NCIMB13064. Only the 13-bp sequence corresponding to the N-terminal part of the dehalogenase was not sequenced since it was encoded by the forward primer. The N-terminal amino acid sequences were identical, however.
FIG. 3.
Proposed pathway for the degradation of trans-1,3-dichloropropene in P. cichorii 170. 1, haloalkane dehalogenase (DhaA); 2, alcohol dehydrogenase; 3, aldehyde dehydrogenase; 4, 3-chloroacrylic acid dehalogenase; 5, decarboxylase. A similar pathway is envisaged for the cis isomer.
Further conversion of the chloroallyl alcohols produced may proceed via oxidation to chloroacrylic acids. This part of the degradation route is known to occur in other organisms. Oxidation of cis- and trans-3-chloroallyl alcohols by cell suspensions of a Pseudomonas strain isolated from soil also led to production of the corresponding chloroacrylic acids (1). Van der Waarde and coworkers (20) showed that in crude extracts of 2-chloroallyl alcohol-grown Pseudomonas cells, 2-chloroallyl alcohol and 3-chloroallyl alcohol are rapidly oxidized to their corresponding chloroacrylic acids without dechlorination taking place.
The cofactor-independent dechlorination of cis- and trans-3-chloroacrylic acid in strain 170 may be catalyzed by enzymes similar to the enzymes present in the gram-positive coryneform bacterial strains CAA2 (6) and FG41 (22). These enzymes are also completely isomer selective and produce malonate semialdehyde as a product of cofactor-independent dehalogenation of 3-chloroacrylic acid. The degradative pathway of the malonate semialdehyde intermediate was studied in detail by Hartmans et al. (6). The results of these workers indicated that a cofactor-independent malonate semialdehyde decarboxylase was involved in the production of acetaldehyde and CO2.
The massive amounts of 1,3-dichloropropene that have been applied worldwide have placed severe selective stress on bacterial populations. This probably led to fast adaptation of microorganisms to this new substrate and may have played an important role in the distribution of dehalogenase genes among different soil bacteria. Our results strongly suggest that horizontal gene transfer between gram-positive and gram-negative organisms occurs under natural conditions and may play a role in the evolution of strains adapted to degrade dichloropropenes. The molecular mechanism underlying the spread of this gene between gram-positive and gram-negative organisms is under investigation. The 1,3-dichloropropene-degrading organisms that have been isolated may have evolved from organisms capable of utilizing allylalcohol or other alcohols. Rapid biodegradation of 3-chloroallyl alcohol has been observed previously, and this compound is a good growth substrate for many bacteria (1, 20). Possibly, the haloalkane dehalogenase gene was transferred to a 3-chloroallyl alcohol-degrading organism, which allowed it to grow directly on 1,3-dichloropropene or made it more resistant to this toxic compound.
ACKNOWLEDGMENTS
This study was supported by the Life Sciences Foundation (SLW), which is subsidized by the Netherlands Organization for Scientific Research (NWO), and by EC Environment and Climate Research Program contract ENV4-CT95-0086.
REFERENCES
- 1.Belser N O, Castro C E. Biodehalogenation—the metabolism of the nematocides cis- and trans-3-chloroallyl alcohol by a bacterium isolated from soil. J Agric Food Chem. 1971;19:23–26. doi: 10.1021/jf60173a047. [DOI] [PubMed] [Google Scholar]
- 2.Bergman J G, Sanik J. Determination of trace amounts of chlorine in naphtha. Anal Chem. 1957;29:241–243. [Google Scholar]
- 3.Castro C E, Belser N O. Hydrolysis of cis- and trans-1,3-dichloropropene in wet soil. J Agric Food Chem. 1966;14:69–70. [Google Scholar]
- 4.Cohen D B, Gilmore D, Fischer B S, Bowes G W. Water quality and pesticides: 1,2-dichloropropane (1,2-D) and 1,3-dichloropropene (1,3-D). Sacramento: California State Water Resources Control Board; 1983. [Google Scholar]
- 5.Curragh H, Flynn O, Larkin M J, Stafford T M, Hamilton J T G, Harper D B. Haloalkane degradation and assimilation by Rhodococcus rhodochrous NCIMB13064. Microbiology. 1994;140:1433–1442. doi: 10.1099/00221287-140-6-1433. [DOI] [PubMed] [Google Scholar]
- 6.Hartmans S, Jansen M W, van der Werf M J, De Bont J A M. Bacterial metabolism of 3-chloroacrylic acid. J Gen Microbiol. 1991;137:2025–2032. doi: 10.1099/00221287-137-8-2025. [DOI] [PubMed] [Google Scholar]
- 7.Innis M A, Gelfand D H. A guide to methods and applications. In: Innes M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press; 1990. pp. 3–11. [Google Scholar]
- 8.Janssen D B, Gerritse J, Brackman J, Kalk C, Jager D, Witholt B. Purification and characterization of a bacterial dehalogenase with activity toward halogenated alkanes, alcohols and ethers. Eur J Biochem. 1988;171:67–72. doi: 10.1111/j.1432-1033.1988.tb13759.x. [DOI] [PubMed] [Google Scholar]
- 9.Janssen D B, Pries F, van der Ploeg J, Kazemier B, Terpstra P, Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol. 1989;171:6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keuning S, Janssen D B, Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985;163:635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulakova A N, Larkin M J, Kulakov L A. The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB13064. Microbiology. 1997;143:109–115. doi: 10.1099/00221287-143-1-109. [DOI] [PubMed] [Google Scholar]
- 12.Rink R, Fennema M, Smids M, Dehmel U, Janssen D B. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 13.Roberts T R, Stoydin G. The degradation of (Z)- and (E)-1,3-dichloropropenes and 1,2-dichloropropane in soil. Pestic Sci. 1976;7:325–335. [Google Scholar]
- 14.Sallis P J, Armfield S J, Bull A T, Hardman D J. Isolation and characterization of a haloalkane halidohydrolase from Rhodococcus erythropolis Y2. J Gen Microbiol. 1990;136:115–120. doi: 10.1099/00221287-136-1-115. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 16.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schanstra J P, Rink R, Pries F, Janssen D B. Construction of an expression and site-directed mutagenesis system of haloalkane dehalogenase in Escherichia coli. Protein Exp Purif. 1993;4:479–489. doi: 10.1006/prep.1993.1063. [DOI] [PubMed] [Google Scholar]
- 18.Scholtz R, Leisinger T, Suter F, Cook A M. Characterization of 1-chlorohexane halidohydrolase, a dehalogenase of wide substrate range from an Arthrobacter sp. J Bacteriol. 1987;169:5016–5021. doi: 10.1128/jb.169.11.5016-5021.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 20.Van der Waarde J J, Kok R, Janssen D B. Degradation of 2-chloroallyl alcohol by a Pseudomonas sp. Appl Environ Microbiol. 1993;59:528–535. doi: 10.1128/aem.59.2.528-535.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Dijk H. Degradation of 1,3-dichloropropenes in the soil. Agro-Ecosystems. 1974;1:193–204. [Google Scholar]
- 22.Van Hylckama Vlieg J E T, Janssen D B. Bacterial degradation of 3-chloroacrylic acid and the characterization of cis- and trans-specific dehalogenases. Biodegradation. 1992;2:139–150. doi: 10.1007/BF00124488. [DOI] [PubMed] [Google Scholar]
- 23.Van Rijn J P, Van Straaten N M, Willems J. Handboek Bestrijdingsmiddelen: gebruik & milieu-effecten. Amsterdam, The Netherlands: VU Uitgeverij; 1995. pp. 629–632. [Google Scholar]
- 24.Verhagen C, Smit E, Janssen D B, van Elsas J D. Bacterial dichloropropene degradation in soil; screening of soils and involvement of plasmids carrying the dhlA gene. Soil Biol Biochem. 1995;27:1547–1557. [Google Scholar]
- 25.Yokota T, Omori T, Kodama T. Purification and properties of haloalkane dehalogenase from Corynebacterium sp. strain m15-3. J Bacteriol. 1987;169:4049–4054. doi: 10.1128/jb.169.9.4049-4054.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]