ABSTRACT
Formins are actin regulators critical for diverse processes across eukaryotes. With many formins in plants and animals, it has been challenging to determine formin function in vivo. We found that the phylogenetically distinct class I integral membrane formins (denoted For1) from the moss P. patens enrich at sites of membrane turnover, with For1D more tightly associated with the plasma membrane than For1A. To probe formin function, we generated formin-null lines with greatly reduced formin complexity. We found that For1A and For1D help to anchor actin near the cell apex, with For1A contributing to formation of cytosolic actin, while For1D contributes to plasma membrane-associated actin. At the cortex, For1A and For1D localized to motile puncta and differentially impacted actin dynamics. We found that class I cortical formin mobility depended on microtubules and only moderately on actin, whereas class II formin (denoted For2) mobility solely depended on actin. Moreover, cortical For2A tightly correlated with the puncta labeled by the endocytic membrane dye FM4-64, and null mutants in class I formins did not affect uptake of a similar dye, FM1-43, suggesting that class I and II formins are involved in distinct membrane trafficking pathways.
Keywords: Actin, Formin, Plant, Endocytosis, Exocytosis, Microtubule
Highlighted Article: Live-cell imaging of endogenously tagged formins coupled with examination of formin-null lines reveals that phylogenetically distinct formins differentially contribute to actin formation in plant cells.
INTRODUCTION
Formins are a diverse family of proteins found throughout eukaryotes that regulate the status of specific actin-based structures within cells (Courtemanche, 2018; Evangelista et al., 2003; Goode and Eck, 2007). Many formins have been characterized as actin-nucleating proteins (Goode and Eck, 2007), but others bundle (Harris et al., 2006) and sever actin filaments as well (Chhabra et al., 2009). Some formins also interact with the microtubule cytoskeleton (Bartolini and Gundersen, 2010; Gaillard et al., 2011), linking actin and microtubule-based functions (Daou et al., 2014; Henty-Ridilla et al., 2016). Together, these activities underlie critical cellular processes, such as establishment of cell polarity, division, adhesion and migration (Campellone and Welch, 2010; Evangelista et al., 1997; Faix and Grosse, 2006; Feierbach and Chang, 2001).
Formins have diverged and expanded throughout the eukaryotic kingdom. For example, humans have 15 formins that group into seven families (Higgs and Peterson, 2005), Arabidopsis has 21 formins in two families (Deeks et al., 2002), and budding yeast has two formins (Bni1 and Bnr1) in one family (Higgs and Peterson, 2005). In yeast, there is strong evidence that formins have distinct functions serving to maintain specific actin-based subcellular structures. For example, the budding yeast Bnr1 generates actin cables in the mother cell from the bud neck to maintain a stable mother–bud axis and to provide transport of vesicles from the mother to the bud, while the other formin, Bni1, generates actin cables to facilitate vesicle transport to varying locations in the bud, ensuring proper bud development (Pruyne et al., 2004). In fission yeast, Cdc12 is essential and generates actin filaments in the contractile ring during cell division (Chang et al., 1997), while For3 is involved in polarized growth and required for actin cable formation and microtubule organization (Feierbach and Chang, 2001). Fission yeast has a third formin, Fus1, which generates a specific actin structure that is critical for yeast cell mating (Dudin et al., 2015). However, in eukaryotes with many formin genes, such as animals and plants, it is not as clear how the various formins participate in generation of specific actin structures.
Plants have evolved a distinct set of formin proteins compared to animals and fungi (Grunt et al., 2008; Higgs and Peterson, 2005). Most seed plants have ∼20 genes that group into two subclasses: class I and II (Grunt et al., 2008). Plant formins are particularly intriguing due to the fact that class I formins are predicted to be integral membrane proteins and thus may be able to link extracellular signals directly to remodeling of the actin cytoskeleton. Because the formin gene family is relatively large in seed plants, it has been challenging to study the family as a whole. While individual formins have been studied in several seed plants including Arabidopsis and rice, single mutants often do not have very strong phenotypes (Diao et al., 2018; Lan et al., 2018; Li et al., 2014, 2018; Liu et al., 2018; Rosero et al., 2016; Sun et al., 2017; van Gisbergen and Bezanilla, 2013). Thus, it has been challenging to determine the role of individual formins and identify the specific subcellular actin structures these formins may be responsible for generating.
The moss, P. patens, has nine formins that group into three phylogenetically distinct classes. Similar to seed plants, P. patens has class I and class II formins (proteins denoted as For1 and For2, respectively), with six and two genes in each class, respectively. P. patens also has a single class III formin, which according to available expression data is only expressed in the archegonia, the female reproductive tissue of the gametophyte (Ortiz-Ramírez et al., 2016). Since class III formins have only been identified in basal land plant lineages, such as mosses, it has been hypothesized that class III formins were lost in seed plants (Grunt et al., 2008).
To study the function of formins in P. patens, Vidali et al. used RNAi to silence the formin gene family and demonstrated that the two class II formins genes are functionally redundant and essential for polarized growth, while class I formins contribute to growth (Vidali et al., 2009b). To pursue the function of class I formins, van Gisbergen et al. used a phylogenetic approach, and discovered that class I formins in bryophytes underwent lineage specific gene duplications (van Gisbergen et al., 2018). Class I formins in P. patens group into three subclasses, with For1D and For1E comprising the most ancestral clade. Moss-specific gene duplications resulted in two additional subclasses, which in P. patens led to the subclass containing For1A, For1B and For1C, and the subclass containing For1F. Interestingly, the gene duplication that gave rise to For1F appears to have stemmed from a gene fusion event resulting in a novel formin. In For1F, a component of the exocyst complex fused to a class I formin resulting in an essential protein that functions in tethering exocytic vesicles as well as a protein that interacts with actin filaments (van Gisbergen et al., 2018).
While mosses evolved this novel formin, they have still retained class I formins that are similar in predicted structure to class I formins in seed plants. Here, we have taken advantage of facile homologous recombination in P. patens to tag the genomic locus of For1A or For1D with sequences encoding a fluorescent protein. We chose these two genes because in juvenile moss tissue (protonemata), For1A and For1D are the highest expressed class I formins in the remaining two class I subclasses (Vidali et al., 2009b). By performing time-lapse live-cell imaging, we analyzed the localization and dynamics of For1A and For1D at the cell cortex, in cell division and during polarized cell expansion.
RESULTS
Formins localize to areas of active membrane turnover
For1A–For1C are predicted to have both a signal peptide and a transmembrane domain (Vidali et al., 2009b). In contrast, For1D and For1E, which belong to the most ancestral bryophyte class I formin subclass (van Gisbergen et al., 2018), have a transmembrane domain but no signal peptide (Vidali et al., 2009b). Using homologous recombination, we tagged For1A and For1D, the two most highly expressed genes in protonemata with three tandem copies of mEGFP (3×mEGFP, henceforth called GFP) at the genomic locus. Correct integration was verified by genotyping (Fig. S1) and full-length expression of the tagged protein was confirmed by immunoblotting (Fig. 1A). To ensure that tagging the locus did not affect plant growth, we measured overall plant area 7 days after regeneration from protoplasts. We found that For1A–GFP and For1D–GFP lines grow indistinguishably from control lines (Fig. S2).
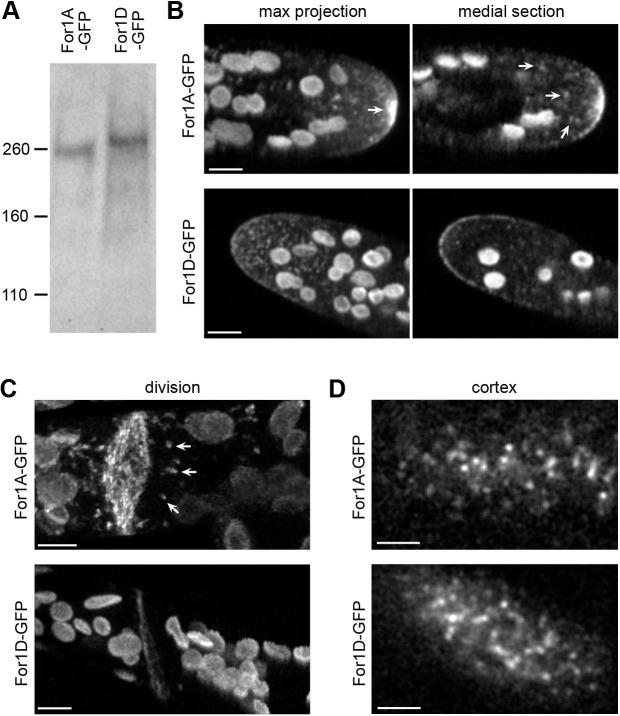
Fig. 1.
Subcellular localization of endogenously tagged For1A and For1D. (A) Immunoblot performed with an antibody to GFP on whole-cell extracts from the For1A or For1D lines where the endogenous locus was tagged in frame with sequences encoding three tandem GFP molecules (GFP). The molecular mass in kDa is indicated. Expected sizes for unmodified proteins are: For1A–GFP, 232 kDa; For1D–GFP, 204 kDa. (B,C) Spinning-disc confocal images of For1A–GFP and For1D–GFP in the apical cell (B) and in a dividing cell (C, For1D–GFP) of a protonemal moss filament. (C, For1A–GFP). Laser scanning confocal images (de-noised with NIS elements software) during cell division. Maximum projections from confocal z-stacks are shown in B and C, and the medial plane is also shown in B. Arrows indicate cytoplasmic For1A–GFP accumulations and puncta. Large globular structures are chloroplasts, which display autofluorescence under these imaging conditions. Scale bars: 5 µm. (D) VAEM images of the cell cortex in the For1A–GFP and For1D–GFP lines. Scale bars: 2 µm. See also Movie 1.
Using confocal microscopy, we observed that For1A–GFP and For1D–GFP are both enriched at the tip of the apical cell (Fig. 1B) and the site of cell division (Fig. 1C). Variable angle epifluorescence microscopy (VAEM) revealed that For1A–GFP and For1D–GFP also localize to dynamic dots at the cell cortex (Fig. 1D, see Movie 1). Interestingly, areas enriched for both For1A–GFP and For1D–GFP are sites of extensive membrane turnover. While both For1A–GFP and For1D–GFP are enriched at the cell apex, their localization patterns differ. For1A–GFP is at the apical plasma membrane and in the apical cytoplasm, forming a focal point behind the tip. Additionally, For1A is found in puncta throughout the cytoplasm. In contrast, For1D–GFP exhibits very few cytoplasmic puncta. The majority of the puncta observed on the maximum projection image of For1D–GFP (Fig. 1B) reside on the cell cortex. Furthermore, imaging the medial section reveals that For1D is more highly enriched along the apical plasma membrane as compared to in the apical cytoplasm.
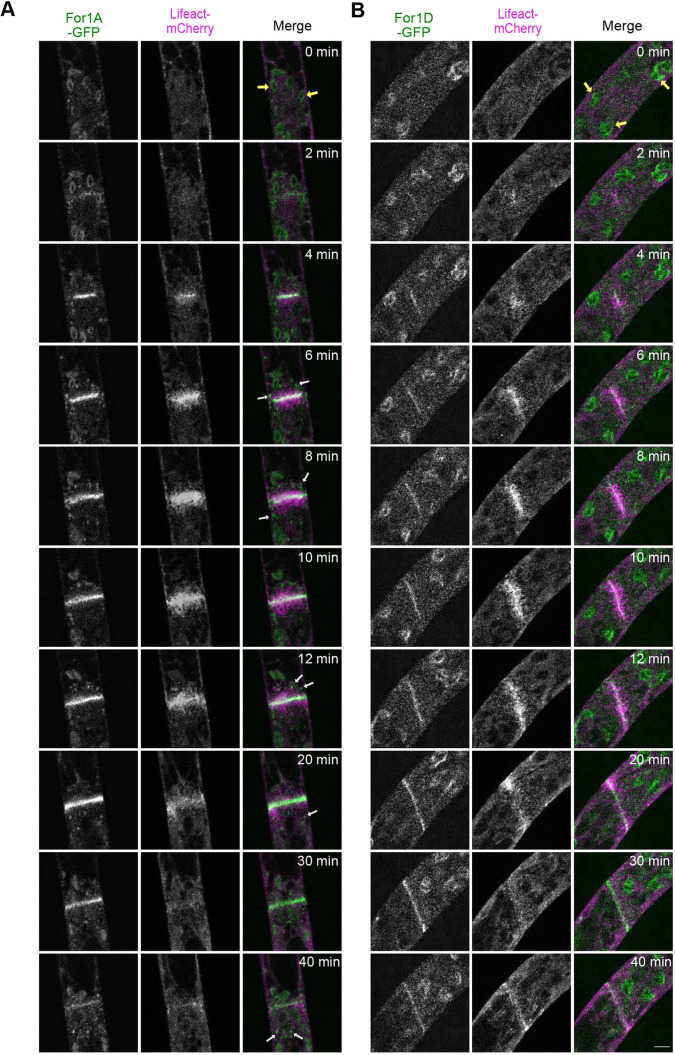
To investigate potential differences between the behavior of For1A–GFP and For1D–GFP during cell division, we monitored For1A–GFP and For1D–GFP in dividing cells (Fig. 2; Movie 2). We found that both For1A–GFP and For1D–GFP localize to the midzone of the phragmoplast before any actin is visible (Fig. 2, 2 min). Both formins remain associated with the midzone region throughout cytokinesis. Later in cytokinesis when actin filaments are found mostly on the leading edge of the forming cell plate (Fig. 2, 20 min), both formins still localize along the entire nascent cell plate. Similar to what we observed near the cell apex (Fig. 1B), For1A–GFP but not For1D–GFP, forms cytoplasmic puncta near the phragmoplast (Fig. 2A, white arrows). Other than the cytoplasmic puncta and a brighter overall signal, For1A–GFP behavior is not remarkably different from For1D–GFP during cell division.
Fig. 2.
Subcellular localization of endogenously tagged For1A and For1D during cell division. Laser-scanning confocal microscope images of For1A–GFP (A) and For1D–GFP (B) with Lifeact–mCherry in dividing cells. Images are a single focal plane acquired every 20 s and deconvolved with NIS elements software (type Richardson–Lucy). White arrows indicate cytoplasmic For1A–GFP accumulations and puncta. Large globular structures are chloroplasts (yellow arrows), which display autofluorescence under these imaging conditions. Scale bar: 5 µm. See also Movie 2.
With distinct localization patterns at the cell apex, we reasoned that For1A–GFP and For1D–GFP might differentially contribute to membrane trafficking or actin dynamics. To probe this, we sought to generate lines that lacked For1A and For1D activity. However, previous RNAi studies demonstrated that class I formins were functionally redundant with respect to growth, and class II formins were redundant with respect to polarized growth (Vidali et al., 2009b). Thus, to remove any possible functional redundancy, we used CRISPR-Cas9 genome editing to generate null alleles in genes encoding the four proteins For1B, For1C, For1E and For2B (Δfor1BCE2B) in a line that also expresses Lifeact–mRuby2 (hereafter referred to as Lifeact–mRuby) (Mallett et al., 2019). We then used CRISPR-Cas9 coupled with homology-directed repair to introduce stop codons downstream of the protospacer target site in For1A and/or For1D. Analyzing the size of plants regenerated from protoplasts, we found that while Δfor1BCE2B is ∼20% smaller than the parental Lifeact–mRuby line, additional deletion of For1A and/or For1D did not further reduce growth (Fig. S3, Table S2), suggesting that For1A and For1D do not significantly contribute to plant size. However, the generated knockout lines provided an opportunity to probe For1A and For1D function with respect to membrane trafficking and actin dynamics in the absence of other highly related class I formins.
Class I formins, unlike class II formins, do not localize to endocytic hotspots
Both class I and class II formins localize to areas of active membrane turnover (Fig. 1, van Gisbergen et al., 2012, 2018; Vidali et al., 2009b), where both exocytosis and endocytosis occur. To investigate whether formin localization correlates with endocytic activity, we stained live cells with FM4-64, a fluorescent lipophilic membrane dye that marks endocytic membranes (van Gisbergen et al., 2008). FM4-64 is enriched at the cell apex, where it is internalized via endocytosis. Both For1A–GFP and For1D–GFP partially overlap with FM4-64 at the plasma membrane (Fig. 3A; Movie 3). To some extent, For1A–GFP also overlaps with internalized FM4-64 in the cytoplasm (Fig. 3A). However, confocal imaging does not readily resolve FM4-64 membrane populations versus cytoplasmic populations.
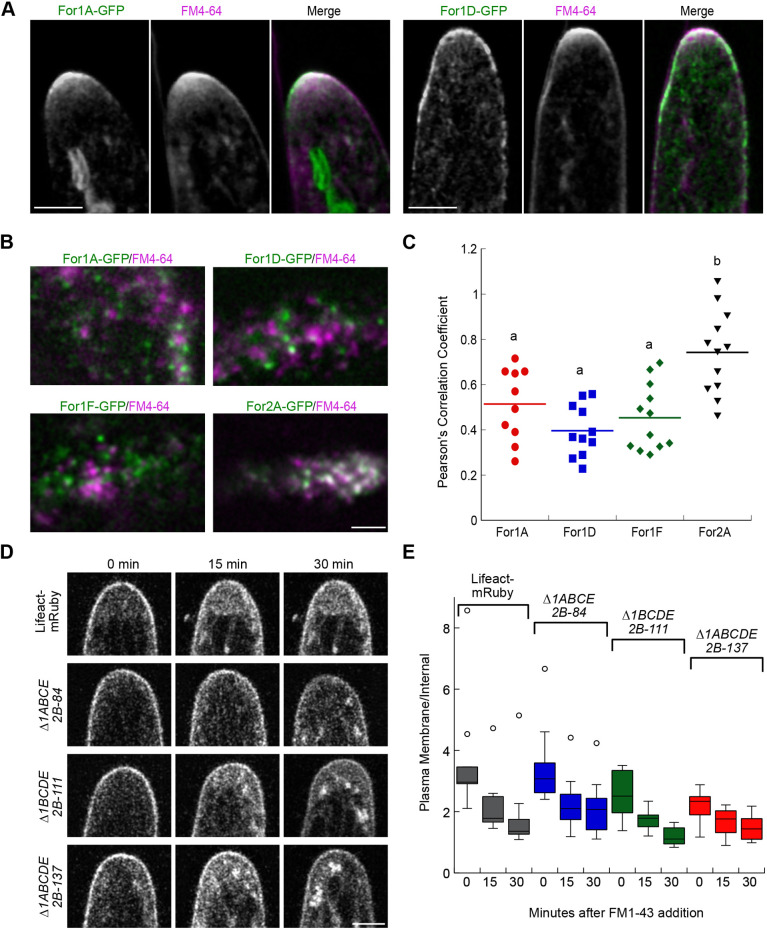
Fig. 3.
Analysis of formin localization with respect to endocytic activity labeled with FM4-64. (A) Laser-scanning confocal images of the medial plane of an apical cell growing in a microfluidic imaging chamber. Images were de-noised with NIS elements software. Large structures in the For1A–GFP image are chloroplasts, which display autofluorescence under these imaging conditions. Scale bars: 5 µm. See also Movie 3. (B) Simultaneous VAEM imaging of endogenously tagged formins as indicated at the cell cortex. See also Movie 4. Scale bar: 2 µm. For all images, the formin is green and FM4-64 is magenta in the merged images. (C) Pearson's correlation coefficient comparing the formin and FM4-64 channels acquired with VAEM. Letters above the bars indicate statistical groups with α<0.05 from one-way ANOVA with a Tukey HSD post hoc test (For1A, n=10 cells; For1D, n=11 cells; For1F, n=12 cells; For2A, n=12 cells). (D) Time course of FM1-43 uptake in control (Lifeact–mRuby) and formin-null lines as indicated. Single focal plane laser scanning confocal images of cells are shown immediately after putting cells in FM1-43 (0 min) and at 15 min intervals. Scale bar: 5 µm. (E) Box plot depicting the quantification of the ratio of the intensity of the plasma membrane to the intensity within the cell within 5–7 µm from the cell tip. The box encloses 50% of the data with the median value drawn as a line. Lines extending from the box mark minimum and maximum values for the data set except for data sets with outliers. Outliers falling outside of the upper quartile are value that are 1.5×interquartile distance, and are depicted as open circles. No significant differences were found at each time point using an one-way ANOVA with a Tukey HSD post hoc test (n=10 cells for all lines).
Since both For1A–GFP and For1D–GFP localize to the membrane, we used VAEM to image the cell cortex to enhance both temporal and spatial resolution. We found that FM4-64 localizes to discrete dots on the cell cortex (Fig. 3B). Simultaneous imaging of For1A–GFP or For1D–GFP with FM4-64 revealed that the formin and FM4-64 puncta are largely non-overlapping (Fig. 3B; Movie 4). We also observed that For1F–GFP and FM4-64 puncta do not overlap (Fig. 3B; Movie 4). Since For1F–GFP was linked to exocytosis and was observed to not overlap with clathrin, which also localizes to discrete cortical puncta (van Gisbergen et al., 2018), these data suggest that FM4-64, like clathrin, labels endocytic hotspots and that class I formins do not associate with these sites.
To test whether class I formins functionally affect endocytosis, we probed endocytosis in formin-knockout lines. Because the formin-knockout lines were generated in a Lifeact–mRuby line, we monitored uptake of FM1-43, a fluorescent lipophilic membrane dye similar to FM4-64 but whose emission profile does not overlap with mRuby. After adding FM1-43, plants were imaged as soon as they were mounted on the microscope (Fig. 3D, 0 min), and then at 15 min intervals. By measuring the ratio of the fluorescence intensity of the plasma membrane to the cytoplasm within 5–7 µm of the cell tip, we found that FM1-43 accumulated within cells. While we observed a trend that Δfor1BCDE2B-111 and Δfor1ABCDE2B-137 accumulated more cytoplasmic FM1-43 than Lifeact–mRuby and Δfor1ABCE2B-84 lines in the first time point, this trend was not statistically significant. Furthermore, uptake at 15 and 30 min was similar across all lines. Thus, these data suggest that class I formins do not significantly contribute to endocytic uptake of FM1-43.
In contrast to what was found for For1A–GFP and For1D–GFP, For2A–GFP, a class II formin, largely overlapped with FM4-64 puncta (Fig. 3B, Movie 4). To quantify the degree of association between formin and FM4-64 signals, we calculated the Pearson's correlation coefficient for all time points in at least ten time-lapse acquisitions. The Pearson's correlation coefficient for FM4-64 with class I formins was significantly lower than FM4-64 with For2A–GFP. Together, these data indicate a possible functional split between class I and class II formins. Namely, class I formins, in particular For1F, are linked to exocytosis, while class II formins associate with presumptive endocytic membranes at the plasma membrane, suggesting that class II formins function during endocytosis.
For1A and For1D differentially affect actin
Formins are known to nucleate and elongate actin filaments (Courtemanche, 2018; Goode and Eck, 2007). Using live-cell imaging, van Gisbergen et al. demonstrated that For2A mediates formation of actin filaments along the plasma membrane (van Gisbergen et al., 2012). To investigate the interaction of For1A and For1D with actin, we used homologous recombination to fuse sequences encoding for three tandem mEGFP molecules in a cell line expressing Lifeact–mCherry, which labels the actin cytoskeleton (van Gisbergen et al., 2012; Vidali et al., 2009a). Proper integration was verified by genotyping (Fig. S1), and we performed growth assays of the lines to ensure that the tagged locus does not alter plant growth (Fig. S2).
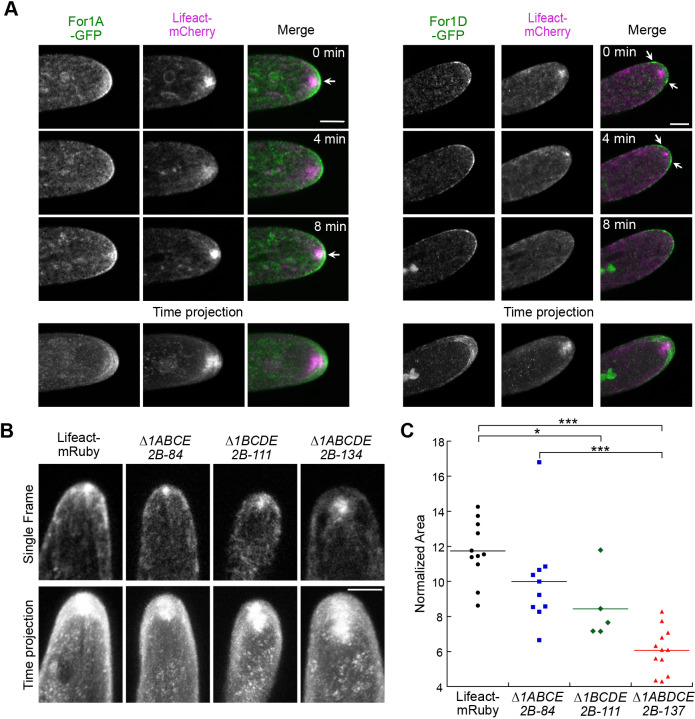
Actively growing cells have an apical actin structure that resides just below the tip of the cell (Fig. 3A; Vidali et al., 2009a). This structure is dynamic, assembling and disassembling in the order of seconds but yet remaining persistent over long time periods below the cell tip where it predicts the site of cell expansion (Furt et al., 2013; Wu and Bezanilla, 2018). Wu et al. demonstrated that enrichment of the class II formin For2A–GFP precedes formation of the apical actin structure (Wu and Bezanilla, 2018). Here, we investigated whether either For1A–GFP or For1D–GFP could also be involved in generating the apical actin structure. Confocal imaging revealed that For1A–GFP strongly accumulated at the apical plasma membrane adjacent to the site nearest the actin focus (Fig. 4A, arrows), which is evident in a maximum intensity projection of frames from the time-lapse acquisition (Fig. 4A, time projection, Movie 5). We also observed frames in the time-lapse (Fig. 4A, 8 min) where cytoplasmic For1A–GFP overlapped with the apical actin structure. Together, these data suggest that For1A–GFP may be involved in generating the cytoplasmic actin focus. In contrast, For1D–GFP accumulates in a larger area along the apical dome, and the points of highest intensity do not correlate with the position of the apical actin structure (Fig. 4A, arrows). A maximum intensity projection of frames from the time-lapse acquisition revealed that the maximum For1D–GFP signal was positioned to either side of the apical actin accumulation, with the majority of For1D–GFP confined to the plasma membrane (Fig. 4A).
Fig. 4.
Simultaneous imaging of For1A–GFP or For1D–GFP with Lifeact–mCherry. (A) Laser-scanning confocal images from the medial plane of an apical cell growing in a microfluidic imaging chamber. Images were de-noised with NIS elements software. Arrows indicate regions of the highest formin signal intensity. Scale bars: 5 µm. See also Movie 5. The time projection is a maximum intensity projection of 24 frames from 8 min of the time-lapse acquisition. (B) Laser-scanning confocal images of Lifeact–mRuby in control and formin-null lines as indicated. The single frame is a timepoint from a time-lapse acquisition, and is a maximum intensity projection of three confocal z-stacks taken near the medial section of the cell. The time projection is a maximum intensity projection of 121 frames from 20 min of the time-lapse acquisition. Scale bar: 5 µm. See also Movie 6. (C) Dot plot of the normalized area of the Lifeact–mRuby signal from the time projections. To normalize, the area was divided by the distance grown over the time-lapse acquisition. An one-way ANOVA with a Tukey HSD post hoc test was performed. *P<0.05; ***P<0.001.
To investigate whether For1A or For1D differentially contribute to actin at the cell apex, we analyzed the actin behavior by monitoring Lifeact–mRuby in formin-knockout lines. We found that the apical actin structure is present in all knockout lines (Fig. 4B). We characterized the dynamics and localization of the structure in cells during growth and discovered that there were consistent differences in the behavior of actin in the formin-null lines. We tracked the behavior of the cytoplasmic actin accumulation using TrackMate in Fiji (Tinevez et al., 2016). To compare across multiple cells and lines, we averaged the absolute value of the change in the estimated size of the cytoplasmic actin focus between each time point in the time-lapse acquisition for each cell (Fig. S4A,B). We found that in the parental line and in cells lacking either For1A or For1D (Δfor1ABCE2B or Δfor1BCDE2B), the size of the cytoplasmic apical accumulation is consistent during growth and the distance between the actin accumulation and the cell tip was less than 1 µm (Fig. S4). In contrast, the apical actin cytoplasmic focus varied dramatically in cells lacking both For1A and For1D (Δfor1ABCDE2B) and the actin accumulation was 1.3 µm further into the cytoplasm (Fig. S4), suggesting that For1A and For1D redundantly contribute to anchoring the apical actin accumulation to the cell membrane.
In addition, actin dynamically associated with the plasma membrane at the cell tip, which was evident in a maximum intensity projection of frames from the time-lapse acquisition (Fig. 4B; Movie 6). Interestingly, cells from mutants lacking For1D (Δfor1BCDE2B and Δfor1ABCDE2B) exhibited a significant reduction in plasma membrane associated Lifeact–mRuby signal. In contrast, cells from the mutant lacking For1A (Δfor1ABCE2B) had similar levels of plasma membrane associated actin as compared to the control line. These data suggest that For1D contributes to actin that is generated near the plasma membrane at the cell tip, consistent with the localization of For1D. To quantify the effect of For1A and/or For1D on apical actin, we measured the total area of the Lifeact–mRuby signal at the cell apex (cytoplasmic apical actin focus and plasma membrane associated signal) in the maximum intensity projections of the frames from the time-lapse acquisitions. We reasoned that loss of actin near the plasma membrane would result in a smaller overall area containing the Lifeact–mRuby signal. Indeed, we found that lines lacking For1D (Δfor1BCDE2B and Δfor1ABCDE2B) had the smallest area (Fig. 4C). Cells with loss of For1A also exhibited a trend for a smaller total area compared to the control line. However, the line lacking both For1A and For1D had the greatest reduction in area (Fig. 4C). These data suggest that For1A and For1D promote actin polymerization at the cell apex, with For1D contributing primarily at the plasma membrane, and For1A in the cytosol near the tip.
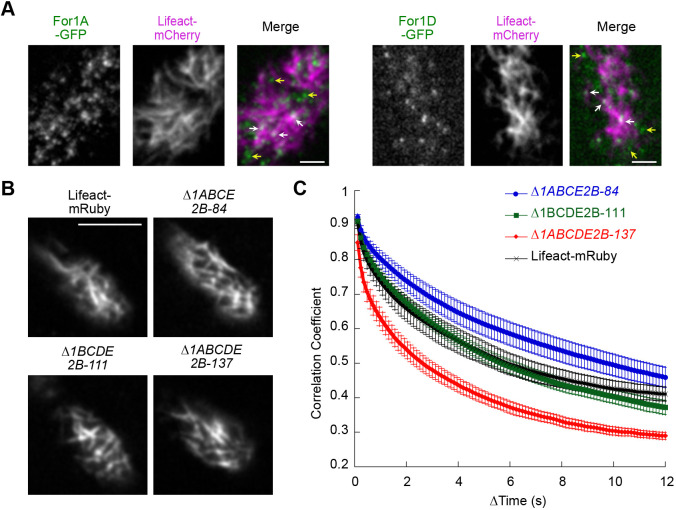
To characterize For1A and For1D behavior with respect to actin filaments at the cell cortex, we used VAEM to simultaneously image For1A–GFP or For1D–GFP and Lifeact–mCherry. In P. patens protonemal cells, the actin network is extremely dense and rapidly remodels (Augustine et al., 2011; Vidali et al., 2010). In addition, cortical For1A–GFP and For1D–GFP signals were weak, making simultaneous VAEM imaging challenging. For both For1A–GFP and For1D–GFP, we observed some overlap between formin puncta and actin filaments (Fig. 5A, arrows; Movie 7), but we also observed a large fraction of puncta not associated with actin filaments (Fig. 5A, yellow arrows; Movie 7). In time-lapse acquisitions (Movie 7), we occasionally observed cortical formin foci move along actin filaments, but these events were very rare, making it difficult to discern any significant patterns from these observations.
Fig. 5.
Characterization of cortical actin in endogenously tagged lines and formin-null lines. (A) Simultaneous VAEM imaging of For1A–GFP or For1D–GFP with Lifeact–mCherry at the cell cortex. Scale bars: 2 µm. See also Movie 7. White arrows indicate examples of formin cortical foci overlapping with actin filaments; yellow arrows indicate examples of formin foci that are not associated with actin filaments. For all images, formin is green and Lifeact–mCherry is magenta in the merged images. (B) VAEM images of cortical actin labeled with Lifeact–mRuby in control and the indicated formin-null lines. Scale bar: 5 µm. See also Movie 8. (C) Quantification of cortical actin dynamics under the indicated conditions. The correlation coefficient between two images was calculated at all possible temporal spacings (time interval). Error bars represent s.e.m. (Lifeact–mRuby, n=10; Δfor1ABCE2B-84, n=13; Δfor1BCDE2B-111, n=14; Δfor1ABCDE2B, n=10).
However, using the knockout formin lines, we had the opportunity to query whether loss of For1A and/or For1D impacted actin dynamics at cell cortex. Using VAEM to image Lifeact–mRuby at the cell cortex, we found that actin filament density was similar across all lines imaged (Fig. 5B; Movie 8). To quantify possible differences in actin dynamics, we measured the correlation coefficient of the intensity of the Lifeact–mRuby signal at all pixel locations for all temporal intervals (Fig. 5C). Over time, as actin filaments change their position due to polymerization or depolymerization, as well as translocation, the correlation coefficient decreases with larger temporal increments (Vidali et al., 2010). A fast decay in the correlation coefficient is indicative of very rapid changes in global actin filament dynamics. Using this method of quantification, we found that loss of For1D does not affect actin filament dynamics at the cell cortex (Fig. 5C). Surprisingly, loss of For1A inhibits actin dynamics, while loss of both For1A and For1D dramatically enhances actin dynamics (Fig. 5C). These data suggest that For1A and For1D differentially affect actin filaments at the cell cortex.
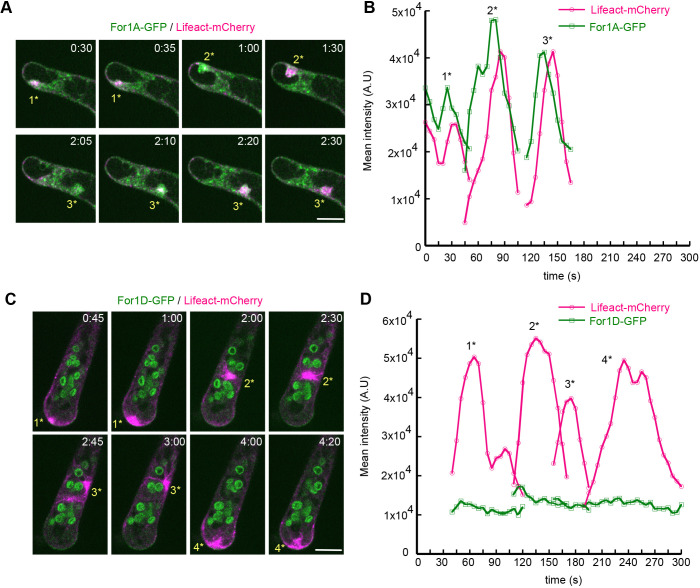
Since imaging at the cell cortex did not readily reveal actin polymerization events associated with either For1A or For1D, we used an assay that induces actin polymerization in the cytoplasm to test whether either class I formin associated with these events. Wu et al. previously observed that, without microtubules, large actin foci form stochastically throughout the cytoplasm, instead of at the cell apex (Wu and Bezanilla, 2018). Furthermore, For2A–GFP intensity accumulated at these ectopic sites of polymerization slightly before actin (Wu and Bezanilla, 2018). To investigate whether microtubules influence For1A or For1D localization and whether either class I formin contributes to generating cytosolic actin foci, we examined the localization of For1A–GFP or For1D–GFP in the Lifeact–mCherry line in the presence of oryzalin, which depolymerizes microtubules. Without microtubules, we observed that For1A–GFP was no longer apically enriched on the plasma membrane. Instead, the cytosolic puncta were enhanced (Fig. 6A). Furthermore, accumulations of For1A–GFP correlated strongly with bursts of actin (Fig. 6A,B; Movie 9). Interestingly, the For1A–GFP intensity peaked just prior to the Lifeact–mCherry signal, suggesting that For1A–GFP contributed to the formation of this structure (Fig. 6B). In contrast to what was seen for For1A–GFP, For1D–GFP intensity did not correlate with the formation of ectopic actin structures (Fig. 6C,D). For1D–GFP resided on the plasma membrane and was not observed in the cytoplasm cell near the actin accumulation (Fig. 6C; Movie 10). These data suggest that For1A, and not For1D, contributes to the formation of the cytosolic portion of the apical actin structure, providing further evidence that For1A and For1D differentially impact actin.
Fig. 6.
For1A and For1D differentially contribute to cytosolic actin structures. Apical cells expressing (A) For1A–GFP (green) or (C) For1D–GFP (green) with Lifeact–mCherry (magenta) growing in microfluidic imaging chambers were treated with 12.5 µM oryzalin and imaged on a laser-scanning confocal microscope. Images are single focal planes acquired every 5 s and deconvolved with NIS elements software (type Richardson–Lucy). The time-lapse images show accumulations of For1A–GFP that closely correlate temporally and spatially with generation of cytosolic actin foci. See also Movie 9. Whereas For1A–GFP strongly accumulates in the cytoplasm in the absence of microtubules, the For1D–GFP signal (remains low in the cytoplasm and mostly stays at the plasma membrane. See also Movie 10. (B,D) Quantification of time-lapse images in A and C. Foci enriched with Lifeact–mCherry were tracked using TrackMate and the mean intensity of For1A–GFP and Lifeact-mCherry in A, and For1D–GFP and Lifeact-mCherry in C were plotted over time. Numbers above the peaks (1*–4*) in the plots correspond to the bursts of actin polymerization indicated in A and C. Time stamps represent min:s. Scale bars: 10 µm.
Class I and II formins are differentially affected by cytoskeletal inhibitors
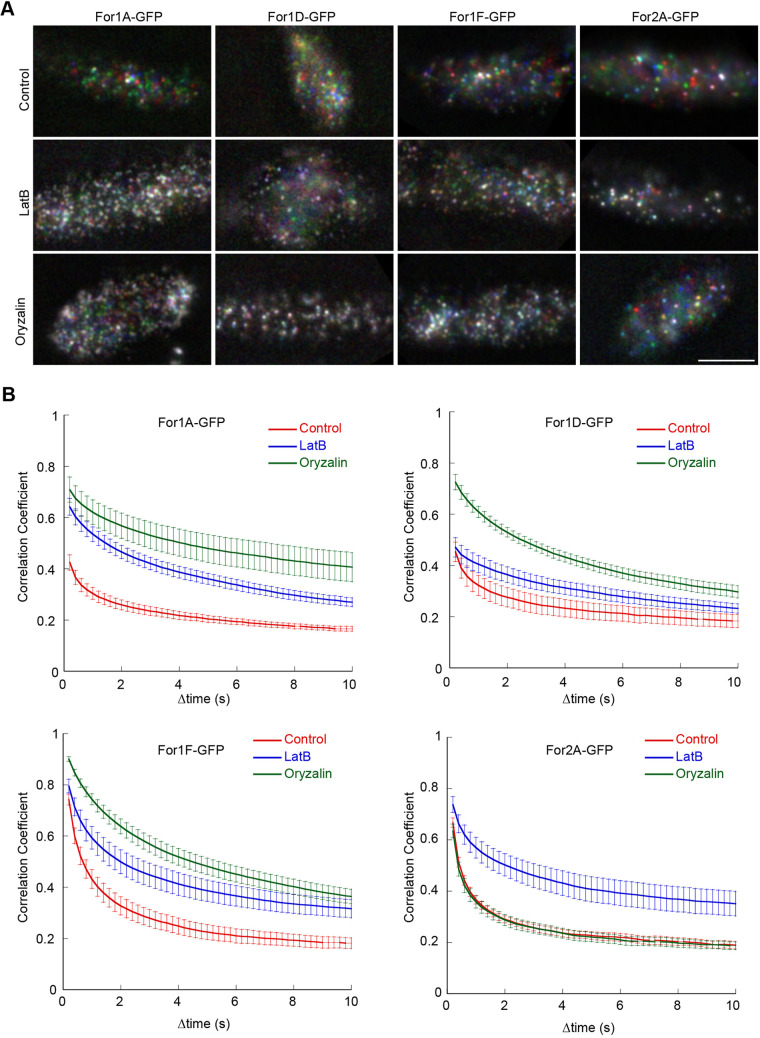
Having observed that microtubules more strongly influence For1A–GFP localization than For1D–GFP, and that For1A–GFP and For1D–GFP have differential activities with respect to actin, we sought to quantify whether microtubules or actin affect the dynamics of either For1A or For1D at the cell cortex. Both For1A–GFP and For1D–GFP localize to dynamic cortical puncta (Fig. 1D; Movie 1), similar to what was observed previously for For1F–GFP (van Gisbergen et al., 2018) and For2A–GFP (van Gisbergen et al., 2012). To quantitatively compare formin cortical dynamics among the three most highly expressed class I formins (For1F, For1A and For1D) (Vidali et al., 2009b) and one of the class II formins (For2A), we used VAEM to image the cell cortex in lines expressing endogenously tagged formins. To determine the effect of the actin and microtubule cytoskeleton on formin dynamics, we used drugs that specifically disassembled either actin filaments (latrunculin B) or microtubules (oryzalin). To visualize dynamics, we assigned color to three consecutive time frames (each 0.2 s apart) as red, green and blue, respectively. Merging these three frames shows that most particles are colored, indicating that they have moved positions in consecutive frames (Fig. 7A). Thus, the majority of particles are motile within 0.2 s in control images for all four formins. Treatment with either latrunculin B or oryzalin results in a significant decrease in dynamics for all class I formins, suggesting that class I formin dynamics are affected by both cytoskeletons (Fig. 7A; Movie 11). In contrast, For2A–GFP dynamics are only altered in cells treated with latrunculin B (Fig. 7A; Movie 11).
Fig. 7.
Formins are differentially affected by cytoskeletal inhibitors. (A) VAEM imaging of endogenously tagged formins at the cell cortex. See also Movie 11. Three frames taken every 200 ms from a time-lapse acquisition were false-colored red, green and blue and then merged into a single image. Movement of cortical dots from one frame to the next appear colored in the merge. If the particle does not move, then the red–green–blue merge results in a white particle. Scale bar: 2 µm. (B) Quantification of cortical formin dynamics under the indicated conditions. The correlation coefficient between two images was calculated at all possible temporal spacings (time interval). Error bars represent s.e.m. (n=10 cells except for For1A control, n=9; For1D LatB, n=8; For2A LatB, n=11).
To quantify these data, we used the correlation coefficient analysis described above for actin filament dynamics (Fig. 5C). Interestingly, loss of actin filaments differentially affected class I formins, with the strongest effects on For1A–GFP and For1F–GFP (Fig. 7B). For1D–GFP, which did not contribute to the formation of ectopic actin foci, was less affected by the absence of actin filaments (Fig. 7B). However, the dynamics of all three class I formins were most dampened in the absence of microtubules. In contrast, the dynamics of the class II formin, For2A–GFP, were only altered in the absence of actin, but not microtubules. These data indicate that formins from distinct groups (class I versus class II) as well as those within a group (class I) are differentially impacted by actin and microtubules.
DISCUSSION
Formins generate actin structures and, in a number of eukaryotes, interact with both the actin and microtubule cytoskeleton (Bartolini and Gundersen, 2010; Daou et al., 2014; Gaillard et al., 2011; Henty-Ridilla et al., 2016; Sun et al., 2017). In P. patens, loss-of-function studies have demonstrated that class I and II formins have distinct functions in cell growth (Vidali et al., 2009b). However, the molecular basis of these functional differences remained to be elucidated. Here, by analyzing formin cortical dynamics, we found that formins representing members from the plant class I and II families differentially respond to pharmacological perturbations of the cytoskeleton, suggesting that sub-functionalization between class I and II may reside in their ability to interact with the cytoskeleton. Our measurements of formin dynamics are an aggregate of appearance and/or disappearance at the plasma membrane, as well as movement along the plasma membrane. However, since cortical formin density is not dramatically altered in the absence of actin or microtubules, we reason that the majority of the differences that we observe in these measurements likely stem from differences in movement of the particles, as opposed to appearance and/or disappearance from the imaging plane. Consistent with this, previous measurements of cortical For2A demonstrated that, in the absence of actin filaments, there was an accumulation of stationary particles, a reduction of particles that exhibit random motility and a complete loss of linear trajectories (van Gisbergen et al., 2012). In our aggregate analysis, we easily detected the reduction of For2A mobility in the absence of actin. However, in the absence of microtubules, we were surprised to find that For2A dynamics were unperturbed, since without microtubules For2A localization is dramatically altered in the cytoplasm (Wu and Bezanilla, 2018). These data suggest that cortical and cytoplasmic pools of these molecules may be driving distinct cellular processes.
In contrast to cortical For2A dynamics, the cortical dynamics of all class I formins measured here (For1A, For1D and For1F) were dampened by treatment with both oryzalin and latrunculin B. Whereas For1D dynamics were only slightly diminished in the absence of actin, loss of actin strongly reduced For1A and For1F dynamics. Intriguingly, all class I formin dynamics were more inhibited by microtubule depolymerization than by actin depolymerization. These data indicate that, at the plasma membrane, class II formin mobility is actin dependent. In contrast, class I formins depend more strongly on microtubules than actin for their dynamics. Future studies utilizing fluorescent fusions of class I and II formins generated in a line containing labeled microtubules will provide further insights into the possible link between formins and microtubules at the cell cortex.
In addition to evidence for functional diversification between the formin classes, our data suggest that class I formin subclasses may have distinct functions. In particular, For1A appears to contribute to formation of the cytoplasmic apical actin accumulation that predicts the site of cell expansion. For1D, which is much more tightly associated with the plasma membrane, appears to contribute to actin at the apical plasma membrane, but not in the cytoplasm in normal cells as well as in cells lacking microtubules. Furthermore, actin dynamics at the cell cortex are differentially affected in the formin knockout lines. Intriguingly, actin dynamics are not altered when For1A and For1F are the two class I formins present (Δfor1BCDE2B). If For1D and For1F are present (Δfor1ABCE2B), then actin dynamics are inhibited. With For1F as the sole class I formin present (Δfor1ABCDE2B), actin dynamics at the cell cortex are dramatically enhanced. Taken together, these data suggest that For1A and, to a greater extent, For1D dampen cortical actin dynamics.
In plants, formins are intimately connected to the membrane, providing a direct link between membrane dynamics and the cytoskeleton (van Gisbergen and Bezanilla, 2013). In seed plants, class I formins are integral membrane proteins, while a subset of class II formins have a phosphatase tensin (PTEN) membrane-binding domain (van Gisbergen and Bezanilla, 2013). In P. patens, with the exception of the moss-specific For1F (van Gisbergen et al., 2018), class I formins are also integral membrane proteins. In P. patens, all class II formins have a PTEN domain, which mediates membrane association by binding to phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2] (van Gisbergen et al., 2012). Formin subcellular localization from previous work (van Gisbergen et al., 2012, 2018; Vidali et al., 2009b) and this study demonstrated that class I and II formins are both found at areas of active membrane remodeling. Here, we provide evidence that formins in phylogenetically distinct clades participate in different membrane activities. We found that none of the class I formins investigated here correlated with FM4-64 particles. The analysis with For1F is consistent with a previous study that demonstrated that For1F does not associate with clathrin particles at the cell cortex (van Gisbergen et al., 2018). In contrast For1F, due to its unique N-terminal Sec10 domain, is part of the exocyst complex and is required for exocytosis (van Gisbergen et al., 2018). Although it is unclear whether the other class I formins participate in exocytosis, loss of For1A and/or For1D did not affect endocytic uptake of FM1-43, suggesting that they are not involved in endocytosis.
In contrast to class I formins, the class II formin, For2A associates closely with presumptive endocytic cortical sites labeled by FM4-64. Furthermore, the apical For2A accumulation in the cytosol also tightly mirrors FM4-64 uptake, both temporally and spatially (van Gisbergen et al., 2012). Silencing class II formins results in a complete loss of polarized cell expansion, resulting in plants composed of spherical cells (Vidali et al., 2009b). Together, these data suggest that For2A generates subcellular actin structures related to endocytic function, which regulates cell polarity. Future studies identifying molecular linkages between formins and their regulators are required to further delineate the functional roles of formins in mediating membrane turnover.
MATERIALS AND METHODS
P. patens propagation, protoplast regeneration and transformation
All P. patens propagation, protoplast regeneration and transformations were performed as previously described (Vidali et al., 2007, 2009b) with minor modifications described as follows. Protoplasts were transformed at a concentration of 2×106 protoplasts/ml. For isolation of endogenously tagged lines, protoplasts were regenerated with top agar [1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 2.7 mM (NH4)2-tartrate, 45 µM FeSO4, 9.93 µM H3BO3, 220 nM CuSO4, 1.966 µM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, 103 nM Na2MoO4, 6% mannitol, 10 mM CaCl2]. To generate null mutants with CRISPR-Cas9-mediated homology directed repair, protoplasts were regenerated with liquid plating medium [1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 2.7 mM (NH4)2-tartrate, 45 µM FeSO4, 9.93 µM H3BO3, 220 nM CuSO4, 1.966 µM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, 103 nM Na2MoO4, 8.5% mannitol, 10 mM CaCl2]. Transformed plants were selected 4 days after transformation on plates containing 15 µg ml−1 hygromycin or 20 µg ml−1 G418. For endogenous tagging, plants were transferred off selection medium for 7–10 days, and then returned to selection medium. Only plants that survived the second selection were selected for genotyping. For CRISPR-Cas9-mediated homology directed repair, plants were transferred off selection medium after 7 days, and grown on medium without selection until they were big enough to genotype using the primers indicated in Table S1.
For protoplast regeneration assays, protoplasts were plated in the liquid plating medium described above. Regenerating plants were moved from the protoplast regeneration medium at day four and transferred to PpNH4 medium [1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 2.7 mM (NH4)2-tartrate, 45 μM FeSO4, 9.93 μM H3BO3, 220 nM CuSO4, 1.966 μM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, 103 nM Na2MoO4]. After 3 days, plants were imaged at room temperature. For endogenously tagged lines, individual plants regenerated from single protoplasts were imaged on a fluorescence stereomicroscope (Leica MZ16FA) equipped with a color camera (Leica DF300FX) using the GFP2 filter set (Leica). For CRISPR-Cas9-mutagenized plants, plants were suspended in a drop of Calcofluor staining solution (0.1 mg ml−1 in water) on top of a slide, and then covered with a coverslip. Images were taken with Nikon SMZ25 dissecting scope using a filter cube (excitation 420/25, dichroic 455, emission 460 longpass 25 nm) with a color camera (Nikon digital sight DS-Fi2). Plant area and morphometric parameters were acquired by an observer who was blind to the experimental conditions and measured as described previously (Vidali et al., 2007). Briefly, a 24-bit RGB image of a 1-week-old plant was manually cropped, and the red channel corresponding to the chlorophyll autofluorescence or the Calcofluor fluorescence pseudo-colored with red was separated. Fluorescence was thresholded using maximum entropy in Fiji software. Total plant area was determined from the thresholded images. Plant area was normalized to the control plants taken on the same day. One-way ANOVA was performed with KaleidaGraph (Synergy).
Plasmid construction
Constructs used to fluorescently tag For1A and For1D were generated as follows: a region 1000 bps directly upstream and downstream of the stop codon was amplified using primers (Table S1) and cloned into pDONR221-P1-P5r and pDONR221-P3-P2, respectively (Invitrogen). For tagging For1A and For1D, these were recombined with pGEM-gate (Vidali et al., 2009b), 3xmEGFP-L5L4 (Vidali et al., 2009b) and R4R3-NOSter-Lox-Hygro-Lox (Augustine et al., 2011). The resulting plasmids were digested with SwaI to make linear targeted pieces for homologous recombination and transformed into protoplasts. Constructs used to induce double-stranded breaks were generated according to Mallett et al. (2019). Briefly, two protospacers were chosen for For1A and For1D genomic sequences using the CRISPOR prediction program (Haeussler et al., 2016). The protospacers were first ligated into entry clones containing the moss U6 promoter sgRNA and subsequently recombined into a final plasmid containing Cas9, creating pMH-For1A-ps21 and pMK-For1D-ps12. Constructs that provided a template for homology-directed repair to knock in a cassette containing stop codons in all three reading frames were generated as follows: a region around 800 bp upstream of the first protospacer-targeting site and a region downstream of the second protospacer-targeting site were amplified using primers (Table S1) and cloned into pDONR221-P1-P4 and pDONR221-P3-P2, respectively, generating two entry clones. These entry clones were recombined with pENTR-R4R3-stop (Mallett et al., 2019) and pGEM-gate. The resulting plasmids, pGEM-For1A-STOP and pGEM-For1D-STOP, were co-transformed into moss protoplasts to induce CRISPR-Cas9-mediated homology directed repair with the corresponding pMH-For1A-ps21 and pMK-For1D-ps12, respectively.
Protein extraction and immunoblotting
For protein extraction from stable moss lines, moss protonemal tissue was dried on a paper towel and weighed. Lysis was performed by grinding in liquid nitrogen. Ground tissue was then resuspended in 400 µl grinding buffer (50 mM HEPES, 150 mM NaCl, 10 mM EDTA, 2 mM DTT, 20 µg/ml leupeptin, 2.5% SDS, and 2 mM PMSF), supplemented with Complete mini EDTA-free protease inhibitor cocktail tablets (1 tablet per 5 ml; Roche) and 0.1% casein. Cell debris was spun down and the supernatant was precipitated with methanol/chloroform. For immunoblotting, the methanol/chloroform-precipitated pellet was first resuspended in 8 M urea with 1% Triton X-100 before adding sample buffer (175 mM Tris base, 2.5% SDS, 80 mM DTT and 7.5% glycerol) and boiling for 10 min. Then, 80% of the sample was loaded and separated on a 6% SDS-PAGE gel, and transferred to a nitrocellulose membrane. The membrane was blocked with 5% nonfat dry milk dissolved in 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween 20 (TBS-T) at 25°C for 1 h. After five washes with TBS-T for 5 min, the membrane was incubated with a polyclonal rabbit anti-GFP antibody (van Gisbergen et al., 2012; 1:5000 dilution with 5% nonfat dry milk in TBS-T) at 4°C for 16 h. After five washes with TBS-T for 5 min, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (1:5000 dilution) at 25°C for 1 h. To remove unbound secondary antibody, samples were washed five times for 5 min with TBS-T. SuperSignal West Femto (Thermo Fisher Scientific) was used according to the manufacturer's recommendations to detect chemiluminescence emission with a gel dock system equipped for chemiluminescence detection (GE Healthcare ImageQuant LAS 500).
Spinning-disc confocal microscopy
One-week old protoplast-regenerated protonemal cells were placed on a 1% agar pad in Hoagland's medium [4 mM KNO3, 2 mM KH2PO4, 1 mM Ca(NO3)2, 89 µM Fe citrate, 300 µM MgSO4, 9.93 µM H3BO3, 220 nM CuSO4, 1.966 µM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, 103 nM Na2MoO4 and 1% sucrose], covered with a glass coverslip, sealed with VALAP (1:1:1 parts of vaseline, lanoline and paraffin) and immediately observed at room temperature. Slides were mounted on a Nikon Ti-E inverted microscope equipped with a Yokogawa CSU-X1 spinning disk head and a 512×512 Andor iXON electron multiplying CCD camera. Images were collected with a Nikon 1.4 NA 60× oil immersion objective at room temperature. 488 and 561 nm laser illumination was used for GFP and FM4-64/mCherry excitation, respectively. Emission filters were 515/30 nm for GFP and 600/32 nm for mCherry/FM4-64. Image acquisition was controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA).
Laser-scanning confocal microscope
One-week old protoplast-regenerated protonemal cells were mounted on agar pads or moss plants were grown in microfluidic imaging devices (Bascom et al., 2016). The microfluidic imaging devices were soaked overnight in Hoagland's medium, after which ground protonemata or plants regenerated from protoplasts were inserted into the imaging chamber. Plants were allowed to recover for 3–7 days, after which the microfluidic devices were imaged. The glass slide or imaging device was mounted on a Nikon A1R laser scanning confocal with a 1.4 NA 60× or 100× oil immersion objective (Nikon) at room temperature. Laser illumination at 488, 514 and 561 nm was used for GFP, FM1-43 and FM4-64/mCherry/mRuby excitation, respectively. Emission filters were 525/50 nm for GFP, 585/65 for FM1-43 and 595/50 for mCherry/FM4-64/mRuby. Image acquisition was controlled by NIS-Elements software (Nikon). For FM4-64 labeling, protonemal cells growing in the imaging chamber were incubated with 20 µM FM4-64 diluted in Hoagland's medium for 5 min and then washed three times with Hoagland's medium to remove excess FM4-64. For FM1-43 labeling, 8-day-old protoplast-regenerated plants were mounted on an agar pad supplied with Hoaglands's medium and 5 µM FM1-43, and covered with a coverslip.
Variable-angle epifluorescence microscopy
Protoplast-regenerated protonemal cells that were 5–7 days old were mounted on agar pads and immediately imaged at room temperature. The slide was mounted on a Nikon Ti-E inverted microscope equipped with a mirror-based Nikon T-FL-TIRF illuminator and imaged with a Nikon 1.49 NA 100× oil immersion TIRF objective. The laser illumination angle was adjusted individually for each sample to achieve the maximum signal to noise ratio. For single-color imaging, GFP was excited with a 488 laser and GFP emission from the specimen was captured with an Andor iXON3 1024×1024 electron-multiplying CCD camera. For simultaneous two-color imaging, images were acquired on a similar system equipped with a Nikon 1.45 NA 60× oil immersion TIRF objective and equipped with a TuCAM (Andor technology) and two Zyla sCMOS cameras (Andor technology). GFP and mCherry/mRuby/FM4-64 were simultaneously excited with 488 and 561 nm lasers, respectively. For cortical imaging of Lifeact–mRuby, images were acquired on a Nikon Ti-E inverted microscope equipped with a TI-TIRF-PAU illuminator, using a Nikon 1.49 NA 100× oil immersion TIRF objective. mRuby was excited with a 561 nm laser, the emission passed through a 610/75 filter and was captured with an Andor 897 EMCCD camera. Image acquisition process was controlled by NIS-Elements software (Nikon).
Image processing and quantification
Unless otherwise noted, images were processed with enhanced contrast, background subtraction (rolling ball diameter of 50) and smoothing using Fiji. All settings were standard. A subset of images, as noted in the figure legends, were de-noised using standard settings in the NIS elements software. Pearson's correlation coefficients were measured using the colocalization test in Fiji. Quantification of cortical actin and formin dynamics was achieved by measuring the decay of the correlation coefficient over all temporal spacings was performed as previously described (Vidali et al., 2010).
Supplementary Material
Acknowledgements
The acquisition of the TIRF microscope at Dartmouth College was funded by an National Institutes of Health (NIH) S10, grant number 1S10OD018046-01.
Footnotes
Author contributions
Conceptualization: P.v.G., S.-Z.W., X.C., K.A.P., M.B.; Methodology: P.v.G., X.C., S.-Z.W., K.A.P., M.B.; Formal analysis: P.v. G., S.-Z.W., M.B.; Investigation: P.v.G., X.C., S.-Z.W., K.A.P.; Writing - original draft: P.v.G., M.B.; Writing - review & editing: P.v.G., X.C., S.-Z.W., M.B.; Visualization: P.v.G., S.-Z.W., X.C.; Supervision: M.B.; Project administration: M.B.; Funding acquisition: M.B.
Funding
P.v.G received support from the Plant Biology Graduate Program at the University of Massachusetts, Amherst and X.C. received support from the Molecular and Cellular Biology Graduate Program at Dartmouth College. This work was supported by National Science Foundation (NSF) grants MCB-1330171 and MCB-1715785 to M.B.
References
- Augustine, R. C., Pattavina, K. A., Tüzel, E., Vidali, L. and Bezanilla, M. (2011). Actin interacting protein1 and actin depolymerizing factor drive rapid actin dynamics in Physcomitrella patens. Plant Cell 23, 3696-3710. 10.1105/tpc.111.090753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini, F. and Gundersen, G. G. (2010). Formins and microtubules. Biochim. Biophys. Acta 1803, 164-173. 10.1016/j.bbamcr.2009.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom, C. S., Wu, S.-Z., Nelson, K., Oakey, J. and Bezanilla, M. (2016). Long-term growth of moss in microfluidic devices enables subcellular studies in development. Plant Physiol. 172, 28-37. 10.1104/pp.16.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone, K. G. and Welch, M. D. (2010). A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237-251. 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F., Drubin, D. and Nurse, P. (1997). cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137, 169-182. 10.1083/jcb.137.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra, E. S., Ramabhadran, V., Gerber, S. A. and Higgs, H. N. (2009). INF2 is an endoplasmic reticulum-associated formin protein. J. Cell. Sci. 122, 1430-1440. 10.1242/jcs.040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche, N. (2018). Mechanisms of formin-mediated actin assembly and dynamics. Biophys Rev. 10, 1553-1569. 10.1007/s12551-018-0468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou, P., Hasan, S., Breitsprecher, D., Baudelet, E., Camoin, L., Audebert, S., Goode, B. L. and Badache, A. (2014). Essential and nonredundant roles for Diaphanous formins in cortical microtubule capture and directed cell migration. Mol. Biol. Cell 25, 658-668. 10.1091/mbc.e13-08-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, M. J., Hussey, P. J. and Davies, B. (2002). Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 7, 492-498. 10.1016/S1360-1385(02)02341-5 [DOI] [PubMed] [Google Scholar]
- Diao, M., Ren, S., Wang, Q., Qian, L., Shen, J., Liu, Y. and Huang, S. (2018). Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. eLife Sci. 7, e36316. 10.7554/eLife.36316.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudin, O., Bendezú, F. O., Groux, R., Laroche, T., Seitz, A. and Martin, S. G. (2015). A formin-nucleated actin aster concentrates cell wall hydrolases for cell fusion in fission yeast. J. Cell Biol. 208, 897-911. 10.1083/jcb.201411124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtine, M. S., Chow, C. J., Adames, N., Pringle, J. R., Peter, M. and Boone, C. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118-122. 10.1126/science.276.5309.118 [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Zigmond, S. and Boone, C. (2003). Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell. Sci. 116, 2603-2611. 10.1242/jcs.00611 [DOI] [PubMed] [Google Scholar]
- Faix, J. and Grosse, R. (2006). Staying in shape with formins. Dev. Cell 10, 693-706. 10.1016/j.devcel.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Feierbach, B. and Chang, F. (2001). Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11, 1656-1665. 10.1016/S0960-9822(01)00525-5 [DOI] [PubMed] [Google Scholar]
- Furt, F., Liu, Y.-C., Bibeau, J. P., Tüzel, E. and Vidali, L. (2013). Apical myosin XI anticipates F-actin during polarized growth of Physcomitrella patens cells. Plant J. 73, 417-428. 10.1111/tpj.12039 [DOI] [PubMed] [Google Scholar]
- Gaillard, J., Ramabhadran, V., Neumanne, E., Gurel, P., Blanchoin, L., Vantard, M. and Higgs, H. N. (2011). Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol. Biol. Cell 22, 4575-4587. 10.1091/mbc.e11-07-0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode, B. L. and Eck, M. J. (2007). Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593-627. 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- Grunt, M., Žárský, V. and Cvrčková, F. (2008). Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 8, 115. 10.1186/1471-2148-8-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler, M., Schönig, K., Eckert, H., Eschstruth, A., Mianné, J., Renaud, J.-B., Schneider-Maunoury, S., Shkumatava, A., Teboul, L., Kent, J., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E. S., Rouiller, I., Hanein, D. and Higgs, H. N. (2006). Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 281, 14383-14392. 10.1074/jbc.M510923200 [DOI] [PubMed] [Google Scholar]
- Henty-Ridilla, J. L., Rankova, A., Eskin, J. A., Kenny, K. and Goode, B. L. (2016). Accelerated actin filament polymerization from microtubule plus ends. Science 352, 1004-1009. 10.1126/science.aaf1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, H. N. and Peterson, K. J. (2005). Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1-13. 10.1091/mbc.e04-07-0565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, Y., Liu, X., Fu, Y. and Huang, S. (2018). Arabidopsis class I formins control membrane-originated actin polymerization at pollen tube tips. PLoS Genet. 14, e1007789. 10.1371/journal.pgen.1007789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Liang, W., Zhang, X., Ren, H., Hu, J., Bennett, M. J. and Zhang, D. (2014). Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc. Natl. Acad. Sci. USA 111, 10377-10382. 10.1073/pnas.1401680111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Yang, X., Zhang, X., Song, Y., Liang, W. and Zhang, D. (2018). Rice morphology determinant-mediated actin filament organization contributes to pollen tube growth. Plant Physiol. 177, 255-270. 10.1104/pp.17.01759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Zhang, Y. and Ren, H. (2018). Actin polymerization mediated by ATFH5 directs the polarity establishment and vesicle trafficking for pollen germination in Arabidopsis. Mol. Plant 11, 1389-1399. 10.1016/j.molp.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Mallett, D. R., Chang, M., Cheng, X. and Bezanilla, M. (2019). Efficient and modular CRISPR-Cas9 vector system for Physcomitrella patens. Plant Direct 3, e00168-e00115. 10.1002/pld3.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramírez, C., Hernandez-Coronado, M., Thamm, A., Catarino, B., Wang, M., Dolan, L., Feijó, J. A. and Becker, J. D. (2016). A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant 9, 205-220. 10.1016/j.molp.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Gao, L., Bi, E. and Bretscher, A. (2004). Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15, 4971-4989. 10.1091/mbc.e04-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosero, A., Oulehlová, D., Stillerová, L., Schiebertová, P., Grunt, M., Žárský, V. and Cvrčková, F. (2016). Arabidopsis FH1 formin affects cotyledon pavement cell shape by modulating cytoskeleton dynamics. Plant Cell Physiol. 57, 488-504. 10.1093/pcp/pcv209 [DOI] [PubMed] [Google Scholar]
- Sun, T., Li, S. and Ren, H. (2017). OsFH15, a class I formin, interacts with microfilaments and microtubules to regulate grain size via affecting cell expansion in rice. Sci. Rep. 7, 6538. 10.1038/s41598-017-06431-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G. M., Reynolds, G. D., Laplantine, E., Bednarek, S. Y., Shorte, S. L. and Eliceiri, K. W. (2016). TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80-90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- van Gisbergen, P. A. C. and Bezanilla, M. E. (2013). Plant formins: membrane anchors for actin polymerization. Trends Cell Biol. 23, 227-233. 10.1016/j.tcb.2012.12.001 [DOI] [PubMed] [Google Scholar]
- van Gisbergen, P. A. C., Esseling-Ozdoba, A. and Vos, J. W. (2008). Microinjecting FM4-64 validates it as a marker of the endocytic pathway in plants. J. Microsc. 231, 284-290. 10.1111/j.1365-2818.2008.02041.x [DOI] [PubMed] [Google Scholar]
- van Gisbergen, P. A. C., Li, M., Wu, S.-Z. and Bezanilla, M. E. (2012). Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J. Cell Biol. 198, 235-250. 10.1083/jcb.201112085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen, P. A. C., Wu, S.-Z., Chang, M., Pattavina, K. A., Bartlett, M. E. and Bezanilla, M. E. (2018). An ancient Sec10-formin fusion provides insights into actin-mediated regulation of exocytosis. J. Cell Biol. 217, 945-957. 10.1083/jcb.201705084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali, L., Augustine, R. C., Kleinman, K. P. and Bezanilla, M. E. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19, 3705-3722. 10.1105/tpc.107.053413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali, L., Rounds, C. M., Hepler, P. K. and Bezanilla, M. E. (2009a). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4, e5744. 10.1371/journal.pone.0005744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali, L., van Gisbergen, P. A. C., Guérin, C., Franco, P., Li, M., Burkart, G. M., Augustine, R. C., Blanchoin, L. and Bezanilla, M. (2009b). Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 106, 13341-13346. 10.1073/pnas.0901170106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali, L., Burkart, G. M., Augustine, R. C., Kerdavid, E., Tüzel, E. and Bezanilla, M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22, 1868-1882. 10.1105/tpc.109.073288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.-Z. and Bezanilla, M. (2018). Actin and microtubule cross talk mediates persistent polarized growth. J. Cell Biol. 217, 3531-3544. 10.1083/jcb.201802039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.