ABSTRACT
The ability of a muscle to break down and reform fibers is vital for development; however, if unregulated, abnormal muscle remodeling can occur, such as in the heart following cardiac infarction. To study how normal developmental remodeling is mediated, we used fluorescently tagged actin, mutant analyses, Ca2+ imaging and controlled Ca2+ release to determine the mechanisms regulating a conspicuous muscle change that occurs in Caenorhabditis elegans males. In hermaphrodites and larval males, the single cell anal depressor muscle, used for waste expulsion, contains bilateral dorsal-ventral sarcomeres. However, prior to male adulthood, the muscle sex-specifically remodels its sarcomeres anteriorly-posteriorly to promote copulation behavior. Although WNT signaling and calcineurin have been implicated separately in muscle remodeling, we unexpectedly found that they participate in the same pathway. We show that WNT signaling through Gαo and PLC-β results in sustained Ca2+ release via IP3 and ryanodine receptors to activate calcineurin. These results highlight the utility of this new model in identifying additional molecules involved in muscle remodeling.
Keywords: G-protein-coupled signaling, Muscle remodeling, WNT/Calcium signaling, WNT/PCP signaling
Summary: Fluorescently tagged actin visualization, mutant analyses and Ca2+-based assays reveal the mechanisms that underpin muscle remodeling under a normal developmental program, which provides insights into how to control pathological remodeling.
INTRODUCTION
Muscle remodeling is an important part of development and disease progression (Freire et al., 2014). In development, muscles must reshape as the organism's needs change. In adults, detrimental events, such as cardiac infarction, can lead to dormant developmental pathways re-activating, which reshapes heart muscle to a pathological degree. To prevent and treat abnormal remodeling, normal well-regulated developmental remodeling must be studied to determine how molecular pathways control this process.
The Caenorhabditis elegans male provides one such opportunity for study as one of its tail muscles undergoes a drastic sex-specific remodeling event during larval to adult development (Jarrell et al., 2012; Sulston et al., 1980; Sulston and Horvitz, 1977). The single cell anal depressor muscle, located in the male tail, goes through a complete remodeling in both structure and function during larval development. In the early larval (L1-L3) male, this muscle contains a dorsal-ventral sarcomere that participates in defecation. During the last larval stage (L4), the muscle remodels. In the adult male, it contains an anterior-posterior sarcomere that positions the copulatory spicule during male mating behavior, facilitating sperm transfer into the hermaphrodite uterus (LeBoeuf and Garcia, 2017). This muscle remodeling takes place concurrently with the morphogenesis of male copulatory structures (Sulston et al., 1980). In contrast, the hermaphrodite anal depressor does not remodel and remains a defecation muscle. We previously reported that the sexual identity of the anal depressor is necessary for developmental remodeling (Chen and García, 2015). However, the pathways regulating this process are unidentified.
Here, we report that multiple WNT pathways are necessary for sarcomere remodeling. During early development, the WNT planar cell polarity (PCP) pathway is required in epithelial cells that neighbor the anal depressor. Later in development, the WNT/calcium pathway has WNT ligands acting directly on the anal depressor, promoting G-protein-coupled signaling that is dependent on Gαo and Gα/GPA-7. This leads to PLC-β activation, which triggers calcium release from internal stores through IP3 and ryanodine receptors. Calcium signaling then promotes the calcium/calmodulin-dependent phosphatase calcineurin to further facilitate the morphogenesis of the anal depressor.
RESULTS
Sarcomere remodeling during male L4 to adult development
Our previous study established that the extensive remodeling of the male anal depressor, which occurs during L4 development, is dependent upon the sexual identity of the muscle (Chen and García, 2015; Jarrell et al., 2012; Garcia et al., 2001; Reiner and Thomas, 1995; Sulston et al., 1980). Problematically, the work used a YFP:actin construct (driven by the unc-103E promoter) that is also expressed in the sex muscles, which partially obscure the adult anal depressor (Chen and García, 2015; Reiner et al., 2006). To visualize the anal depressor more clearly in adults, we generated an integrated YFP:actin construct that uses the aex-2 promoter, which is expressed in the anal depressor, sphincter, intestinal muscles and the motor neuron DVB, but not in the male sex muscles (Fig. 1) (Mahoney et al., 2008). The Paex-2:YFP:actin construct allowed clearer dissection of anal depressor remodeling.
Fig. 1.
Visualization of male anal depressor rearrangement using YFP:actin. (A,A′,E-I′) DIC (A,E,F,G,H,I) and fluorescent (A′,E′,F′,G′,H′,I′) images of the male tail. The arrows in the fluorescent images point to the ventral slit that denotes the anterior and posterior domains. (B) Diagram representing the transverse view of the hermaphrodite and larval male anal depressor (adp). The sarcomere orientation is indicated by the gray shading. There are two dorsal attachments, left and right. D, dorsal; R, right. (C-D′) DIC (C,D) and fluorescent (C′,D′) images of the hermaphrodite tail. (J) Diagram of the male anal depressor representing both the actin (red lines) and cell shape (in gray). The location of the nucleus is in yellow. For all images except B, dorsal is up and anterior is to the left. Scale bars: 10 µm.
In both hermaphrodites and larval males, the single cell anal depressor had left and right sarcomeres that spanned from the dorsal cuticle to the ventral anus (Fig. 1A,B). The sarcomeres contract during the last step of defecation, allowing expulsion of the gut contents (Thomas, 1990). Although similar in form and function, the larval male anal depressor did not expand like the hermaphrodite muscle (Fig. 1C-D′) (Chen and García, 2015). During the last larval stage (L4), the sarcomeres in the male anal depressor underwent a dramatic reorganization, so that the cell transformed from a dorsal-ventral defecation muscle to an anterior-posterior auxiliary mating muscle. First, during late L3 to early L4 larval stage, a ventral slit formed along the rectal attachment, demarking the anterior and posterior domains (Fig. 1E,E′). The domain anterior to the slit elongated along the neighboring epithelia to attach to the developing spicule protractor muscles (Fig. 1F; Fig. S1). While this occurs, the dorsal-ventral sarcomere in the posterior domain was present and was not fully disassembled until the end of L4 (Fig. 1G-I′). The fluorescently tagged actin from the Paex-2:YFP:actin integration showed a similar disassembly process to the Punc-103E:YFP:actin transgene (Fig. 1) (Chen and García, 2015). Interestingly, we noted that each left and right dorsal-ventral sarcomere was reorganized into two partially overlapping anterior-posterior sarcomeres, a feature that was not seen with the Punc-103E:YFP:actin construct (Fig. 1J; Fig. S2).
WNT ligands and receptor regulate anal depressor rearrangement
To determine which developmental pathways are used for C. elegans male anal depressor rearrangement, we generated an integrated transgenic line expressing cytoplasmic YFP from the aex-2 promoter. YFP expression was diffuse throughout the cell, showing both the ventrally located muscle attachments and the dorsally located nucleus (Fig. 2A). We crossed this marker to strains defective in different developmental pathways, including EGF/LIN-3 (Fig. 2B), notch/LIN-12 (Fig. 2C), TGFβ/SMA-6 (Fig. 2D) and WNT ligands (EGL-20, LIN-44). Only mutations in the WNT pathway ligands showed anal depressor rearrangement defects (Fig. 2E,F), suggesting that WNT signaling is involved in anal depressor remodeling.
Fig. 2.
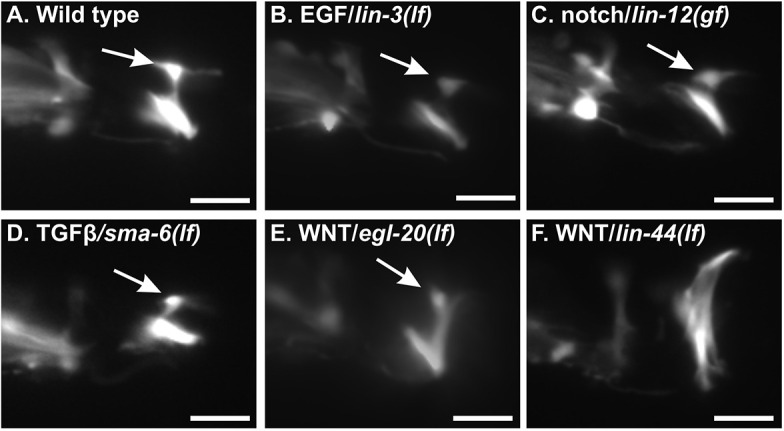
Anal depressor rearrangement defects in developmental mutants. (A-F) YFP fluorescent images of the anal depressor muscle in wild-type (A) and mutant (B-F) adult male tails. Dorsal is up and posterior is to the right. Arrows point to the location of the cell body. In F, the cell body is hidden in the abnormal anal depressor. Scale bars: 10 μm.
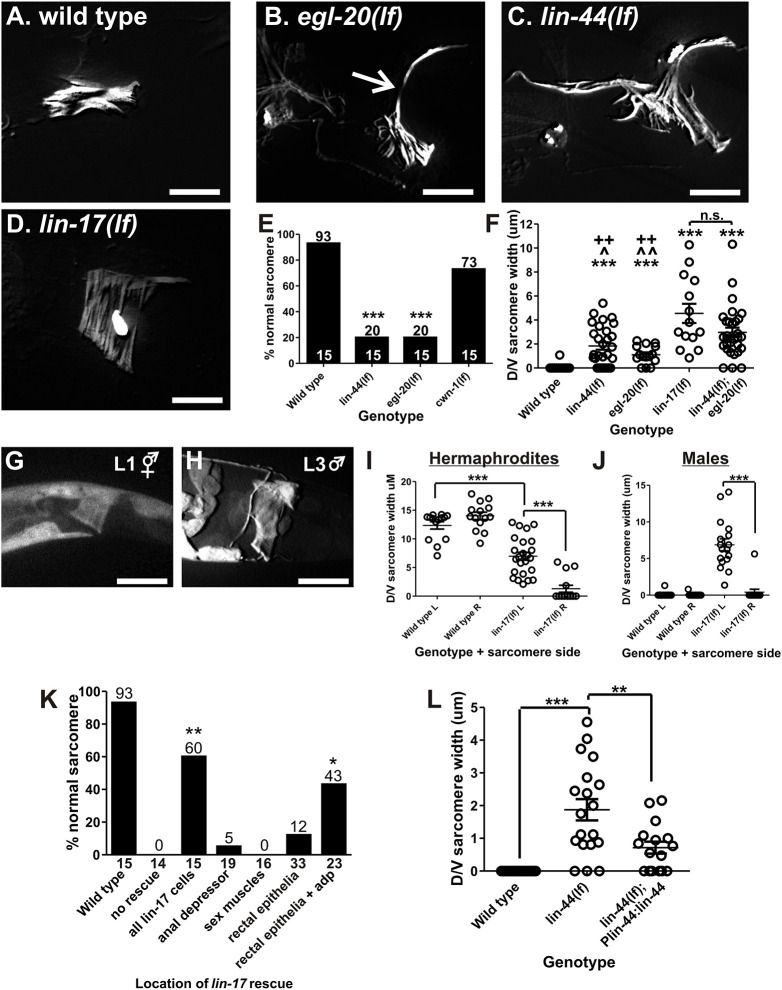
The cytoplasmic YFP only shows the overall morphology of the anal depressor, not sarcomere-specific defects. As depicted in Fig. 1, the entire dorsal-ventral sarcomere of the developing male tail is remodeled into anterior-posterior sarcomeres, necessitating sarcomere disassembly. To determine the state of the anal depressor sarcomere in WNT mutants, we crossed the Paex-2:YFP:actin integration into WNT mutant backgrounds. The dorsal-ventral sarcomere in early wild-type L4 males, immediately before disassembly begins, is ∼9 µm, whereas the hermaphrodite anal depressor around the same developmental stage is ∼14 µm (Chen and García, 2015). However, by the time the males are adults, no dorsal-ventral sarcomere should be present (Fig. 3A), meaning that any remaining dorsal-ventral sarcomere indicates incomplete disassembly. Mutant males could have minor defects, where most of the dorsal-ventral sarcomere disassembles but some remained (Fig. 3B), to more severe defects, where a significant portion of the dorsal-ventral sarcomere did not disassemble (Fig. 3C), and finally to a complete lack of disassembly (Fig. 3D). We counted the number of males with normal and defective sarcomeres (Fig. 3E). Any male that had even minor defects was considered mutant. In addition, we also measured the dorsal-ventral sarcomere width (Fig. 3F), and reported that measurement, where necessary, to highlight differences in severity between mutants.
Fig. 3.
WNT mutant anal depressor defects. (A-D) Fluorescent image of YFP:actin in the wild-type (A), egl-20(lf) (B), lin-44(lf) (C) and lin-17 (D) anal depressors. The arrow in B indicates dorsal-ventral sarcomere width measured in F,I,J and L. (E) Graph depicting the percentage of males with normal sarcomeres in the indicated mutants. Numbers above the bars give the percentage and numbers above the x-axis represent the total sample number for each line. ***P<0.0001, Fisher's exact test, compared with wild type. (F) Graph depicting the severity of the remodeling defects. Each circle represents the dorsal/ventral sarcomere width of either the left or right dorsal attachment in one male (see arrow in B). Data are mean±s.e.m. ***P<0.0001, Mann–Whitney t-test compared with wild type. ^P<0.05, ^^P<0.005, Mann–Whitney t-test compared with lin-44(lf);egl-20(lf). ++P<0.005, Mann–Whitney t-test compared with lin-17(lf). n.s., not significant. (G,H) Fluorescent images of YFP expressed from the lin-17 promoter. The anal depressor is at the center of the image. (I,J) Left (L) and right (R) dorsal-ventral sarcomere width in adult anal depressors. Each circle represents the width of the sarcomere on the indicated side of an individual animal. ***P<0.0001, Mann–Whitney t-test. (K) Graph depicting the percentage of males with normal sarcomeres. The x-axis indicates where wild-type lin-17 cDNA is expressed. The numbers on top of the bars represent the percentages. The number below the x-axis represents the total sample number for each location. *P<0.05, **P<0.005, Fisher's exact test, compared with no rescue. (L) lin-44(lf) anal depressor defects are rescued with a transgene. Each circle represents the dorsal/ventral sarcomere width of either the left or right dorsal attachment in one male (see arrow in C). Data are mean±s.e.m. **P<0.005, ***P<0.0001, Mann–Whitney t-test. For all images, dorsal is up and anterior is to the left. Scale bars: 10 µm.
Three WNT ligands are expressed in the tail: LIN-44, EGL-20, and CWN-1 (Pan et al., 2006; Whangbo and Kenyon, 1999; Herman et al., 1995). Whereas cwn-1 loss-of-function (lf) mutant males showed occasional minor defects, defects in lin-44(lf) and egl-20(lf) mutant males were more severe (Fig. 3E). Some sarcomere disassembly occured but was not always completed, and the anterior-posterior sarcomere was formed but was not always wild type (Fig. 3B,C). Thus, lin-44 and egl-20-encoded ligands are probably the primary drivers of WNT-regulated anal depressor rearrangement in the developing male tail.
WNT ligands EGL-20 and LIN-44 both activate the frizzled receptor, which is encoded by lin-17 (Sawa et al., 1996). LIN-17 functions in early male tail development (Wu and Herman, 2007, 2006), therefore it might be involved in anal depressor rearrangement. When we expressed YFP:actin in lin-17(lf) mutants, we found that they displayed more severe defects than any of the WNT ligands alone (Fig. 3D,F). In many males, no disassembly occurred, and the anterior-posterior sarcomere was never formed (Fig. 3D,F). Thus, lin-17 is a WNT receptor involved in anal depressor rearrangement. As the mutant frizzled receptor phenotype is more severe than lin-44(lf) or egl-20(lf), we created a double WNT ligand mutant and observed that lin-44(lf); egl-20(lf) males displayed similar defects to lin-17(lf) males (Fig. 3F). Any differences in disassembly defects between the lin-44(lf); egl-20(lf) double mutant and lin-17(lf) single mutant might be accounted for by minor compensation from cwn-1. Thus, LIN-44 and EGL-20 promote sarcomere disassembly through LIN-17.
We examined the location of lin-17 expression using the lin-17 promoter to drive YFP. We determined that lin-17 was expressed in both hermaphrodite and male anal depressors from an early stage (Fig. 3G,H, Fig. S3A). Because of its expression pattern, we investigated whether lin-17 also plays a role in the hermaphrodite anal depressor. We measured the dorsal-ventral sarcomere width of wild-type and mutant hermaphrodites, and determined that lin-17(lf) mutant hermaphrodites lacked anal depressor symmetry (Fig. 3I); this asymmetry was also seen in males (Fig. 3J). Thus, lin-17 is also required in both sexes for maintaining anal depressor symmetry during development.
To confirm that a reduction in the number of of frizzled/LIN-17 receptors leads to the defective anal depressor, we generated a lin-17 cDNA rescuing construct. We used the endogenous lin-17 promoter to drive expression in male tail cells, including the anal depressor and the epithelial cells (the B cell and its descendants) that the muscle attaches to as it remodels (Sawa et al., 1996). These B-derived cells produce the male copulatory spicules, associated neurons and the proctodeum. Some of these cells also provide the substrate to which the copulatory muscles, including the anal depressor, attach (Sulston et al., 1980). We injected the Plin-17:lin-17 construct into lin-17(lf) mutant worms and rescued the anal depressor defects (Fig. 3K, Fig. S3B), confirming that LIN-17 is involved in male anal depressor rearrangement.
We next investigated where in the male tail lin-17 expression was important for sarcomere disassembly. To address this question, we expressed lin-17 in a variety of tissues in the lin-17(lf) mutant male tail, including the sex muscles (using the Punc-103E promoter), the anal depressor (using the Paex-2 promoter) and the rectal epithelial cells that neighbor the anal depressor (using the Ppop-1 promoter). At the L1 stage, the pop-1 promoter expresses in the B cell, its descendants B.a and, to a lesser extent, B.p (Fig. S3C-G) (Wu and Herman, 2006); the pop-1 promoter does not drive expression in the anal depressor. Expressing lin-17 in the sex muscles or anal depressor had no impact on the lin-17(lf) phenotype (Fig. 3K, Fig. S3B). However, expressing lin-17 in the rectal epithelial B-cell descendants did reduce the severity of the defect severity but not the penetrance (Fig. 3K, Fig. S3B). As restoring lin-17 in the rectal epithelial cells partially rescued the anal depressor defect, we checked whether co-expressing lin-17 in multiple locations could improve rescue. Indeed, we found that expressing lin-17 in both the rectal epithelial cells and the anal depressor of lin-17 mutants produced a better rescue than either one alone (Fig. 3K). This result indicates that lin-17 functions both in the anal depressor and the neighboring rectal epithelial cells to promote sarcomere disassembly.
We suggest that the reason the Ppop-1:lin-17 transgene can rescue the lin-17(lf) disassembly defects is due to it restoring the patterning of the B cell descendants. When these epithelial cells are patterned correctly, they can provide the proper structural substrate necessary for the anal depressor to rearrange. The B cell does not divide in hermaphrodites and this is the reason that their anal depressor does not remodel. In males, LIN-17 is required for early asymmetrical cell divisions that establish the fates of these epithelial cells (Wu and Herman, 2007, 2006). The epithelial cells are all descendants of the B cell, a large cell in the male tail during the L1 developmental stage (Fig. S3C-E). This cell divides into anterior and posterior cells (B.a and B.p), with B.a being larger than B.p. This asymmetry is dependent on lin-17 (Wu and Herman, 2006). Following this cell division, the anal depressor is located over B.p (Fig. S3E,H) (Barr et al., 2018). During the L2 and L3 stages, the B.p cell divides further, with the anal depressor eventually maintaining contact with the B.paa and B.pap descendants (Barr et al., 2018) (Fig. S3I).
To further support a role for the B cell descendants in anal depressor rearrangement, we laser-ablated the B cell descendants that directly contact the anal depressor (B.pa or B.paa and B.pap) in wild-type larval males. Although these B cell descendants should be patterned by the time of ablation, the anal depressor in a significant number of operated males failed to disassemble (Fig. S3J). We could not analyze the results of operations conducted at an earlier developmental time, as ablating the B or B.p cells results in the intestine herniating through the rectum. Nonetheless, our results indicate a role for the B cell descendants in anal depressor remodeling.
Previous work established that LIN-17 on the B cell descendant membranes was stimulated by the WNT ligand LIN-44 (Wu and Herman, 2006). Our analysis of lin-44(lf) mutants indicated that the ligand promotes proper anal depressor rearrangement (Fig. 3E,F). To confirm this result, in lin-44(lf) mutants we rescued lin-44 expression using the genomic sequence and its own promoter. This construct was able to significantly reduce the sarcomere rearrangement defect (Fig. 3L). However, although the defects were not as severe, they were still present. This was probably a result of either mosaicism of the transgene or over-expression of WNT. Both over-expression and reduction of WNT signaling are capable of causing mutant phenotypes (Kirszenblat et al., 2011; Pénigault and Félix, 2011; Pan et al., 2006).
Cell-autonomous membrane-tethered egl-20/WNT can promote sarcomere disassembly
Previous work established that WNT/LIN-44 activates frizzled/LIN-17 on the developing posterior rectal epithelial cells to establish male-specific structures (Wu and Herman, 2006). However, as lin-44(lf) mutant defects are not as severe as lin-17(lf) mutant defects, an additional WNT ligand must also function in the male tail. The other WNT ligand is EGL-20, as the severity of the lin-44(lf); egl-20(lf) double mutant is similar to that of the lin-17(lf) mutant. Additionally, we showed that the lin-17(lf) muscle remodeling defect is best rescued through expression of wild-type lin-17 in the epithelial cells and the anal depressor. As LIN-44 is the WNT ligand that specifies B-cell descendant asymmetry, EGL-20 might be the WNT ligand acting on the anal depressor.
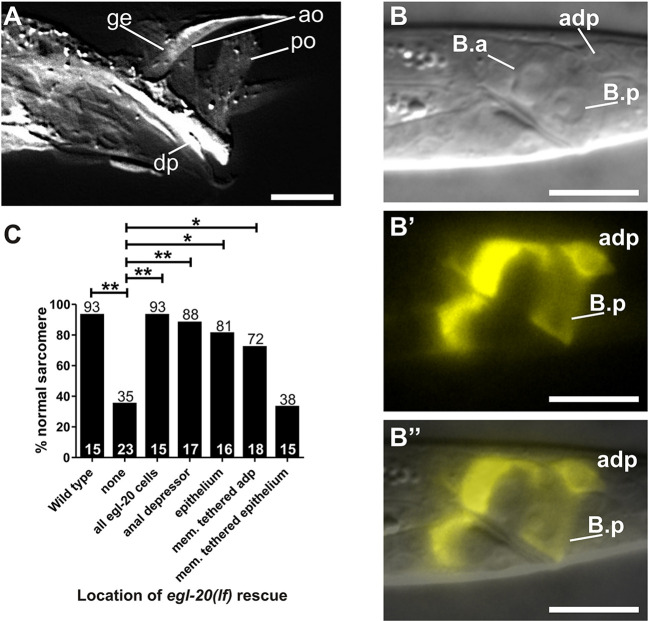
To determine where egl-20 is functionally expressed, we made a reporter construct containing the genomic egl-20 locus that starts at the ATG, ends after the 3′ UTR, and contains an SL2 trans-spliced red fluorescent protein (RFP). Expression of egl-20 occurs in the anal depressor, the rectal epithelial cells neighboring the anal depressor and the sex muscles in the male tail (Harterink et al., 2011; Whangbo and Kenyon, 1999) (Fig. 4A-B″). We placed the egl-20 promoter in front of our egl-20 genomic construct and injected this DNA into egl-20(lf) mutants; this DNA successfully rescued the disassembly defects (Fig. 4C). Expressing egl-20 in either the anal depressor (via the aex-2 promoter) or the rectal epithelial cells (via the pop-1 promoter) also rescued the egl-20(lf) phenotype (Fig. 4C). Thus, egl-20 can be provided by multiple male tail cells to regulate anal depressor remodeling.
Fig. 4.
WNT/EGL-20 ligand functions in the tail to regulate anal depressor rearrangement. (A-B″) egl-20 expression pattern in males. (A) Confocal DsRed1-E5 fluorescent image of an adult male. DIC L2 male (B) and egl-20-expressed RFP fluorescent images (B′) are shown separately. B″ shows these images stacked on top of one another, to highlight where egl-20 is expressed. (C) Graph depicting the percentage (numbers on top of the basr) of males with normal sarcomeres. The x-axis indicates where wild-type egl-20 genomic DNA is expressed. The numbers above the x-axis represent the total sample number for each location. *P<0.05, **P<0.005, Fisher's exact test. For all images, dorsal is up and anterior is to the left. adp, anal depressor; ge, gubernaculum erector; ao, anterior oblique; po, posterior oblique; dp, dorsal protractor. Scale bars: 10 µm.
WNT is primarily known to function as a secreted ligand (Pani and Goldstein, 2018); however, WNT can rescue many WNT-deficient defects as a membrane-tethered ligand (Alexandre et al., 2014). To determine whether WNT has to be secreted or can act as a membrane-tethered ligand in anal depressor remodeling, we fused a membrane-spanning domain of PAT-3 integrin to the C-terminus of EGL-20 (Gettner et al., 1995). We then expressed this construct in the anal depressor (via the aex-2 promoter) of the egl-20(lf) mutants and determined that it is capable of rescuing disassembly defects (Fig. 4C). Expressing membrane-tethered WNT in the adjacent epithelial cells (via the pop-1 promoter) failed to rescue these defects (Fig. 4C). Therefore, although secreted WNT from multiple sources can promote sarcomere remodeling, rescue of the egl-20(lf) phenotype by membrane-tethered WNT can only be accomplished if it is expressed in the anal depressor itself. Although this experiment cannot address how WNT functions normally (as a secreted ligand, membrane-tethered ligand or both), the failure of epithelial-expressed membrane-tethered WNT to rescue egl-20(lf) defects suggests that WNT must be in a form that can be internalized in the anal depressor. Unlike the membrane-tethered anal depressor WNT, which can bind to its cell-autonomous receptor and be internalized, anchoring WNT to the surrounding cells probably obstructs its proper interaction with the muscle-expressed LIN-17 and subsequent internalization.
WNT/β-catenin pathways are involved in anal depressor rearrangement
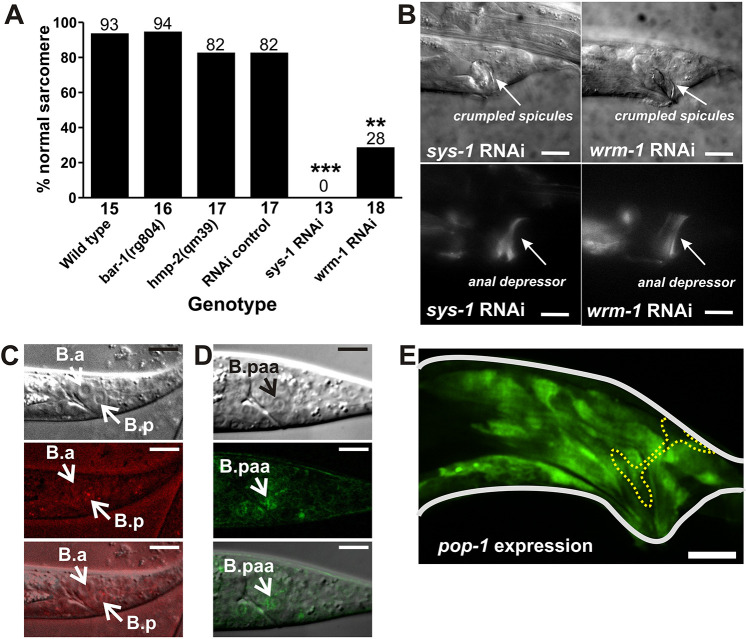
Canonical WNT signaling through β-catenin is involved in many C. elegans developmental processes (Sawa and Korswagen, 2013). Thus, we investigated whether β-catenin participates in anal depressor remodeling. C. elegans has four β-catenin genes: bar-1, hmp-2, sys-1 and wrm-1. Owing to the bar-1 locus being on the same chromosome (X) as our integrated Paex-2:YFP:actin reporter construct, we could not easily use canonical bar-1 mutations. Instead, we used CRISPR/Cas9 to generate a new bar-1 allele, rg804, in our integrated Paex-2:YFP:actin background. This allele has a 4 bp deletion that results in the loss of a lysine at amino acid 45, creating a frame shift that leads to a stop codon 15 amino acids downstream of the mutation. This allele produces defects associated with bar-1 loss-of-function mutants in hermaphrodites, such as uncoordinated movement and reduced egg-laying behavior. Despite this, bar-1(rg804), as well as a mutation in hmp-2, did not cause any anal depressor defects (Fig. 5A).
Fig. 5.
β-Catenin functions in cells outside the anal depressor to regulate disassembly. (A) Graph depicting the percentage of males with normal sarcomeres (complete dorsal-ventral disassembly). Numbers above the bars indicate the percentage and the numbers below the x-axis represent the total sample number for each genotype. **P<0.005, ***P<0.0001, Fisher's exact test. (B) DIC (top) and fluorescent (bottom) images of YFP:actin-expressing male tails developed in sys-1 and wrm-1 RNAi-expressing bacteria. Arrows indicate the spicules (DIC images) or the anal depressor (fluorescent images). (C) DIC (top) and fluorescent (middle) images of SYS-1:mcherry in the nucleus of B.p of a late L1 male. Bottom image is the top and middle images combined. Arrows indicate B.a and B.p. (D) DIC (top) and fluorescent (middle) images of WRM-1:GFP in the B.paa cell (arrow) of a L3 male. Bottom image is the top and middle images combined. (E) Fluorescent image of YFP expressed by the pop-1 promoter. Adult male tail outlined in white. Anal depressor outlined by a yellow dashed line. Scale bars: 10 µm.
WRM-1 and SYS-1 are involved in the regulation of daughter cell asymmetry during embryonic and larval cell division (Sawa and Korswagen, 2013). We used the RNAi-feeding method to knock down sys-1 or wrm-1 expression in developing L1 to adult males. RNAi of these genes individually induced major spicule developmental defects and interfered with normal anal depressor rearrangement (Fig. 5A,B). We then investigated whether sys-1 and wrm-1 are expressed in the anal depressor during rearrangement using direct protein fusions with mcherry and GFP, respectively (Baldwin and Phillips, 2014; Takeshita and Sawa, 2005). We did not find expression in the anal depressor, suggesting that β-catenin does not function in this cell. However, we did observe nuclear localization of SYS-1 in B.p after the first asymmetric B cell division in late L1 (Fig. 5C), suggesting this β-catenin is important in patterning the B cell descendants. Additionally, we observed WRM-1 expression and nuclear localization in the B.paa cell, which is produced through two divisions of B.p (Fig. 5D, Fig. S3I). Previous research established a role for sys-1, but not for wrm-1, in asymmetric B cell division (Wu and Herman, 2006). However, our observations suggest that these β-catenins could have roles in establishing cell differences during development.
Consistent with the idea that sys-1 and wrm-1 function in the B cell descendants and not the anal depressor, is the observation that the downstream effector of β-catenin TCF/LEF, encoded by pop-1, is not expressed in the anal depressor (Fig. 5E, Fig. S3F,G). Taken together, our results suggest SYS-1 and WRM-1 act with LIN-17 in the B cell descendants to promote anal depressor remodeling.
Calcium WNT pathways are involved in anal depressor rearrangement
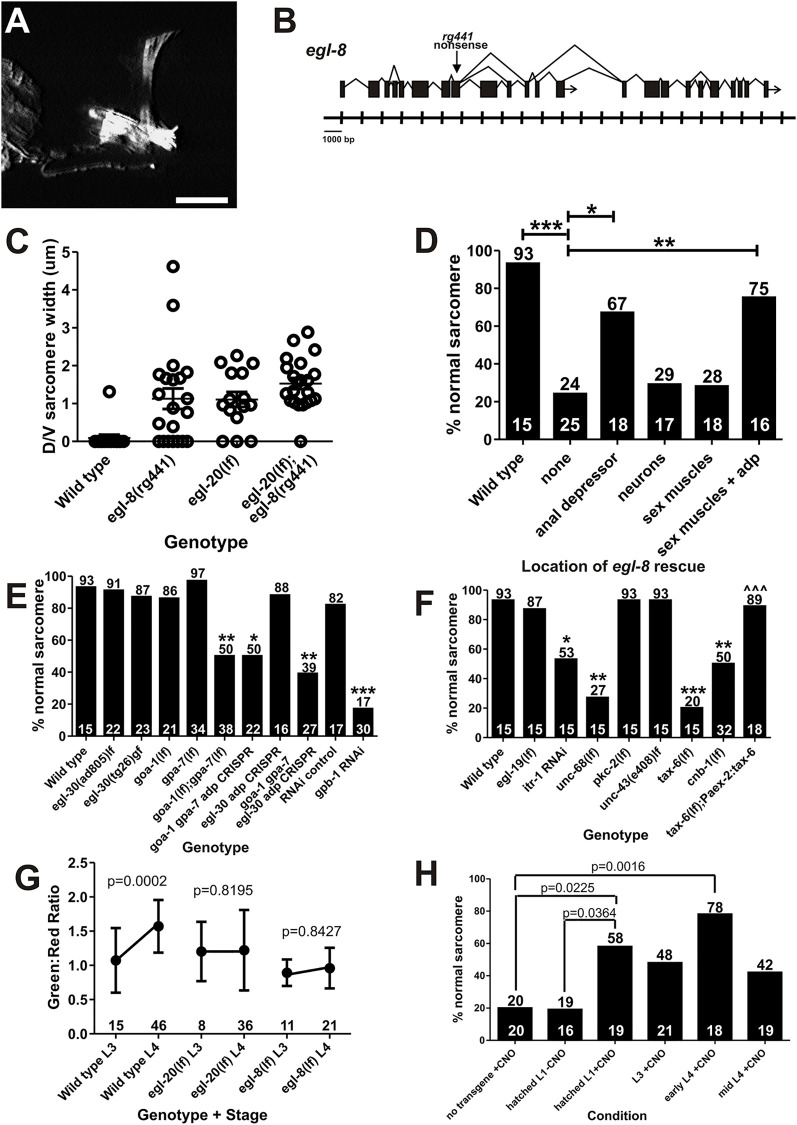
We performed a mutant screen to identify additional signaling components that might participate with WNT-mediated remodeling. We exposed worms to ethyl methanesulfonate (EMS) and screened their progeny for sarcomere rearrangement defects that resemble the egl-20(lf) phenotype; consequently, we identified the mutant rg441 (Fig. 6A). Using single-nucleotide polymorphism (SNP) mapping and whole-genome sequencing, we identified a nonsense mutation in the middle of PLC-β/egl-8 (Fig. 6B) (Wicks et al., 2001; Lackner et al., 1999; Miller et al., 1999). PLC-β (phospholipase C-β) cleaves PIP2 into IP3 and DAG, resulting in Ca2+ release from the IP3 receptor (Walker et al., 2009; Gower et al., 2005). Although PLC has not been shown to be associated with WNT signaling in C. elegans, it is downstream of WNT signaling in vertebrate systems (McQuate et al., 2017).
Fig. 6.
PLC-β/EGL-8 promotes calcium release from internal stores to regulate anal depressor sarcomere disassembly. (A) Fluorescent image of egl-8 mutant adult male anal depressor expressing YFP:actin. Dorsal is up and anterior is to the left. Scale bar: 10 µm. (B) Genomic structure and splice variants of egl-8 with the location of the rg441 allele in exon 8 indicated by the downward pointing arrow. Tick marks denote DNA base pairs in thousands. (C) Graph depicting the severity of disassembly defect. Each circle represents the dorsal-ventral sarcomere width of either the left or right dorsal attachment in one male. Data are mean±s.e.m. (D) Graph depicting the percentage (numbers on top of bars) of males with normal sarcomeres. The x-axis indicates where the wild-type egl-8 minigene is expressed. adp, anal depressor. The numbers above the x-axis represent the total sample number for each location. *P<0.05, **P<0.005, ***P<0.0005, Fisher's exact test. (E,F) Graphs depicting the percentage (numbers on top of bars) of males with normal sarcomeres. The numbers above the x-axis represent the total sample number for each genotype/stage. *P<0.05, **P<0.005, ***P<0.0001, Fisher's exact test, compared with wild type. itr-1 RNAi and pkc-2(lf) were scored using an Punc-103E:YFP:actin expressing transgene (Chen and García, 2015). (G) Calcium transients in the anal depressor monitored by measuring changes in G-CaMP fluorescence (green) normalized to an unchanging RFP control (red). Each dot indicates a time point where males were averaged±s.d. The x-axis represents genotype and developmental stage. The numbers above the x-axis represent the total sample number for each genotype/stage. P-values were determined using the Mann-Whitney t-test. (H) Percentage of adult ceDREADD-expressing egl-20(lf) males with a fully remodeled anal depressor. Males other than the ‘no transgene +CNO’ control have Paex-2:ceDREADD expressed from a transgene. x-axis indicates the developmental stage during which the males were first exposed to CNO. The numbers on top of the bars indicate the percentage of adult males with a normal sarcomere. The numbers above the x-axis indicate the total sample number for each condition. P-values were determined using Fisher's exact test.
To determine whether egl-8 acts with WNT signaling in the C. elegans male, we made a double mutant with the WNT ligand egl-20. The egl-8 and egl-20 mutants share additional phenotypes, including egg retention and the inability to sire progeny. If egl-8 acts in an independent genetic pathway from egl-20, the severity of the double mutant phenotype should be greater than either single mutant. However, if they promote the same remodeling process, the double mutant phenotype should be similar to either single mutant. We found no statistical difference in the spectrum of sarcomere disassembly defects between single mutants and double mutants (Fig. 6C). This suggests that egl-8 and egl-20 participate in the same process. However, compared with the single mutants, fewer adult double mutant males had wild-type anal depressors (Fig. S4A). As EGL-8 has roles in other pathways, we do not rule out the possibility that EGL-8 also provides parallel WNT-independent contributions to anal depressor remodeling.
We next checked where PLC-β/egl-8 functions to regulate sarcomere rearrangement. The egl-8 expression pattern in C. elegans males includes the sex muscles, ray neurons, additional neurons in the tail and the anal depressor (Gower et al., 2005). We created an egl-8 minigene using egl-8 cDNA for exons 1-14 fused to genomic DNA for the remainder of the gene. Because the normal expression pattern of egl-8 is determined by introns between exons 1-8 (Gower et al., 2005), we did not use its own promoter to drive expression. Instead, we used a series of tissue-specific promoters, including the aex-2 promoter (anal depressor), the rab-3 promoter (pan-neuronal), the hlh-8 promoter [early M cell, from which sex muscles are descended (Harfe et al., 1998)] and the unc-103E promoter (sex muscles and anal depressor). We found that expressing egl-8 in neurons or sex muscles did not rescue egl-8(lf) defects (Fig. 6D, Fig. S4B). However, expressing egl-8 in either the sex muscles and anal depressor, or the anal depressor alone, rescued the sarcomere disassembly defects (Fig. 6D, Fig. S4B). Thus, EGL-8 functions in the anal depressor to regulate sarcomere disassembly during male development.
In C. elegans, egl-8 is an effector of Gαq/EGL-30 (Xu and Chisholm, 2011; Lackner et al., 1999; Miller et al., 1999). We analyzed mutant males containing both loss- and gain-of-function mutations in egl-30 and did not find any anal depressor defects (Fig. 6E). Thus, for anal depressor rearrangement, Gαq is either not required or functions redundantly with other Gα proteins.
We examined which other Gα proteins could be upstream of PLC-β. In the Drosophila WNT/Ca2+ pathway, the frizzled receptor is genetically upstream of Gαo (Katanaev et al., 2005). In C. elegans, Gαo is encoded by the broadly expressed goa-1 (Mendel et al., 1995). Another possibility is gpa-7, a Gα present in the anal depressor (Hunt-Newbury et al., 2007). Similar to our egl-30 mutant analysis, we did not find anal depressor defects in either goa-1(lf) or gpa-7(lf) single mutants (Fig. 6E). However, when we generated the goa-1(lf); gpa-7(lf) double mutant, we found that 50% of males had remodeling defects (Fig. 6E, Fig. S4C). In addition, some males also had an abnormal tail morphology (Fig. S4C). Thus, these Gα proteins redundantly promote anal depressor remodeling.
If the Gα proteins are functioning upstream of PLC-β/egl-8, they should also be functioning in the anal depressor. We attempted to address this issue by rescuing goa-1 in the goa-1(lf); gpa-7(lf) strain but the double mutant produces too few viable progeny for maintaining transgenic lines. To address where these proteins function, we used tissue-specific CRISPR/Cas9. We designed guide RNAs to specific G-protein components and expressed Cas9 in the enteric muscles, using the aex-2 promoter (Dickinson et al., 2013; Mahoney et al., 2008). We then expressed our tissue-specific CRISPR/Cas9 constructs in wild-type Paex-2:YFP:actin-expressing animals. Tissue-specific CRISPR/Cas9 should disrupt the gene only in the specific cells of interest. If males display mutant anal depressors, we can conclude that these cells use the wild-type genes for muscle remodeling.
We first checked whether our tissue-specific CRISPR/Cas9 Gα protein constructs were working by analyzing the goa-1; gpa-7 combination. Similar in proportion to the genetic goa-1(lf); gpa-7(lf) double mutants, the goa-1; gpa-7 anal depressor-targeted CRISPR/Cas9 animals displayed defective anal depressor remodeling (Fig. 6E), consistent with a cell-autonomous site of action. We then investigated whether egl-30 might act redundantly with goa-1 and gpa-7 to promote remodeling. Triple mutants of goa-1, gpa-7 and egl-30 were not viable, so we generated egl-30 single- and goa-1; gpa-7; egl-30 triple tissue-specific CRISPR/Cas9 lines. However, we found that males containing egl-30 single CRISPR/Cas9 transgenes were wild type, and males containing the guide RNAs to the three G-proteins had a phenotype that was no more severe than that in males containing tissue specific goa-1; gpa-7 CRISPR/Cas9 transgenes (Fig. 6E). In contrast to the canonical pathway in other C. elegans tissues, there is a lack of evidence that Gαq/egl-30 is upstream of PLC-β/egl-8 signaling in male anal depressor remodeling (Xu and Chisholm, 2011; Lackner et al., 1999; Miller et al., 1999); this could be because of incomplete penetrance of the CRISPR constructs or because egl-30 is not involved in the developmental process.
In addition to Gα proteins, PLC-β can be activated by the βγ subunits (Park et al., 1993). Using RNAi, we knocked down the expression of C. elegans Gβ gpb-1 to examine whether perturbing other G protein members can disrupt anal depressor remodeling. Males grown with gpb-1 RNAi were severely defective in terms of anal depressor remodeling during development (Fig. 6E, Fig. S4C). Therefore, functioning heterotrimeric complexes of Gβγ and GOA-1 or GPA-7 are needed to promote sarcomere disassembly.
The calcium released as a result of PLC-β/EGL-8 activation could come from internal or external stores. To determine the calcium source, we examined mutant worms defective in the cell membrane-located L-type voltage-gated Ca2+ channel EGL-19, and the ER-located IP3 receptor ITR-1 and ryanodine receptor UNC-68 (Baylis et al., 1999; Lee et al., 1997; Maryon et al., 1996). We determined that, although the anal depressors of egl-19(lf) males appear normal, both itr-1 RNAi and unc-68(lf) resulted in sarcomere disassembly defects (Fig. 6F, Fig. S4E). Thus, the calcium required for sarcomere disassembly is mobilized from internal stores.
WNT-promoted calcium release activates calcium-dependent kinases, including protein kinase C (PKC) and calcium/calmodulin-dependent kinase II (CaMKII) (Kuhl et al., 2000; Sheldahl et al., 1999; Slusarski et al., 1997). We examined the sarcomere structure in C. elegans males mutant in pkc-2 (PKC) and unc-43 (CaMKII), and determined that these effectors are not part of the remodeling pathway in the anal depressor (Fig. 6F, Fig. S4E).
Another known WNT effector is calcineurin, a calcium/calmodulin-dependent phosphatase that primarily works through NFAT (Ando et al., 2016; Saneyoshi et al., 2002). Although C. elegans does not have an NFAT homolog, the worm does have two genes that encode different calcineurin subunits: the phosphatase subunit tax-6 and the regulatory subunit cnb-1 (Bandyopadhyay et al., 2004; Kuhara et al., 2002). The tax-6 gene is expressed in the anal depressor (Lee et al., 2005). We examined the anal depressor in males lacking either subunit and found that, in both cases, sarcomere rearrangement was impaired; this defect was rescued in tax-6(lf) males by enteric muscle-specific tax-6 (Fig. 6F, Fig. S4E). Therefore, one effector for the increase in calcium in the anal depressor is calcineurin.
If increasing calcium signaling results in sarcomere rearrangement through WNT signaling, then removing WNT signaling should result in decreased calcium levels in the anal depressor. To measure this, we expressed the calcium-sensing fluorescent protein G-CaMP6 in the anal depressor of wild-type, WNT/egl-20(lf) and PLC-β/egl-8(lf) mutant hermaphrodites and males (Correa et al., 2012; Tian et al., 2009; Nakai et al., 2001). We then recorded calcium changes from L2 to adult. Individual worms were placed on agar pads and the G-CaMP6 (calcium sensor) and RFP (control) fluorescent levels were measured at the developmental stage indicated (Fig. S4F,G, Fig. S5). Owing to the amount of data collected, we displayed the average and standard deviation for each sex and developmental stage (Fig. 6G, Fig. S4G). Calcium levels in the hermaphrodite anal depressor did not change during development (Fig. S4F,G). In contrast, the wild-type male showed an increase in calcium transients between L3 and L4, when rearrangement occurs (Fig. 6G, Fig. S4F,G), but not when the males were deficient in egl-20(lf) or egl-8(lf) (Fig. 6G, Fig. S5). These data support the idea that calcium release is correlated with muscle disassembly, and further indicate a role for WNT signaling through PLC-β.
Our experimental results suggest that WNT signaling works upstream of G-protein-coupled signaling to increase calcium levels in the anal depressor. Therefore, we investigated whether we could bypass the need for the WNT/EGL-20 ligand via artificially activating intracellular calcium signaling in the anal depressor. We used the C. elegans version of DREADD (ceDREADD), which is a modified gar-3-encoded G-protein-coupled receptor that is normally activated by acetylcholine. ceDREADD is stimulated by clozapine-N-oxide (CNO) (Prömel et al., 2016). The activated ceDREADD should promote intracellular calcium mobilization through signaling to PLC-β/EGL-8 but via Gαq/EGL-30 instead of GOA-1. We expressed ceDREADD in the anal depressor of egl-20(lf) mutant males using the aex-2 promoter and then exposed these males to CNO to activate G-protein-driven signaling. Neither egl-20(lf) mutant males expressing ceDREADD without the CNO nor growing egl-20(lf) mutant males on CNO rescued the anal depressor defects (Fig. 6H). In contrast, when the ceDREADD-expressing egl-20(lf) males were placed on CNO plates at different time points, including hatched L1, L3, early L4 and mid L4, they developed normal adult anal depressors. ceDREADD-expressing males exposed to CNO as early L4s, when the anal depressor rearrangement begins, showed the largest developmental response to G-protein-driven calcium increase (Fig. 6H). Therefore, in wild-type males, the start of L4 is probably when the EGL-20/WNT ligand is activating G-protein-driven calcium increase to promote sarcomere disassembly.
DISCUSSION
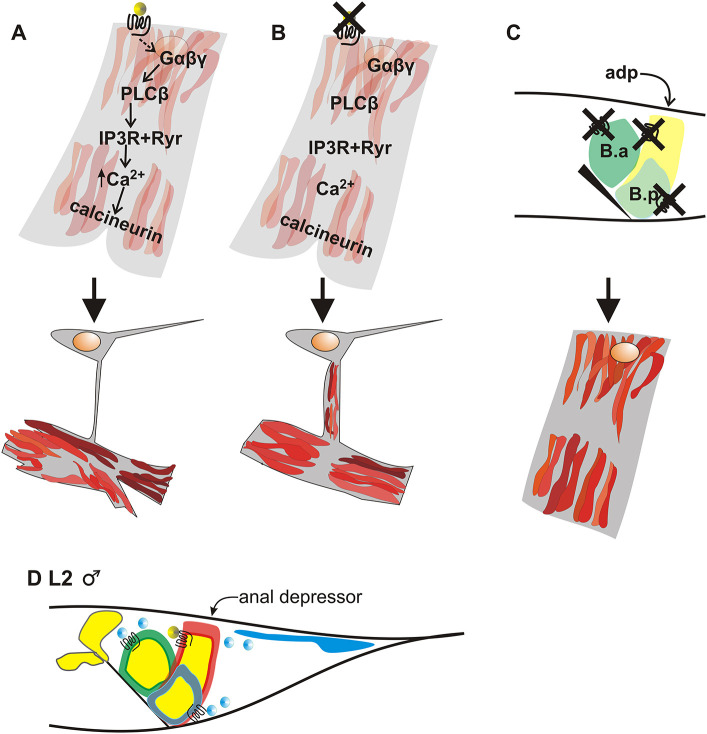
Regulation of calcium levels is important in both development and disease progression (Paudel et al., 2018). There are different types of calcium signaling in muscle; one type is the brief spikes and subsequent calcium removal that control muscle contraction, and another is sustained calcium release that promotes longer-term changes. Altering the calcium handlers, which control the contractile machinery, can activate the pathways leading to changes in sarcomere structure and potential disease states (Shimizu et al., 2017). Here, we showed how and why a muscle activates sustained calcium release to disassemble a sarcomere (Fig. 7). Determining the regulation of these pathways is vital for addressing how to compensate for their dysfunction.
Fig. 7.
A model for programmed muscle remodeling in the anal depressor. (A-C) Cartoon of cell signaling and anal depressor rearrangement. The outline of the cell is in gray. The actin component of the sarcomere is in red. The yellow circle is the WNT/EGL-20 ligand, which binds to the frizzled/LIN-17 receptor. The orange circle shows the location of the nucleus. (A) Wild-type anal depressor signaling and rearrangement. The top images represent early L4 stage and the bottom images represent the adult stage. (B) Inhibiting the WNT/Ca2+ pathway during development of the anal depressor results in incomplete sarcomere disassembly. (C) Inhibiting the WNT/Ca2+ pathway in the anal depressor (adp) and the WNT/PCP pathway of the surrounding epithelial B cell descendants (B.a and B.p) results in the adult male anal depressor resembling the larval muscle. The top image is of an L2 male tail. The black triangle depicts the anus. (D) Cartoon of an L2 male tail. adp, red outline; B.a cell, green outline; B.p cell, gray outline; WNT/LIN-44 ligand expression, blue; WNT/LIN-44 ligand, blue circles; WNT/EGL-20 expression, yellow; ligand itself, yellow circles; anus, black line.
Sustained calcium release can be a consequence of the WNT signaling pathway (McQuate et al., 2017). In the developing male anal depressor, sustained calcium release is accomplished through PLC-β (Fig. 7A). Similarly, in C. elegans adult wound closure, PLC-β is used to activate sustained calcium release, in this instance through a melastatin family (TRPM) channel and Gαq (Xu and Chisholm, 2011). Wound closure also requires cytoskeleton remodeling, indicating that sustained calcium release controlled by PLC-β is a preferred method to regulate programmed remodeling events.
Excitable cells need to separate sustained calcium release from the transient Ca2+ spikes, which are the basis for muscle contraction. In muscle, one enzyme known for responding to sustained but not transient calcium is the calcium/calmodulin-dependent serine/threonine phosphatase calcineurin (Dolmetsch et al., 1997). We showed that calcineurin is an important part of the signal transduction pathway activated by calcium levels in the anal depressor (Fig. 7A). Similar to our study, work in mice kidneys showed that calcineurin is activated via WNT signaling (independent of β-catenin), whereas CaMKII is not (Ando et al., 2016). Calcineurin has been reported downstream of WNT/Ca2+ signaling in other contexts, including promoting aquaporin localization to kidney cell membranes (Ando et al., 2016) and patterning in Xenopus embryos (Saneyoshi et al., 2002).
What the potential downstream targets of calcineurin are is an open question, as C. elegans lacks a common phosphatase target, the transcription factor NFAT (Graef et al., 2001). Known C. elegans targets of calcineurin include the transcription factor FOXO/daf-16, the transcriptional co-activator CRTC-1 and a gene of unknown function, cnp-1 (Tao et al., 2013; Jee et al., 2012; Mair et al., 2011). However, whether these potential targets are expressed in the anal depressor is not known. Remodeling events often include transcription regulation, therefore calcineurin targets might include molecules that regulate transcription. Additionally, CRTC-1 is important in monitoring energy status, and C. elegans might find linking the high-energy-demand remodeling process with the energy status of the organism beneficial (Burkewitz et al., 2015).
Calcineurin is also activated in response to elevated calcium levels that lead to the removal of excess neuronal synapses during development (Miller-Fleming et al., 2016). This removal might be dependent on a member of the cell death pathway, ced-4, which activates a protein that cleaves F-actin (Meng et al., 2015). Although these studies focused on neurons, C. elegans muscle might use a similar mechanism for cytoskeletal remodeling. The effectors of calcineurin in the anal depressor could include molecules involved in the disassembly process and/or transcription-associated genes that regulate genes that participate in remodeling.
Mutating components of the WNT/Ca2+ pathway in the anal depressor results in males with adult sarcomeres that were only partially disassembled during development (Fig. 7B). However, mutating components of the WNT pathway in the surrounding epithelial cells (the B cell descendants) along with components in the anal depressor results in a complete lack of larval sarcomere disassembly (Fig. 7C). Noncanonical WNT/PCP signaling is important for determining anterior-posterior cell identity, as the B cell descendants go through several rounds of asymmetrical divisions (Wu and Herman, 2006; Sulston et al., 1980). In this paper, we also show a role for β-catenin-like proteins WRM-1 and SYS-1 in B cell descendant patterning. We propose that these cell divisions result in epithelial progeny that the anal depressor uses as instructors for its rearrangement. Although the anal depressor does not rearrange until the male reaches the L4 developmental stage, the male anal depressor probably receives competence information from the B cell descendants before remodeling occurs (Fig. 7D) (Chen and García, 2015).
Drastic muscle remodeling is found in humans in disease states, such as pathological heart muscle remodeling following cardiac damage (Parra and Rothermel, 2017). These pathological models show similar molecular mechanisms to those identified in the C. elegans anal depressor. However, the anal depressor presents an advantage, as this is not a pathological state, but a normal deterministic program. We are able to discover how these pathways function under normal biological conditions, which can provide insight into how they are co-opted to produce disease states. Separately, WNT and calcineurin signaling are known to be involved in the pathological cardiac remodeling that takes place following injury, although these two signaling pathways are rarely explicitly linked in this context (Fu et al., 2019; Parra and Rothermel, 2017; Freire et al., 2014; Yoshida et al., 2004). Our work demonstrates a direct link between WNT and calcineurin signaling in programmed muscle remodeling, highlighting the need to focus on how these pathways integrate to coordinate this process. Furthermore, the calcineurin effector NFAT seems to be primarily involved in the pathological response to increased calcium signaling following cardiac injury, whereas the effectors activated as a result of physiological responses remain unknown (Zhao et al., 2019; Parra and Rothermel, 2017; Balakumar and Jagadeesh, 2010). As C. elegans does not have an NFAT homolog, and we are studying a non-pathological remodeling event, this system provides a unique opportunity to determine additional calcineurin targets, thereby separating pathological and physiological responses.
MATERIALS AND METHODS
Experimental model and subject details
Caenorhabditis elegans and bacterial strains
C. elegans strains were cultured at 20°C and manipulated following standard protocols (Brenner, 1974). Strains listed as ‘Wild type’ contained him-5(e1490) on LG V (Hodgkin et al., 1979). Bacteria strains used to feed worms include Escherichia coli OP50 for nearly all experiments and HT115(DE3) for RNAi experiments.
The egl-8(rg441) allele described in this study was isolated from an EMS screen that selected for males that have anal depressor sarcomere disassembly defects. The strain CG941, which carries the integrated transgene rgIs3 [Plev-11:RFP; Plev-11:G-CaMP], was mutagenized to generate mutant lines. The rg441 strain was identified and outcrossed four times to eliminate background mutations. Whole-genome sequencing was conducted by BGI Americas Corporation, in combination with SNP mapping (Wicks et al., 2001), to locate the rg441 allele within the egl-8 locus on LG V. The rg441 mutation changed the wild-type sequence CTTGACCAAT to the mutant sequence CTTGACTAAT (Gln to Ochre).
The bar-1(rg804) allele described in this study was isolated using CRISPR/Cas9 as described by Dickinson et al. (2013). Plasmid construction is described below. Additional alleles and strains, as well as primers, resources and reagents, used in this study are listed in Table S1.
Method details
Plasmid construction, transgenic strain and integrated line generation
Construction of Paex-2:YFP:actin integration
We removed the introns from the genomic actin sequence with YFP fused at the N terminus that we used previously in Guo et al. (2012) and Chen and García (2015) to create the plasmid pXC22. This plasmid contained an Invitrogen Gateway Reading Frame Cassette (RfC), allowing for recombination with pBL348 [plasmid contains the aex-2 promoter (LeBoeuf and Garcia, 2017)] to create pBL397. pBL397 was injected with pLR361(Pges-1:RFP) and pUC18 into N2 hermaphrodites. The ges-1 promoter drives expression in the intestine and we used it as a marker for the generation of transgenic lines (Egan et al., 1995). A transgenic line was selected for UV-irradiation-based integration (Mello and Fire, 1995) to create strain CG1640 that contains the rgIs15 integration. rgIs15 was mapped to LGX. This integration was then combined with other mutations to visualize anal depressor sarcomere morphology. ‘Wild type’ in this paper refers to strain CG1722, which is the rgIs15 integration with the him-5(e1490) mutation.
Nearly all strains in which the anal depressor sarcomere was visualized contain rgIs15. A few strains, indicated when used, contain a transgene with Punc-103E:YFP:genomic actin, which was created as described by Chen and García (2015).
Construction of Paex-2:YFP integration
The plasmid containing Paex-2 (pBL348) was recombined with pGW77YFP, which contains an RfC and YFP to generate pBL351. pBL351 was injected with pLR361(Pges-1:RFP) and pUC18 into N2 hermaphrodites. A transgenic line was selected for UV-irradiation-based integration to create strain CG1647 that contains the rgIs21 integration. rgIs21 was mapped to LGII. This integration was then combined with other mutations to visualize anal depressor morphology.
Construction of lin-17 cDNA plasmids
The full length lin-17 cDNA was PCR-amplified using primers flin-17 and lin-17r. This PCR product was ligated to linearized pBL405 (see section on construction of egl-20 plasmids for further details) using Takara Bio In-Fusion (linearizing primers: DownRmvsdha and UPremegl20) to create pBL423. This plasmid was sequenced and a point mutation was fixed using primers fpBL423a and pBL423ar to create pBL426. pBL426 was recombined with pBL348 (Paex-2), pXC11 (Plin-17) and pLR21 (Punc-103E) (Reiner et al., 2006) to create pBL428, pBL429 and pBL427, respectively. pXC11 was created using primers attB1-Plin-17-Forward and attb2-Plin-17-Reverse to amplify the 6.5 kb region upstream of the lin-17 ATG from genomic DNA, and then recombining that PCR product with pDG15 (Reiner et al., 2006). pXC11 was recombined with pGW322YFP to create pXC12 (Plin-17:YFP). The RFP in pBL426 was replaced with cyan fluorescent protein (CFP) using primers fpBL412CFP and pBL412CFPr to amplify CFP, and fpBL412 and pBL412r to linearize pBL426 minus RFP. An In-Fusion reaction combined the two PCR products to create pBL444. pBL444 was recombined with pBL455 (Ppop-1) to create pBL457. pBL455 was created using primers FPpop-1 and Ppop-1R to amplify the 2.6 kb region upstream of the pop-1 ATG from genomic DNA and then recombining that PCR product with pDG15. pBL427, pBL428, pBL429 and pBL457 were injected with pUC18 and pBX1 (pha-1) (Schnabel and Schnabel, 1990) as a marker for transgenic lines into the strain CG1721.
The pop-1 promoter expression pattern in males was determined by recombining pBL455 with pGW77YFP to create pBL462. This plasmid was injected into CG531 males with pBX1 and pUC18 to create CG1753.
Construction of lin-44 genomic plasmids
Genomic lin-44 DNA was PCR amplified using primers RVL44gnmJG44lnks and FL44pxc99lnks. This PCR product was ligated to linearized pBL405 using Takara Bio In-Fusion to create pBL407. pBL407 was recombined with pXC108 (Plin-44) to create pBL409. pXC108 was created using primers Flin-44pro and lin-44proR to amplify the 2.7 kb region upstream of the lin-44 ATG from genomic DNA and then recombining that PCR product with pDG15. pBL409 was injected with pUC18 and Pmyo-2:GFP as a marker for transgenic lines into the strain CG1692.
Construction of egl-20 plasmids
Genomic egl-20 sequence (2.3 kb) from ATG to the last stop codon was PCR amplified using the primers egl-20-coding-F and egl-20-coding-R. These two primers contain homology to the sequences that flank the G-CaMP3-coding sequence in pLR279. The primers linear-pLR306-formab-5-for and linear-pLR306-formab-5re were used to linearize pLR279 and remove the G-CaMP-coding sequence. The two primers contain homology to the 3′ and 5′ sequence of the egl-20 genomic sequence, respectively. The egl-20 genomic sequence was ligated into the SL2:DsRed plasmid backbone using In-Fusion to make pXC99 (the gateway ccdB cassette:egl-20:SL2:RFP). A loxP site was added in front of the egl-20 ATG using primers UpForloxPegl20 and RevLOXPegl20 to PCR amplify pXC99. The primers contain the loxP site; after amplification, the original plasmid was removed by DpnI digestion, leaving only the amplified plasmid with loxP sites, which was ligated using NEB Quick Ligase. This process created plasmid pBL404. The process was repeated using primers Fordwnxc99loxP and Revdwnxc99loxP to add loxP sites after RFP to create plasmid pBL405.
We then added the egl-20 3′UTR to the plasmid. The plasmid itself was amplified using primers fpBL405sl2 and pBL405egl-20r. The egl-20 3′UTR was amplified using primers Fegl-20 3′UTR and Egl-20 3′UTRr. These PCR products were ligated together using In-Fusion to create plasmid pBL412 (RfC:loxP:egl-20:3′UTR:SL2:RFP:loxP). This plasmid was recombined with pXC33 (Pegl-20), pBL348 (Paex-2) and pXC108 (Plin-44) to create pBL415, pBL416 and pBL422, respectively. These plasmids were injected along with pUC18 and Pmyo-2:GFP into the strain CG1673 to generate transgenic lines.
pXC33 contains the 7 kb promoter region of egl-20 and was cloned using primers Pegl-20-For-1 and Pegl-20-Re-1-imme-beforeATG recombined with pDG15 (Whangbo and Kenyon, 1999). We created a plasmid to determine egl-20 expression in males by recombining pXC33 with pLR186 (RfC:DsRed1-E5) (LeBoeuf et al., 2011) to make pXC35. pXC35 was injected into him-5(e1490) hermaphrodites with Pmyo-2:GFP as an injection marker. A transgenic line was selected for UV-irradiation-based integration to create strain CG1656 that contains the rgIs23 integration. rgIs23 was mapped to LGV. This integration was then used to visualize egl-20 expression patterns in males.
We replaced the RFP in pBL412 with CFP using primers fPBL412 and pBL412r to amplify pBL412 without RFP, and amplified CFP using primers fpBL412CFP and pBL412CFPr. We ligated the two PCR products together using In-Fusion to create plasmid pBL436. pBL426 was recombined with pBL455 (Ppop-1) to create plasmid pBL476. pBL476 was injected with pUC18 and Pmyo-1:GFP into the strain CG1673 to generate the transgenic line CG1762.
Genomic egl-20 sequence (2.3 kb) from ATG to the codon before the stop codon was PCR-amplified using primers Fegl-20pPD122 and egl-20pPD122R. The primers contained homology for pPD122-39, which contains two transmembrane domains from pat-3 integrin tagged with YFP (Gettner et al., 1995). pPD122-39 was amplified using primers FpPD122-39 and pPD122-39R. The two PCR products were combined using In-Fusion to create plasmid pBL481. We then added a Gateway Reading Frame Cassette (RfC) to pBL481 using primers pBL481f and RpBL481 to amplify the plasmid and primers ForRfC and RfcRev to amplify the RfC. We ligated the two PCR products together to create plasmid pBL484, which we recombined with pBL455 (Ppop-1) and pBL348 (Paex-2) to create pBL486 and pBL487. pBL486 and pBL487 were injected along with pUC18 and Pmyo-2:GFP into strain CG1652 to create strains CG1812 and CG1801, respectively.
Construction of the egl-8 minigene
Owing to the size of the egl-8 locus (23 kb), a minigene was created. Primers Fegl-8atg and egl-8midR were used to PCR amplify the first half of the egl-8 cDNA. This product was ligated to plasmid pBL405 (ccdB:loxP:egl-20:SL2:RFP:loxP) that had been linearized using primers DownRmvsdha and UPremegl20 to remove egl-20 and add egl-8 in its place using an In-Fusion Dry-Down PCR Cloning Kit to create plasmid pBL424. Next, the second half of the egl-8 cDNA was amplified from pXC78, which contains egl-8 genomic sequences. pXC78 was constructed using primers e8-12-16ex-2nd-for and e8-12-16ex-2nd-re to amplify the egl-8 genomic region from exon 12 to exon 16. This PCR product was recombined with pDG15 using BP clonase to make pXC53. We then cloned the genomic region from exon 17 to the genomic sequence that is 3.6 kb downstream of egl-8 using the primers egl-8-17th-down-for and egl-8-17th-down-re. The primers contain homology to the 3′ end of exon 16. pXC53 was linearized using primers lin-e8-12-16pdg15-f17-d-for and lin-e8-12-16pdg15-f17-d-re. The PCR product of exon 17 to the downstream sequence was ligated into pXC53 using In-Fusion to make pXC54 (egl-8 genomic sequence from exon 12 to the 3.6 kb sequence downstream of egl-8). The insertion mutation contained within pXC54 was corrected by single-site mutagenesis to make pXC78 (egl-8 genomic sequence from exon 12 to the 3.6 kb sequence downstream of egl-8 with the mutation corrected).
From pXC78, egl-8 was PCR-amplified using primers Fegl-8exon14 and Egl-8 3UTR. This was ligated using an In-Fusion reaction with linearized pBL424, linearized using primers DownRmvsdha and pBL424r, to create plasmid pBL425. pBL425 was sequenced, revealing two point mutations in the genomic region of egl-8. These two mutations were fixed using primers (Finfegl-8ex14 and Infegl-8ex17R) that did not contain the point mutations to amplify a PCR product that was then ligated using In-Fusion to pBL425 linearized using primers pBL425egl-8r and FpBL425egl-8 to create plasmid pBL432. pBL432 was sequenced and found to contain a mutation in one exon. This mutation was fixed by amplifying the area containing it with the primers FEgl-8exon15a and egl-8exon20R and then combined using In-Fusion with a linearized pBL432 using primers FEgl-8mini and egl-8miniR to create pBL447. pBL447 contains the egl-8 minigene with no mutations.
To create egl-8 expression constructs, pBL447 was recombined using LR Clonase with pBL348 (Paex-2), pBL363 [Prab-3 (LeBoeuf and Garcia, 2017)] and pBL458 [Punc-103E (smaller version)] to create plasmids pBL448, pBL449 and pBL459, respectively. These plasmids were injected along with pUC18 and Pmyo-2:GFP into the strain CG1697 to create transgenic lines.
pBL458 contains a shorter version of Punc-103E that was originally published by Reiner et al. (2006). The original Punc-103E, at 5.4 kb, was deemed too large to work with the 16.2 kb pBL447. Since the publication by Reiner et al. (2006), another unc-103 exon 1 was identified within the original Punc-103E (LeBoeuf and Garcia, 2017). This exon and promoter were removed to create a shorter version (2.4 kb) of Punc-103E using primers FPunc-103E-J and Punc-103E-JR. This promoter was recombined with pDONR using BP clonase to create pBL458.
As the hlh-8 promoter is too large to put in front of the egl-8 minigene, the pXC27 plasmid (Chen and García, 2015) containing Phlh-8 was co-injected with pBL447. Functional transgenes were confirmed by fluorescent imaging of the RFP tag.
Construction of the tax-6 genomic DNA plasmid
The full-length genomic DNA of tax-6 was PCR amplified from purified C. elegans genomic DNA using primers Ftax-6lowcopy and tax-6R. In-Fusion was used to combine this PCR product with pBL436 linearized with primers FCFPATG and RpBL436wRfCB to create plasmid pWD5(RfC:tax-6:CFP). There were two mutations in tax-6, one in exon 6 and one in exon 11. We used primers that fixed these mutations, Ftax-6ex6 and tax-6ex11R, to create a 3665 bp PCR product. We used primers FpWD5 and pWD5R to amplify the rest of the plasmid. We then used In-Fusion to put these two PCR products together, creating plasmid pBL498(RfC:tax-6:CFP). We used LR Clonase to recombine this plasmid with pBL348(Paex-2) to generate plasmid pBL499.
Construction of Paex-2:G-CaMP6M:SL2:RFP integration
To reliably measure differences in calcium levels in the anal depressor across strains, we generated a G-CaMP integrated line. We injected pBL352 (Paex-2:G-CaMP6M:SL2:RFP) (LeBoeuf and Garcia, 2017) into N2 with Pacr-10:YFP as an injection marker. A transgenic line was selected for UV irradiation-based integration to create strain CG1644 that contains the rgIs19 integration. rgIs19 was mapped to LGII. This integration was then used to visualize calcium levels in the anal depressor in egl-20 and control males.
DREAD expression
The C. elegans designer receptors exclusively activated by designer drugs (ceDREADD) has been described previously and extensively characterized by Prömel et al. (2016). In short, the muscarinic G-protein sequence of gar-3:YFP in the plasmid pLR344 contains Y146C and A237G substitutions, which make the expressed G-protein-coupled receptor insensitive to acetylcholine but sensitive to clozapine-N-oxide (CNO). The aex-2 promoter from the plasmid pBL348 was recombined in front of the ceDREADD sequence using LR clonase, to make the plasmid pLR345.
Paex-2:ceDREADD::YFP was introduced into C. elegans by injecting 20 ng/µl pLR348, 20 ng/µl pPD132.102 (Pmyo-2:GFP) and 160 ng/µl of pUC18 into the germline of CG1673 hermaphrodites to generate the extrachromosomal array rgEx841 in strain CG1806.
The DREADD ligand CNO (Sigma-Aldrich) was dissolved in dimethyl sulfoxide (DMSO) to make a 20 mM stock solution. The CNO stock solution was further diluted in M9 buffer and added to NGM plates, so that the final drug concentration in the plates was 100 µM. OP50 was then spotted onto the CNO-containing plates and allowed to grow overnight. The next day, five or six CG1806 hermaphrodites were placed on the CNO- or DMSO blank-plates. The anal depressor of their transgenic adult male progeny was then analyzed by microscopy. L3, early L4 and mid L4 CG1806 males grown under normal conditions (without CNO) were identified and transferred to CNO plates. Their anal depressors were analyzed by microscopy once they were adults.
bar-1 plasmid construction
We generated a CRISPR/Cas9 plasmid targeting bar-1 using primers bar-1sgRNA1 and sgRNA-Rev-common to create pBL468. We injected pBL468 into CG1640, isolating CG1780. We sequenced CG1780 and determined that in the sequence 5′ CACCGCAAGGTGG the four underlined bases were deleted. TGG in the sequence is the PAM site we chose, indicating the Cas9 enzyme cut exactly where it was designed to.
The 6.3 kb promoter region of bar-1 was cloned using the primers bar-1-promot-F and bar-1-promot-R. The primer set contains the ATTB site, therefore the promoter region was recombined, using BP clonase, into pDG15 to generate pXC67. pXC67 was recombined with pGW322YFP to make pXC70 (Pbar-1:YFP). The genomic sequence of bar-1 from ATG to codon 761 was PCR-amplified using the primers bar1-coding-F and bar1-coding-R. The primers contain homology to the 3′ end of bar-1 promoter region and 5′ end of YFP. pXC70 (Pbar-1:YFP) was linearized using primers linPbY-For2nd-infbar1-761 and lin-PbY-infcod-R. The primer set contains homology to the 3′ end and 5′ end of cloned bar-1 genomic sequence, respectively. The bar-1 genomic region was infused with the linearized vector pXC70 using In-Fusion to make pXC87 (Pbar-1:bar-1:YFP). pXC67 (Pbar-1) was also recombined with pGW77C (plasmid containing Gateway ATTR site in front of CFP) to make pXC93 (Pbar-1:CFP). pXC93 and pXC87 were injected with pBX1 and pUC18 into CG531 to make CG1572.
RNAi
itr-1
rrf-3(pk1426); him-5(e1490) L4 hermaphrodites carrying the transgene rgEx497 (Punc-103E: YFP::actin) were fed with bacteria producing double-stranded RNAs to target the open reading frame of itr-1. Bacteria with the L4440 (control) or double-T7 vector including exons of target genes were grown and induced by isopropylthio-β-D-galactoside (IPTG) using a standard protocol (Kamath et al., 2001). Carbenicillin was used at the concentration of 10 mg/ml to increase the level of plasmid maintenance and therefore the effectiveness of RNAi. Exons 14 to 19 of itr-1 were cloned into the double-T7 vector. Males that displayed the corresponding mutant phenotype were picked for anal depressor imaging and defects detection.
wrm-1 and sys-1
The Gateway ATTP sequence was PCR-amplified from the Gateway entry vector pDONR221 (Invitrogen) and inserted in the NheI site, between the double T7 promoters of the L4440 vector (Addgene), to generate the plasmid pMB3. Fragments (300-400 bp) of sys-1 and wrm-1 exon 3 were PCR amplified with the primer pairs: Attb1sys1, Attb2sys1, and Attb1wrm1, Attb2wrm1, respectively. The PCR fragments were recombined into pMB3 using BP clonase (Invitrogen) to generate the plasmids pLR390 (sys-1 RNAi) and pLR391 (wrm-1 RNAi). pLR390 and pLR391 were transformed into the T7 RNA polymerase-expressing strain HT115(DE3).
A drop of solution containing four parts bleach and three parts 4 N NaOH was added to groups of gravid rgIs15 hermaphrodites that were placed on bacteria-free NG plates. The lack of food stalled the development of L1 worms that hatched from the bleach-resistant eggs. The next day, HT115(DE3) cells containing pLR390 or pLR391 were grown in a shaking incubator at 37°C in Luria-Bertani broth containing 100 µg/ml of carbenicillin. When the bacterial culture reached mid-log density, IPTG was added at a final concentration of 100 µM. Cells were grown for an additional hour, centrifuged for 5 min at 21,000 g, concentrated 10× and spotted on a nematode growth (NG) plate that contained 100 µg/ml of carbenicillin and 100 µM IPTG. The overnight-synchronized starved L1 animals were then added to the HT115(DE3)-containing plates. Adult males were scored using fluorescence microscopy.
CRISPR/Cas9-mediated disruption of goa-1, gpa-7, egl-30 and gpb-1 in the anal depressor
We replaced the eft-3 promoter, which drives Cas9 in pDD162 [the Cas9-expressing empty sgRNA plasmid (Dickinson et al., 2013)], with the Gateway ccdB destination cassette A (Invitrogen) to generate pYW42. To drive Cas9 expression in the anal depressor, LR clonase (Invitrogen) was used to recombine the aex-2 promoter from pBL348 into pYW42 to generate pLR392. To generate the goa-1 CRISPR/Cas9 guide RNA plasmid pLR393, the 19 base pair guide sequence to exon 2 of goa-1 (5′ AGCCATGAGCAACTTAGGC 3′) was added to pLR392 using PCR and the primers goa1gRNA and sgRNA(universal)REV. To generate the gpb-1 CRISPR/Cas9 guide RNA plasmid pLR394, the 19 bp guide sequence to exon 8 of gpb-1 (5′ GGCTGGTCACGATAACCGA 3′) was added to pLR392 using PCR and the primers gpb1gRNA and sgRNA(universal)REV. To generate the gpa-7 CRISPR/Cas9 guide RNA plasmid pLR397, the 19 base pair guide sequence to exon 3 of gpa-7 (5′ AGCGTGACTCCCAGTTAGT 3′) was added to pLR392 using PCR and the primers Gpa7gRNA and sgRNA(universal)REV. To generate the egl-30 CRISPR/Cas9 guide RNA plasmid pLR400, the 19 base pairs guide sequence to exon 2 of egl-30 (5′ GACTTGTCTATCAGAACGT 3′) was added to pLR392 using PCR and the primers Egl30gRNA and sgRNA(universal)REV.
Injection mixes containing: 160 ng/µl pUC18, 20 ng/µl pBL65 (Pges-1::CFP) and 20 ng/µl pLR394 (Paex-2:Cas9; gpb-1 gRNA); 160 ng/µl pUC18, 20 ng/µl pBL65 and 20 ng/µl pLR400 (Paex-2:Cas9; egl-30 gRNA); 140 ng/µl pUC18, 20 ng/µl pBL65 and 20 ng/µl each of pLR393 (Paex-2:Cas9; goa-1 gRNA) and pLR397 (Paex-2:Cas9; gpa-7 gRNA); or 120 ng/µl pUC18, 20 ng/µl pBL65 (Pges-1::CFP) and 20 ng/µl each of pLR395, pLR397 and pLR400 were microinjected into the germline of rgIs15 hermaphrodites.
The anal depressors of transgenic males, which express the co-injected Pges-1::CFP in the intestine, were scored using fluorescence microscopy. Transgenic males, which contain pLR394 (Paex-2:Cas9; gpb-1gRNA) and died as L1 larvae, were constipated or were obviously mosaic for the transgene and superficially wild type; transgenic males that were constipated at L1 to L3 larval stages were picked for inspection. Transgenic males that contain either Paex-2:Cas9; egl-30 gRNA or Paex-2:Cas9; goa-1, gpa-7 gRNA or Paex-2:Cas9; goa-1, gpa-7 and egl-30 gRNA, transmit the extrachromosomal array well; all adult males that expressed a CFP-fluorescent intestine were inspected.
Imaging and measurements
To facilitate imaging, the worms were mounted on agar pads made from a melted agar solution (Sulston and Horvitz, 1977). The agar solution was made by adding M9 to Difco Noble agar. The concentration of the agar pads for larval males and hermaphrodites was 2%. We used abamectin (Sigma-Aldrich) to immobilize the worms without interfering with development. Treatments such as NaN3, levamisole, arecoline, nicotine and dopamine killed the worms, wore off after a period of time or stalled development. Abamectin opens glutamate-gated chloride channels, which are located on the ventral cord neurons, leading to paralysis (Hart et al., 1995; Maricq et al., 1995). However, the larval anal depressor does not seem to have abamectin-sensitive receptors. We made a stock of 50 mg/ml in DMSO and then added 5 µl of stock solution to 1 ml of M9. This solution was then used to pick the worms via a glass needle from the plates they were grown on. After a few minutes, the worms were paralyzed and transferred to the agar pads. This same procedure was used for adult males, as abamectin did not change the structure of the tail, allowing for clear visualization of tail structures. Cover slips were then put on the top of the worm. All the images were taken using an Olympus IX81 microscope fitted with a Yokogawa CSU-X1 Spinning Disk Unit (Andor Technology). The two outermost lateral sides of the worm tail were set up as the starting and ending z stack for optical sectioning. Series of confocal images were then collected from one lateral side of the worm tail to the other. For each animal, the image that best showed the dorsal-ventral sarcomere width was selected. If no single image gave a good area for measurement, images of the individual attachment (either the left or the right) of the anal depressor were overlaid together using MetaMorph software (version 7.8.0.0, Molecular Devices) to create a flattened image. A line that had been calibrated for the magnification used was then drawn to determine the dorsal-ventral sarcomere width.
Calcium imaging
Males at the mid-L4 stage were placed on agar pads that were made from 2.5% noble agar dissolved in S-basal solution. Abamectin was added to the agar solution to immobilize the worm. The final concentration was 0.25 mg/ml. The G-CaMP and DsRed fluorescence signals at the male tail were recorded simultaneously using a dual view simultaneous image splitter (Photometrics) and a Hamamatsu ImagEM Electron multiplier (EM) CCD camera, with an exposure time of 1 s. The worms were imaged using the 40× objective, and the imaging continued until the worm reached the L4-molt stage. The Ca2+ data were analyzed using the Hamamatsu SimplePCI (version 6.6.0.0) software and Microsoft Excel, as previously described (LeBoeuf et al., 2011).
Cell ablations
CG1722 L2 or L3 males were mounted on 2% noble agar pads dissolved in M9 plus 12 mM NaN3. Cell ablations were performed as described by Bargmann and Avery (1995) using a Micropoint 337-NDS-USAS laser (Photonic Instruments) attached to an Olympus BX51 microscope.
Quantification and statistical analysis
Graphs and statistical analysis were constructed and carried out using GraphPad Prism 5.
Supplementary Material
Acknowledgements
We thank the Caenorhabditis Genetics Center for providing strains [funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440)]; Bryan Phillips for the BTP105 strain; and Kyoungsun Rha, Jimmy Goncalves, Yufeng Wan, Paola Correa, Liusou Zhang, Changhoon Jee and Daisy Gualberto for discussion and manuscript review.
Footnotes
Author contributions
Conceptualization: B.L., X.C., L.R.G.; Methodology: B.L., X.C., L.R.G.; Validation: B.L., X.C., L.R.G.; Formal analysis: B.L., X.C., L.R.G.; Investigation: B.L., X.C., L.R.G.; Resources: B.L., X.C., L.R.G.; Writing - original draft: B.L.; Writing - review & editing: X.C., L.R.G.; Visualization: B.L., X.C., L.R.G.; Project administration: L.R.G.; Funding acquisition: L.R.G.
Funding
This work was supported by a gift from the Howard Hughes Medical Institute awarded to L.R.G.
Contributor Information
Brigitte LeBoeuf, Department of Biology, Texas A&M University, College Station, TX 77843, USA.
Xin Chen, Department of Biology, Texas A&M University, College Station, TX 77843, USA.
Luis Rene Garcia, Department of Biology, Texas A&M University, College Station, TX 77843, USA.
References
- Alexandre, C., Baena-Lopez, A. and Vincent, J.-P. (2014). Patterning and growth control by membrane-tethered Wingless. Nature 505, 180-185. 10.1038/nature12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, F., Sohara, E., Morimoto, T., Yui, N., Nomura, N., Kikuchi, E., Takahashi, D., Mori, T., Vandewalle, A., Rai, T., et al. (2016). Wnt5a induces renal AQP2 expression by activating calcineurin signalling pathway. Nat. Commun. 7, 13636. 10.1038/ncomms13636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakumar, P. and Jagadeesh, G. (2010). Multifarious molecular signaling cascades of cardiac hypertrophy: can the muddy waters be cleared? Pharmacol. Res. 62, 365-383. 10.1016/j.phrs.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Baldwin, A. T. and Phillips, B. T. (2014). The tumor suppressor APC differentially regulates multiple β-catenins through the function of axin and CKIα during C. elegans asymmetric stem cell divisions. J. Cell Sci. 127, 2771-2781. 10.1242/jcs.146514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay, J., Lee, J. and Bandyopadhyay, A. (2004). Regulation of calcineurin, a calcium/calmodulin-dependent protein phosphatase, in C. elegans. Mol. Cells 18, 10-16. [PubMed] [Google Scholar]
- Bargmann, C. I. and Avery, L. (1995). Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48, 225-250. 10.1016/S0091-679X(08)61390-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, M. M., García, L. R. and Portman, D. S. (2018). Sexual dimorphism and sex differences in Caenorhabditis elegans neuronal development and behavior. Genetics 208, 909-935. 10.1534/genetics.117.300294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylis, H. A., Furuichi, T., Yoshikawa, F., Mikoshiba, K. and Sattelle, D. B. (1999). Inositol 1,4,5-trisphosphate receptors are strongly expressed in the nervous system, pharynx, intestine, gonad and excretory cell of Caenorhabditis elegans and are encoded by a single gene (itr-1). J. Mol. Biol. 294, 467-476. 10.1006/jmbi.1999.3229 [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz, K., Morantte, I., Weir, H. J. M., Yeo, R., Zhang, Y., Huynh, F. K., Ilkayeva, O. R., Hirschey, M. D., Grant, A. R. and Mair, W. B. (2015). Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell 160, 842-855. 10.1016/j.cell.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. and García, L. R. (2015). Developmental alterations of the C. elegans male anal depressor morphology and function require sex-specific cell autonomous and cell non-autonomous interactions. Dev. Biol. 398, 24-43. 10.1016/j.ydbio.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Correa, P., LeBoeuf, B. and Garcia, L. R. (2012). C. elegans dopaminergic D2-like receptors delimit recurrent cholinergic-mediated motor programs during a goal-oriented behavior. PLoS Genet. 8, e1003015. 10.1371/journal.pgen.1003015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D. J., Ward, J. D., Reiner, D. J. and Goldstein, B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028-1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch, R. E., Lewis, R. S., Goodnow, C. C. and Healy, J. I. (1997). Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386, 855-858. 10.1038/386855a0 [DOI] [PubMed] [Google Scholar]
- Egan, C. R., Chung, M. A., Allen, F. L., Heschl, M. F. P., Van Buskirk, C. L. and McGhee, J. D. (1995). A gut-to-pharynx/tail switch in embryonic expression of the Caenorhabditis elegans ges-1 gene centers on two GATA sequences. Dev. Biol. 170, 397-419. 10.1006/dbio.1995.1225 [DOI] [PubMed] [Google Scholar]
- Freire, A. G., Resende, T. P. and Pinto-do-Ó, P. (2014). Building and repairing the heart: what can we learn from embryonic development? BioMed Res. Int. 2014, 679168. 10.1155/2014/679168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W.-B., Wang, W. E. and Zeng, C.-Y. (2019). Wnt signaling pathways in myocardial infarction and the therapeutic effects of Wnt pathway inhibitors. Acta Pharmacol. Sin. 40, 9-12. 10.1038/s41401-018-0060-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, L. R., Mehta, P. and Sternberg, P. W. (2001). Regulation of distinct muscle behaviors controls the C. elegans male's copulatory spicules during mating. Cell 107, 777-788. 10.1016/S0092-8674(01)00600-6 [DOI] [PubMed] [Google Scholar]
- Gettner, S. N., Kenyon, C. and Reichardt, L. F. (1995). Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J. Cell Biol. 129, 1127-1141. 10.1083/jcb.129.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower, N. J. D., Walker, D. S. and Baylis, H. A. (2005). Inositol 1,4,5-trisphosphate signaling regulates mating behavior in Caenorhabditis elegans males. Mol. Biol. Cell 16, 3978-3986. 10.1091/mbc.e05-02-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef, I. A., Gastier, J. M., Francke, U. and Crabtree, G. R. (2001). Evolutionary relationships among Rel domains indicate functional diversification by recombination. Proc. Natl. Acad. Sci. USA 98, 5740-5745. 10.1073/pnas.101602398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Navetta, A., Gualberto, D. G. and García, L. R. (2012). Behavioral decay in aging male C. elegans correlates with increased cell excitability. Neurobiol. Aging 33, 1483.e5-1483.e23. 10.1016/j.neurobiolaging.2011.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., Vaz Gomes, A., Kenyon, C., Liu, J., Krause, M. and Fire, A. (1998). Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 12, 2623-2635. 10.1101/gad.12.16.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, A. C., Sims, S. and Kaplan, J. M. (1995). Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 378, 82-85. 10.1038/378082a0 [DOI] [PubMed] [Google Scholar]
- Harterink, M., Kim, D. H., Middelkoop, T. C., Doan, T. D., van Oudenaarden, A. and Korswagen, H. C. (2011). Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development 138, 2915-2924. 10.1242/dev.064733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, M. A., Vassilieva, L. L., Horvitz, H. R., Shaw, J. E. and Herman, R. K. (1995). The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83, 101-110. 10.1016/0092-8674(95)90238-4 [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., Horvitz, H. R. and Brenner, S. (1979). Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91, 67-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury, R., Viveiros, R., Johnsen, R., Mah, A., Anastas, D., Fang, L., Halfnight, E., Lee, D., Lin, J., Lorch, A., et al. (2007). High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5, e237-e237. 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell, T. A., Wang, Y., Bloniarz, A. E., Brittin, C. A., Xu, M., Thomson, J. N., Albertson, D. G., Hall, D. H. and Emmons, S. W. (2012). The connectome of a decision-making neural network. Science 337, 437-444. 10.1126/science.1221762 [DOI] [PubMed] [Google Scholar]
- Jee, C., Choi, T.-W., Kalichamy, K., Yee, J. Z., Song, H.-O., Ji, Y. J., Lee, J., Lee, J. I., L'Etoile, N. D., Ahnn, J., et al. (2012). CNP-1 (ARRD-17), a novel substrate of calcineurin, is critical for modulation of egg-laying and locomotion in response to food and lysine sensation in Caenorhabditis elegans. J. Mol. Biol. 417, 165-178. 10.1016/j.jmb.2012.01.012 [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. and Ahringer, J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, Research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katanaev, V. L., Ponzielli, R., Sémériva, M. and Tomlinson, A. (2005). Trimeric G protein-dependent frizzled signaling in Drosophila. Cell 120, 111-122. 10.1016/j.cell.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Kirszenblat, L., Pattabiraman, D. and Hilliard, M. A. (2011). LIN-44/Wnt directs dendrite outgrowth through LIN-17/Frizzled in C. elegans Neurons. PLoS Biol. 9, e1001157. 10.1371/journal.pbio.1001157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara, A., Inada, H., Katsura, I. and Mori, I. (2002). Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron 33, 751-763. 10.1016/S0896-6273(02)00607-4 [DOI] [PubMed] [Google Scholar]
- Kuhl, M., Sheldahl, L. C., Malbon, C. C. and Moon, R. T. (2000). Ca(2+)/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 275, 12701-12711. 10.1074/jbc.275.17.12701 [DOI] [PubMed] [Google Scholar]
- Lackner, M. R., Nurrish, S. J. and Kaplan, J. M. (1999). Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24, 335-346. 10.1016/S0896-6273(00)80848-X [DOI] [PubMed] [Google Scholar]
- LeBoeuf, B. and Garcia, L. R. (2017). Caenorhabditis elegans Male Copulation Circuitry Incorporates Sex-Shared Defecation Components To Promote Intromission and Sperm Transfer. G3 7, 647-662. 10.1534/g3.116.036756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBoeuf, B., Guo, X. and García, L. R. (2011). The effects of transient starvation persist through direct interactions between CaMKII and ether-a-go-go K+ channels in C. elegans males. Neuroscience 175, 1-17. 10.1016/j.neuroscience.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. Y. N., Lobel, L., Hengartner, M., Horvitz, H. R. and Avery, L. (1997). Mutations in the alpha1 subunit of an L-type voltage-activated Ca2+ channel cause myotonia in Caenorhabditis elegans. EMBO J. 16, 6066-6076. 10.1093/emboj/16.20.6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Song, H.-O., Jee, C., Vanoaica, L. and Ahnn, J. (2005). Calcineurin regulates enteric muscle contraction through EXP-1, excitatory GABA-gated channel, in C. elegans. J. Mol. Biol. 352, 313-318. 10.1016/j.jmb.2005.07.032 [DOI] [PubMed] [Google Scholar]
- Mahoney, T. R., Luo, S., Round, E. K., Brauner, M., Gottschalk, A., Thomas, J. H. and Nonet, M. L. (2008). Intestinal signaling to GABAergic neurons regulates a rhythmic behavior in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 105, 16350-16355. 10.1073/pnas.0803617105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, W., Morantte, I., Rodrigues, A. P. C., Manning, G., Montminy, M., Shaw, R. J. and Dillin, A. (2011). Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature 470, 404-408. 10.1038/nature09706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricq, A. V., Peckol, E., Driscoll, M. and Bargmann, C. I. (1995). Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378, 78-81. 10.1038/378078a0 [DOI] [PubMed] [Google Scholar]
- Maryon, E. B., Coronado, R. and Anderson, P. (1996). unc-68 encodes a ryanodine receptor involved in regulating C. elegans body-wall muscle contraction. J. Cell Biol. 134, 885-893. 10.1083/jcb.134.4.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuate, A., Latorre-Esteves, E. and Barria, A. (2017). A Wnt/Calcium signaling cascade regulates neuronal excitability and trafficking of NMDARs. Cell Rep. 21, 60-69. 10.1016/j.celrep.2017.09.023 [DOI] [PubMed] [Google Scholar]
- Mello, C. and Fire, A. (1995). DNA transformation. Methods Cell Biol. 48, 451-482. 10.1016/S0091-679X(08)61399-0 [DOI] [PubMed] [Google Scholar]
- Mendel, J. E., Korswagen, H. C., Liu, K. S., Hajdu-Cronin, Y. M., Simon, M. I., Plasterk, R. H. and Sternberg, P. W. (1995). Participation of the protein Go in multiple aspects of behavior in C. elegans. Science 267, 1652-1655. 10.1126/science.7886455 [DOI] [PubMed] [Google Scholar]
- Meng, L., Mulcahy, B., Cook, S. J., Neubauer, M., Wan, A., Jin, Y. and Yan, D. (2015). The cell death pathway regulates synapse elimination through cleavage of Gelsolin in Caenorhabditis elegans neurons. Cell Rep. 11, 1737-1748. 10.1016/j.celrep.2015.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. G., Emerson, M. D. and Rand, J. B. (1999). Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron 24, 323-333. 10.1016/S0896-6273(00)80847-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Fleming, T. W., Petersen, S. C., Manning, L., Matthewman, C., Gornet, M., Beers, A., Hori, S., Mitani, S., Bianchi, L., Richmond, J., et al. (2016). The DEG/ENaC cation channel protein UNC-8 drives activity-dependent synapse removal in remodeling GABAergic neurons. eLife 5, e14599. 10.7554/eLife.14599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, J., Ohkura, M. and Imoto, K. (2001). A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137-141. 10.1038/84397 [DOI] [PubMed] [Google Scholar]
- Pan, C.-L., Howell, J. E., Clark, S. G., Hilliard, M., Cordes, S., Bargmann, C. I. and Garriga, G. (2006). Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10, 367-377. 10.1016/j.devcel.2006.02.010 [DOI] [PubMed] [Google Scholar]
- Pani, A. M. and Goldstein, B. (2018). Direct visualization of a native Wnt in vivo reveals that a long-range Wnt gradient forms by extracellular dispersal. eLife 7, e38325. 10.7554/eLife.38325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D., Jhon, D. Y., Lee, C. W., Lee, K. H. and Rhee, S. G. (1993). Activation of phospholipase C isozymes by G protein beta gamma subunits. J. Biol. Chem. 268, 4573-4576. [PubMed] [Google Scholar]
- Parra, V. and Rothermel, B. A. (2017). Calcineurin signaling in the heart: the importance of time and place. J. Mol. Cell. Cardiol. 103, 121-136. 10.1016/j.yjmcc.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel, S., Sindelar, R. and Saha, M. (2018). Calcium signaling in vertebrate development and its role in disease. Int. J. Mol. Sci. 19, 3390. 10.3390/ijms19113390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pénigault, J. B. and Félix, M. A. (2011). High sensitivity of C. elegans vulval precursor cells to the dose of posterior Wnts. Dev. Biol. 357, 428-438. 10.1016/j.ydbio.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Prömel, S., Fiedler, F., Binder, C., Winkler, J., Schöneberg, T. and Thor, D. (2016). Deciphering and modulating G protein signalling in C. elegans using the DREADD technology. Sci. Rep. 6, 28901. 10.1038/srep28901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, D. J. and Thomas, J. H. (1995). Reversal of a muscle response to GABA during C. elegans male development. J. Neurosci. 15, 6094-6102. 10.1523/JNEUROSCI.15-09-06094.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, D. J., Weinshenker, D., Tian, H., Thomas, J. H., Nishiwaki, K., Miwa, J., Gruninger, T., Leboeuf, B. and Garcia, L. R. (2006). Behavioral genetics of Caenorhabditis elegans unc-103-encoded erg-like K(+) channel. J. Neurogenet. 20, 41-66. 10.1080/01677060600788826 [DOI] [PubMed] [Google Scholar]
- Saneyoshi, T., Kume, S., Amasaki, Y. and Mikoshiba, K. (2002). The Wnt/calcium pathway activates NF-AT and promotes ventral cell fate in Xenopus embryos. Nature 417, 295-299. 10.1038/417295a [DOI] [PubMed] [Google Scholar]
- Sawa, H. and Korswagen, H. C. (2013). Wnt signaling in C. elegans. WormBook 1-30. 10.1895/wormbook.1.7.2. [DOI] [PMC free article] [PubMed]
- Sawa, H., Lobel, L. and Horvitz, H. R. (1996). The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila frizzled protein. Genes Dev. 10, 2189-2197. 10.1101/gad.10.17.2189 [DOI] [PubMed] [Google Scholar]
- Schnabel, H. and Schnabel, R. (1990). An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science 250, 686-688. 10.1126/science.250.4981.686 [DOI] [PubMed] [Google Scholar]
- Sheldahl, L. C., Park, M., Malbon, C. C. and Moon, R. T. (1999). Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr. Biol. 9, 695-698. 10.1016/S0960-9822(99)80310-8 [DOI] [PubMed] [Google Scholar]
- Shimizu, H., Langenbacher, A. D., Huang, J., Wang, K., Otto, G., Geisler, R., Wang, Y. and Chen, J.-N. (2017). The Calcineurin-FoxO-MuRF1 signaling pathway regulates myofibril integrity in cardiomyocytes. eLife 6, e27955. 10.7554/eLife.27955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarski, D. C., Corces, V. G. and Moon, R. T. (1997). Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature 390, 410-413. 10.1038/37138 [DOI] [PubMed] [Google Scholar]
- Sulston, J. E. and Horvitz, H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., Albertson, D. G. and Thomson, J. N. (1980). The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. 78, 542-576. 10.1016/0012-1606(80)90352-8 [DOI] [PubMed] [Google Scholar]
- Takeshita, H. and Sawa, H. (2005). Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes Dev. 19, 1743-1748. 10.1101/gad.1322805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, L., Xie, Q., Ding, Y.-H., Li, S.-T., Peng, S., Zhang, Y.-P., Tan, D., Yuan, Z. and Dong, M.-Q. (2013). CAMKII and calcineurin regulate the lifespan of Caenorhabditis elegans through the FOXO transcription factor DAF-16. eLife 2, e00518. 10.7554/eLife.00518.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. H. (1990). Genetic analysis of defecation in Caenorhabditis elegans. Genetics 124, 855-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., Hires, S. A., Mao, T., Huber, D., Chiappe, M. E., Chalasani, S. H., Petreanu, L., Akerboom, J., McKinney, S. A., Schreiter, E. R., et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6, 875-881. 10.1038/nmeth.1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D. S., Vázquez-Manrique, R. P., Gower, N. J. D., Gregory, E., Schafer, W. R. and Baylis, H. A. (2009). Inositol 1,4,5-trisphosphate signalling regulates the avoidance response to nose touch in Caenorhabditis elegans. PLoS Genet. 5, e1000636. 10.1371/journal.pgen.1000636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo, J. and Kenyon, C. (1999). A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol. Cell 4, 851-858. 10.1016/S1097-2765(00)80394-9 [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., Yeh, R. T., Gish, W. R., Waterston, R. H. A. and Plasterk, R. H. (2001). Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160-164. 10.1038/88878 [DOI] [PubMed] [Google Scholar]
- Wu, M. and Herman, M. A. (2006). A novel noncanonical Wnt pathway is involved in the regulation of the asymmetric B cell division in C. elegans. Dev. Biol. 293, 316-329. 10.1016/j.ydbio.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Wu, M. and Herman, M. A. (2007). Asymmetric localizations of LIN-17/Fz and MIG-5/Dsh are involved in the asymmetric B cell division in C. elegans. Dev. Biol. 303, 650-662. 10.1016/j.ydbio.2006.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S. and Chisholm, A. D. (2011). A Gαq-Ca2+ signaling pathway promotes actin-mediated epidermal wound closure in C. elegans. Curr. Biol. 21, 1960-1967. 10.1016/j.cub.2011.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y., Kim, S., Chiba, K., Kawai, S., Tachikawa, H. and Takahashi, N. (2004). Calcineurin inhibitors block dorsal-side signaling that affect late-stage development of the heart, kidney, liver, gut and somitic tissue during Xenopus embryogenesis. Dev. Growth Differ. 46, 139-152. 10.1111/j.1440-169X.2004.00733.x [DOI] [PubMed] [Google Scholar]
- Zhao, G., Qiu, Y., Zhang, H. M. and Yang, D. (2019). Intercalated discs: cellular adhesion and signaling in heart health and diseases. Heart Fail. Rev. 24, 115-132. 10.1007/s10741-018-9743-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.