ABSTRACT
Continual spermatogenesis relies on the actions of an undifferentiated spermatogonial population that is composed of stem cells and progenitors. Here, using mouse models, we explored the role of RNA-binding proteins (RBPs) in regulation of the biological activities of this population. Proteins bound to polyadenylated RNAs in primary cultures of undifferentiated spermatogonia were captured with oligo (dT)-conjugated beads after UV-crosslinking and profiled by proteomics (termed mRBPome capture), yielding a putative repertoire of 473 RBPs. From this database, the RBP TRIM71 was identified and found to be expressed by stem and progenitor spermatogonia in prepubertal and adult mouse testes. Tissue-specific deletion of TRIM71 in the male germline led to reduction of the undifferentiated spermatogonial population and a block in transition to the differentiating state. Collectively, these findings demonstrate a key role of the RBP system in regulation of the spermatogenic lineage and may provide clues about the influence of RBPs on the biology of progenitor cell populations in other lineages.
Keywords: mRBPome capture, RNA-binding protein, TRIM71, Spermatogonia, Differentiation, Spermatogenesis
Highlighted Article: mRBPome capture reveals that the RNA-binding protein TRIM71 is expressed by mouse undifferentiated spermatogonial, and conditional knockout of Trim71 uncovers a key role in regulation of spermatogonial differentiation.
INTRODUCTION
Post-transcriptional regulation of gene expression plays a central role in intracellular processes and depends on the function of RNA-binding proteins (RBPs), which regulate pre-mRNA splicing, mRNA nuclear export, and transcript stability (Glisovic et al., 2008; Hentze et al., 2018). The fate or function of RNAs is influenced greatly by interaction with translational machinery, and disruption of RNA-protein complex formation is associated with a variety of diseases (Cooper et al., 2009; Hentze et al., 2018). A classic example of RBP-mediated influence on cellular functions is the regulation of pluripotent stem cells. In particular, by acting on specific RNA species, LIN28 shapes the cell metabolome to influence reprogramming of differentiated somatic cells and maintenance of a pluripotent state (Shyh-Chang et al., 2013; Zhang et al., 2016). At present, the scope of RBP involvement in fate decisions of tissue-specific stem cell-based systems remains largely unknown, although there are some studies focusing on the role of RBPs in germline cells.
Continual spermatogenesis represents one of the most productive regeneration systems in the body, relying on the balance of self-renewal and differentiation of an undifferentiated spermatogonial population consisting of stem and transit-amplifying progenitor subsets (Brinster, 2007). A minor subset of the undifferentiated spermatogonial population possesses regenerative capacity; these cells are generally referred to as spermatogonial stem cells (SSCs). Progenitor spermatogonia arise from the SSC pool and transiently amplify in number before transitioning to a differentiating state as A1 spermatogonia in response to a pulse of retinoic acid (RA) (Griswold et al., 2012; Lord et al., 2018; Rooij and Russell, 2000). Previous studies revealed that the RBP NANOS2, through interaction with dead end 1 (DND1), plays a key role in establishment of the undifferentiated spermatogonial population in both mice and pigs (Niimi et al., 2019; Park et al., 2017; Sada et al., 2009; Zhou et al., 2015).
Beyond the role of NANOS2, understanding of the regulatory role for RBPs in spermatogenic lineage function is rudimentary. Previous studies yielded a systematic, unbiased and comprehensive method for profiling RBP repertoires in cells termed ‘mRNA interactome capture’ or ‘mRBPome capture’ (Castello et al., 2012, 2013). Using this methodology, several comprehensive atlases of RBPs with canonical and noncanonical RNA-binding domains (RBDs) have been identified in human HeLa cells (Castello et al., 2012), mouse embryonic stem cells (ESCs) (Kwon et al., 2013) and fly embryonic tissue (Wessels et al., 2016). Overall, an estimated 1914 RBP-encoding genes are present in the mouse genome (Hentze et al., 2018). In the present study, we profiled the repertoire of RBPs in mouse undifferentiated spermatogonia using mRBPome capture, and from the database we identified TRIM71 (also known as LIN41) as a potential novel regulator of spermatogonial functions. Conditional knockout (cKO) of Trim71 revealed a key role in promoting expansion of the transit-amplifying progenitor population, as well as transition to a differentiating state. Taken together, these findings provide an advance in understanding of the influence that the RBP system has on regulation of mammalian spermatogonial biology, which may be relevant to stem and progenitor cell populations of other tissues.
RESULTS
mRBPome of the mouse undifferentiated spermatogonial population
To profile the repertoire of proteins that bind directly to mRNAs in undifferentiated spermatogonia (Fig. 1A, Fig. S1A), we generated primary cultures from postnatal day (P) 6 to 8 C57BL/6 mice as described previously (Wang et al., 2017). These cultures are known to contain a functional SSC pool as well as abundant progenitor spermatogonia (Helsel et al., 2017; Oatley et al., 2009; Takashima et al., 2015). To validate the cultures used for mRBPome analyses, we performed transplantation experiments with aliquots of cells that were derived from B6;129S-Gt (ROSA) 26Sor/J (also known as Rosa-lacZ) or Gt (ROSA)26Sortm4(ACTB-tdTomato,-EGFP)LUO/J) (also known as Rosa-tdTomato) transgenic mice to confirm the existence of an SSC pool (Fig. S1B). Next, for each RBP capture experiment, approximately 40 million cultured undifferentiated spermatogonia (Fig. S1B1) were exposed to UV radiation at 254 nm (Fig. 1A) to crosslink RBP-mRNA complexes. The mRNA interactome was captured through oligo(dT) magnetic bead-mediated pull-down of polyadenylated RNAs (Fig. 1A). Proteins bound to mRNAs were released by RNase treatment and subjected to mass spectrometry (MS) analysis (Fig. 1A). Silver staining was used for visualization of the proteins in gels after electrophoresis to check whether there were enriched bands in the UV-crosslinked samples compared with the non-UV-crosslinked controls. The enrichment of distinct protein bands with oligo(dT) captured in lysates of UV-crosslinked cells confirmed the success of mRBPome capture (Fig. 1B). Using quantitative real-time RT PCR, we found that polyadenylated transcripts, such as glyceraldehyde 3-phosphate dehydrogenase (Gapdh) and beta-actin (Actb), were enriched in the oligo(dT) captured lysate, whereas 18S ribosomal RNA (rRNA) was not (Fig. 1C). Western blot analysis confirmed that known canonical RBPs, IGF2BP1 and LIN28A, were present in the UV-crosslinked samples but absent in non-UV-crosslinked controls (Fig. 1D). As expected, common contaminants, such as cytoskeleton proteins (e.g. tubulin and ACTB), were not detectable by oligo(dT) capture (Fig. 1D). These results confirmed enrichment of RBPs from undifferentiated spermatogonia. In total, we identified 587 proteins, of which 415 (70.7%) were present in all three biological replicates, and 488 proteins (83.1%) were detected in at least two of the three replicate cultures (Fig. 1E, Table S1) (accession number: PXD017377, ProteomeXchange via the PRIDE database, a public database platform improved by Perez-Riverol et al., 2019).
Fig. 1.
mRBPome profiling of mouse undifferentiated spermatogonia. (A) Schematic of the experimental approach. mRNA-protein complexes in primary cultures of undifferentiated spermatogonia were UV-crosslinked and captured using oligo(dT) magnetic beads. (B) Representative image of a silver-stained SDS-PAGE gel confirming enrichment of protein bands in UV-crosslinked (CL; +) compared with non-UV-crosslinked control (non-CL; −) input and oligo(dT)-captured samples. (C) Relative levels of 18S rRNA, Actb and Gapdh mRNAs measured by quantitative real-time RT-PCR analysis. Data are mean±s.e.m. for n=3 different cultures. (D) Representative image of western blot analysis confirming enrichment of the known RBPs IGF2BP1 and LIN28A in UV-crosslinked samples subjected to oligo(dT)-capture (+). (E) Venn diagram analysis of RBPs identified by mass spectrometry analysis; data are from three independent primary cultures.
Characteristics of RBPs in undifferentiated spermatogonia
Next, we aimed to screen the mRBPome dataset to generate a focused list of potential novel candidates that impact the biology of undifferentiated spermatogonia. First, we excluded 21 proteins that were captured in both UV-crosslinked (experimental) and non-UV-crosslinked (control) samples, and only considered candidate proteins that were detected in at least two biological replicates, yielding 473 proteins as the putative repertoire of RBPs in undifferentiated spermatogonia (Fig. 2A, Table S1). Second, to exclude technical bias from MS analysis that can occur with complex proteomes (Brunner et al., 2007; Schrimpf et al., 2009), we compared the distribution of isoelectric points and the ratio of disordered residues among three groups of proteins: global proteins predicted from whole mouse genome (all), RBPs previously reported in gene ontology (GO) annotation (RNA binding), and the repertoire of captured RBPs in undifferentiated spermatogonia (captured RBPs, cRBPs). Compared with all proteins and known RBPs, cRBPs exhibited a shift towards alkaline isoelectric points (Fig. 2B) and a higher ratio of disordered residues (Fig. 2C), suggesting that our oligo(dT) cRBPs from undifferentiated spermatogonia were predominantly basic proteins, and enriched for intrinsically disordered regions. These properties are consistent with features of RBPs identified in previous large-scale screens, in which the physicochemical properties were found to be distinct from those of whole cell lysates (Castello et al., 2012). Third, we performed GO enrichment analysis for the cRBPs and found that most of the enriched functional terms for molecular processes, cellular components and biological processes were directly associated with RNA-binding activities, as well as ribonucleoprotein complexes and RNA biology-related functions and processes (Fig. 2D-F, Table S1). The majority of captured proteins were functionally annotated as RBPs (83.7%, 396 of 473), with only 23 proteins being annotated as RNA-unrelated and 44 proteins lacking annotation (Fig. 2G). The proteins annotated as RBPs were then classified according to their domains, which revealed that a large proportion contained either classical RBDs (172) or non-classical RBDs (114) (Fig. 2H). Using the Pfam protein domain database, we found that multiple classical (PRM, dead box helicase, KH) and non-classical (helicase-C, zinc finger CCHC) protein domains were enriched in RBPs from undifferentiated spermatogonia (Fig. 2I, Table S1, sheet 2, columns H-J, red). Lastly, the putative non-classical RBDs identified in undifferentiated spermatogonia were compared with RBPome datasets from other mouse cell populations (Boucas et al., 2015; He et al., 2016; Kwon et al., 2013; Liao et al., 2016; Liepelt et al., 2016). Of the 114 candidate RBPs with non-classical RBDs, 107 (93.9%; Fig. S2A) are also found in other cell types, thereby validating the mRBPome profile generated for mouse undifferentiated spermatogonia (Table S1, sheet 2, columns K-S, purple).
Fig. 2.
Characteristics of mRBPome-captured proteins in undifferentiated spermatogonia. (A) Venn diagram of oligo(dT)-captured proteins identified by mass spectrometry analysis in the UV-crosslinked (experimental) versus non-UV-crosslinked (control) samples (587 total candidates were obtained from three independent experiments, data are presented in Fig. 1E). After removing the 21 proteins that were also detected in control, 473 captured RBPs (cRBPs) were present in at least two biological replicates. (B,C) Distribution of isoelectric point and disordered residues among total proteins (magenta), undifferentiated spermatogonial mRBPome-captured proteins (blue) and known RBPs (green). (D-F) Significantly enriched Gene Ontology (GO) terms for mRBPome-captured proteins based on molecular processes (D), cellular components (E) and biological processes (F). (G,H) Classification of mRBPome-captured proteins according to the GO annotations of RNA-associated functions (G) and predefined domains (H). (I) Top ten enriched Pfam domains among mRBPome-captured proteins.
Transcripts encoding mRBPome-defined RBPs are upregulated in mouse undifferentiated spermatogonia
Proliferation of undifferentiated spermatogonia is stimulated, at least in part, by glial cell line-derived neurotrophic factor (GDNF) (Oatley et al., 2006; Wang et al., 2017), whereas the transition to a differentiating state is triggered by RA signaling (Endo et al., 2015). To assess the extent to which the profiled undifferentiated spermatogonial mRBPome is subject to regulation by these external signals, we mined databases of GDNF- and RA-regulated gene expression in mouse undifferentiated spermatogonia generated by previous studies (GSE66998 and GSE99147, Gene Expression Omnibus database) (Li et al., 2016; Oatley et al., 2006). Of the 473 candidate RBP genes identified in the mRBPome, 27 (5.7%) were regulated by GDNF signaling (Fig. 3A), including Lin28a, Esrp1 and Mov10; whereas 29 (6.1%) were regulated by RA (Fig. 3B), including Col1a1, Ybx3, Rhpn1 and Rbms3. A small number of RBP genes, including Bc048507 and Rbm24, were regulated by both GDNF and RA. These findings indicate that a majority of RBP-encoding genes in mouse undifferentiated spermatogonia are not regulated by extrinsic signals and may instead be autonomously expressed.
Fig. 3.
Expression profiles of mRBPome-captured proteins in undifferentiated spermatogonia. (A,B) Scatter plots of gene expression for candidate RBPs in undifferentiated spermatogonia in response to GDNF (A) or RA (B) signaling as measured by microarray analysis (taken from Wang et al., 2017). Each dot represents individual gene identifiers with upregulated genes shown in yellow/red and downregulated genes shown in blue. (C) Venn diagram comparing undifferentiated spermatogonial mRBPome-captured proteins (labeled as SSC here) with mRBPome-captured proteins in MEFs (taken from Boucas et al., 2015) and mouse ESCs (Kwon et al., 2013; He et al., 2016). (D) Heatmap depiction of gene expression levels for candidate undifferentiated spermatogonia mRBPome-captured proteins in different mouse testicular cell types.
Next, to narrow down the full list to a few top candidates for further study, the undifferentiated spermatogonial mRBPome dataset of 473 RBPs was compared with mRBPome datasets for mouse ESCs (representative of a stem cell dataset) and mouse embryonic fibroblasts (MEFs, representative of a somatic cell dataset) that were generated by previous studies (Boucas et al., 2015; He et al., 2016; Kwon et al., 2013). As presented in Fig. 3C, 103 RBP-encoding genes were found to be uniquely expressed in undifferentiated spermatogonia (class I core RBPs) compared with ESCs and MEFs, and 147 genes were expressed in undifferentiated spermatogonia and ESCs but not MEFs (class II core RBPs). In comparison with the curated RNA interactome database generated by Hentze et al. (2018), we found that 90.2% of the RBPs expressed in mouse undifferentiated spermatogonia were also present in subsets of other cell lineages (Fig. S2B).
We next queried a well-defined transcriptome database of testis cell populations (www.ebi.ac.uk/arrayexpress, accession number E-TABM-130) for the expression patterns of the 250 RBP genes (103 class I core RBPs and 147 class II core RBPs) in spermatogonia, spermatocytes, spermatids and Sertoli cells. A majority of these RBP genes (216 of 250) were represented, with distinct expression patterns in germline cells versus Sertoli cells (Table S1). Of these, 15 genes were preferentially expressed in mouse spermatogonia, including seven that are unique to undifferentiated spermatogonia (Ythdf1, Unkl, Drosha, Dnmt1, Nlrp4c, Dnd1 and Nxf2) and eight that are expressed in both undifferentiated spermatogonia and ESCs (Mki67, Mex3a, Noc2l, Trim71, Dazl, Ddx10, Lin28a and Esrp1) (Fig. 3D, Fig. S2C). Comparison of expression patterns in stem cells (ESC and undifferentiated spermatogonia) versus somatic cells (STO) revealed that TRIM71 and ESRP1 were only expressed in the former, whereas MEX3A was also expressed in somatic cells (Fig. S2D), suggesting that further experimental validation of expression is warranted for these RBPs.
Knockdown of TRIM71, ESRP1 and MEX3A impairs proliferation and survival of primary undifferentiated spermatogonial cultures
Among the 15 RBPs found to be preferentially expressed in spermatogonia, six (Nxf2, Dnd1, Dnmt1, Lin28a, Dazl and Drosha) have been shown previously to influence spermatogonial biology (Chakraborty et al., 2014; Pan et al., 2009; Schrans-Stassen et al., 2001; Wu et al., 2012; Yamaji et al., 2017); whereas three of the RBPs (Trim71, Esrp1 and Mex3a) had not been studied in the mammalian germline, although a role in somatic stem and progenitor cells was demonstrated previously (Barriga et al., 2017; Bebee et al., 2015; Nguyen et al., 2017). To address this gap in knowledge, we first localized expression of TRIM71, ESRP1 and MEX3A in the mouse testis by immunostaining of cross-sections and found that all are expressed by germ cells at P3 (Fig. 4A) and then spermatogonia at P10 (Fig. 4A). In adult (P70) mouse testis, TRIM71 immunostaining was detectable in spermatogonia, whereas staining for ESRP1 and MEX3A was observed in a subset of spermatogonia but also in spermatocytes and spermatids (Fig. 4A). Within the spermatogonial population, expression of TRIM71, ESRP1 and MEX3A colocalized with immunostaining for the undifferentiated marker ZBTB16 (also known as PLZF; Buaas et al., 2004; Costoya et al., 2004) in P10 testes. In the adult testis, only TRIM71 colocalized with ZBTB16 but ESRP1 and MEX3A were also expressed by advanced germ cells (Fig. 4B).
Fig. 4.
Localization of TRIM71, ESRP1 and MEX3A expression in mouse testes. (A) Representative images of immunohistochemical staining for TRIM71, ESRP1 and MEX3A in cross-sections of mouse testes during neonatal (P3), prepubertal (P10) and adult (P70) stages of development. Insets show magnification of the area where the positive cells are located. Arrowheads indicate the positive cells. (B) Whole-mount immunofluorescence staining for TRIM71, ESRP1 and MEX3A expression in undifferentiated spermatogonia based on colocalization with the marker ZBTB16. Insets show magnification of the area where the positive cells are located. (C) Representative images of western blot analysis for knockdown efficiency of TRIM71, ESRP1 and MEX3A levels 7 days after gene-specific shRNA treatment compared with non-targeting shRNA control (shVector). ACTB level was assessed as a loading control. (D) Quantification of total cell number in primary cultures of undifferentiated spermatogonia at 0, 4 and 7 days after treatment with Trim71, Esrp1 or Mex3a shRNA to knock down expression. (E) Quantification of apoptotic and dead cells in primary cultures of undifferentiated spermatogonia at 7 days after treatment with Trim71, Esrp1 or Mex3a shRNA. All data are mean±s.e.m. for n=3 different cultures. *P<0.05, **P<0.01 (Student's t-test). Scale bars: 20 μm.
Next, we aimed to determine whether TRIM71, ESRP1 and MEX3A have an important role in regulation of undifferentiated spermatogonial biology. To achieve this, primary cultures were treated with small hairpin RNAs (shRNAs) designed to knockdown mRNA levels for each factor. We found that the in vitro expansion rate of undifferentiated spermatogonial populations treated with shRNA-mediated knockdown of TRIM71 or MEX3A was significantly reduced compared with cells treated with control shRNA (P<0.01; Fig. 4C,D, Fig. S3). In addition, the percentage of cells undergoing apoptosis was significantly increased following knockdown of TRIM71 compared with treatment with non-targeting control shRNA (P<0.01; Fig. 4E). However, although numerically different, the level of apoptosis in cultures subjected to knockdown of ESRP1 or MEX3A was not significantly altered compared with control shRNA-treated cultures (P>0.05). Taken together, these findings suggested that RBPs, such as TRIM71 and MEX3A, may play important roles in maintenance of the undifferentiated spermatogonial population.
The two conserved RBPs, LIN28A and TRIM71, were first identified in Caenorhabditis elegans as key regulators of cell fate decisions in stem cells (Ambros and Horvitz, 1984; Reinhart et al., 2000) where they work in concert to suppress Let-7 miRNA function in somatic cells. Thus, we examined whether there was cooperative binding in mouse undifferentiated spermatogonia. TRIM71 was found to colocalize with LIN28A, a well-known marker of undifferentiated spermatogonia, in both pre-pubertal and adult testes (Fig. S4A). However, based on co-immunoprecipitation (co-IP) analysis using a validated anti-LIN28A antibody and either MS or western blot analysis, the presence of TRIM71 could not be detected (Fig. S4B,C). These findings indicate that although colocalized in undifferentiated spermatogonia, LIN28A and TRIM71 are not binding partners.
TRIM71 regulates spermatogonial differentiation in vivo
Outcomes of TRIM71 knockdown in primary cultures of undifferentiated spermatogonia suggested a role in the promotion of proliferation and survival. To extend this finding to the population in vivo, we generated a Trim71 cKO mouse model using the Cre/loxP system. First, a novel line was generated using a gene-targeting approach to insert loxP sites flanking exon 4 of the Trim71 coding sequence (Fig. S5A). Second, mice homozygous for the floxed allele (Trim71fl/fl) were backcrossed with Ddx4-Cre mice to produce animals with germ cell-specific inactivation of Trim71 (designated hereafter as Trim71 cKO males) (Fig. S5B,C). The efficiency of Trim71 floxed allele recombination was assessed by co-immunostaining for ZBTB16 and TRIM71 in cross-sections of testes from control and Trim71 cKO mice at P10 and outcomes revealed complete loss of TRIM71 expression (Fig. S5D). At adulthood, testes from Trim71 cKO males were strikingly smaller compared with control littermates with one or two functionally intact Trim71 alleles (Fig. 5A). Histological assessment of cross-sections from testes of adult Trim71 cKO males revealed severe disruption of spermatogenesis (Fig. 5B). In addition, sperm were absent in cross-sections of epididymis from adult Trim71 cKO males (Fig. 5B). Quantification of total germ cell number (based on immunostaining for GCNA1+ cells) revealed no difference in the prospermatogonial population (precursors to the undifferentiated spermatogonial population) between Trim71 cKO and control males at P1 (Fig. 5C,H). hereafter, the number of undifferentiated spermatogonia (i.e. ZBTB16+ cells) in testes of Trim71 cKO males was found to be significantly (P<0.01) reduced to 48% and 13% of wild-type controls at P10 (ZBTB16+ cells/tubule) and P56 [number of ZBTB16+ cells × seminiferous tubule diameter (µm)], respectively (Fig. 5D,E,I,J).
Fig. 5.
Impact of Trim71 conditional knockout in the male germline. (A) Representative images of testis and epididymis from adult (P56) Trim71 conditional knockout (cKO) and control mice. Ruler shows 1 mm per division. (B) Representative images of Hematoxylin and Eosin (H&E)-stained cross-sections of seminiferous tubules and epididymis from P56 control and Trim71 cKO mice. (C-G) Representative images of immunofluorescence staining for the pan germ cell marker GCNA1 at P1 (C), the undifferentiated spermatogonial marker ZBTB16 and primitive spermatogonial marker GFRA1 at P10 (D) and P56 (E), and the undifferentiated spermatogonial marker ZBTB16 and differentiating marker STRA8 at P10 (F) and P56 (G) in cross-sections of testes from control and Trim71 cKO mice. (H) Quantification of total germ cell number (based on GCNA1+ cells) in seminiferous tubule cross-sections from control and Trim71 cKO mice at P1. (I,J) Quantification of undifferentiated spermatogonial number [based on ZBTB16+ cells/seminiferous tubule at P10 or number (#) of ZBTB16+ cells × seminiferous tubule diameter (µm) at P56], percentage of the population that is the most primitive subset (based on GFRA1+ among ZBTB16+ cells), and percentage of the population that has transitioned to a differentiating state (based on STRA8+ among ZBTB16+ cells) in cross-sections of testes from control and Trim71 cKO mice at prepubertal (P10) (I) and adult (P56) (J) age points. Data in H-J are mean±s.e.m. for n=3 different mice, dots represent average values of individual mice, **P<0.01 (Student's t-test). Scale bars: 20 μm.
To clarify further the impact of Trim71 deficiency on spermatogenesis, we immunostained for GFRA1, which marks the primitive subset of the undifferentiated population (Nakagawa et al., 2010), including SSCs, and ZBTB16 in cross-sections of testes from control and Trim71 cKO mice. Quantitative assessment revealed that the percentage of GFRA1+ spermatogonia within the ZBTB16+ population was not different at P10 but was significantly (P<0.01) greater at P56 in Trim71 cKO testes compared with controls (Fig. 5D,E,I,J). Additional co-staining revealed that all GFRA1+ spermatogonia co-express TRIM71 (Fig. S5E), indicating that the higher percentage of GFRA1+/ZBTB16+ in Trim71 cKO testes at P56 was not caused by the original lack of TRIM71 expression in wild-type GFRA1+ spermatogonia and instead revealed that the differentiation of undifferentiated spermatogonia was blocked by TRIM71 inactivation. Interestingly, the first round of spermatogenesis that is known to derive independent of the undifferentiated spermatogonial population was impaired in Trim71 cKO mice, evidenced by the smaller testes and lack of elongating spermatids at P35 (Fig. S6A,B) and significant (P<0.01) reduction in the number of STRA8+ spermatogonia, a marker of spermatogonial differentiation, compared with controls at P10 (Fig. 5F,I). However, the gross nuclear morphology of spermatogonia in cross-sections of testes appeared to be similar between control and Trim71 cKO mice (Fig. S6B). In adult testes, STRA8+ spermatogonia (i.e. ZBTB16+) were not evident in cross-sections of testes from Trim71 cKO (Fig. 5G,J, Fig. S6C,D) at either P35 or P56. Furthermore, immunostaining for the meiotic marker SYCP3 revealed that spermatocytes failed to form in testes of Trim71 cKO mice (Fig. S6E). Collectively, these findings indicate that Trim71 deficiency in spermatogonia impairs the capacity for transit-amplifying progenitors to expand in number and transition to a differentiating state.
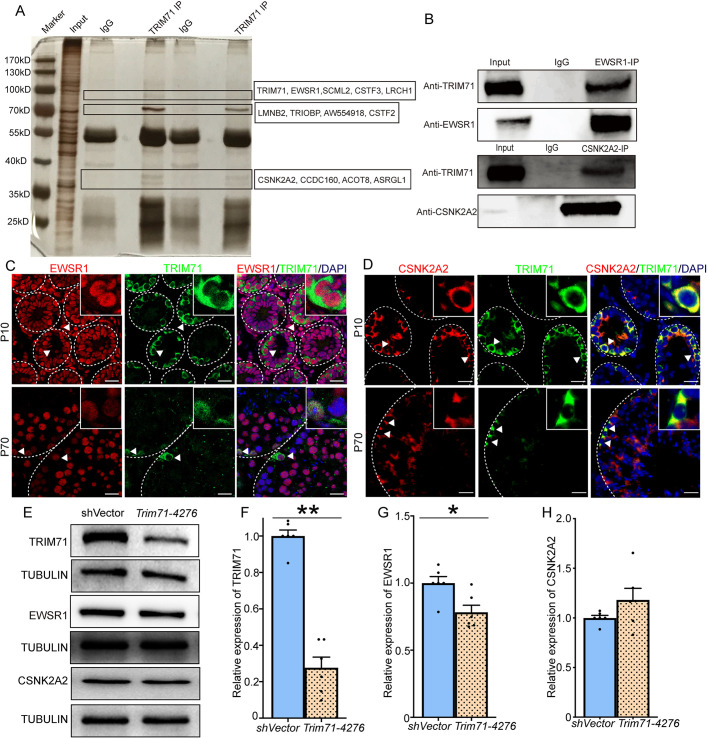
EWSR1 is a binding partner protein of TRIM71
Lastly, we explored whether there are protein binding partners of TRIM71 in undifferentiated spermatogonia. To achieve this, an anti-TRIM71 antibody was generated using two antigen peptide sequences: KFGAQGSGFGQMDRPSGI (Chang et al., 2012) and CVRAHQRVRLTKDHYIER (Worringer et al., 2014). The antibody was then used for IP to isolate TRIM71-containing protein complexes from testes of P10 mice. The IP proteins were then subjected to MS analysis (Fig. 6A, Fig. S7A, representative IP data). Candidate proteins, such as the RNA-binding protein EWS (EWSR1), casein kinase II subunit alpha (CSNK2A2), sex comb on midleg-like 2 (SCML2) and Acyl-CoA thioesterase 8 (ACOT8), were identified (Table S2). Interestingly, EWSR1 is known to regulate hematopoietic stem cell senescence, and inactivation of EWSR1 leads to arrest of pre-B cell differentiation and male infertility (Cho et al., 2011; Li et al., 2007). Also, CSNK2A2 deficiency causes male sterility (Xu et al., 1999). Thus, EWSR1 and CSNK2A2 were selected for validation as TRIM71 binding partners. In corroboration with the MS data, TRIM71 was detected in the IP of EWSR1 and CSNK2A2 by western blot analysis (Fig. 6B). In addition, immunostaining revealed that both EWSR1 and CSNK2A2 colocalize with TRIM71 in cross-sections of testes from P10 and P70 mice (Fig. 6C,D). Paradoxically, EWSR1 appeared to be nuclear whereas TRIM71 was mainly detectable in the cytoplasm. To address this, we isolated the cytoplasmic and nuclear fractions from testis lysates and detected TRIM71 protein in both (Fig. S7B,C). Finally, we used shRNA treatment to knock down Trim71 levels in cultures of undifferentiated spermatogonia in order to assess whether TRIM71 regulates the expression of EWSR1 or CSNK2A2. Although the level of CSNK2A2 was unaffected in Trim71 knockdown cells; the expression of EWSR1 was significantly (P<0.05) decreased compared with control (Fig. 6E-H). Collectively, these findings suggest that EWSR1 levels in undifferentiated spermatogonia are influenced post-transcriptionally by interaction with TRIM71.
Fig. 6.
EWSR1 is a protein-binding partner of TRIM71 in mouse testes. (A) Silver staining of proteins isolated from TRIM71 IP of P10 testis lysate and identified by MS. (B) Enrichment of TRIM71 in EWSR1 and CSNK2A2 IP using P10 mouse testes. (C,D) Co-immunofluorescence of TRIM71 and EWSR1 (C) or CSNK2A2 (D) in cryostat sections from P10 and adult testes. (E-H) Western blot (E) and quantification of relative expression levels of TRIM71 (F), EWSR1 (G) and CSNK2A2 (H) after TRIM71 knockdown by shRNA treatment. Data in F-H are mean±s.e.m. for three different primary cultures and two separate knockdowns. *P<0.05, **P<0.01 (Student's t-test). Scale bars: 20 μm.
DISCUSSION
In eukaryotic cells, RBPs bind thousands of target RNAs and control their respective fate by regulating pre-RNA splicing, polyadenylation, transport, translation and cellular localization (Glisovic et al., 2008; Hentze et al., 2018). Identification and characterization of the role of these proteins in RNA-associated ribonucleoprotein complexes have provided valuable insights into the highly dynamic RNA-protein networks in living cells. Traditionally, RBPs are thought to interact with sequences or structural motifs of RNAs via RBDs, and the search for novel RBPs has been based largely on this relationship. However, this conventional approach is not suitable for identifying unknown RBPs with noncanonical RBDs and decoding their functions in a specific cell lineage or tissue type. To overcome these limitations, several approaches have been developed for the systematic search for RBPs (Scherrer et al., 2011), including a methodical, unbiased and comprehensive method termed mRNA interactome capture that has been successfully applied to immortalized cell lines (e.g. HeLa and HEK293T) and ESCs to profile RBP pools (Castello et al., 2012, 2013; Kwon et al., 2013). This methodology has resulted in the identification of large numbers of RBPs, including proteins that were not previously annotated as RBPs or that lacked classical RBDs.
Tissue-specific stem cells and transit-amplifying progenitors exist in most, if not all, cell lineages and are crucial for the maintenance of organ homeostasis and regeneration (Biteau et al., 2011; Miyajima et al., 2014). To date, mRNA interactome capture has not been applied to tissue-specific stem cell or progenitor populations, probably because of the difficulty in obtaining a sufficient number of cells for analysis. In the male germline, stem cell and transit-amplifying progenitor pools reside in the undifferentiated spermatogonial population, and methods have been devised that support long-term expansion of mouse cells as a primary culture. Here, we were able to use this platform to generate a sufficient number of cells for application of mRNA interactome capture. The outcomes produced an atlas of 473 candidate RBPs that provide invaluable information to aid in elucidating a role for the RBP system in regulation of undifferentiated spermatogonial biology.
Of the 473 captured proteins, the majority are annotated as RBPs (396, 83.7%) based on current databases. Over half of the proteins (431, 90.2%) in the current and previous studies (Hentze et al., 2018) are found in multiple cell types. However, we also identified an RBP pool of 103 proteins that might be expressed uniquely in undifferentiated spermatogonia and 147 that seemed to be expressed in both undifferentiated spermatogonia and ESCs. These proteins may represent the core mammalian mRNA interactome in undifferentiated spermatogonia. Recent studies have provided evidence of RBPs playing crucial roles in undifferentiated spermatogonial cell fate decisions by regulating RNA transport, alternative splicing patterns, and stability. A prominent example is NANOS2, a zinc-finger RBP that has been identified as a key regulator of spermatogonial stem cell biology. A NANOS2-containing messenger ribonucleoprotein complex defines the post-transcriptional profile in undifferentiated spermatogonia by recruiting and repressing specific transcripts (Sada et al., 2009; Zhou et al., 2015). Another example is DND1, a vertebrate-conserved RBP that controls the mRNA destabilization that is required for survival of mouse spermatogonial precursors (i.e. prospermatogonia) (Yamaji et al., 2017). Interestingly, DND1 is a binding partner of NANOS2 that facilitates interaction with target RNAs in undifferentiated spermatogonia (Suzuki et al., 2016). In our mRBPome capture, we also identified DND1 as one of 15 RBPs preferentially expressed in both undifferentiated spermatogonia and ESCs. We were not able to identify NANOS2, which may be due to low peptide abundance in the current capture from 40 million undifferentiated spermatogonia or the bias of mRBPome to enrich for proteins that are bound to transcripts with U-rich motifs. However, in a previous MS screen involving more source material (∼300 million spermatogonia) NANOS2 was identified as a GDNF-regulated protein (Wang et al., 2017).
Interestingly, six of the 15 RBPs identified in our screen as being preferentially expressed in spermatogonia have known roles; these are LIN28A, NXF2, DNMT1, DAZL, DND1 and ribonuclease type III DROSHA. The pluripotency factor LIN28A was previously identified as an undifferentiated spermatogonial marker (Zheng et al., 2009), and regulates proliferation of the undifferentiated spermatogonial population in mouse testes (Chakraborty et al., 2014). Deletion of the nuclear RNA export factor NXF2 in mouse testes leads to age-dependent depletion of undifferentiated spermatogonia (Pan et al., 2009). Also, depletion of DNMT1 uncouples DNA methylation and inactivation of both DNMT1 and p53 causes upregulation of SOX2, which in turn bolsters OCT4 (POU5F1) levels and eventually drives undifferentiated spermatogonia into a pluripotent state (Takashima et al., 2013, 2009). In addition, DAZL deficiency blocks the differentiation from transit-amplifying progenitor to A1 states in spermatogonia thereby leading to sterility (Schrans-Stassen et al., 2001). Furthermore, inactivation of Drosha in the mouse testes causes severe spermatogenic disruption, caused by disruption of miRNA biogenesis (Wu et al., 2012).
Based on the known roles for six of the 15 RBPs with preferential expression in undifferentiated spermatogonia, we speculated that the remaining nine candidates – YTHDF1, UNKL, NIRP4C, MKI67, NOC2L, DDX10, MEX3A, ESRP1 and TRIM71 – may influence spermatogonial biology as well. The cellular function for YTHDF1, UNKL, NIRP4C, MKI67, NOC2L and DDX10 has not been defined in any stem cell type. However, MEX3A has been shown to mark a small population of slowly dividing LGR5+ intestinal stem cells that are resistant to chemotherapy (Barriga et al., 2017). Through an interaction with the transcription factor CDX2, MEX3A regulates intestinal stem/progenitor differentiation, polarity and regeneration (Pereira et al., 2013). ESRP1, an epithelial-specific alternative splicing factor, is responsible for establishing a proper skin barrier. Deletion of ESRP1 leads to neonatal death due to disruption in alternative splicing of target genes including Fgfr1, Fgfr3 and Enah, and impairment of epidermal development (Bebee et al., 2015). TRIM71 is known to be regulator of neural progenitor and ESC maintenance, and loss of function in mice results in embryonic lethality due to neural tube closure defects (Maller Schulman et al., 2008). In the current study, we characterized expression of TRIM71, MEX3A and ESRP1 by colocalization with well-known markers of undifferentiated spermatogonia in mouse testis. Importantly, all were found to be expressed by undifferentiated spermatogonia at both neonatal (P3) and the prepubertal (P10) stages of development. However, MEX3A and ESRP1 were found to also be expressed by advanced spermatogenic subtypes in adulthood. In contrast, TRIM71 expression was restricted to undifferentiated spermatogonia even in adult mice.
Although well studied in invertebrate models as a regulator of Let-7 miRNA (Reinhart et al., 2000; Slack et al., 2000), TRIM71 function in the mammalian germline is unknown. In the current study, we found that conditional inactivation in the spermatogenic lineage led to ablation of spermatogenesis. In addition to the undifferentiated spermatogonial population being significantly impacted in a Trim71-deficient state, the ability to transition to a differentiating state and therefore initiate a round of spermatogenesis was also impaired. Previous studies identified that in addition to expression by undifferentiated spermatogonia, ZBTB16 is also expressed by the first layer of differentiating spermatogonia (Niedenberger et al., 2015). Thus, at P10 when the first round of spermatogenesis commences, the decrease in ZBTB16+ cells could be due to deficiency in the first cohort of differentiating spermatogonia. However, the fact that the GFRA1+ spermatogonia population appeared to be unaffected following TRIM71 inactivation at P10 suggests that the SSC pool was intact. Furthermore, outcomes of co-IP analysis revealed another possible role for TRIM71 via physical interaction with EWSR1. Previous studies showed that EWSR1 influences XY bivalent formation and meiotic recombination, and regulates expression of colony stimulating factor 1 receptor (CSF1R) expression, which is enriched in THY1+ cells and is an extrinsic stimulator for self-renewal of mouse SSCs (Hume et al., 2008; Li et al., 2007; Oatley et al., 2009). Thus, TRIM71 may also be acting to inhibit meiotic entry and promote transition from a stem cell to progenitor state in spermatogonia.
Together, our present study describes the first set of RBPs captured by mRBPome from undifferentiated spermatogonia, identifying regulators of mRNA fate in these cells. We demonstrate that one of the RBP molecules, TRIM71, may through acting with EWSR1, and plays a key role in spermatogonia promoting the transition from an undifferentiated to a differentiating state.
MATERIALS AND METHODS
Animals
All animals were maintained under specific pathogen-free conditions at 20-26°C and 40-70% humidity in the animal core facility of Nanjing Medical University and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (ID: IACUC1403044). The Vasa-Cre (Ddx4-Cre) mouse line was maintained on a mixed genetic background (129/C57BL/6×FVB/N) (J006954, Jackson Laboratory). To generate a conditional Trim71 allele, intron 3 and the 3′ UTR were flanked by LoxP sites to delete exon 4. Male Trim71fl/fl or Trim71fl/+ mice were crossed with Ddx4-Cre transgenic mice to obtain male mice carrying Trim71fl/+ and Ddx4-CreTg/+ alleles, which were further crossed with female Trim71fl/fl mice to obtain mice with germ cell-specific Trim71−/− alleles. To prevent global recombination of Trim71 floxed alleles, only young (<7 weeks old) Ddx4-CreTg/+; Trim71f/+ males were used for breeding. Genotyping for Trim71 floxed alleles was performed by PCR analysis with tail genomic DNA and specific primer pairs listed in Table S3.
Undifferentiated spermatogonia used for each of the three mRBPome capture experiments were derived from C57BL/6 mice and cells used for transplantation were derived from B6;129S-Gt (ROSA) 26Sor/J transgenic mice (002073, The Jackson Laboratory), or Gt (ROSA)26Sortm4(ACTB-tdTomato, -EGFP)LUO/J transgenic mice (007576, The Jackson Laboratory).
Primary cultures of mouse undifferentiated spermatogonia
In order to provide sufficient cell numbers for mRBPome profiling, primary cultures of mouse undifferentiated spermatogonia were established as described previously (Wang et al., 2017; Wu et al., 2009). Briefly, THY1 (CD90.2)-positive cells were enriched from single cell suspensions of testes of 6-8 days postpartum C57BL/6J mice by magnetic-activated cell sorting. Cultures were maintained at 37°C in a humidified incubator in 5% CO2 with mitotically inactivated STO feeder cell monolayer and in chemically defined serum-free medium (SFM) consisting of MEMalpha (12561-049, Gibco) supplemented with 0.2% bovine serum albumin (BSA; A3803, Sigma-Aldrich), 10 μg/ml transferrin (T1283, Sigma-Aldrich), 50 μM free fatty acid mixture, 30 nM Na2SeO3 (481815, Sigma-Aldrich), 2 mM L-glutamine (25030, Gibco), 50 μM 2-mercaptoethanol (M7522, Sigma-Aldrich), 5 μg/ml insulin (I5500, Sigma-Aldrich), 10 mM HEPES (0887, Sigma-Aldrich), 60 μM putrescine (5780, Sigma-Aldrich), 20 ng/ml recombinant human GDNF (212-GD/CF, R&D Systems), 150 ng/ml rat GFRA1 (560-GR/CF, R&D Systems) and 1 ng/ml basic fibroblast growth factor (354060, Corning). Medium was changed every 2 days and cells were passaged onto fresh feeder cells at a ratio of 1:2 or 1:3 every 6-7 days.
Oligo-dT mRBPome capture
To systematically identify RBPs in undifferentiated spermatogonia, we performed mRNA interactome capture combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses as described previously (Castello et al., 2012, 2013) but with slight modifications. For each capture experiment, approximately 40,000,000 cultured undifferentiated spermatogonia were used. Following removal of medium from the culture dish, cells were rinsed three times with pre-warmed HBSS and placed on ice immediately before cross-linking at a distance of 10 cm from the UV source (CL-1000 Crosslinkers, UVP). Cells were irradiated by 15 mJ/cm2 for 1 min at 254 nm UV light for cross-linking as experimental groups; cells not irradiated by UV light were used as controls. Next, cells were collected in 1 ml of ice-cold HBSS per well using gentle pipetting, followed by centrifugation at 600 g for 5 min at 4°C, and placed in lysis buffer followed by rapid pipetting. The lysate was then homogenized by passing through a needle syringe (gauge 0.5 mm diameter, 301805, BD Biosciences) four times and incubated for 10 min at 4°C.
Lysates were incubated with 1 ml of oligo (dT)25 magnetic beads (S1419S, New England Biolabs) for 1 h at 4°C by gentle rotation and captured using a magnetic bead separation rack (50 ml Magnetic Separation Rack, New England Biolabs) at 4°C until beads were completely captured. mRNAs were captured by two rounds of enrichment with oligo (dT)25 beads according to the manufacturer's protocol.
To recover proteins, 100 μl of 10× RNase buffer and 1 μg of RNase A (ST576, Beyotime) were added to the remaining pooled eluate, and incubated for 1 h at 37°C. Proteins were purified using an Amicon Ultra-0.5 Centrifugal Filter Unit with Ultracel-3 membrane (UFC500324, Millipore) and eluted proteins were identified by LC-MS/MS. Aliquots (10 μl) of the final sample were used for protein quality control using silver staining and western blotting.
LC-MS/MS analysis
Liquid separation and mass spectrometry analysis were performed on a Nano CHIP-1D-LC system (Eksigent) and a TripleTOF 5600 mass spectrometer (AB Sciex), respectively. The LC gradient was formulated with MS buffer A (2% acetonitrile and 0.1% formic acid in HPLC water) and buffer B (2% water and 0.1% formic acid in acetonitrile). The analytical column (75 μm×15 cm) was used in the chip LC with 3 μm 150 Å Magic C18 resin (Eksigent). Each sample was injected and separated with a linear gradient of 2-35% buffer B over 120 min at a flow rate of 0.3 μl/min. The column was then flushed with 90% buffer B for 5 min, and re-equilibrated with 2% buffer B for 10 min. For shotgun experiments, the TripleTOF mass spectrometer was operated with a ‘top30’ method. From the 500 ms survey scan (TOF-MS), the top 30 most intense precursor ions were selected for subsequent automated MS/MS in measurements where each MS/MS event consisted of a 150 ms scan. The selection criteria for parent ions included intensity, where ions had to be greater than 200 counts per second, a charge state greater than 2+. The precursors selected for fragmentation were added to a dynamic exclusion list for 24 s. In the instances for which there were ≤20 parent ions per survey scan that met the selection criteria, ions were subjected to extended MS/MS accumulation time to maintain a constant total cycle of 3. The raw data were uploaded into ProteomeXchange via the PRIDE database (Perez-Riverol et al., 2019).
Database search and screening of candidate RNA-binding proteins
The raw data generated by the LC-MS/MS analyses were processed using MaxQuant software (version 1.5.2.8) (Cox and Mann, 2008), and searched against the reference mouse proteome from the UniProt database (release 2016_01, 58239 protein sequences) (Magrane and Uniport Consortium, 2011). Enzyme specificity was considered to be fully cleaved by trypsin, and two maximum missed cleavage sites were permitted. The minimum peptide length required was six. Carbamidomethyl (C) was set as fixed modification and variable modifications included Acetylation (Protein N-term) and Oxidation (M). The mass tolerance for precursor ions was set to 0.1 Da, and the mass tolerance for fragment ions was set to 40 ppm. At least one unique peptide was required for identification of a protein. The false discovery rate (FDR) of the identification was estimated by searching against databases with reverse protein sequences. The FDR cut-offs for site, peptide and protein were set to 0.05.
Bioinformatics analysis
All statistical plotting was conducted using R software (version 3.0.3). Protein accessions or probe set IDs of downloaded microarray data were mapped to Ensembl gene identities for functional annotation using BioMart (Ensembl version 83). Functional category enrichment (including GO and Pfam domains) was determined using ToppGene suite (release 2016_04) software. The whole genome was set as background and a FDR (Benjamini and Hochberg method) of less than 0.05 was required for determining significantly enriched terms. Classical and non-classical RBDs were defined according to previously reported criteria (Kwon et al., 2013; Lunde et al., 2007). Raw data of previous microarray studies were mined to investigate the expression patterns of candidate RBPs among different testis cell types (including spermatogonia, spermatocytes, spermatids and Sertoli cells) (Chalmel et al., 2007). We further classified gene expression patterns for the candidate RBPs into categories of spermatogonial specific (>0.9), enriched (∼0.7-0.9), high (∼0.5-0.7) and not high (<0.5) using the algorithm of Pattern Gene Finder (PaGeFinder, local version: 2014_03, http://bioinf.xmu.edu.cn/PaGeFinder/download.jsp) (Pan et al., 2012).
RNA isolation and quantitative real-time RT-PCR
RNAs were extracted using Trizol (15596-018, Invitrogen) and reverse transcribed using the PrimeScript RT reagent kit (RR036A, Takara Bio). Analysis of 18s, Actb, Gapdh, Trim71, Esrp1 and Mex3a RNA levels was performed in a StepOne Plus real-time RT-PCR system (ABI StepOnePlus, Life Technologies) using SYBR green reagent (Q141-02, Takara Bio) and specific primer pairs (Table S3). Each sample was amplified in triplicate and amplicon specificity was verified by dissociation curve analysis and agarose gel electrophoresis.
Western blotting
Cultured undifferentiated spermatogonia were lysed in RIPA buffer (P0013C, Beyotime) containing protease inhibitor cocktail (11697498001, Roche). Protein lysates were separated on 12% SDS-PAGE gels followed by electrotransfer (300 mA, 90 min) onto 0.22 μM PVDF membranes (162-0177, Bio-Rad). The blots were blocked for 2 h with 5% nonfat milk in Tris-buffered saline with 0.1% (v/v) Tween 20 (TBST) and then incubated overnight at 4°C with primary antibody (see Table S4). HRP-conjugated secondary antibody (see Table S4) and ECL reagent (NEL105001EA, PerkinElmer) were used to visualize immunopositive bands.
SDS-PAGE and silver staining
Silver staining was performed as described previously (Castello et al., 2012). Briefly, SDS-PAGE gels were fixed by incubating in a solution of 40% ethyl alcohol (Sinopharm Chemical Reagent Company) and 10% acetic acid (Sinopharm Chemical Reagent Company) for 1 h at room temperature. Fixed gels were subsequently dipped in sensitizing agent containing 50% ethyl alcohol, 0.2% sodium thiosulfate (Shanghai Lingfeng Chemical Reagent Company) and 6.8% anhydrous sodium acetate (Sinopharm Chemical Reagent Company) for 30 min at room temperature by gentle rocking. The gels were washed three times in distilled water for 5 min each and then dipped in silver solution for 20 min in the dark. Silver solution was prepared by dissolving 0.25 g silver nitrate (M122, Amresco) and 20 μl 37% formaldehyde in 100 ml distilled water. Following washing in distilled water for 5 min, proteins were visualized by incubation in 2.5% sodium carbonate (Sinopharm Chemical Reagent Company) and 0.74% formaldehyde at room temperature for 5-15 min. The reaction was terminated by addition of 0.4% glycine (YiFei Xue Biotechnology, China) and kept in 10% ethyl alcohol at 4°C for later experiments.
Immunofluorescence and immunohistochemistry
To prepare frozen sections, testes were fixed in 4% paraformaldehyde (PFA; P6148, Sigma-Aldrich) at 4°C for 24 h, 5% (w/v) sucrose/PBS for 3 h at room temperature and 30% (w/v) sucrose/PBS for 4°C overnight. Testes were then embedded with optimal cutting temperature (4583, Sakura) compound and stored at −80°C. Sections (5 μm) were cut using a cryostat microtome (Cryotome FSE, Thermo Fisher Scientific) and washed three times in 0.01 M PBS before incubation in 3% hydrogen peroxide, followed by boiling for 15 min in antigen retrieval buffers (P0083, Beyotime). After cooling to room temperature, sections were incubated with primary antibody (see Table S4) at 4°C overnight. TRITC-conjugated secondary antibody and FITC-conjugated secondary antibody for immunofluorescence or biotinylated secondary antibody (see Table S4) and diaminobenzidine (DAB; ZLI9017, Zhongshan Jinqiao Biotechnology) were used for visualization of immunostaining. Images were captured and processed using confocal (N-SIM, ZEISS) or compound (Axio Scope A1, ZEISS or LEICA DMi8) microscopy.
Whole-mount immunostaining
Seminiferous tubules were prepared as described previously but with minor modifications (Wu et al., 2009). Tubules from P10 mice were separated using sharp needle-nosed tweezers. Tubules from P70 (adult) mice were dispersed by incubation in a solution of 1.5 ml DNase I (7 mg/ml, DN25, Sigma-Aldrich) and 0.5 ml collagenase (1 mg/ml, C5138, Sigma-Aldrich) at 37°C incubator for 3-4 min. Tubules were fixed in 4% PFA for 3 h at 4°C, washed twice with PBS, and serially dehydrated with 25%, 50%,75% and 100% methanol in PBS with 0.1% (v/v) Tween 20 (PBST) for 5 min on ice with gentle rocking. After storage at −20°C for at least 2 h, tubules were rehydrated, incubated in 0.2% Triton X-100 and 0.9% hydrogen peroxide for 1 h, washed for 5 min in PBS and blocked by incubation in 5% fetal bovine serum, 2% BSA and 0.1% Triton X-100 in PBS for 3 h on ice with gentle rocking. The tubules were then incubated with primary antibody (see Table S4) in PBS with 2% BSA and 0.1% Triton X-100 at 4°C overnight, washed three times in PBST for 10 min each, and incubated with TRITC, FITC-conjugated secondary antibody and Hoechst 33342 (H3570, Thermo Fisher Scientific) or DAPI (D9542, Sigma-Aldrich) to stain DNA for 2 h at room temperature. Images were captured and processed by confocal microscopy (LSM700, Zeiss).
IP and protein MS analysis
TRIM71 antibodies (peptide antigens: KFGAQGSGFGQMDRPSGI and CVRAHQRVRLTKDHYIER) were generated as described previously (Chang et al., 2012; Worringer et al., 2014). Briefly, the peptide fragments (purity >99%) were synthesized by Sangon Biotech (China) and dissolved in sterile PBS (antigen storage, concentration: 1 mg/ml) followed by immunizing rabbits with 0.50 mg of antigen in complete Freund's Adjuvant (F5881, Sigma-Aldrich). Booster immunizations were performed 15 days apart for three times [0.25 mg of antigen in incomplete Freund's Adjuvant (F5506, Sigma-Aldrich)] and the rabbits were sacrificed 15 days after the last booster to collect serums.
For IP, whole testes from P10-P14 mice were lysed in RIPA buffer (50 mM Tris-Hcl, 150 mM NaCl, 1% NP40, 1 mM DTT, 0.5% sodium deoxycholate; 0.05% SDS, 1 mM EDTA) containing protease inhibitor cocktail (11697498001, Roche). Protein lysates were then blocked by incubation with Protein A Dynabeads (10002D, Invitrogen) for 2 h at 4°C followed by incubation with primary antibody (see Table S4) or control IgG (normal rabbit or mouse IgG, sc-2027, sc-2025, Santa Cruz Biotechnology) overnight at 4°C. The lysates were then incubated with Protein A Dynabeads for 2 h at 4°C and proteins eluted in SDS-PAGE buffer for 10 min at 100°C after stringent washing in lysis buffer. The protein profiles were determined by MS analysis.
Lentivirus-mediated knockdown
shRNA for TRIM71, ESRP1 and MEX3A knockdown (see Table S3 for sequences) were designed using the Genetic Perturbation Platform (https://portals.broadinstitute.org/gpp/public/), and plasmids constructed according to the manufacturer's guidelines (http://www.addgene.org/tools/protocols/plko/). Lentiviral treatments were conducted as described previously (Wang et al., 2017). Briefly, cells were exposed to lentiviral particles for 12 h and then seeded into 24-well plates containing STO feeder at a concentration of 2.0×105 cells/well in 1 ml of SFM. The pLKO.1 shRNA lentivirus vector and lentivirus packaging plasmids (pmd-REV and pmd-1G/pmd-LG) were generous gifts from the Dr Dahua Chen (State Key Laboratory of Reproductive Biology, Beijing, China).
Cell quantification and apoptosis analysis
Primary cultures of undifferentiated spermatogonia were harvested by trypsin-EDTA digestion 4 or 7 days after lentiviral transduction and pelleted by centrifugation (600 g 7 min at 4°C). Cells were then resuspended in 1 ml SFM and 10 μl of suspension was loaded into hemocytometer.
Apoptosis was assessed using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) detection kit (BD556547, BD Biosciences) according to the manufacturer's instructions. Briefly, cells were collected 7 days after lentiviral transduction by incubation in EDTA-free trypsin (25200-114, Gibco) at 37°C for 5 min. Cells were then washed in ice-cold HBSS, and resuspended in 110 μl buffer that contained 5 μl Annexin V-FITC and 5 μl PI. Samples were then incubated at room temperature in the dark for 15 min followed by addition of 200 μl of binding buffer. Flow cytometric analysis (BD FACS Verse flow cytometer, BD Biosciences) was used for determining the percentage of apoptotic cells in each sample.
Purification of cytoplasmic and nuclear fractions
To separate cellular fractions, 100 mg of testis tissue was homogenized in 1 ml fractionation buffer A (250 mM sucrose, 10 mM pH 8.0 Tris-HCl, 10 mM MgCl2, 1 mM EGTA, 1× protease inhibitor cocktail) and incubated at 4°C for 30 min with gentle rocking. Lysates were then passed through a 40 µm cell strainer followed by centrifugation at 300 g for 5 min at 4°C. Approximately 100 µl of supernatant was recovered as the cytoplasmic portion and the remaining cell pellet was further homogenized using 200 µl Cell Fractionation Buffer (from PARISTM Kit, AM1921, Life Technologies) followed by incubation on ice for 10 min and then centrifugation at 500 g for 5 min at 4°C. After washing three times in fractionation buffer A, 100 µl of ice-cold cell disruption buffer (from PARIS Kit, AM1921, Life Technologies) was added to the pellet and vortexed vigorously to homogenize the sample. Samples were again incubated on ice for 10 min followed by centrifugation at 21,130 g for 5 min at 4°C. The resulting supernatant was harvested as the nuclear fraction. Unfractioned lysate, cytoplasmic or nuclear fractions were then analyzed by western blotting to detect the expression of TRIM71. Blotting for GAPDH or H3K4me3 was used as positive controls for cytoplasmic and nuclear fractions, respectively.
Quantification and statistics
For quantification of cell number in testis cross-sections, images were captured using confocal microscopy at 10× magnification, and at least two random fields of view were used with samples from P35 and P56 mice. For P1 and P10 mice, images were captured either 10× (one field per cross-section) or 20× (two fields per cross-section) magnification. Only round seminiferous tubule cross-sections were used for analysis. Quantification of ZBTB16+ cells in cross-sections of P56 mice was normalized based on diameter of the seminiferous tubule analyzed. All quantitative data presentation was carried out using GraphPad Prism (version 8) software. Statistical differences between mean value was determined using Student's t-test function of SPSS software (IBM Corporation, version 19.0), and P-value <0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank CAM-SU International Cooperation Center, Soochow University, China for help in generation of Trim71 floxed mice. We thank Dr S. Eckardt for help with manuscript preparation.
Footnotes
Author contributions
Conceptualization: J.Z.L., J.M.O., X. Wu; Methodology: G.D., X. Wang, X. Wu; Software: T.Z.; Validation: G.D., X. Wang, W.X., M.W., L.Y., L.L., L.C.; Formal analysis: G.D., T.Z.; Investigation: G.D., M.W.; Resources: M.L.; Data curation: X. Wu; Writing - original draft: G.D.; Writing - review & editing: G.D., P.J.W., J.M.O., X. Wu; Supervision: J.M.O., X. Wu; Project administration: X. Wu; Funding acquisition: J.Z.L., X. Wu.
Funding
This study was supported by the National Basic Research Program of China (973 Program) (2018YFC1003302 to X. Wu); the National Natural Science Foundation of China (31872844, 31571537 to X. Wu; 31601195 to L.L., 31771588 to M.L.; 81471079, 31771307 to J.Z.L.); a key project grant from the State Key Laboratory of Reproductive Medicine (SKLRM-2018B1 to X. Wu); the Natural Science Foundation of Jiangsu Province for Youth (BK20150991 to L.L.); and project (C084) to G.D. from International Cooperation and Exchanges of Nanjing Medical University.
Data availability
The raw mRBPome data are available in the ProteomeXchange via the PRIDE database with accession number PXD017377.
References
- Ambros, V. and Horvitz, H. R. (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409-416. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- Barriga, F. M., Montagni, E., Mana, M., Mendez-Lago, M., Hernando-Momblona, X., Sevillano, M., Guillaumet-Adkins, A., Rodriguez-Esteban, G., Buczacki, S. J. A., Gut, M., et al. (2017). Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell 20, 801-816.e807. 10.1016/j.stem.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee, T. W., Park, J. W., Sheridan, K. I., Warzecha, C. C., Cieply, B. W., Rohacek, A. M., Xing, Y. and Carstens, R. P. (2015). The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. eLife 4, e08954. 10.7554/eLife.08954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau, B., Hochmuth, C. E. and Jasper, H. (2011). Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9, 402-411. 10.1016/j.stem.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucas, J., Fritz, C., Schmitt, A., Riabinska, A., Thelen, L., Peifer, M., Leeser, U., Nuernberg, P., Altmueller, J., Gaestel, M., et al. (2015). Label-free protein-RNA interactome analysis identifies Khsrp signaling downstream of the p38/Mk2 kinase complex as a critical modulator of cell cycle progression. PLoS ONE 10, e0125745. 10.1371/journal.pone.0125745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster, R. L. (2007). Male germline stem cells: from mice to men. Science 316, 404-405. 10.1126/science.1137741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, E., Ahrens, C. H., Mohanty, S., Baetschmann, H., Loevenich, S., Potthast, F., Deutsch, E. W., Panse, C., de Lichtenberg, U., Rinner, O., et al. (2007). A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576. 10.1038/nbt1300 [DOI] [PubMed] [Google Scholar]
- Buaas, F. W., Kirsh, A. L., Sharma, M., McLean, D. J., Morris, J. L., Griswold, M. D., de Rooij, D. G. and Braun, R. E. (2004). Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36, 647-652. 10.1038/ng1366 [DOI] [PubMed] [Google Scholar]
- Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C., Davey, N. E., Humphreys, D. T., Preiss, T., Steinmetz, L. M., et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393-1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Castello, A., Horos, R., Strein, C., Fischer, B., Eichelbaum, K., Steinmetz, L. M., Krijgsveld, J. and Hentze, M. W. (2013). System-wide identification of RNA-binding proteins by interactome capture. Nat. Protoc. 8, 491. 10.1038/nprot.2013.020 [DOI] [PubMed] [Google Scholar]
- Chakraborty, P., Buaas, F. W., Sharma, M., Snyder, E., de Rooij, D. G. and Braun, R. E. (2014). LIN28A marks the spermatogonial progenitor population and regulates its cyclic expansion. Stem Cells 32, 860-873. 10.1002/stem.1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel, F., Rolland, A. D., Niederhauser-Wiederkehr, C., Chung, S. S. W., Demougin, P., Gattiker, A., Moore, J., Patard, J.-J., Wolgemuth, D. J., Jégou, B., et al. (2007). The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl. Acad. Sci. USA 104, 8346-8351. 10.1073/pnas.0701883104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H. M., Martinez, N. J., Thornton, J. E., Hagan, J. P., Nguyen, K. D. and Gregory, R. I. (2012). Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 3, 923. 10.1038/ncomms1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Lai, F. and Niswander, L. (2012). The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 26, 803-815. 10.1101/gad.187641.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J., Shen, H., Yu, H., Li, H., Cheng, T., Lee, S. B. and Lee, B. C. (2011). Ewing sarcoma gene Ews regulates hematopoietic stem cell senescence. Blood 117, 1156-1166. 10.1182/blood-2010-04-279349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, T. A., Wan, L. and Dreyfuss, G. (2009). RNA and disease. Cell 136, 777-793. 10.1016/j.cell.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya, J. A., Hobbs, R. M., Barna, M., Cattoretti, G., Manova, K., Sukhwani, M., Orwig, K. E., Wolgemuth, D. J. and Pandolfi, P. P. (2004). Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 36, 653-659. 10.1038/ng1367 [DOI] [PubMed] [Google Scholar]
- Cox, J. and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Endo, T., Romer, K. A., Anderson, E. L., Baltus, A. E., de Rooij, D. G. and Page, D. C. (2015). Periodic retinoic acid–STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc. Natl. Acad. Sci. USA 112, E2347-E2356. 10.1073/pnas.1505683112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic, T., Bachorik, J. L., Yong, J. and Dreyfuss, G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977-1986. 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold, M. D., Hogarth, C. A., Bowles, J. and Koopman, P. (2012). Initiating meiosis: the case for retinoic acid. Biol. Reprod. 86, 35, 31-37. 10.1095/biolreprod.111.096610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C., Sidoli, S., Warneford-Thomson, R., Tatomer, D. C., Wilusz, J. E., Garcia, B. A. and Bonasio, R. (2016). High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell 64, 416-430. 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel, A. R., Yang, Q.-E., Oatley, M. J., Lord, T., Sablitzky, F. and Oatley, J. M. (2017). ID4 levels dictate the stem cell state in mouse spermatogonia. Development 144, 624-634. 10.1242/dev.146928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M. W., Castello, A., Schwarzl, T. and Preiss, T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. 19, 327-341. 10.1038/nrm.2017.130 [DOI] [PubMed] [Google Scholar]
- Hume, D. A., Sasmono, T., Himes, S. R., Sharma, S. M., Bronisz, A., Constantin, M., Ostrowski, M. C. and Ross, I. L. (2008). The Ewing sarcoma protein (EWS) binds directly to the proximal elements of the macrophage-specific promoter of the CSF-1 receptor (csf1r) gene. J. Immunol. Res. 180, 6733-6742. 10.4049/jimmunol.180.10.6733 [DOI] [PubMed] [Google Scholar]
- Kwon, S. C., Yi, H., Eichelbaum, K., Föhr, S., Fischer, B., You, K. T., Castello, A., Krijgsveld, J., Hentze, M. W. and Kim, V. N. (2013). The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122. 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- Li, H., Watford, W., Li, C., Parmelee, A., Bryant, M. A., Deng, C., O'Shea, J. and Lee, S. B. (2007). Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Invest. 117, 1314-1323. 10.1172/JCI31222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Wang, M., Wang, M., Wu, X., Geng, L., Xue, Y., Wei, X., Jia, Y. and Wu, X. (2016). A long non-coding RNA interacts with Gfra1 and maintains survival of mouse spermatogonial stem cells. Cell Death Dis. 7, e2140. 10.1038/cddis.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Castello, A., Fischer, B., Leicht, S., Föehr, S., Frese, C. K., Ragan, C., Kurscheid, S., Pagler, E., Yang, H., et al. (2016). The cardiomyocyte RNA-binding proteome: links to intermediary metabolism and heart disease. Cell Rep. 16, 1456-1469. 10.1016/j.celrep.2016.06.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepelt, A., Naarmann-de Vries, I. S., Simons, N., Eichelbaum, K., Föhr, S., Archer, S. K., Castello, A., Usadel, B., Krijgsveld, J., Preiss, T., et al. (2016). Identification of RNA-binding proteins in macrophages by interactome capture. Mol. Cell. Proteomics 15, 2699-2714. 10.1074/mcp.M115.056564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, T., Oatley, M. J. and Oatley, J. M. (2018). Testicular architecture is critical for mediation of retinoic acid responsiveness by undifferentiated spermatogonial subtypes in the mouse. Stem Cell Rep. 10, 538-552. 10.1016/j.stemcr.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde, B. M., Moore, C. and Varani, G. (2007). RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. 8, 479. 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane, M. and Uniport Consortium . (2011). UniProt Knowledgebase: a hub of integrated protein data. Database 2011, bar009. 10.1093/database/bar009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller Schulman, B. R., Liang, X., Stahlhut, C., DelConte, C., Stefani, G. and Slack, F. J. (2008). The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7, 3935-3942. 10.4161/cc.7.24.7397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima, A., Tanaka, M. and Itoh, T. (2014). Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14, 561-574. 10.1016/j.stem.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., Sharma, M., Nabeshima, Y.-I., Braun, R. E. and Yoshida, S. (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62-67. 10.1126/science.1182868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, D. T. T., Richter, D., Michel, G., Mitschka, S., Kolanus, W., Cuevas, E. and Wulczyn, F. G. (2017). The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ. 24, 1063. 10.1038/cdd.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenberger, B. A., Busada, J. T. and Geyer, C. B. (2015). Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction 149, 329. 10.1530/REP-14-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi, Y., Imai, A., Nishimura, H., Yui, K., Kikuchi, A., Koike, H., Saga, Y. and Suzuki, A. (2019). Essential role of mouse Dead end1 in the maintenance of spermatogonia. Dev. Biol. 445, 103-112. 10.1016/j.ydbio.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Oatley, J. M., Avarbock, M. R., Telaranta, A. I., Fearon, D. T. and Brinster, R. L. (2006). Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. USA 103, 9524-9529. 10.1073/pnas.0603332103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley, J. M., Oatley, M. J., Avarbock, M. R., Tobias, J. W. and Brinster, R. L. (2009). Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 136, 1191-1199. 10.1242/dev.032243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J., Eckardt, S., Leu, N. A., Buffone, M. G., Zhou, J., Gerton, G. L., McLaughlin, K. J. and Wang, P. J. (2009). Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev. Biol. 330, 167-174. 10.1016/j.ydbio.2009.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J.-B., Hu, S.-C., Wang, H., Zou, Q. and Ji, Z.-L. (2012). PaGeFinder: quantitative identification of spatiotemporal pattern genes. Bioinformatics 28, 1544-1545. 10.1093/bioinformatics/bts169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K.-E., Kaucher, A. V., Powell, A., Waqas, M. S., Sandmaier, S. E. S., Oatley, M. J., Park, C.-H., Tibary, A., Donovan, D. M., Blomberg, L. A., et al. (2017). Generation of germline ablated male pigs by CRISPR/Cas9 editing of the NANOS2 gene. Sci. Rep. 7, 40176. 10.1038/srep40176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, B., Sousa, S., Barros, R., Carreto, L., Oliveira, P., Oliveira, C., Chartier, N. T., Plateroti, M., Rouault, J.-P., Freund, J.-N., et al. (2013). CDX2 regulation by the RNA-binding protein MEX3A: impact on intestinal differentiation and stemness. Nucleic Acids Res. 41, 3986-3999. 10.1093/nar/gkt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., Inuganti, A., Griss, J., Mayer, G., Eisenacher, M., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442-D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R. and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Rooij, D. G. and Russell, L. D. (2000). All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 21, 776-798. [PubMed] [Google Scholar]
- Sada, A., Suzuki, A., Suzuki, H. and Saga, Y. (2009). The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 325, 1394-1398. 10.1126/science.1172645 [DOI] [PubMed] [Google Scholar]
- Scherrer, T., Femmer, C., Schiess, R., Aebersold, R. and Gerber, A. P. (2011). Defining potentially conserved RNA regulons of homologous zinc-finger RNA-binding proteins. Genome Biol. 12, R3. 10.1186/gb-2011-12-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrans-Stassen, B. H. G. J., Saunders, P. T. K., Cooke, H. J. and de Rooij, D. G. (2001). Nature of the spermatogenic arrest in Dazl−/− mice. Biol. Reprod. 65, 771-776. 10.1095/biolreprod65.3.771 [DOI] [PubMed] [Google Scholar]
- Schrimpf, S. P., Weiss, M., Reiter, L., Ahrens, C. H., Jovanovic, M., Malmström, J., Brunner, E., Mohanty, S., Lercher, M. J., Hunziker, P. E., et al. (2009). Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7, e1000048. 10.1371/journal.pbio.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang, N., Zhu, H., De Soysa, T. Y., Shinoda, G., Seligson, M. T., Tsanov, K. M., Nguyen, L., Asara, J. M., Cantley, L. C. and Daley, G. Q. (2013). Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155, 778-792. 10.1016/j.cell.2013.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, F. J., Basson, M., Liu, Z., Ambros, V., Horvitz, H. R. and Ruvkun, G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659-669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Suzuki, A., Niimi, Y., Shinmyozu, K., Zhou, Z., Kiso, M. and Saga, Y. (2016). Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep. 17, 37-46. 10.15252/embr.201540828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima, S., Takehashi, M., Lee, J., Chuma, S., Okano, M., Hata, K., Suetake, I., Nakatsuji, N., Miyoshi, H., Tajima, S., et al. (2009). Abnormal DNA methyltransferase expression in mouse germline stem cells results in spermatogenic defects. Biol. Reprod. 81, 155-164. 10.1095/biolreprod.108.074708 [DOI] [PubMed] [Google Scholar]
- Takashima, S., Hirose, M., Ogonuki, N., Ebisuya, M., Inoue, K., Kanatsu-Shinohara, M., Tanaka, T., Nishida, E., Ogura, A. and Shinohara, T. (2013). Regulation of pluripotency in male germline stem cells by Dmrt1. Genes Dev. 27, 1949-1958. 10.1101/gad.220194.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima, S., Kanatsu-Shinohara, M., Tanaka, T., Morimoto, H., Inoue, K., Ogonuki, N., Jijiwa, M., Takahashi, M., Ogura, A. and Shinohara, T. (2015). Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Rep. 4, 489-502. 10.1016/j.stemcr.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Guo, Y., Wang, M., Zhou, T., Xue, Y., Du, G., Wei, X., Wang, J., Qi, L., Zhang, H., et al. (2017). The glial cell-derived neurotrophic factor (GDNF)-responsive phosphoprotein landscape identifies raptor phosphorylation required for spermatogonial progenitor cell proliferation. Mol. Cell. Proteomics 16, 982-997. 10.1074/mcp.M116.065797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, H.-H., Imami, K., Baltz, A. G., Kolinski, M., Beldovskaya, A., Selbach, M., Small, S., Ohler, U. and Landthaler, M. (2016). The mRNA-bound proteome of the early fly embryo. Genome Res. 26, 1000-1009. 10.1101/gr.200386.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer, K. A., Rand, T. A., Hayashi, Y., Sami, S., Takahashi, K., Tanabe, K., Narita, M., Srivastava, D. and Yamanaka, S. (2014). The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14, 40-52. 10.1016/j.stem.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Schmidt, J. A., Avarbock, M. R., Tobias, J. W., Carlson, C. A., Kolon, T. F., Ginsberg, J. P. and Brinster, R. L. (2009). Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc. Natl. Acad. Sci. USA 106, 21672-21677. 10.1073/pnas.0912432106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q., Song, R., Ortogero, N., Zheng, H., Evanoff, R., Small, C. L., Griswold, M. D., Namekawa, S. H., Royo, H., Turner, J. M., et al. (2012). The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J. Biol. Chem. 287, 25173-25190. 10.1074/jbc.M112.362053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Toselli, P. A., Russell, L. D. and Seldin, D. C. (1999). Globozoospermia in mice lacking the casein kinase II α′ catalytic subunit. Nat. Genet. 23, 118. 10.1038/12729 [DOI] [PubMed] [Google Scholar]
- Yamaji, M., Jishage, M., Meyer, C., Suryawanshi, H., Der, E., Yamaji, M., Garzia, A., Morozov, P., Manickavel, S., McFarland, H. L., et al. (2017). DND1 maintains germline stem cells via recruitment of the CCR4–NOT complex to target mRNAs. Nature 543, 568. 10.1038/nature21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Ratanasirintrawoot, S., Chandrasekaran, S., Wu, Z., Ficarro, S. B., Yu, C., Ross, C. A., Cacchiarelli, D., Xia, Q., Seligson, M., et al. (2016). LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell 19, 66-80. 10.1016/j.stem.2016.05.009 [DOI] [PubMed] [Google Scholar]
- Zheng, K., Wu, X., Kaestner, K. H. and Wang, P. J. (2009). The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev. Biol. 9, 38. 10.1186/1471-213X-9-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Shirakawa, T., Ohbo, K., Sada, A., Wu, Q., Hasegawa, K., Saba, R. and Saga, Y. (2015). RNA binding protein Nanos2 organizes post-transcriptional buffering system to retain primitive state of mouse spermatogonial stem cells. Dev. Cell 34, 96-107. 10.1016/j.devcel.2015.05.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



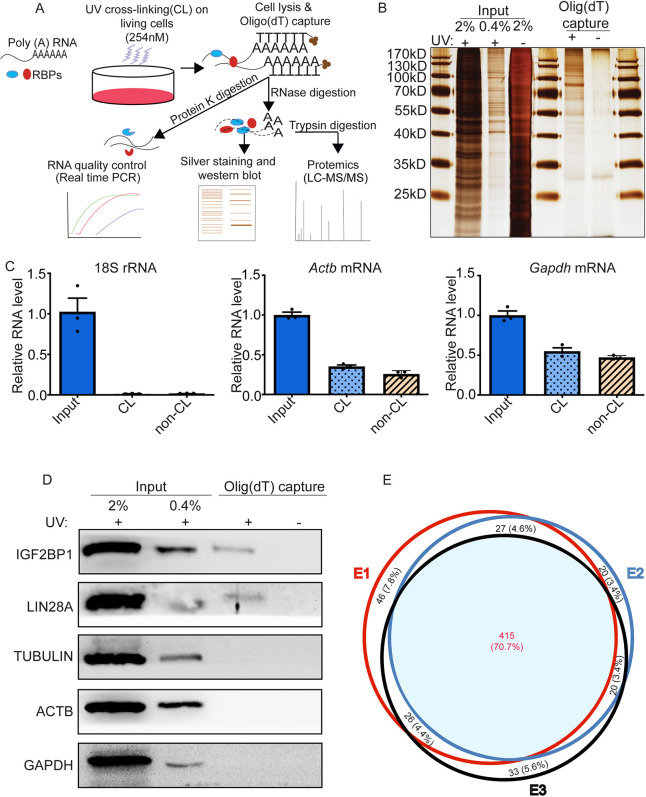
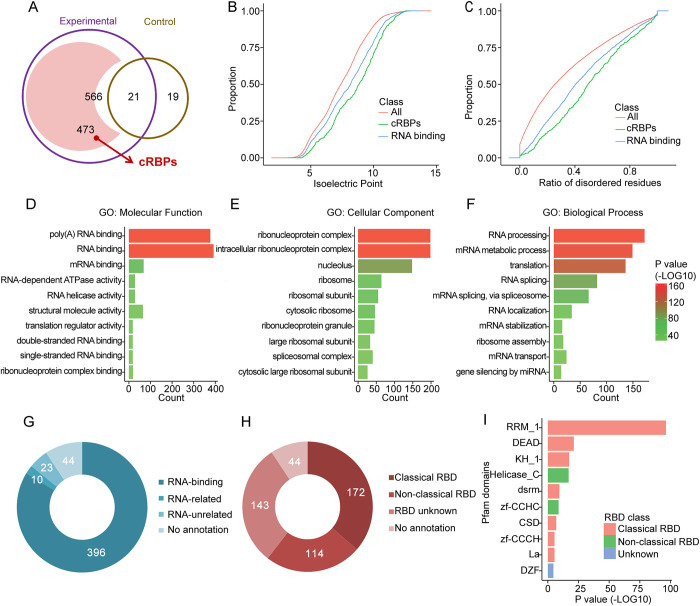
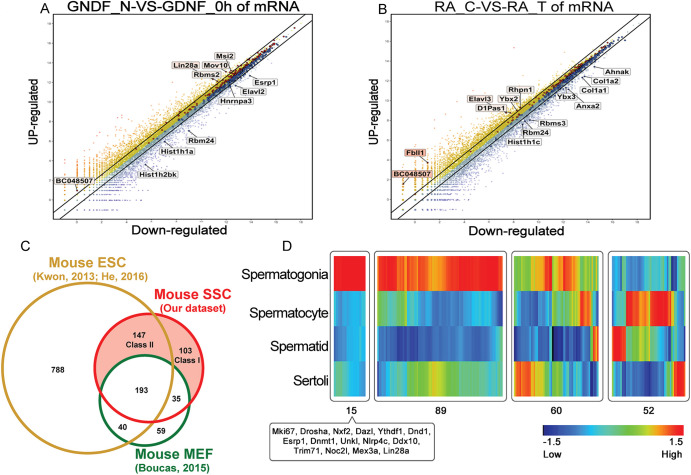
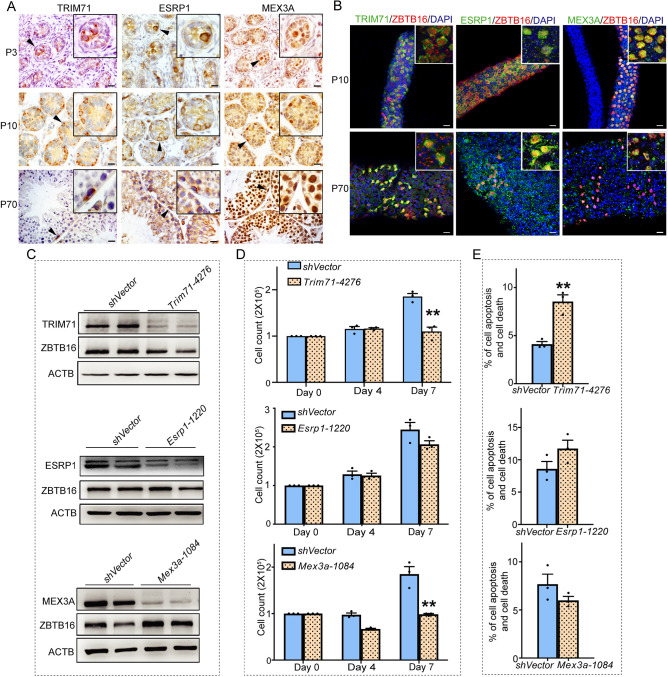
![Fig. 5. Impact of Trim71 conditional knockout in the male germline. (A) Representative images of testis and epididymis from adult (P56) Trim71 conditional knockout (cKO) and control mice. Ruler shows 1 mm per division. (B) Representative images of Hematoxylin and Eosin (H&E)-stained cross-sections of seminiferous tubules and epididymis from P56 control and Trim71 cKO mice. (C-G) Representative images of immunofluorescence staining for the pan germ cell marker GCNA1 at P1 (C), the undifferentiated spermatogonial marker ZBTB16 and primitive spermatogonial marker GFRA1 at P10 (D) and P56 (E), and the undifferentiated spermatogonial marker ZBTB16 and differentiating marker STRA8 at P10 (F) and P56 (G) in cross-sections of testes from control and Trim71 cKO mice. (H) Quantification of total germ cell number (based on GCNA1+ cells) in seminiferous tubule cross-sections from control and Trim71 cKO mice at P1. (I,J) Quantification of undifferentiated spermatogonial number [based on ZBTB16+ cells/seminiferous tubule at P10 or number (#) of ZBTB16+ cells × seminiferous tubule diameter (µm) at P56], percentage of the population that is the most primitive subset (based on GFRA1+ among ZBTB16+ cells), and percentage of the population that has transitioned to a differentiating state (based on STRA8+ among ZBTB16+ cells) in cross-sections of testes from control and Trim71 cKO mice at prepubertal (P10) (I) and adult (P56) (J) age points. Data in H-J are mean±s.e.m. for n=3 different mice, dots represent average values of individual mice, **P<0.01 (Student's t-test). Scale bars: 20 μm.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/5f82/10679512/f7370b9ac5e5/develop-147-184655-g5.jpg)