Summary
Mechanisms that underlie homeostatic plasticity have been extensively investigated at single-cell levels in animal models, but are less well understood at the network level. Here, we used microelectrode arrays to characterize neuronal networks following induction of homeostatic plasticity in human induced pluripotent stem cell (hiPSC)-derived glutamatergic neurons co-cultured with rat astrocytes. Chronic suppression of neuronal activity through tetrodotoxin (TTX) elicited a time-dependent network re-arrangement. Increased expression of AMPA receptors and the elongation of axon initial segments were associated with increased network excitability following TTX treatment. Transcriptomic profiling of TTX-treated neurons revealed up-regulated genes related to extracellular matrix organization, while down-regulated genes related to cell communication; also astrocytic gene expression was found altered. Overall, our study shows that hiPSC-derived neuronal networks provide a reliable in vitro platform to measure and characterize homeostatic plasticity at network and single-cell levels; this platform can be extended to investigate altered homeostatic plasticity in brain disorders.
Keywords: hiPSC, homeostatic plasticity, human neuronal networks
Highlights
-
•
TTX treatment elicits time-dependent re-arrangement of hiPSC-derived neuronal networks
-
•
Homeostatic plasticity involves increased mEPSC amplitude and elongation of AIS
-
•
TTX treatment induces transcriptional changes in neurons and astrocytes
In this article, Yuan et al. describe a human in vitro human neuronal model for studying homeostatic plasticity at the network level, and gain insight into how synaptic strength and intrinsic excitability cooperate to stabilize neuronal network activity. They demonstrate that chronic deprivation of neuronal activity with TTX elicits a time-dependent re-arrangement of neuronal networks and transcriptional changes in neurons and astrocytes, which may underlie the network re-arrangement.
Introduction
In the healthy brain, neuronal activity adapts dynamically to a changing environment. Two principal mechanisms accommodate such adaptive behavior: Hebbian plasticity and homeostatic plasticity. Traditional forms of Hebbian plasticity, such as long-term potentiation (LTP) and long-term depression (LTD), induce changes in the strength of individual synaptic connections and constitute the biological substrate of learning and memory consolidation. However, without effective negative feedback regulation, their effects could cause a destabilization of neuronal networks (Fox and Stryker, 2017). Homeostatic plasticity is a negative feedback mechanism that stabilizes neuronal network activity by adjusting synaptic strength and intrinsic properties of the neurons in response to activity perturbations during learning and development. Eventually, the homeostatic changes that occur through these mechanisms prevent the neuronal networks from becoming hypo- or hyper-active (Tien and Kerschensteiner, 2018; Wefelmeyer et al., 2016; Turrigiano, 2012; Watt and Desai, 2010). As such, homeostatic plasticity mechanisms play a critical role in the correct functioning of the nervous system (Antoine et al., 2019). Previous studies identified disrupted homeostatic plasticity in rodent models for different neurodevelopmental disorders (NDDs), such as Fragile X syndrome, Kleefstra syndrome, Rett syndrome, and tuberous sclerosis, suggesting that altered or insufficient homeostatic plasticity during development contributes to cognitive and behavioral impairments that characterize NDDs (Bulow et al., 2019; Lee et al., 2018; Benevento et al., 2016; Dani et al., 2005; Zeng et al., 2007; Antoine et al., 2019).
While homeostatic plasticity mechanisms have been well characterized at the single-cell level, using rodent dissociated-cell cultures and organotypic slice cultures (Moulin et al., 2020; Debanne et al., 2019; Debanne and Russier, 2019), homeostatic plasticity at the network level is poorly understood. In addition, it remains unclear how intrinsic properties and synaptic strength cooperate to stabilize neuronal networks in response to changes in activity levels (Antoine et al., 2019; Turrigiano, 2011). Thus, establishing a framework to assess homeostatic plasticity at the network level in a human model is essential for a better understanding of human neurodevelopment and neuronal function, both in normal and pathological conditions.
Human induced pluripotent stem cell (hiPSC)-derived neuronal models allow for the generation of networks of controllable cellular composition that contain spontaneously active and synaptically connected neurons in a patient-specific background (Frega et al., 2017; Bardy et al., 2016). They are increasingly used as a model system to understand the pathophysiology of brain disorders, primarily by studying spontaneous activity under basal conditions (McCready et al., 2022; van Hugte and Nadif Kasri, 2019). However, despite the overwhelming implication of synaptic and homeostatic plasticity deficits in brain disorders, only a handful of studies have investigated mechanisms of plasticity in hiPSC-derived neurons (Cordella et al., 2022; Pre et al., 2022; Meijer et al., 2019; Zhang et al., 2018). In particular, no attention has been given to the study of homeostatic plasticity at the network level in hiPSC-derived neurons.
Here, we established a model of tetrodotoxin (TTX)-induced homeostatic plasticity in co-cultures of hiPSC-derived glutamatergic neurons on microelectrode arrays (MEAs), which we have previously shown to facilitate non-invasive, reproducible, real-time, and multidimensional measurement of activity in hiPSC-derived neuronal networks (Mossink et al., 2021). We characterized single-cell and network changes induced with this form of homeostatic plasticity, together with changes in gene expression that may underlie them. We show that hiPSC-derived neuronal networks form a reliable platform to measure and characterize homeostatic plasticity, which can also be harnessed to investigate homeostatic plasticity in human models for brain disorders at the network and single-cell level.
Results
TTX-induced homeostatic plasticity leads to re-arrangement of neuronal networks
Populations of hiPSC-derived neurons cultured on MEAs generate highly synchronous network bursting activity within a few days in vitro (DIV) (Frega et al., 2019; Frega et al., 2017). During the initial 2 weeks of differentiation, the activity of the control glutamatergic neuronal network primarily consisted of random spikes (isolated asynchronous spikes) and bursts (high frequency action potentials). As differentiation progressed, these bursts organized into network bursts (rhythmic, synchronous events). Throughout maturation, the networks displayed an increase in mean firing rate (MFR) and (network) burst rate (NBR/BR), and a decrease in (network) burst duration (NBD/BD), and percentage of random spikes (PRS). Starting from DIV 27, these parameters reached a plateau, indicating stabilization of neuronal network activity.
To investigate the effect of chronic activity perturbation on neuronal network dynamics, we co-cultured glutamatergic neurons, derived from three independent control hiPSC lines (Ctrl1, 2, and 3), with rat astrocytes on MEAs and treated them with TTX, a sodium channel blocker, or with a control vehicle (Figure 1A). This procedure allows for testing if neuronal networks adapt to changes in neuronal activity by means of homeostatic plasticity. At both DIV 30 and DIV 49, the presence of rhythmic and synchronous network bursts, integrated by many spikes and involving most of the channels (Figures 1B, 1C, S1A, S1B, and S1E), indicated that control hiPSC-derived glutamatergic neurons had organized into synaptically connected and spontaneously active neuronal networks (Frega et al., 2017).
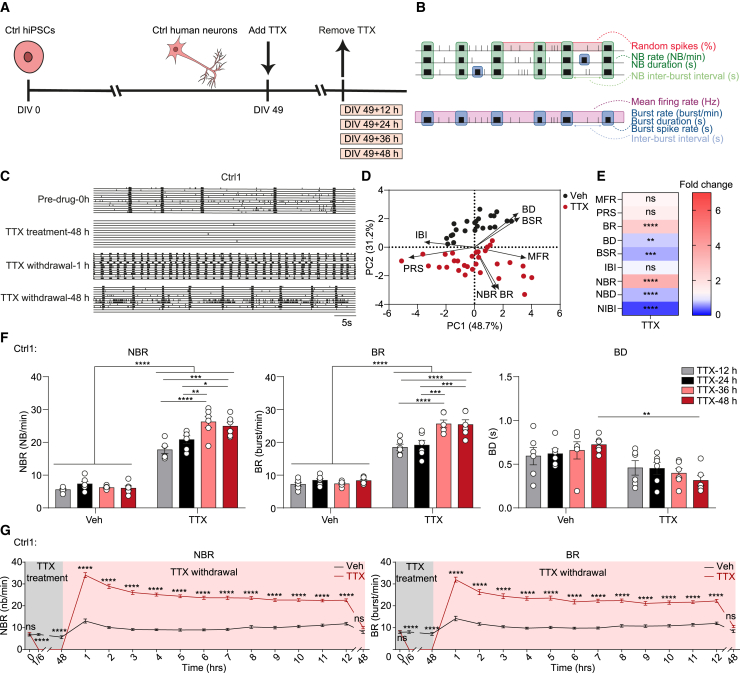
Figure 1.
Tetrodotoxin (TTX)-treated neurons show a time-dependent re-arrangement of the networks
(A) Schematic representation of human induced pluripotent stem cell (hiPSC) differentiation and TTX treatment workflow.
(B) Schematic overview of extracted parameters from microelectrode arrays (MEAs) recordings. NB = network burst.
(C) Representative raster plots showing 1 min of spontaneous activity from hiPSC-derived neuronal networks (Ctrl1) before and after 1 μM TTX treatment, including before addition of TTX (Pre-drug-0 h), 48 h after addition of TTX (TTX treatment-48 h), 1 h after TTX withdrawal (TTX withdrawal-1 h), and 48 h after TTX withdrawal (TTX withdrawal-48 h).
(D) Principal-component analysis (PCA) plot displaying PC1 and PC2 of all nine analyzed MEA parameters for vehicle-treated (Veh) and 48-h TTX-treated (TTX) neurons (Ctrl1 and Ctrl2). All MEA parameters were measured 1 h after TTX withdrawal. MFR = mean firing rate, PRS = percentage of random spike, BR = mean burst rate, BD = mean burst duration, BSR = burst spike rate, IBI = inter-burst interval, NBR = network burst rate, NBD = network burst duration, NIBI = Network burst IBI. n = number of MEA wells/batches: Veh n = 23/5, TTX n = 31/5 (Veh: n = 15/3 from Ctrl1, n = 8/2 from Ctrl2; TTX: n = 21/3 from Ctrl1, n = 10/2 from Ctrl2).
(E) Heatmap showing fold changes of all nine analyzed MEA parameters after TTX treatment (Ctrl1 and Ctrl2). All MEA parameters were measured at 1 h after TTX withdrawal. n = number of MEA wells/batches: Veh n = 23/5, TTX n = 31/5 (Veh: n = 15/3 from Ctrl1, n = 8/2 from Ctrl2; TTX: n = 21/3 from Ctrl1, n = 10/2 from Ctrl2).
(F) Bar graphs showing the effect of 12 h, 24 h, 36 h, and 48 h TTX treatment on the NBR, BR, and BD for Ctrl1 neuronal networks. All MEA parameters were measured 1 h after TTX withdrawal. n = number of MEA wells/batches: Veh n = 6/2 for each group, TTX n = 6/2 for each group.
(G) Quantification of NBR and BR over time for vehicle-treated (Veh) and 48-h TTX-treated (TTX) neurons (Ctrl1). n = number of MEA wells/batches: Veh n = 15/3, TTX n = 21/3. Data represent means ± SEM. ns: not significant, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001. For (E), unpaired Student’s t test was performed between two groups. For (F) and (G), two-way ANOVA test followed by a post hoc Bonferroni correction was performed between conditions. All means, SEMs, and test statistics are listed in Table S3.
TTX treatment for 48 h on DIV 49 networks completely abolished neuronal activity (Figure 1C). Removing TTX after the 48-h treatment led to a significant increase in network bursting activity, compared with pre-drug and vehicle-treated control conditions (Figure 1C, S1B, S1E, and S1F). To obtain a full picture of the network changes, we performed a principal-component analysis (PCA), including nine independent MEA parameters (Mossink et al., 2021). PCA indicated a clear separation between TTX- and vehicle-treated neuronal networks (Figure 1D). The analysis identified mean NBR, BR, BD, and burst spike rate (BSR) as the primary parameters responsible for the observed changes between the two treatment groups (Figure 1D). NBR and BR were increased, while BD, BSR, NBD, and network inter-burst interval (NIBI) were decreased following TTX exposure compared with the vehicle-treated condition. In contrast, we found no change in MFR, PRS, and inter-burst interval (IBI) after TTX exposure (Figure 1E). Moreover, upon TTX treatment, the cross correlation within the entire neuronal network was increased. Specifically, neuronal networks subjected to TTX treatment showed increased link weight, while the number of links remained unchanged (Figure S1I). Together, these results suggest that the induction of homeostatic plasticity by prolonged TTX exposure leads to re-arrangement of neuronal networks, without an increase in network activity (as measured by MFR).
Previous studies in rodents have shown that homeostatic plasticity response to TTX at the single-cell level is dependent on the duration of TTX treatment (Benevento et al., 2016; Ibata et al., 2008). We tested if this was also the case at the network level in human neuronal networks by applying TTX for 12, 24, 36, and 48 h. Following the withdrawal of TTX, we observed a significant increase in both NBR and BR already after only 12 h of treatment. These parameters continued to increase with longer treatment durations, peaking at 36 to 48 h of TTX treatment before reaching a plateau. However, BD only decreased after 48 h of TTX exposure, and not with shorter TTX exposures (Figures 1F and S1C). Of interest, when we recorded the network activity on an hourly basis to investigate if the changes in network activity persisted after TTX withdrawal, we found that 48-h TTX-treated neuronal networks gradually returned to pre-drug levels within 48 h (Figures 1G and S1D). This confirms the continuous nature of homeostatic plasticity in human neuronal networks.
GluA2-lacking AMPA receptors are expressed in an early stage of homeostatic plasticity in hiPSC-derived neurons
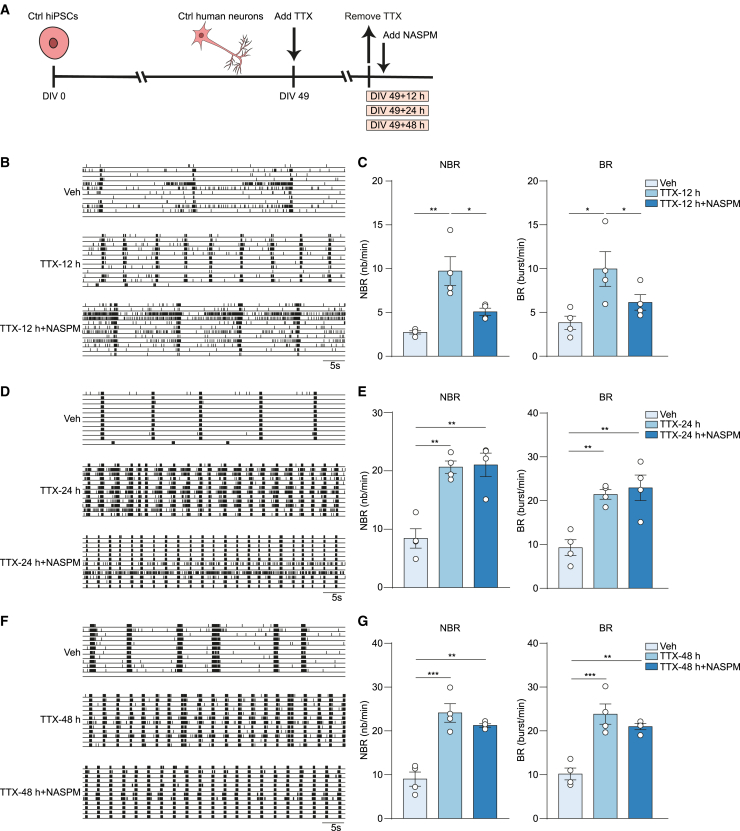
Membrane insertion of GluA2-lacking AMPA receptors (calcium-permeable AMPA receptors) is required for the induction of homeostatic plasticity. GluA2-lacking AMPA receptors are subsequently replaced with GluA2-containing AMPA receptors (Sutton et al., 2006; Hou et al., 2008). These mechanisms regulate synaptic strength and are known to underlie synaptic scaling mechanisms (Man, 2011; Soares et al., 2013). To investigate if these mechanisms are also contributing to homeostatic plasticity in this human model system, we first tested if activity in control hiPSC-derived neuronal networks is driven by AMPA receptors, by adding a selective antagonist of AMPA receptors (NBQX). NBQX completely abolished the network bursting activity compared with vehicle-treated conditions (Figures S2C and S2D) (Frega et al., 2019). Next, we applied 1-naphthyl acetyl spermine trihydrochloride (NASPM), a specific antagonist for GluA2-lacking AMPA receptors, to hiPSC-derived neuronal networks that were already exposed to TTX for 12, 24, and 48 h (Figure 2A). We found that NASPM prevented the TTX-induced increase in NBR and BR (Figures 2B and 2C) in the neuronal networks treated for 12 h with TTX. Importantly, NASPM had no effect on NBR and BR before TTX application (Figures S2A and S2B), confirming that the expression of GluA2-lacking AMPA receptors was exclusively induced by neuronal activity suppression with TTX. After longer TTX exposure, i.e., 24 h and 48 h, NASPM did not affect the increase in NBR and BR (Figures 2D–2G), suggesting that GluA2-lacking AMPA receptors had already been replaced by GluA2-containing AMPA receptors at these time points. Taken together, these results indicate that GluA2-lacking AMPA receptors do contribute to network re-arrangement in hiPSC-derived neuronal networks, where they are incorporated into synapses in an early stage of TTX-induced homeostatic plasticity (0–12 h).
Figure 2.
GluA2-lacking AMPA receptors are expressed in an early stage of homeostatic plasticity
(A) Schematic representation of tetrodotoxin (TTX) treatment and 1-naphthyl acetyl spermine trihydrochloride (NASPM) treatment workflow.
(B) Representative raster plots showing 1 min of spontaneous activity from Ctrl2 neuronal networks grown on microelectrode arrays (MEAs). Where indicated, the neurons were vehicle-treated (Veh), or neurons were first treated with 1 μM TTX for 12 h and then TTX was removed (TTX-12 h), or neurons were treated with 10 μM NASPM after TTX was removed (TTX-12 h+NASPM).
(C) Bar graphs showing the quantification of mean network burst rate (NBR) and mean burst rate (BR) for (B). n = number of MEA wells/batches: Veh n = 4/1, TTX-12 h n = 4/1, TTX-12 h+NASPM n = 4/1.
(D) Representative raster plots showing 1 min of spontaneous activity from Ctrl1 neuronal networks grown on MEAs. Where indicated, the neurons were vehicle-treated (Veh), or neurons were first treated with 1 μM TTX for 24 h and then TTX was removed (TTX-24 h), or neurons were treated with 10 μM NASPM after TTX was removed (TTX-24 h+NASPM).
(E) Bar graphs showing the quantification of NBR and BR for (D). n = number of MEA wells/batches: Veh n = 4/1, TTX-24 h n = 4/1, TTX-24 h+NASPM n = 4/1.
(F) Representative raster plots showing 1 min of spontaneous activity from Ctrl1 neuronal networks grown on MEAs. Where indicated, the neurons were vehicle-treated (Veh), or neurons were first treated with 1 μM TTX for 48 h and then TTX was removed (TTX-48 h), or neurons were treated with 10 μM NASPM after TTX was removed (TTX-48 h+NASPM).
(G) Bar graphs showing the quantification of NBR and BR for (F). n = number of MEA wells/batches: Veh n = 4/1, TTX-48 h n = 4/1, TTX-48 h+NASPM n = 4/1. Data represent means ± SEM. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, one-way ANOVA test followed by a post hoc Bonferroni correction was performed between conditions. All means, SEMs, and test statistics are listed in Table S3.
Homeostatic plasticity involves increased miniature excitatory postsynaptic current amplitude and elongation of axon initial segments in hiPSC-derived neurons
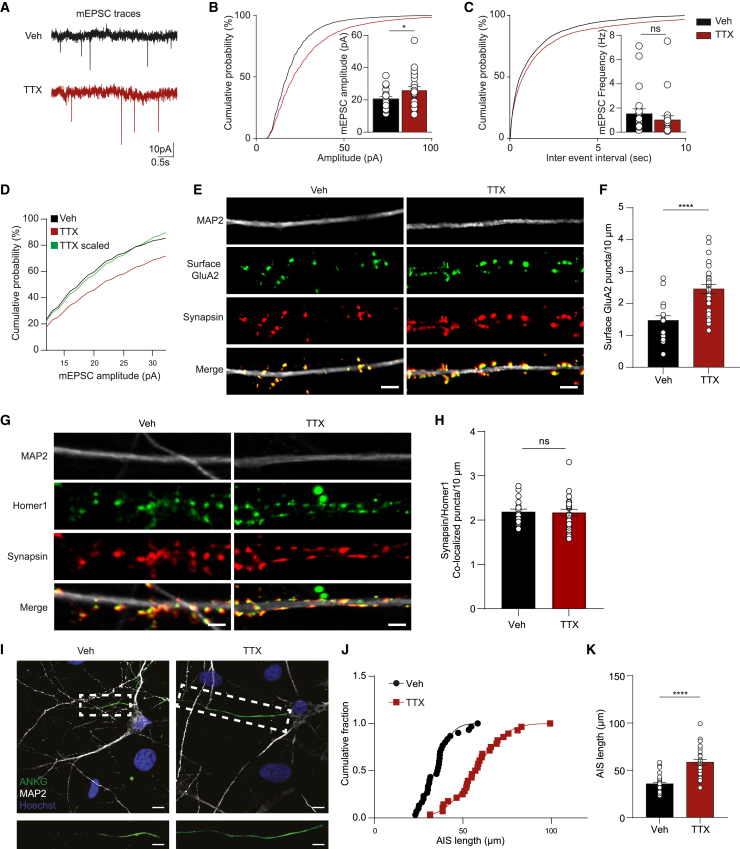
We next investigated presynaptic and postsynaptic contributions to synaptic scaling following TTX treatment at the single-cell level. With whole-cell electrophysiological recordings of hiPSC-derived neurons treated with vehicle or TTX for 48 h, we found that TTX treatment induced an increase in miniature excitatory postsynaptic current (mEPSC) amplitude without affecting mEPSC frequency (Figures 3A–3C and S3A–S3C). The changes in mEPSC amplitude were scalable, as they were observed across different mEPSC amplitudes (Figures 3D and S3D), which is a major property of synaptic scaling (Moulin et al., 2020; Turrigiano et al., 1998). This functional readout suggests that neuronal activity suppression results in increased postsynaptic AMPA receptor expression. To corroborate these results, we quantified the surface expression of AMPA receptors GluA1 and GluA2 at the postsynaptic membrane after TTX treatment. Analysis of GluA2 surface expression indeed revealed an increase in the number of surface GluA2 puncta after 48 h of TTX treatment (Figures 3E and 3F), which is in line with our functional data at single-cell and neuronal network levels. However, TTX did not cause changes in the number of surface GluA1 puncta (Figures S3H and S3I). In addition, we found no difference in the density of Synapsin/Homer1 co-localized puncta between vehicle- and TTX-treated neurons (Figures 3G and 3H), suggesting that the TTX treatment did not induce any alteration in the number of functional synapses. However, an increased density of Homer1 puncta was noticed following TTX treatment, while no changes were observed in the intensity of Homer1 or the density of Synapsin (Figures S3E–S3G).
Figure 3.
Increased miniature excitatory postsynaptic current (mEPSC) amplitudes and elongation of axon initial segment (AIS) following TTX treatment
(A–D) Representative mEPSC traces of vehicle-treated (Veh) and 48-h TTX-treated (TTX) neurons (Ctrl1; A). Quantification of the amplitude (B) and frequency of mEPSCs (C) in Veh- and TTX-treated neurons. n = number of cells/batches: Veh n = 21/3, TTX n = 23/3. Rescaled cumulative mEPSCs amplitude (Scaling = TTX values ∗ 1.21; D).
(E) Representative images of vehicle-treated (Veh) and 48 h TTX-treated (TTX) neurons (Ctrl1) stained for MAP2 (gray), surface GluA2 (green), and Synapsin 1/2 (red) (scale bar, 5 μm).
(F) Quantification of GluA2 puncta (number per 10 μm). n = number of cells/batches: Veh n = 18/2, TTX n = 28/2.
(G) Representative images of vehicle-treated (Veh) and 48-h TTX-treated (TTX) neurons (Ctrl1) stained for MAP2 (gray), Homer1 (green), and Synapsin 1/2 (red) (scale bar, 5 μm).
(H) Quantification of Synapsin/Homer1 co-localized puncta (number per 10 μm). n = number of cells/batches: Veh n = 20/1, TTX n = 25/1.
(I) Representative images of vehicle-treated (Veh) and 48 h TTX-treated (TTX) neurons (Ctrl1) stained for MAP2 (gray), ANKG (green), and Hoechst (blue) at DIV 49 (scale bar, 5 μm).
(J and K) Plots showing cumulative fraction and quantification of the length axon initial segment (AIS; μm). n = number of cells/batches: Veh n = 14/2, TTX n = 14/2. Data represent means ± SEM. ns: not significant, ∗p < 0.05, ∗∗∗∗p < 0.0001, unpaired Student’s t test was performed between two groups. All means, SEMs, and test statistics are listed in Table S3.
In addition to synaptic scaling, neurons also respond to altered activity by modifying their intrinsic excitability. Changes in neuronal intrinsic excitability can occur through a variety of mechanisms, including changes in structural characteristics of the axon initial segment (AIS). For instance, the elongation of the AIS has been shown to increase neuronal excitability and to facilitate action potential generation (Kuba et al., 2010, 2015). TTX treatment (48 h) significantly increased AIS length compared with vehicle-treated neurons (Figures 3I–3K). This confirms that, besides changes in functional synaptic properties, alterations in the structural characteristics of axons also occur following neuronal activity suppression with TTX and may contribute to the increase in network excitability.
Transcriptional changes in neurons following TTX treatment are associated with homeostatic plasticity
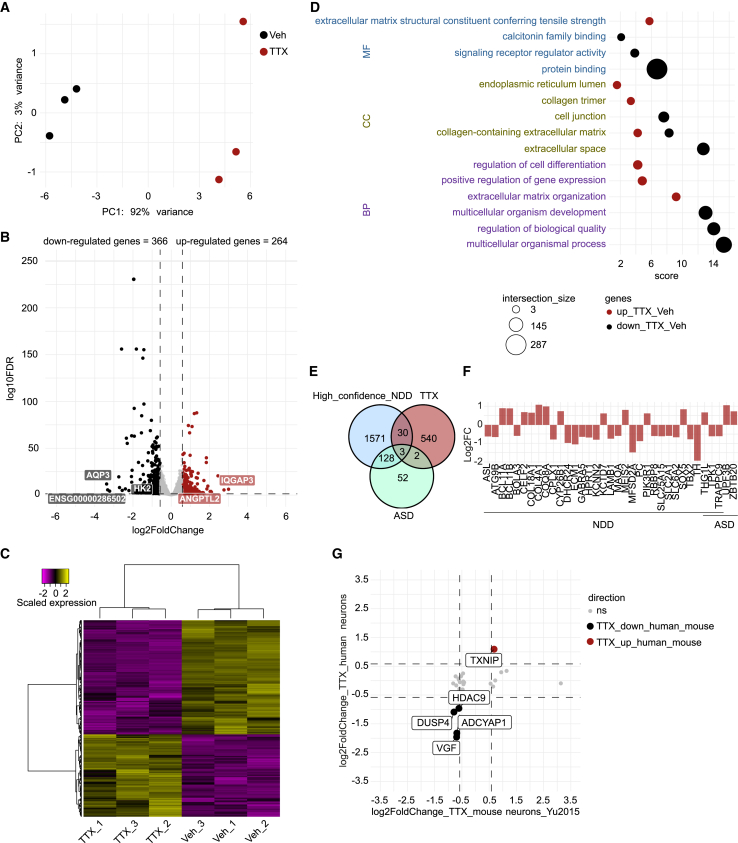
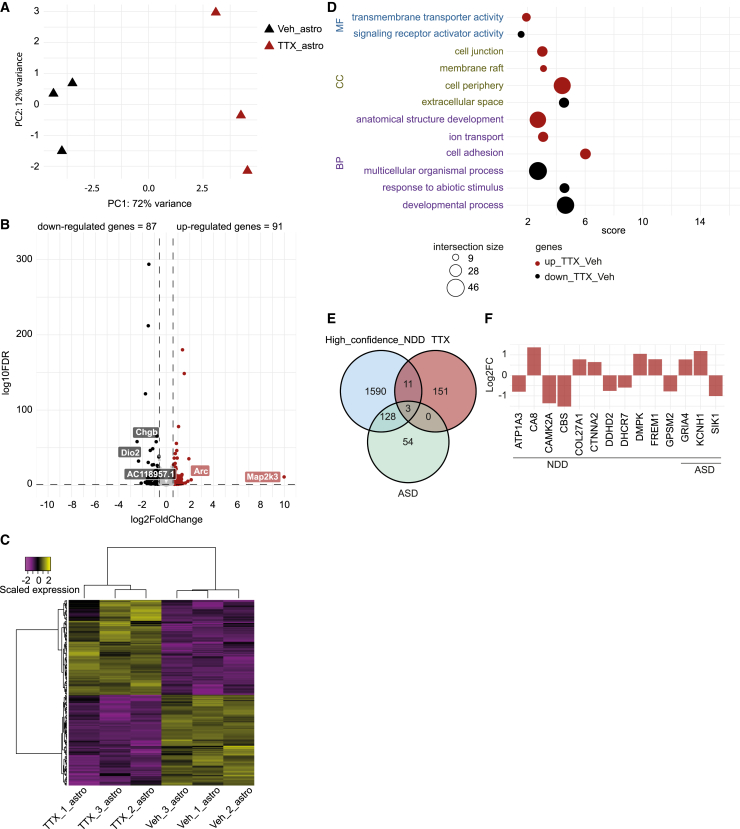
The increase in AMPA receptor levels can be seen as early as 4–6 h after activity suppression. The increase in synaptic strength continues for up to 24 or 48 h (Ibata et al., 2008). This time frame of homeostatic plasticity-related processes suggests that changes in the transcription program of neurons could mediate such processes (Ibata et al., 2008; Schaukowitch et al., 2017). In order to further understand the molecular changes associated with a later stage of TTX-induced homeostatic plasticity, in both neurons and astrocytes, we performed RNA-sequencing (RNA-seq) of vehicle- and 48-h TTX-treated DIV 49 neuron-astrocyte co-cultures. hiPSCs-neuronal networks highly expressed SLC17A6, as well as genes coding for AMPA (e.g., GRIA2), Kainate (e.g., GRIK5), and NMDA (e.g., GRIN3A and GRIN2D; Figure S4E) receptor subunits, confirming the glutamatergic identity of our cultures. PCA analysis segregated TTX- and vehicle-treated neurons and astrocytes, confirming TTX treatment-related changes in the transcriptional activity of the neuronal networks (Figures 4A and 5A).
Figure 4.
Transcriptional changes in human neurons associated with homeostatic plasticity
(A) Principal-component analysis (PCA) of RNA-sequencing data of six samples, three TTX- and three vehicle-treated hiPSC-derived neurons samples (Ctrl1). Colors indicate the received treatment (red = TTX, and black = vehicle).
(B) Volcano plot depicting differential gene expression between TTX- and vehicle-treated neurons. Colored dots indicate differentially expressed genes (DEGs; absolute log2 fold change (Log2FC) > 0.58 and adjusted p value <0.05). Up-regulated genes in TTX-treated neurons are shown in red, down-regulated genes are depicted in black. The five genes with the greatest change in expression activity between the two conditions are indicated.
(C) Heatmap depicting scaled expression of DEGs in the six samples.
(D) Scatterplot of the enrichment analysis results depicting the top three significant gene ontology (GO) terms associated with up-regulated (red dots) and down-regulated genes (black dots) in neurons exposed to TTX treatment as compared with vehicle-treated neurons. Terms are ordered per ontological category (MF: molecular function; CC: cellular component; BP: biological process). The size of the dots indicates the number of genes observed in the list of DEGs and the list of genes associated with the particular term. Score: negative log10 of the adjusted p value resulting from the enrichment analysis.
(E) Venn diagram depicting the number of shared genes between the list of DEGs in TTX-treated neurons (red circle, TTX) and genes previously related to neurodevelopmental disorders (NDDs; light-blue circle, High_confidence_NDD) (Leblond et al., 2021) and highly confident autism-related genes (light-green circle, ASD) (Fu et al., 2022).
(F) Bar plot depicting the Log2FC value of the 35 shared genes observed in (E). Fold change was calculated by normalizing gene expression level of TTX-treated condition to vehicle-treated condition.
(G) Four-way volcano plot depicting expression changes in hiPSC-derived neurons and mouse neurons treated with TTX (absolute Log2FC > 0.58 and adjusted p value <0.05) (Yu et al., 2015). Each dot represents a gene. Red dots represent genes up-regulated in both hiPSC-derived neurons and mouse neurons in response to TTX, black dots represent down-regulated genes in both hiPSC-derived neurons and mouse neurons exposed to TTX.
Figure 5.
Transcriptional changes are induced by suppression of neuronal activity in rat astrocytes
(A) PCA plot of RNA-sequencing data of six samples, three TTX- and three vehicle-exposed rat astrocytes derived from co-cultures with hiPSC-derived neurons (Ctrl1). Colors indicate the received treatment (red = TTX, and black = vehicle).
(B) Volcano plot depicting differential gene expression between TTX- and vehicle-exposed astrocytes. Colored dots indicate DEGs (Absolute Log2FC > 0.58 and adjusted p value <0.05). Up-regulated genes in TTX-exposed astrocytes are shown in red, down-regulated genes are depicted in black. The five genes with the greatest change in expression activity between the two conditions are indicated.
(C) Heatmap depicting scaled expression of DEGs in the six astrocyte samples.
(D) Scatterplot depicting the top three significant GO terms, per ontological category (molecular function: MF, cellular component: CC and biological process: BP), associated with up-regulated (red dots) and down-regulated genes (black dots) in astrocytes exposed to TTX treatment, as compared with vehicle-exposed astrocytes. The size of the dots indicates the number of intersected genes between the list of DEGs and the genes associated with the particular term. Score: negative logarithm10 of the adjusted p value resulting from the enrichment analysis.
(E) Venn diagram depicting the number of shared genes between the list of DEGs in TTX-exposed astrocytes (red circle, TTX) and genes previously related to NDDs (light-blue circle, High_confidence_NDD) (Leblond et al., 2021) and highly confident autism-related genes (light-green circle, ASD) (Fu et al., 2022).
(F) Bar plot depicting the Log2FC value of the 14 shared genes observed in (E). Fold change was calculated by normalizing gene expression level of TTX-treated condition to vehicle-treated condition.
With an absolute log2 fold change (FC) >0.58 and multiple testing-adjusted p value <0.05, we identified 366 down-regulated and 264 up-regulated genes in TTX-treated neuronal networks, compared with vehicle-treated ones (Figure 4B and Table S1). These transcriptional changes were consistent across samples within each condition (Figure 4C). The largest changes in gene expression were observed for IQGAP3, ANGPTL2, AQP3, ENSG00000286502, and HK2 (Figure 4B and Table S1).
Among genes up-regulated by TTX treatment, gene ontology (GO) analysis showed “extracellular matrix organization” to be the most strongly enriched term, including members of the Disintegrin and Metalloproteinase with Thrombospondin motifs (ADAMTS) family, such as ADAMTS17, ADAMTS8, and ADAMTS18 (Figures 4D and S4A, and Table S1). Additionally, the term “positive regulation of gene expression” was also enriched, including several transcription factors, such as ATF3 and HOXA5 (Figure S4A and Table S1).
Genes down-regulated by TTX treatment, including ADAMTSL4, ADAMTS9, NPTX1, BDNF, VGF, NR4A1, and TNC (Figure S4A and Table S1), were generally related to cell communication; enriched terms included “response to stimulus”, “signaling”, “transport”, “vesicle”, “extracellular space”, and “cell junction” (Figure 4D and Table S1). Neuronal pentraxin-1 (NPTX1) and brain-derived neurotrophic factor (BDNF) have been previously implicated in homeostatic plasticity. While chronic network deprivation leads to decreased BDNF release (Karpova et al., 2010; Castren et al., 1992), increased Nptx1 expression was observed 6 h after TTX treatment in mouse hippocampal cultures (Schaukowitch et al., 2017). We observed reduced BDNF and NPTX1 expression in the TTX-treated networks, which was corroborated with quantitative polymerase chain reaction (Figure S4B). The dissimilarity in NPTX1 expression level between our study and the previous finding (Schaukowitch et al., 2017) might be attributed to different stages of homeostatic plasticity being examined. Our RNA-seq analysis focused on a later stage (48 h) of homeostatic plasticity, during which we exclusively observed a decrease in NPTX1 expression, potentially leading to an enhancement in intrinsic excitability (Figueiro-Silva et al., 2015). We did not observe changes in the expression of genes coding for AMPA or NMDA receptors following TTX treatment (Figure S4E), indicating that the observed alterations in AMPA receptors occur exclusively at the postsynaptic membrane surface and not at the transcriptional level. By assessing the function of TTX-induced differentially expressed genes (DEGs) in the synaptic compartment through SynGO ontologies and annotations (Koopmans et al., 2019), we observed that DEGs were mostly associated with synaptic organization, synaptic signaling, and presynaptic and postsynaptic processes (Figures S4C and S4D, and Table S1).
Considering that alterations in homeostatic plasticity during development may contribute to the pathophysiology of different NDDs (Antoine et al., 2019; Bulow et al., 2019; Lee et al., 2018; Benevento et al., 2016; Zeng et al., 2007; Dani et al., 2005), we tested whether the TTX-induced transcriptional changes were associated with genes previously related to NDDs (Fu et al., 2022; Leblond et al., 2021). While we did not identify a significant association between the list of TTX-induced DEGs and lists of genes previously related to NDDs (Table S1), we still observed some overlap (5.6%; 35 of 630; Figures 4E and 4F), including MAOA, TH, and TRAPPC9 (Figure 4F and Table S1). This result suggests that altered expression of these genes could affect homeostatic plasticity in some NDDs.
Finally, to identify potential conserved molecular mechanisms involved in homeostatic plasticity between human and mouse neurons, we compared our list of TTX-induced DEGs with previously identified TTX-induced DEGs in mouse primary hippocampal neurons (Yu et al., 2015). The overlap with previously identified up-regulated (0.4%; 1 of 264) and down-regulated DEGs was, however, low (1%; 4 of 366; Figure 4G and Table S1). Overlapping genes were TXNIP, HDAC9, DUSP4, ADCYAP1, and VGF (Figure 4G and Table S1). This may suggest that changes in gene expression activity contributing to homeostatic plasticity differ between mice and humans. However, differences in the experimental design, including different studied brain regions (mouse primary hippocampal neurons versus co-cultures of human cortical neurons and rat astrocytes), or different stages of homeostatic plasticity (4-h TTX treatment versus 48-h TTX treatment) could also be contributing to the lack of consistency between the species.
Overall, these results identify multiple genes that likely contribute to homeostatic plasticity. Suppression of neuronal activity may induce reduction in the expression of genes involved in cell communication, while changes in the extracellular matrix might contribute to increased neuronal synchronized activity observed in homeostatic plasticity.
Suppression of neuronal activity induces transcriptional changes in astrocytes
Astrocytes are sensitive to changes in neuronal activity and are known regulators of homeostatic plasticity (Lines et al., 2020; Perez-Catalan et al., 2021). We identified 91 up-regulated and 87 down-regulated astrocytic genes in response to neuronal activity suppression with TTX (Figure 5B and Table S2). Transcriptional changes in astrocytes were consistently seen in the same conditions (Figure 5C and Table S2). Arc, Map2k3, Dio2, Chgb, and AC118957.1 were identified as the genes with the strongest expression changes between vehicle- and TTX-exposed astrocytes (Figure 5B and Table S2).
GO enrichment analysis indicated significant association between the up-regulated astrocytic genes and ontological terms likely related to neuronal function (Figure 5D and Table S2), such as “cell junction”, “ion transport”, “neuron projection”, and “transmembrane transporter activity”. Particularly, the expression of Arc, Egr3, Avpr1a, and Astn2 was increased in TTX-exposed astrocytes (Figure S5A and Table S2). Together, these observations suggest that upon suppression of neuronal activity, astrocytes increase the expression of genes that could modulate synaptic activity. In contrast, down-regulated genes in astrocytes exposed to TTX showed significant association with terms related to “response to abiotic stimulus”, “response to external stimulus”, and “signaling receptor activity”, suggesting that a reduced expression of genes related to response to stimuli in astrocytes may be a consequence of neuronal activity suppression, which could decrease the stimulation of the astrocytes in culture. We further assessed the role of astrocytic DEGs induced by neuronal activity suppression in the synaptic compartment using SynGO ontologies and annotations (Koopmans et al., 2019); we observed that astrocytic DEGs were related to both presynaptic and postsynaptic processes (Figures S5C and S5D, and Table S2).
Similar to our analysis for the hiPSC-derived neurons, we compared the list of DEGs in astrocytes exposed to neuronal activity suppression with lists of genes previously related to NDDs (Fu et al., 2022; Leblond et al., 2021). We found that 7.9% (14 of 178) of DEGs were known NDD genes (Figure 5E and Table S2), such as Camk2a (Figure 5F and Table S2). Their observed roles in NDDs might thus be related to an impaired astrocytic contribution to homeostatic plasticity.
Finally, to identify potentially shared mechanisms between neurons and astrocytes that could contribute to homeostatic plasticity in our co-cultures, we compared the transcriptional changes induced by TTX treatment in the two cell types. To this end, we converted rat gene symbols to human gene symbols and combined the transcriptional profiles of all sequenced samples, including astrocytes and neurons. PCA showed cell type to be the major source of variation in gene expression. Nonetheless, samples were also segregated by treatment, suggesting some shared transcriptional changes in neurons and astrocytes after neuronal activity suppression (Figure S5B). When intersecting the list of DEGs by TTX treatment in neurons and astrocytes, we identified 11 commonly down-regulated genes, including VEGFA, SCG2, and CCK, while the expression of ATF3 and COL13A1 was increased in both cell types. We also identified nine genes with opposite changes in transcriptional activity between neurons and astrocytes, including SPON1, IGFBP3, and DISP3 (Figure S5E). These oppositely regulated genes, of which several are involved in lipid and energy metabolism, may be involved in crosstalk mechanisms between neurons and astrocytes that contribute to homeostatic plasticity.
Concluding, our transcriptional analysis of astrocytes in the neuronal networks provides evidence corroborating their role in the regulation of homeostatic plasticity and identifies candidates for underlying genes and biological pathways.
Discussion
In this study, we describe a human in vitro neuronal model for studying homeostatic plasticity at the network level. Using this model, we provide insight into how synaptic strength and intrinsic properties of neurons cooperate to stabilize neuronal network activity in response to activity suppression. We demonstrated that chronic deprivation of neuronal activity through the inhibition of voltage-gated sodium channels with TTX elicited a time-dependent re-arrangement of neuronal networks. This re-arrangement was characterized by significant increase in the synchronized network activity following TTX treatment, as indicated by metrics such as mean NBR, mean BR, and cross correlation, while exhibiting no changes in global activity, as determined by MFR. TTX-induced modifications in neuronal network properties were accompanied by increased surface expression of postsynaptic AMPA receptors, as well as by elongation of axon initial segments. Additionally, we identified transcriptional changes induced by suppression of neuronal activity in neurons and astrocytes, which may underlie the network re-arrangement.
At the network level, we identified an increase in the mean NBR and mean BR, along with decreased mean BD, BSR, mean NBD, and NIBI following treatment with TTX. Previous studies have demonstrated that both AMPA and NMDA receptors play a crucial role in driving network bursting activity (Odawara et al., 2016; Suresh et al., 2016). However, our previous study demonstrated that in control hiPSC-derived neuronal networks, network bursting activity is mainly mediated by AMPA receptors (Frega et al., 2019). This was evidenced by the inhibition of AMPA receptors with NBQX completely abolished network bursting activity, whereas the blockade of NMDA receptors (D-AP5) only slightly reduced this form of activity, and mostly reduced NBD. In the present work, we observed increased expression of surface AMPA receptors following TTX treatment. An increase in AMPA receptors may facilitate excitatory neurotransmission (Watson et al., 2017; Niedringhaus et al., 2013), which could be translated into increased network bursts. Taken together, these findings suggest that changes in bursting activity, by means of increased NBR and BR, and reduced NIBI, can primarily be attributed to the up-regulation of surface AMPA receptors. Moreover, AMPA receptor-driven bursts have shorter durations, while NMDA receptor-driven bursts have comparatively longer durations (Odawara et al., 2016; Suresh et al., 2016). Therefore, the decrease in BD, NBD, and BSR may also be explained by increased expression of AMPA receptors after adding TTX. Further investigation is necessary to elucidate this relationship in greater detail.
Increased synchronized activity following neuronal activity suppression has been described in a rodent model of homeostatic plasticity, where short-term treatment with TTX induced synchronized burst oscillations (Zhou et al., 2009). Our study in hiPSC-derived neurons verified earlier reported presence of homeostatic plasticity mechanisms regulating neuronal excitability at the network and single-cell levels (Cordella et al., 2022; Zhang et al., 2018). We found that in the hiPSC-derived neurons, the early increase (0–12 h) in synchronized network activity observed after suppression of neuronal activity was dependent on the expression of GluA2-lacking AMPA receptors and, in a later stage (24–48 h), was associated with increased expression of GluA2-containing AMPA receptors. These findings align with previous studies in rodents showing involvement of GluA2-lacking AMPA receptors in the induction of homeostatic plasticity, particularly in the early initiation phase (Hou et al., 2008; Sutton et al., 2006). Furthermore, a study of cultured rat hippocampal neurons demonstrated that just 1 h blockade of NMDA receptors leads to an up-regulation of GluA2-lacking AMPA receptors. However, it is important to note that GluA2-lacking receptors do not exert influence during the late phase of homeostatic plasticity, occurring 2 days after the suppression of firing rate through Kir2.1 transfection (Hou et al., 2008) and after 24–48 h of TTX in our in vitro human neurons. Moreover, we confirmed that the network re-arrangement following TTX treatment was associated with increased mEPSC amplitudes and elongation of the AIS, matching expected changes in structural properties of AIS and postsynaptic AMPA receptors after suppression of neuronal activity (Chater and Goda, 2014; Grubb and Burrone, 2010). Taken together, these results indicate that we provide a valid model of homeostatic plasticity.
Using our model system, we observed that increased synchronized activity gradually returned to pre-drug level within 48 h after TTX removal, consistent with the notion that homeostatic plasticity, as induced by TTX, will restore network activity from a hyper-active level to a set-point level. In mammalian synapses, pharmacological block of postsynaptic AMPA receptors or genetic knockout of GluA4 AMPA receptors is known to trigger a presynaptic form of homeostatic plasticity (Delvendahl et al., 2019; Jakawich et al., 2010; Lindskog et al., 2010). Thus in our model, we may hypothesize that the accumulation of postsynaptic AMPA receptors, as induced by the prolonged TTX exposure, might cause an increase in miniature excitatory postsynaptic potential amplitude, which then induces a homeostatic decrease in presynaptic vesicle release. Future studies are needed to test this hypothesis.
Previous studies in rodents suggested that transcriptional changes contribute to TTX-induced homeostatic plasticity (Valakh et al., 2023; Schaukowitch et al., 2017; Benevento et al., 2016; Yu et al., 2015). We noticed that both Valakh et al. and Schaukowitch et al. highlighted the modulation of transcriptional factor gene expression levels during TTX-induced homeostatic plasticity. Valakh et al. specifically emphasized that the activation of transcription factors from the PAR bZIP family (e.g., Hlf and Tef) plays a role in restraining the homeostatic up-regulation of network activity in response to activity suppression, particularly following a 5-day TTX treatment in mouse brain slice cultures. Similarly, Schaukowitch et al. demonstrated the up-regulation of two other transcription factors (Elk1 and Srf) after a 6-h TTX treatment in primary mouse cortical cultures, and they could mediate TTX-dependent Nptx1 induction. In our current study, we also observed an up-regulation of genes associated with “positive regulation of gene expression” in neurons following TTX treatment. Among these genes, certain transcription factors such as ATF3 and HOXA5 (Figure S4A) have been previously demonstrated to regulate synaptic transmission and synaptic plasticity (Ahlgren et al., 2014; Lizen et al., 2017). Therefore, these transcription factors could potentially act as significant modulators of the homeostatic responses. In our previous study (Benevento et al., 2016) conducted on rat primary cortical neurons, we observed that genes exhibiting down-regulation following TTX treatment were associated with various biological processes, such as cholesterol biosynthesis and potassium ion channels. However, in our present study, we found that the down-regulated neuronal genes in response to TTX treatment were primarily related to cell communication. It is important to note that changes in gene expression related to cell communication may be a mere consequence of suppressing neuronal activity and might not directly connect to the underlying molecular mechanisms of homeostatic plasticity. For instance, the expression of NR4A1 is activity-dependent (Jeanneteau et al., 2018; Hawk and Abel, 2011), and we observed reduced NR4A1 in TTX-treated neurons. Feedback loops may be hypothesized, by which the detection of a reduced expression of genes related to cell communication could contribute to activating homeostatic plasticity mechanisms. In our comparative analysis between the DEGs resulting from TTX treatment (48 h), as observed in our study, and the ones reported by Yu et al., a subset of genes was found to be shared, including TXNIP, HDAC9, DUSP4, ADCYAP1, and VGF. Notably, Yu et al. focused on investigating the effects of TTX treatment on mouse primary hippocampal neurons at very early time points (4 h), indicating that these overlapping genes potentially hold significance in both stages of homeostatic plasticity. However, we have to highlight that our culture system lacks inhibitory input, which can have implications for modeling homeostatic plasticity in neuronal networks and may contribute to divergent observations across studies.
In addition, the transcriptional program induced by neuronal activity suppression implicated increased expression of neuronal genes related to “extracellular matrix organization.” Extracellular matrix (ECM) molecules are synthesized by neurons as well as by non-neural cells, and are secreted into the extracellular space (Dityatev and Schachner, 2003). Reorganization of the ECM can modulate neuronal connectivity and plays a role in synaptic plasticity and homeostasis (Bikbaev et al., 2015; Frischknecht and Gundelfinger, 2012; Dityatev et al., 2010). Members of the ADAMTS family, such as ADAMTS17, ADAMTS8, and ADAMTS18, exhibited increased expression in response to TTX (Figure S4A). These proteins are involved in the degradation of the ECM during development, and play an essential role in neuroplasticity (Ferrer-Ferrer and Dityatev, 2018; Gottschall and Howell, 2015). Based on our results, the reorganization of the ECM also appears important in homeostatic plasticity. Other ECM molecules also were altered in their expression by suppression of neuronal activity, such as tenascin C (TNC), which exhibited reduced neuronal expression in response to TTX (Figure S4A). Reduced TNC might cause the increase of mEPSC amplitude we observed by reducing L-type voltage-gated calcium channel-mediated signaling (Evers et al., 2002; Wang et al., 2011).
Our in vitro model of network plasticity allows us to explore not only neuronal mechanisms of homeostatic plasticity but also the potential role of astrocytes in this process. In our study, we observed significant transcriptional changes in astrocytes exposed to neuronal activity suppression. For example, Arc (Figure S5A), an immediate-early gene (IEG), was up-regulated in its expression. In neurons, Arc expression is dynamically regulated by neuronal activity, and it modulates synaptic strength by controlling endocytosis of AMPA receptors at the synapse (Shepherd and Bear, 2011; Shepherd et al., 2006). It has previously been reported that Arc and other IEGs are also expressed in astrocytes (Rodriguez et al., 2008; Rubio, 1997; Kato et al., 1995). We speculate that Arc was transcribed and may be released from the astrocytes into the synapse, where it may modulate synaptic plasticity, as described for other astrocyte-secreted factors (Perez-Catalan et al., 2021; Wang et al., 2021). We also observed increased expression of Egr3, Avpr1a, and Astn2 in TTX-exposed astrocytes (Figure S5A). Avpr1a belongs to the subfamily of G-protein-coupled receptors that bind arginine vasopressin (AVP), and it promotes LTP induction (Namba et al., 2016); Egr3 is important for normal hippocampal LTD (Gallitano-Mendel et al., 2007); Astn2 is a transmembrane protein that also modulates synaptic function by trafficking of neuroligins and synaptic proteins in Purkinje cells (Behesti et al., 2018). Together, these observations suggest that astrocytes increase the expression of genes that could modulate synaptic activity upon suppression of activity of (neighboring) neurons. Such findings indicate that the astrocytic contribution to homeostatic plasticity might be more pronounced than previously considered and should be studied more intensively.
Some limitations should be taken into account in the interpretation of our results. Considering that some sub-types of astrocytes exhibit sensitivity to TTX (McNeill et al., 2021; Sontheimer and Waxman, 1992), we cannot discriminate transcriptional changes directly related to suppression of neuronal activity and homeostatic plasticity mechanisms, from changes induced by TTX effects on astrocytes (that may or may not influence homeostatic plasticity). Comparing the gene expression profile of our TTX-treated neuron-astrocyte co-cultures with pure astrocyte cultures treated with TTX may help identify genes and processes induced by TTX treatment in astrocytes and provide a basis for testing their role in homeostatic plasticity. Also, the application of other methods to induce homeostatic plasticity, more complex co-cultures (incorporating inhibitory neurons), and setups using human astrocytes in future studies could provide a more comprehensive understanding of homeostatic plasticity in human neuronal networks.
To conclude, we provide evidence that hiPSC-derived neuronal networks display homeostatic plasticity at the network and single-cell levels. Our in vitro model of network plasticity is versatile and allows investigation of human homeostatic plasticity mechanisms. It may also provide a platform for (high-throughput) drug screening, to explore whether specific compounds can be used to rescue altered or insufficient homoeostatic plasticity in the context of brain disorders, such as NDDs.
Experimental procedures
Resource availability
Corresponding author
Nael Nadif Kasri: n.nadif@donders.ru.nl.
Materials availability
This study did not generate new unique reagents.
Human iPSC cell culture
In this study, we used three independent control hiPSC lines, including Ctrl1, Ctrl2, and Ctrl3. All of them were obtained by reprogramming skin fibroblasts and have been described in detail previously (Frega et al., 2019; Mossink et al., 2021). More details also can be found in our supplemental experimental procedures. HiPSCs were cultured in Essential 8 Flex medium (Gibco, A2858501) supplemented with 0.1 mg/mL Primocin (Invivogen, ant-pm-2) on 6-well plates pre-coated with Matrigel (1:15 diluted in DMEM/F12 medium; Matrigel: Corning, #356231; DMEM/F12 medium: Gibco, 11320074) at 37°C, 5% CO2. See supplemental experimental procedures for full details.
Neuronal differentiation
At DIV 0, hiPSCs were dissociated with Accutase (Sigma-Aldrich, A6964) to generate single cells. HiPSCs were then plated on microelectrode array (MEA) plates (20,000 cells/well) or glass, nitric-acid-treated coverslips (20,000 cells/well) in Essential 8 medium (Gibco, A1517001) supplemented with Primocin (0.1 mg/mL, Invivogen, ant-pm-2), RevitaCell (1:100, Gibco, A2644501), and doxycycline (4 μg/mL). MEA plates and coverslips were pre-coated with 50 μg/mL poly-L-ornithine (Sigma-Aldrich, P4957) for 3 h at 37°C, 5% CO2, and 20 μg/mL human recombinant laminin (BioLamina, LN521) overnight at 4°C. See supplemental experimental procedures for full details.
Procedure for TTX treatment and TTX withdrawal on MEA plates
The effect of TTX treatment was exclusively evaluated in neurons at DIV 49 due to the observed sensitivity of younger neurons to the washing out process. This sensitivity was reflected by increased neuronal network activity at DIV 30, but not at DIV 49 after washing out TTX (Figure S1A). Nonetheless, similar to DIV 49 neurons, DIV 30 neuronal networks exhibited an increase in both mean NBR and mean BR following TTX treatment (Figures S1G and S1H). Additional information regarding procedure of TTX treatment and washing out is provided in the details in the supplemental experimental procedures.
MEA recordings and data analysis
All MEA recordings were performed using the 24-well MEA system (Multichannel Systems, MCS GmbH, Reutlingen, Germany). Recordings and data analysis procedures have been described previously in detail (Frega et al., 2019; Mossink et al., 2021). See supplemental experimental procedures for full details.
Immunocytochemistry
Cells cultured on coverslips were fixed with 4% paraformaldehyde supplemented with 4% sucrose for 15 min at room temperature, followed by permeabilization with 0.2% Triton in PBS for 10 min at room temperature. Nonspecific binding sites were blocked by incubation in blocking buffer (PBS, 5% normal goat serum, 1% bovine serum albumin, 0.2% Triton) for 1 h at room temperature. Cells were incubated in a primary antibody solution wherein antibodies were diluted in blocking buffer overnight at 4°C. Secondary antibodies, conjugated to Alexa-fluorochromes, were diluted in blocking buffer and applied for 1 h at room temperature. Hoechst 33342 (Molecular Probes) was used to stain the nucleus before the cells were mounted using DAKO fluorescent mounting medium (DAKO). More details can be found in the supplemental experimental procedures.
RNA-seq
Cells were treated with or without 1 μM TTX (Tocris, 1069) at DIV 49 and harvested at DIV 51. In all conditions, hiPSC-derived neurons were co-cultured with rat astrocytes. For RNA-seq, the prepared samples were sequenced on an Illumina NovaSeq 6000 S1 platform at an average depth of >50 million reads per sample using a read length of 2∗100 base pairs. See supplemental experimental procedures for full details.
RNA-seq data processing
RNA-seq reads were mapped against the hybrid human and rat reference genome (GRCh38+ Rnor6.0) with STAR/2.7.8a (Dobin et al., 2013) using ENCODE parameters. Gene quantification was performed with RSEM/1.3.0 (Li and Dewey, 2011) using the human gencode39 and rat ensembl104 annotations. See supplemental experimental procedures for full details.
Quantification and statistical analysis
The statistical analysis of the data was performed using GraphPad Prism 9 (GraphPad Software, Inc., CA, USA). We first determined whether data were normally distributed. Significance analysis was done for different experimental conditions by one-way ANOVA or two-way ANOVA with post hoc Bonferroni correction when different cell lines, time lines, and drug-treated samples were included. Analysis was done using unpaired Student’s t tests when comparing two conditions at a single time point. Results with p values <0.05 were considered as significantly different (∗), p < 0.005 (∗∗), p < 0.0005 (∗∗∗), p < 0.0001 (∗∗∗∗). Data are shown as mean ± standard error of the mean (SEM). All means, SEM, and test statistics are listed in Table S3.
Acknowledgments
The work was supported by funding from the European Community’s Horizon 2020 Programme (H2020/2014–2020) under grant agreement no. 728018 (Eat2beNICE) (to B.F.); ERA-NET NEURON-102 SYNSCHIZ grant (NWO) 013-17-003 4538 (to D.S.); China Scholarship Council 201906100038 (to X.Y.); ISCIII /MINECO (PT17/0009/0019) and FEDER (to A.E.C.); and M.M. was supported by an internal grant from the Donders Centre for Medical Neurosciences of the Radboud University Medical Center.
Author contributions
X.Y., B.F., and N.N.K. conceived the hypothesis and designed the experiments. N.N.K., B.F., and D.S. supervised the study. X.Y., J.R.V.R., and U.C. performed the experiments. X.Y., S.P., J.R.V.R., U.C., and M.F. analyzed the data. A.E.C., M.M., S.R., and E.J.H.v.H. assisted in data analysis. A.O. and C.S. assisted in experiments. X.Y., S.P., and J.R.V.R drafted the manuscript. All authors reviewed and edited the draft manuscript.
Declaration of interests
B.F. has received educational speaking fees from Medice.
Published: October 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.09.011.
Supplemental information
Tab 1, Results from differential expression analysis for human induced pluripotent stem cell (hiPSC)-derived neurons treated with TTX for 48 h, related to Figure 4. Tab 2, Results from gene ontology (GO) enrichment analysis for hiPSC-derived neurons treated with TTX for 48 h, related to Figure 4. Tab 3 and 4, Results from comparison between differentially expressed genes (DEGs) in hiPSC-derived neurons induced by TTX treatment and genes related to neurodevelopmental disorders (NDDs) as well as confident autism-related genes, related to Figure 4. Tab 5, Results from SynGO analysis for human induced pluripotent stem cell (hiPSC)-derived neurons treated with TTX for 48 h, related to Figure 4.
Tab 1, Results from differential expression analysis for rat astrocytes treated with TTX for 48 h, related to Figure 5. Tab 2, Results from GO enrichment analysis for rat astrocytes treated with TTX for 48 h, related to Figure 5. Tab 3 and 4, Results from comparison between DEGs in rat astrocytes induced by TTX treatment and genes related to NDDs as well as confident autism-related genes, related to Figure 5. Tab 5, Results from SynGO analysis for rat astrocytes treated with TTX for 48 h, related to Figure 5.
Data and code availability
The GEO accession number for the RNA-seq data in this paper is GSE225761.
References
- Ahlgren H., Bas-Orth C., Freitag H.E., Hellwig A., Ottersen O.P., Bading H. The nuclear calcium signaling target, activating transcription factor 3 (ATF3), protects against dendrotoxicity and facilitates the recovery of synaptic transmission after an excitotoxic insult. J. Biol. Chem. 2014;289:9970–9982. doi: 10.1074/jbc.M113.502914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine M.W., Langberg T., Schnepel P., Feldman D.E. Increased Excitation-Inhibition Ratio Stabilizes Synapse and Circuit Excitability in Four Autism Mouse Models. Neuron. 2019;101:648–661.e4. doi: 10.1016/j.neuron.2018.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy C., van den Hurk M., Kakaradov B., Erwin J.A., Jaeger B.N., Hernandez R.V., Eames T., Paucar A.A., Gorris M., Marchand C., et al. Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology. Mol. Psychiatr. 2016;21:1573–1588. doi: 10.1038/mp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behesti H., Fore T.R., Wu P., Horn Z., Leppert M., Hull C., Hatten M.E. ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins. Proc. Natl. Acad. Sci. USA. 2018;115:E9717–E9726. doi: 10.1073/pnas.1809382115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevento M., Iacono G., Selten M., Ba W., Oudakker A., Frega M., Keller J., Mancini R., Lewerissa E., Kleefstra T., et al. Histone Methylation by the Kleefstra Syndrome Protein EHMT1 Mediates Homeostatic Synaptic Scaling. Neuron. 2016;91:341–355. doi: 10.1016/j.neuron.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Bikbaev A., Frischknecht R., Heine M. Brain extracellular matrix retains connectivity in neuronal networks. Sci. Rep. 2015;5:14527. doi: 10.1038/srep14527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulow P., Murphy T.J., Bassell G.J., Wenner P. Homeostatic Intrinsic Plasticity Is Functionally Altered in Fmr1 KO Cortical Neurons. Cell Rep. 2019;26:1378–1388.e3. doi: 10.1016/j.celrep.2019.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrén E., Zafra F., Thoenen H., Lindholm D. Light regulates expression of brain-derived neurotrophic factor mRNA in rat visual cortex. Proc. Natl. Acad. Sci. USA. 1992;89:9444–9448. doi: 10.1073/pnas.89.20.9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater T.E., Goda Y. The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Front. Cell. Neurosci. 2014;8:401. doi: 10.3389/fncel.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordella F., Ferrucci L., D'Antoni C., Ghirga S., Brighi C., Soloperto A., Gigante Y., Ragozzino D., Bezzi P., Di Angelantonio S. Human iPSC-Derived Cortical Neurons Display Homeostatic Plasticity. Life. 2022;12:1884. doi: 10.3390/life12111884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani V.S., Chang Q., Maffei A., Turrigiano G.G., Jaenisch R., Nelson S.B. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debanne D., Inglebert Y., Russier M. Plasticity of intrinsic neuronal excitability. Curr. Opin. Neurobiol. 2019;54:73–82. doi: 10.1016/j.conb.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Debanne D., Russier M. The contribution of ion channels in input-output plasticity. Neurobiol. Learn. Mem. 2019;166:107095. doi: 10.1016/j.nlm.2019.107095. [DOI] [PubMed] [Google Scholar]
- Delvendahl I., Kita K., Müller M. Rapid and sustained homeostatic control of presynaptic exocytosis at a central synapse. Proc. Natl. Acad. Sci. USA. 2019;116:23783–23789. doi: 10.1073/pnas.1909675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev A., Schachner M. Extracellular matrix molecules and synaptic plasticity. Nat. Rev. Neurosci. 2003;4:456–468. doi: 10.1038/nrn1115. [DOI] [PubMed] [Google Scholar]
- Dityatev A., Schachner M., Sonderegger P. The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat. Rev. Neurosci. 2010;11:735–746. doi: 10.1038/nrn2898. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers M.R., Salmen B., Bukalo O., Rollenhagen A., Bösl M.R., Morellini F., Bartsch U., Dityatev A., Schachner M. Impairment of L-type Ca2+ channel-dependent forms of hippocampal synaptic plasticity in mice deficient in the extracellular matrix glycoprotein tenascin-C. J. Neurosci. 2002;22:7177–7194. doi: 10.1523/JNEUROSCI.22-16-07177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Ferrer M., Dityatev A. Shaping Synapses by the Neural Extracellular Matrix. Front. Neuroanat. 2018;12:40. doi: 10.3389/fnana.2018.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiro-Silva J., Gruart A., Clayton K.B., Podlesniy P., Abad M.A., Gasull X., Delgado-García J.M., Trullas R. Neuronal pentraxin 1 negatively regulates excitatory synapse density and synaptic plasticity. J. Neurosci. 2015;35:5504–5521. doi: 10.1523/JNEUROSCI.2548-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K., Stryker M. Integrating Hebbian and homeostatic plasticity: introduction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20160413. doi: 10.1098/rstb.2016.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frega M., Linda K., Keller J.M., Gümüş-Akay G., Mossink B., van Rhijn J.R., Negwer M., Klein Gunnewiek T., Foreman K., Kompier N., et al. Neuronal network dysfunction in a model for Kleefstra syndrome mediated by enhanced NMDAR signaling. Nat. Commun. 2019;10:4928. doi: 10.1038/s41467-019-12947-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frega M., van Gestel S.H.C., Linda K., van der Raadt J., Keller J., Van Rhijn J.R., Schubert D., Albers C.A., Nadif Kasri N. Rapid Neuronal Differentiation of Induced Pluripotent Stem Cells for Measuring Network Activity on Micro-electrode Arrays. <J. Vis. Exp. 2017;119:e54900. doi: 10.3791/54900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht R., Gundelfinger E.D. The brain's extracellular matrix and its role in synaptic plasticity. Adv. Exp. Med. Biol. 2012;970:153–171. doi: 10.1007/978-3-7091-0932-8_7. [DOI] [PubMed] [Google Scholar]
- Fu J.M., Satterstrom F.K., Peng M., Brand H., Collins R.L., Dong S., Wamsley B., Klei L., Wang L., Hao S.P., et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022;54:1320–1331. doi: 10.1038/s41588-022-01104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallitano-Mendel A., Izumi Y., Tokuda K., Zorumski C.F., Howell M.P., Muglia L.J., Wozniak D.F., Milbrandt J. The immediate early gene early growth response gene 3 mediates adaptation to stress and novelty. Neuroscience. 2007;148:633–643. doi: 10.1016/j.neuroscience.2007.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschall P.E., Howell M.D. ADAMTS expression and function in central nervous system injury and disorders. Matrix Biol. 2015;44–46:70–76. doi: 10.1016/j.matbio.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb M.S., Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 2010;465:1070–1074. doi: 10.1038/nature09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibata K., Sun Q., Turrigiano G.G. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Hawk J.D., Abel T. The role of NR4A transcription factors in memory formation. Brain Res. Bull. 2011;85:21–29. doi: 10.1016/j.brainresbull.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q., Zhang D., Jarzylo L., Huganir R.L., Man H.Y. Homeostatic regulation of AMPA receptor expression at single hippocampal synapses. Proc. Natl. Acad. Sci. USA. 2008;105:775–780. doi: 10.1073/pnas.0706447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakawich S.K., Nasser H.B., Strong M.J., McCartney A.J., Perez A.S., Rakesh N., Carruthers C.J.L., Sutton M.A. Local presynaptic activity gates homeostatic changes in presynaptic function driven by dendritic BDNF synthesis. Neuron. 2010;68:1143–1158. doi: 10.1016/j.neuron.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneteau F., Barrère C., Vos M., De Vries C.J.M., Rouillard C., Levesque D., Dromard Y., Moisan M.P., Duric V., Franklin T.C., et al. The Stress-Induced Transcription Factor NR4A1 Adjusts Mitochondrial Function and Synapse Number in Prefrontal Cortex. J. Neurosci. 2018;38:1335–1350. doi: 10.1523/JNEUROSCI.2793-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova N.N., Rantamäki T., Di Lieto A., Lindemann L., Hoener M.C., Castrén E. Darkness reduces BDNF expression in the visual cortex and induces repressive chromatin remodeling at the BDNF gene in both hippocampus and visual cortex. Cell. Mol. Neurobiol. 2010;30:1117–1123. doi: 10.1007/s10571-010-9544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Kogure K., Araki T., Itoyama Y. Induction of Jun-like immunoreactivity in astrocytes in gerbil hippocampus with ischemic tolerance. Neurosci. Lett. 1995;189:13–16. doi: 10.1016/0304-3940(95)11437-2. [DOI] [PubMed] [Google Scholar]
- Koopmans F., van Nierop P., Andres-Alonso M., Byrnes A., Cijsouw T., Coba M.P., Cornelisse L.N., Farrell R.J., Goldschmidt H.L., Howrigan D.P., et al. SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron. 2019;103:217–234.e4. doi: 10.1016/j.neuron.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba H., Oichi Y., Ohmori H. Presynaptic activity regulates Na(+) channel distribution at the axon initial segment. Nature. 2010;465:1075–1078. doi: 10.1038/nature09087. [DOI] [PubMed] [Google Scholar]
- Kuba H., Yamada R., Ishiguro G., Adachi R. Redistribution of Kv1 and Kv7 enhances neuronal excitability during structural axon initial segment plasticity. Nat. Commun. 2015;6:8815. doi: 10.1038/ncomms9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond C.S., Le T.L., Malesys S., Cliquet F., Tabet A.C., Delorme R., Rolland T., Bourgeron T. Operative list of genes associated with autism and neurodevelopmental disorders based on database review. Mol. Cell. Neurosci. 2021;113:103623. doi: 10.1016/j.mcn.2021.103623. [DOI] [PubMed] [Google Scholar]
- Lee K.Y., Jewett K.A., Chung H.J., Tsai N.P. Loss of fragile X protein FMRP impairs homeostatic synaptic downscaling through tumor suppressor p53 and ubiquitin E3 ligase Nedd4-2. Hum. Mol. Genet. 2018;27:2805–2816. doi: 10.1093/hmg/ddy189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindskog M., Li L., Groth R.D., Poburko D., Thiagarajan T.C., Han X., Tsien R.W. Postsynaptic GluA1 enables acute retrograde enhancement of presynaptic function to coordinate adaptation to synaptic inactivity. Proc. Natl. Acad. Sci. USA. 2010;107:21806–21811. doi: 10.1073/pnas.1016399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines J., Martin E.D., Kofuji P., Aguilar J., Araque A. Astrocytes modulate sensory-evoked neuronal network activity. Nat. Commun. 2020;11:3689. doi: 10.1038/s41467-020-17536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizen B., Moens C., Mouheiche J., Sacré T., Ahn M.T., Jeannotte L., Salti A., Gofflot F. Conditional Loss of Hoxa5 Function Early after Birth Impacts on Expression of Genes with Synaptic Function. Front. Mol. Neurosci. 2017;10:369. doi: 10.3389/fnmol.2017.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man H.Y. GluA2-lacking, calcium-permeable AMPA receptors--inducers of plasticity? Curr. Opin. Neurobiol. 2011;21:291–298. doi: 10.1016/j.conb.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready F.P., Gordillo-Sampedro S., Pradeepan K., Martinez-Trujillo J., Ellis J. Multielectrode Arrays for Functional Phenotyping of Neurons from Induced Pluripotent Stem Cell Models of Neurodevelopmental Disorders. Biology. 2022;11:316. doi: 10.3390/biology11020316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill J., Rudyk C., Hildebrand M.E., Salmaso N. Ion Channels and Electrophysiological Properties of Astrocytes: Implications for Emergent Stimulation Technologies. Front. Cell. Neurosci. 2021;15:644126. doi: 10.3389/fncel.2021.644126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M., Rehbach K., Brunner J.W., Classen J.A., Lammertse H.C.A., van Linge L.A., Schut D., Krutenko T., Hebisch M., Cornelisse L.N., et al. A Single-Cell Model for Synaptic Transmission and Plasticity in Human iPSC-Derived Neurons. Cell Rep. 2019;27:2199–2211.e6. doi: 10.1016/j.celrep.2019.04.058. [DOI] [PubMed] [Google Scholar]
- Mossink B., Verboven A.H.A., van Hugte E.J.H., Klein Gunnewiek T.M., Parodi G., Linda K., Schoenmaker C., Kleefstra T., Kozicz T., van Bokhoven H., et al. Human neuronal networks on micro-electrode arrays are a highly robust tool to study disease-specific genotype-phenotype correlations in vitro. Stem Cell Rep. 2021;16:2182–2196. doi: 10.1016/j.stemcr.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin T.C., Rayêe D., Williams M.J., Schiöth H.B. The Synaptic Scaling Literature: A Systematic Review of Methodologies and Quality of Reporting. Front. Cell. Neurosci. 2020;14:164. doi: 10.3389/fncel.2020.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T., Taniguchi M., Murata Y., Tong J., Wang Y., Okutani F., Yamaguchi M., Kaba H. Activation of arginine vasopressin receptor 1a facilitates the induction of long-term potentiation in the accessory olfactory bulb of male mice. Neurosci. Lett. 2016;634:107–113. doi: 10.1016/j.neulet.2016.09.056. [DOI] [PubMed] [Google Scholar]
- Niedringhaus M., Chen X., Conant K., Dzakpasu R. Synaptic potentiation facilitates memory-like attractor dynamics in cultured in vitro hippocampal networks. PLoS One. 2013;8:e57144. doi: 10.1371/journal.pone.0057144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odawara A., Katoh H., Matsuda N., Suzuki I. Physiological maturation and drug responses of human induced pluripotent stem cell-derived cortical neuronal networks in long-term culture. Sci. Rep. 2016;6:26181. doi: 10.1038/srep26181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Catalan N.A., Doe C.Q., Ackerman S.D. The role of astrocyte-mediated plasticity in neural circuit development and function. Neural Dev. 2021;16:1. doi: 10.1186/s13064-020-00151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pre D., Wooten A.T., Biesmans S., Hinckley S., Zhou H., Sherman S.P., Kakad P., Gearhart J., Bang A.G. Development of a platform to investigate long-term potentiation in human iPSC-derived neuronal networks. Stem Cell Rep. 2022 doi: 10.1016/j.stemcr.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez J.J., Davies H.A., Errington M.L., Verkhratsky A., Bliss T.V.P., Stewart M.G. ARG3.1/ARC expression in hippocampal dentate gyrus astrocytes: ultrastructural evidence and co-localization with glial fibrillary acidic protein. J. Cell Mol. Med. 2008;12:671–678. doi: 10.1111/j.1582-4934.2007.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio N. Interferon-gamma induces the expression of immediate early genes c-fos and c-jun in astrocytes. Immunology. 1997;91:560–564. doi: 10.1046/j.1365-2567.1997.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K., Reese A.L., Kim S.K., Kilaru G., Joo J.Y., Kavalali E.T., Kim T.K. An Intrinsic Transcriptional Program Underlying Synaptic Scaling during Activity Suppression. Cell Rep. 2017;18:1512–1526. doi: 10.1016/j.celrep.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J.D., Bear M.F. New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J.D., Rumbaugh G., Wu J., Chowdhury S., Plath N., Kuhl D., Huganir R.L., Worley P.F. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares C., Lee K.F.H., Nassrallah W., Béïque J.C. Differential subcellular targeting of glutamate receptor subtypes during homeostatic synaptic plasticity. J. Neurosci. 2013;33:13547–13559. doi: 10.1523/JNEUROSCI.1873-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer H., Waxman S.G. Ion channels in spinal cord astrocytes in vitro. II. Biophysical and pharmacological analysis of two Na+ current types. J. Neurophysiol. 1992;68:1001–1011. doi: 10.1152/jn.1992.68.4.1001. [DOI] [PubMed] [Google Scholar]
- Suresh J., Radojicic M., Pesce L.L., Bhansali A., Wang J., Tryba A.K., Marks J.D., van Drongelen W. Network burst activity in hippocampal neuronal cultures: the role of synaptic and intrinsic currents. J. Neurophysiol. 2016;115:3073–3089. doi: 10.1152/jn.00995.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M.A., Ito H.T., Cressy P., Kempf C., Woo J.C., Schuman E.M. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Tien N.W., Kerschensteiner D. Homeostatic plasticity in neural development. Neural Dev. 2018;13:9. doi: 10.1186/s13064-018-0105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G. Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harbor Perspect. Biol. 2012;4:a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu. Rev. Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
- Turrigiano G.G., Leslie K.R., Desai N.S., Rutherford L.C., Nelson S.B. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Valakh V., Wise D., Zhu X.A., Sha M., Fok J., Van Hooser S.D., Schectman R., Cepeda I., Kirk R., O'Toole S.M., Nelson S.B. A transcriptional constraint mechanism limits the homeostatic response to activity deprivation in mammalian neocortex. Elife. 2023;12:e74899. doi: 10.7554/eLife.74899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hugte E., Nadif Kasri N. Modeling Psychiatric Diseases with Induced Pluripotent Stem Cells. Adv. Exp. Med. Biol. 2019;1192:297–312. doi: 10.1007/978-981-32-9721-0_15. [DOI] [PubMed] [Google Scholar]
- Wang H.L., Zhang Z., Hintze M., Chen L. Decrease in calcium concentration triggers neuronal retinoic acid synthesis during homeostatic synaptic plasticity. J. Neurosci. 2011;31:17764–17771. doi: 10.1523/JNEUROSCI.3964-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fu W.Y., Cheung K., Hung K.W., Chen C., Geng H., Yung W.H., Qu J.Y., Fu A.K.Y., Ip N.Y. Astrocyte-secreted IL-33 mediates homeostatic synaptic plasticity in the adult hippocampus. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020810118. e2020810118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.F., Ho H., Greger I.H. Synaptic transmission and plasticity require AMPA receptor anchoring via its N-terminal domain. Elife. 2017;6:e23024. doi: 10.7554/eLife.23024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt A.J., Desai N.S. Homeostatic Plasticity and STDP: Keeping a Neuron's Cool in a Fluctuating World. Front. Synaptic Neurosci. 2010;2:5. doi: 10.3389/fnsyn.2010.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefelmeyer W., Puhl C.J., Burrone J. Homeostatic Plasticity of Subcellular Neuronal Structures: From Inputs to Outputs. Trends Neurosci. 2016;39:656–667. doi: 10.1016/j.tins.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Su Y., Shin J., Zhong C., Guo J.U., Weng Y.L., Gao F., Geschwind D.H., Coppola G., Ming G.L., Song H. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat. Neurosci. 2015;18:836–843. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.H., Ouyang Y., Gazit V., Cirrito J.R., Jansen L.A., Ess K.C., Yamada K.A., Wozniak D.F., Holtzman D.M., Gutmann D.H., Wong M. Abnormal glutamate homeostasis and impaired synaptic plasticity and learning in a mouse model of tuberous sclerosis complex. Neurobiol. Dis. 2007;28:184–196. doi: 10.1016/j.nbd.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Marro S.G., Zhang Y., Arendt K.L., Patzke C., Zhou B., Fair T., Yang N., Südhof T.C., Wernig M., Chen L. The fragile X mutation impairs homeostatic plasticity in human neurons by blocking synaptic retinoic acid signaling. Sci. Transl. Med. 2018;10:eaar4338. doi: 10.1126/scitranslmed.aar4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Li X., Liu M., Zhao Y., Zhu G., Luo Q. Homeostatically regulated synchronized oscillations induced by short-term tetrodotoxin treatment in cultured neuronal network. Biosystems. 2009;95:61–66. doi: 10.1016/j.biosystems.2008.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab 1, Results from differential expression analysis for human induced pluripotent stem cell (hiPSC)-derived neurons treated with TTX for 48 h, related to Figure 4. Tab 2, Results from gene ontology (GO) enrichment analysis for hiPSC-derived neurons treated with TTX for 48 h, related to Figure 4. Tab 3 and 4, Results from comparison between differentially expressed genes (DEGs) in hiPSC-derived neurons induced by TTX treatment and genes related to neurodevelopmental disorders (NDDs) as well as confident autism-related genes, related to Figure 4. Tab 5, Results from SynGO analysis for human induced pluripotent stem cell (hiPSC)-derived neurons treated with TTX for 48 h, related to Figure 4.
Tab 1, Results from differential expression analysis for rat astrocytes treated with TTX for 48 h, related to Figure 5. Tab 2, Results from GO enrichment analysis for rat astrocytes treated with TTX for 48 h, related to Figure 5. Tab 3 and 4, Results from comparison between DEGs in rat astrocytes induced by TTX treatment and genes related to NDDs as well as confident autism-related genes, related to Figure 5. Tab 5, Results from SynGO analysis for rat astrocytes treated with TTX for 48 h, related to Figure 5.
Data Availability Statement
The GEO accession number for the RNA-seq data in this paper is GSE225761.