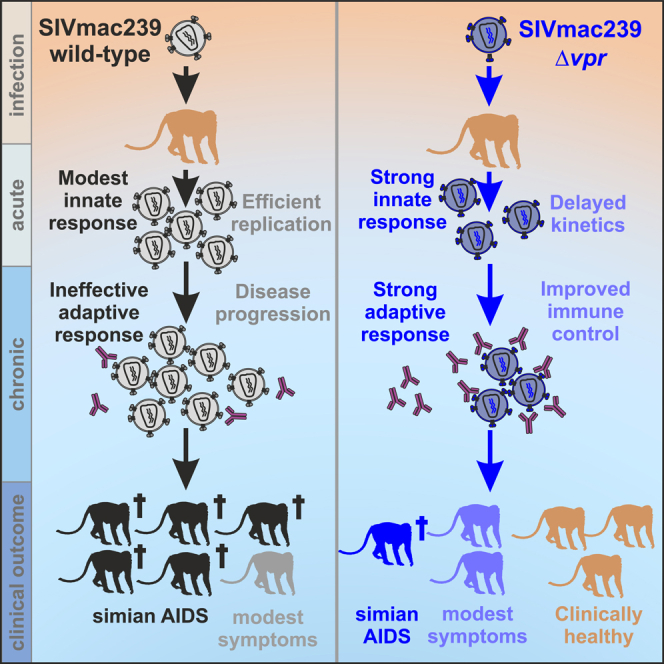
Summary
The accessory viral protein R (Vpr) is encoded by all primate lentiviruses. Vpr counteracts DNA repair pathways, modulates viral immune sensing, and induces cell-cycle arrest in cell culture. However, its impact in vivo is controversial. Here, we show that deletion of vpr is associated with delayed viral replication kinetics, rapid innate immune activation, development and maintenance of strong B and T cell responses, and increased neutralizing activity against SIVmac239 in rhesus macaques. All wild-type SIVmac239-infected animals maintained high viral loads, and five of six developed fatal immunodeficiency during ∼80 weeks of follow-up. Lack of Vpr was associated with better preservation of CD4+ T cells, lower viral loads, and an attenuated clinical course of infection in most animals. Our results show that Vpr contributes to efficient viral immune evasion and the full pathogenic potential of SIVmacin vivo. Inhibition of Vpr may improve humoral immune control of viral replication.
Subject areas: Immunology, Immunity, Immune response
Graphical abstract
Highlights
-
•
Deletion of vpr delays SIV replication and increases immune control in vivo
-
•
Lack of Vpr is associated with increased neutralization
-
•
Vpr leads to generation of lower-avidity antibodies against the p27 antigen
-
•
Vpr contributes to the full pathogenic potential of SIV in rhesus macaques
Immunology; Immunity; Immune response
Introduction
Viral protein R (Vpr) is encoded by all primate lentiviruses and currently perhaps the most enigmatic accessory factor.1 While numerous in vitro activities have been reported,2,3,4 additional cellular targets and functions most likely remain to be discovered. In cell culture Vpr promotes infection of macrophages,5 causes cell-cycle arrest,6,7 modulates nuclear factor κB (NF-κB) activity,8,9,10 up-regulates natural killer (NK) activating ligands,11 modulates cellular gene expression,12,13 and targets key enzymes in DNA repair pathways for proteasome-dependent degradation.14 Specifically, Vpr hijacks the CRL4A-DCAF1 E3 ubiquitin ligase complex to promote ubiquitination and degradation of the cellular uracil DNA glycosylase 2 (UNG2), the crossover junction endonuclease Mus81, the helicase-like transcription factor (HLTF), and exonuclease 1 (Exo1) for proteasomal degradation.15,16,17,18,19,20 It has also been reported that Vpr induces premature activation of the SLX4 complex to promote G2/M arrest and escape from innate immune sensing.21 Others found, however, that Vpr activates the DNA damage response through an as-yet-unknown SLX4-independent mechanism.22 In addition, it has been reported that Vpr causes mitochondrial damage and apoptosis23,24,25 and promotes immune activation.26 Thus, the role of Vpr is multi-layered and its importance for viral replication, immune evasion, and pathogenicity in vivo is poorly understood.
Simian immunodeficiency virus (SIV) infection of macaques closely mimics the pathogenesis and immunology of HIV-1 infection and is one of the best models for acquired immunodeficiency syndrome (AIDS) in humans and vaccine development.27 SIVmac originated from SIVsmm naturally infecting sooty mangabeys,28 which has also been transmitted to humans on at least nine independent occasions, giving rise to HIV-2 groups A–I.29,30 It is established that SIVsmm is non-pathogenic in its natural host.31 In comparison, HIV-2 can cause AIDS but is less pathogenic than HIV-1.32 The SIVmac239 molecular clone replicates to high levels in rhesus macaques and typically causes fatal simian AIDS within one year after experimental infection.33 Early studies in the SIVmac/macaque model suggested that Vpr might be dispensable for efficient viral replication and pathogenesis in vivo.34,35 However, the number of animals was low and the studies provided limited information on the replication kinetics and immunological outcome of vpr-deleted and wild-type (wt) SIVmac239 infection. Here, we performed in-depth analyses of the impact of Vpr on the virological, immunological, and clinical course of infection in SIVmac-infected rhesus macaques. Our results show that Vpr promotes viral immune evasion and contributes to full viral replication fitness and pathogenic potential in vivo.
Results
Lack of vpr delays and attenuates viral replication in vivo
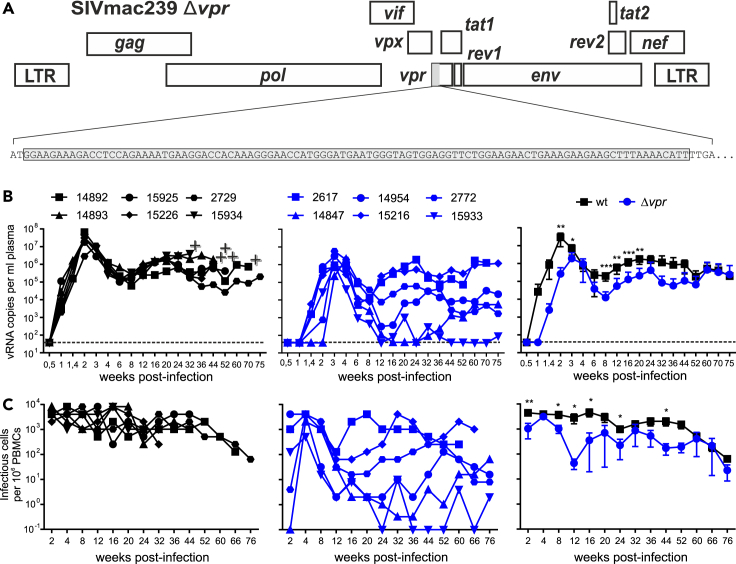
To examine the relevance of Vpr in vivo, we used a mutant SIVmac239 construct containing a large deletion of 101 nucleotides eliminating the vpr gene (Figure 1A).35,36 It has been previously reported that the Δvpr virus replicates with similar or slightly delayed kinetics and efficiency as the wt SIVmac239 construct in CEMx174 cells, rhesus peripheral blood mononuclear cells (PBMCs), and alveolar macrophages.35,36 In addition, deletion of vpr did not impair expression of the Vpx protein.35 A total of twelve rhesus macaques (six animals each) were exposed intravenously to wt or Δvpr SIVmac, and all became productively infected. Lack of vpr was associated with delayed viral replication kinetics and ∼15-fold lower average peak levels of viral RNA in the blood of infected monkeys (3.0x107 ± 8.5x106 vs. 1.9x106 ± 8.5x106 copies per mL plasma; p = 0.009; mean values ± SEM) during acute infection (Figure 1B). After the peaks at two and three weeks post-infection (wpi), respectively, the average levels of viral RNA declined by about two orders of magnitude in both wt and Δvpr SIVmac infection. During the chronic phase, all six wt-infected rhesus macaques showed high plasma viral RNA loads ranging from ∼105 to ∼106 copies/mL from 12 wpi until the end of follow-up at ∼89 wpi (Figure 1B). In contrast, the levels of viral RNA in Δvpr SIVmac-infected monkeys were highly variable ranging from undetectable (i.e., ≤40 copies/mL) to >106 copies per mL blood plasma (Figure 1B). In agreement with the levels of viremia, the cell-associated viral loads were generally high in the wt group but varied in the Δvpr group of animals (Figure 1C). Together, these results showed that Vpr is critical for efficient SIVmac replication during acute infection and for consistent development and maintenance of high viral loads in the chronic phase.
Figure 1.
Effect of vpr on SIVmac replication in vivo
(A) Schematic of the SIVmac239 genome and the deleted DNA sequence of the vpr gene highlighted in brackets.
(B) Viral RNA loads in plasma of rhesus macaques infected with wt or Δvpr SIVmac239. The left and middle panels show values obtained for individual animals, and the right panel shows mean viral RNA loads (±SD) in the two groups. The detection limit for viral RNA was 40 copies/mL plasma. +, indicates death of the infected animal.
(C) Cell-associated viral loads. Shown are the numbers of infectious cells per 1 million PBMCs isolated from the blood of macaques infected with wt (left) or Δvpr SIVmac239 (right). p values: ∗, <0.05; ∗∗, <0.01; ∗∗∗, <0.001.
Lack of vpr is associated with efficient antiviral gene expression in viremic animals
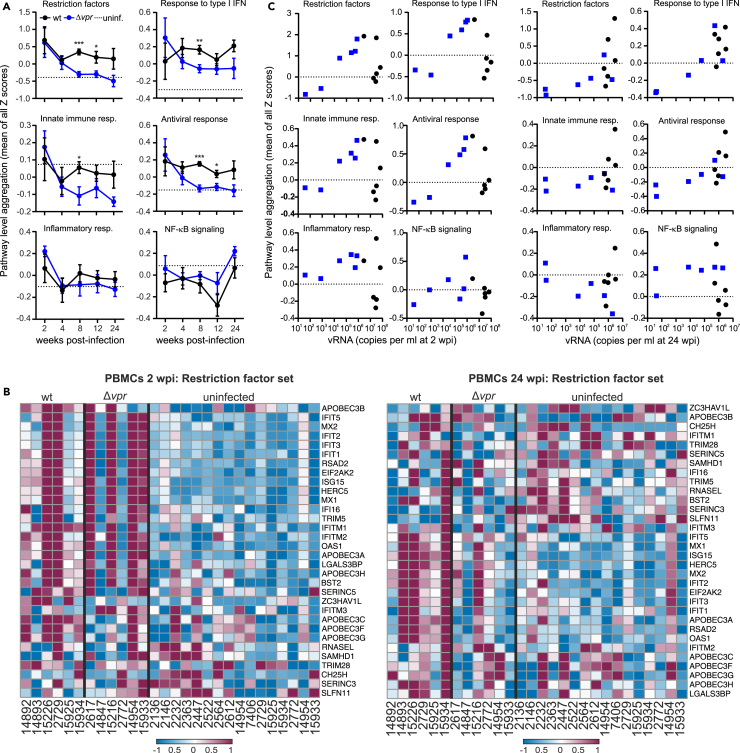
To determine whether Vpr affects the host response to infection in vivo, we performed RNA sequencing (RNA-seq) of sequentially isolated PBMC samples from all 12 infected and 16 uninfected control animals. Normalized principal-component analysis (PCA) showed that wt-infected animals formed a distinct cluster at 2 wpi, while the Δvpr group clustered with uninfected animals (Figure S1A). This is conceivable since lack of vpr was associated with 112-fold lower average viral RNA loads at this time point (Figure 1B). To assess the impact of Vpr on the response to virus infection, we performed comprehensive gene set enrichment analyses (GSEAs) (Figure S1B). Despite lower viral loads, four of six Δvpr-infected animals showed an early and strong induction of innate immune genes (Figures 2A and S1C). The two exceptions were Macaca mulatta (Mm)14847 and Mm2772, which showed undetectable or very low viral RNA loads (Figure 1B) and little induction of innate antiviral responses, as well as expression of restriction factors at 2 wpi (Figures 2B and S1C). The remaining four Δvpr-infected macaques, however, showed strong activation of interferon (IFN)-stimulated genes (ISGs), including members of the IFIT (IFN-induced proteins with tetratricopeptide repeats) family, ISG15 (interferon-stimulated gene 15), OAS1 (2′-5′-oligoadenylate synthetase 1), MX1 (myxovirus resistance protein 1), MX2, tetherin (BST2), and IFI16 (Interferon Gamma Inducible Protein 16) (Figures 2B and S1D). Notably, the levels of innate antiviral gene expression at 2 wpi correlated well with the corresponding viral loads in Δvpr SIVmac infection, while most wt-infected animals showed low levels despite high viral RNA loads (Figure 2C). Altogether, the levels of innate immune activation did not differ significantly between the Δvpr and wt groups at 2 wpi since the two Δvpr-infected macaques that showed little if any viremia also showed little innate immune activation. Despite ∼75-fold lower average viral loads compared to wt-infected macaques, however, all four animals with significant viremia in the Δvpr group showed high levels of innate antiviral gene expression at the earliest time point investigated. After acute infection, the levels of innate immune gene expression declined in the Δvpr group but remained elevated in wt infected-animals (Figure 2A). At 24 wpi, expression of restriction factors was typically higher in wt compared to Δvpr infection (Figures 2B and S1D). This was most likely due to the higher levels of viral replication since the two Δvpr-infected animals with the highest viral loads at 24 wpi showed expression levels of type I IFN and restriction factors similar to wt-infected animals at this time point (Figure 2C). Altogether, these results indicated that lack of vpr is typically associated with efficient antiviral gene expression during acute SIVmac infection.
Figure 2.
Impact of vpr on innate immune activation
(A) Relative activation or inhibition state of the indicated pathways in uninfected rhesus macaques (n = 16) compared to animals infected with wt or Δvpr SIVmac239. Shown are mean values of Z scores obtained for all animals (±SEM). The dotted line indicates the average value obtained for uninfected animals. p values: ∗, <0.05; ∗∗, <0.01; ∗∗∗, <0.001. Individual values are shown in Figure S1D.
(B) Heat maps of wt versus Δvpr in PBMC dataset for the restriction factor gene sets. The datasets were obtained from PBMC samples collected at 2 or 24 wpi, respectively. The color scale is shown at the bottom, with lowest to highest gene expression across all animals represented by the blue to red color gradient.
(C) Correlation between the mean pathway level aggregation scores and the viral RNA loads at 2 (left) and 24 (right) wpi (see also Figure S1).
Low levels of pro-inflammatory cytokines during acute Δvpr SIVmac infection
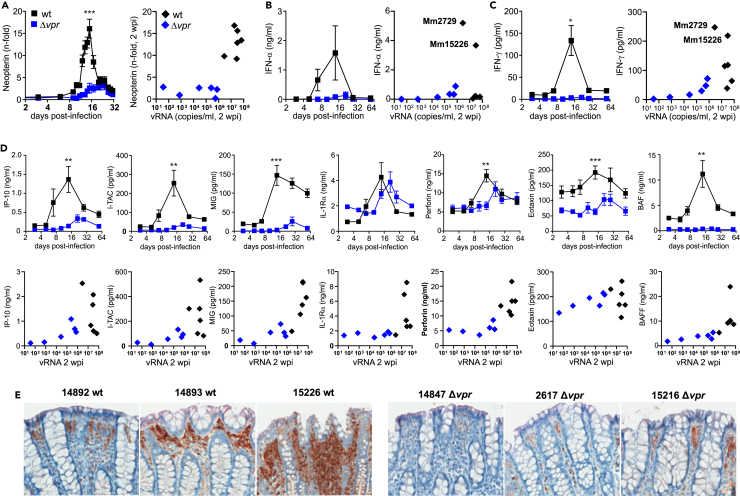
The transcriptomics analyses revealed rapid innate immune activation in the four viremic animals infected with Δvpr SIVmac. To further determine the impact of Vpr on the innate immune response to infection, we measured the levels of various indicators of inflammation during acute infection. Induction of urinary neopterin representing a marker of cellular immune activation37 correlated with viral loads and increased ∼15-fold after wt infection but only up to 3-fold in the Δvpr group (Figure 3A). Similarly, the peak levels of IFN-α and IFN-γ in the wt group were much higher compared to the Δvpr group (Figures 3B and 3C). However, especially IFN-α levels varied enormously in wt SIVmac infections. They were very high in Mm15226 and Mm2729 (Figure 3B) in line with strong induction of restriction factors (Figure 2B) and the highest levels of IFN-γ (Figure 3C). Despite higher viral loads, the plasma levels of these IFNs in the remaining wt-infected animals were similar to Δvpr-infected macaques. In parallel with the viral RNA loads, pro-inflammatory cytokines such as IFN-γ induced protein 10 (IP-10 or CXCL10), IFN-inducible T cell alpha chemoattractant (I-TAC or CXCL11), and monokine induced by IFN-γ (MIG or CXCL9) were induced more rapidly and to higher levels in wt- compared to Δvpr SIVmac-infected animals (Figure 3D). Perforin, a pore-forming protein released by cytotoxic T lymphocytes (CTLs) or NK cells, also peaked earlier in the wt compared to the Δvpr group (Figure 3D). Finally, eotaxin (CXCL11), a potent chemoattractant for eosinophils, and the B cell activating factor (BAFF) were strongly induced in wt- but not in Δvpr SIVmac-infected animals (Figure 3D). In line with higher inflammation and viral loads in wt-infected animals, the MX1, representing a key indicator of the IFN-induced antiviral response, was expressed at high levels in colon biopsies obtained at 24 wpi from wt-infected animals (Figure 3E). Altogether, delayed and attenuated replication of SIVmac239 Δvpr was associated with fast and efficient acute induction of antiviral gene expression but low levels of pro-inflammatory cytokines in blood plasma.
Figure 3.
Expression of pro-inflammatory cytokines and Mx1 in wt and Δvpr SIV infection
(A) Levels of urinary neutering in animals infected with wt or Δvpr SIVmac. Shown are mean fold increases (±SEM) of neutering/creatinine ratios over the mean ratios determined prior to infection for both groups. The right panel shows the correlation by between increases in neutering and viral RNA levels at 2 wpi for individual animals.
(B and C) Levels of IFN-α (B) and IFN-γ (C) in the plasma of animals infected with wt or Δvpr SIVmac. The left panel indicates mean values (±SEM) obtained at the indicated days post-infection, and the right panels indicate the correlation between IFN and vRNA levels at 2 wpi. For clarity, the two wt-infected animals showing strong innate immune activation (Mm2779 and Mm15226) are indicated.
(D) Detection of pro-inflammatory cytokines in plasma of the 12 rhesus macaques infected with wt or Δvpr SIVmac239. Cytokines were measured by Luminex at the indicated time points. The upper panel shows mean values (±SEM), and the lower panel shows the levels of the indicated cytokines and vRNA at 2 wpi. Asterisks in all panels indicate significant differences between the groups (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
(E) Expression of Mx1 in colon biopsies of the indicated animals infected with wt or Δvpr SIVmac239 at 24 wpi (see also Figure S2).
Vpr affects expression of pathways associated with cell cycling and oxidative stress in vivo
Cell culture studies suggest that Vpr causes cell-cycle arrest,6,7 mitochondrial damage, and oxidative stress,23,25,38 inhibits DNA repair pathways,15,16,20,21,22 affects cellular transcription,12,13 modulates NF-κB activity,8,9,39 and induces structural changes in cellular DNA.40 In agreement with these in vitro findings, pathways related to mitochondrial function, cell cycle transition, and DNA replication were among the top 20 enriched in wt SIVmac infection at 2 wpi (Figure S2A). Numerous genes involved in DNA templated DNA replication were strongly enriched in wt-infected animals during acute infection (Figures S2B). Mitochondrial and cell cycle pathways were also induced in wt vs. Δvpr SIVmac infection at 24 wpi. Strikingly, 13 of the top 20 pathways enriched in wt SIVmac infection at this later time point were linked to respiratory and oxidative stress, electron transport, and ATP synthesis (Figures S2A and S2C). Thus, infection with SIVmac expressing Vpr caused substantial oxidative stress and changes in the metabolism and induced, e.g., pathways involved in “antioxidant activity” and “detoxification” to limit these damaging effects. Altogether, these results suggest that some effects reported for Vpr in vitro are recapitulated in the SIVmac model in vivo. SIVmac Vpr was also reported to inhibit NF-κB activity.41 Despite lower viral loads and innate gene expression, lack of vpr was (on average) associated with enrichment of NF-κB target gene expression at later time points (8, 12, and 24 wpi), although the differences to the wt group failed to reach significance (Figure 2A). NF-κB governs numerous cellular processes, and modulation of its activity by Vpr may contribute to some of the aforementioned effects.
Vpr suppresses humoral immune responses and generation of neutralizing antibodies
Our results indicated that Vpr promotes viral replication and innate immune evasion during acute SIVmac infection. To determine whether Vpr also affects the adaptive cellular response to SIVmac infection, we examined the IFN-γ ELISpot responses against viral Gag peptides and whole antigen (AT-2 inactivated SIV), the latter preferentially stimulating CD4+ cells.42 CD8+ T-cell-mediated Gag responses were highly variable in both groups of macaques (Figure S3A). All animals infected with the Δvpr SIVmac239 construct showed significant and persisting CD4+ T cell responses against AT-2 inactivated SIV that contains all of the virion proteins but is non-infectious. In contrast, animals infected with the wt virus showed only transient and modest responses against SIV AT-2 during the early phase of infection (Figure S3B). The overall levels of CD4+ and CD8+ lymphocyte proliferation (Ki67), activation (CD69, CD25, HLA-DR), exhaustion (PD-1) and chemokine (CCR6 and CCR7) receptor expression varied throughout the course of infection but did not differ significantly between the wt and Δvpr groups (Figure S3C). Altogether, lack of vpr was associated with persistent IFN-γ responses to whole SIV antigen suggesting stable levels of virus-specific CD4+ cells.
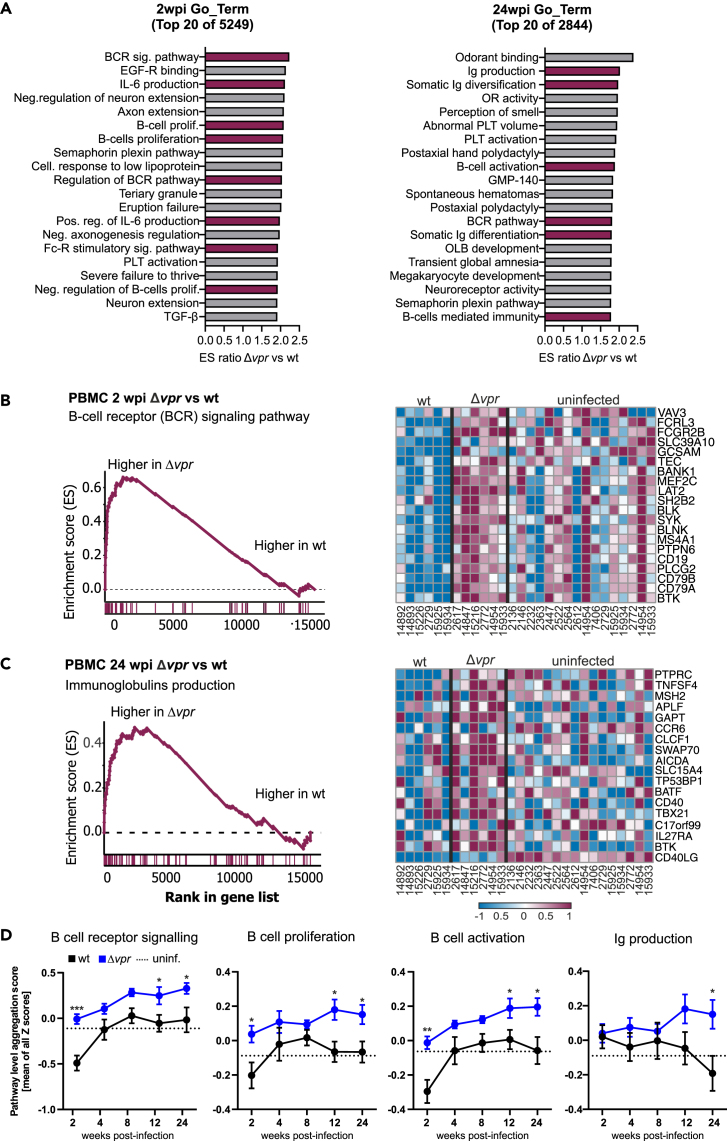
Despite substantially lower viral loads and BAFF induction, pathways involved in B cell receptor (BCR) signaling, B cells activation, and proliferation were induced at higher levels in Δvpr- compared to wt SIVmac-infected macaques at 2 wpi (Figure 4A, left). In agreement with the transcriptomics data, activated CD80+ B cells during acute infection increased faster in the Δvpr group than in the wt group of infected animals (Figure S4A). As previously reported,43 wt SIVmac infection induced a transient loss of peripheral activated memory B cells (CD21− CD27+) at 2 wpi, while their frequencies were unaltered in Δvpr-infected macaques (Figure S4A). Later during infection (24 wpi) the lack of vpr was preferably associated with the induction of pathways associated with immunoglobulin (Ig) production, activation, and somatic diversification (Figure 4A, right). GSEA plots and heatmaps confirmed that genes involved in BCR signaling were strongly enriched during acute Δvpr SIVmac infection (Figures 4B and S4B). During later stages of infection genes associated with antibody production were enriched in the absence of Vpr (Figures 4C and S4C). Despite delayed and attenuated replication kinetics, BCR signaling as well as B cell proliferation and activation pathways associated genes were expressed to higher levels in the Δvpr group at all time points from 2 to 24 wpi, while Ig production was only significantly higher at 12 and 24 wpi (Figure 4D). It has recently been reported that HIV-1 cell-to-cell spread induces CD69 and Blimp1 (PRDM1) expression and promotes productive infection of resting memory CD4+ T cells in a Vpr-dependent manner.12 This study also showed RhoA signaling as the top pathway that is differentially enriched in the absence of Vpr in HIV-1-infected resting memory CD4+ T cells. On average, Blimp1 expression was reduced and RhoA signaling induced in the Δvpr group (Figure S4D). While the differences failed to reach significance for most time points, these results suggest that some of the effects of HIV-1 Vpr on transcription in human T cells are recapitulated in SIVmac-infected rhesus macaques.
Figure 4.
Lack of vpr is associated with increased B cell activation
(A) Top 20 Go term pathways enriched in Δvpr compared to wt SIVmac239 infection of rhesus macaques. Pathways involved in B cell activation and proliferation or antibody production are highlighted in magenta.
(B) GSEA plot (left) and heatmap (right) of Δvpr vs. wt in PBMC collected at 2 wpi for the B cell receptor signaling gene set (FDR <0.001). The running enrichment score (y axis) is indicated for each gene ordered by their rank in the whole dataset for that specific comparison (shown by the bars below the x axis). The right panel shows a heatmap for the leading-edge genes of the B cell receptor (BCR) signaling gene set. The color scale is shown at the bottom, with lowest to highest gene expression across all animals represented by the blue to red color gradient.
(C) GSEA plot (left) and heatmap (right) of Δvpr vs. wt in PBMC collected at 24 wpi for the Ig production gene set (FDR = 0.22). Refer to panel B for detail.
(D) Relative activation of B cell pathways in uninfected rhesus macaques (n = 16) compared to animals infected with wt or Δvpr SIVmac239. Shown are mean values of Z scores obtained for all animals (±SEM) at the indicated time points. Individual values are shown in Figure S4C. The dotted line indicates the average values obtained for uninfected animals. p values: ∗, <0.05; ∗∗, <0.01; ∗∗∗, <0.001 (see also Figures S3 and S4).
Increased neutralizing antibody responses in Δvpr infection
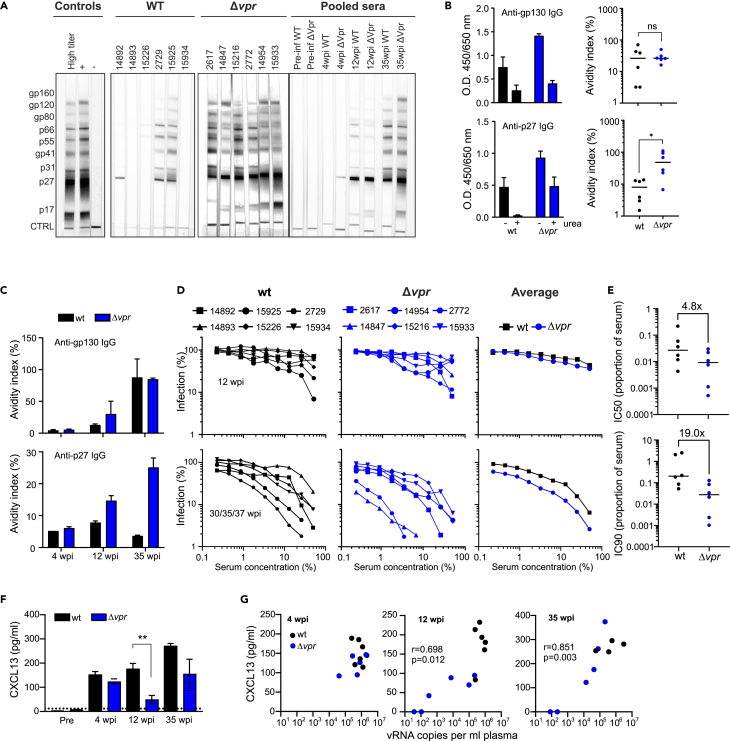
To investigate the impact of Vpr on B cell antibody production, we investigated the humoral response to SIV antigens. Western blot analyses of sera obtained at 35 wpi (33 wpi for Mm 15934) revealed higher levels of antibodies against viral proteins in all animals from the Δvpr group (Figure 5A). Antibody levels were generally lower in sera from wt-infected macaques, and three animals (Mm 14893, 15226, and 15934) showed little if any reactivity. ELISA assays showed that the levels of antibodies against the p27 capsid antigen varied strongly and declined after the first 8 to 12 wpi in the six animals infected with wt SIVmac (Figure S5A). In agreement with the western blot data, macaques infected with Δvpr SIVmac showed stronger and stable anti-p27 Gag responses. Three of six wt- and all Δvpr-infected animals showed persistent humoral immune responses against the gp130 Env glycoprotein (Figure S5B). Altogether, these results demonstrated increased humoral immune responses to the Gag and Env gp130 antigens in the Δvpr compared to the wt group of SIVmac-infected macaques.
Figure 5.
Antibody production and neutralizing activity in wt and Δvpr SIVmac239 infection
(A) Detection of SIV-specific antibodies in sera from the indicated animals at 35 wpi and pooled sera of wt or Δvpr SIVmac239-infected macaques at different time points post-infection. SIV-specific proteins are indicated on the left. Pre-inf, pre-infection.
(B) Detection of gp130 Env and p27 capsid antibodies in sera of animals infected with wt or Δvpr SIVmac239 at 35 wpi determined by ELISA. The assay was performed under stand conditions and in the presence of 8 M urea and the avidity index (right) calculated by normalizing the values obtained in presence of urea to those in absence of urea (100%) for the same serum dilution. ∗, p < 0.05.
(C) Pooled sera obtained at the indicated wpi from both groups of animals were analyzed and the avidity index determined as described in panel B.
(D) Neutralization of SIVmac239 by sera obtained from wt- or Δvpr-infected animals at 12 or 35/37 wpi. Serially diluted sera were used to determine the impact of Vpr on the induction of neutralizing Abs in vivo. The left and middle panels show values obtained for individual animals, and the right panel shows the mean viral neutralization curves obtained for the two groups.
(E) Concentrations of plasma from wt- and Δvpr-infected animals at 35 or 37 wpi required to inhibit SIVmac239 infection by 50% and 90%.
(F) CXCL13 concentration in plasma obtained at the indicated time points determined by ELISA. ∗∗, p < 0.01.
(G) Correlation between plasma levels of CXCR13 and viral RNA (see also Figure S5).
To examine whether Vpr affects not only the strength but also the quality of the humoral immune response, we examined IgG binding to gp130 and p27 antigens by ELISA under standard conditions and in the presence of 8 M urea. This approach allows to measure the avidity of antibodies elicited by viral infection.44 Testing sera obtained from the 12 animals at 35 wpi under non-saturating conditions (dilution 1:200) revealed that lack of Vpr significantly increased the avidity of antibodies for the p27 antigen (Figure 5B). In contrast, no significant difference was observed for anti-gp130 IgG, although it is noteworthy that two wt- but none of Δvpr-infected animals show avidity indexes <10%. Analyses of pooled sera from the 4, 12, and 35 wpi time points confirmed that those from the Δvpr group showed stronger binding to the p27 antigen (Figure 5C).
To further address the functional consequences of the differences in B cell antibody generation, we determined the impact of Vpr on the neutralizing activity of monkey sera against SIVmac239 which is known to be difficult to neutralize.45,46 Analyses of sera from individual monkeys showed that their neutralizing activities differed substantially in both groups of animals and predictably increased substantially from 12 to 35/37 wpi (Figure 5D). On average, however, the concentrations of plasma derived from monkeys infected with Δvpr SIVmac239 required to achieve 50% and 90% inhibition were 4.8- and 19.0-fold lower compared to those obtained from wt-infected macaques at the latter time points (Figure 5E). Analyses of pooled sera confirmed that those obtained from the Δvpr group at 35 or 37 wpi showed higher neutralizing activity compared to those derived from the wt group (Figure S5C). It has been reported that plasma levels of CXCL13 (chemokine [C-X-C motif] ligand 13) are a marker of germinal center activity in humans and macaques and positively correlated with the magnitude of broadly neutralizing antibodies in infected humans.47 We hypothesized that effective replication of wt SIVmac might impair germinal center function and hence effective humoral immune responses.48 However, the plasma levels of CXCL13 were usually higher in wt-infected animals (Figure 5F) and correlated with the viral RNA loads (Figure 5G). Altogether, these results showed that Vpr attenuates B cell function and reduces the production of neutralizing antibodies but does not disrupt germinal center activity.
Lack of vpr attenuates viral pathogenicity
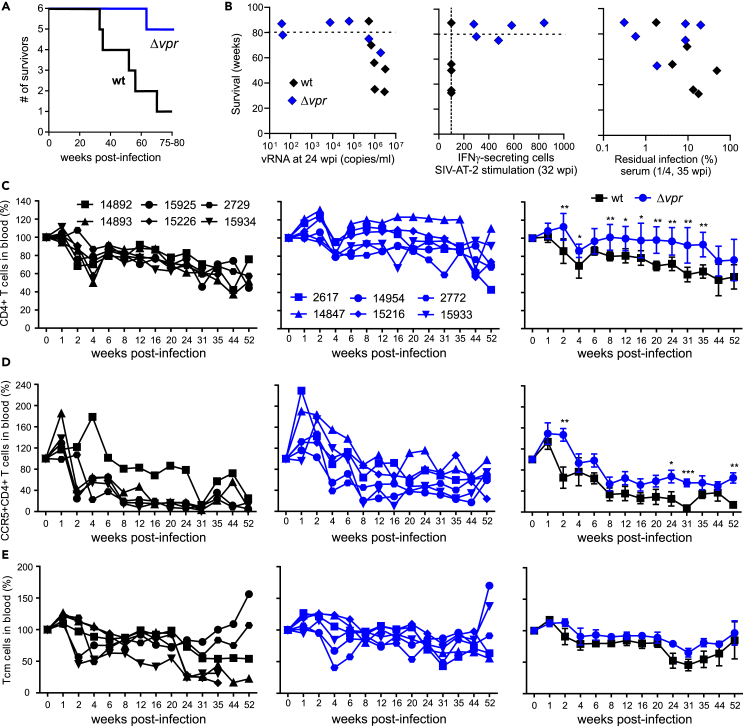
Five of the six animals infected with wt SIVmac developed simian AIDS and had to be euthanized within 80 weeks of follow-up (Figure 6A; Table 1). In contrast, only one animal (Mm2617) infected with Δvpr SIVmac developed severe disease and had to be euthanized before the end of the investigation period. Long survival in the Δvpr group was associated with low viral RNA loads, IFN-γ responses to AT-2 inactivated SIV, and efficient antibody neutralization (Figure 6B). Predictably, disease progression was associated with declining numbers of CD4+ T cells. Flow cytometric analyses revealed that the percentage of total CD4+ T cells in blood decreased by ∼40% in wt- and ∼20% in Δvpr SIVmac-infected animals during the first year after infection (Figure 6C). Declines in CCR5+/CD4+ T cells were more pronounced and occurred more rapidly. Again, they were stronger (∼70%) in wt-infected macaques compared to animals that received the Δvpr virus (∼40%; Figure 6D). In nine of the 12 animals, the percentages of central memory T cells (Tcm) remained stable or declined only slightly during the first year after infection (Figure 6E). The exceptions were three wt-infected macaques (Mm14893, Mm15226, and Mm15934) which showed strong declines in Tcm cells (∼75%) from 24 wpi. These animals also showed weak antibody responses (Figure 5) and more rapid progression to simian AIDS compared to all remaining macaques and had to be euthanized after 33, 35, and 51 wpi, respectively (Table 1).
Figure 6.
Lack of vpr increases survival rates and attenuated CD4+ T cell depletion
(A) Kaplan-Meier survival curves for the wt and Δvpr groups of SIV-infected macaques with time measured from the date of infection.
(B) Correlations between the survival time of animals infected with wt and Δvpr and the viral loads, T cell response, and neutralizing efficiency. The dashed line indicates the beginning of termination of the study and the dotted line the detection limit.
(C–E) Levels of (C) CD4+, (D) CCR5+CD4+ T cells, and (E) central memory T cells in blood of infected macaques compared to baseline (100%). The right panels show mean values (±SEM) measured for the wt (black) and Δvpr groups, and the middle and left panels show values obtained for individual animals. Asterisks indicate significant differences between the groups (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Table 1.
Clinical outcome and necropsy findings of wild-type and Δvpr SIVmac239 infection
| Animal no. | SIV construct | Death at wpia | Diagnosis and clinical signs | Main necropsy findings |
|---|---|---|---|---|
| 14892 | wt | 70 | Simian AIDS; reduced feed intake 3 days before necropsy, mild diarrhea, moderate to severe anemia by wk 60, severe thrombocytopenia before necropsy | Erosive-ulcerative transmural jejunitis, multiple extramedullary hematopoiesis foci (indicating impaired hematopoiesis), generalized hyperplasia of lymphatic system |
| 14893 | wt | 51 | Simian AIDS; reduced feed intake for 1 wk before necropsy, coughing and dyspnea by wk 45, diarrhea by wk 50 | Lymphatic system depleted, interstitial pneumonia with intranuclear inclusion bodies, suspect of CMV-induced pneumonia, subacute colitis |
| 15226 | wt | 35 | Simian AIDS; persisting diarrhea by wk 25, loss of appetite by wk 31 | Depletion of lymphatic system, severe chronic-active bacterial and parasitic gastroenteritis induced by protozoa |
| 2729 | wt | 89 | Moderate to severe thrombocytopenia by wk 44, otherwise clinically healthy | Chronic endocardiosis of mitral valve, generalized moderate to severe activation of lymphatic system including GALT and BALT system and kidney |
| 15925 | wt | 56 | Simian AIDS; recurrent diarrhea by wk 53, reduced appetite shortly before necropsy | Moderate chronic-active gastroenteritis, generalized activation of the lymphatic system |
| 15934 | wt | 33 | Simian AIDS; mild diarrhea by wk 26, moderate amount of stool protozoa, reduced feed intake and dyspnea by wk 32 | Severe diffuse pneumocystosis of the lung |
| 2772 | Δvpr | 89 | Clinically healthy | Mild activation of the lymphatic system |
| 14954 | Δvpr | 88 | Clinically healthy | Mild oligofocal follicular hyperplasia of the spleen |
| 15933 | Δvpr | 87 | Clinically healthy | none |
| 2617 | Δvpr | 64 | Simian AIDS; loss of appetite; watery diarrhea; severe anemia; moderate dyspnea | Focal thrombotic endocardiosis of the right atrium with lung embolism, heart insufficiency with abdominal and pericardial effusions, focal mild interstitial pneumonia |
| 14847 | Δvpr | 78 | Mild transient diarrhea by wk 76 | Generalized severe activation of the lymphatic system with transition to lymphoid depletion; chronic colitis |
| 15216 | Δvpr | 75 | Simian AIDS; mild diarrhea by wk 73 | Focally severe, partially erosive chronic gastroenteritis, severe diffuse pneumocystosis of the lung, generalized severe hyperplasia of the lymphatic system with transition to lymphoid depletion |
wt, wild-type SIV mac239; Δvpr, SIVmac239vpr deletion mutant; wk, week; wpi, weeks post-infection.
Animals were euthanized at the end of the investigation period or when they developed severe SIV-induced immunodeficiency.
In agreement with the high levels of plasma viremia, persistently high cell-associated viral loads, and declines in CD4+ T cells, most wt SIVmac239-infected animals developed clinical symptoms, such as diarrhea and thrombocytopenia. Postmortem examination revealed generalized hyperplasia and/or depletion of the lymphatic tissues as well as opportunistic infections in wt-infected animals (Table 1). In comparison, three of the six animals infected with the Δvpr SIVmac construct remained clinically healthy and one showed only mild diarrhea toward the end of the observation period. Only Mm2617 and Mm15216 showed viral RNA and cell-associated viral loads as high as those observed in wt-infected animals (Figure 1) and both developed simian AIDS and had to be euthanized at 64 and 75 wpi, respectively (Table 1). Postmortem examination revealed focal thrombotic endocardiosis and mild interstitial pneumonia in Mm2617 and severe hyperplasia to depletion of the germinal centers of the lymph nodes, severe multiorgan vasculitis and perivasculitis with evidence of cytomegalovirus infection, and severe pneumocystosis of the lung in Mm15216 (Table 1). In comparison, the three Δvpr-infected animals (Mm2772, Mm14954, and Mm15933) with low to intermediate virus loads (Figure 1) remained clinically healthy and showed little if any activation of the lymphoid system upon necropsy (Table 1). Altogether, these results show that lack of Vpr is associated with improved immune control of virus replication and delayed disease progression.
Discussion
In this study, we show that lack of an intact vpr gene is associated with improved innate and adaptive immune responses, delayed and attenuated levels of SIVmac replication during acute infection, variable outcomes during chronic infection, and significantly reduced viral pathogenicity. Our finding that Vpr plays a role in viral immune evasion and the ability to maintain high levels of replication agrees with its general conservation among primate lentiviruses. Like all accessory factors of human and SIVs, Vpr is multi-functional.1,2 Our finding that Vpr is associated with changes in DNA replication, RhoA signaling, mitochondrial function, respiratory stress, and cell cycling in SIVmac-infected rhesus macaques suggests that various activities established in cell culture are relevant in vivo. Most likely, several of these Vpr activities cooperate to suppress stable and effective cellular and humoral immune responses thereby allowing persistently high levels of viral replication.
Early results suggested that deletion of vpr has little to no impact on SIVmac replication, CD4 declines, and progression to simian AIDS in rhesus monkeys.35 However, just four animals infected with Δvpr SIVmac239 were analyzed and the impact of Vpr on viral replication efficiencies, transcriptional changes, and the immunological response to infection remained largely elusive. Our findings agree with the accumulating evidence that Vpr exerts numerous activities promoting viral evasion of the immune system.8 Despite delayed and attenuated replication, Δvpr SIVmac induced high levels of innate immune gene expression during acute infection in viremic animals. This agrees with reports showing that Vpr inhibits innate immunity by degrading and/or inhibiting nuclear transport of IFN regulatory factor 3 (IRF3) and NF-κB that are important for efficient induction of type I IFN and antiviral gene expression.49,50 It is conceivable that the rapid and efficient induction of antiviral factors contributed to the delayed replication and lower peak viral loads in the absence of Vpr. However, the levels of IFN and other inflammatory cytokines were substantially lower during acute Δvpr infection. While the induction of antiviral genes in the absence of Vpr correlated well with the viral loads, it seemed to be actively suppressed in four of the six wt SIVmac-infected macaques (Figure 2C). Altogether, the results suggest that rapid innate immune responses might be sufficient to achieve significant control of virus replication, while the expression of high levels of IFNs and other cytokines is driven by high viral loads. However, the impact of Vpr varied substantially between individual animals. Two wt SIVmac-infected macaques also showed strong innate immune activation, and in one case (Mm2729) this was associated with delayed replication kinetics and long-term survival similar to Δvpr infection.
We found that both cellular and humoral adaptive immune responses are clearly enhanced and more stable in the absence of Vpr. This agrees with previous findings that Vpr suppresses cellular immune responses and reduces the effectiveness of anti-SIV vaccines in mouse models.51,52 Our transcriptomics data revealed that the lack of vpr is associated with marked increases in BCR signaling and activation during acute infection, while expression of genes associated with Ig production and somatic differentiation pathways were enriched at later time points. Functional analyses confirmed that IgG production was higher in Δvpr-infected monkeys. Importantly, we found that, on average, sera derived from macaques infected with Δvpr SIVmac showed higher neutralizing activity compared to those obtained from animals infected with the wt virus. In addition, the avidity of p27 Gag but not of gp130 envelope antibodies was significantly higher in Δvpr-infected animals (Figure 5). Vpr might affect antibody production and quality by several mechanisms. It is established that HIV-1 infection frequently leads to disturbances of immune cell subsets that are not permissive to infection, including B cells.53 Many steps of the B cell life cycle and activation are dependent on functional T cells. Thus, indirect effects of Vpr on B cells may be mediated by Vpr-induced G2 cell-cycle arrest, inhibition of cell proliferation, and induction of apoptosis in infected, dysfunctional T cells. For example, upon binding of an antigen presented on B cells by the T cell receptor the T cells produce costimulatory factors such as CD40L, BAFF, interleukin (IL)-4, and IL-21, leading to B cell activation.54 Activated B cells undergo initial proliferation and Ig class switching before they enter lymphoid follicles to form a germinal center where they proliferate and undergo Ig class switch and somatic hypermutation, processes that are again aided by T follicular helper (TFH) cells. As a consequence, Vpr-mediated cell-cycle arrest or depletion of T cells, especially TFH cells, thus may lead to reduced B cell activation in WT virus-infected monkeys. In line with this, previous data showed decreased affinity maturation of B cells in wt SIV-infected monkeys.55 In agreement with previous data on HIV-1,12 our results suggest that Vpr may suppress RhoA signaling, which is a key regulator of innate and adaptive immune responses.56 Our transcriptomics data and the finding that B cell activity and neutralizing antibodies are increased in animals infected with Δvpr SIVmac further support that Vpr exerts various activities that compromise B cell activity and contribute to humoral immune escape of HIV-1.
The present data agree with previous reports showing that Vpr is not absolutely required for AIDS progression in SIVmac239-infected rhesus macaques.34,35 It also demonstrates, however, that lack of Vpr substantially delays and attenuates viral replication as well as progression to simian AIDS. Thus, while Vpr is not generally required for efficient viral replication, it is clearly advantageous for the virus. This is consistent with previous data showing that disruptive point mutations in the vpr gene reverted in SIVmac-infected macaques,34 and in a lab worker who became accidentally infected with an HIV-1 variant containing a frameshift mutation in vpr.57 Wild-type HIV-1 and SIVmac infection results in high viral loads and progression to immunodeficiency in the majority of infected individuals and animals, respectively. A minority of infected hosts, however, mount particularly effective immune responses that control viral replication and delay or even prevent disease progression even after infection with intact wt viruses. Conversely, lack of Vpr is associated with enhanced immune responses, but some infected individuals still fail to mount protective immune responses and progress to disease. The determinants underlying the highly divergent outcome of vpr-defective SIVmac infection are most likely complex but clearly warrant further study.
SIVmac is closely related to HIV-2, and both originate from SIVsmm naturally infecting sooty mangabeys without causing disease.30,31,58 One feature that distinguishes these viruses from HIV-1 is the presence of a vpx gene. It is thought that vpx originated from duplication or recombination of a vpr gene.59,60 Thus, some HIV-1 Vpr functions might be split between SIVmac Vpr and Vpx and Vpr be more important for HIV-1 and other primate lentiviruses lacking Vpx. Indeed, it has been shown that combined mutation of vpr and vpx attenuates SIVmac replication and pathogenicity more severely than lack of vpr alone.61 However, SIVmac and HIV-2 Vpx proteins also exert activities that HIV-1 Vpr is lacking, such as counteraction of the restriction factor SAM domain and HD domain-containing protein 1 (SamHD1) and the human silencing hub (HUSH) complex.62,63,64 In comparison, several functions, such as induction of cell-cycle arrest, induction of apoptosis, and modulation of DNA repair pathways are conserved between HIV and SIV Vpr proteins.65,66,67,68 Similar to the present findings, it has been shown that Vpr accelerates HIV-1 replication during acute infection in a human hematopoietic stem cell-transplanted humanized mouse model.69 Altogether, most observations made in the present study are most likely relevant for both HIV-1 and HIV-2 infection.
In summary, we show that lack of Vpr affects the systemic immune response and the virological and clinical outcomes of SIV infection in rhesus macaques. Specifically, our data demonstrate that lack of Vpr clearly delays and attenuates viral replication during acute infection and allows most animals to mount efficient and persisting immune responses and higher levels of neutralizing antibodies. Thus, Vpr contributes to viral immune evasion and replication fitness in primate lentiviruses. Further studies on the impact of Vpr on antibody-dependent cell-mediated cytotoxicity (ADCC), size of the viral reservoirs, and selective pressures driving Env evolution will be of interest. Finally, further elucidation of the underlying mechanisms of Vpr function and their relative contribution to viral immune evasion and pathogenesis may help to develop more effective vaccines against HIV-1.
Limitations of the study
The virologic and clinical outcome of infection in the six rhesus macaques that received Δvpr SIVmac239 was highly variable. Thus, some findings are suggestive, but further studies are needed for definitive conclusions and to clarify which host factors govern the differential outcome of vpr-defective virus infection. The initial studies on the generation and characterization of the Δvpr SIVmac239 construct reported that the deletion has no significant effect on viral replication in rhesus monkey PBMCs and macrophages.35,36 However, slightly reduced and delayed replication kinetics compared to wt SIVmac239 were observed in some experiments.35,36 Thus, it is possible that modest differences in replicative fitness rather than effects on innate immune functions contributed to the delayed in vivo replication kinetics of Δvpr SIVmac. Similarly, higher levels of replication and enhanced immune damage may have contributed to impaired B cell function in the wt group of infected animals. However, we observed increased B cell signaling and proliferation in the Δvpr group already early during infection prior to the development of immunodeficiency in any of the twelve infected animals. Finally, it came as surprise that pathways related to mitochondrial function, cell cycle transition, and DNA replication were enriched in acute wt SIVmac infection since Vpr should mainly affect infected cells representing a minority in vivo. This systemic impact of Vpr on cellular gene expression clearly warrants further study. Since Vpr is found in virions and exerts some activities prior to proviral integration, it is tempting to speculate that it may affect cells that become abortively infected. Finally, Vpr may be more important for replication and pathogenicity of HIV-1 since unlike SIVmac and HIV-2 it lacks a vpx gene.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 (clone SP34-2, Alexa Fluor 700) | BD Bioscience | Cat#557943; RRID:AB_396952 |
| CD4 (clone L200; Horizon V450) | BD Bioscience | Cat#560811; RRID:AB_2033927 |
| CD21 (clone B-ly4, FITC) | BD Bioscience | Cat#561372; RRID:AB_10895576 |
| CD28 (clone CD28.2, FITC) | BD Bioscience | Cat#555728; RRID:AB_396071 |
| CD45 (clone D058-1283, Horizon V500) | BD Bioscience | Cat#561489; RRID:AB_10683313 |
| CD95 (clone Dx2, PE-Cy7) | BD Bioscience | Cat#561636; RRID:AB_10896323 |
| CD195 (clone 3A9, PE) | Biolegend | Cat#550632; RRID:AB_2072548 |
| CD8 (clone RPA-T8, PerCP-Cy5.5) | Biolegend | Cat#301032; RRID:AB_893422 |
| CD8 (clone SK1, APC-Cy7) | Biolegend | Cat#344714; RRID:AB_2044006 |
| CD20 (clone 2H7, PE-Cy7) | Biolegend | Cat#302312; RRID:AB_314260 |
| CD25 (clone M-A251, APC-Fire) | Biolegend | Cat#356136; RRID:AB_2616723 |
| CD27 (clone M-T271, APC) | Biolegend | Cat#356410; RRID:AB_2561957 |
| CD196 (clone G034E3, PE-Dazzle) | Biolegend | Cat#353430; RRID:AB_2564233 |
| CD197 (clone G043H7, APC, Brilliant Violet 421) | Biolegend | Cat#353208; RRID:AB_11203894 |
| HLA-DR (clone L243, APC-Cy7) | Biolegend | Cat#307618; RRID:AB_493586 |
| Ki67 (clone Ki67, Brilliant Violet 605) | Biolegend | Cat#151215; RRID:AB_2876504 |
| PD-1 (clone EH12.2H7, PE-Cy7) | Biolegend | Cat#329918; RRID:AB_2159324 |
| MX1 (M143) | Pr. Kochs, Freiburg University | N/A |
| Peroxidase-conjugated Goat-Anti-Human-IgG | Jackson Bio-Lab | Cat#109-036-088 |
| Goat-anti-monkey-IgA-PO | Nordic | Cat#109-036-088 |
| Bacterial and virus strains | ||
| XL2-Blue MRF’ TM Ultracompetent cells | Agilent Technologies | Cat#200151 |
| Chemicals, peptides, and recombinant proteins | ||
| L-Glutamine | Pan Biotech | Cat#P04-80100 |
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | Cat#41965039 |
| Penicillin-Streptomycin | ThermoFisher | Cat#15140122 |
| FCS | Gibco | Cat#10270106 |
| SIV Gag peptides | NIBSC | Cat#EVA7066.1-16 |
| SIV AT-2 | NIBSC | Cat#ARP1018.1 |
| SIV p27 | NIBSC | Cat#EVA643.2 |
| SIV gp130 | NIBSC | Cat#100841 |
| Critical commercial assays | ||
| IFN-γ ELISpot | Mabtech | Cat#3421M |
| GalScreen | Applied Bioscience | Cat#T1027 |
| SIV Western Blot assay | ZeptoMetrix | Cat#0801500 |
| Paxgene RNA purification kit | Qiagen | Cat#762164 |
| QiaAmp viral RNA isolation mini kit | Qiagen | Cat#52906 |
| TruSeq Stranded mRNA Library Prep | Illumina | Cat#20020595 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131-1096 |
| RBC Lysis/Fixation Solution (10X) | Biolegend | Cat#422401 |
| FOXP3 Fix/Perm Buffer Set | Biolegend | Cat#421403 |
| Rhesus Macaque BLC ELISA Kit | Invitrogen | Cat#EP3RB |
| TaKaRa Prime-Script-One-Step-RT-PCR kit | Takara | Cat#RR014A |
| Deposited data | ||
| RNA-seq data | Gene Expression Omnibus (GEO) repository | Accession number GSE229310 |
| Experimental models: Cell lines | ||
| Human: HEK293T cells | ATCC | Cat#CRL-3216 RRID: CVCL_0063 |
| Human: TZM-bl cells | NIH AIDS Reagent Program | Cat#8129 RRID: CVCL_B478 |
| Experimental models: Organisms/strains | ||
| Rhesus macaques | German Primate Center | N/A |
| Oligonucleotides | ||
| gag forward (5′-ACCCAGTACAACAAATAGGTGGTAACT-3′) | This paper | N/A |
| gag reverse (5′-TCAATTTTACCCAG-GCATTTAATGT-3′) | This paper | N/A |
| gag probe (5′-6FAM(6-carboxyfluorescein)-TGTCCACCT-GCCATTAAGCCCGAG-TAMRA(6-carboxytetramethylrhodamine-3′) | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pBR322_SIVmac 239 | F. Kirchhoff (Ulm University) | N/A |
| Plasmid: pBR322_SIVmac 239 ΔVpr | F. Kirchhoff (Ulm University) | N/A |
| Software and algorithms | ||
| Corel DRAW 2021 | Corel Corporation | https://www.coreldraw.com/ |
| GraphPad Prism Version 10 | GraphPad Software, Inc. |
https://www.graphpad.com RRID: SCR_002798 |
| Rstudio Rx64 4.1.14 | Posit | https://posit.co/downloads/ |
| Aperio image scope version 12.3.1.5011 | Leica | https://aperio-imagescope.software.informer.com/12.3/ |
| DESeq2 version 1.22.1 R | R Package | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| GSEA desktop module 4.2.3 | Broad Institute | https://www.gsea-msigdb.org/gsea/index.jsp |
| FACS DIVA software 6.1.3 | BD Bioscience | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Frank Kirchhoff (frank.kirchhoff@uni-ulm.de)
Materials availability
All unique reagents generated in this study are listed in the key resources table and available from the lead contact.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Transcriptomics data have been submitted to the GEO repository and assigned the accession number GSE229310. the code will be provided once available. They are available for review at: to https://nam11.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.ncbi.nlm.nih.gov%2Fgeo%2Fquery%2Facc.cgi%3Facc%3DGSE229310&data=05%7C01%7Cgktharp%40emory.edu%7C641c7cb43f3e45afb4cd08db39eee4c8%7Ce004fb9cb0a4424fbcd0322606d5df38%7C0%7C0%7C638167470424595022%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=XGvUcza83IJRR0qGpqq90DmE8KjBHoE1PbZVOVuXPIQ%3D&reserved=0; enter token yvsxuoyepvcjrqj into the box. The data will be made public upon acceptance.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Ethical statement for animal experiments
The rhesus monkeys (Macaca mulatta) from this study were cared for by qualified staff of the German Primate Center (DPZ) and housed according to the German Animal Welfare Act which complies with the European Union guidelines on the use of non-human primates for biomedical research and the Weatherall report. The study was approved by the Lower Saxony State Office for Consumer Protection and Food Safety and performed with the project licences 33.19-42502-04-12/0758 and 33.19-42502-04-15/2001. The DPZ has the permission to breed and house non-human primates under license number 392001/7 granted by the local veterinary office and conforming with § 11 of the German Animal Welfare act.
Experimental animals
Twelve purposed-bred young adult rhesus macaque, 10 males and 2 females, were obtained from the DPZ breeding colony. When allocated to the experiment, they were 3.8-7.25 years old with a bodyweight between 4.1-7.6 kg. All monkeys were seronegative for Simian Retrovirus Typ D, SIV, and Simian T-lymphotropic Virus. They were also negative for major histocompatibility complex MHC class I Mamu-A1∗001:01, and -B∗017:01 alleles known to be associated with slow disease progression. Typing was performed as previously described.70 Animals were randomly distributed to two groups with six monkeys each.
Animal care
Socially compatible monkeys were housed in groups of two by combining cages. If animals had to be caged individually they had constant visual, olfactory and acoustic contact to their roommates and could still groom their neighbours through small mash inserts in the separating side walls. Each cage was equipped with a perch. The animals had water access ad libitum and were fed with dry monkey biscuits containing adequate carbohydrate, energy, fat, fiber (10%), mineral, protein, and vitamin content twice daily. The feed was supplemented by fresh fruit or vegetables and other edible objects like nuts, cereal pulp and different seeds to offer variety to the diet. Moreover, for environmental enrichment monkeys were provided feeding puzzles boards, alternate toys and wood sticks for gnawing. During the study, animals were assessed by experienced keepers twice a day for any signs of distress, pain or sickness by checking water and feed intake, feces consistency and the general condition. In case of any abnormal presentation, animals were attended by veterinarians.
Ethical statement for human samples
The use of established cell lines (HEK293T and TZM-bl cells) did not require the approval of the Institutional Review Board.
Cell lines
All cells were cultured at 37°C in a 5% CO2 atmosphere. HEK293T (Human embryonic kidney) cells (ATCC: #CRL3216; derived from a femal fetus in 1973) and TZM-bl (NIH: ARP5011, HeLa cell line; derived from an adenocarcinoma of cervix of a 31-year-old female in 1951) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), L-glutamine (2 mM), streptomycin (100 μg/ml) and penicillin (100 U/ml). TZM-bl cells were provided and authenticated by the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc (Platt et al., 1998). TZM-bl are derived from HeLa cells, which were isolated from a 30-year-old female. Purchased cell lines were authenticated by the respective vendor and were not validated further in our laboratory. All cell lines used in this study were regularly checked for the presence of mycoplasma by PCR.
Method details
Virus stocks and transductions
To generate virus stocks to perform the neutralization assays, HEK293T cells were transfected with the proviral SIVmac239 using LT1 following the manufacturer protocol (Mirus) Two days post-transfection, supernatants containing infectious virus was harvested and concentrated 10 times using the Amicon filter tubes. Viral stocks were tittered on TZM-bl cells.
Virus inoculation and specimen collection
Infections were carried out at two time-points, about 1.5 years apart. In each of the two experiments, three animals were infected with wt and three animals with Δvpr SIVmac239. Blood was collected from the femoral vein using the BD Vacutainer system (BD, Heidelberg) and 22G BD Vacutainer® Eclipse™ Blood Collection Needles. For virus inoculation, medical interventions like removal of peripheral lymph nodes, and as premedication for euthanasia, all for which a deeper anesthesia was required, animals were administered i.m. a mixture of 5-10 mg ketamine, 1-2 mg xylazine, and 0.01-0.02 mg atropine per kg BW. Each animal of the first experimental group (five males and one female) was inoculated intravenously with 1 ml of cell-free culture supernatant containing 500 median tissue culture infectious doses of wt or Δvpr SIVmac239. The animals of the other group (five males and one female) received SIVmac239 wild type at the same dose and by the same route as for the mutant virus. Blood samples were collected four to five times before infection, two to three times up to day 7, thereafter at weeks 2, 3, and four, followed by 2-4-week intervals until 24 wpi and thereafter around every 8 weeks until euthanasia because of the development of AIDS-like disease or the end of the study (weeks 82-89). Colon biopsies were obtained by endoscopy two times before infection and up to seven times between 2 and 66 wpi. Peripheral lymph nodes were surgically removed once or twice before infection and up to six times between 2 and 66 wpi. At necropsy, animals were euthanized by an overdose of 160-240 mg sodium pentobarbital per kg BW injected into the circulation.
RNA purification
Total RNA extraction was performed using the Paxgene RNA purification kit (Qiagen, Hilden, Germany). Briefly, 2.5 ml of whole blood was collected in Paxgene blood RNA tubes and total RNA extraction was performed using the Paxgene RNA purification kit according to manufacturer’s specifications; on-column DNAse digestion was also performed to remove Genomic DNA. RNA integrity of the extracted RNA was assessed by Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) capillary electrophoresis on a Bioanlayzer RNA nanochip.
Viral RNA loads
Viral RNA was extracted from 200 μl plasma using QiaAmp viral RNA isolation mini kit. For qRT-PCR of viral RNA, 8.5 μl RNA-solution were reverse transcribed and amplified using TaKaRa Prime-Script-One-Step-RT-PCR kit (TaKaRa Bio Europe) using primers gag forward (5′-ACCCAGTACAACAAATAGGTGGTAACT-3′), gag reverse (5′-TCAATTTTACCCAG-GCATTTAATGT-3′) and a fluorescent probe (5′-6FAM(6-carboxyfluorescein)-TGTCCACCT-GCCATTAAGCCCGAG-TAMRA(6-carboxytetramethylrhodamine-3′). Reverse transcription was performed at 45°C for 5 min, amplification was started by an initial denaturation step at 95°C for 10 s, followed by 45 cycles of denaturation for 5 s at 95°C and annealing and elongation at 60°C for 30 s using the Rotor-Gene Q apparatus and software (Qiagen).
Cell-associated viral load
The cell-associated virus load in peripheral blood was determined by a limiting dilution co-cultivation assay as described.71,72 Instead of testing supernatants of cell cultures for SIV p27, SIV positive cultures were identified by immunoperoxidase staining for intracellular viral proteins with a SIV-hyperimmune serum from an infected macaque as reported.73 To increase adherence of indicator cells, plates used for staining were coated with Concanavalin A.74
T cell ELISpot
To determine the SIV-specific T-cell response in blood of the infected animals, an IFN-γ ELISpot assay was performed using commercially available reagents (Mabtech), as previously reported.42,75,76 Purified PBMCs were seeded at 1×105 cells per well in triplicate wells of 96-well plates (MLLIPORE, MAIP S4510). Cells were stimulated as reported42 with SIV Gag peptides (EVA7066.1-16, Centre for AIDS Reagents, NIBSC, United Kingdom) at 2 μg/ml each or inactivated whole SIV (aldrithiol-2-inactivated whole SIVmac251 particles [SIV AT-2], ARP1018.1, kindly provided by Jeff Lifson, National Cancer Institute [NCI], Frederick, MD, USA, and distributed through the Centre for AIDS Reagents, NIBSC, United Kingdom). As peptide pool background control SIV-unrelated hepatitis C virus NS3 peptides were included.76 Microvesicles (ARP1018.2, see source above) were used as background control for viral particle stimulation.42 Spots were counted by means of a Bioreader-3000 ELISPOT reader (Bio-Sys) and values calculated for 106 cells. Samples were considered positive if yielding both >100 Spot Forming Cells (SFC)/106 PBMC minus background and exceeding twice the values of the respective controls.
Serological testing
To analyse the SIV-specific humoral immune response in blood, a standard ELISA was performed using recombinant SIV p27 (EVA643.2) and SIV gp130 (cat. # 100841) kindly provided by the Centre for AIDS Reagents (National Institute for Biological Standards and Control), UK. Antigens were coated at 50 ng protein per well. Antibody levels were assessed using 1:200-diluted plasma.
Neopterin
Urinary neopterin was analysed by high performance liquid chromatography (HPLC) as described previously.77 Urinary samples were collected pre and up to day 31 post-infection in the morning. To equalize physiological differences in urinary concentration creatinine was used as an internal standard.
Transcriptome analysis
The collection of blood PBMC from wt SIVmac239 and Δvpr infected animals was carried out in two sets. Set A consists of PBMC samples from wt (n=3) and Δvpr (n=3) infected animals at only post-infection time points. In addition to PBMC samples from infected animals, PBMC samples in Set A were also collected from uninfected control animals (n=10). Set B consists of PBMC from wt (n=3) and Δvpr (n=3) infected animals before and post-infection. RNA was extracted from these samples using RNeasy Mini kits (Qiagen, CA) with DNase digest and QIAcube automation stations. Quantification of extracted RNA was done on NanoDrop 2000 spectrophotometer (Thermo Scientific Inc. Wilmington, DE) and the quality was assessed by Bioanalyzer analysis (Agilent Technologies, Santa Clara, CA). For Set A samples, libraries were prepared using the Illumina (Illumina Inc. San Diego, Ca, USA) TruSeq mRNA stranded kit, as per manufacturer’s instructions, with 400 ng of total RNA as input. For Set B samples, 10 ng of total RNA was used as input for cDNA amplification using 5′ template-switch PCR with the Clontech SMART-Seq v4 Ultra Low Input RNA kit. Further, for this set, amplified cDNA was fragmented and appended with dual indexed bar codes using Illumina NexteraXT DNA Library Prep kits. The amplified libraries from both sets were validated by capillary electrophoresis on the Agilent 4200 TapeStation. The libraries were normalized, pooled and sequenced on the Illumina HiSeq 3000 system employing a single-end 101 cycles run at average read depths of 16 million reads/sample (Set A) and 25 million reads/sample (Set B). Infections were performed at two time points ∼2.5 years apart each involving six animals (3x wt and 3x Δvpr). The sequencing data was demultiplexed using Illumina bcl2fastq version 2.20.0.422. The quality of raw reads was assessed with FastQC version 0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were mapped to the MacaM version 7 assembly of the Indian rhesus macaque genomic reference78 assembly available at: http://www.unmc.edu/rhesusgenechip/index.htm#NewRhesusGenome using STAR version 2.5.2b with default alignment parameters.79 Abundance estimation of raw read counts per transcript was done internally with STAR using the htseq-count algorithm.80 DESeq2 version 1.22.1 R package was used to produce normalized read counts and a regularized log expression table.81 PCA showed that each experiment rather than the different groups of animals clustered separately prior to normalization (Figure S1A). To enable comparison of the two datasets they were normalized with the mean of all of the values set to 0 and the standard deviation to 1 (Z-score). After normalization, set A and B samples were combined for further analysis. Uninfected dataset consists of PBMC samples collected from control animals and those from infected animals at 4 weeks before infection.
Gene set enrichment analysis and heatmaps
Gene set enrichment analysis (GSEA) was performed on the PBMC normalized counts expression data using the Broad Institute GSEA desktop module 4.2.3 (http://www.broadinstitute.org/gsea/). Enrichment analysis was performed for Δvpr vs wt datasets for PBMC and wt vs Δvpr. Heatmaps were generated to visualize the expression patterns of the leading-edge genes in the indicated gene sets for that enrichment analysis. For PBMC regularized log expression data, the expression for each gene was normalized by the mean expression across all samples (Δvpr, wt and uninfected samples). These were then plotted based on a gradient color-scale.
Multiplex cytokine profile analysis
A total of 15 cytokines/chemokines were measured in macaque plasma by Luminex at baseline, days 5, 7, and 10, and weeks 2, 3 4, and 8 post SIV infection. The analytes included: B-cell activating factor (BAFF), eotaxin (CCL11), interferon alpha (IFN-α) and gamma (IFN-γ), IL-1 receptor antagonist (IL-1Ra), monokine induced by IFN-gamma (MIG/CXCL9), macrophage inflammatory protein 1- beta (MIP-1β/CCL4), perforin. The multiplex assay was conducted as previously described.82,83
Neutralization assay
To determine the neutralizing activities in both groups of animals, 10,000 TZM-bl cells per well were seeded in 96 well plates. The next day, cells were treated with 1:2 serial dilutions of the sera and mixed with SIV239mac. Three days post-infection, cells were lysed and β-galactosidase reporter gene expression was determined using the GalScreen Kit (Applied Bioscience) according to the manufacturer’s instructions with an Orion microplate luminometer (Berthold). To perform the neutralizing assay from pooled sera, equal amounts of serum from each monkey were combined before dilutions.
Avidity and CXCL13 ELISA
To measure the binding avidity of antibodies present in the serum, a modified SIV p27 and SIV gp130 ELISA was performed by the addition of a chaotropic agent, 8M urea, to the wash buffer. ELISA was performed using recombinant SIV p27 (EVA643.2) and SIV gp130 (cat. # 100841) kindly provided by the Centre for AIDS Reagents (National Institute for Biological Standards and Control), UK. Antigens were coated at 50 ng protein per well. The avidity index is calculated by normalizing the values obtained in presence of urea to those in absence of urea for the same serum dilution. The serum was diluted to reach linear range in the normal ELISA (OD<1.5) and the values from the according dilution in the condition with urea were kept (here, 1:50 for p27 and 1:1000 for gp130). To measure the plasma levels of CXCL13, the Rhesus Macaque BLC ELISA Kit (Cat# EP3RB, Invitrogen) was used according to protocol of the manufacturer. In brief, pre-coated plates were incubated with 100 ul of plasma or standard overnight and calculation of the CXCL13 concentrations were done using a 4PL sigmoidal standard curve with prior subtraction of blanks.
Immunohistochemical detection of Mx1 in colon biopsies
Colon biopsies and lymph nodes planned for immunohistochemistry (IHC) were fixed in 4% formaldehyde. The samples were embedded in paraffin and sectioned at 3 μm. For Mx1 detection we used the monoclonal antibody M143 (kindly provided from Professor Kochs, Freiburg University). IHC was performed with an automated immunostaining system (Discovery XT, Roche Diagnostics GmbH, Mannheim, Germany) using the SABC (streptavidin–biotin complex) method and DAB (diaminobenzidine tetrahydrochloride) for signal detection (DAB Map Kit, Roche Diagnostics GmbH, Mannheim, Germany). Slides were scanned using the Aperio ScanScope, at a high magnification (400x). The enumeration of positive immunohistochemical signals was performed using the Positive-Pixel-Count algorithm of the Aperio image scope version 12.3.1.5011.
Immunoblotting of SIV antibodies
To determine to which SIV antibodies were present in the sera of monkeys infected with either wt or Δvpr SIVmac239, we performed the SIV Western Blot assay (ZeptoMetrix Cat#0801500) following the manufacturer protocol. Briefly, the antigen-coated strips were incubated with the serum at 37°C for two hours before washing and incubation with a conjugated secondary antibody. Bands are revealed by addition of 5-bromo-4chloro-3indolyl-phosphate (BCIP) substrate and nitroblue tetrazolium (NBT). Pictures were taken with Biorad GelDoc XR+ imaging system.
Flow cytometry
50μL of whole blood were stained for 30 min at room temperature in the dark with different mixtures of monoclonal antibodies at optimal pretitrated concentrations. The mixtures included CD3 (clone SP34-2, Alexa Fluor 700), CD4 (clone L200; Horizon V450), CD21 (clone B-ly4, FITC), CD28 (clone CD28.2, FITC), CD45 (clone D058-1283, Horizon V500), CD95 (clone Dx2, PE-Cy7), CD195 (clone 3A9, PE), all from BD Biosciences, Heidelberg, Germany; CD8 (clone RPA-T8, PerCP-Cy5.5; clone SK1, APC-Cy7), CD20 (clone 2H7, PE-Cy7), CD25 (clone M-A251, APC-Fire), CD27 (clone M-T271, APC), CD196 (clone G034E3, PE-Dazzle), CD197 (clone G043H7, APC, Brilliant Violet 421), HLA-DR (clone L243, APC-Cy7), Ki67 (clone Ki67, Brilliant Violet 605), PD-1 (clone EH12.2H7, PE-Cy7) all from BioLegend, San Diego, USA and CD69 (clone TP1.55.3, ECD) from Beckman Coulter, Krefeld, Germany. Lysis of residual RBCs (red blood cells) and fixation was performed by incubation with 1mL RBC lysis/fixation solution from BioLegend for 15min. For intranuclear staining of Ki67 the FOXP3 Fix/Perm Buffer set from BioLegend was used according to the manufacturer's instructions. Briefly, following surface staining of whole blood and RBC lysis, cells were incubated with fix/perm solution and subsequently perm buffer. Cells were then stained with Ki67 antibody diluted in perm buffer for 30min. Cells were acquired using a LSRII cytometer (BD Biosciences) equipped with three lasers. Compensation was calculated by FACS DIVA software 6.1.3 using appropriate single antibody labeled compensation beads from SpheroTech, Lake Forest, USA. Analysis was performed using FlowJo 9.6.4 (Treestar, Ashland, OR, USA).
Quantification and statistical analysis
Statistical analyses were performed with GraphPad PRISM (GraphPad Software) and Microsoft Excel. P-values were calculated using the two-tailed unpaired Student’s-t-test. Correlations were calculated with the linear regression module. Unless otherwise stated, all in vitro experiments were performed in three technical replicates for each animal and the data are shown as mean ± SEM. No methods were used to determine whether the data met assumptions of the statistical approach. Significant differences are indicated as: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Statistical parameters are specified in the figure legends.
Acknowledgments
We thank Dré van der Merwe, Kerstin Regensburger, Daniela Krnavek, Martha Mayer, Regina Burger, Sandra Heine, and Nicole Leuchte for technical assistance and Matthias Mietsch for excellent and comprehensive veterinary support and Andrés Finzi (Montreal) for helpful advice and support. This work was supported by grants from the German Research Foundation (DFG CRC 1279 and SPP 1923). The Emory NPRC Genomics Core is supported in part by NIH P51 OD011132 and S10 OD026799. This investigation used resources supported by the Southwest National Primate Research Center grant P51 OD011133 from the Office of Research Infrastructure Programs, National Institutes of Health and Biomedical Primate Research Centre (BPRC; Rijswijk, Netherlands). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
A.L., C.P.B., and V.H. performed most of the in vitro analyses. S.J. also provided experimental data and generated mutant vpr constructs. U.S., B.R., A.K., M.D., K.M.-R., N.S.-L., M.D., D.F., and C.S.-H. performed the vivo analyses in the SIV/macaque model. G.K.T., S.A.N., P.M.G., and S.E.B. performed the transcriptome analyses, and L.P. and L.G. determined the multiplex cytokine profiles. A.L. and C.P.B. performed gene expression analyses supported by V.H. and K.M.J.S. S.E.B., L.G., G.S., K.M.J.S., C.S.-H., and F.K. conceived and designed the experiments and analyzed the data. F.K. wrote the initial draft manuscript. All authors have seen, edited, and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108351.
Supplemental information
References
- 1.Guenzel C.A., Hérate C., Benichou S. HIV-1 Vpr-a still “enigmatic multitasker.”. Front. Microbiol. 2014;5:127. doi: 10.3389/fmicb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabryova H., Strebel K. Vpr and Its Cellular Interaction Partners: R We There Yet? Cells. 2019;8:1310. doi: 10.3390/cells8111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nodder S.B., Gummuluru S. Illuminating the Role of Vpr in HIV Infection of Myeloid Cells. Front. Immunol. 2019;10:1606. doi: 10.3389/fimmu.2019.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauter D., Kirchhoff F. Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins. Cytokine Growth Factor Rev. 2018;40:3–12. doi: 10.1016/j.cytogfr.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Mashiba M., Collins D.R., Terry V.H., Collins K.L. Vpr overcomes macrophage-specific restriction of HIV-1 Env expression and virion production. Cell Host Microbe. 2014;16:722–735. doi: 10.1016/j.chom.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jowett J.B., Planelles V., Poon B., Shah N.P., Chen M.L., Chen I.S. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 1995;69:6304–6313. doi: 10.1128/JVI.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Planelles V., Jowett J.B., Li Q.X., Xie Y., Hahn B., Chen I.S. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 1996;70:2516–2524. doi: 10.1128/JVI.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kogan M., Deshmane S., Sawaya B.E., Gracely E.J., Khalili K., Rappaport J. Inhibition of NF-κB activity by HIV-1 Vpr is dependent on Vpr binding protein. J. Cell. Physiol. 2013;228:781–790. doi: 10.1002/jcp.24226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu R., Lin Y., Jia R., Geng Y., Liang C., Tan J., Qiao W. HIV-1 Vpr stimulates NF-κB and AP-1 signaling by activating TAK1. Retrovirology. 2014;11:45. doi: 10.1186/1742-4690-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu R., Tan J., Lin Y., Jia R., Yang W., Liang C., Geng Y., Qiao W. HIV-1 Vpr activates both canonical and noncanonical NF-κB pathway by enhancing the phosphorylation of IKKα/β. Virology. 2013;439:47–56. doi: 10.1016/j.virol.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 11.Richard J., Sindhu S., Pham T.N.Q., Belzile J.-P., Cohen E.A. HIV-1 Vpr up-regulates expression of ligands for the activating NKG2D receptor and promotes NK cell-mediated killing. Blood. 2010;115:1354–1363. doi: 10.1182/blood-2009-08-237370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuschl A.-K., Mesner D., Shivkumar M., Whelan M.V.X., Pallett L.J., Guerra-Assunção J.A., Madansein R., Dullabh K.J., Sigal A., Thornhill J.P., et al. HIV-1 Vpr drives a tissue residency-like phenotype during selective infection of resting memory T cells. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauby H., Ward C.C., Hugh-White R., Swanson C.M., Schulz R., Goujon C., Malim M.H. HIV-1 Vpr Induces Widespread Transcriptomic Changes in CD4+ T Cells Early Postinfection. mBio. 2021;12 doi: 10.1128/mBio.01369-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brégnard C., Benkirane M., Laguette N. DNA damage repair machinery and HIV escape from innate immune sensing. Front. Microbiol. 2014;5:176. doi: 10.3389/fmicb.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrecka K., Hao C., Shun M.-C., Kaur S., Swanson S.K., Florens L., Washburn M.P., Skowronski J. HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc. Natl. Acad. Sci. USA. 2016;113:E3921–E3930. doi: 10.1073/pnas.1605023113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J., Shun M.-C., Hao C., Zhang Y., Qian J., Hrecka K., DeLucia M., Monnie C., Ahn J., Skowronski J. HIV-1 Vpr Reprograms CLR4DCAF1 E3 Ubiquitin Ligase to Antagonize Exonuclease 1-Mediated Restriction of HIV-1 Infection. mBio. 2018;9:e017322-18–e1818. doi: 10.1128/mBio.01732-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahouassa H., Blondot M.-L., Chauveau L., Chougui G., Morel M., Leduc M., Guillonneau F., Ramirez B.C., Schwartz O., Margottin-Goguet F. HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc. Natl. Acad. Sci. USA. 2016;113:5311–5316. doi: 10.1073/pnas.1600485113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X., DeLucia M., Hao C., Hrecka K., Monnie C., Skowronski J., Ahn J. HIV-1 Vpr protein directly loads helicase-like transcription factor (HLTF) onto the CRL4-DCAF1 E3 ubiquitin ligase. J. Biol. Chem. 2017;292:21117–21127. doi: 10.1074/jbc.M117.798801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan N., Regalado-Magdos A.D., Stiggelbout B., Lee-Kirsch M.A., Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J., Shun M.-C., Zhang Y., Hao C., Skowronski J. HIV-1 Vpr counteracts HLTF-mediated restriction of HIV-1 infection in T cells. Proc. Natl. Acad. Sci. USA. 2019;116:9568–9577. doi: 10.1073/pnas.1818401116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laguette N., Brégnard C., Hue P., Basbous J., Yatim A., Larroque M., Kirchhoff F., Constantinou A., Sobhian B., Benkirane M. Premature activation of the slx4 complex by vpr promotes g2/m arrest and escape from innate immune sensing. Cell. 2014;156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Fregoso O.I., Emerman M. Activation of the DNA Damage Response Is a Conserved Function of HIV-1 and HIV-2 Vpr That Is Independent of SLX4 Recruitment. mBio. 2016;7 doi: 10.1128/mBio.01433-16. e014333-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C.-Y., Chiang S.-F., Lin T.-Y., Chiou S.-H., Chow K.-C. HIV-1 Vpr triggers mitochondrial destruction by impairing Mfn2-mediated ER-mitochondria interaction. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart S.A., Poon B., Jowett J.B., Chen I.S. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J. Virol. 1997;71:5579–5592. doi: 10.1128/JVI.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacotot E., Ravagnan L., Loeffler M., Ferri K.F., Vieira H.L., Zamzami N., Costantini P., Druillennec S., Hoebeke J., Briand J.P., et al. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J. Exp. Med. 2000;191:33–46. doi: 10.1084/jem.191.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vermeire J., Roesch F., Sauter D., Rua R., Hotter D., Van Nuffel A., Vanderstraeten H., Naessens E., Iannucci V., Landi A., et al. HIV Triggers a cGAS-Dependent, Vpu- and Vpr-Regulated Type I Interferon Response in CD4+ T Cells. Cell Rep. 2016;17:413–424. doi: 10.1016/J.CELREP.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Johnson P.R., Hirsch V.M. SIV infection of macaques as a model for AIDS pathogenesis. Int. Rev. Immunol. 1992;8:55–63. doi: 10.3109/08830189209056641. [DOI] [PubMed] [Google Scholar]
- 28.Chakrabarti L., Guyader M., Alizon M., Daniel M.D., Desrosiers R.C., Tiollais P., Sonigo P. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature. 1987;328:543–547. doi: 10.1038/328543a0. [DOI] [PubMed] [Google Scholar]
- 29.Ayouba A., Akoua-Koffi C., Calvignac-Spencer S., Esteban A., Locatelli S., Li H., Li Y., Hahn B.H., Delaporte E., Leendertz F.H., Peeters M. Evidence for continuing cross-species transmission of SIVsmm to humans: Characterization of a new HIV-2 lineage in rural Côte d’Ivoire. AIDS. 2013;27:2488–2491. doi: 10.1097/01.aids.0000432443.22684.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chahroudi A., Bosinger S.E., Vanderford T.H., Paiardini M., Silvestri G. Natural SIV Hosts: Showing AIDS the Door. Science. 2012;335:1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell-Yesufu O.T., Gandhi R.T. Update on Human Immunodeficiency Virus (HIV)-2 Infection. Clin. Infect. Dis. 2011;52:780–787. doi: 10.1093/cid/ciq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regier D.A., Desrosiers R.C. The Complete Nucleotide Sequence of a Pathogenic Molecular Clone of Simian Immunodeficiency Virus. AIDS Res. Hum. Retrovir. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 34.Hoch J., Lang S.M., Weeger M., Stahl-Hennig C., Coulibaly C., Dittmer U., Hunsmann G., Fuchs D., Müller J., Sopper S. vpr deletion mutant of simian immunodeficiency virus induces AIDS in rhesus monkeys. J. Virol. 1995;69:4807–4813. doi: 10.1128/JVI.69.8.4807-4813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbs J.S., Lackner A.A., Lang S.M., Simon M.A., Sehgal P.K., Daniel M.D., Desrosiers R.C. Progression to AIDS in the absence of a gene for vpr or vpx. J. Virol. 1995;69:2378–2383. doi: 10.1128/JVI.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbs J.S., Regier D.A., Desrosiers R.C. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 1994;10:607–616. doi: 10.1089/aid.1994.10.607. [DOI] [PubMed] [Google Scholar]
- 37.Higham J.P., Kraus C., Stahl-Hennig C., Engelhardt A., Fuchs D., Heistermann M. Evaluating noninvasive markers of nonhuman primate immune activation and inflammation. Am. J. Phys. Anthropol. 2015;158:673–684. doi: 10.1002/ajpa.22821. [DOI] [PubMed] [Google Scholar]
- 38.Deshmane S.L., Mukerjee R., Fan S., Del Valle L., Michiels C., Sweet T., Rom I., Khalili K., Rappaport J., Amini S., Sawaya B.E. Activation of the Oxidative Stress Pathway by HIV-1 Vpr Leads to Induction of Hypoxia-inducible Factor 1α Expression. J. Biol. Chem. 2009;284:11364–11373. doi: 10.1074/jbc.M809266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langer S., Hammer C., Hopfensperger K., Klein L., Hotter D., De Jesus P.D., Herbert K.M., Pache L., Smith N., van der Merwe J.A., et al. Hiv-1 vpu is a potent transcriptional suppressor of nf-kb-elicited antiviral immune responses. Elife. 2019;8 doi: 10.7554/eLife.41930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iijima K., Kobayashi J., Ishizaka Y. Structural alteration of DNA induced by viral protein R of HIV-1 triggers the DNA damage response. Retrovirology. 2018;15:8. doi: 10.1186/s12977-018-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hotter D., Krabbe T., Reith E., Gawanbacht A., Rahm N., Ayouba A., Van Driessche B., Van Lint C., Peeters M., Kirchhoff F., Sauter D. Primate lentiviruses use at least three alternative strategies to suppress NF-κB-mediated immune activation. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauermann U., Radaelli A., Stolte-Leeb N., Raue K., Bissa M., Zanotto C., Krawczak M., Tenbusch M., Überla K., Keele B.F., et al. Vector Order Determines Protection against Pathogenic Simian Immunodeficiency Virus Infection in a Triple-Component Vaccine by Balancing CD4+ and CD8+ T-Cell Responses. J. Virol. 2017;91 doi: 10.1128/JVI.01120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Titanji K., Velu V., Chennareddi L., Vijay-Kumar M., Gewirtz A.T., Freeman G.J., Amara R.R. Acute depletion of activated memory B cells involves the PD-1 pathway in rapidly progressing SIV-infected macaques. J. Clin. Invest. 2010;120:3878–3890. doi: 10.1172/JCI43271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tauzin A., Gendron-Lepage G., Nayrac M., Anand S.P., Bourassa C., Medjahed H., Goyette G., Dubé M., Bazin R., Kaufmann D.E., Finzi A. Evolution of Anti-RBD IgG Avidity following SARS-CoV-2 Infection. Viruses. 2022;14:532. doi: 10.3390/v14030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson W.E., Sanford H., Schwall L., Burton D.R., Parren P.W.H.I., Robinson J.E., Desrosiers R.C. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 2003;77:9993–10003. doi: 10.1128/jvi.77.18.9993-10003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson W.E., Lifson J.D., Lang S.M., Johnson R.P., Desrosiers R.C. Importance of B-cell responses for immunological control of variant strains of simian immunodeficiency virus. J. Virol. 2003;77:375–381. doi: 10.1128/jvi.77.1.375-381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Havenar-Daughton C., Lindqvist M., Heit A., Wu J.E., Reiss S.M., Kendric K., Bélanger S., Kasturi S.P., Landais E., Akondy R.S., et al. CXCL13 is a plasma biomarker of germinal center activity. Proc. Natl. Acad. Sci. USA. 2016;113:2702–2707. doi: 10.1073/pnas.1520112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moukambi F., Rabezanahary H., Rodrigues V., Racine G., Robitaille L., Krust B., Andreani G., Soundaramourty C., Silvestre R., Laforge M., Estaquier J. Early Loss of Splenic Tfh Cells in SIV-Infected Rhesus Macaques. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan H., Sumner R.P., Rasaiyaah J., Tan C.P., Rodriguez-Plata M.T., Van Tulleken C., Fink D., Zuliani-Alvarez L., Thorne L., Stirling D., et al. HIV-1 Vpr antagonizes innate immune activation by targeting karyopherin-mediated NF-κB/IRF3 nuclear transport. Elife. 2020;9 doi: 10.7554/eLife.60821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okumura A., Alce T., Lubyova B., Ezelle H., Strebel K., Pitha P.M. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology. 2008;373:85–97. doi: 10.1016/j.virol.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayyavoo V., Muthumani K., Kudchodkar S., Zhang D., Ramanathan P., Dayes N.S., Kim J.J., Sin J.-I., Montaner L.J., Weiner D.B. HIV-1 viral protein R compromises cellular immune function in vivo. Int. Immunol. 2002;14:13–22. doi: 10.1093/intimm/14.1.13. [DOI] [PubMed] [Google Scholar]
- 52.Muthumani K., Bagarazzi M., Conway D., Hwang D.S., Ayyavoo V., Zhang D., Manson K., Kim J., Boyer J., Weiner D.B. Inclusion of Vpr accessory gene in a plasmid vaccine cocktail markedly reduces Nef vaccine effectiveness in vivo resulting in CD4 cell loss and increased viral loads in rhesus macaques. J. Med. Primatol. 2002;31:179–185. doi: 10.1034/j.1600-0684.2002.02004.x. [DOI] [PubMed] [Google Scholar]
- 53.Moir S., Fauci A.S. B-cell exhaustion in HIV infection: the role of immune activation. Curr. Opin. HIV AIDS. 2014;9:472–477. doi: 10.1097/COH.0000000000000092. [DOI] [PubMed] [Google Scholar]
- 54.Crotty S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015;15:185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong J.J., Amancha P.K., Rogers K., Ansari A.A., Villinger F. Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J. Immunol. 2012;188:3247–3256. doi: 10.4049/jimmunol.1103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bros M., Haas K., Moll L., Grabbe S. RhoA as a Key Regulator of Innate and Adaptive Immunity. Cells. 2019;8:733. doi: 10.3390/cells8070733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goh W.C., Rogel M.E., Kinsey C.M., Michael S.F., Fultz P.N., Nowak M.A., Hahn B.H., Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 58.Sauter D., Kirchhoff F. Key Viral Adaptations Preceding the AIDS Pandemic. Cell Host Microbe. 2019;25:27–38. doi: 10.1016/j.chom.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Tristem M., Marshall C., Karpas A., Hill F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 1992;11:3405–3412. doi: 10.1002/j.1460-2075.1992.tb05419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharp P.M., Bailes E., Stevenson M., Emerman M., Hahn B.H. Gene acquisition in HIV and SIV. Nature. 1996;383:586–587. doi: 10.1038/383586a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Desrosiers R.C., Lifson J.D., Gibbs J.S., Czajak S.C., Howe A.Y., Arthur L.O., Johnson R.P. Identification of Highly Attenuated Mutants of Simian Immunodeficiency Virus. J. Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chougui G., Munir-Matloob S., Matkovic R., Martin M.M., Morel M., Lahouassa H., Leduc M., Ramirez B.C., Etienne L., Margottin-Goguet F. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat. Microbiol. 2018;3:891–897. doi: 10.1038/s41564-018-0179-6. [DOI] [PubMed] [Google Scholar]
- 63.Hrecka K., Hao C., Gierszewska M., Swanson S.K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M.P., Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller S.M., Lang S.M. The first HxRxG motif in simian immunodeficiency virus mac239 Vpr is crucial for G(2)/M cell cycle arrest. J. Virol. 2002;76:11704–11709. doi: 10.1128/jvi.76.22.11704-11709.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Y., Gelbard H.A., Roshal M., Pursell S., Jamieson B.D., Planelles V. Comparison of cell cycle arrest, transactivation, and apoptosis induced by the simian immunodeficiency virus SIVagm and human immunodeficiency virus type 1 vpr genes. J. Virol. 2001;75:3791–3801. doi: 10.1128/JVI.75.8.3791-3801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casey L., Wen X., de Noronha C.M.C. The functions of the HIV1 protein Vpr and its action through the DCAF1.DDB1.Cullin4 ubiquitin ligase. Cytokine. 2010;51:1–9. doi: 10.1016/j.cyto.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fletcher T.M., Brichacek B., Sharova N., Newman M.A., Stivahtis G., Sharp P.M., Emerman M., Hahn B.H., Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 69.Sato K., Misawa N., Iwami S., Satou Y., Matsuoka M., Ishizaka Y., Ito M., Aihara K., An D.S., Koyanagi Y. HIV-1 Vpr accelerates viral replication during acute infection by exploitation of proliferating CD4+ T cells in vivo. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sauermann U., Siddiqui R., Suh Y.-S., Platzer M., Leuchte N., Meyer H., Mätz-Rensing K., Stoiber H., Nürnberg P., Hunsmann G., et al. Mhc class I haplotypes associated with survival time in simian immunodeficiency virus (SIV)-infected rhesus macaques. Gene Immun. 2008;9:69–80. doi: 10.1038/sj.gene.6364448. [DOI] [PubMed] [Google Scholar]
- 71.Stahl-Hennig C., Dittmer U., Nisslein T., Petry H., Jurkiewicz E., Fuchs D., Wachter H., Mätz-Rensing K., Kuhn E.M., Kaup F.J., et al. Rapid development of vaccine protection in macaques by live-attenuated simian immunodeficiency virus. J. Gen. Virol. 1996;77:2969–2981. doi: 10.1099/0022-1317-77-12-2969. [DOI] [PubMed] [Google Scholar]
- 72.Stahl-Hennig C., Voss G., Nick S., Petry H., Fuchs D., Wachter H., Coulibaly C., Lüke W., Hunsmann G. Immunization with Tween-ether-treated SIV adsorbed onto aluminum hydroxide protects monkeys against experimental SIV infection. Virology. 1992;186:588–596. doi: 10.1016/0042-6822(92)90025-K. [DOI] [PubMed] [Google Scholar]
- 73.Norley S.G., Löwer J., Kurth R. Insufficient inactivation of HIV-1 in human cryo poor plasma by beta-propiolactone: results from a highly accurate virus detection method. Biologicals. 1993;21:251–258. doi: 10.1006/biol.1993.1082. [DOI] [PubMed] [Google Scholar]
- 74.Gundlach B.R., Reiprich S., Sopper S., Means R.E., Dittmer U., Mätz-Rensing K., Stahl-Hennig C., Uberla K. Env-independent protection induced by live, attenuated simian immunodeficiency virus vaccines. J. Virol. 1998;72:7846–7851. doi: 10.1128/jvi.72.10.7846-7851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suh Y.S., Park K.S., Sauermann U., Franz M., Norley S., Wilfingseder D., Stoiber H., Fagrouch Z., Heeney J., Hunsmann G., et al. Reduction of viral loads by multigenic DNA priming and adenovirus boosting in the SIVmac-macaque model. Vaccine. 2006;24:1811–1820. doi: 10.1016/j.vaccine.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 76.Stahl-Hennig C., Kuate S., Franz M., Suh Y.S., Stoiber H., Sauermann U., Tenner-Racz K., Norley S., Park K.S., Sung Y.C., et al. Atraumatic oral spray immunization with replication-deficient viral vector vaccines. J. Virol. 2007;81:13180–13190. doi: 10.1128/JVI.01400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schroecksnadel K., Winkler C., Fuchs D. Method for urinary neopterin measurements by HPLC. J. Biochem. Biophys. Methods. 2006;66:99–100. doi: 10.1016/j.jbbm.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 78.Zimin A.V., Cornish A.S., Maudhoo M.D., Gibbs R.M., Zhang X., Pandey S., Meehan D.T., Wipfler K., Bosinger S.E., Johnson Z.P., et al. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol. Direct. 2014;9:20. doi: 10.1186/1745-6150-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giavedoni L.D. Simultaneous detection of multiple cytokines and chemokines from nonhuman primates using luminex technology. J. Immunol. Methods. 2005;301:89–101. doi: 10.1016/j.jim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 83.Woollard S.M., Olwenyi O.A., Dutta D., Dave R.S., Mathews S., Gorantla S., Johnson N., Giavedoni L., Norgren R.B., Byrareddy S.N. Preliminary Studies on Immune Response and Viral Pathogenesis of Zika Virus in Rhesus Macaques. Pathogens. 2018;7:70. doi: 10.3390/pathogens7030070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Transcriptomics data have been submitted to the GEO repository and assigned the accession number GSE229310. the code will be provided once available. They are available for review at: to https://nam11.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.ncbi.nlm.nih.gov%2Fgeo%2Fquery%2Facc.cgi%3Facc%3DGSE229310&data=05%7C01%7Cgktharp%40emory.edu%7C641c7cb43f3e45afb4cd08db39eee4c8%7Ce004fb9cb0a4424fbcd0322606d5df38%7C0%7C0%7C638167470424595022%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=XGvUcza83IJRR0qGpqq90DmE8KjBHoE1PbZVOVuXPIQ%3D&reserved=0; enter token yvsxuoyepvcjrqj into the box. The data will be made public upon acceptance.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.