Abstract
Low pathogenic influenza A viruses (IAVs) have shown promising oncolytic potential in lung cancer–bearing mice. However, as replication-competent pathogens, they may cause side effects in immunocompromised cancer patients. To circumvent this problem, we genetically engineered nonreplicating IAVs lacking the hemagglutinin (HA) gene (ΔHA IAVs), but reconstituted the viral envelope with recombinant HA proteins to allow a single infection cycle. To optimize the therapeutic potential and improve immunomodulatory properties, these replication-incompetent IAVs were complemented with a murine interferon-gamma (mIFN-γ) gene. After intratracheal administration to transgenic mice that develop non-small cell lung cancer (NSCLC), the ΔHA IAVs induced potent tumor destruction. However, ΔHA IAVs armed with mIFN-γ exhibited an even stronger and more sustained effect, achieving 85% tumor reduction at day 12 postinfection. In addition, ΔHA-mIFN-γ viruses were proven to be efficient in recruiting and activating natural killer cells and macrophages from the periphery and in inducing cytotoxic T lymphocytes. Most important, both viruses, and particularly IFN-γ-encoding viruses, activated tumor-associated alveolar macrophages toward a proinflammatory M1-like phenotype. Therefore, replication-incompetent ΔHA-mIFN-γ-IAVs are safe and efficient oncolytic viruses that additionally exhibit immune cell activating properties and thus represent a promising innovative therapeutic option in the fight against NSCLC.
Keywords: replication-incompetent influenza A viruses, hemagglutinin gene-deficiency, single-cycle viruses, interferon gamma, oncolytic viruses, non-small cell lung cancer, tumor immune environment, cancer cell destruction, activation of tumor-associated macrophages, TAMs
Graphical abstract

Replication-incompetent influenza viruses armed with an interferon-γ gene appeared to be safe and efficient oncolytic agents against non-small cell lung carcinomas in mice. In addition, they attract cells of the innate and adaptive immune system to the tumor-bearing lung and mediate their activation.
Introduction
Despite advances in cancer therapy, lung cancers remain the leading cause of cancer deaths, accounting for 1.8 million worldwide.1 Non-small cell lung cancer (NSCLC) is the most common form and accounts for 85% of all lung cancers.2 The delayed diagnosis at an advanced stage continues to be a challenge for the treatment of NSCLC. In addition, this type of tumor is relatively insensitive to classical chemotherapy and radiotherapy,1,2 and not every tumor is amenable to treatment with immune checkpoint inhibitors (ICIs) or targeted therapy with tyrosine kinase blockers.3 Moreover, like most solid tumors, NSCLC promotes the development of an immunosuppressed milieu consisting of anti-inflammatory immune cells such as tumor-associated macrophages (TAMs) that support growth rather than destruction of the tumor.4,5 Therefore, new cancer therapies that target tumor cells while redirecting a patient’s immune system to fight cancer would be a breakthrough in the treatment of lung tumors. Oncolytic viruses (OVs) appear to be a promising new option for lung cancer therapy.
Viral replication is highly dependent on cellular signaling cascades, some of which are known to be functionally activated in transformed cancer cells as well.6 This renders tumor cells highly permissive to viral replication, leading to increased viral amplification and tumor cell death.7 Importantly, OVs not only infect and destroy tumor cells but they also abrogate the immunosuppressed environment and redirect immune cells to the tumor, supporting the lysis of uninfected tumor cells.8,9
Influenza A viruses (IAVs) are of particular interest for targeting NSCLC. NSCLC cells are not only primary targets for IAVs but they also exhibit an oncogene-activated Raf/MEK/ERK signaling pathway that strongly supports IAV replication and renders them highly susceptible to oncolytic IAVs.10,11 Consistent with this, we have demonstrated strong tropism of IAVs for Raf-transformed lung cancer cells in vivo.12,13 Furthermore, our investigation of the general molecular mechanisms underlying the activation and recruitment of subtypes of innate and adaptive immune cells to the lung after acute IAV infection has shown that infection with low doses of IAVs lead to immunomodulation of the lung environment toward a proinflammatory state.13,14,15 Furthermore, we demonstrated potent oncolytic efficacy of intranasally applied IAVs in a preclinical immunocompetent mouse model of NSCLCs whose tumorigenicity was due to oncogenic dysregulation of the Raf/MEK/ERK pathway.13 Interestingly, the oncolytic efficacy was not only based on the viral infection itself but was also complemented by the restoration of immunocompetence of TAMs in the tumor microenvironment (TME).
Influenza viruses are invasive pathogens and rather unattractive for clinical use against lung tumors, however. Safety and selectivity of OVs that prevent the spread of the virus in tissues are critical preconditions for clinical use. This usually requires attenuation of the viruses, which often compromises their lytic ability. Hence, preclinical studies aim to translate the promising potential of oncolytic IAVs into human cancer therapy by improving the safety profile of IAVs.16,17
Here, we describe the generation of replication-incompetent IAVs that lack the hemagglutinin (HA) gene, which is essential for the infectivity of the virus. Instead, recombinant HA proteins were provided in trans during the virus production process, allowing a single cycle of infection. These ΔHA viruses were able to infect epithelial cells, undergo the first replication cycle, and induce apoptosis. To enhance the local immune response after IAV infection, we engineered HA gene–deficient viruses to harbor a mouse interferon gamma (mIFN-γ) gene sequence. Finally, both single-cycle IAV types were evaluated for their oncolytic potential and ability to abrogate immunosuppression in the lung tumor environment using an NSCLC mouse model.
Results
Generation and in vitro characterization of single-cycle infectious IAVs
To generate recombinant IAVs with defective replication, we used the 8-plasmid transfection system described by Hoffmann et al. in 2000,18 which capitalizes on the segmented genome of IAVs and has the advantage that deletions or mutations could be experimentally introduced within the viral genome. The entire coding sequence (measuring 1,382 nt) of the HA gene of the H1N1 strain A/Puerto Rico/8/34, hereafter referred to as PR8, with the exception of 165 and 151 nt at the 5′ and 3′ ends, respectively, was replaced by either a noncoding stuffer sequence (166 nt) or the coding sequence of the murine IFN-γ gene (468 nt). A stuffer sequence was used to keep a certain length of the HA segment to allow sufficient packaging and to integrate restriction sites for further cloning processes. The packaging sequence, including the 5′ and 3′ UTRs, were left intact to ensure successful segment packaging and the formation of ΔHA virus particles.19,20,21 Finally, to exclude any translation of the HA sequence, the remaining start codon ATG was replaced with the nucleotides CTA (Figure 1A).
Figure 1.
Generation of replication-incompetent single-cycle ΔHA IAVs
(A) Design of viral vector constructs (based on pHW2000 plasmids). The 5′ and 3′ end UTRs as well as 165- and 151-nt packaging sequences at the corresponding ends of the HA gene were left intact, whereas the rest was replaced with either a placeholder (Stuffer, 166 nt) or an mIFN-γ sequence (468 nt) to generate ΔHA or ΔHA-mIFN-γ IAVs, respectively. (B and C) HEK293 and MDCK cell lines were transduced with retroviruses comprising the HA gene of the PR8 IAV, and the number of HA-expressing cells was analyzed by flow cytometry. MDCK cells infected with WT PR8 IAV served as positive controls, and nontransduced/noninfected cells were included as negative controls (Mock). The histograms shown in (B) representatively depict the gating to determine HA+ cells, and dashed lines indicate nonspecific binding of the fluorophore-labeled secondary antibody. Independent experiments verify the stable HA expression in both cell lines (i.e., MDCK-HA and HEK-HA) (C). (D) MDCK and MDCK-HA (PR8) cells were infected with PR8 WT, ΔHA, and ΔHA-mIFN-γ IAVs, and virus titers were determined in a plaque assay. (E) MDCK cells were infected with IFN-γ-armed viruses, and secretion of virus-derived IFN-γ was measured at the indicated time points in a cytokine release assay (LEGENDplex). (F and G) B16-F10 melanoma cells were infected with an MOI of 5 of PR8 WT, ΔHA, or ΔHA-mIFN-γ IAVs. At 10 h PI, cell lysates were prepared and RNA was isolated to perform qRT-PCR on the expression level of the apoptosis marker genes Fas (F) and Tnfsf10 (G). (C–G) Data are expressed as mean values ± SD of 3 independent experiments. ∗p < 0.05, ∗∗p < 0.0052, ∗∗∗p < 0.0001. (D and E) Unpaired t test. (F and G) Ordinary 1-way ANOVA followed by Tukey’s multiple comparison test. ctrl., control; h.p.i., hours postinfection; n.d., not determined; ns, not significant; Pos., positive.
Nonreplicating IAVs with HA gene deficiency require the HA protein on their surface to be infectious. Therefore, to generate these viruses, stable expression of the HA protein of PR8 was achieved on the surface of the human embryonic kidney 293 (HEK293) and Madin-Darby canine kidney (MDCK) cell lines by retroviral transduction. The first cell line has a high transfection rate and the second a high infection rate. The technical details of the virus rescue are described in the Materials and methods section. As shown in Figures 1B and 1C, almost all of the transduced MDCK and HEK293 cells express the recombinant HA protein at levels similar to those observed after the infection of MDCK cells with PR8 wild-type (WT) viruses. Using the established cell lines (hereafter referred to as MDCK-HA and HEK-HA), two recombinant HA gene–deficient IAVs, henceforth called ΔHA and ΔHA-mIFN-γ, were obtained, purified, and concentrated by ultracentrifugation.
When cultured on MDCK-HA cells, both ΔHA and ΔHA-mIFN-γ IAVs reached infectious virus titers above 107 plaque-forming units (PFU)/mL, which was almost as high as the titer of PR8 WT viruses (i.e., 108 PFU/mL; Figure 1D). However, neither of the two HA gene–deficient virus types produced infectious progeny in MDCK cells that did not express the recombinant HA protein (Figure 1D). However, both HA gene–deficient viruses were infectious, and MDCK cells infected with ΔHA-mIFN-γ IAVs produced and secreted high levels of virus-based IFN-γ (Figure 1E), and both virus types were capable of inducing apoptosis of infected cells, as was the nonmutated WT IAV (Figures 1F and 1G). Strikingly, the ΔHA-mIFN-γ viruses were more efficient in apoptosis induction than ΔHA or WT viruses, as shown by the expression of apoptosis marker genes. Taken together, these results show that attenuated ΔHA viruses are infectious and induce apoptosis but do not produce infectious progeny.
A single dose of replication-incompetent IAVs resulted in tumor reduction in vivo
In a previous study, we showed that the infection of Raf-BxB mice with 500 PFU of WT PR8 IAVs was well tolerated by the animals and resulted in 50% tumor reduction on day 3 postinfection and viral titers of ≥106 PFU/mL in the lungs.13 In contrast, single-cycle HA gene–deficient viruses are expected to produce only noninfectious viral particles and are unable to spread within the lung. However, to achieve efficient oncolysis, a high number of cancer cells must be infected. Based on these assumptions, we treated cancer-bearing mice with a dose of 0.8 × 107 PFU replication-incompetent IAVs per animal (i.e., with a number of virus particles approximately equal to that produced by WT IAVs during the infection period). To prevent the loss of viral particles in the upper respiratory tract, HA gene–deficient viruses were administered directly into the lung of Raf-BxB mice by intratracheal intubation, and the oncolytic potential was analyzed at indicated time points (Figure 2A). Both ΔHA and ΔHA-mIFN-γ IAVs were evaluated, and the same viral dose was used for both virus types. Control mice received PBS via the same route, and body weight changes of the animals were used as an indicator of disease severity, monitored over a 12-day period after infection (Figure 2B).
Figure 2.
Infection with IFN-γ-armed ΔHA IAVs destroyed 85% of the lung tumor
(A) Time schedule of ΔHA virus infection. Mice were infected with 0.8 × 107 PFU of either unarmed (ΔHA) or armed (ΔHA-mIFN-γ) viruses. At indicated time points, mice were sacrificed and lungs were analyzed. (B) Body weight loss of mice infected with either of the two virus subtypes. Control mice received PBS via the same intratracheal route. (C) Tumor reduction in the lungs of Raf-BxB mice after infection with ΔHA or ΔHA-mIFN-γ IAVs. The Raf-BxB mRNA expression was determined by TaqMan qRT-PCR. Mean values of noninfected control mice were taken as unity. (D) The percentage of tumor tissue in the lung was calculated as the ratio between Raf-BxB+ tumor area and total lung area after specific days of infection. (C and D) Unpaired t test. (E) Correlation between Raf-BxB mRNA expression level and tumor area. (F) Paraffin-embedded lung sections of mice infected with ΔHA (upper row) or ΔHA-mIFN-γ IAVs (center row) were stained for human Raf-BxB protein (red), representing oncogene-expressing tumor cells. Nuclei were counterstained with hematoxylin. Uninfected mice (day 0 [d0]) received PBS only. Scale bar, 1,000 μm. For better visualization, enlarged images (20× and 40× magnification) of certain areas (black rectangle) of noninfected (a) as well as of infected lungs (b, 6 DPI) are presented in the lower row (scale bar, 20 μm). Mean values ± SEM of 5–6 animals per group are depicted. ∗p = 0.0101, ∗∗∗p = 0.0001.
As expected, the control mice tolerated the intratracheal intubation very well and showed no abnormal symptoms or changes in body weight over time. In contrast, all of the infected mice showed mild but still influenza-like symptoms (e.g., sleepiness) and moderate weight loss, which peaked at day 2 of infection. Mice infected with ΔHA-mIFN-γ viruses lost slightly more weight than did the mice infected with unarmed ΔHA viruses, although all of the mice gained weight by day 3 postinfection (PI) and fully recovered from infection on days 4–5.
The antitumor effect of replication-incompetent viruses was evaluated by two different methods: by determining the transcript levels of the oncogenic human Raf-BxB gene in the lung (Figure 2C) and by evaluating the lung tumor area in relation to the lung area in histological sections after staining with a specific anti-human Raf-BxB antibody (Figure 2D). Although distinct variation between individual animals was observed (Figure S1), both assessment methods showed very similar tumor-reduction kinetics, and the amount of Raf-BxB mRNA estimated by qRT-PCR significantly correlated with the histologically evaluated lung tumor area (Figure 2E), which was in accordance with the observations made in previous studies.13 Remarkably, lung tissue showed a significant reduction in the hRaf-BxB mRNA and hRaf-BxB+ cells after infection with both replication-incompetent virus types (Figures 2C and 2D), indicating the destruction of Raf-transformed tumor cells. For both virus types, the maximum tumor loss rate was observed within the first 3 days of infection and slowed down thereafter. The oncolytic activity of the unarmed ΔHA viruses was very strong, reducing the tumor over 65% by day 6 PI. Afterward, however, the tumor loss seemed not to proceed but was still slightly over 50% of the initial value on day 12 PI. It is striking that the overall oncolytic effect of ΔHA-mIFN-γ viruses was much stronger than that of ΔHA IAVs and reached a worth seeing value of more than 85% of the initial lung tumor mass on day 12 PI, highlighting the effective oncolytic capacity of the IFN-γ-armed ΔHA viruses.
Closer histological analysis of infected Raf-BxB lungs with a specific anti-human Raf-BxB antibody confirmed the oncolytic capacity of both ΔHA IAVs (Figure 2F). Due to ubiquitous expression of the recombinant hRaf-BxB transgene in type II pneumocytes, the mice develop multiple tumor foci distributed throughout the lung.22 In uninfected mice, they exhibited a well-structured and dense shape represented by purple spots. However, the lungs infected with either of the two replication-incompetent virus types revealed a significantly lower amount of tumor tissue, and the remaining cancerous areas showed a more diffuse and less dense tumor structure compared to uninfected mice. Although infection with unarmed ΔHA viruses resulted in remarkable oncolysis on day 6 PI, the lung tissue from IFN-γ virus-infected mice revealed a tremendous reduction of the tumor area as early as day 3 PI (Figure 2F). Furthermore, hardly any tumor foci were detectable 6 and 12 days PI with the IFN-γ-armed viruses. In addition, the infection with nonreplicating unarmed or armed ΔHA viruses attracted immune cells to the lungs and caused inflammation in the tumor environment. Strikingly, the areas with massive immune cell infiltration hardly had any tumor cells left (Figure 2F, enlarged images).
Replication-incompetent IAVs initiated proinflammatory immune responses in tumor-containing lung tissue
Infection with IAVs is usually accompanied by the upregulation of proinflammatory cytokines in the lung, which attract immune cells to the site of infection and initiate immune responses to clear the viruses.23,24,25 Therefore, we wondered whether replication-incompetent ΔHA IAVs are capable of eliciting a proinflammatory cytokine response and whether infection with ΔHA-mIFN-γ-armed IAVs differs in its effectiveness from unarmed ΔHA IAVs. For this purpose, bronchoalveolar lavage fluid (BALF) was collected 3, 6, and 12 days PI (DPI), and the cytokine profile was characterized using the flow cytometry–based cytoplex assay. As shown by the data in Figure 3A, both replication-incompetent ΔHA virus types were able to induce proinflammatory cytokines in a manner similar to that of WT IAVs in terms of quantity and time intervals.13,26,27 The only difference between unarmed and armed replication-incompetent viruses was that we detected very early secretion of IFN-γ after infection with ΔHA-mIFN-γ viruses, confirming the expression of IFN-γ by the genetically modified viruses. In particular, already on day 3 of infection, high levels of IFN-γ could be measured, which persisted until day 6, when the secretion of IFN-γ increased upon infection with unarmed IAVs. Remarkably, IFN-γ levels were already very strong at day 3 after ΔHA-mIFN-γ virus infection, which was usually not the case in our previous experiments with WT IAV infection.14,28 However, the IFN-γ secretion on day 6 PI with either replication-incompetent virus was similar to the IFN-γ induction after WT IAV infection.
Figure 3.
Single-cycle ΔHA IAVs induced infiltration of innate immune cells into the TME
BALF from NSCLC-bearing mice at different times of infection with 0.8 × 107 PFU of either unarmed (ΔHA, open symbols) or cytokine-armed (ΔHA-mIFN-γ, filled black symbols) IAVs was analyzed according to the schedule shown in Figure 2A. (A) Cytokines released during viral infection were measured using a multiplex cytokine release assay. Individual and mean values ± SEM (columns) per group and time point are presented. Mean values of uninfected tumor-bearing mice (PBS, filled gray symbols) were arbitrarily set to 1. (B–F) BALF-derived immune cells were analyzed for different surface and intracellular markers of peripheral macrophages and NK cells by flow cytometry. Total numbers of leukocytes (CD45.2+) as well as monocytes/granulocytes (CD11b+ CD11c−) and NK cells (CD3- CD49b+) as parts of CD45.2+ are shown. (B) The percentage of peripheral macrophages (F4/80+) among CD11b+ CD11c− cells and the percentage of MHC class II–expressing peripheral macrophages infiltrating the lung are depicted. (C) Representative dot-plot images highlighting the gating strategy for MHC class II+ peripheral macrophages infiltrating the lung 12 days after ΔHA (left image) or ΔHA-mIFN-γ (center image) IAV infection and immunoglobulin G (IgG)-control (right image) are shown. (D) Geometric mean fluorescence intensities (GeoMFI) are depicted exemplarily for MHC class II+ peripheral macrophages on day 6 PI. Individual and mean values ± SEM per group are shown. (E) Infiltration kinetics of activated NK cells (CD3− CD49b+ NKp46+) into the lung and percentages of IFN-γ+ and MHC class II+ cells among them are presented. (F) Representative dot-plot images showing the gating strategy for NKp46+ (left) among NK cells (CD3− CD49b+) and Granzyme B+ among CD49b+ NKp46+ activated NK cells (right) at day 6 PI with either ΔHA IAVs (left image) or ΔHA-mIFN-γ IAVs (center image) and IgG/FMO-control (right image). Means ± SEM of 5–6 mice per group are shown. ∗p < 0.05, ∗∗p < 0.006, ∗∗∗p < 0.0009, ∗∗∗∗p < 0.0001. (A, Kruskal-Wallis test with Dunn’s multiple comparisons test; B and E, ordinary 1-way ANOVA followed by Tukey’s multiple comparison test). d.p.i., days postinfection; IP-10, interferon gamma-induced protein-10; MCP-1, monocyte chemoattractant protein-1.
The strong induction of proinflammatory cytokines and the kinetics of their expression imply that the replication-defective IAVs are also capable of inducing infiltration and activation of immune cells into the infected lung. To address this question, we first estimated changes in the total amount of CD45+ leukocytes in the BALF of infected lungs. No significant difference was observed between ΔHA and ΔHA-mIFN-γ virus-infected mice. The number of immune cells infiltrating the bronchoalveolar space profoundly increased, peaked on day 6 of infection, and decreased by day 12 after both infections. This kinetic is very similar to the kinetics following infection with WT PR8 virus, except that it appears to attenuate more rapidly.13,15,28,29
As expected, both types of replication-incompetent viruses attracted cells of the innate immune system, peripheral macrophages (CD11b+ CD11c− F4/80+) and natural killer (NK) cells (CD3− CD49b+), but their infiltration kinetics differed depending on the type of virus used. Infection with ΔHA-mIFN-γ viruses led to an earlier infiltration of innate immune cells compared to unarmed ΔHA viruses. Similar to WT PR8 viruses, infection with ΔHA viruses resulted in the maximal attraction of both innate immune cell types on day 6, with a subsequent decline. However, after infection with ΔHA-mIFN-γ viruses, the absolute number of these cells in the lungs increased sharply by day 3 PI and remained high, until day 6 or later (Figures 3B and 3E). Most of the infiltrating macrophages exhibited a proinflammatory M1-like phenotype, as reflected by an increased proportion of major histocompatibility complex class II (MHC class II)–expressing cells and the amount of the receptor on their surface (Figures 3B and 3D). NK cells were also highly activated and exhibited the cytolytic marker NKp46 (CD3− CD49b+ NKp46+), and the NKp46+ cells secreted large amounts of the proapoptotic granzyme B as well as the proinflammatory IFN-γ, further confirming their activated state.
Thus, the most striking finding was that infection with IFN-γ-armed viruses resulted in an earlier infiltration of innate immune cells and a significantly greater activation of recruited macrophages and NK cells compared to unarmed viruses (Figures 3B–3F).
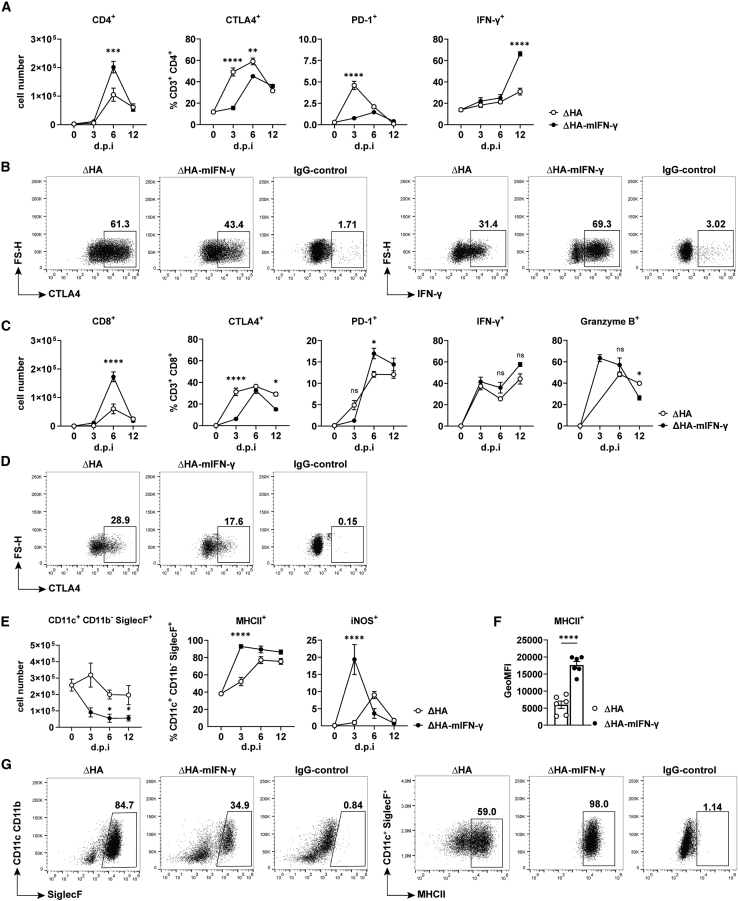
The infiltration of the IAV-infected lung with cells of the innate immune system typically is followed by adaptive T lymphocytes. The adaptive immune response is very specific and strong due to the high antigenicity of influenza viruses. It must be newly developed, however; thus, infiltration of the infected lung with T cells is delayed compared to cells of the innate immune system. However, once unfolded, the cytotoxic T cells are highly efficient in clearing the IAV infection. Therefore, we wondered whether the replication-incompetent viruses could also elicit an adaptive immune response and whether their kinetics and activity differ after the infection of Raf-BxB mice. Helper (CD3+ CD4+) as well as cytotoxic (CD3+ CD8+) T cells were analyzed (Figures 4A–4C).
Figure 4.
Infection with single-cycle ΔHA IAVs activated adaptive T cell responses and led to a significant reduction of TAMs
Raf-BxB mice were intratracheally infected with HA gene–deficient viruses and BALF was analyzed by flow cytometry for adaptive T lymphocyte populations and alveolar macrophages 3, 6, and 12 DPI. (A) Total numbers of CD3+ CD4+ T helper lymphocytes (left) as parts of CD45.2+ leukocytes (see Figure 3B) and percentages of CTLA4+ (second left) as well as PD-1+ (second right) and IFN-γ-expressing (right) cell populations among CD3+ CD4+ cells. (B) The gating strategy is exemplified by CTLA4 (at day 6 PI, left) and IFN-γ (on day 12 PI, right) as well as IgG-control (right image) on T helper cells from mice infected with either ΔHA IAVs (left image) or ΔHA-mIFN-γ IAVs (center image). (C) Absolute number of CD3+ CD8+ T cytotoxic lymphocytes (left) and percentage of CTLA4+ (second left) and PD-1+ (center) cell populations among them and their activation status expressed by IFN-γ+ (second right) and Granzyme B+ (right) over the time of infection. (D) The gating strategy is exemplarily demonstrated for the immune checkpoint CTLA4 (at day 12 PI) and the corresponding IgG-control (right image) on CD8+ T cells during infection with either ΔHA IAVs (left image) or ΔHA-mIFN-γ IAVs (center image), respectively. (E) Absolute numbers of alveolar macrophages (CD11c+ CD11b− SiglecF+, left) and the percentages of MHC class II+ and iNOS+ alveolar macrophages. (F) GeoMFI are depicted exemplarily for MHC class II+ alveolar macrophages at day 3 PI. Individual values and mean values (columns) ± SEM per group are shown. (G) Representative dot-plot images emphasizing the gating strategy, including FMO controls for quantification of parameters shown in (E), for SiglecF (6 DPI, left) and MHC class II (3 DPI, right). Mean values ± SEM with n = 5–6 animals per group are shown. ∗p < 0.03, ∗∗p < 0.002, ∗∗∗p < 0.0002, ∗∗∗∗p < 0.0001. (A, C, and E) Ordinary 1-way ANOVA followed by Tukey’s multiple comparison test. FS-H, forward scatter-height; PD-1, programmed cell death protein-1.
Both cell types strongly infiltrated the lung after infection with ΔHA and ΔHA-mIFN-γ replication-defective IAVs, peaking on day 6 PI. It is most remarkable that the onset and course of adaptive immune responses were comparable for both viruses, whereas the total number of infiltrating helper T cells (CD3+ CD4+) and cytotoxic T cells (CD3+ CD8+) was significantly higher on day 6 PI with IFN-γ-expressing viruses compared to infection with the unarmed viruses (2 and 3 times, respectively). In addition, adaptive immune cell activation was higher and occurred earlier in mice infected with ΔHA-mIFN-γ viruses, as shown by the analysis of IFN-γ+ and granzyme B+ CD4+ and CD8+ T cells (Figures 4A–4D). Interestingly, although the adaptive immune response was stronger in mice infected with ΔHA-mIFN-γ viruses, the immune checkpoint receptor CTLA4 was significantly weaker expressed on CD4+ as well as on CD8+ T cells than after infection with unarmed ΔHA IAVs (Figure 4A).
Infection with IFN-γ-armed ΔHA IAVs accelerated the activation of TAMs
Finally, we wanted to know whether the infection of tumor-bearing Raf-BxB mice with replication-incompetent IAVs could, similar to WT IAVs, activate TAMs. These kind of immune cells are generally the most abundant resident immune cell type in the lung and have been shown to be converted to the tumor-supportive M2 state by NSCLCs,13 which is consistent with the finding of immunosuppressed TAMs in human NSCLC patients.
In good agreement with previously published data that alveolar macrophages undergo apoptotic death after IAV infection, their numbers also decreased after infection with replication-incompetent IAVs (Figure 4E). It was surprising that the decrease was much more pronounced after infection with IFN-γ-encoding IAVs than with unarmed ΔHA IAVs. More important, as infection progressed, the remaining alveolar macrophages exhibited markers of the activated M1 state such as MHC class II and inducible nitric oxide synthase (iNOS) (Figure 4E). Although iNOS expression appeared to be transient, MHC class II expression was more sustained. In mice infected with IFN-γ-armed ΔHA IAVs, alveolar macrophage activation was not only more intense but also occurred earlier upon infection. In particular, already on day 3 PI with ΔHA-mIFN-γ viruses, more than 90% of all cells expressed high levels of the MHC class II receptor and almost 20% of the cells were iNOS+, compared to only 50% and 1%, respectively, in the ΔHA virus-infected lungs without additional secretion of virus-based IFN-γ (Figures 4E–4G). Along this line, the level of MHC class II receptor expression per macrophage (CD11c+ CD11b− SiglecF+) on day 3 PI was also higher in mice infected with IFN-γ-armed viruses than in mice infected with unarmed viruses (Figure 4F). In summary, the present study demonstrates that lung tumors are efficiently destroyed in vivo after infection with replication-incompetent oncolytic IAVs. Furthermore, ΔHA viruses armed with IFN-γ induce strong but controlled immune responses, resulting in the modulation of the lung immune environment in mice harboring lung cancer.
Discussion
In this study, we demonstrated that nonreplicating HA gene-deficient IAVs (i.e., ΔHA IAVs) are effective oncolytic agents that initiate tremendous tumor reduction in a mouse model of NSCLC. The oncolytic effect was even higher when the ΔHA viruses were genetically modified to express IFN-γ upon infection. These ΔHA-mIFN-γ viruses caused an 85% abrogation of Raf-BxB oncogene expression at day 12 PI (compared to 50% for unarmed ΔHA viruses), indicating the destruction of Raf-transformed tumor cells. In addition, infection with either of the two virus types resulted in the reconversion of a suppressed to a highly activated lung immune environment and caused a strong infiltration of the lung with activated innate and adaptive immune cells. The abrogation of the immunosuppressed lung environment as well as the infiltration of the lung with immune cells were earlier and much stronger when virus-derived IFN-γ was present early PI.
Despite the progress made in the development of immunotherapies against solid tumors, treatment remains challenging. The main obstacle remains the immunosuppressed environment caused by various tumor entities, which promotes tumor growth and persistence. In NSCLC patients, the accumulation of TAMs correlates with poor prognosis. Moreover, ICIs, which should enhance antitumor immunity, are effective in only 20% of NSCLC patients, mainly due to the suppression of reactive T cells by the TME and the absence of tumor-associated antigens, which are required for the production of reactive antitumor T lymphocytes.
Interestingly, OVs have been shown to be effective in overcoming the immunosuppressed TME and are being intensively studied as a standard therapy or in combination with existing treatment regimens, and several OVs are in clinical trials against NSCLCs.30,31 However, IAVs have not yet made it into clinical trials as OVs against solid tumors, despite preclinical evidence of their oncolytic properties against lung cancer.15,32 Furthermore, lung-cancer patients hospitalized for influenza infection were found to have lower overall lung cancer mortality.32 These findings altogether render IAV as a suitable oncolytic candidate.
An explanation for why IAVs are not further investigated as OVs in clinical approaches is most likely the fear of potential complications when even low-pathogenic but replicating viruses are administered to immunocompromised cancer patients. To improve the feasibility of IAVs as OVs for the treatment of lung cancer in humans, preclinical studies are focusing on the development of highly attenuated oncolytic IAVs that have a lower risk of severe side effects.17 In the present study, we generated nonreplicating HA gene–deficient viruses from the PR8 strain to completely prevent the risk of viral spread in lung tissue.
It has been shown that arming OVs with coding sequences for immune mediators enhances the virus-induced antitumoral immunity.33 The type II IFN-γ is a pleiotropic cytokine that orchestrates cells of the innate as well as of the adaptive immune system.34 Furthermore, it is known that IFN-γ plays a pivotal role in inhibiting tumor progression and dissemination, since solid tumors grew much faster and established earlier metastases in IFN-γ knockout mice.35,36 Due to its antiproliferative and proapoptotic functions IFN-γ is evaluated alone or as an immunotherapy adjuvant in preclinical and clinical trials against cancer.37 However, local expression of IFN-γ is essential to elicit its antitumoral functions and the short half-life of IFN-γ in serum limits a systemic application. To achieve intratumoral, and hence, local, expression of IFN-γ, we inserted the sequence for the mIFN-γ gene into the viral genome of our ΔHA IAVs and analyzed the potential to boost proinflammatory immune responses.
Both replication-incompetent viruses (i.e., ΔHA and IFN-γ-armed ΔHA) accomplished a single cycle of infection due to a substituted recombinant HA-entry protein in the viral envelope (Figures 1C and 1D). Cells infected with ΔHA IAVs produced noninfectious viral progeny in vitro (Figure 1D) and in vivo (Figure S2), which enormously increase safety concerns. With nonreplicating viruses, viral spread within lung tissue can be excluded, giving HA gene–deficient IAVs a significant advantage over NS1 gene–deficient viruses that may replicate in tumor cells in which the IFN response is downregulated.16
The infection of MDCK cells with IFN-γ-encoding ΔHA-IAVs resulted in the high secretion of IFN-γ, indicating successful transgene expression from the viral genome (Figure 1E). As expected, both replication-incompetent virus types induced apoptosis of tumor cells, as evidenced by the upregulation of the apoptosis marker genes Fas and Tnfsf10 in infected cells. Of note, the presence of virus-based IFN-γ upregulated the apoptotic marker genes more strongly than just single-infection cycle ΔHA or WT viruses (Figures 1F and 1G). These results are in line with studies describing the induction of Fas-mediated apoptosis by IFN-γ38 and the death of lung cancer cells triggered by IFN-γ.39 Most important, we compared the oncolytic activity of unarmed and IFN-γ-armed ΔHA IAVs in mice developing NSCLC (i.e., in Raf-BxB mice). Due to the targeted expression of the constitutively active truncated human Raf-BxB kinase in type II pneumocytes, Raf-BxB mice spontaneously develop NSCLC, but because of the obligatory expression of the Raf-BxB oncogene in epithelial cells, the resulting tumor foci are distributed throughout the lung. This is a limitation of the Raf-BxB model, because it does not fully reflect the rather nodular growth characteristics of human lung tumors. However, the advantage of the model is that these mice are fully immunocompetent and that type II pneumocytes are the natural host cells of IAVs; and it seems plausible to consider the whole lung tissue as the tumor environment in this specific case. Using the Raf-BxB mice, we were able to show that both virus types caused a reduction in tumor tissue and that the modification with IFN-γ impressively destroyed 85% of Raf-induced tumors (Figures 2C and 2D). Viral infection not only reduced the overall tumor area in the lung (Figures 2D and 2F) but also caused the disintegration of encapsulated tumor foci and opened the tumor tissue to infiltration by immune cells (Figure 2F).
Virus-infected epithelial cells produce cytokines as an early viral response that attract innate immune cells such as macrophages and NK cells to the side of infection and trigger their phagocytic and cytolytic functions.23,40 The lungs of mice infected with the WT IAV PR8 indeed secrete early viral response cytokines such as type I IFNs, tumor necrosis factor (TNF)-α, and interleukin (IL)-6.41 In our study, a sublethal dose of 0.8 × 107 PFU of nonreplicating IAVs was sufficient to induce a cytokine release comparable to WT virus infection. Analysis of BALF from mice infected with either unarmed or IFN-γ-armed ΔHA IAVs revealed elevated levels of TNF-α and IL-6 (Figure 3A), which corresponds to increased numbers of peripheral macrophages and NK cells infiltrating the lung from day 3 PI onward (Figures 3B–3D). Infection with IFN-γ-armed viruses attracted innate immune cells to the infected lung tissue much earlier as compared with unarmed viruses. Furthermore, the infiltrating cells showed higher and stronger activation, indicating that virus-released IFN-γ enhances proinflammatory immune responses in the tumor-bearing lung. These observations are in agreement with other studies elucidating the beneficial effect of OVs engineered with an IFN-γ gene. Infection of a mouse model for breast cancer with a vesicular stomatitis virus (VSV) genetically modified to express IFN-γ showed an enhanced activation of innate immune cells in contrast to the unarmed parental virus. Furthermore, these IFN-γ-armed VSVs reduced the tumor size more efficiently compared to unarmed VSVs.42
In contrast to type I interferons, which can be produced by any nucleated cell, immune cells such as macrophages,43 NK cells,44 and T lymphocytes45 are the main source of IFN-γ. In mice infected with single-cycle unarmed ΔHA viruses, IFN-γ secretion increased on day 6 PI to levels comparable to IFN-γ levels after infection with fully replicating WT PR8 viruses.15 As we expected, infection with IFN-γ-armed ΔHA viruses resulted in an earlier secretion of IFN-γ, which was expressed from the modified viral genome. In detail, the concentration of IFN-γ measured in BALF on day 3 PI was spectacularly 1,000-fold higher compared to levels of unarmed ΔHA virus-infected mice (Figure 3A). This result confirmed the successful expression and secretion of virus-encoded IFN-γ upon in vivo infection. Interestingly, on day 6 PI, IFN-γ levels were similar for unarmed as well as for cytokine-armed ΔHA viruses and did not increase further in mice infected with ΔHA-mIFN-γ viruses. This correlates with the finding that infectious virus particles were no longer present 3 days PI, thus abolishing virus-based IFN-γ production (Figure S2). Both NK cells and T lymphocytes contributed to increased IFN-γ secretion, with the number of IFN-γ+ cells being highest on days 6 and 12 PI, respectively (Figures 3E, 4A, and 4C). Cytotoxic T lymphocytes are essential for the recognition and killing of both virus-infected and transformed tumor cells. The infection with either ΔHA virus type induced migration of adaptive T cells into the lung on day 6 PI. Remarkably, the number of infiltrating CD4+ and CD8+ T cells was 2-fold higher in IFN-γ-armed infected lungs compared to unarmed virus infection and was indeed as high as observed after WT IAV infection (Figures 4A and 4C).14 Furthermore, T cell activation was also higher after ΔHA-mIFN-γ compared to unarmed ΔHA virus infection, consistent with the greater activation of innate immune cells in IFN-γ virus-infected mice. In particular, most CD8+ T killer cells expressed IFN-γ and cytolytic granzyme B, reflecting a highly activated effector phenotype (Figure 4C). Although we did not explicitly analyze tumor-specific T cell immune responses in this study, we would not rule out this scenario, because massive virus-induced tumor cell destruction releases sufficient tumor-specific antigens, if available. Also, the greater and sustained tumor reduction after IFN-γ-virus infection may suggest T cell involvement in tumor cell destruction in addition to NK cells and macrophages. However, this question needs to be clarified in detail in future experiments. It is interesting to note that the unarmed HA gene–deficient viruses elicited an effective immune response in the tumor-bearing lung, in contrast to recently published replication-incompetent oncolytic IAVs, which elicit a strong immune response only after conjugation with an immunostimulatory peptide.17 Different routes of viral administration and different viral doses could account for the discrepancy. For example, Ji et al. infected mice intranasally, which we believe is less effective than intratracheal administration of nonreplicating IAVs, and administered much fewer viral particles.17
Tumor-bearing Raf-BxB mice show an accumulation of TAMs resembling the phenotype of the TME in human lung cancer patients. TAMs are a main source of granulocyte macrophage-colony-stimulating factor (GM-CSF), and it has been reported that certain NSCLCs from human patients as well secrete elevated amounts of GM-CSF.46 It is worth noting that GM-CSF was the only cytokine in the cytoplex assay that showed lower levels in infected compared to noninfected mice (Figure 3A). This observation was consistent with the decrease in both TAMs and tumor cells following the infection with replication-incompetent viruses and is in accordance with previous findings that mRNA transcript levels of GM-CSF decreased upon the infection of Raf-BxB mice with WT IAVs.13 Again, the decline in TAMs following ΔHA-mIFN-γ infection was earlier and much greater than the decline following infection with unarmed ΔHA viruses (Figure 4E). In particular, almost all of the remaining alveolar macrophages (CD11c+ CD11b− SiglecF+) were activated (MHC class II+) and showed a stable M1-like phenotype throughout the observation period (i.e., 12 DPI). Furthermore, lung-resident macrophages showed iNOS expression in nearly 20% of cells on day 3 PI (compared to less than 1% in unarmed ΔHA virus-infected lungs at the same time point). Both MHC class II expression and NOS induction are known to be stimulated in response to IFN-γ,47,48 which is in good agreement with the increased levels of virus-derived IFN-γ on day 3, as shown in Figure 3A.
The effective oncolytic dose of 0.8 × 107 PFU of unarmed or IFN-γ-armed ΔHA IAVs was well tolerated by the infected animals. This is because the viruses are unable to replicate after a single cycle of infection. All of the mice recovered from day 3 PI onward, as depicted in Figure 2B, and no immunopathology was observed. In particular, cytokine levels measured 12 DPI were back to the levels of noninfected mice, and the number of lung-infiltrated immune cells decreased 12 DPI, indicating a termination of the inflammation and controlled immune responses after tumor destruction (Figures 3A, 3E, 4A, and 4C).
Due to the transgenic nature of the Raf-BxB mouse model, this type of lung tumor cannot be eradicated completely. It will always recur and regrow as long as type II pneumocytes are present, even with a stronger induction of immune responses by the virus-based IFN-γ. The constant appearance of new tumor cells is a disadvantage of the mouse model, and it would be important to investigate the oncolytic activity in a future study in an additional mouse model.
We have conclusively demonstrated for the first time that HA gene–deficient single-cycle IAVs are potent OVs in NSCLC-bearing mice. Virus-derived IFN-γ even accelerated the tumor-reducing capacity of ΔHA IAVs. Moreover, IFN-γ-armed viruses shifted the formerly suppressed lung immune environment into a highly active state. By activating proinflammatory immune responses within the lung and an efficient reduction of the tumor area, replication-incompetent ΔHA-mIFN-γ IAVs can serve as promising candidates for the development of a safe and efficient virotherapy in humans with NSCLC.
Materials and methods
Plasmids for virus generation
The bidirectional plasmid pHW2000 harboring the HA gene segment of the virus strain PR8 was genetically modified to abrogate HA viral ribonucleic acid (vRNA) formation and HA protein translation. In particular, the WT HA segment (including 5′ and 3′ UTRs) comprise 1,775 nt, from which 1,382 nt were deleted, whereas packaging sequences at the 5′ (197 nt) and 3′ (196 nt) ends were maintained. The start codon ATG was mutated to CTA to exclude translation, and a noncoding placeholder (stuffer, 166 nt) with a multicloning site was inserted into the truncated HA segment. This ΔHA construct had a length of 559 nt (197 nt + 166 nt + 196 nt) and was synthesized by VectorBuilder (Chicago, IL) on a provided pHW2000-HA-WT plasmid backbone. Afterward, we cloned the cDNA of mIFN-γ (468 nt) into the pHW2000-ΔHA vector using HindIII-HF and SpeI (NEB, Frankfurt am Main, Germany). The ΔHA-mIFN-γ construct comprises 936 nt (197 nt + 7 nt Kozak consensus sequence + 468 nt + 68 nt remaining from stuffer sequence +196 nt). Primer sequences are listed in Table S1.
Stable HA-expressing cell lines
Madin-Darby canine kidney cells (MDCK) and human embryonic kidney 293 (HEK293) cells were cultured in Eagle’s minimum essential medium (EMEM; Merck, Darmstadt, Germany) supplemented with 5% heat-inactivated fetal bovine serum (FBS; Capricorn Scientific, Ebsdorfergrund, Germany) and DMEM (PAN-Biotech, Aidenbach, Germany) with 10% FBS, respectively, at 37°C and 5% CO2. To generate MDCK and HEK293 cells stably expressing the viral HA protein from PR8 IAV, the coding sequence of HA was obtained by PCR using a pcDNA3-HA (PR8) plasmid (VectorBuilder) as a template and cloned into the retroviral vector pEGZ-Zeocin using BamHI (NEB) restriction sites. To generate retroviral particles expressing the HA gene of PR8, Phoenix-AMPHO (ATCC, Manassas, VA) cells were seeded with 2.5 × 105 cells/6-well 48 h before transfection with 2 μg of the pEGZ-HA vector. Two days later, cells were transferred into a 10-cm dish, and selected for zeocin resistance (250 μg/mL) for 12 days; the medium was changed every 3–4 days. After culturing a dense monolayer of these cells overnight, the retrovirus-containing supernatant was harvested, filtered through a 0.45-μm nylon membrane, mixed with polybrene (8 μg/mL), and transferred to MDCK and HEK293 cells for 24 h. Transduced MDCK and HEK293 cells were cultured in the presence of 500 and 250 μg/mL zeocin, respectively, to obtain cell populations uniformly expressing the recombinant HA protein. Zeocin-resistant clones of MDCK-HA (PR8) and HEK293-HA (PR8) cells were then expanded, and the HA expression was verified by flow cytometry using an anti-HA (PR8-specific) antibody (11684-T56, [1:100], SinoBiological, Eschborn, Germany). The antibodies are listed in Table S2.
Generation of recombinant viruses
HA gene–deficient IAVs of the H1N1 strain PR8 were generated using a DNA plasmid transfection system, as previously described, with appropriate modifications. Briefly, HEK-HA (PR8) cells were simultaneously transfected with 7 pHW2000 plasmids encoding WT segments of the PR8 virus (i.e., NP, PA, PB1, PB2, M, NS1, NA), and either the modified pHW2000-ΔHA or pHW2000-ΔHA-mIFN-γ plasmid using TransIT-LT1 transfection reagent (Mirus Bio, Madison, WI). MDCK-HA (PR8) cells suspended in DMEM containing 10% FBS and tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)–treated trypsin (1:8,000) were seeded on top of HEK293-HA cells 24 h after transfection. Trypsin must be added to the medium because, unlike lung epithelial cells, MDCK cells do not possess this endoprotease. However, proteolytic cleavage is essential to activate viral HA and allow sufficient viral replication. Another 48 h later, the supernatant of cocultured cells was harvested and transferred to freshly plated MDCK-HA cells to further amplify ΔHA or ΔHA-mIFN-γ IAVs. After 48 h, the virus-containing supernatant was collected and a plaque assay on strain-matched MDCK-HA cells was performed to determine viral titers as previously described.49 Single clones were plaque purified, and virus stocks were grown on MDCK-HA cells using standard methods. Recombinant WT PR8 viruses were generated from 8 pHW2000 plasmids encoding 8 WT genomic segments using the same method, but with HEK293 and MDCK cells not expressing the viral HA protein.
In vitro infection experiments
To determine the number of HA+ cells after infection with WT IAVs, MDCK cells were infected with a multiplicity of infection (MOI) of 5 of PR8 viruses, and 12 h later, HA-expressing cells were analyzed by flow cytometry.
For the verification of the replication incompetence of HA gene–deficient viruses, MDCK and MDCK-HA cells were seeded 24 h before the infection with PR8 WT, ΔHA, or ΔHA-mIFN-γ IAVs at a rate of 1 × 106 cells/6-well in EMEM plus 10% FBS without antibiotics. Cells were then washed once with PBS and infected with viruses diluted in EMEM containing 0.2% BSA, 1 mM MgCl2, 0.9 mM CaCl2, 100 U mL−1 penicillin, and 0.1 mg mL−1 streptomycin and TPCK-treated trypsin (1:8,000) and incubated at 37°C for 24 h. Supernatants were harvested, filtered through a 0.45-μm nylon membrane, and analyzed for infectious virus particles in a plaque assay using MDCK or MDCK-HA cells.
The functionality of the IFN-γ transgene expression from the viral genome was verified by IFN-γ detection in the supernatant of infected cells. MDCK cells (5 × 105 cells/12-well) were infected with an MOI of 1 of ΔHA-mIFN-γ IAVs and incubated at 37°C for 30 min. Afterward, the virus inoculum was aspirated and the cells were washed once with PBS and cultured in EMEM (containing 0.2% BSA, 1 mM MgCl2, 0.9 mM CaCl2, 500 μg/mL gentamicin, and TPCK-treated trypsin [1:8,000]). Supernatants were collected at 8, 12, 24, and 48 h PI, and IFN-γ levels were measured in a flow cytometry–based LEGENDPlex assay using a custom single-plex panel (BioLegend, Koblenz, Germany). Cells incubated in PBS only served as controls.
To assess the apoptosis induction by HA gene–deficient IAVs, murine melanoma B16-F10 cells (1 × 106 cells/6-well) were infected with an MOI of 5 of either PR8 WT, ΔHA, or ΔHA-mIFN-γ IAVs, cell lysates were prepared 10 h PI, and RNA was isolated for subsequent gene expression analysis by qRT-PCR. Primer sequences are listed in Table S3.
Animal studies
All of the animal experiments were approved by the local ethics committee and performed in strict accordance with the German regulations of the Society for Laboratory Animal Science and the European Health Law of the Federation of Laboratory Animal Science Associations. The protocols were approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, Germany.
All of the in vivo experiments were performed with 4- to 6-month-old homozygous transgenic Raf-BxB mice17 of both genders maintained under specific pathogen-free conditions, and 5–6 animals were included per experimental group. Mice were anesthetized by isoflurane inhalation and intratracheally intubated to deliver 0.8 × 107 PFU of either ΔHA or ΔHA-mIFN-γ IAVs diluted in 50 μL PBS directly into the lungs. Mice of the control group received PBS via the same route of application. The health status of the mice was checked daily, including monitoring of body weight loss. None of the animals lost ≥20% of their original body weight during the entire experimental period. At indicated time points (i.e., 3, 6, and 12 DPI), mice were sacrificed and BALF was collected to analyze the immune cell status and cytokine content in the lungs. Afterward, the right main bronchus was clamped at the bifurcation and the four right lung lobes were excised and used for total RNA isolation, while the left lobe was fixed in situ with 4% paraformaldehyde (PFA) in PBS, extracted, and further processed for immunohistochemistry (IHC) staining.
RNA isolation and qRT-PCR
Four right lung lobes per mouse were homogenized in 2.0 mL lysing matrix A tubes (MP Biomedicals, Santa Ana, CA) and total RNA extraction was performed with 1.0 mL of peqGOLD TriFast (VWR Life Science, Darmstadt, Germany) according to the manufacturer’s instructions. Purified RNA was transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific, Schwerte, Germany). RNA expression levels of the human c-Raf-BxB kinase were determined by TaqMan qRT-PCR on a LightCycler 480II machine (Roche, Mannheim, Germany). Each cDNA sample was analyzed in triplicate and mean Cp values were normalized to the signal of the housekeeping gene transcripts of glyceraldehyde 3-phosphate dehydrogenase and cytochrome c. Primer-sequences and TaqMan probes used for qRT-PCR are listed in Table S3.
Immunofluorescence and IHC staining
The left lung lobe of each mouse was fixed with 0.7 mL of 4% PFA in PBS for 1 h at room temperature. The fixed tissue was dehydrated in ascending isopropanol dilutions for a maximum of 24 h, followed by incubation in histol for 5 h; lungs were then embedded in paraffin and sections of 4 μm were analyzed. Tumor area of two different sections (approximately 400 μm apart) from each mouse and from 4 mice per group and time point were quantified. Briefly, tissue sections were rehydrated and heat-mediated antigen retrieval was performed using 10 mM citric acid buffer (pH 6.0) for 20 min. To prevent unspecific antibody binding, lung specimens were blocked with 10% FBS and 0.1% Triton X-100 for 30 min. Lung tissue was then stained with a primary rabbit anti-human Raf-BxB kinase antibody (SP-63, dilution 1:100) for 1 h at room temperature, followed by a 40-min incubation with an Alexa Fluor 647-conjugated secondary antibody. The fluorescence-positive areas of the samples were determined using the Celigo Image Cytometer (Nexcelom Biosciences, Lawrence, MA), classified as tumor area, and their relative area to the total tissue area was calculated per sample using Gimp software. IHC staining of lung specimens was performed exactly as described above, except that a species-specific biotinylated secondary antibody (Vector Laboratories, Newark, CA) was used and positive signals were visualized using the Vectastain ABC-AP kit (Vector Laboratories) according to the manufacturer’s instructions. Lung sections were then stained with hematoxylin for 2–3 min to visualize cell nuclei. Images of the entire lung were obtained using the Biorevo BZ-9000 microscope (Keyence, Neu-Isenburg, Germany) and Keyence BZ Analyzer software. Tissue images of higher magnification were acquired with a Carl Zeiss microscope (Axiovert 200M; Zeiss, Oberkochen, Germany) using AxioVision software.
Analysis of the lung immune status
BAL was performed to obtain immune cells and cytokines from the lungs for further investigation. In particular, mice were euthanized, the trachea was exposed, and a cannula with an atraumatic catheter was inserted into the trachea. The cannula was removed, and the lungs were gently washed 5 times with 600 μL PBS containing 2 mM EDTA via the remaining catheter. BALF from the first wash was collected separately for the determination of viral particles and analysis of cytokine levels. The cell pellet from the first BAL was pooled with the cells of the subsequent lavages and centrifuged at 350 × g for 10 min at 4°C. The erythrocytes were lysed, and the percent viability of the cells was determined by the trypan blue exclusion method using the Cell Countess System. Equal amounts of viable cells were then stimulated for 5 h with eBioscience Cell Stimulation Cocktail (Thermo Fisher Scientific) to simultaneously block protein transport and secretion. Cells were then detached from the 6-well dish by incubation in 10 mM EDTA on ice for 20 min (repeated twice). Then, cells were incubated with an anti-CD16/CD32 antibody (BD Pharmingen, Heidelberg, Germany) to block Fc receptors and subsequently stained for multicolor flow cytometric analyses. Extracellular staining of cells for multicolor flow cytometric analyses was performed by incubation with the appropriate fluorophore-labeled antibodies on ice for 30 min. Cells were then washed with PBS and fixed and permeabilized for intracellular staining using the Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific), following the procedure described in the manufacturer’s protocol. Fixed and stained cells were subjected to flow cytometry (Gallios, Beckmann Coulter, Krefeld, Germany), and the data obtained were quantified using FlowJo software version 10 (Becton Dickinson, Ashland, OR) based on the fluorescence minus one (FMO) strategy. The total number of immune cells was determined by single-cell discrimination, followed by normalization to the absolute number of cells counted in the BALF of each mouse. The principal gating strategy to distinguish immune cell subsets and their respective activation markers was applied as previously described23 and is exemplified in Figures 3 and 4. Fluorophore-labeled antibodies are listed in Table S4.
Quantification of secreted cytokines
The collected first BALF was analyzed for quantities of released cytokines by the flow-based LEGENDplex 13-plex mouse antivirus response panel kit (BioLegend) according to the manufacturer’s recommendations. Flow cytometry files were analyzed with BioLegend’s Cloud-based LEGENDplex data analysis software.
Statistical analysis
All of the analyses were performed with GraphPad Prism software, version 8. The statistical tests are indicated in the respective figure legends.
Data and code availability
The data that support the findings of this study are available from the corresponding authors upon request.
Acknowledgments
We thank Seema Lakdawala and Peter Palese for helpful discussions on genomic segment packaging of the IAV strain PR8. We further thank Tanja Schied for excellent technical assistance. In addition, we thank Michael Schoefbaenker and Sriram Kumar for their helpful advice. This work was supported by the Cusanuswerk, a sponsorship organization financed by the Federal Ministry of Education and Research (Germany), granted to R.M., and the Interdisciplinary Center of Clinical Research of the Medical Faculty of the University of Münster under grant no. Lud2/013/21.
Author contributions
R.M., V.W., F.P.W., J.C.J., T.R., D.M., and Y.B. performed and analyzed the experiments. R.M., V.W., D.M., B.G.H., and S.L. designed the study. R.M. and V.W. wrote the paper. V.W. and S.L. supervised the project. All of the authors critically read and approved the manuscript text.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2023.100741.
Contributor Information
Viktor Wixler, Email: vwixler@uni-muenster.de.
Stephan Ludwig, Email: ludwigs@uni-muenster.de.
Supplemental information
References
- 1.International agency for research on cancer Global Cancer Observatory. 2020. https://gco.iarc.fr/
- 2.Herbst R.S., Heymach J.V., Lippman S.M. Lung cancer. N. Engl. J. Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 4.Anichini A., Perotti V.E., Sgambelluri F., Mortarini R. Immune Escape Mechanisms in Non Small Cell Lung Cancer. Cancers. 2020;12 doi: 10.3390/cancers12123605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quatromoni J.G., Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am. J. Transl. Res. 2012;4:376–389. [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig S., Planz O., Pleschka S., Wolff T. Influenza-virus-induced signaling cascades: targets for antiviral therapy? Trends Mol. Med. 2003;9:46–52. doi: 10.1016/s1471-4914(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 7.Masemann D., Boergeling Y., Ludwig S. Employing RNA viruses to fight cancer: novel insights into oncolytic virotherapy. Biol. Chem. 2017;398:891–909. doi: 10.1515/hsz-2017-0103. [DOI] [PubMed] [Google Scholar]
- 8.Marelli G., Howells A., Lemoine N.R., Wang Y. Oncolytic Viral Therapy and the Immune System: A Double-Edged Sword Against Cancer. Front. Immunol. 2018;9:866. doi: 10.3389/fimmu.2018.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin D.H., Nguyen T., Ozpolat B., Lang F., Alonso M., Gomez-Manzano C., Fueyo J. Current strategies to circumvent the antiviral immunity to optimize cancer virotherapy. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2020-002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pleschka S., Wolff T., Ehrhardt C., Hobom G., Planz O., Rapp U.R., Ludwig S. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 2001;3:301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber A., Boff L., Anhlan D., Krischuns T., Brunotte L., Schuberth C., Wedlich-Söldner R., Drexler H., Ludwig S. Dissecting the mechanism of signaling-triggered nuclear export of newly synthesized influenza virus ribonucleoprotein complexes. Proc. Natl. Acad. Sci. USA. 2020;117:16557–16566. doi: 10.1073/pnas.2002828117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olschläger V., Pleschka S., Fischer T., Rziha H.J., Wurzer W., Stitz L., Rapp U.R., Ludwig S., Planz O. Lung-specific expression of active Raf kinase results in increased mortality of influenza A virus-infected mice. Oncogene. 2004;23:6639–6646. doi: 10.1038/sj.onc.1207883. [DOI] [PubMed] [Google Scholar]
- 13.Masemann D., Köther K., Kuhlencord M., Varga G., Roth J., Lichty B.D., Rapp U.R., Wixler V., Ludwig S. Oncolytic influenza virus infection restores immunocompetence of lung tumor-associated alveolar macrophages. Oncoimmunology. 2018;7:e1423171. doi: 10.1080/2162402x.2017.1423171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masemann D., Meissner R., Schied T., Lichty B.D., Rapp U.R., Wixler V., Ludwig S. Synergistic anti-tumor efficacy of oncolytic influenza viruses and B7-H3 immune- checkpoint inhibitors against IC-resistant lung cancers. Oncoimmunology. 2021;10 doi: 10.1080/2162402x.2021.1885778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sitnik S., Masemann D., Leite Dantas R., Wixler V., Ludwig S. PD-1 IC Inhibition Synergistically Improves Influenza A Virus-Mediated Oncolysis of Metastatic Pulmonary Melanoma. Mol. Ther. Oncolytics. 2020;17:190–204. doi: 10.1016/j.omto.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hock K., Laengle J., Kuznetsova I., Egorov A., Hegedus B., Dome B., Wekerle T., Sachet M., Bergmann M. Oncolytic influenza A virus expressing interleukin-15 decreases tumor growth in vivo. Surgery. 2017;161:735–746. doi: 10.1016/j.surg.2016.08.045. [DOI] [PubMed] [Google Scholar]
- 17.Ji D., Zhang Y., Sun J., Zhang B., Ma W., Cheng B., Wang X., Li Y., Mu Y., Xu H., et al. An engineered influenza virus to deliver antigens for lung cancer vaccination. Nat. Biotechnol. 2023 doi: 10.1038/s41587-023-01796-7. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann E., Neumann G., Kawaoka Y., Hobom G., Webster R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii Y., Goto H., Watanabe T., Yoshida T., Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. USA. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe T., Watanabe S., Noda T., Fujii Y., Kawaoka Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 2003;77:10575–10583. doi: 10.1128/jvi.77.19.10575-10583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh G.A., Hatami R., Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 2007;81:9727–9736. doi: 10.1128/jvi.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerkhoff E., Fedorov L.M., Siefken R., Walter A.O., Papadopoulos T., Rapp U.R. Lung-targeted expression of the c-Raf-1 kinase in transgenic mice exposes a novel oncogenic character of the wild-type protein. Cell Growth Differ. 2000;11:185–190. [PubMed] [Google Scholar]
- 23.Julkunen I., Melén K., Nyqvist M., Pirhonen J., Sareneva T., Matikainen S. Inflammatory responses in influenza A virus infection. Vaccine. 2000;19(Suppl 1):S32–S37. doi: 10.1016/s0264-410x(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 24.van Reeth K. Cytokines in the pathogenesis of influenza. Vet. Microbiol. 2000;74:109–116. doi: 10.1016/s0378-1135(00)00171-1. [DOI] [PubMed] [Google Scholar]
- 25.Iwasaki A., Pillai P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peper R.L., van Campen H. Tumor necrosis factor as a mediator of inflammation in influenza A viral pneumonia. Microb. Pathog. 1995;19:175–183. doi: 10.1006/mpat.1995.0056. [DOI] [PubMed] [Google Scholar]
- 27.Jewell N.A., Vaghefi N., Mertz S.E., Akter P., Peebles R.S., Bakaletz L.O., Durbin R.K., Flaño E., Durbin J.E. Differential type I interferon induction by respiratory syncytial virus and influenza a virus in vivo. J. Virol. 2007;81:9790–9800. doi: 10.1128/jvi.00530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Köther K., Nordhoff C., Masemann D., Varga G., Bream J.H., Gaestel M., Wixler V., Ludwig S. MAPKAP kinase 3 suppresses Ifng gene expression and attenuates NK cell cytotoxicity and Th1 CD4 T-cell development upon influenza A virus infection. FASEB J. 2014;28:4235–4246. doi: 10.1096/fj.14-249599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masemann D., Leite Dantas R., Sitnik S., Schied T., Nordhoff C., Ludwig S., Wixler V. The Four-and-a-Half LIM Domain Protein 2 Supports Influenza A Virus–Induced Lung Inflammation by Restricting the Host Adaptive Immune Response. Am. J. Pathol. 2018;188:1236–1245. doi: 10.1016/j.ajpath.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Villalona-Calero M.A., Lam E., Otterson G.A., Zhao W., Timmons M., Subramaniam D., Hade E.M., Gill G.M., Coffey M., Selvaggi G., et al. Oncolytic reovirus in combination with chemotherapy in metastatic or recurrent non-small cell lung cancer patients with KRAS-activated tumors. Cancer. 2016;122:875–883. doi: 10.1002/cncr.29856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pol J.G., Acuna S.A., Yadollahi B., Tang N., Stephenson K.B., Atherton M.J., Hanwell D., El-Warrak A., Goldstein A., Moloo B., et al. Preclinical evaluation of a MAGE-A3 vaccination utilizing the oncolytic Maraba virus currently in first-in-human trials. Oncoimmunology. 2019;8 doi: 10.1080/2162402x.2018.1512329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman J.H., Chesson C.B., Herzog N.L., Bommareddy P.K., Aspromonte S.M., Pepe R., Estupinian R., Aboelatta M.M., Buddhadev S., Tarabichi S., et al. Intratumoral injection of the seasonal flu shot converts immunologically cold tumors to hot and serves as an immunotherapy for cancer. Proc. Natl. Acad. Sci. USA. 2020;117:1119–1128. doi: 10.1073/pnas.1904022116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andtbacka R.H.I., Ross M., Puzanov I., Milhem M., Collichio F., Delman K.A., Amatruda T., Zager J.S., Cranmer L., Hsueh E., et al. Patterns of Clinical Response with Talimogene Laherparepvec (T-VEC) in Patients with Melanoma Treated in the OPTiM Phase III Clinical Trial. Ann. Surg. Oncol. 2016;23:4169–4177. doi: 10.1245/s10434-016-5286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder K., Hertzog P.J., Ravasi T., Hume D.A. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 35.Pulaski B.A., Smyth M.J., Ostrand-Rosenberg S. Interferon-gamma-dependent phagocytic cells are a critical component of innate immunity against metastatic mammary carcinoma. Cancer Res. 2002;62:4406–4412. [PubMed] [Google Scholar]
- 36.duPre' S.A., Redelman D., Hunter K.W. Microenvironment of the murine mammary carcinoma 4T1: endogenous IFN-gamma affects tumor phenotype, growth, and metastasis. Exp. Mol. Pathol. 2008;85:174–188. doi: 10.1016/j.yexmp.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Jorgovanovic D., Song M., Wang L., Zhang Y. Roles of IFN-γ in tumor progression and regression: a review. Biomark. Res. 2020;8:49. doi: 10.1186/s40364-020-00228-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H.-J., Kim J.Y., Park J.E., Yoon Y.-D., Tsang B.K., Kim J.-M. Induction of Fas-Mediated Apoptosis by Interferon-γ is Dependent on Granulosa Cell Differentiation and Follicular Maturation in the Rat Ovary. Dev. Reprod. 2016;20:315–329. doi: 10.12717/dr.2016.20.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang C., Weng T., Hu S., Yuan Z., Xiong H., Huang B., Cai Y., Li L., Fu X. IFN-γ-induced ER stress impairs autophagy and triggers apoptosis in lung cancer cells. Oncoimmunology. 2021;10 doi: 10.1080/2162402x.2021.1962591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sládková T., Kostolanský F. The role of cytokines in the immune response to influenza A virus infection. Acta Virol. 2006;50:151–162. [PubMed] [Google Scholar]
- 41.Hennet T., Ziltener H.J., Frei K., Peterhans E. A kinetic study of immune mediators in the lungs of mice infected with influenza A virus. J. Immunol. 1992;149:932–939. [PubMed] [Google Scholar]
- 42.Bourgeois-Daigneault M.-C., Roy D.G., Falls T., Twumasi-Boateng K., St-Germain L.E., Marguerie M., Garcia V., Selman M., Jennings V.A., Pettigrew J., et al. Oncolytic vesicular stomatitis virus expressing interferon-γ has enhanced therapeutic activity. Mol. Ther. Oncolytics. 2016;3 doi: 10.1038/mto.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darwich L., Coma G., Peña R., Bellido R., Blanco E.J.J., Este J.A., Borras F.E., Clotet B., Ruiz L., Rosell A., et al. Secretion of interferon-gamma by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. 2009;126:386–393. doi: 10.1111/j.1365-2567.2008.02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perussia B. Lymphokine-activated killer cells, natural killer cells and cytokines. Curr. Opin. Immunol. 1991;3:49–55. doi: 10.1016/0952-7915(91)90076-d. [DOI] [PubMed] [Google Scholar]
- 45.Sad S., Marcotte R., Mosmann T.R. Cytokine-induced differentiation of precursor mouse CD8+ T cells into cytotoxic CD8+ T cells secreting Th1 or Th2 cytokines. Immunity. 1995;2:271–279. doi: 10.1016/1074-7613(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 46.Uemura Y., Kobayashi M., Nakata H., Kubota T., Bandobashi K., Saito T., Taguchi H. Effects of GM-CSF and M-CSF on tumor progression of lung cancer: roles of MEK1/ERK and AKT/PKB pathways. Int. J. Mol. Med. 2006;18:365–373. [PubMed] [Google Scholar]
- 47.Steimle V., Siegrist C.A., Mottet A., Lisowska-Grospierre B., Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science (New York, N.Y.) 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 48.Zhao W., Cha E.N., Lee C., Park C.Y., Schindler C. Stat2-dependent regulation of MHC class II expression. J. Immunol. 2007;179:463–471. doi: 10.4049/jimmunol.179.1.463. [DOI] [PubMed] [Google Scholar]
- 49.Hrincius E.R., Wixler V., Wolff T., Wagner R., Ludwig S., Ehrhardt C. CRK adaptor protein expression is required for efficient replication of avian influenza A viruses and controls JNK-mediated apoptotic responses. Cell. Microbiol. 2010;12:831–843. doi: 10.1111/j.1462-5822.2010.01436.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.