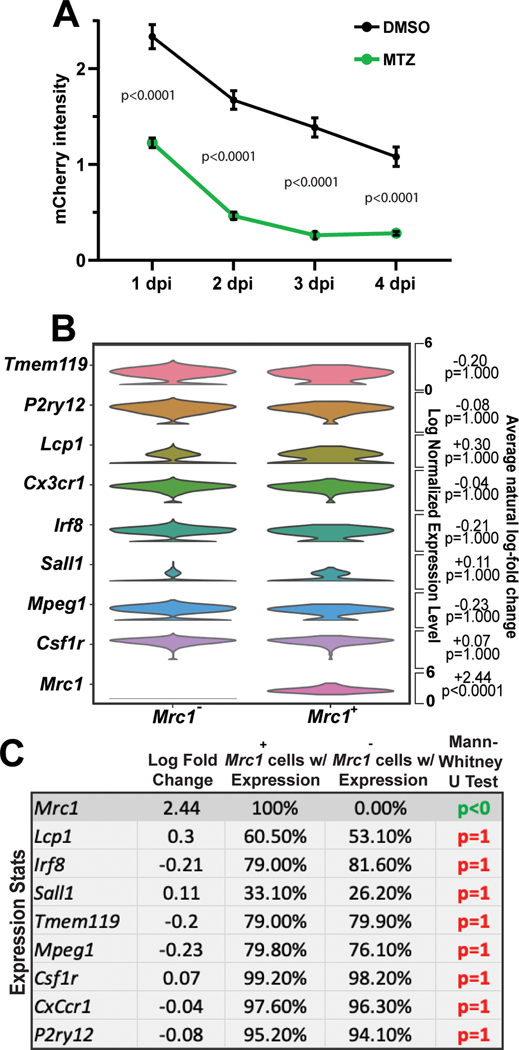
Abstract
Microglia are the resident macrophages of the central nervous system that serve critical roles in brain construction. Although human brains contain microglia by 4 weeks gestation, an understanding of the earliest microglia that seed the brain during its development remains unresolved. Using timelapse imaging in zebrafish, we discovered a mrc1a+ microglia precursor population that seeds the brain before traditionally described microglia. These early microglia precursors are dependent on lymphatic vasculature that surrounds the brain and are independent of pu1+ yolk sac-derived microglia. Single-cell RNA sequencing datasets reveal Mrc1+ microglia in the embryonic brain of mice and humans. We then show in zebrafish that these early mrc1a+ microglia precursors preferentially expand during pathophysiological states in development. Taken together, our results identify a critical role of lymphatics in the microglia precursors that seed the early embryonic brain.
INTRODUCTION:
Microglia have substantial roles in brain construction1–9. We know microglia precursors can be detected in the human embryonic brain as early as 4 weeks gestation10–13, before peak stages of neurogenesis, gliogenesis and synaptogenesis14. Colonization is believed to occur over several weeks in humans, with peak microglia density reached around 20 weeks15. Microglia colonize the mouse around E9.5 with substantial expansion by E10.516. We know fate mapping with tamoxifen-inducible Runx1-Cre mice activated at E7.5 demonstrates labeling of 30% of microglia at E10.516. In zebrafish, we also know the yolk sac generates microglia precursors that seed the brain in early development between 2–3 dpf (days post-fertilization), although L-plastin+ cells can be identified at 35 hpf17,18. Studies in mice and zebrafish support the hypothesis that microglia may also have additional sources besides the yolk sac19–22. Still, relatively little is known about the first microglia, or pioneer microglia, to colonize the embryonic brain.
Here we utilize timelapse imaging to show a population of mrc1a+ microglia-like cells that colonizes the embryonic zebrafish brain before traditionally described pu1+ microglia populations and prior to peak stages of synaptic and glia genesis. Analysis of scRNA sequencing datasets from both mouse and human fetal tissue demonstrate mammalian embryonic microglia also express MRC1/Mrc1. We show mrc1a+ microglia precursors develop independently of the traditional pu1+ yolk sac-derived microglia lineage in zebrafish. Intravital imaging revealed that mrc1a+ cells within and associated with lymphatic vessels that surround the brain, leave the vessels and colonize the brain. Colonization of mrc1a+ microglia in zebrafish is reduced with multiple manipulations that disrupt lymphangiogenesis. Finally, we show mrc1a+ microglia are the primary responding microglia population during a developmental injury. Together, these data implicate brain-border lymphatic vessels as central to the early colonization of microglia precursors.
RESULTS:
Microglia-like cells express mrc1a
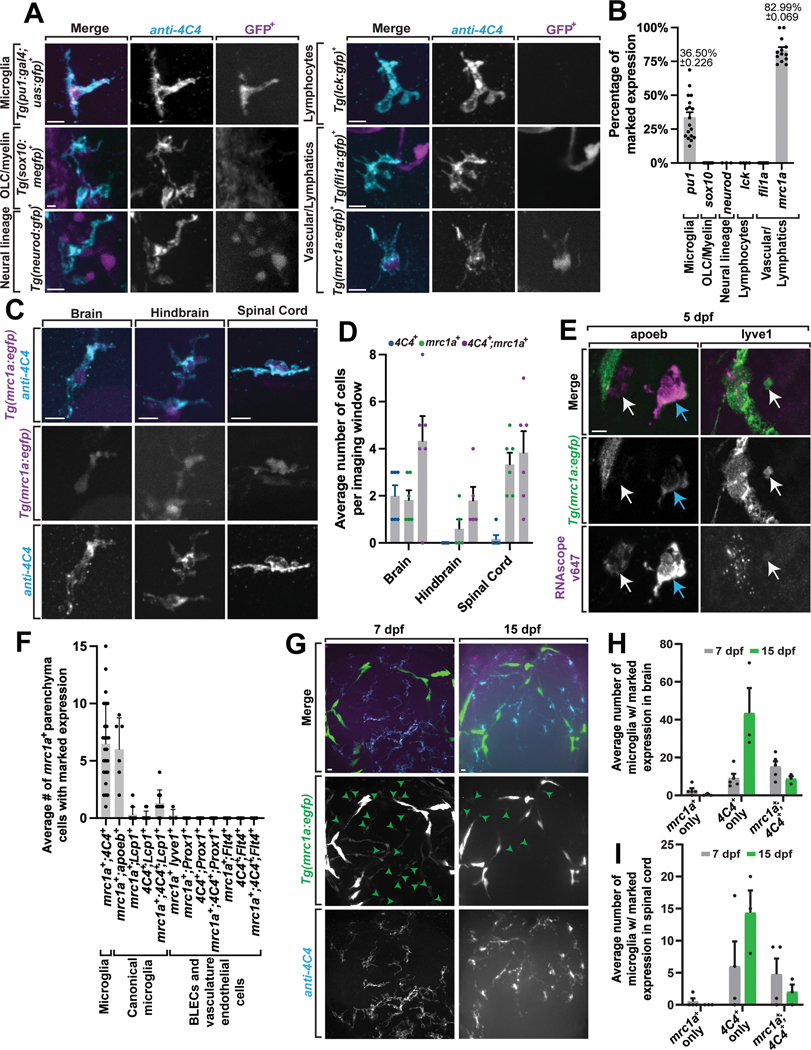
To explore embryonic microglia, we initially scored the abundance of microglia in the 5 dpf zebrafish brain. To do this we used the 4C4 antibody which specifically labels microglia in zebrafish23. We then co-localized 4C4 with Tg(pu1:Gal4;UAS:gfp) which labels yolk sac-derived microglia/macrophage populations24. This analysis identified that 36.5 ± 0.226% of zebrafish microglia could be labeled by the pu1 transgene. 4C4 also marked 28.1% of Eos+ cells from Tg(pu1:eos) animals, consistent with the idea that Tg(pu1:Gal4;UAS:gfp) animals were marking the portion of 4C4+ cells that were also pu1+ (Extended Data Figure 1A).
To next explore the possibility that additional embryonic populations label or contribute to the pool of microglia, we screened transgenes labeling major cell types and lineages in the brain or periphery, including oligodendrocyte lineage cells or neural crest (sox10), neurons (neurod), lymphocytes (lck), vasculature (fli1a), and venous vasculature/lymphatics (mrc1a)25. We did not detect 4C4 immunolabeling in sox10, neurod, lck, or fli1 cells. In contrast, mrc1a labeled 82.99 ±0.069% of 4C4+ cells in the brain (Figure 1A–1B), indicating a potential contribution of mrc1a+ cells to the embryonic microglia pool. For simplicity’s sake, below we refer to GFP+ cells in Tg(mrc1a:egfp) animals as mrc1a+.
Figure 1. Microglia-like cells express mrc1a.
(A) Confocal z-projection of Tg(sox10:megfp), Tg(neurod:gfp), Tg(lck:gfp), Tg(fli1a:gfp), Tg(pu1:gfp), and Tg(mrc1a:egfp) animals stained with 4C4 at 5 dpf. (B) The percentage of 4C4+ cells that co-express GFP. Quantifications represent all CNS regions of animals (n=59 animals). (C) Confocal z-projection of Tg(mrc1a:egfp) animals stained with 4C4 at 5 dpf labeling microglia-like cells in different CNS regions. (D) The average number of cells located within different CNS regions. Imaging window for the brain equals 0.0027 mm3, hindbrain equals 0.0027 mm3, and spinal cord equals 0.0108 mm3 (n=6 animals). (E) Confocal z-plane images of Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) animals hybridized with RNAscope probes apoeb and lyve1b at 5 dpf. White arrows: mrc1a+ only microglia. Cyan arrows: mrc1a+;apoeb+ microglia. (F) The average number of mrc1a+ parenchyma cells with co-labeled marked expression across various lymphatic, BLECs, vascular endothelial cells, and canonical cell markers (n=31 animals). (G) Confocal z-projection of Tg(mrc1a:egfp) animals stained with 4C4 at 7 dpf and 15 dpf. (H) The average number of mrc1a+ only, 4C4+ only, and mrc1a+;4C4+ microglia in the brain imaging window (n=8 animals). (I) The average number of mrc1a+ only, 4C4+ only, and mrc1a+;4C4+ microglia in the spinal cord imaging window (n=8 animals). Imaging window equals a 0.0027 mm3 region of the brain (A-I). Scale bar equals 10μm (A,C,E, G). Error bars denote ± SEM (B, D, F, H-I).
Microglia are located throughout the CNS, so we next quantified whether mrc1a+;4C4+ cells are present across various CNS regions. To do this we imaged the brain, hindbrain, and spinal cord of Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) animals, stained with 4C4, and scored the abundance of mrc1a+;4C4+ cells in each CNS region (Figure 1C–1D). Co-labeled cells were located in all areas of the CNS. We confirmed this localization with a second transgenic line that marked the parenchyma with a panneuronal-expressing dsred, Tg(mrc1a:egfp);Tg(nbt:dsred) (Extended Data Figure 1B–1E). Unlike vessel associated cell populations like brain lymphatic endothelial cells, FGPs, and brain-border macrophages that would be expected to interact with vessels outside of the CNS parenchyma26–28, mrc1a+;4C4+ cells were located within the CNS proper (Figure 1C–1D). We further confirmed that these mrc1a+ cells in the parenchyma are separate and distinguishable from stationary lymphatic vessel endothelium by timelapse imaging motile mrc1a+ cells in the embryonic brain (Supplementary Video 1). They are also present in the spinal cord, where FGP cells (which also label with mrc1a) are not present (Figure 1C–1D)27. These data are consistent with the hypothesis that mrc1a+ cells may represent a microglia-like precursor population that seeds the embryonic brain.
We would expect microglia or their precursors to be labeled with apoeb and L-plastin (Lcp1)29. Therefore, we first stained Tg(mrc1a:egfp) animals at 5 dpf with Lcp1 and 4C4. In this analysis, we identified mrc1a+;Lcp1+;4C4+ cells in the zebrafish brain (Figure 1E–1F, Extended Data Figure 1F–1G). In situ hybridization demonstrated that mrc1a+ cells in the parenchyma also expressed apoeb. (Figure 1E–F, Extended Data Figure 1C). We also tested if they were marked by lyve1b, Flt4, or Prox1, which are expressed in vascular endothelial populations (Figure 1E–F; Extended Data Figures 1F–G)25,30,31. However, mrc1a+ cells in the parenchyma did not label with those markers (Figure 1E–F; Extended Data Figure 1C). These data are consistent with the hypothesis that a mrc1a+ microglia population seeds the zebrafish embryonic brain.
To determine if this is a transient population that is present only during embryogenesis, we scored the abundance of mrc1a+;4C4+ cells in Tg(mrc1a:egfp) brains at 7 and 15 dpf (Figure 1G–1I). Most microglia at both time points are only labeled with 4C4. However, mrc1a+;4C4+ microglia could also be detected at 7 and 15 dpf and thus persist at least into the juvenile zebrafish brain.
mrc1a+ microglia function like traditional microglia
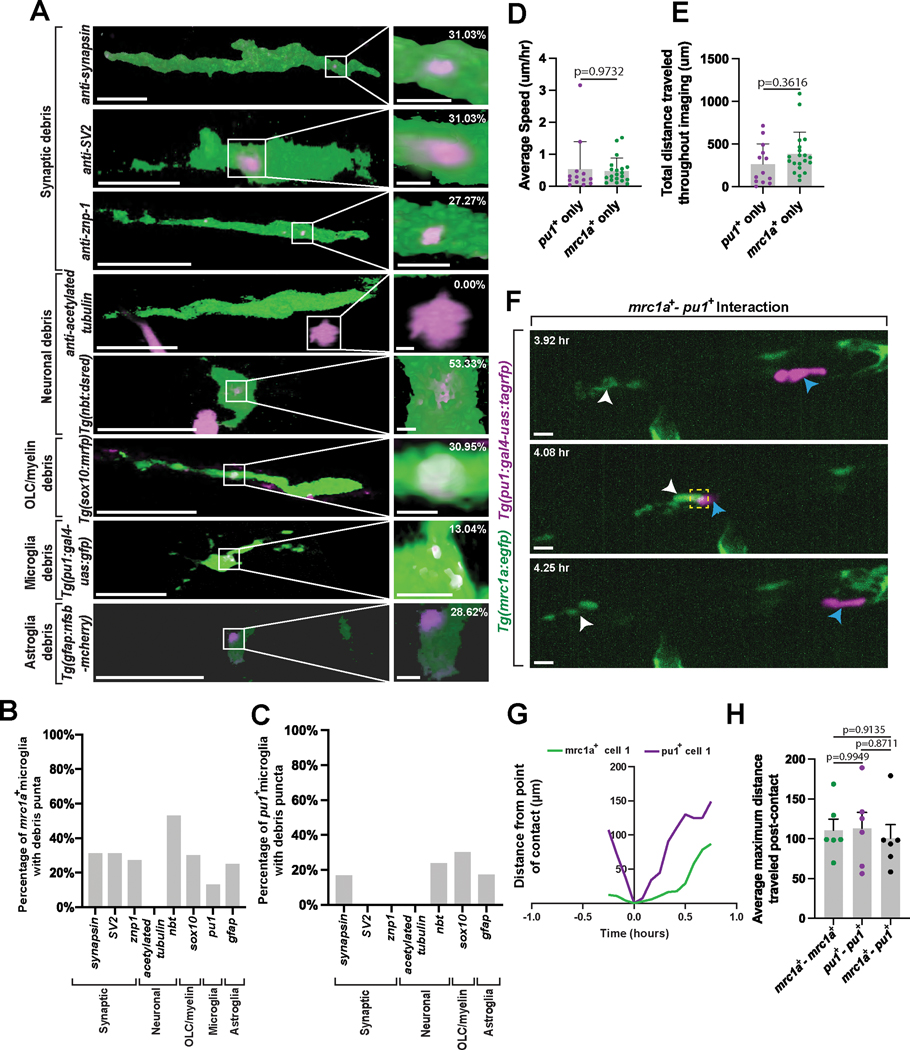
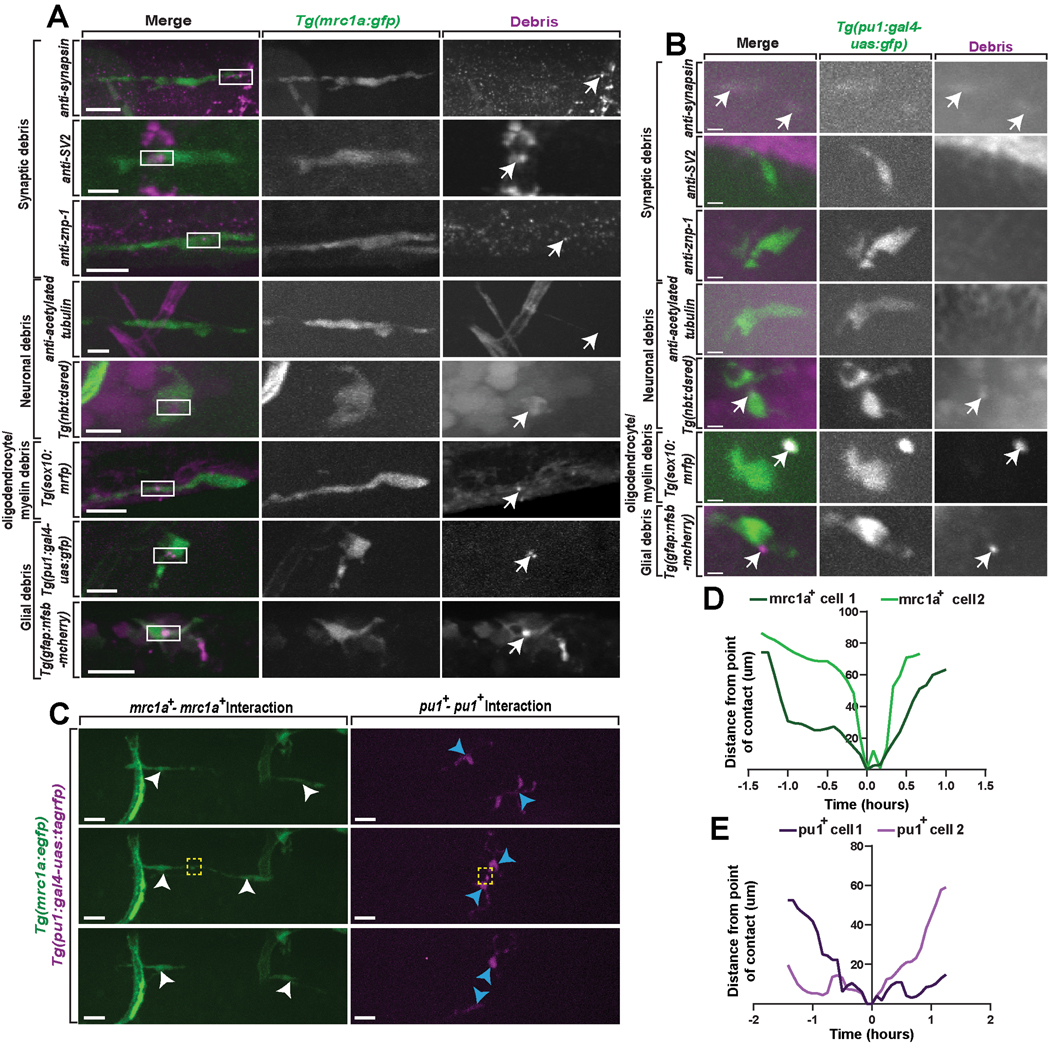
Microglia clear debris in the developing CNS1–9. To test if mrc1a+ microglia also clear parenchymal debris, we used 5 dpf Tg(mrc1a:egfp) animals immunostained with 4C4 and a combination of transgenic animals and antibody staining to label synaptic (synapsin, SV2, and znp-1), neuronal (acetylated tubulin, nbt), oligodendrocyte lineage cell (sox10), astroglial (GFAP), and microglia (pu1) debris. We used IMARIS to confirm and visualize the engulfment of labeled debris by mrc1a+ microglia(Figure 2A, Extended Data Figure 2A). This revealed that mrc1a+ microglia phagocytose most neural debris in the developing CNS (Figure 2B). mrc1a+ cells contained debris in higher proportions than pu1+ microglia for some types of debris (Figure 2C, Extended Data Figure 2B). Thus like typical microglia, mrc1a+ microglia clear multiple types of debris in the parenchyma.
Figure 2. mrc1a+ microglia function like traditional microglia.
(A) IMARIS 3D surface renderings of 5 dpf Tg(mrc1a:egfp) animals stained with antibodies or other transgenic animals to label debris from synaptic, neuronal, oligodendrocyte, microglia, or astroglia populations. White boxes: magnified region of engulfed debris puncta (right column). (B) The percentage of types of labeled debris cleared by mrc1a+ microglia (n=32 animals). (C) The percentage of the types of labeled debris cleared by pu1+ microglia (n=53 animals). (D) The average speed of pu1+ only, and mrc1a+ only microglia from 4 dpf to 5 dpf (Ordinary one-way ANOVA/Tukey’s multiple comparisons tests: pu1+ vs. mrc1a+ p=0.9732, Mean diff.=0.0571, DF=42, q=0.3141, SE of diff.=0.2571)(n=32 cells; n=7 animals). (E) The total distance traveled by pu1+ only, and mrc1a+ only microglia from 4 dpf to 5 dpf (Ordinary one-way ANOVA/Tukey’s multiple comparisons tests: pu1+ vs. mrc1a+ p=0.3616, Mean diff.=−118.3, DF=42, q=1.948, SE of diff.=85.88)(n=32 cells; n=7 animals). (F) Confocal z-projections of Tg(mrc1a:egfp);Tg(pu1:gal4-uas:tagrfp) animals showing homotypic interactions between pu1+ and mrc1a+ microglia populations from 4 dpf to 5 dpf. White arrowheads: mrc1a+ microglia, blue arrowheads: pu1+ microglia. (G) The migration path of individual pu1+ microglia and mrc1a+ microglia traveled pre and post contact (n=3 animals). (H) The average maximum distance pu1+ only, mrc1a+ only, and pu1+;mrc1a+ microglia traveled post-contact (Ordinary one-way ANOVA/Tukey’s multiple comparisons test: mrc1a-mrc1a vs. pu1-pu1 p=0.9949, Mean diff.=−2.328, DF=15, q=0.1362, SE of diff.=24.17; mrc1a-mrc1a vs. mrc1a-pu1 p=0.9135, Mean diff.=9.824, DF=15, q= pu1-pu1 vs. mrc1a-pu1 p=0.8711, Mean diff. = 12.15, DF=15, q=0.711, SE of diff=24.17)(n=7 animals). Imaging window equals 0.0027 mm3 (A-C), 0.0081 mm3 (D-H). Scale bar equals 10μm (A), 100μm (F). Error bars denote ± SEM (D-E, H).
We next asked if mrc1a+ microglia migrate like traditional microglia by tracking individual microglia labeled with pu1 and mrc1a across three imaging regions in the CNS from 4 dpf to 5 dpf. We did not observe a difference in the average speed or total distance traveled by migrating microglia regardless of their pu1+ or mrc1a+ identity (Figure 2D,E). Previous work shows homotypic microglia-microglia interactions are repulsive while macrophage-microglia interactions are not repulsive32. To test whether mrc1a+ microglia also exhibit repulsion following interactions with other microglia, we tracked pu1+ and mrc1a+ microglia throughout three 0.0027 mm3 imaging windows for 24 hours and observed three unique microglia interactions: pu1-pu1, mrc1a-mrc1a, and mrc1a-pu1. Each interaction resulted in contact-dependent repulsion (Figure 2F–2H; Extended Data Figures 2C–2E), consistent with a microglia identity of the mrc1a+ cells.
Mrc1a+ cells colonize the brain early in development
Microglia populations can be identified in the human brain as early as 4 weeks gestation14. To ask whether mrc1a+ microglia might contribute to pioneer microglia-like cells, we explored the earliest seeding of microglia precursors in the brain. We compared this to pu1+ and mpeg1+ cells, traditional markers for microglia precursors that seed the zebrafish brain at 2 to 3 dpf18–20. To explore this, we imaged Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) and Tg(mrc1a:egfp); Tg(mpeg1:mCherry) in the zebrafish brain at 36 hpf and 48 hpf. Such ages correspond with the neurogenesis stages where human microglia can first be identified. We fixed and stained these animals with GFAP to mark the CNS radial glial boundary and confirm a cell was present within the brain parenchyma and away from the mrc1a+ lymphatic/venous vessels that surrounded the brain (Figure 3A–3E). We could not detect pu1+ or mpeg1+ cells present in the brain at 36 hpf. An average of 0.429 ± 0.202 pu1+ and 0.444 ± 0.242 mpeg1+ cells were present in the brain imaging window at 48 hpf, consistent with previous reports that pu1+ precursors seed the brain after 2 dpf (Figure 3F)18. In contrast, an average of 1.077 ± 0.211 mrc1a+ cells were present within the brain imaging window at 36 hpf and mrc1a+ cells were more abundant in the brain at 48 hpf, with higher relative abundance than pu1+ and mpeg+ cells. Cells that label with both mrc1a+;4C4+ were detected at 48 hpf, consistent with the hypothesis that the early mrc1a+ cells persist throughout the embryonic brain (Figure 3F). We could not detect any 4C4+ cells at 48 hpf that also label with pu1 or mpeg1.
Figure 3. Mrc1a+ cells colonize the brain early in development.
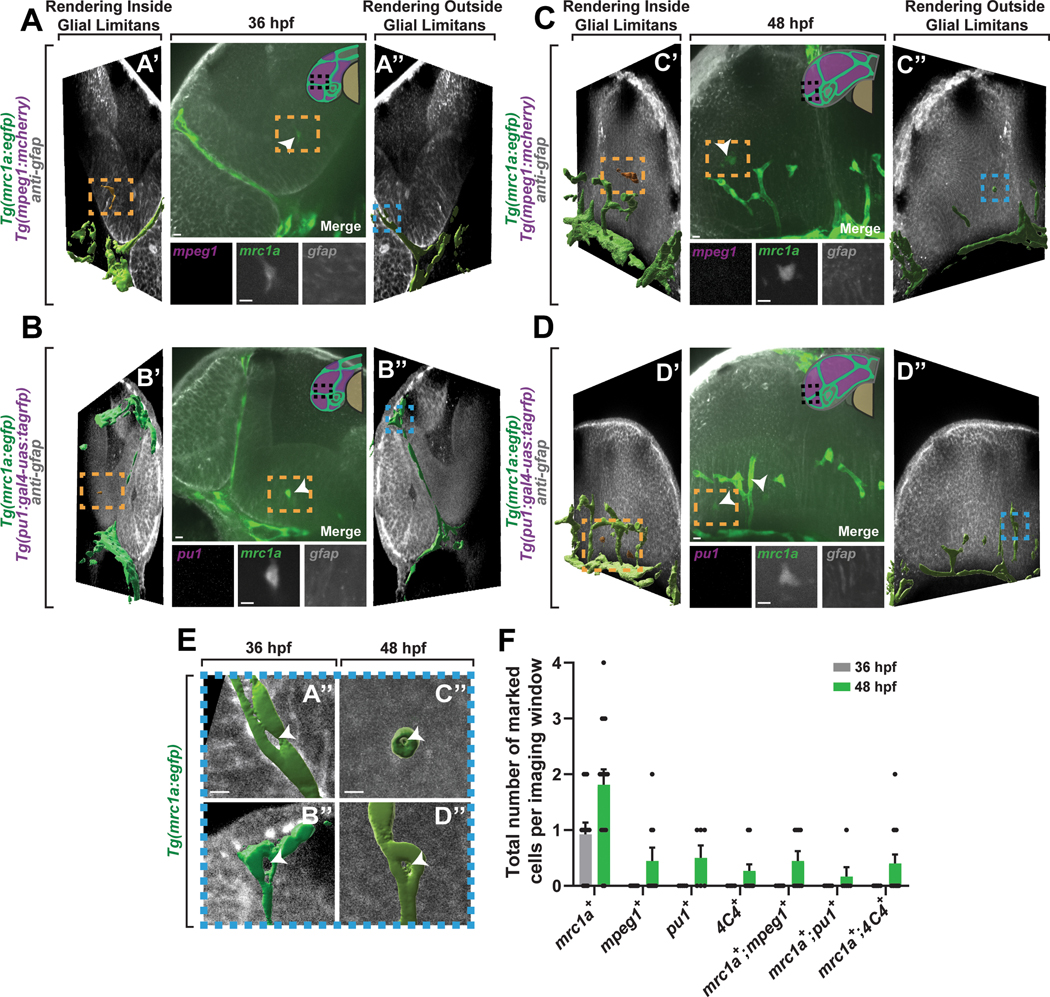
(A) Confocal z-projection of Tg(mrc1a:egfp);Tg(mpeg1:mCherry) animals stained with GFAP at 36 hpf (middle). (A’-A”) IMARIS 3D surface rendering of vessels (green) and mrc1a+ microglia (orange) combined with a 10μm confocal z-projection slice stained with GFAP to label the glial limitans. The combined 3D surface rendering and confocal image were rotated 45 in the negative (A’) and positive (A”) orthogonal z-plane to confirm the presence of mrc1a+ cells inside the glial limitans (A’) and absence of mrc1a+ cells outside the glial limitans (A”). (B) Confocal z-projection of Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) animals stained with GFAP at 36 hpf (middle). B’-B” represent the same views described in A’-A”. (C) Confocal z-projection of Tg(mrc1a:egfp);Tg(mpeg1:mCherry) animals stained with GFAP at 48 hpf (middle). C’-C” represent the same views described in A’-A”. (D) Confocal z-projection of Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) animals stained with GFAP at 48 hpf (middle). D’-D” represent the same views described in A’-A”. Dashed orange boxes: mrc1a+ microglia-like cells within the GFAP+ limitans (A-D, A’-D’). White arrowheads: mrc1a+ microglia-like cells (A-D). Dashed blue boxes: vessel cross sections from A”-D” of insets represented in (E). Graphical illustrations of the embryonic zebrafish brain (magenta) and vessels (green) at 36 hpf (A’ and B’ middle upper right corner) and 48 hpf (C’ and D’ middle upper right corner). (E). Magnified insets of Tg(mrc1a:egfp) vessel cross sections highlighted by the dashed blue boxes in A”-D”. White arrowheads: hollow vessel cores. (F) The total number of marked cells inside the glial limitans at 36 hpf and 48 hpf in Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) or Tg(mrc1a:egfp);Tg(mpeg:dsred) animals (A-F) (n=29 animals). Imaging window equals a 0.0027 mm3 region of the brain. Scale bar equals 10μm (A-E). Error bars denote ± SEM (F).
Microglia in the mammalian embryonic brain express Mrc1
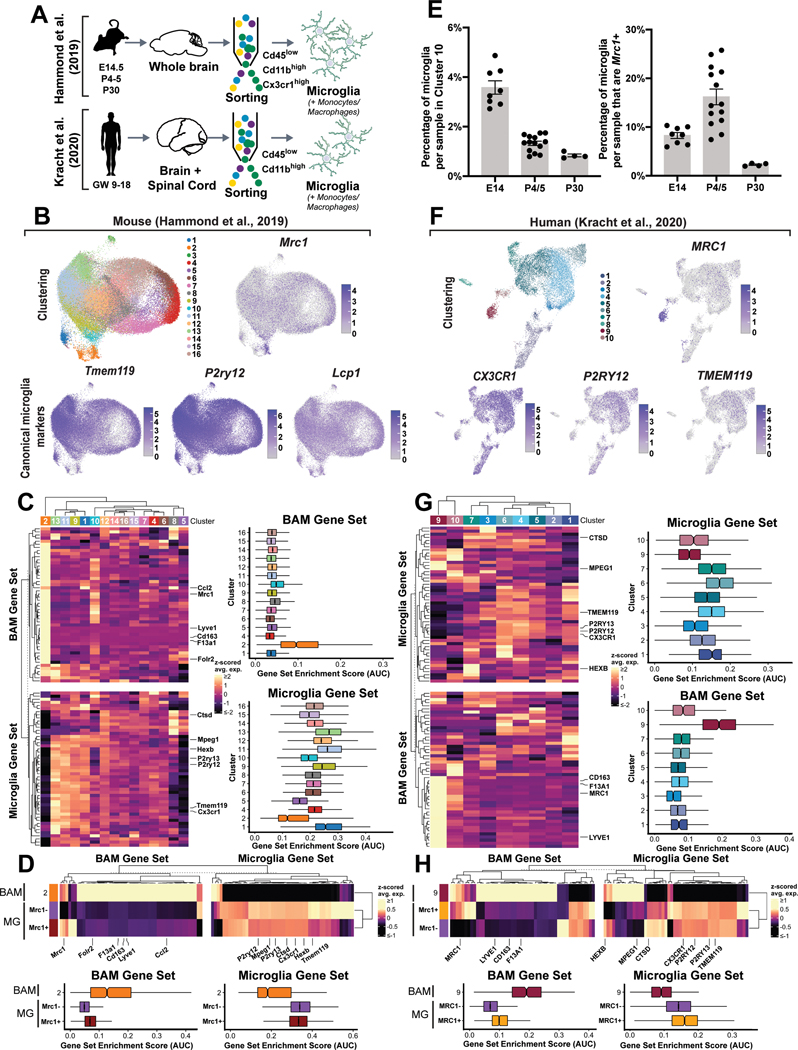
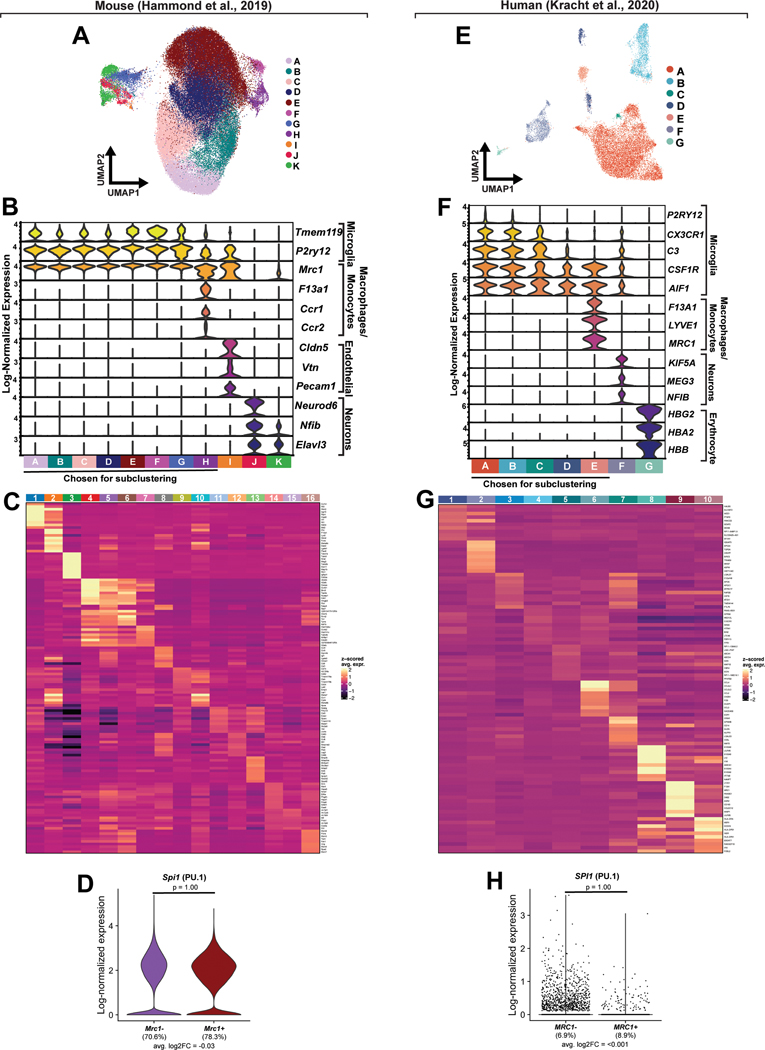
We next examined the transcriptomes of murine microglia in early development using previously published single-cell RNA sequencing data to determine if Mrc1 is expressed in mammalian microglia precursors, in addition to the expression that has already been described in border-associated macrophages (BAMs)29,33(Figure 4A). We first analyzed approximately 70,000 transcriptomes of microglia and macrophages from E14.5, P4/5, and P30 mice. E14.5 captures embryonic microglia and P4/P5 captures post-embryonic, early-juvenile microglia ages that would correspond with the myelination states that are present in 5 dpf zebrafish when we can detect mrc1a+ microglia-like cells. P30 mice were included for an adult reference. We utilized hierarchical clustering to remove contaminating neurons and endothelial cells, and then re-clustered the remaining cells (Extended Data Figure 3A–B), which revealed 16 distinct clusters (Figure 4B; Extended Data Figure 3C). While cluster 3 expressed neuronal markers, the remaining clusters all expressed macrophage genes (Figure 4B; Extended Data Figure 3C). To determine whether these clusters were composed of microglia or BAMs, we created sets of marker genes which were differentially expressed in microglia and BAMs in published RNA-sequencing data from embryonic mice34. We then examined expression of these gene sets across the 15 macrophage clusters. Cluster 2 exhibited notably higher expression of genes in the BAM gene set – such as Mrc1, F13a1, Lyve1, and Cd163 – compared to all other clusters (Figure 4C). The fourteen other clusters expressed genes in the microglia gene set – such as Tmem119, P2ry12, and Hexb35–38,37–40 – more highly than Cluster 2 cells (Figure 4C). Based on expression of the two gene sets, we classified Cluster 2 as BAMs and the remaining clusters as microglia. Several of these clusters replicated previously described subtypes29, including an Ms4a7+ cluster highly enriched in Mrc1 (Cluster 10), which was most highly prevalent at E14.5 compared to later ages, supporting the hypothesis that Mrc1 may label a subset of embryonic microglia in mammals (Figure 4C & E). Interestingly, we observed 11.6% of the cells in the fourteen microglia clusters expressed detectable Mrc1. To determine whether these Mrc1+ cells were microglia-like and not BAMs, we examined expression of the microglia and BAM gene sets between Mrc1+ and Mrc1− cells from the 14 putative microglia clusters, including Cluster 2 cells as a reference for BAMs (Figure 4D). We find that the Mrc1+ cells exhibited minimal differences in expression of these gene sets compared to the Mrc1− cells (Figure 4D; Supplementary Table 1). We further found that Mrc1+ microglia exhibit higher enrichment of the microglia gene set and lower enrichment of the BAM gene set than Cluster 2 BAMs. The small differences in microglia gene expression are likely due to developmental age, as Mrc1+ microglia were more highly prevalent in younger animals (E14.5 and P4–5) and were markedly reduced at P30 (Figure 4E).
Figure 4. Mrc1 is expressed in developmental microglia in the mammalian brain.
(A) Schematic depicting Hammond et al. (2019) and Kracht et al. (2020) isolations. (B) UMAP of subclustering of E14.5, P4/5, and P30 cells from Hammond et al, upper left, and log-normalized gene expression of Mrc1 and canonical microglia. (C) (left) Heatmap of expression of BAM and microglia genes from Utz et al. (2020)34 across microglia and macrophage clusters from Hammond et al. (right) Box plots of single-cell gene set enrichment scores for the BAM and microglia gene sets across clusters. Enrichment scores are reported as AUC (area under curve), calculated using AUCell. (D) (top) Heatmap of expression of BAM and microglia signature genes across Mrc1+ and Mrc1− microglia and Cluster 2 BAMs. (bottom) Box plots of single-cell gene set enrichment scores for the BAM and microglia gene sets across the Mrc1+ and Mrc1− microglia and Cluster 2 BAMs. (E) (left) Percentage of microglia per sample in Mrc1-enriched Cluster 10 versus sample age. Error bars denote ± SEM. P-values correspond to two-sided Tukey’s multiple comparisons test (E14.5 vs. P4/5: p < 0.0001; E14.5 vs. P30: p < 0.0001; P4/5 vs. P30: p= 0.2071) following ANOVA (p < 0.0001). (right) Percentage of microglia per sample that are Mrc1+ (E14.5: n = 8; P4/5: n = 14; P30: n = 4). P-values correspond to Tukey’s multiple comparisons tests (E14.5 vs. P4/5: p = 0.0023; E14.5 vs. P30: p = 0.1102; P4/5 vs. P30: p < 0.0001) following ANOVA (p < 0.0001). (F) UMAP plot of clustering of immune cells from Kracht et al, (upper left). (upper right and lower row) log-normalized gene expression of MRC1 and canonical microglia markers overlaid on UMAP plots. (G) (left) Heatmap of expression of BAM and microglia genes, human orthologs of the genes identified from Utz et al, across clusters. (right) Box plots of gene set enrichment scores for the BAM and microglia gene sets across clusters. (H) (top) Heatmap of expression of BAM and microglia genes across MRC1+ and MRC1− microglia and Cluster 9 BAMs. (bottom) Box plots of single-cell gene set enrichment scores for the BAM and microglia gene sets across the MRC1+ and MRC1− microglia and Cluster 9 BAMs. For box plots in (C-D, G-H), the transecting line indicates median. Notches indicate 95% confidence interval surrounding median. Box boundaries indicate interquartile range (25% to 75%). Whiskers indicate minima and maxima.
We next asked whether MRC1-expressing microglia are present in human development by exploring a dataset of over 13,000 microglia from human fetal tissue33 (Figure 4A). Using hierarchical clustering, we first removed contaminating populations of neurons and erythrocytes, and then re-clustered the remaining cells, yielding 10 final clusters (Extended Data Figure 3E–F). One of these clusters (Cluster 8), was enriched in leukocyte genes like S100A9 and LILRA5, while the other clusters expressed typical macrophage genes (Extended Data Figure 3G; Supplementary Table 1). Examining microglia versus BAM gene sets showed Cluster 9 exhibited the highest expression of BAM genes and the lowest expression of microglia genes (Figure 4G). The remaining clusters more highly expressed the microglia gene set and did not display notable enrichment of the BAM gene set. Clusters 3 & 10 exhibited lower expression of microglia genes but also minimal enrichment of BAM genes, which may indicate these cells are less mature microglia. We further discovered that, of the 13,430 cells in the potential microglia clusters, 8.6% expressed MRC1. The MRC1+ cells expressed the microglia gene set more highly and the BAM gene set lower than Cluster 2 BAMs, and expressed both these gene sets comparably to MRC1− microglia (Figure 4H; Supplementary Table 1). We thus conclude that these MRC1+ cells are indeed microglia. Like the mouse scRNA sequencing analysis, Spi1/Pu.1 was detected in a portion of both MRC1+ and MRC1− microglia (Extended Data Figure 3D,H). Although microglia in both mice and humans express Mrc1/MRC1 during development, further work will be needed to confirm the existence of these populations in mammals, and to determine whether this population shares other characteristics with the embryonic zebrafish microglia.
mrc1a+ microglia are dependent on lymphangiogenesis
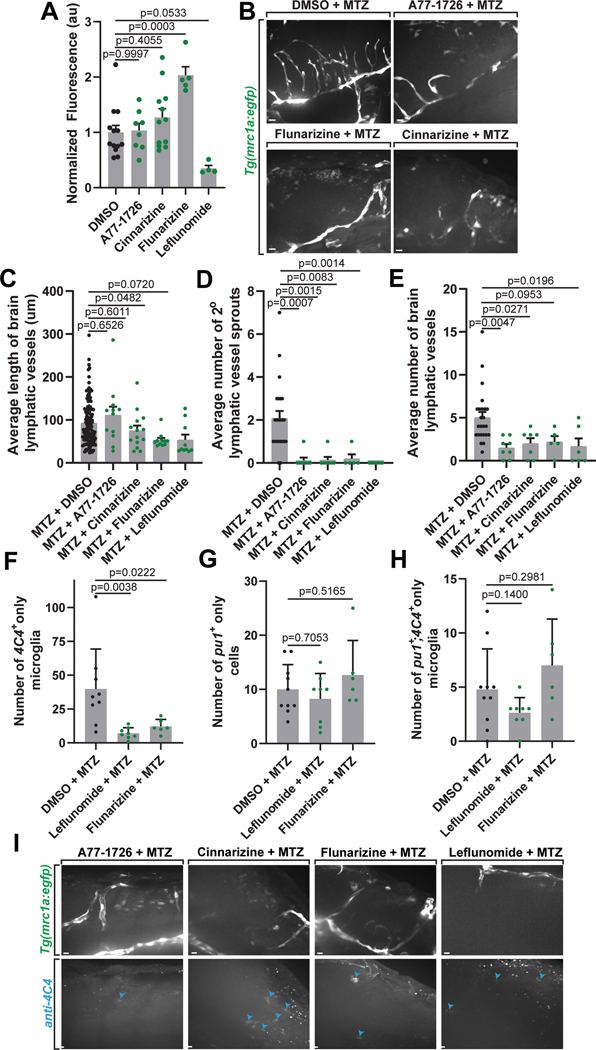
We next explored the developmental ontogeny of mrc1a+ microglia in zebrafish. In addition to expression in parenchyma-located microglia, mrc1a/Mrc1 is expressed in lymphatic and venous vessels, brain-border macrophages, FGPs and brain lymphatic endothelial cells (BLECs)26,27,39. These mrc1a+ vessels surrounding the brain uptake Qdot705 when it is injected into the head, consistent with their classification as lymphatic vasculature26. Light sheet microscopy revealed that lymphatic vessels surround the brain in the embryo, right outside the gfap+ boundary (Figure 5A–5B). To investigate the potential role of these embryonic brain-border lymphatic vessels in mrc1a+ microglia, we timelapsed the brain-border mrc1a+ vessel in the anterior region of the head and its surrounding brain region for 24 hrs. Orthogonal rotations of these images demonstrate a luminal space in the center of the vessel (Figure 5C). In timelapse movies, we noted two observations that showed an interaction with migratory mrc1a+ cells and mrc1a+ lymphatic vessels. First, mrc1a+ cells could be seen migrating along and circling mrc1a+ lymphatic vessels (Figure 5D–5E, Supplementary Video 1). We also identified mrc1a+ cells within the mrc1a+ lymphatic vessels that extended processes outside of the vessel and then eventually exited the vessel (Supplementary Video 2,3). Rotational images in IMARIS confirmed the intra-vessel location throughout the movie. While exiting, they displayed an hourglass shape, indicative of cells leaving confined barriers32,40. These migratory mrc1a+ vessels then traveled away from the vessel, unlike FGP cells that remain associated or close to the lymphatic vessels. Together these data are consistent with the possibility that mrc1a+ microglia precursors could utilize the lymphatic vessels for brain colonization, placing the brain-border lymphatics as central to embryonic microglia colonization.
Figure 5. mrc1a+ microglia are dependent on lymphangiogenesis.
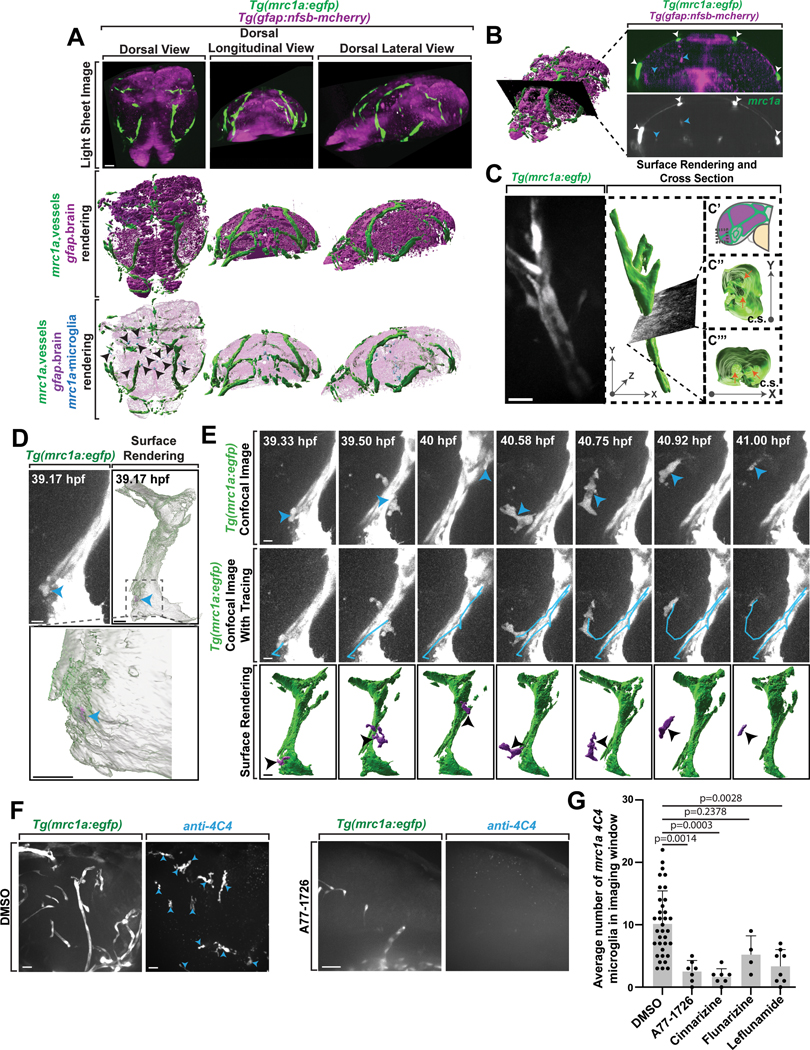
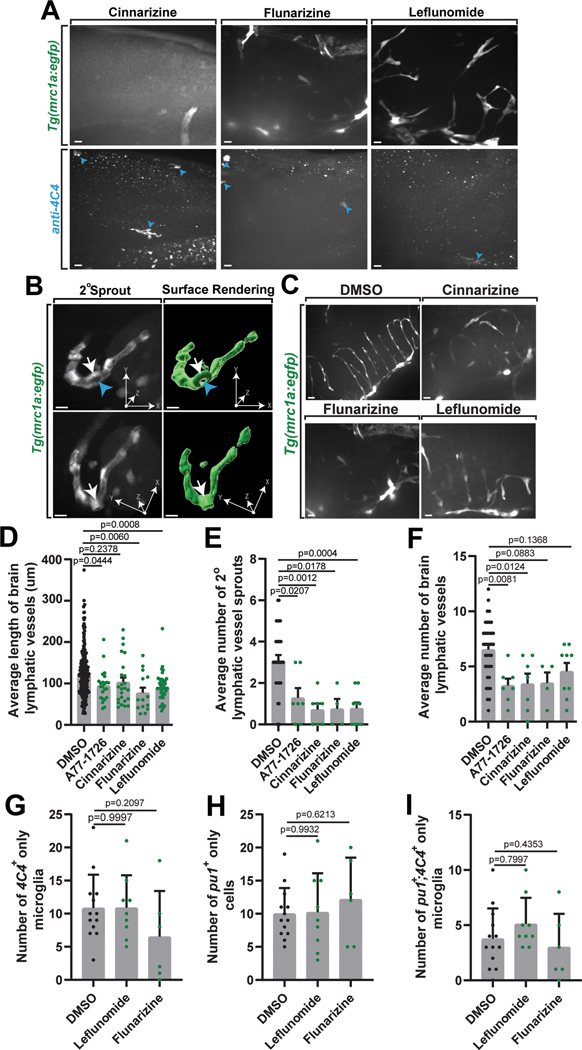
(A) Z-projection of images from light sheet microscopy (top row) and IMARIS surface renderings (bottom two rows) of the brain and surrounding mrc1a+ vessels at 3 dpf in Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) animals. Black arrowheads: pseudocolored cyan mrc1a+ microglia inside the gfap+ boundary. (B) IMARIS surface rendering (left) of the 3 dpf brain and surrounding mrc1a+ vessels with a 2.3 μm cross sectional slice through the center (right). White arrowheads: mrc1a+ vessels outside the gfap+ boundary. Blue arrowheads: mrc1a+ microglia inside the gfap+ boundary. (C) Confocal z-projection (left) and IMARIS 3D surface rendering (middle) of dorsal lymphatic vessels outside the glial limitans. C’ graphical illustration of the embryonic zebrafish brain and lymphatic vessels at 48 hpf. Grey box: dorsal lymphatic vessel location of confocal images depicted in (D). (C”-C”’) dorsal and lateral cross section (c.s.) views of the vessel. Orange arrows: hollow cores of vessel cross sections. (D) Confocal image (left) and surface rendering (right) from a 24-hour timelapse movie starting at 34 hpf showing an mrc1a+ cell (blue arrowheads) located within an mrc1a+ lymphatic vessel at 39.17 hpf. Black dashed box: magnified view of mrc1a+ cell inside the vessel (bottom). (E) Images (top), overlay of the cell’s migration path (middle), and surface rendering (bottom) from a 24-hour timelapse movie starting at 34 hpf in Tg(mrc1a:egfp) animals showing an mrc1a+ cell (blue and black arrowheads) exiting and interacting with a lymphatic vessel. (F) Confocal z-projection of DMSO- vs lymphatic inhibitor (A77–1726)-treated Tg(mrc1a:egfp) animals stained with 4C4. Blue arrowheads: mrc1a+;4C4+ microglia. (G) The percentage of microglia per imaging window that are mrc1a+ in DMSO- vs lymphatic inhibitor (A71–1726, cinnarizine, flunarizine, or leflunomide)-treated animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. A77–1726 p=0.0014, Mean diff.=7.269, DF=69, q=3.825, SE of diff=1.994; DMSO vs. cinnarizine p=0.0003, Mean diff.=8.486, DF=69, q=4.255, SE of diff.=1.994; DMSO vs. flunarizine p=0.02378, Mean diff.=4.95, DF=69, q=1.947, SE of diff.=2.542, DMSO vs. leflunamide p=0.0028, Mean diff.=6.825, DF=69, q=3.616, SE of diff=1.687) (n=61 animals). Scale bar equals 10μm (B-E), 100μm (A). Error bars denote ± SEM (G).
Therefore, we next asked if mrc1a+ microglia colonization was dependent on lymphatic vessels. To test this, we first replicated a drug screen that revealed small molecules that impede lymphangiogenesis41. Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) animals were dosed with individual inhibitors daily from 48 hpf to 120 hpf and fixed and immunostained with 4C4 at 120 hpf. We confirmed inhibition of lymphangiogenesis by measuring the average length and number of lymphatic vessels and secondary sprouts (Extended Data Figures 4A–4F). To determine whether mrc1a+ microglia are dependent on lymphatic vessels, we next quantified the average percentage of 4C4+ microglia which were mrc1a+. We observed a significant reduction in the number of microglia in the brain which were mrc1a+ across the lymphatic inhibitor treatment groups compared to DMSO (Figure 5F–5G), consistent with the hypothesis that mrc1a+ microglia could be dependent on lymphangiogenesis.
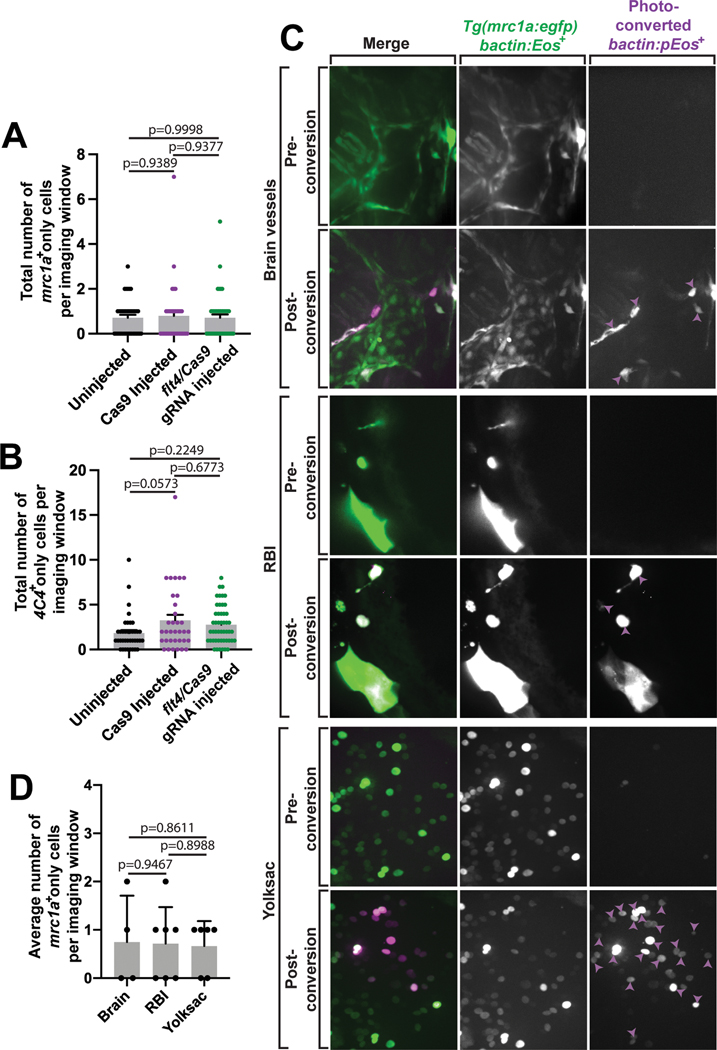
As a complementary approach, we also inhibited lymphatic vessel formation genetically using two flt4 sgRNAs. Flt4 is required for the production of lymphatic vessels25,28,42. We first confirmed that injection of flt4 sgRNA/Cas9 reduced the lymphatic vessels in Tg(mrc1a:egfp) animals, but not in Cas9 only or uninjected controls(Figure 6A). With T7E1 we identified that the animals with perturbed lymphatic vessels had indels at the flt4 locus. To determine if mrc1a+ microglia were reduced after flt4 sgRNA injection, mrc1a+ cells in the parenchyma were immunostained with 4C4. We noted that mrc1a+;4C4+ cells were reduced after injection with flt4 sgRNA (Figure 6B–6C, Extended Data Figures 5A–5B). These injections did not reduce the number of 4C4+ only cells in the brain or those that were labeled with only mrc1a. Although it remains a possibility that mrc1a+ microglia require flt4 for differentiation, together these results with the pharmacological manipulations of lymphatic vessels are most consistent with the hypothesis that mrc1a+;4C4+ microglia are dependent on lymphatic vessels.
Figure 6. mrc1a+ microglia are dependent on lymphangiogenesis and lymphatics in the head.
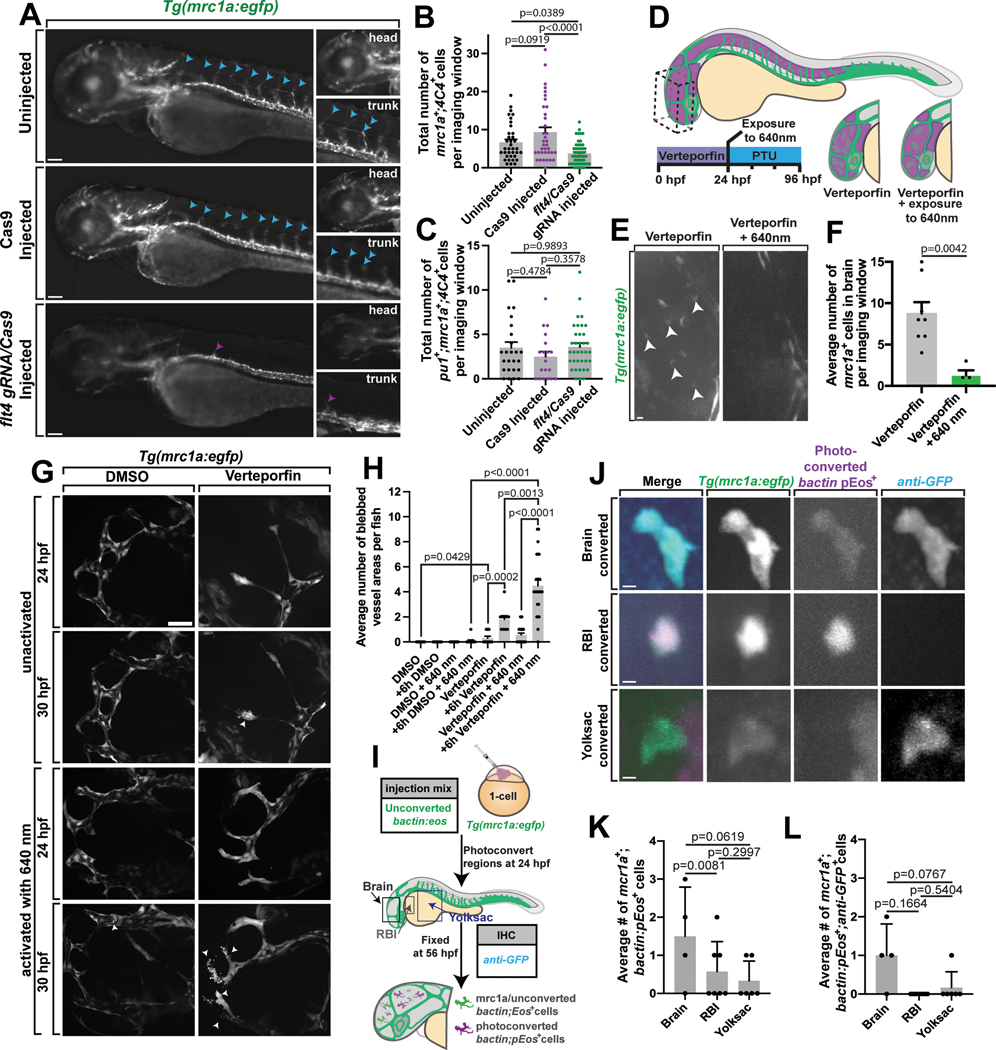
(A) Tg(mrc1a:egfp) at 5 dpf. Blue arrowheads: developed lymphatic vessels. Purple arrowheads: disrupted lymphatic vessels. (B-C) Total number of microglia in uninjected, Cas9 only, and flt4 gRNA/Cas9-injected animals that are mrc1a+;4C4+ (B) (one-way ANOVA/Tukey’s multiple comparisons: uninjected vs. Cas9 only p=0.0919, Mean diff = −2.667, DF=119, q=2.986, SE of diff=1.263, uninjected vs. flt4 gRNA/Cas9 p=0.0389, Mean diff=2.899, DF=119, q=3.5, SE of diff=1.171, Cas9 only vs. flt4 gRNA/Cas9 p<0.0001, Mean diff=5.566, DF=119, q=6.719, SE of diff=1.171)(n=122 animals) or pu1+;mrc1a+;4C4+ (C) (one-way ANOVA/Tukey’s multiple comparisons: uninjected vs. Cas9 only injected p=0.4784, Mean diff=1.006, DF=77, q=1.646, SE of diff=0.8648; uninjected vs. flt4 gRNA/Cas9 injected p=0.9893, Mean diff=−0.1033, DF=77, q=0.1975, SE of diff=0.7398; Cas9 only injected vs. flt4 gRNA/Cas9 injected p=0.3578, mean diff=−1.11, DF=77, q=1.948, SE of diff=0.8058)(n=80 animals). (D) Schematic illustrating Verteporfin treatment. (E) Verteporfin- or Verteporfin + 640nm-treated Tg(mrc1a:egfp) animals at 24 hpf. White arrowheads: microglia within the brain imaging window. (F) Average number of mrc1a+ cells in the brain imaging window (t-test: Verteporfin vs. Verteporfin + 640nm p=0.0042, DF=10, two-tailed) (n=12). (G) Unactivated and activated DMSO- or Verteporfin-treated Tg(mrc1a:egfp) animals. White arrowheads: blebbing vessels. (H) Average number of blebbed vessel areas (t-test: DMSO vs. Verteporfin p=0.0420, Verteporfin vs. +6h Verteporfin p=0.0002, +6h DMSO + 640 nm vs. +6h Verteporfin + 640 nm p<0.0001, +6h Verteporfin vs. +6h Verteporfin + 640 nm p=0.0013, Verteporfin + 640 nm vs. +6h Verteporfin + 640 nm p<0.0001; all DF=20 and all two-tailed)(n=108). (I) Schematic outlining the injection, photoconversion of bactin:eos, and IHC staining in Tg(mrc1a:egfp) animals. (J) bactin:eos-injected animals photoconverted in various regions at 24 hpf and stained with GFP at 56 hpf. (K) Average number of mrc1a+;pEos+ cells converted in various regions (t-test: brain vs. RBI p=0.0081, RBI vs. yolksac p=0.2997, brain vs. yolksac p=0.0619; all two-tailed)(n=17). (L) Average number of mrc1a+;pEos+;anti-GFP+ cells (t-test: brain vs. RBI p=0.1664, RBI vs. yolksac p=0.5404, brain vs. yolksac p=0.0767; all two-tailed)(n=17). Imaging window equals 0.0027 mm3 (E,G,J). Scale bars: 10μm (E), 50μm (A,G). Error bars denote ± SEM (B-C, F, H, K-L).
We next asked more specifically if lymphatic endothelial vessels or cells around or in the brain are required for colonization of mrc1a+ microglia precursors. To test this, we disrupted mrc1a+ endothelial cells in and around the brain with Verteporfin at 24 hpf, 12 hours before we initially visualize mrc1a+ cell migration from lymphatic vessels, and then scored the number of free-roaming mrc1a+ cells in the brain at 4 dpf (Figure 6D–6H). Verteporfin is a photodynamic drug that when exposed to 640 nm light causes specific perturbation of lymphatic endothelial cells, thereby providing precise spatiotemporal control of the perturbation43,44. As a control, we exposed animals to 1 μM verteporfin but not 640 nm light. We first confirmed via timelapse imaging that the exposure to Verteporfin and 640 nm light caused mrc1a+ lymphatic vessels outside the brain to swell and degrade by 3.21±1.32 hours post 640 nm exposure (n=4 animals) (Figure 6G–6H). Three days post treatment, Verteporfin-treated animals exposed to 640 nm in the head exhibited less mrc1a+ cells in the brain (1.25 cells on average in the parenchyma per 0.0027 mm3 imaging window) compared to non-photoactivated control animals (on average 8.75 mrc1a+ cells; p=0.0042; Figure 6E–6F). These data support the hypothesis that mrc1a+ microglia may be dependent on brain-border lymphatic endothelial vessels or cells.
Our data supports the hypothesis that mrc1a+ microglia colonize the brain at a distinct time from pu1+ microglia (Figure 2F). To ask if these pu1+ microglia also utilize lymphatics distinctly to colonize the brain, we repeated two of the lymphatic inhibitor treatments with strong reduction in the mrc1a+ microglia (Figure 5D–5E), in Tg(pu1:Gal4;UAS:gfp); Tg(gfap:nfsb-mCherry) animals immunostained with 4C4. The number of pu1+ cells did not change upon treatment with leflunomide and flunarizine compared to DMSO-treated animals (Extended Data Figure 4G–4I), consistent with the hypothesis that brain lymphatics have a role in mrc1a+ microglia colonization–a role that is not shared with pu1+ yolk sac-derived microglia.
Traditional pu1+ microglia migrate from the rostral blood islands (RBI) in the yolk sac to the brain16,19. To narrow the potential direct source of mrc1a+ microglia, we fate-mapped cells with the photoconvertible fluorescent protein, Eos. In this paradigm, bactin:eos was injected into Tg(mrc1a:egfp) at the one-cell stage, photoconverted with a diffraction-limited laser in specific regions of interest at 24 hpf, grown for 24 hours post-conversation and then fixed for anti-GFP staining (Figure 6I). Photoconversion of Eos in the yolk sac at 24 hpf, resulted in few mrc1a+;p-Eos+ cells in the brain at 48 hpf, although we could detect mrc1a+ cells and p-Eos+ cells in the brain (Figure 6J–6L). Photoconversion of the RBI at 24 hpf also did not produce mrc1a+;p-Eos+ cells in the brain at 48 hpf. These results are inconsistent with the hypothesis that the yolk sac or RBI directly produces the majority of the mrc1a+ cells that are located in the parenchyma. Given that mrc1a+ microglia are reduced following perturbation of brain-border lymphatics, we next tested if precursor cells in the head at 24 hpf produced mrc1a+ microglia by photoconverting Eos in the entirety of the head. At 48 hpf, we scored mrc1a+;pEos+ cells in the brain in 75% of the animals (Figure 6J–6L, Extended Data Figures 5C–5D). While it is still possible that mrc1a+ microglia are derived at some point from other sources, these data support the hypothesis that mrc1a+ microglia at 48 hpf originate from cells that were present in the head at 24 hpf.
mrc1a+ microglia are distinct from pu1+ microglia
We next sought to address if mrc1a+ microglia could be further distinguished from traditional pu1+ microglia. Traditional yolk sac-derived microglia express and are dependent on pu1 (spi1b)19,35. To determine whether mrc1a+ microglia are independent of traditionally described yolk sac-derived microglia, we first imaged Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) animals immunostained with 4C4 at 5 dpf and scored the average number of cells with varying mrc1a, pu1, and 4C4 marked expression (Figure 7A). We identified seven marked expression patterns, including small populations of 4C4+ only, 4C4+;pu1+, and pu1+;mrc1a+ cells, and larger populations of pu1+ only and mrc1a+ only cells within the CNS imaging window. Most notably, there is an abundant cell population that is mrc1a+;4C4+ that does not express pu1+ (Figure 7B).
Figure 7. mrc1a+;4C4+ microglia are distinct from yolk sac-derived microglia.
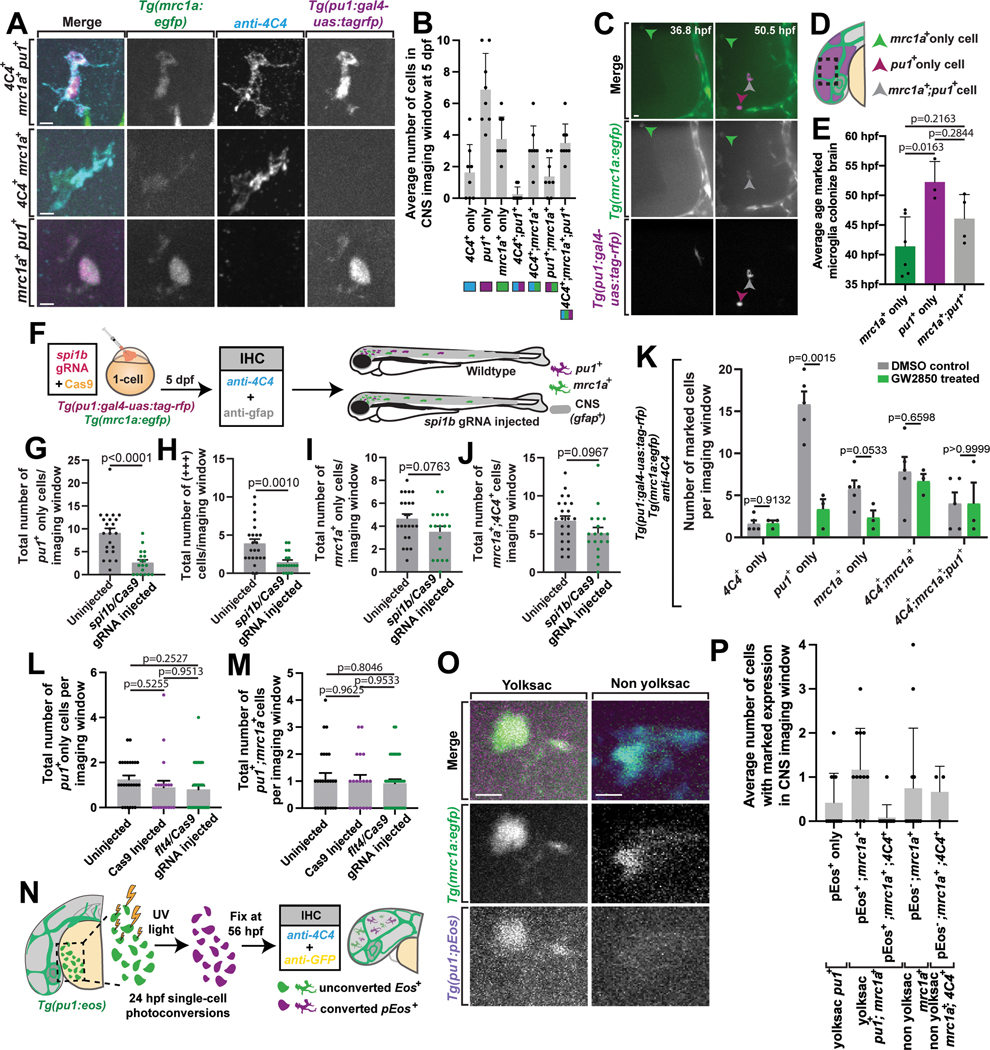
(A) 5 dpf Tg(mrc1a:egfp);Tg(pu1:gal4-uas:tagrfp) animals stained with 4C4. (B) Average number of cells in the CNS imaging window with varied expression. Imaging window represents 0.0027 mm3 of the brain. Colored rectangles underneath graph labels correspond to expression patterns in (A)(n=9) (C) Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) animals 36 hpf-56 hpf. (D) Anatomical schematic of the imaging window in (C). (E) Average age various types of microglia colonize the brain (one-way ANOVA/Dunnett’s multiple comparisons: mrc1a+ only vs. pu1+ only p=0.0163, pu1+ only vs. mrc1a+;pu1+ p=0.2844, mrc1a+ only vs. mrc1a+;pu1+ p=0.2163). (F) Graphical illustration outlining our CRISPR/Cas9 spi1b single-gRNA injection experiment. (G-J) The total number of pu1+ (G), pu1+;mrc1a+;4C4+ (+++) (H), mrc1a+ (I), and mrc1a+;4C4+ (J) cells in spi1b sgRNA injected animals compared to uninjected controls (t-test, spi1b sgRNA injected vs. uninjected: pu1+ only p<0.0001, +++ p=0.0010, mrc1a+ only p=0.0763, mrc1a+;4C4+ p=0.0967; all two-tailed)(n=41 G-J). (K) Average number of marked variants per imaging window across treatment groups (t-test, DMSO vs. GW2850: 4C4+ only p=0.9132, pu1+ only p=0.0015, mrc1a+ only p=0.0533, 4C4+;mrc1a+ p=0.06598, 4C4+;mrc1a+;pu1+ p>0.9999; all two-tailed)(n=8 animals). (L-M) Average number of microglia in uninjected and Cas9 only animals compared to flt4 gRNA injected animals that were pu1+ only (L) (one-way ANOVA/Tukey’s multiple comparisons: uninjected vs. Cas9 injected p=0.5255, Mean diff =0.3453, DF=77, q=1.536, SE of diff=0.3179; uninjected vs. flt4 gRNA/Cas9 injected p=0.2527, Mean diff=0.2719, DF=77, q=2.26, SE of diff=0.2719; Cas9 injected vs. flt4 gRNA/Cas9 injected p=0.9513, Mean diff=0.08919, DF=77, q=0.4259, SE of diff=0.2962) (n=80) or pu1+;mrc1a+ (M) (one-way ANOVA/Tukey’s multiple comparisons: uninjected vs. Cas9 injected p=0.9625, Mean diff=0.08, DF=77, q=0.3727, SE of diff=0.3035; uninjected vs. flt4 gRNA/Cas9 injected p=0.9533, Mean diff=0.08333, DF=77, q=0.4167, SE of diff=0.2828; Cas9 injected vs. flt4 gRNA/Cas9 injected p=0.8046, Mean diff=0.1633, DF=77, q=0.8696, SE of diff=0.2596)(n=80). (N) Schematic illustrating the single-cell photoconversion and IHC in (O) and (P). (O) Tg(mrc1a:egfp);Tg(pu1:eos) animals at 56 hpf. (P) Average number of pEos+ cells with marked expression at 56 hpf (n=12). Imaging window equals 0.0027 mm3 of the brain (A-P). Scale bar equals 10μm (A, C, O). Error bars denote ± SEM (B, E, G-M, P).
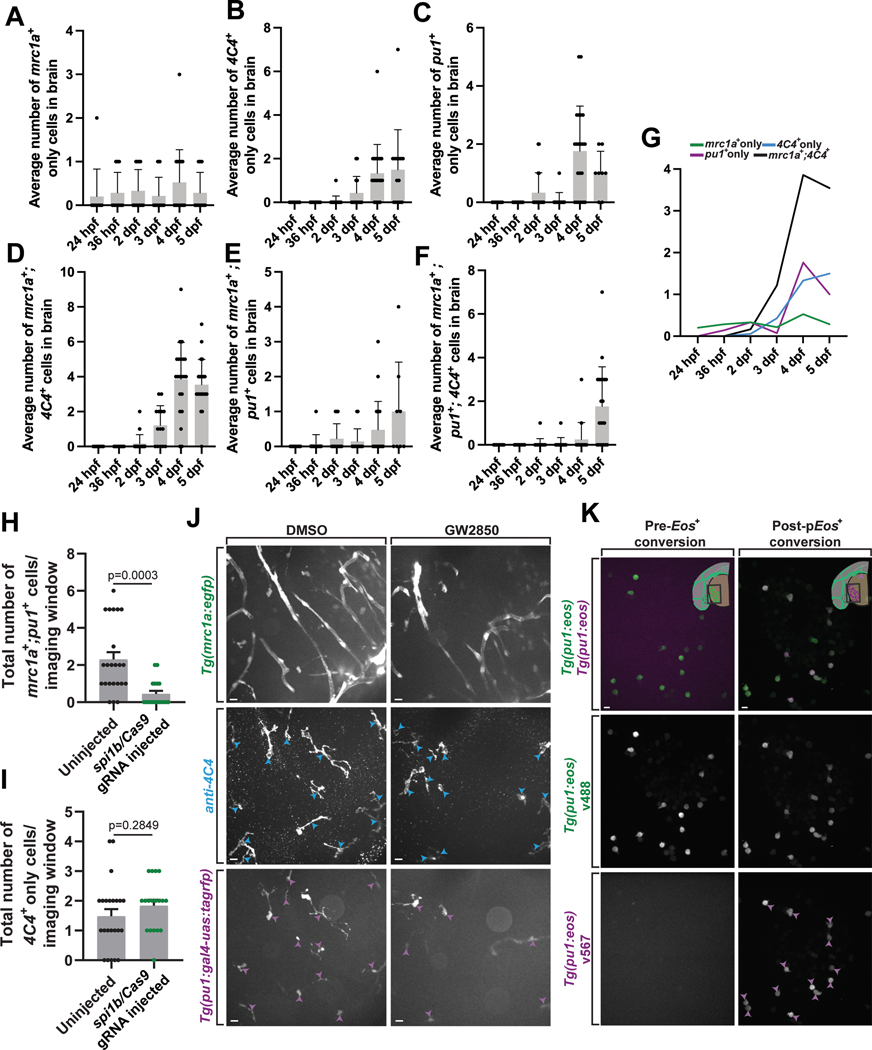
To further explore how mrc1a+ microglia differed from the pu1+, and pu1+;mrc1a+ cells, we assayed their development across embryonic ages from 1–5 dpf in Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) animals (Extended Data Figures 6A–6G). We also co-stained with 4C4 to identify their mature microglia identity. mrc1a+ cells in the parenchyma could first be identified at 24 and 36 hpf ((Extended Data Figures 6A). The mrc1a+;4C4+ population could first be detected at 2 dpf and expanded after (Extended Data Figure 6D). pu1+ cells arrived to the brain at 2 dpf, consistent with previous reports of yolk sac-derived microglia populations (Extended Data Figures 6A–6G). As development progressed, so did the populations labeled with 4C4, consistent with the idea that the early seeding of these populations produces a 4C4+ mature microglia population (Extended Data Figures 6A–6G). To get a precise time of infiltration, we complemented this analysis with timelapse imaging of Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) animals from 36–56 hpf, a time point which corresponds with the arrival times from the above staining. In these movies, mrc1a+ cells were first to arrive in the brain, followed by mrc1a+;pu1+ and then lastly pu1+ cells (Figure 7C–7E). Together, these data indicate mrc1a+ microglia precursors arrive before both the pu1+ and mrc1a+;pu1+ population.
To further investigate how mrc1a+;4C4+ were different from other populations of microglia, we used CRISPR/Cas9 to target the pu1 genomic locus without disrupting the pu1 transgenes. We injected the single synthetic-gRNA and Cas9 into Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) single-cell embryos, fixed and stained with 4C4 and GFAP at 5 dpf, and scored the number of mrc1a+, pu1+, and 4C4+ cells (Figure 7F). Embryos that were not injected were used as controls. The number of cells in all pu1+ categories was reduced in crispant animals compared to uninjected animals. These populations include the pu1+ only, pu1+;mrc1a+;4C4+, and pu1+;mrc1a+ cell-types (Figure 7G–7J, Extended Data Figure 6H). In contrast, the total number of mrc1a+ only, and mrc1a+;4C4+, and 4C4+ only cells were not reduced in crispant animals compared to uninjected animals (Figure 7I–7J, Extended Data Figure 6H–6I). Together, these data demonstrate a successful reduction of all pu1+ populations but not mrc1a+ cells, again consistent with the hypothesis that mrc1a+ microglia precursors likely arise independent of pu1+ microglia.
To determine if mrc1a+;4C4+ were different from other populations of microglia with a second complementary approach, we treated Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) animals with GW2850, a Csf1r inhibitor, since traditional microglia require Csf1r signaling for development and survival32,45,46. Animals were treated from 1 to 5 dpf (Extended Data Figure 6J). We then fixed and immunostained with 4C4 at 5 dpf and quantified that in Csf1r inhibitor-treated animals, the pu1+ only cells were significantly reduced from an average of 15.8 cells to 3.3 cells per animal (p=0.0015), while the mrc1a+;4C4+ microglia did not change between GW2850-treated (average: 7.8 cells) and DMSO-treated (average: 6.7 cells) animals (p=0.6598; Figure 7K). These data demonstrate that mrc1a+;4C4 microglia develop independently of pu1 and are resistant to Csf1r inhibition, again distinct from pu1+ microglia.
Analysis and manipulations of mrc1a+;4C4+ cells indicated a population of cells that were also pu1+;mrc1a+. This pu1+;mrc1a+ migrated at an average of 0.486 μm/hr and traveled 287.63 μm, both similarly to pu1+ and mrc1a+ microglia (pu1+;mrc1a+ vs. pu1+, p=0.999 and pu1+;mrc1a+ vs mrc1a+, p=0.971). We could not test the repulsive nature of the microglia with other microglia because we did not detect contact between mrc1a+;pu1+ cells and other microglia. Our data indicate that the pu1+;mrc1a+ population was also reduced by injection of spi1b gRNA but not reduced by the Csf1r inhibition (Figure 7K, Extended Data Figure 6H). Further, the pu1+;mrc1a+ population was present in the brain after the mrc1a+ population (Figure 7C–7E). There were three likely hypotheses regarding the pu1+;mrc1a+ population: 1. a subset pu1+ cells from the yolk sac are mrc1a+, 2. mrc1a+ lymphatic-dependent cells become pu1+ and 3. pu1+;mrc1a+ are a distinct third population. To determine whether the pu1+;mrc1a+ was related to the mrc1a+ population we first injected flt4 sgRNA into Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) animals and scored the presence of the pu1+;mrc1a+ population. These results indicated pu1+;mrc1a+ or pu1+ cells were not reduced (Figure 7L–7M). However, mrc1a+;4C4+ cells were reduced (Figure 6B) and thus indicated the pu1+;mrc1a+ is not dependent on lymphatics. Further, if mrc1a+ cells upregulate pu1, we may expect to see mrc1a+ cells in timelapse movies become RFP+ in Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) animals. However, we were not able to detect the cells transition to label as pu1+;mrc1a+. These results indicate again that mrc1a+;4C4+ cells are distinct from other microglia subpopulations.
We further tested this relationship by lineage tracing pu1+ cells from the yolk sac. To do this, we created Tg(pu1:eos) animals that express the photoconvertible protein Eos with pu1 regulatory regions. Then, in Tg(pu1:eos); Tg(mrc1a:egfp) animals, we photoconverted single yolk sac pu1+ cells at 24 hpf, grew the animals to 56 hpf, and fixed and stained for GFP (Figure 7N, Extended Data Figure 6K). If a portion of pu1+ cells from the yolk sac are mrc1a+, we would expect mrc1a+;pEos+ cells in the brain. We detected two populations of mrc1+ cells in the brain, mrc1+;pEos+ or mrc1+;pEos−. The mrc1+;pEos+ population is consistent with a yolk sac origin (Figure 7O). However, mrc1+;pEos− or mrc1+;pEos−;4C4+ cells are consistent with a non-yolk sac origin (Figure 7P). We also detected a third population that was pEos+;mrc1−, likely indicating the pu1+ cells we detect in the brain (Figure 7P). The simplest explanation for these results is that mrc1a+;4C4+ microglia are distinct from other subpopulations of microglia that express pu1 and that originate from the 24 hpf yolk sac.
mrc1a+ microglia expand in response to CNS injury
The response and expansion of microglia after CNS injury is a hallmark of the cell-type47. We thus sought to compare the responses of mrc1a+-lymphatic dependent and pu1+ populations to pathophysiological conditions in development. To do this, we chemogenetically ablated radial glia using Tg(gfap:nfsb-mCherry) animals, which express a nitroreductase-tagged mCherry under the gfap promoter48. When bathed in the prodrug metronidazole (MTZ), nitroreductase metabolizes MTZ into cytotoxic products to induce radial glia death. Treatment with MTZ from 48 to 72 hpf induced rapid loss of radial glia, which was assessed by mCherry fluorescence in the hindbrain and spinal cord versus uninjured DMSO-treated animals (Figure 8A–8B, Extended Data Figure 7A). Death of radial glia was observed just 24 hours after treatment and continued until 120 hpf, 3 days post injury (dpi) (Extended Data Figure 7A).
Figure 8. mrc1a+;4C4+ microglia expand in response to CNS injury.
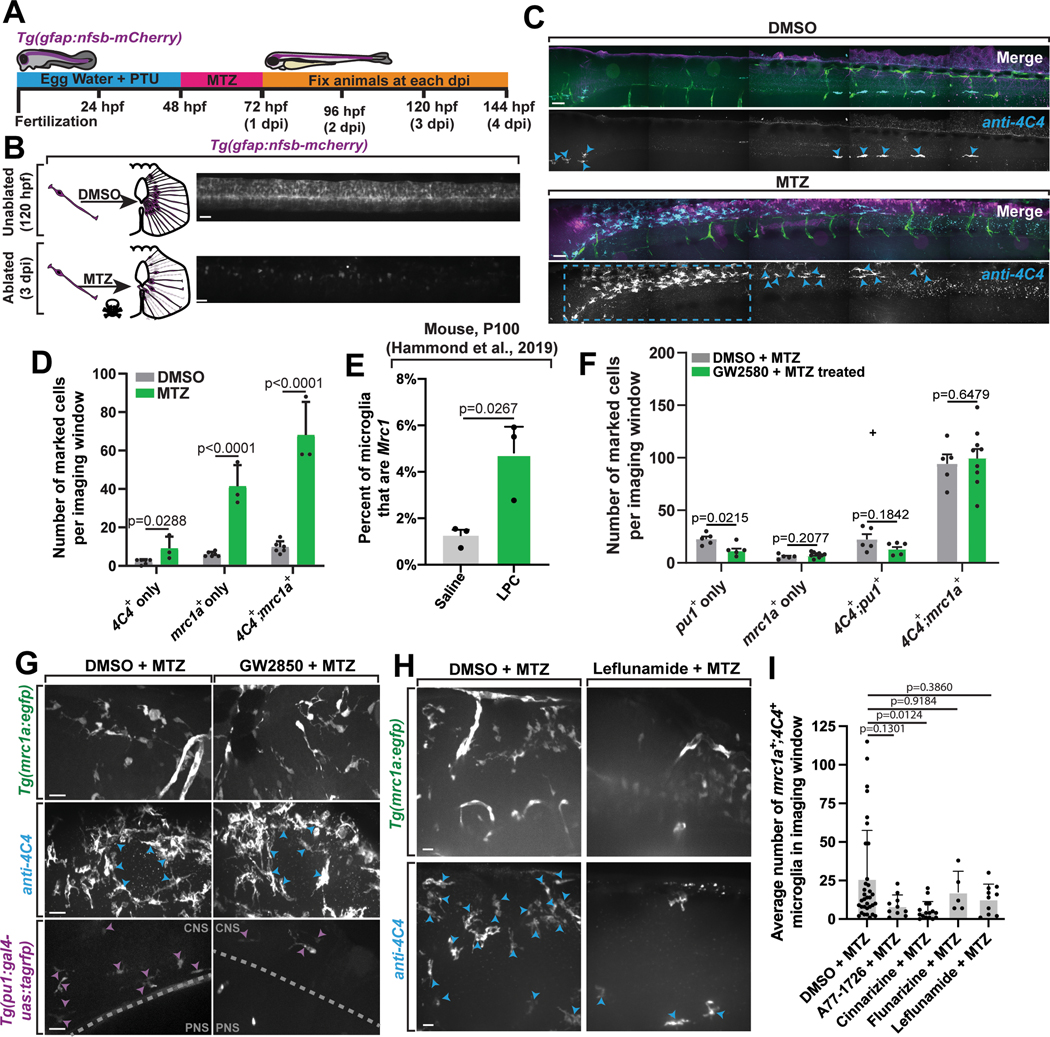
(A) Graphical illustration of MTZ drug treatment and staining. (B) Graphical illustration (left) and confocal z-projection (right) of the injury paradigm in 5 dpf animals. (C) Tiled confocal z-projection from a DMSO control and MTZ-treated 5 dpf Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) zebrafish stained with 4C4 representing the entire animal. Individual mrc1a+;4C4+ microglia (blue arrowheads) and a larger, expanded cluster (dashed blue box) can be seen. (D) Number of cells with marked expression in DMSO- vs MTZ-treated animals per imaging window (t-test, DMSO vs. MTZ: 4C4+ onlyp=0.0288, mrc1a+ only p<0.0001, 4C4+;mrc1a+ p<0.0001; all two tailed)(n=9). (E) Percentage of microglia that express at least one read of Mrc1 from saline control- vs. LPC-injected P100 mice (data from Hammond et al., 2019) (t-test: saline vs. LPC p=0.0267; two tailed)(n=6). (F) The average number of marked variants per imaging window in GW2580 + MTZ-treated vs. DMSO + MTZ-treated control animals(t-test, DMSO + MTZ vs. GW2850 + MTZ: pu1+ only p=0.0215, mrc1a+ only p=0.2077, 4C4+;pu1+ only p=0.1842, 4C4+;mrc1a+ only p=0.6479; all two-tailed)(n=24). (G) Representative confocal z-projection of Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) and Tg(pu1:gal4-uas:tagrfp);Tg(gfap:nfsb-mCherry) animals stained with 4C4. (H) Representative confocal z-projections of 5 dpf Tg(mrc1a:egfp);Tg:gfap:nfsb-mCherry) animals stained with 4C4. Blue arrowheads: mrc1a+;4C4+ microglia. Grey dashed line: glial limitans, microglia above dashed line (purple arrowheads) are inside the CNS and cells below the dashed line are pu1+ macrophages. (I) The percentage of mrc1a+ microglia in lymphatic inhibitor-treated animals compared to DMSO + MTZ control animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO + MTZ vs. A77–1726 + MTZ p=0.1301, Mean diff=17.59, DF=72, q=2.232, SE of diff=7.884; DMSO + MTZ vs. cinnarizine + MTZ p=0.0124, Mean diff=20.79, DF=72, q=3.13, SE of diff=6.644; DMSO + MTZ vs. flunarizine + MTZ p=0.9184, Mean diff=8.694, DF=72, q=0.8282, SE of diff=10.5; DMSO + MTZ vs. leflunomide + MTZ p=0.3860, MEan diff=13.19, DF=72, q=1.673, SE of diff=7.884)(n=75). Imaging windows: six 0.0027 mm3 regions per animal, or 3000μm quantified to include the entire CNS of each animal (A-D, F), one 0.0027 mm3 region per animal (F-I). Scale bars: 10μm (G, H) 100μm (B,C). Error bars denote ± SEM (D-F, I).
To assess whether mrc1a+ microglia expand in response to this injury, we conducted the injury paradigm in Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) animals and immunolabeled microglia with 4C4. By 3 dpi, mrc1a+;4C4+ microglia had expanded seven-fold relative to uninjured DMSO-treated animals (p < 0.0001) and comprised on average 88% of the responding 4C4+ microglia (Figure 8C–8D). mrc1a−;4C4+ microglia also expanded relative to uninjured controls (p = 0.029), but only four-fold and comprised only an average of 12% of the responding microglia. We also observed a significant expansion of mrc1a+ only cells in response to injury (p < 0.0001) (Figure 8C–8D). Thus, mrc1a+ microglia exhibit microgliosis in response to damage to the CNS.
To next examine the possibility that Mrc1+ microglia are present in mammals after injury, we again analyzed scRNA sequencing data, in which microglia from adult mice were isolated from a focal demyelinating lesion caused by LPC injection or from the same region in saline injected control mice29. In LPC-injected mice, Mrc1+ cells significantly increased in number compared to saline injected mice (Figure 8E; p = 0.0267). Comparing canonical microglia marker expression in Mrc1+ and Mrc1− cells revealed no significant differences in expression of those genes(Extended Data Figures 7B–7C). Although the increase in Mrc1+ microglia in the scRNA sequencing data of mouse injury does not distinguish between the upregulation of Mrc1 in previously Mrc1− microglia or the expansion of an existing Mrc1+ microglia population, it does indicate that Mrc1+ microglia are more present after injury to the mammalian brain.
mrc1a+ microglia are independently responsive to injury
Given both mrc1a+ and pu1+ microglia populations are present during development and injury, we next investigated whether mrc1a+ and pu1+ microglia injury responses are independent, by treating Tg(mrc1a:egfp); Tg(gfap:nfsb-mCherry) or Tg(pu1:Gal4;UAS:gfp); Tg(gfap:nfsb-mCherry) animals with the Csf1r inhibitor, GW2850, from 24 to 120 hpf, and MTZ from 48 to 72 hpf to induce injury (Figure 8F). Treatment with the inhibitor reduced pu1+ cells but not mrc1a+;4C4+ microglia or mrc1a+ only cells (Figure 8F, 8G), again supporting the conclusion that mrc1a⁺ microglia are independent of the pu1+ subpopulation.
To test this hypothesis with a second approach, we treated Tg(mrc1a:egfp); Tg(gfap:nfsb-mCherry) animals with inhibitors of lymphangiogenesis from 48 to 120 hpf and MTZ from 48 to 72 hpf to create the injury (Figure 8H, 8I)41. All four inhibitors successfully inhibited the development of lymphatic vessels (Extended Data Figures 8I). Inhibitor-treated animals also exhibited a reduction in the abundance of mrc1a+;4C4+ (Figure 8E–8F), demonstrating the response of mrc1a+ microglia to the injury is altered when animals are treated with drugs that reduce lymphatic vessels.
To ask if pu1+ microglia also require lymphatics to respond to the injury, we repeated two of the lymphatic inhibitor treatments in Tg(pu1:Gal4;UAS:gfp); Tg(gfap:nfsb-mCherry) animals dosed with MTZ from 48 to 72 hpf and immunostained with 4C4. Treatment with leflunomide and flunarizine did not cause a change in the number of pu1+ cells (p=0.7053, p=0.5165) or pu1+;4C4+ microglia (p = 0.14, p = 0.2981), indicating pu1+ microglia injury response is not disrupted when the animal is exposed to drugs that inhibit lymphatics (Extended Data Figures 8F–8I). Together these data indicate that mrc1a+ and pu1+ microglia respond independently to injury and thereby further establish that these populations are distinct.
DISCUSSION:
Here we demonstrate that the zebrafish brain is seeded by a mrc1a+ microglia population that is dependent on lymphatic vessels. We also show that induction of a pathophysiological state in embryonic animals results in expansion of mrc1a+ microglia. This expansion in disease states is disrupted by perturbations that alter lymphatic vessels.
Beyond microglia, the brain is populated with brain lymphatic endothelial cells (BLECs), fluorescent granular perithelial cells (FGPs), brain-border and perivascular macrophages, and Mato cells26,27. Unlike these populations, the mrc1a+ cells in the parenchyma display distinguishing features expected of microglia. They are located in the parenchyma, clear debris that is largely localized to the parenchyma, demonstrate heterotypic recognition with other microglia, and label with the microglia specific-antibody 4C4, as well as Lcp1 and apoeb. Distinct from FGPs, these mrc1a+ cells in zebrafish express lower levels of mrc1a:egfp, are not labeled with Flt4, Prox1 or lyve1b, and are located in the parenchyma and are present in the spinal cord where lymphatic vessels and FGPs have yet to form and colonize25. Unlike macrophage populations, mrc1a+ microglia display contact-dependent repulsion away from other microglia populations in zebrafish.
We know that precursors from extraembryonic yolk sac blood islands give rise to primitive macrophages which then colonize the brain and differentiate into mature microglia16,35. Other waves of hematopoiesis occurring outside of the yolk sac may also generate microglia progenitors in both mice and zebrafish19,20,22. Interestingly, the earliest microglia colonize the human CNS (at ~4–5 gestational weeks) prior to the establishment of active circulation12,13,49. Avian chimera studies have similarly described early colonization prior to brain vascularization49; however, lineage tracing demonstrates that yolk sac-derived microglia precursors require active circulation to colonize the brain in mice16. Our results may help bridge the gap between these disparate findings: the earliest colonizing microglia precursors may arise from or depend on lymphatic endothelium, while a later wave of yolk sac-derived progenitors colonizes the brain after circulation.
There are several indications that lymphatic endothelial vessels could be a site of cytogenesis. In the developing embryo, much of the nascent vasculature is hemogenic50. In mice, primitive hematopoiesis that produces myeloid cells can occur through a hemogenic endothelial intermediate located in vasculature30,51. Later waves of hematopoiesis generate blood cells from hemogenic endothelium in the wall of the dorsal aorta52–54, and in zebrafish this region gives rise to an additional wave of microglia colonization in the adult animal19,20. Which vascular endothelium is hemogenic is unresolved but blood cells can be generated from vascular endothelium in the heart and cranium51,55–57. The possibility that lymphatic vessels are required for a subset of early microglia progenitors and that cells in the head could produce these mrc1a+ microglia is surprising but logical given the previously demonstrated ability of intraembryonic vascular endothelium to generate immune-like cells27,56,57. An alternative possibility is that lymphatic vessels serve as migration routes for early microglia precursors. Lastly, recently the bone marrow of the skull of mice, has been shown to be hematopoietic in order to produce macrophages that can respond to injury of the brain58. Our experiments do not distinguish between these possibilities.
Since the rediscovery of the brain lymphatic system, it has been implicated in disease states59,60. Because of its recent characterization, it is unlikely that its potential role in microglia seeding of the brain would have been investigated. The mature brain lymphatics are also thought to develop postnatally in mice and at later larval stages of zebrafish61, after Mrc1+ progenitors colonize the zebrafish brain interfaces. Whether the mammalian brain has similar embryonic lymphatics surrounding the brain, like in zebrafish (Figure 5A,B), is unclear. The use of timelapse imaging in zebrafish was essential to visualize cells within the lymphatic vessels and departure through vessels, but does not exclude the possibility that mrc1a+ microglia are at some point derived from yolk sac cells. Our data only supports the conclusion that the more direct source of mrc1a+ microglia in zebrafish is likely the head. Nonetheless, these data place lymphatics at the epicenter of mrc1a+ microglia colonization. The role and necessity of pioneer microglia and lymphatics are intriguing avenues for future studies.
METHODS:
Data Availability
Data that supports the findings of this study are available in the Source Data tables. All data collected for the study are included in the figures. All code for the single-cell RNA sequencing data analysis in this manuscript can be accessed at https://github.com/michael-r-odea/Green_ODea_2022.
Experimental Model and Subject Details:
All animal studies were approved by the University of Notre Dame Institutional Animal Care and Use Committee. Zebrafish strains used for this study include: Tg(mrc1a:egfp)25, Tg(pu1:Gal4;UAS:tagrfp)62, Tg(pu1:Gal4;UAS:gfp)62, Tg(gfap:nfsb-mCherry)48, Tg(sox10:mrfp)63, Tg(sox10:megfp)64, Tg(neurod:gfp)65, Tg(neurod:rfp)66, Tg(lck:gfp)67, Tg(fli1a:gfp)68, Tg(nbt:dsred)69, Tg(pu1:eos) (generated here) and Tg(mpeg1:mCherry)69. Pairwise matings produced embryos and embryos were raised at 28°C in egg water in total darkness. Animals were staged by hours or days post fertilization (hpf and dpf)70. Stable germline transgenic lines were used. Embryos of either sex were used for all experiments.
Method Details
In vivo imaging
Animals were anesthetized with 3-amino-benzoic acid ester (Tricaine), enveloped in 0.8% low-melting point agarose and mounted accordingly for best imaging results. Animals were flat mounted on their right side, back mounted, or mounted dorsally in glass-bottomed 35 mm Petri dishes32,71. Images were acquired on a spinning disk confocal microscope custom built by 3i technology© as previously described71,72. Images in timelapse microscopy were collected every 5 min for 18–48 hours depending on the experiment. Adobe Illustrator, IMARIS, and ImageJ were used to process images. Brightness and contrast were enhanced in presented images.
Light sheet microscopy
The 3 dpf embryonic brain, surrounding lymphatic vessels, and mrc1a+ cells inside the gfap+ labeled brain of Tg(mrc1a:egfp);Tg(gfap:nfsb-mCherry) animals were imaged using light sheet microscopy. Animals were anesthetized with Tricaine, enveloped in 1% low-melting point agarose, and mounted in a 100 μL glass capillary tube (Blaubrand, Germany). Light sheet microscopy was performed using a MuVi-SPIM system (Bruker) equipped with dual detection, 16x water-immersion objective lenses (N.A. 0.8, Nikon) and Orca Flash 4.0 V3 sCMOS cameras (Hamamatsu). Images were captured at 32X magnification using a 2x lens magnification changer. Image processing was performed using LuxControl software (version 3.4.0, Bruker). Further 3D surface reconstructions of the light sheet images of the gfap+ brain and mrc1a+ labeled vessels and cells inside gfap+ brain were created using IMARIS.
Immunohistochemistry
The primary antibodies used in this study include 4C4 (1:50, mouse, Seiger, Becker and Becker Labs) (Ohnmacht et al., 2016)23, GFP (1:500, chicken, Kerafast, EMU101), Lcp1 (1:500, rabbit, GeneTex, GTX134697), Prox1 (1:500, rabbit, AngioBio, 11–002), Flt4 (1:500, rabbit, Kerafast, ES1002), GFAP (1:500, rabbit, ZIRC, AB_1−−13806), acetylated tubulin (1:250, mouse, Sigma-Aldrich catalog #T7451), Synapsin 1/2 (1:1000, rabbit, Synaptic Systems, catalog #106 102), SV2A (1:500, mouse, DHSB), and Znp-1 (1:500, mouse, DHSB). The secondary antibodies used in this study include Alexa Fluor 405, goat anti-rabbit (1:600, ThermoFisher, catalog #: A-11034), Alexa Fluor 647, goat anti-rabbit (1:600, ThermoFisher, catalog #: R3121), Alexa Fluor 647 goat anti-chicken (1:600, ThermoFisher, catalog #: A21449), and Alexa Fluor 647, goat anti-mouse (1:600, ThermoFisher, catalog #: A-21235). Staining was performed using the protocol in Nichols & Smith (2019)72. Larvae were fixed at 36 hpf, 48 hpf, 72 hpf, 4 dpf, 5 dpf, 6 dpf, 7 dpf and 15 dpf in fresh 4% PFA in 0.1% PBS Triton-X.
RNAscope in situ hybridization
The probes used were: apoeb (1:50, 80 μL, C3, ACD) and lyve1b (1:50, 80 μL, C2, ACD). RNAScope was performed using the protocol detailed in Kikel-Coury et al., (2020,2021)73,74. Larvae were fixed at 5 dpf in 4% PFA at 25°C for 30 minutes.
Single-cell RNA sequencing analysis
Data Sources
Single-cell RNA-sequencing data from Hammond et al. (2019)29 and Kracht et al. (2020)33 was obtained from the NCBI GEO under the access numbers GSE121654 and GSE141862, respectively.
Hammond et al. (2019) Analysis
Raw UMI count data was obtained from the NCBI GEO database for E14.5, P4, P5, and P30 samples. Analysis was performed using R (v4.1) and Seurat (v4.0.4)75,76. Cells expressing fewer than 400 or greater than 3,000 genes, cells with greater than 10,000 transcripts, and cells with greater than 3% of their transcripts mapping to the mitochondrial genome were excluded from further analysis to eliminate shallow reads, multiplets, and apoptotic cells, respectively. Expression data was log-normalized (UMIs per cell were divided by total UMIs per cell, natural-log transformed, and multiplied by a scale factor of 10,000) and integrated using Harmony77. Dimensionality reduction was performed via UMAP using 40 Harmony-corrected principal components, and hierarchical clustering was performed using HGC78. After removing 11 singlets, and cutting the resulting dendrogram at k = 11, we identified 11 clusters (7 microglia, 1 monocyte/macrophage, 1 endothelial, and 2 neuronal). We then selected the 7 microglia clusters and the monocyte/macrophage cluster for sub-clustering to identify microglial and macrophage subpopulations. We again batch corrected using Harmony and performed dimensionality reduction using UMAP (using 50 principal components), and performed sub-clustering on the resultant 69,862 cells using HGC. After removing 67 singlets which did not cluster, we cut the cluster dendrogram at k = 21 and merged terminal sibling branches when one sibling cluster included less than 600 cells to avoid focusing analysis on very rare cell subtypes. This yielded 16 clusters. One cluster, cluster 3, was identified as neurons on the basis of expression of canonical markers such as Meg3, and Neurod6. The remaining clusters expressed typical macrophage lineage markers. To identify these clusters as either microglia or border-associated macrophages (BAMs) in an unbiased manner, we first identified embryonic microglia and BAM gene signatures from bulk RNA-sequencing data from Utz et al. (2020)34. FASTQ files were obtained from the NCBI GEO database (GSE146928), trimmed with TrimGalore (v0.6.6)79, and mapped with Salmon (v1.5.2)80. DESeq281 was used to identify genes differentially expressed between microglia and BAMs across all embryonic ages. A log2-fold change threshold of ±1 and a multiple comparisons-adjusted p-value threshold of 0.05 were used to determine statistical significance. Significant differentially expressed genes were sorted by the baseMean expression to identify the genes most likely to be detected via scRNA-seq. The top 50 significant microglia markers and top 50 BAM markers by baseMean expression detected in the Hammond et al. (2019)29 single-cell data were used to form the microglia and BAM gene signature sets. Genes in the microglia gene set were excluded if less than 20% of cells expressed the gene, to eliminate genes poorly detected in the dataset. Z-scored average log-normalized expression of genes in these sets across the macrophage-lineage clusters was visualized via heatmaps using the ComplexHeatmap package82, and differential expression testing was performed using Seurat. Per-cell gene set enrichment scores for the microglia and BAM sets were calculated using AUCell83 using default parameters. Clusters 1, and 4–16 were identified as microglia given their higher expression of the microglia gene set and lower expression of the BAM gene set. Cluster 2 was highly enriched in BAM genes and was classified as BAMs. Differential expression testing was performed using Seurat via Wilcoxon rank-sum test, and significance thresholds were set at ± 0.5 log2-fold change and p < 0.01. Microglia (from clusters 1, and 4–16) were further subsetted into Mrc1+ (log-normalized expression > 0) and Mrc1- (log-normalized expression = 0) groups and compared to the cluster 2 BAMs with the same previously used microglia and BAM gene sets. P100 LPC-injected and saline-injected control mouse raw UMI data was also obtained from the NCBI GEO database. Counts were log-normalized, and the proportion of Mrc1+ (log-normalized expression > 0) cells per sample was calculated. Cells from LPC-injected mice were then divided into Mrc1+ (log-normalized expression > 0) and Mrc1- groups and differential expression testing was performed using Seurat and a list of canonical microglia genes.
Kracht et al., (2020) Analysis
UMI count data was obtained from the NCBI GEO database, and analysis was performed using R (v4.1) and Seurat (v4.0.4)75,76. ENSEMBL gene IDs were converted to gene symbols using the EnsDB.Hsapiens.v7984 Bioconductor package. Cells with fewer than 200 or greater than 3000 genes detected, greater than 90,000 reads per cell, or greater than 10% of reads mapping to the mitochondrial genome were removed from downstream analysis. The count data was log-normalized and integrated across sub-samples (separately sequenced runs from each sample) using Harmony77. Dimensionality reduction was performed via UMAP using 15 Harmony-corrected principal components, and hierarchical clustering was performed using HGC78. The clustering dendrogram was cut at k = 7, resulting in four microglia clusters, one neuronal cluster, one erythrocytic cluster, and one macrophage/monocyte cluster, classified by examining cluster-specific differentially expressed genes. The microglia and macrophage clusters were selected for subclustering, reintegrated using Harmony, and dimensionality reduction was again performed via UMAP using 10 corrected principal components followed by hierarchical clustering using HGC. The dendrogram was cut at k = 10, yielding one leukocyte cluster and nine macrophage-lineage clusters. Murine microglia and BAM gene sets were identified from Utz et al. (2020)34, and converted to human orthologs using the biomaRt package85. The top 50 microglia and top 50 BAM genes that were detectable in the Kracht et al. data were retained for gene sets. Gene set expression and enrichment analysis was performed as described above. Clusters 1–7 & 10 were identified as microglia on the basis of higher microglia gene set expression and lower expression of the BAM gene set, while cluster 9 was identified as BAMs and was highly enriched in BAM genes. Microglia (from clusters 1–7 & 10) were divided into Mrc1+ (log-normalized expression > 0) and Mrc1- (log-normalized expression = 0) partitions and DE testing was again performed using Seurat and the microglia and BAM gene sets as described above.
All data plots were generated using the Seurat, ggplot2, ggpubr, or ComplexHeatmap packages in R76,82,86,87.
Chemical Treatments
Lymphatic inhibitor treatments
The chemical reagents used in these experiments were A77–1726 (Millpore Sigma Cat. 100128) at 2μM, cinnarizine (Alfa Aesar Cat. J64568) at 28uM, leflunomide (Alfa Aesar Cat. J65917) at 4μM, and flunarizine (Alfa Aesar Cat. J62969) at 5μM41. Stock solutions of each drug were stored at −20°C with concentrations of 1% DMSO for A77–1726 and cinnarizine. For drugs with additional vehicles, stock solutions were stored at −20°C with concentrations of 20% 2-hydroxypropyl-B-cyclodextrin for flunarizine and 1.5% carboxymethylcellulose (Alfa Aesar Cat A181105) for leflunomide. All treated embryos were dechorionated at 24 hpf and then dosed each day from 24 hpf to 120 hpf. Control animals were also incubated with 1% DMSO in egg water (A77–1726, cinnarizine), 20% 2-hydroxypropyl-B-cyclodextrin in egg water (flunarizine), and 1.5% carboxymethylcellulose in egg water (leflunomide) daily from 24 hpf to 120 hpf. All drug values were selected and all lymphatic treatments were performed following a previous anti-lymphatic drug screen protocol41. Treated and control animals were then fixed and stained with 4C4 at 5 dpf following the protocol in Nichols & Smith (2019)88.
Verteporfin (MedChemExpress, HY-B0146/CS-1950) was reconstituted in 100% DMSO and 10mM stock solutions were stored at −20C. All treated embryos were incubated with 1μM Verteporfin (0.0001% DMSO) from 0–24 hpf and PTU from 24–96 hpf. Verteporfin+640nm animals were dechorionated at 24 hpf, exposed to 640nm light (laser power 2, 100 ms exposure) in a 30μm stack at a step size of 1μm in the forebrain, midbrain, and regions just outside the brain, and then grown until 96 hpf. Exposure to 640nm light was achieved by dorsally mounting dechorionated, Verteporfin-treated animals and angling the animals’ tails upward so that the forebrain and midbrain of the animals were flush with the bottom of the glass dish and therefore exposed to the laser. Positioning for the 30μm stack was achieved by setting a midpoint 15μm into the animal from the most dorsal view of the intracranial lymphatic vessels surrounding the midbrain.
Csf1r inhibitor treatments
The chemical reagent used in this experiment is GW2580 (ApexBio). Stock solutions of 10μM were stored at −20C with concentrations of 1% in DMSO32. All embryos were dechorionated at 24 hpf and incubated with 3 mL egg water until desired treatment time. Animals were treated daily starting at 24 hpf until 144 hpf. Control animals were incubated daily with 1% DMSO in egg water from 24 hpf to 144 hpf.
Generation of transgenics and plasmids
bactin:eos
Gateway cloning was used to generate bactin:eos. The p5e-bactin89, pMe-eos90, 3pe-pA89 and 395 destination vectors were recombined with LR clonase to produce pCS-DFD14 (bactin:eos)89. In one cell Tg(mrc1a:gfp) animals, 1 nl of inject mix composed of 25 ng/ul of bactin:eos and 75 ng/ul of RNA transposase. F0 animals were used for all bactin:eos experiments.
Tg(pu1:eos)
Gateway cloning was used to generate pu1:eos. The p5e-pu162, pMe-eos90, p3e-pA89 and 395 destination vectors were recombined with LR clonase to produce pu1:eos89. One cell AB animals were then injected with 1 nl of injection mix composed of 12.5 ng/ul of pu1:eos and 75 ng/ul of RNA transposase89. F0 animals were screened as Eos+ and then grown to adulthood. F0 animals were crossed to AB and Eos+ progeny were identified then grown to adulthood as the F1 population. All Tg(pu1:eos) experiments were completed on stable transgenic animals that were outcrossed for at least 2 generations.
Photoconversion Experiments
bactin:eos region photoconversions
Tg(mrc1a:egfp) embryos were injected with a bactin:eos construct at the one-cell stage. Embryos were then grown to 24 hpf. Confocal z-stack images of the green v488 and red v561 channels were to confirm positive Eos expression pre-conversion and confirm negative expression in the red channel prior to photoconversion. bactin:eos cells were then photoconverted using 5 ms bursts of v405 nm laser exposure to specified regions on our confocal microscope using mVector. Photoconversions were performed in three groups with one distinct region of the animal converted; the yolk sac, the RBI, and the brain vessels. The brain vessel converted animals involved photoconverting the bactin:eos cells lining the vessels of the brain using the line tool in Slidebook software. Single bactin:eos cells were photoconverted in the RBI and yolk sac regions using the cursor tool in Slidebook software. The RBI region represents the early rostral blood islands located in between the developing head and yolk sac. The bactin:eos cells located along the outside of the yolk sac represent the yolk sac region. Following photoconversion, we took post-photoconversion images in the green v488 and red v561 channels to confirm successful conversion of the photoconverted Eos protein from green v488 to red v561. Animals were then grown to 56 hpf, fixed, and stained with GFP (v647) and GFAP (v405) following the immunohistochemistry protocol in Nichols & Smith (2019). Confocal z-stack images were taken of Eos+ cells in the laterally mounted right side of the brain 0.0027 mm3 imaging window in stained 56 hpf animals.
Tg(pu1:eos) single-cell photoconversions
We created Tg(pu1:eos) animals and crossed them to Tg(mrc1a:egfp) animals. Tg(pu1:eos);Tg(mrc1a:egfp) embryos were grown to 24 hpf. Pre-conversion confocal z-stack images were taken in the green v488 and red v561 channels to confirm there was no non-specific photoconversion. Single pu1+ cells were then photoconverted using a 5 ms v405 nm laser pulse in the embryonic yolk sac at 24 hpf following the single-cell photoconversion protocol in Green and Smith, 201891. Following photoconversion, post-conversion confocal z-stack images were taken in the green v488 and red v561 channels to confirm expression of the converted Eos+ protein. Photoconverted animals were then grown to 56 hpf and then fixed and stained with 4C4 and GFP following the immunohistochemistry protocol in Nichols & Smith (2019)72,88. Confocal z-stack images were taken of photoconverted Eos+ cells in the laterally mounted right side of the brain 0.0027 mm3 imaging window in stained 56 hpf animals. We scored the different expression profiles of mrc1a+,p-Eos+, and 4C4+ cells.
Genetic Perturbation
flt4 genetic perturbation
Two synthetic gRNA duplexes, crRNA;trRNA92, (crRNA - altR system, IDT) were created (5’-CGTTAGCGTTAATCACAAGC-3’) and (5’-AATAACCCGAGTCATTGGCC-3’) that targeted the translated region of flt4 at exon 2 and 3. Tg(mrc1a:egfp) and Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) embryos were injected with a sgRNA at the one-cell stage and imaged at 5 dpf. The sequences of primers for genotyping of Tg(mrc1a:egfp) only animals injected with flt4 gRNAs were: 5’-TAGGTCTGGTGAATGGGTTTTC-3’ (forward, exon 2) and 5’- AAAAGCTCTGTGCTGTGACAAA-3’ (reverse, exon 2); 5’-CTGGCCTGAAGAGTCTTTGAGT-3’ (forward, exon 3) and 5’- CACAGCTCTTACCTCGAACAAA-3’ (reverse, exon 3). Following genomic DNA amplification, PCR products were purified and a T7 endonuclease I digestion (New England Biolabs, Cat# M0302L) to detect mutation was performed. F0 mutants were confirmed by the presence of two bands, an uncut (288bp) and cut (<200bp) band. Wildtype animals were identified by a single uncut (288bp) band. Uninjected and Cas9 only injected Tg(mrc1a:egfp) animals served as controls. Animals were genotyped after scoring to ensure blinded quantification.
spi1b genetic perturbation
A synthetic gRNA duplex, crRNA;trRNA92, (crRNA - IDT) was created (5’ - TGCATCCGTACAGAATGGAGGGG - 3’) that targeted the translated region of spi1b. Tg(mrc1a:egfp); Tg(pu1:Gal4;UAS:rfp) embryos were injected with a sgRNA at the one-cell stage and imaged at 5 dpf. The sequences of primers for genotyping of spi1b were: ACAGTTTTGAAAGCCCTTGAGA (5’ - forward) and TCAAACGCAAAATAATGCAAAC (3’ - reverse). Following genomic DNA amplification, PCR products were purified and a T7 endonuclease I digestion (New England Biolabs, Cat# M0302L) to detect mutation was performed. F0 mutants were confirmed by the presence of two bands, an uncut (288bp) and cut (<200bp) band. Wildtype animals were identified by a single uncut (288bp) band. Mutants in F0 larvae were confirmed by Sanger sequencing of amplified genomic DNA. Uninjected Tg(mrc1a:egfp) animals served as a control. Animals were genotyped after scoring to ensure blinded quantification.
MTZ Ablation
To perform gfap-specific cell ablations, Tg(gfap:nfsb-mCherry) animals tagged with nitroreductase under a gfap promoter were used48,93. A 20mM Metronidazole (MTZ, Fisher Scientific, Cat# AC210340050) stock solution was made and protected from light. All embryos were dechorionated at 24 hpf and incubated with 3 mL egg water until desired treatment time48. Animals were dosed with 10mM MTZ in 0.0003% PTU and 1% DMSO in egg water and then incubated starting at 48 hpf. MTZ was removed at 72 hpf and replaced with 0.0003% PTU and 1% DMSO in egg water until desired imaging time. Control animals were incubated with 0.0003% PTU and 1% DMSO in egg water starting at 24 hpf until desired imaging time.
Quantification and statistical analysis
3i Slidebook software (Denver, CO) was used to generate composite z-images of microglia. All individual z-images were sequentially observed. IMARIS (Notre Dame Imaging Core) was used to create 3D surface renderings of microglia. All graphical data represent both the mean and individual values used in each experiment unless otherwise noted. All quantifications were performed using various plugins available in ImageJ (Bethesda, MD) and Microsoft Excel (Seattle, WA). GraphPad Prism (San Diego, CA) software was used to perform all statistical analysis. Full list of sample sizes, central tendency and variance, statistical tests, and p-values can be found in the Source Data tables.
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications9,32,63,64. All statistical tests were run with biological replicates, not technical replicates. Healthy animals were randomly selected for experiments. No data points were excluded from analysis. Data distribution was assumed to be normal, but this was not formally tested. It is indicated where data collection and analysis performed were blinded. If not indicated, data collection and analysis were not performed blind to the conditions of the experiments. Each experiment was repeated at least twice with similar results.
Quantification of debris puncta
To visualize debris puncta, confocal images were taken of several Tg(mrc1a:egfp) and Tg(pu1:Gal4;UAS:gfp) animals stained with 4C4 and with other debris types labeled in magenta. 3D surface renderings were created of individual mrc1a+ microglia using IMARIS (Notre Dame Imaging Core). To confirm the presence of debris within or outside an mrc1a+ microglia, rotated the images up to 270 degrees in the orthogonal z-plane were used. Magenta puncta present inside green mrc1a+ microglia were considered a type of debris cleared by the microglia population. If magenta puncta were not present inside green mrc1a+ they were considered a type of debris not cleared by the microglia population.
Quantification of cell migration
To create the migration plots and calculate the distance mrc1a+ and pu1+ cells travel post-heterotypic or homotypic contact with another mrc1a+ or pu1+ cell, we timelapse imaged Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) animals from 36–56 hpf, 48–72hpf, and 72–96 hpf. We used the MTrackJ plugin in ImageJ to track the individual paths that mrc1a+ only, pu1+ only and mrc1a+;pu1+ cells traveled prior to contacting another marked cell throughout a 24-hour imaging window. We set the MTrackJ X,Y coordinates of the point of contact between the two cells as the origin for the migration plot. We also used the MTrackJ plugin in ImageJ to track the individual paths that mrc1a+ only, pu1+ only and mrc1a+;pu1+ cells traveled post-contact with another marked cell. The distance equation in Excel was used to measure the distance between the X, Y coordinates of the maximum distance each cell traveled post-contact and the X, Y coordinates of the origin (point of contact).
Quantification of lymphatic vessels
Lymphatic vessel number, length, and number of secondary sprouting event quantifications were represented following the quantitative methods listed in Astin et al. (2014)41. We used the freehand tracing tool in ImageJ to trace and measure the raw length of each lateral lymphatic vessel present in the brain of each DMSO control and lymphatic inhibitor treated animal. All further quantifications were performed using Excel and all statistical analyses were performed using GraphPad Prism.
Quantification of GFAP fluorescence
The mean gray value of mCherry fluorescence in the brain and spinal cord was measured by tracing the brain and spinal cord boundaries using the freehand selection tool in ImageJ. Then, the mean gray value of the background of each individual z-stack image was measured by drawing a small circle (or multiple small circles and averaging between them) in the upper right corner with the freehand selection tool. This data for each image was normalized by subtracting the background mean gray value from the mean gray value inside the brain or spinal cord boundary. When treating with both lymphatic inhibitors and MTZ, fluorescence was normalized to the DMSO control group.
Extended Data
Extended Data Fig. 1. mrc1a+ microglia are distinct from other BLEC and vascular endothelial cells.
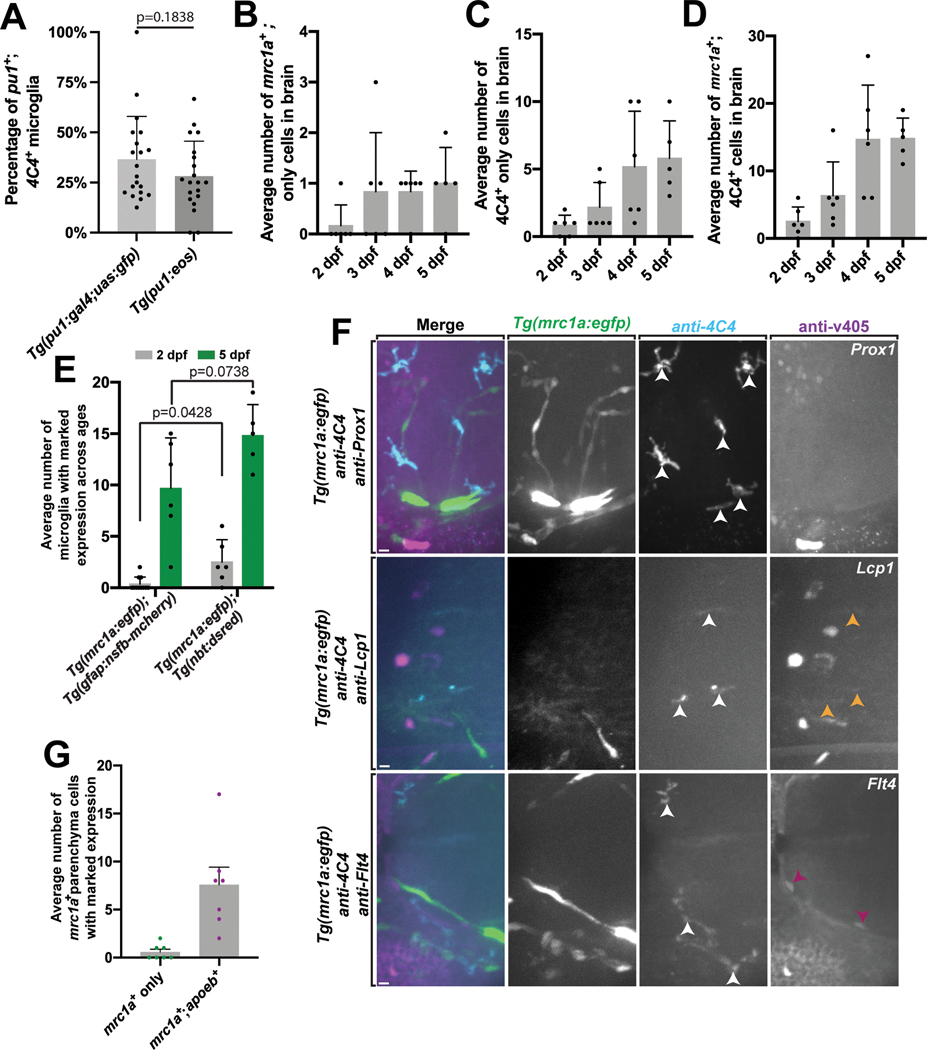
(A) Quantification of the percentage of pu1+;4C4+ microglia in Tg(pu1:Gal4;UAS:rfp) compared to Tg(pu1:eos) animals (t-test: Tg(pu1:gal4-uas:tagrfp) vs Tg(pu1:eos) p=0.1838; two-tailed) (n=40 animals). (B) Quantification of the average number of mrc1a+ only cells in Tg(mrc1a:egfp);Tg(nbt:dsred) animals from 2 dpf - 5 dpf (n=20 animals). (C) Quantification of the average number of 4C4+ only cells in Tg(mrc1a:egfp);Tg(nbt:dsred) animals from 2 dpf - 5 dpf (n=20 animals). (D) Quantification of the average number of mrc1a+;4C4+ cells in Tg(mrc1a:egfp);Tg(nbt:dsred) animals from 2 dpf - 5 dpf (n=20 animals). (E) Quantification of the average number of microglia with marked expression in Tg(mrc1a:egfp);Tg(gfap:nsfb-mCherry) and Tg(mrc1a:egfp);Tg(nbt:dsred) animals at 2 dpf and 5 dpf (t-test: 2 dpf Tg(mrc1a:egfp);Tg(gfap:nsfb-mcherry) vs. Tg(mrc1a:egfp);Tg(nbt:dsred) p=0.0428, 5 dpf Tg(mrc1a:egfp);Tg(gfap:nsfb-mcherry) vs. Tg(mrc1a:egfp);Tg(nbt:dsred) p=0.0738; all two-tailed)(n=28 animals). (F) Confocal z-projection images of Tg(mrc1a:egfp) animals at 5 dpf stained with 4C4, Lcp1, Prox1, and Flt4. White arrowheads indicate 4C4+ microglia. Orange arrowheads indicate a small number of 4C4+;Lcp1+ microglia. Purple arrowheads indicate Flt4+ cells located along the mrc1a+ vessel endothelium that are not 4C4+. (G) Quantification of the average number of mrc1a+ parenchyma cells with mrc1a+ only expression compared to mrc1a+;apoeb+ microglia (n=7 animals). Imaging window represents one 0.0027 mm3 region per animal (A-G). Scale bar equals 10μm (B).
Extended Data Fig. 2. mrc1a+ microglia function like traditional microglia.
(A) Confocal z-projection images of 5 dpf Tg(mrc1a:egfp) animals stained with antibodies or other transgenic animals to label debris from synaptic, neuronal, oligodendrocyte, microglia, or astroglia populations. White boxes indicate regions of engulfed debris puncta. Arrows indicate individual debris puncta within mrc1a+ microglia. (B) Confocal z-projection images of 5 dpf Tg(pu1:Gal4;UAS:gfp) animals stained with same antibodies /transgenic animals represented in (A). Arrows indicate individual debris puncta within mrc1a+ microglia. (C) Confocal z-projection still images from a 24 hour timelapse of Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) animals from 4 dpf to 5 dpf showing homotypic interactions between mrc1a+ and mrc1a+ microglia and pu1+ and pu1+ microglia populations. White arrowheads indicate mrc1a+ microglia and blue arrowheads indicate pu1+ microglia. Dashed yellow box indicates contact point for two microglia. White arrowheads indicate mrc1a+ microglia. Blue arrowheads indicate pu1+ microglia. (D) Quantification of the migration path two individual mrc1a+ microglia traveled pre and post contact (n=7 animals). (E) Quantification of the migration path two individual pu1+ microglia traveled pre and post contact (n=7 animals). Imaging window equals 0.0027 mm3 (A,B), 0.0081 mm3 (C-E). Scale bar equals 10μm (A,B), 100μm (C).
Extended Data Fig. 3. Mrc1 is expressed in developmental microglia in the mammalian brain.
Panels A-D refer to analysis of data from Hammond et al. (2019)36. (A) UMAP of initial clustering of all cells from E14, P4 & P5, and P30 from Hammond et al (2019). Clusters A-G represent microglia; cluster H is macrophages/monocytes; cluster I is endothelial cells; and clusters J and K are neuronal. (B) Violin plot of log-normalized expression of Mrc1, microglia markers (Tmem119, P2ry12), macrophage/monocyte markers (F13a1, Ccr1, Ccr2), endothelial markers (Cldn5, Vtn, Pecam1), and neuronal markers (Neurod6, Nfib, Elavl3). Clusters A-H were chosen for subclustering to identify microglial subpopulations. (C) Heatmap of z-scored average expression of the top 10 differentially expressed genes for each of the 16 clusters identified in the second round of clustering of microglia and macrophages. (D) Violin plot comparing expression of Spi1 (the gene encoding the PU.1 transcription factor) in Mrc1+ and Mrc1− microglia (both aggregated from clusters 1 & 4–16). Panels E-H refer to analysis of data from Kracht et al., (2020)41. (E) UMAP of initial clustering of all cells from Kracht et al. (2020). Clusters A-D are microglia; cluster E is monocytes/macrophages; cluster F is neurons; and cluster G is erythrocytes. (F) Violin plots of log-normalized expression of canonical microglia, macrophage/monocyte, neuronal, and erythrocytic genes across the seven initial clusters. Clusters A-E were chosen for subclustering to identify microglia and macrophage subpopulations. (G) Heatmap of z-scored average expression of top 10 differentially expressed genes per cluster across all 10 clusters identified in the sub-clustering of microglia and macrophages from Kracht et al. (2020). (H) Violin plot comparing expression of SPI1 (the gene encoding the PU.1 transcription factor) in MRC1+ and MRC1− microglia (both aggregated from clusters 1 & 4–16). Points were added for single cells as a low proportion of cells in both groups expressed detectable MRC1. For (D) and (H) P-value refers to a Wilcoxon rank-sum test with Bonferroni correction for multiple comparisons. “Avg. log2FC” refers to the average log2-fold change in expression between Mrc1/MRC1+ and Mrc1/MRC1− microglia. Positive values refer to higher expression in Mrc1/MRC1+ microglia. The percentage of cells in each group expressing at least one detected read of Mrc1/MRC1 is shown in parentheses. Statistical significance in differential expression testing was determined by Wilcoxon rank-sum test (ɑ = 0.05, with Bonferroni correction) and an average log fold-change threshold of ±0.5.
Extended Data Fig. 4. mrc1a+ microglia are dependent on lymphangiogenesis.
(A) Representative images of two orthogonal rotations of confocal z-projection (left) and IMARIS 3D surface rendering (right) of 6 dpf Tg(mrc1a:egfp) animals showing a secondary sprout of a growing lymphatic vessel. White arrows indicate vessel secondary sprout site. Blue arrowheads indicate hollow vessel center. (B) Representative confocal z-projections of Tg(mrc1a:egfp) animals stained with 4C4 showing the reduction of mrc1a+;4C4+ microglia in cinnarizine, flunarizine, and leflunomide treated animals compared to DMSO control animals. Blue arrowheads represent mrc1a+;4C4+ microglia. (C) Representative confocal z-projections of 5 dpf Tg(mrc1a:egfp) animals showing the disruption of vessel growth and development in animals treated with A77–1726, cinnarizine, flunarizine, or leflunomide compared to control DMSO animals. (D) Quantification showing the reduced average length of brain lymphatic vessels in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. A77–1726 p=0.0444, Mean diff=22.94, DF-358, q=2.622, SE of diff=8.749; DMSO vs. cinnarizine p=0.2378, Mean diff=16.62, DF=358, q=1.937, SE of diff=8.592; DMSO vs. flunarizine p=0.0060, Mean diff=35.94, DF=358, q=3.263, SE of diff=11.02; DMSO vs. leflunomide p=0.0008, Mean diff=25.83, DF=358, q=3.81, SE of diff=6.78) (n=80 animals) (E) Quantification showing the reduced average number of secondary sprouts in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. A77–1726 p=0.0207, mean diff =1.8, DF=63, q=2.966, SE of diff=0.6068; DMSO vs. cinnarizine p=0.0012, Mean diff=2.371, DF=63, q=3.908, SE of diff=0.6068; DMSO vs. flunarizine p=0.0178, Mean diff=2.336, DF=63, q=3.019, SE of diff=0.7736; DMSO vs. leflunomide p=0.0004, Mean diff=2.308, DF=63, q=4.213, SE of diff=0.5478)(n=80 animals). (F) Quantification showing the reduced average number of lymphatic vessels surrounding the brain in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. A77–1726, DMSO vs. cinnarizine p=0.0081, Mean diff=3.242, DF=64, q=3.29, SE of diff=0.9854; DMSO vs. cinnarizine p=0.0124, Mean diff=3.099, DF=64, q=3.145, SE of diff=0.9854; DMSO vs. flunarizine p=0.0883, Mean diff=3.028, DF=64, q=2.408, SE of diff=1.257; DMSO vs. leflunomide p=0.1368, Mean diff=1.972, DF=64, q=2.218, SE of diff=0.889)(n=80 animals). (G) Quantification of the number of 4C4+ only microglia in DMSO control animals compared to leflunomide and flunarizine treated animals (t-test: DMSO vs. leflunomide, DMSO vs. flunarizine (Ordinary one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. leflunamide p=0.9997, Mean diff=−0.04274, DF=25, q=0.01816, SE of diff=2.353; DMSO vs. flunarizine p=0.2097, Mean diff=4.346, DF=25, q=1.623, SE of diff=2.678)(n=29 animals). (H) Quantification of the number of pu1+ only microglia in DMSO control animals compared to leflunomide and flunarizine treated animals (Ordinary one-way ANOVA/Dunnett’s multiple comparisonst: DMSO vs. leflunamide p=0.9932, Mean diff=−0.2222, DF=36, q=0.09986, SE of diff=2.225; DMSO vs. flunarizine p=0.6213, Mean diff=−2.167, DF=25, q=0.8554, SE of diff=2.533)(n=29 animals). (I) Quantification of the number of pu1+;4C4+ microglia in DMSO control animals compared to leflunomide and flunarizine treated animals (Ordinary one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. Leflunamide p=0.7997, Mean diff=−1.342, DF-25, q=1.19, SE of diff=1.167; DMSO vs. Flunarizine p=0.4353, Mean diff=0.7692, DF=25, q=0.5789, SE of diff=1.329)(n=29 animals). Imaging window equals 0.0027 mm3 (A-F), 0.0081 mm3 or 3000μm (G-I). Scale bar equals 10μm (A-C).
Extended Data Fig. 5. mrc1a+ microglia are dependent on lymphangiogenesis.
(A) Quantification of the total number of mrc1a+ only microglia in uninjected and Cas9 only animals compared to flt4 gRNA injected animals (one-way ANOVA/Dunnett’s multiple comparisons: uninjected vs. Cas9 only injected p=0.9389, Mean diff==0.08359, DF=18, q=0.4783, SE of diff=0.2472; Cas9 only injected vs. flt4 gRNA/Cas9 injected p=0.9377, Mean diff=−0.003759, DF=118, q=0.02349, SE of diff=0.2263; uninjected vs. flt4 gRNA/Cas9 injected p=0.9998, Mean diff=0.07983, DF=118, q=0.4831, SE of diff=0.2337)(n=122 animals). (B) Quantification of the total number of 4C4+ only microglia in uninjected and Cas9 only animals compared to flt4 gRNA injected animals (one-way ANOVA/Dunnett’s multiple comparisons: uninjected vs. Cas9 only injected p=0.0573, Mean diff=−1.423, DF=118, q=3.277, SE of diff=0.614; Cas9 only injected vs. flt4 gRNA/Cas9 injected p=0.6773, Mean diff=−0.9334, DF=118, q=2.348, SE of diff=0.5622; uninjected vs. flt4 gRNA/Cas9 injected p=0.2249, Mean diff=0.4892, DF=118, q=1.192, SE of diff=0.5805)(n=122 animals). (C) Confocal z-projections of the brain, RBI, and Yolksac regions in Tg(mrc1a:egfp) animals injected with bactin:eos pre and post-photoconversion. Purple arrowheads indicate successfully photoconverted pEos+ cells. (D) Quantification of the average number of mrc1a+ only cells in the brain at 56 hpf following photoconversion of bactin:eos in the brain vessels, RBI, and yolk sac (t-test: bra(n=17 animals) in vs. RBI p=0.9467, RBI vs. yolksac p=0.8988, brain vs. yolksac p=0.8611;all two-tailed). Imaging window equals 0.0027 mm3 (A-D). Scale bar equals 10μm (C).
Extended Data Fig. 6. mrc1a+ microglia are distinct from yolk-sac derived microglia.
(A) Quantification of the average number of mrc1a+ only cells in the brain imaging window over time (n=69 animals). (B) Quantification of the average number of 4C4+ only cells in the brain imaging window over time. (C) Quantification of the average number of pu1+ only cells in the brain imaging window over time (n=69 animals). (D) Quantification of the average number of mrc1a+;4C4+ cells in the brain imaging window over time (n=69 animals). (E) Quantification of the average number of mrc1a+;pu1+ cells in the brain imaging window over time (n=69 animals). (F) Quantification of the average number of mrc1a+;pu1+;4C4+ cells in the brain imaging window over time (n=69 animals). (G) Quantification of compiled average number of mrc1a+ only, 4C4+ only, pu1+ only, and mrc1a+;4C4+ cells over time. (H) Quantification of the total number of mrc1a+;pu1+ cells per imaging window in uninjected animals compared to spi1b sgRNA injected animals (t-test: uninjected vs. spi1b gRNA/Cas9 injected p=0.0003’two-tailed)(n=41 animals). (I) Quantification of the total number of 4C4+ only cells per imaging window in uninjected animals compared to spi1b sgRNA injected animals (t-test: uninjected vs. spi1b/Cas9 injected p=0.2849;two-tailed)(n=41 animals). (J) Representative confocal z-projection images of Tg(mrc1a:egfp); Tg(gfap:nsfb-mCherry) and Tg(pu1:Gal4;UAS:rfp);Tg(gfap:nsfb-mCherry) animals stained with 4C4 showing the reduction of pu1+ microglia and no change in the mrc1a+;4C4+ microglia in the GW2580 treated animals compared to DMSO control animals. Blue arrowheads: represent mrc1a+;4C4+ microglia. Purple arrowheads represent pu1+ microglia. (K) Confocal z-projections of single pu1 cells in the embryonic yolksac of Tg(mrc1a:egfp);Tg(pu1:eos) animals at 24 hpf pre and post-photoconversion. Purple arrowheads indicate successfully photoconverted pEos+ cells. Imaging window equals 0.0027 mm3 (A-G, J-K), 0.0081 mm3 or 3000μm (H-I). Scale bar equals 10μm (J,K).
Extended Data Fig. 7. Injury paradigms alter mrc1a+ microglia and expression of mammalian Mrc1 during injury.
(A) Quantification of spinal cord mCherry intensity in MTZ-treated animals across time from 1 to 4 dpi compared to DMSO treated zebrafish animals (t-test, two-tailed; multiple comparisons corrected: 1 dpi DMSO vs. MTZ p<0.0001, 2 dpi DMSO vs. MTZ p<0.0001, 3 dpi DMSO vs. MTZ p<0.0001, 4 dpi DMSO vs. MTZ p<0.0001; all two-tailed)(n=70 animals). (B) Stacked violin plot of microglia from Hammond et al. (2019) comparing canonical microglia marker gene expression between Mrc1+ and Mrc1− microglia from P100 LPC-injected animals. Microglia were subsetted as Mrc1+ if log normalized expression of Mrc1 was > 0. (C) Differential expression testing results table comparing expression of canonical microglia markers in Mrc1+ versus Mrc1− microglia from Hammond et al. (2019). In (C) “Log Fold Change” refers to natural log fold-change, with positive values indicating higher expression in Mrc1+ microglia versus Mrc1− microglia. “Mrc1+ microglia with expression” and “Mrc1- microglia with expression” report the percentage of Mrc1+ and Mrc1− cells, respectively, with at least one read of the gene detected. Statistical significance was determined by Wilcoxon rank-sum test and an average log fold-change threshold of ±0.5. Imaging window equals 0.0027 mm3 (A).
Extended Data Fig. 8. mrc1a+ microglia are dependent on lymphangiogenesis during injury.
(A) Quantification of the normalized fluorescence value of DMSO control animals compared to lymphatic inhibitor treated animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77–1726 p=0.997, DMSO vs. cinnarizine p=0.4055, DMSO vs. flunarizine p=0.0003, DMSO vs. leflunomide p=0.0533)(n=32 animals). (B) Representative confocal z-projections of 5 dpf Tg(mrc1a:egfp) animals showing the disruption of vessels in animals treated with A77–1726 + MTZ, cinnarizine + MTZ, or flunarizine + MT compared to control DMSO + MTZ control animals. (C) Quantification of the average length of brain lymphatic vessels in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77–1726 p=0.6526,Mean diff=1.962, DF=44, q=4.081, SE of diff=0.4807; DMSO vs. cinnarizine p=0.6011, Mean diff=3.845, DF=44, q=3.845, SE of diff=0.5056; DMSO vs. flunarizine p=0.0482, Mean diff=1.887, DF=44, q=3.265, SE of diff=0.5779; DMSO vs. leflunomide p=0.0702, Mean diff=2.087, DF=44, q=3.887, SE of diff=0.5369)(n=72 animals). (D) Quantification showing the reduced average number of secondary sprouts (Figure S4B) in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77–1726 p=0.0007, Mean diff=3.542, DF=45, q=3.459, SE of diff=.024; DMSO vs. cinnarizine p=0.0015, Mean diff=1.944, DF=45, q=3.845, SE of diff=0.5056; DMSO vs. flunarizine p=0.0083, Mean diff=2.842, DF=45, q=2.305, SE of diff=1.233; DMSO vs. leflunomide p=0.0014, Mean diff=3.375, DF=45, q=2.948, SE of diff=1.145)(n=72 animals). (E) Quantification of the average number of lymphatic vessels that surround the brain in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77–1726 p=0.0047, DMSO vs. cinnarizine p=0.0271, DMSO vs. flunarizine p=0.0953, DMSO vs. leflunomide p=0.0196)(n=72 animals). (F) Quantification of the number of 4C4+ only microglia in DMSO + MTZ control animals compared to leflunomide + MTZ and flunarizine + MTZ treated animals (One-way ANOVA Dunnett’s multiple comparisons: DMSO vs. leflunomide p=0.0038, DMSO vs. flunarizine p=0.0222)(n=29 animals). (G) Quantification of the number of pu1+ only microglia in DMSO + MTZ control animals compared to leflunomide + MTZ and flunarizine + MTZ treated animals (One-way ANOVA Dunnett’s multiple comparisons: DMSO vs. leflunomide, DMSO vs. flunarizine p=0.7053, Mean diff=32.76, q= 3.554, SE of diff=9.22; DMSO vs. flunarizine p=0.5165, Mean diff=27.72, q=2.772, SE of diff=10)(n=29 animals). (H) Quantification of the number of pu1+;4C4+ microglia in DMSO + MTZ control animals compared to leflunomide + MTZ and flunarizine + MTZ treated animals (One-way ANOVA Dunnett’s multiple comparisons: DMSO vs. leflunomide p=0.1400, Mean diff=1.75, q=0.723, SE of diff=2.421; DMSO vs. flunarizine p=0.2981, Mean diff=−2.667, q=1.012, SE of diff=2.635) (n=29 animals) (I) Representative confocal z-projections of 6 dpf Tg(mrc1a:egfp);Tg(gfap:nsfb-mCherry) animals stained with 4C4 showing the reduction of mrc1a+;4C4+ microglia in A77–1726 + MTZ, cinnarizine + MTZ, flunarizine + MTZ, and leflunomide + MTZ, treated animals compared to DMSO + MTZ control animals. Blue arrowheads represent mrc1a+4C4+ microglia. Imaging window equals 0.0027 mm3 (A-E,I), 3000μm (F-H). Scale bar equals 10μm (B,I).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brant Weinstein (NIH) for sending us the Tg(mrc1a:egfp) animals, Dirk Seiger for p5e-pu1, and Wilson Clements for Tg(lck:gfp) and Tg(fli1:gfp) animals. We thank Beth Stevens, Tim Hammond, Kelly Monk, Chris Bennett, and Siyuan Zhang for their helpful comments and reagent guidance. We also thank Brent Redford, Sam Connell and 3i for imaging related questions, Sara Cole in the NDiiF Optical Microscopy Core for help with light sheet imaging (OMC/NDIIF and the National Science Foundation-Major Research Instrumentation Program 1919832) and IMARIS analysis, and Deborah Bang, Karen Heed, and Brittany Gervais for zebrafish housing and upkeep. This work was supported by the University of Notre Dame, the Elizabeth and Michael Gallagher Family, Centers for Zebrafish Research and Stem Cells and Regenerative Medicine at the University of Notre Dame, the Indiana Spinal Cord and Brain Injury Research with the Indiana State Board of Health(CJS), the Alfred P. Sloan Foundation(CJS), and the NIH (DP2NS117177)(CJS). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
Supplemental Data
All supplemental data are provided in Source Data files for each main and Extended Data figure that contain all raw n-values, data values, and statistical data exported from GraphPad Prism (version 8). These data also contain results for normality tests. For all datasets that did not pass normality tests, secondary unpaired nonparametric t-test analysis were used and yielded similar p-values.
REFERENCES:
- 1.Miyamoto A. et al. Microglia contact induces synapse formation in developing somatosensory cortex. Nat. Commun. 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schafer D. et al. Microglia sculpt postnatal neuronal circuits in an activivty and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paolicelli RC et al. Synaptic pruning by microglia is necessary for normal brain development. Science (80-. ). 333, 1456–1458 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Cunningham CL, Martinez-Cerdeno V. & Noctor SC Microglia Regulate the Number of Neural Precursor Cells in the Developing Cerebral Cortex. J. Neurosci. 33, 4216–4233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marín-Teva JL et al. Microglia Promote the Death of Developing Purkinje Cells. Neuron 41, 535–547 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Wakselman S. et al. Developmental neuronal death in hippocampus requires the microglial CD11b integrin and DAP12 immunoreceptor. J. Neurosci. 28, 8138–8143 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueno M. et al. Layer v cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 16, 543–551 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y. & Sato K. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 34, 2231–2243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes Alexandria N. & Appel Bruce. Microglia phagocytose myelin sheaths to modify developmental myelination. Nat. Neurosci. (2020). doi: 10.1016/j.expneurol.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi BH Hematogenous cells in the central nervous system of developing human embryos and fetuses. J. Comp. Neurol. 196, 683–694 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Andjelkovic AV, Nikolic B, Pachter JS & Zecevic N. Macrophages/microglial cells in human central nervous system during development: An immunohistochemical study. Brain Res. 814, 13–25 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Monier A. et al. Entry and distribution of microglial cells in human embryonic and fetal cerebral cortex. J. Neuropathol. Exp. Neurol. 66, 372–382 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Verney C, Monier A, Fallet-Bianco C. & Gressens P. Early microglial colonization of the human forebrain and possible involvement in periventricular white-matter injury of preterm infants. J. Anat. 217, 436–448 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menassa DA & Gomez-Nicola D. Microglial dynamics during human brain development. Front. Immunol. 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monier A, Evrard P, Gressens P. & Verney C. Distribution and Differentiation of Microglia in the Human Encephalon during the First Two Trimesters of Gestation. J. Comp. Neurol. 499, 565–582 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Ginhoux F. et al. Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science (80-. ). 330, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbom P, Thisse B. & Thisse C. Zebrafish Early Macrophages Colonize Cephalic Mesenchym e and D eveloping Brain, Retina, and Epiderm is through a M-CSF Receptor-D ependent Invasive Process. Dev. Biol. 288, 274–288 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Casano AM, Albert M. & Peri F. Developmental Apoptosis Mediates Entry and Positioning of Microglia in the Zebrafish Brain. Cell Rep. 16, 897–906 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Xu J. et al. Temporal-Spatial Resolution Fate Mapping Reveals Distinct Origins for Embryonic and Adult Microglia in Zebrafish. Dev. Cell 34, 632–641 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Ferrero G. et al. Embryonic Microglia Derive from Primitive Macrophages and Are Replaced by cmyb-Dependent Definitive Microglia in Zebrafish. Cell Rep. 24, 130–141 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Chen SK et al. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell 141, 775–785 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H. et al. Fate mapping via CCR2-CreER mice reveals monocyte-to-microglia transition in development and neonatal stroke. 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohnmacht J. et al. Spinal motor neurons are regenerated after mechanical lesion and genetic ablation in larval zebrafish. Development 143, 1464–1474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peri F. & Nüsslein-Volhard C. Live Imaging of Neuronal Degradation by Microglia Reveals a Role for v0-ATPase a1 in Phagosomal Fusion In Vivo. Cell 133, 916–927 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Jung HM et al. Development of the larval lymphatic system in zebrafish. Development 144, 2070–2081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Lessen M. et al. Intracellular uptake of macromolecules by brain lymphatic endothelial cells during zebrafish embryonic development. Elife 6, 1–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galanternik MV et al. A novel perivascular cell population in the zebrafish brain. Elife 6, 1–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin M. et al. Vegfc acts through ERK to induce sprouting and differentiation of trunk lymphatic progenitors. Dev. 144, 531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond TR et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plein A, Fantin A, Denti L, Pollard JW & Ruhrberg C. Erythro-myeloid progenitors contribute endothelial cells to blood vessels. Nature 562, 223–228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koltowska K. et al. Vegfc Regulates Bipotential Precursor Division and Prox1 Expression to Promote Lymphatic Identity in Zebrafish. Cell Rep. 13, 1828–1841 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Green LA, Nebiolo JC & Smith CJ Microglia exit the CNS in spinal root avulsion. PLOS Biol. 17, e3000159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kracht L. et al. Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science (80-. ). 369, 530–537 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Utz SG et al. Early Fate Defines Microglia and Non-parenchymal Brain Macrophage Development. Cell 181, 557–573.e18 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Kierdorf K. et al. Microglia emerge from erythromyeloid precursors via Pu. 1- and Irf8-dependent pathways. Nat. Publ. Gr 16, 273–280 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Bennett ML et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. 113, E1738–E1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butovsky O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbomel P, Thisse B. & Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Goldmann T. et al. Origin, fate and dynamics of macrophages at CNS interfaces HHS Public Access. Nat Immunol 17, 797–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith CJ, Morris AD, Welsh TG & Kucenas S. Contact-Mediated Inhibition Between Oligodendrocyte Progenitor Cells and Motor Exit Point Glia Establishes the Spinal Cord Transition Zone. PLoS Biol. 12, e1001961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astin JW et al. An in vivo antilymphatic screen in zebrafish identifies novel inhibitors of mammalian lymphangiogenesis and lymphatic-mediated metastasis. Mol. Cancer Ther. 13, 2450–2462 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Zhang L. et al. VEGFR-3 ligand-binding and kinase activity are required for lymphangiogenesis but not for angiogenesis. Cell Res. 20, 1319–1331 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Tammela T. et al. Photodynamic ablation of lymphatic vessels and intralymphatic cancer cells prevents metastasis. Sci. Transl. Med. 3, (2011). [DOI] [PubMed] [Google Scholar]
- 44.Kilarski WW et al. Optimization and regeneration kinetics of lymphatic-specific photodynamic therapy in the mouse dermis. Angiogenesis 17, 347–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elmore MRP et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erblich B, Zhu L, Etgen AM, Dobrenis K. & Pollard JW Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One 6, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q. & Barres BA Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. (2017). doi: 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- 48.Smith CJ, Johnson K, Welsh TG, Barresi MJF & Kucenas S. Radial glia inhibit peripheral glial infiltration into the spinal cord at motor exit point transition zones. Glia 64, 1138–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuadros MA, Martin C, Coltey P, Almendros A. & Navascués J. First appearance, distribution, and origin of macrophages in the early development of the avian central nervous system. J. Comp. Neurol. 330, 113–129 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Gritz E. & Hirschi KK Specification and function of hemogenic endothelium during embryogenesis. Cell. Mol. Life Sci. 73, 1547–1567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lancrin C. et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457, 892–895 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swiers G. et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 4, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boisset JC et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Jaffredo T, Gautier R, Eichmann A. & Dieterlen-Lièvre F. Intra-aortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 125, 4575–4583 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Jordan HE Evidence of hemogenic capacity of endothelium. Anat. Rec. 10, 417–420 (1916). [Google Scholar]
- 56.Nakano H. et al. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat. Commun. 4, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z. et al. Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell 11, 663–675 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Cugurra A, Mamuladze T, Rustenhoven J, Dykstra T, Beroshvili G, Greenberg ZJ, Baker W, Papadopoulos Z, Drieu A, Blackburn S, Kanamori M, Brioschi S, Herz J, Schuettpelz LG, Colonna M, Smirnov I, Skull JK and vertebral bone marrow are myeloid reservoirs for the meninges and CNS parenchyma. Science (80-. ). in press, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Da Mesquita S. et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature 560, 185–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Louveau A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castranova D. et al. Live Imaging of Intracranial Lymphatics in the Zebrafish. Circ. Res. 128, 42–58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sieger D, Moritz C, Ziegenhals T, Prykhozhij S, Peri F. Long-Range Ca2+ Waves Transmit Brain-Damage Signals to Microglia. Dev Cell. 2012;22: 1138–1148. doi: 10.1016/j.devcel.2012.04.012 [DOI] [PubMed] [Google Scholar]
- 63.Kucenas S, Wang W-D, Knapik EW, Appel B. A Selective Glial Barrier at Motor Axon Exit Points Prevents Oligodendrocyte Migration from the Spinal Cord. J Neurosci. 2009;29: 15187–15194. doi: 10.1523/JNEUROSCI.4193-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9: 1506–1511. doi: 10.1038/nn1803 [DOI] [PubMed] [Google Scholar]
- 65.Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251: 45–58. doi: 10.1006/dbio.2002.0820 [DOI] [PubMed] [Google Scholar]
- 66.McGraw HF, Snelson CD, Prendergast A, Suli A, Raible DW. Postembryonic neuronal addition in Zebrafish dorsal root ganglia is regulated by Notch signaling. Neural Dev. 2012;7: 23. doi: 10.1186/1749-8104-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JPD, Kanki JP, et al. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A. 2004;101: 7369–7374. doi: 10.1073/pnas.0402248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248: 307–318. doi: 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- 69.Rosenberg AF, Wolman MA, Franzini-Armstrong C, Granato M. In Vivo Nerve-Macrophage Interactions Following Peripheral Nerve Injury. J Neurosci. 2012;32: 3898–3909. doi: 10.1523/JNEUROSCI.5225-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203: 253–310. doi: 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 71.Nichols EL, Green LA, Smith CJ. Ensheathing cells utilize dynamic tiling of neuronal somas in development and injury as early as neuronal differentiation. Neural Dev. Neural Development; 2018;13: 19. doi: 10.1186/s13064-018-0115-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nichols EL, Smith CJ. Pioneer axons employ Cajal’s battering ram to enter the spinal cord. Nat Commun. Springer US; 2019;10: 562. doi: 10.1038/s41467-019-08421-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kikel-Coury NL, Brandt JP, Correia IA, O’Dea MR, DeSantis DF, Sterling F, et al. Identification of astroglia-like cardiac nexus glia that are critical regulators of cardiac development and function. PLoS Biol. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kikel-Coury NL, Green LA, Nichols EL, Zellmer AM, Pai S, Hedlund SA, et al. Pioneer Axons Utilize a Dcc Signaling-Mediated Invasion Brake to Precisely Complete Their Pathfinding Odyssey. J Neurosci. 2021;41: 6617–6636. doi: 10.1523/JNEUROSCI.0212-21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. In: Vienna, Austria: [Internet]. 2021. Available: https://www.r-project.org/ [Google Scholar]
- 76.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. Cell; 2021;184: 3573–3587.e29. doi: 10.1016/J.CELL.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. Springer US; 2019;16: 1289–1296. doi: 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zou Z, Hua K, Zhang X. HGC: fast hierarchical clustering for large-scale single-cell data. Bioinformatics. Oxford Academic; 2021;37: 3964–3965. doi: 10.1093/BIOINFORMATICS/BTAB420 [DOI] [PubMed] [Google Scholar]
- 79.Krueger F, James F, Ewels P, Afyounian E, Schuster-Boeckler B. FelixKrueger/TrimGalore: v0.6.7 - DOI via Zenodo. 2021; doi: 10.5281/ZENODO.5127899 [DOI] [Google Scholar]
- 80.Patro R. Salmon provides fast and bias-aware quantification of transcript expression. 2017; doi: 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. BioMed Central Ltd; 2014;15: 1–21. doi: 10.1186/S13059-014-0550-8/FIGURES/9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. doi: 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 83.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017;14: 1083–1086. doi: 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bioconductor - EnsDb.Hsapiens.v79 [Internet]. [cited 29 Apr 2022]. Available: https://bioconductor.org/packages/release/data/annotation/html/EnsDb.Hsapiens.v79.html
- 85.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. 2009; doi: 10.1038/nprot.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wickham H. ggplot2. Cham: Springer International Publishing; 2016; doi: 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- 87.Kassambara A. ggpubr: “ggplot2” based publication ready plots. [Internet]. 2021. Available: https://rdrr.io/cran/ggpubr/ [Google Scholar]
- 88.Nichols EL, Smith CJ. Synaptic-like Vesicles Facilitate Pioneer Axon Invasion. Curr Biol. Elsevier Ltd; 2019; 1–13. doi: 10.1016/j.cub.2019.06.078 [DOI] [PubMed] [Google Scholar]
- 89.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: A multisite gateway-based construction Kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236: 3088–3099. doi: 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- 90.Prendergast A, Linbo TH, Swarts T, Ungos JM, McGraw HF, Krispin S, et al. The metalloproteinase inhibitor Reck is essential for zebrafish DRG development. Development. 2012;139: 1141–1152. doi: 10.1242/dev.072439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Green L, Smith CJ. Single-cell Photoconversion in Living Intact Zebrafish. J Vis Exp. 2018; e57024–e57024. doi: 10.3791/57024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoshijima K, Jurynec MJ, Klatt Shaw D, Jacobi AM, Behlke MA, Grunwald DJ. Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev Cell. Elsevier Inc; 2019;51: 645–657.e4. doi: 10.1016/j.devcel.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson K, Barragan J, Bashiruddin S, Smith CJ, Tyrrell C, Parsons MJ, et al. Gfap-positive radial glial cells are an essential progenitor population for later-born neurons and glia in the zebrafish spinal cord. Glia. 2016;64: 1170–1189. doi: 10.1002/glia.22990 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that supports the findings of this study are available in the Source Data tables. All data collected for the study are included in the figures. All code for the single-cell RNA sequencing data analysis in this manuscript can be accessed at https://github.com/michael-r-odea/Green_ODea_2022.