Abstract
The management of metastatic estrogen receptor (ER) positive HER2 negative breast cancer (ER+) has improved; however, therapeutic resistance and disease progression emerges in majority of cases. Using unbiased approaches, as expected PI3K and MTOR inhibitors emerge as potent inhibitors to delay proliferation of ER+ models harboring PIK3CA mutations. However, the cytostatic efficacy of these drugs is hindered due to marginal impact on the expression of cyclin D1. Different combination approaches involving the inhibition of ER pathway or cell cycle result in durable growth arrest via RB activation and subsequent inhibition of CDK2 activity. However, cell cycle alterations due to RB loss or ectopic CDK4/cyclin D1 activation yields resistance to these cytostatic combination treatments. To define means to counter resistance to targeted therapies imparted with RB loss; complementary drug screens were performed with RB-deleted isogenic cell lines. In this setting, RB loss renders ER+ breast cancer models more vulnerable to drugs that target DNA replication and mitosis. Pairwise combinations using these classes of drugs defines greater selectivity for RB deficiency. The combination of AURK and WEE1 inhibitors, yields synergistic cell death selectively in RB-deleted ER+ breast cancer cells via apoptosis and yields profound disease control in vivo. Through unbiased efforts the XIAP/CIAP inhibitor birinapant was identified as a novel RB-selective agent. Birinapant further enhances the cytotoxic effect of chemotherapies and targeted therapies used in the treatment of ER+ breast cancer models selectively in the RB-deficient setting. Using organoid culture and xenograft models, we demonstrate the highly selective use of birinapant based combinations for the treatment of RB-deficient tumors. Together, these data illustrate the critical role of RB-pathway in response to many agents used to treat ER+ breast cancer, whilst informing new therapeutic approaches that could be deployed against resistant disease.
INTRODUCTION
Estrogen receptor (ER) positive HER2 negative (ER+) breast cancer accounts for more than 70% of all the breast cancer cases and metastatic disease represents a leading cause for cancer-related death worldwide [1, 2]. The ER signaling pathway is a major driver for tumorigenesis and it has been exploited therapeutically as a treatment option for the ER+ breast cancers [3, 4]. Different strategies impacting the ER signaling were clinically approved that includes selective ER modulators (tamoxifen), selective ER down-regulators (fulvestrant), and aromatase inhibitors to inhibit estrogen biosynthesis (letrozole) [3–7]. Despite clear clinical efficacy, a sub-population of patients may not respond to first-line endocrine therapy or experience tumor recurrence due to acquired resistance. There are several underlying mechanisms that impart resistance, including ER mutations, “cross talk” between ER and other mitogenic signaling pathways, and cell cycle perturbations [8]. Prognostic biomarkers used in the management of ER+ breast cancer (e.g. OncotypeDx or PAM50) largely monitor basal cell cycle status as a determinant of response to adjuvant endocrine therapy [9–11].
Previous studies reported that the aberrant activation of PI3K/ mTOR signaling pathway, which is dysregulated in breast cancers due to mutations in the PIK3CA gene and other mechanisms can be targeted therapeutically [12, 13]. While these pathways contribute to resistance to endocrine therapy, potent inhibitors against PI3K (e.g. alpelisib) and MTOR (e.g. everolimus) have been developed [14]. Much like endocrine therapy, PI3K and MTOR inhibitors enhance cytostatic activity in the metastatic setting, however, tumor progression on treatment occurs, even in selected patient population (e.g. PIK3CA mutated) [14]. The basis for the adaptive resistance to these inhibitors remains poorly understood but can involve pathway cross talk and likely other mechanisms that enable proliferation [15]. Therefore, understanding fundamental proliferative networks may be critical in elucidating resistance mechanisms and developing potent treatment options for tumors that have progressed on these targeted therapeutic regimens.
Cell cycle regulatory processes are deregulated in ER+ breast cancer via a variety of mechanisms. Activation of CDK4 or CDK6 kinases via the binding of cyclin D1 initiates cell cycle progression by phosphorylating the retinoblastoma tumor suppressor (RB) [16]. The CDK-mediated phosphorylation of RB inactivates its transcriptional repressive function, thereby enhancing the expression of multiple target genes required for cell division [17]. Aberrant CDK4/6 kinase activity frequently occurs in breast cancers due to the amplification of cyclin D1 as well as a host of deregulated signaling pathways [18]. Owing to its oncogenic function, pharmacological CDK4/6 inhibitors (e.g. palbociclib) were developed that potently suppresses cellular proliferation of ER+ breast cancers in preclinical studies [19, 20]. Additionally, CDK4/6 inhibitors as monotherapy and in combination with endocrine therapies are FDA-approved for the treatment of metastatic ER+ breast cancer [19, 21–23]. While these agents are effective, acquired resistance evolves in tumor sub-populations due to cell cycle deregulation [24, 25]. The mechanisms of resistance are varied but can include aberrant oncogenic signaling (e.g. KRAS mutation), cell cycle deregulation due to amplification of cyclin E1 loss of the RB tumor suppressor [26–29].
It is clinically relevant to elucidate the major cell cycle regulatory networks that can drive cross-resistance or modulate sensitivity to PI3K, MTOR, and CDK4/6 inhibitors as patients can be treated sequentially with all three drug classes [30, 31]. Interestingly, coordinate targeting of multiple pathways could elicit more durable responses as illustrated by synergism between PI3K and MTOR inhibitors with CDK4/6 inhibitors in ER+ breast cancer models [32]. As more potent cytostatic regimens are emerging, it is expected that deregulation of cell cycle via RB loss or other mechanisms (e.g. Cyclin E amplification) will become more frequent, which is observed in the context of standard disease progression in metastatic ER+ breast cancer [29, 33, 34]. To circumvent this issue, several studies have evaluated synthetic vulnerabilities of RB-deficient tumors, albeit in tumors that are generally fast-growing and frequently harbor intrinsic RB loss (e.g. triple-negative breast cancer and small cell lung cancer) [35–37]. These studies illustrated increased sensitivities to drugs that target aurora kinases, other cell cycle regulatory proteins, or specific chemotherapies [37, 38]. Unfortunately, the therapeutic window of such agents maybe only modestly expanded with RB loss due to the pan-essentiality of such genes, suggesting that improved synthetic strategies or combination approaches could be most relevant in targeting RB loss. Here we employed a series of unbiased screens, isogenic models, and xenograft models to interrogate features of the therapeutic resistance and new vulnerabilities in breast cancer models.
RESULTS
Cellular response to different molecular targeted drugs as single agents
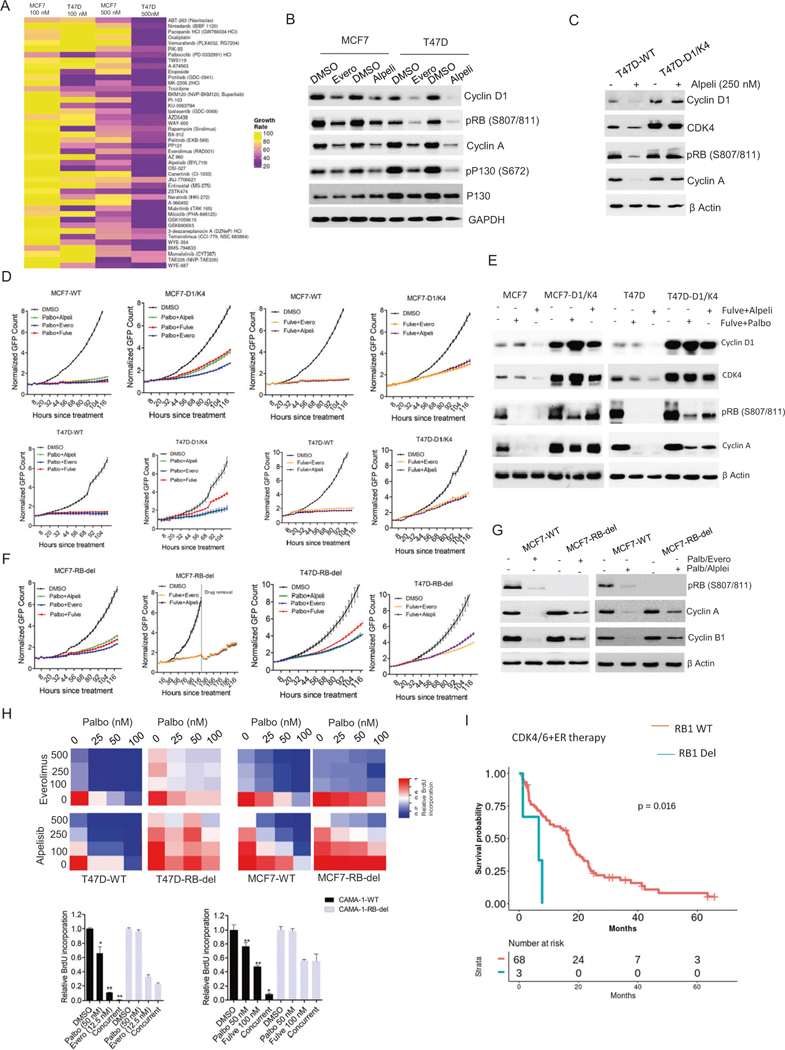
To investigate the response of ER+ breast cancer models to multiple targeted agents, MCF7 and T47D cell lines that harbor PIK3CA mutation were subjected to drug screening analysis using live-cell imaging (Fig. S1) [39]. Cells were labeled to stably express H2B-GFP and exposed to a 305-drug library at two different concentrations (100 nM and 500 nM) for 5 days (Fig. S2). Time lapse imaging of the H2B signals was used to determine relative growth rate of the cells in each well over the period of time as a measure of drug efficacy. A scatter plot based on the relative growth rate from two independent experiments indicated a significant correlation thereby validating the reproducibility of the drug screen (Fig. S2). Unbiased principal component analysis (PCA) was used to cluster drugs according to their anti-proliferative effects (Fig. S2). Initially, we focused on Cluster 1, which was highly enriched for drugs that target PI3K, AKT and mTOR pathways and inhibited the growth of MCF7 and T47D cells (Fig. 1A, Fig. S2). Biochemical analysis was carried out in MCF7 and T47D cells using 2 different inhibitors from drug screen, alpelisib and everolimus that target PI3K and mTOR respectively to investigate the impact on cell cycle machinery. Based on the phosphorylation status of RB1 and p130, as well as cyclin A expression, it was clear that alpelisib had a stronger inhibitory effect selectively in T47D cells as compared to that in MCF7 cells (Fig. 1B). Moreover, the IC50 of alpelisib in T47D cells is significantly lower than that in MCF7 cells, which is consistent with previous report (Fig. S2) [40]. The cellular effects of PI3K and mTOR inhibitors correlated with their ability in downregulating cyclin D1 expression, which integrates the mitogenic signals to cell cycle progression [41], suggesting that the residual levels of cyclin D1 following the treatment with PI3K/mTOR inhibitors might be sufficient to drive the cell cycle progression (Fig. 1B). This concept is supported by the finding that depletion of CCND1 gene resulted in a durable growth arrest in both MCF7 and T47D cells (Fig. S3). Conversely, ectopic overexpression of cyclin D1 and CDK4 in T47D cells (T47D-D1/K4), could mitigate the inhibitory effect of alpelisib on cell cycle proteins (Fig. 1C). Since the clinical use of PI3K/mTOR inhibitors involves combination with endocrine therapy or CDK4/6 inhibitors we evaluated the impact of Cyclin D1/CDK4 overexpression on the response to combination therapies in both MCF7 and T47D cells. Following different pairwise combination treatments that include fulvestrant, palbociclib and PI3K/mTOR inhibitors, MCF7-D1/K4 and T47D-D1/K4 cells continued proliferation following the combination treatments while the wild-type cells underwent growth arrest (Fig. 1D). Similar to PI3K/mTOR inhibitors, the combination of fulvestrant with two different Akt inhibitors yielded growth arrest in MCF7-WT cells while the MCF7-D1/K4 cells were partially resistant (Fig. S3). Biochemical analysis further confirmed that the inhibitory effect on cell cycle induced by the combination treatments could be alleviated by overexpressing cyclin D1/CDK4 that retains RB phosphorylation and cyclin A expression in both MCF7-D1/K4 and T47D-D1/K4 cells (Fig. 1E, Fig. S3).
Fig. 1. RB-mediated cell cycle arrest by standard care of therapies.
A Heat map representing the relative growth rate of MCF7 and T47D cells in the presence of drug library at two different concentrations (100 nM and 500 nM) from cluster 1. B Western blot analysis on the indicated proteins from MCF7 and T47D cells that were treated with everolimus (250 nM) and alpelisib (250 nM) up to 48 H. C Western blot analysis to investigate the effect of alpelisib (250 nM) on cell cycle proteins from T47D and T47D-D1/K4 that were treated up to 48 H. D Live cell imaging to monitor the growth of MCF7-WT, MCF7-D1/K4, T47D-WT, and T47D-D1/K4 cells treated with different pairwise combination of palbociclib (50 nM), fulvestrant (100 nM), everolimus (125 nM) and alpelisib (125 nM). Error bars represent mean and SD from triplicates. Experiments were done at two independent times. E Western blot on the indicated proteins from MCF7-WT, MCF7-D1/K4, T47D-WT and T47D-D1/K4 that were treated with palbociclib (50 nM) + fuvlestrant (100 nM) and alpelisib (125 nM) + fulvestrant (100 nM) for 48 h. F Growth curves illustrating the effect of indicated combination treatments involving palbociclib (50 nM), fulvestrant (100 nM), everolimus (125 nM) and alpelisib (125 nM) in MCF7-RB-del and T47D-RB-del cells. Mean and SD were calculated from triplicates. G Western blot analysis to demonstrate the differential effect of palbociclib (50 nM) + Everolimus (125 nM) and palbociclib (50 nM)+ alpelisib (125 nM) on cell cycle proteins. H Heat map to demonstrate the synergistic effect between the combination of palbociclib with everolimus (Palb/Evero) and alpelisib (Palb/Alpe) on BrdU incorporation at the indicated doses in MCF-WT, MCF7-RB-del, T47D-WT, and T47D-RB-del cells. The values in heat map represent the mean from triplicates. BrdU incorporation was determined in CAMA-1 WT and CAMA-1-RB-del cells treated with the palbociclib (palbo) in combination with everolimus (evero) and fulvestrant (fulve) at indicated concentrations for 72 h. Error bars represent SD from triplicates (*p < 0.05, **p < 0.01 as determined by two-way ANOVA). I PFS of patients receiving CDK4/6i in combination with endocrine therapy with tumors harboring WT RB (n = 68) and RB-deletion mutation (n = 3). All p values were indicated.
Since RB activation status appears to determine the impact of the combination treatments on cell cycle progression, we examined how RB expression modulates the efficacy of those targeted therapies in ER+ breast cancer models. We generated two isogenic cells lines, MCF7-RB-del and T47D-RB-del that harbor CAS-mediated RB deletion and monitored the proliferation in the presence of different combination treatments [42]. RB deletion in MCF7 cells rendered them partially resistant to palbociclib in combination with fulvestrant, alepelisib and everolimus as the cells continued proliferation (Fig. 1F). Although, the combination of fulvestrant with alpelisib and everolimus robustly delayed the proliferation of MCF7-RB-del cells, the effect was reversible because the cellular outgrowth following the drug removal occurred selectively in RB-del cells as compared to the MCF7-WT cells (Figs. 1F, S3). However, the T47D-RB-del cells continued to proliferate with all the combination treatments that were tested (Fig. 1F). To define the mechanistic impact of combination targeted therapy gene expression analysis was carried out in MCF7 cells treated with palbociclib and everolimus. Although more genes were significantly downregulated by palbociclib and everolimus in MCF7-RB-del cells as compared to single agent treatment, the magnitude of repression was significantly higher in RB-proficient cells (Fig. S4, Table S2). Interestingly, these genes are involved in cell cycle pathway (e.g. Cyclin A, cyclin B1, CDK1 and FOXM1) whose transcriptional activation is regulated through E2F, suggesting the direct role of RB in mediating the cellular response (Fig. S4). Biochemical analysis confirmed the selective suppression of cyclin A and cyclin B1 following the treatment with palbo/evero in both MCF7-WT and T47D-WT cells (Fig. 1G and Fig. S4). Moreover, BrdU incorporation revealed that the synergism between palbociclib and everolimus (palb/evero) or alpelisib (palb/alpel) was significantly attenuated in the RB-deleted cells, indicating therapeutic resistance (Fig. 1H and Fig. S5). While, MCF7 and T47D cells harbor PIK3CA mutation, we included another ER+ breast cancer cell line, CAMA-1 that harbors wild-type PIK3CA and mutant PTEN and are highly resistant to lapels (Fig. S5) [39]. Like MCF7 and T47D cells, RB deletion in CAMA-1 cells resulted in resistance to the synergistic effect induced by palbociclib in combination with fulvestrant and everolimus based on BrdU incorporation, indicating that RB loss renders resistance to the standard cytostatic treatments irrespective of the PIK3CA status (Fig. 1H). To investigate the clinical relevance of RB loss and its association with therapeutic outcome to CDK4/6 inhibition, we explored a patient cohort with 222 metastatic ER+ breast cancer patients (NCT04526587). To determine the RB1 mutation, genetic testing was performed on 71 patients, which revealed that 3 patients (4.2%) harbored RB1-deletion mutation (Fig. S6). Recurrent disease was observed in patients with wild-type RB1 (n = 45) and all the patients with RB1-deletion mutation (n = 3) who had prior treatments either with hormone therapy or endocrine therapy (Fig. S6). Among the 71 patients, PIK3CA mutation was observed in both wild-type (n = 28) and RB-deleted (n = 1) patient groups (Fig. S6). Consistent with previous clinical data, patients with RB-loss mutation had a significant decrease in progression-free survival (PFS) as compared to those with wild-type RB following the treatment with CDK4/6 inhibitor in combination with endocrine therapy (Fig. 1I) [29, 34, 43]. Overall these data indicate that RB deficiency is a genetic aberration that associates with poor clinical outcome with standard of care therapies.
RB loss-mediated molecular alterations render ER+ models vulnerable to selective drug classes
To understand the mechanism of resistance to cytostatic therapies, gene expression data was further investigated, which revealed that RB loss does not prominently alter the basal expression of genes involved in ER/PI3K pathway and cell cycle except for increased CCNE1 expression in both MCF7 and T47D RB-deficient cells (Fig. S7). Moreover, RB loss could only slightly enhance the growth rate of both MCF7 and T47D cells. To confirm the gene expression data, the differential expression of representative proteins was evaluated by western blotting, which revealed that T47D-RB-del cells have lower mTOR activity based on the phosphorylation status of S6 (Fig. S7). However, this phenomenon was not observed in MCF7-RB-del cells (Fig. S7). The upregulation of cyclin E1 due to RB loss occurred in both MCF7 and T47D RB-del cells, which is consistent with the gene expression data, indicating that it might assist in maintaining the CDK2 kinase activity to evade the cell cycle arrest induced by targeted therapies (Fig. S7). In accordance to this observation, following the palbociclib and everolimus treatment the RB-deficient cells (MCF7 and T47D) retained the cyclin E1/CDK2 and cyclin A/CDK2 complexes to maintain an active CDK2 kinase as determined by in vitro kinase assay and a live cell sensor that tracks CDK2 activity as described in our previous study (Fig. S8) [44].
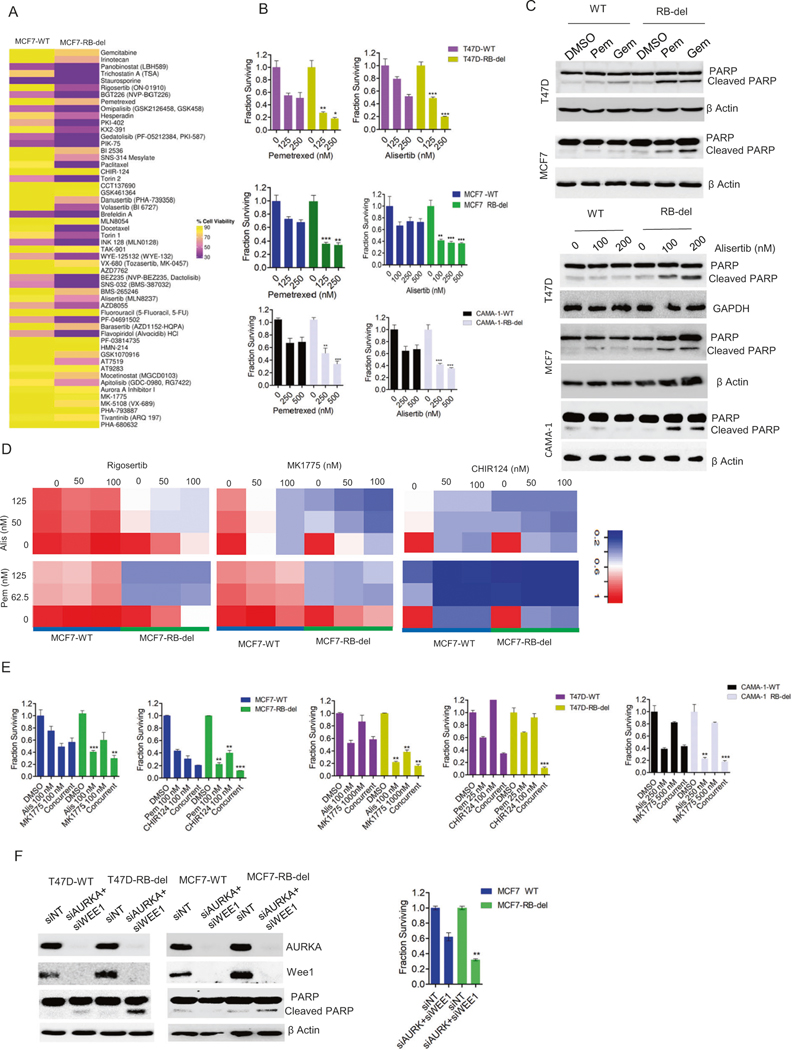
To define a therapeutic strategy for RB-deficient ER+ breast cancer models, we further investigated the drug screen analysis, which revealed that RB deletion rendered both the MCF7 and T47D cells to be more vulnerable to drugs, targeting AURK, nucleotide synthesis, PLK and multiple CDKs (Fig. S9). Since the inhibition of these targets could induce cell death, we repeated the drug screen in MCF7-WT and RB-del cells and examined the cytotoxic effect of the drugs using a cell-titer-glow (CTG) assay. Consistent with the primary drug screen, there was more cell killing by these classes of drugs in RB-deficient cells as compared to the wild-type cells (Fig. 2A). The differential effect on cell viability between the MCF7-WT and RB-del cells were significantly correlated between two independent experiments (Fig. S9). To validate the drug screen, we individually examined the drug efficacy at different concentrations in MCF7, T47D and CAMA-1 cells. We chose one representative drug that target AURK (Alisertib) and nucleotide synthesis (Pemetrexed), which showed enhanced cytotoxic effect in RB-deficient MCF7, T47D, and CAMA-1. (Fig. 2B). Moreover, the cell death caused by these drugs in all the RB-deficient cell lines was associated with enhanced PARP cleavage, indicative of apoptosis (Fig. 2C).
Fig. 2. Vulnerability due to RB loss.
A Heat map representing the differential effect of drugs (250 nM) from cluster 2 on the viability of MCF7-WT and RB-del cells as determined by CTG assay. B Differential effects of alisertib and pemetrexed at the indicated concentrations on the RB-proficient and RB-deficient MCF7, T47D and CAMA-1 cells following 6 days of exposure. Bars represent mean and SD from triplicates. Experiments were performed at 2 independent times (**p < 0.01, ***p < 0.001 as determined by 2-way ANOVA). C Western blot analysis to demonstrate the effect of pemetrexed (Pem) (200 nM) and gemcitabine (Gem) (100 nM) on PARP cleavage in RB-proficient and RB-deficient MCF7 and T47D cells after 72 h treatment. Effect of alisertib at two different concentrations on cleaved PARP in MCF7, T47D and CAMA-1 WT and RB-del cells following 72 h treatment. D Heat maps to demonstrate the synergistic effect on the inhibition of cell viability by different pairwise combination of the indicated drugs in MCF-WT and MCF7-RB-del cells following 6 days of exposure. E CTG assay on MCF7, T47D and CAMA-1 WT/RB-del cell lines following the treatment with the indicated drugs at indicated concentrations as single agents and combination for up to 6 days. Mean and SD were calculated from triplicates (**p < 0.01, ***p < 0.001 as determined by 2-way ANOVA). F Western blot analysis on MCF7-WT, MCF7-RB-del, T47D-WT and T47D-RB-del cells that were reverse transfected with RNAis to silence WEE1 and AURKA. Viability of MCF7-WT and RB-del cells were determined by CTG assay following the knockdowns of WEE1 and AURKA up to 6 days. Bars represent mean and SD from triplicates. Experiments were performed at two independent times (**p < 0.01 as determined by two-way ANOVA). Western blots represent replicates from two independent experiments.
Although the identified drugs are selective against RB-deficient cells as single agents these agents target genes that are considered universally required for proliferation. We reasoned that low dose combinations could be utilized to enhance selectivity for RB-deficient cells. Using the identified drugs, we carried out different pairwise drug combinations in MCF7-WT and RB-del cells, including additional inhibitors that target CHK1 (CHIR124) and WEE1 (MK1775) that are associated with DNA replication and mitosis respectively. Interestingly all the combination treatments yielded selective killing in RB-deficient cells (Fig. 2D). The combination of alisertib and MK1775 yielded a synergistic interaction in selectively killing the RB-deficient cells (Fig. S10). Similarly, pemetrexed in combination with different inhibitors (rigosertib, MK1775, and CHIR124) yielded an enhanced cytotoxic effect, which is more potent in the RB-deleted MCF7 cells (Fig. 2D, Fig. S10). The combination analyses were further validated by repeating the treatment at single concentration of each drug in the RB-deficient MCF7, T47D, and CAMA-1 cells. In all the cell lines tested the combination treatment showed significantly enhanced cell killing selectively in RB-deficient setting (Fig. 2E). Since, AURK and WEE1 are critical targets for RB-dependent lethality, we depleted their expressions using RNAis, which showed that RB loss partially sensitizes both T47D and MCF7 cells to loss of these proteins and renders them to undergo apoptosis and cell death as determined by PARP cleavage (Fig. 2F, Fig. S10).
Mechanistic impact of RB loss on sensitivity to AURK and WEE1 inhibitors
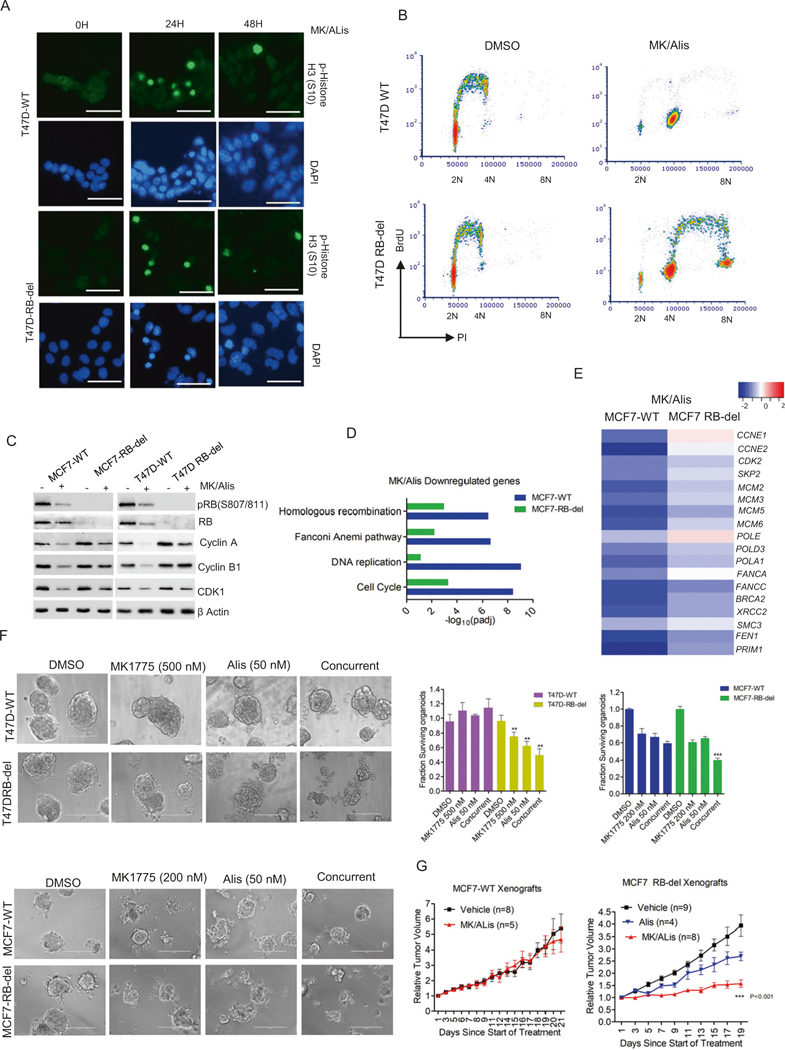
To decipher the cellular mechanisms that renders RB-deficient models sensitive to AURK and WEE1 inhibitors, the impact on mitotic stress was determined in the presence of these agents. We examined the nuclei of MCF7-WT and RB-del cells in the presence of Alisertib in combination with MK1775, which revealed that the phosphorylation of Ser10-Histone H3 (pHH3) occurred as an acute response and eventually diminished over the period in both MCF7 and T47D cells (Fig. 3A, Fig. S11). This indicates that the cells undergo a transient mitotic arrest but exit from mitotic state. This phenomenon occurred in an RB-independent manner (Fig. 3A). Further cell cycle analysis illustrated that RB-deficient MCF7 and T47D cells possessed high-level of DNA replication and progressed to higher 8 N ploidy in the presence of MK1775 and alisertib as observed by the BrdU incorporation (Fig. 3B, Fig. S11). In contrast, in RB-proficient MCF7 and T47D cells, the replication of 4 N cells was inhibited in the presence of the drug combination (Fig. 3B, Fig. S11). Biochemical analysis on both MCF7 and T47D cells indicated that alisertib +MK1775 suppressed the expressions of cyclin A, CDK1, and cyclin B1 selectively in RB-proficient cells through RB dephosphorylation (Fig. 3C). These molecular events indicate that the accumulation of cells with 4 N DNA content is a consequence of pseudo-G1 arrest, while in RB-deleted cells the 4 N population continues cell division [45]. RNA sequencing on MCF7-WT and RB-del cells following the exposure to alisertib+MK1775 revealed significant downregulation of a large collection of genes associated with DNA replication selectively in RB-proficient cells (Fig. 3D). The downregulated genes were highly enriched for E2F-target genes, confirming the effect of alisertib+MK1775 on RB activation and the direct role of RB in modulating their expression (Fig. 3E). We also evaluated the impact of RB loss on the cellular responses to pemetrexed in combination with CHIR124. Since, these classes of drugs are known to induce DNA damage, we evaluated the γH2AX foci formation in T47D cells [46]. Based on immunofluorescence and western blotting pemetrexed + CHIR124 prominently increased the phosphorylation of H2AX in both RB-proficient and deficient cells, suggesting that RB does not modulate the cellular DNA damage induced in response to this combination treatment (Fig. S11). However, PARP cleavage occurred selectively in RB-deficient cells, suggesting that RB loss primes the cells to be more vulnerable to DNA damage or replication stress and activate apoptotic pathways (Fig. S11).
Fig. 3. Impact of RB loss on AURK and WEE1 inhibition.
A Immunofluorescence staining of p-Histone H3 (S10) on T47D-WT and RB-del cells treated with MK1775 (200 nM) in combination with Alisertib (200 nM) at different time points. Scale bar represents 50 μm. B Representative flow cytometry analysis of T47D-WT and RB-del cells treated with Alisertib (200 nM) and MK1775 (200 nM) for 48 h. X and Y axis represent DNA content and BrdU incorporation respectively. C Western blot analysis on the indicated proteins from RB-proficient and deficient MCF7 and T47D cells that were treated with Alisertib (200 nM) and MK1775 (200 nM) up to 72 H. D ENRICHR analysis to represent the top gene ontology sets that were significantly suppressed in MCF7-WT and MCF7-RB-del cells treated with MK1775 (200 nM) in combination with Alisertib (200 nM) based on RNA sequencing analysis, which was done in triplicates. E Heat map indicating the fold change of selected genes in MCF7WT and RB-deficient cells treated with MK/Alis. F Representative images of organoids derived from T47D-WT, T47D- RB-del, MCF7-WT, and MCF7-RB-del cells that were treated with Alisertib and MK1775 at indicated concentrations for 6 days. The viability of the organoids was determined by CTG assay. Graphs represent mean and SD from triplicates. Experiments were done at two independent times (**p < 0.01, ***p < 0.001 as determined 2-way ANOVA). G Mice bearing MCF7-WT and RB-del xenografts were randomized for treatment with vehicle and MK1775 (30 mg/kg) in combination with Alisertib (10 mg/kg) for 3 weeks and the tumor growth was monitored regularly. Data show mean SEM (***p < 0.001 as determined two-way ANOVA).
To further interrogate that combination approaches are more potent in inducing selective lethality in RB-deficient models, we employed organoid models derived from MCF7-WT/RB-del and T47D-WT/RB-del cells and treated them with alisertib and MK1775. While the single agents had limited effect on the organoids, the combination of alisertib + MK1775 selectively impacted the viability of organoids derived from both MCF7-RB-del T47D-RB-del cells as determined by CTG assay (Fig. 3F). To interrogate whether the impact of RB loss on the effects of alisertib + MK1775 could be translated in vivo, MCF7-WT and RB-del xenograft models were deployed. Western blotting and immunofluorescence on the tumor tissues confirmed the RB status of the xenograft models (Fig. S12). In accordance with the cell-culture and organoid data, MCF7-WT tumors did not respond to alisertib +MK1775 treatment and the tumor continued to progress over the course of treatment (Fig. 3G). In contrast, the progression of MCF7-RB-del tumors was partially inhibited in the presence of alisertib, while the combination treatment alisertib+MK1775 exerted a significant effect in controlling the tumor growth, further confirming the selective vulnerability imparted by RB-depletion (Fig. 3G, Fig. S12).
RB loss and apoptosis
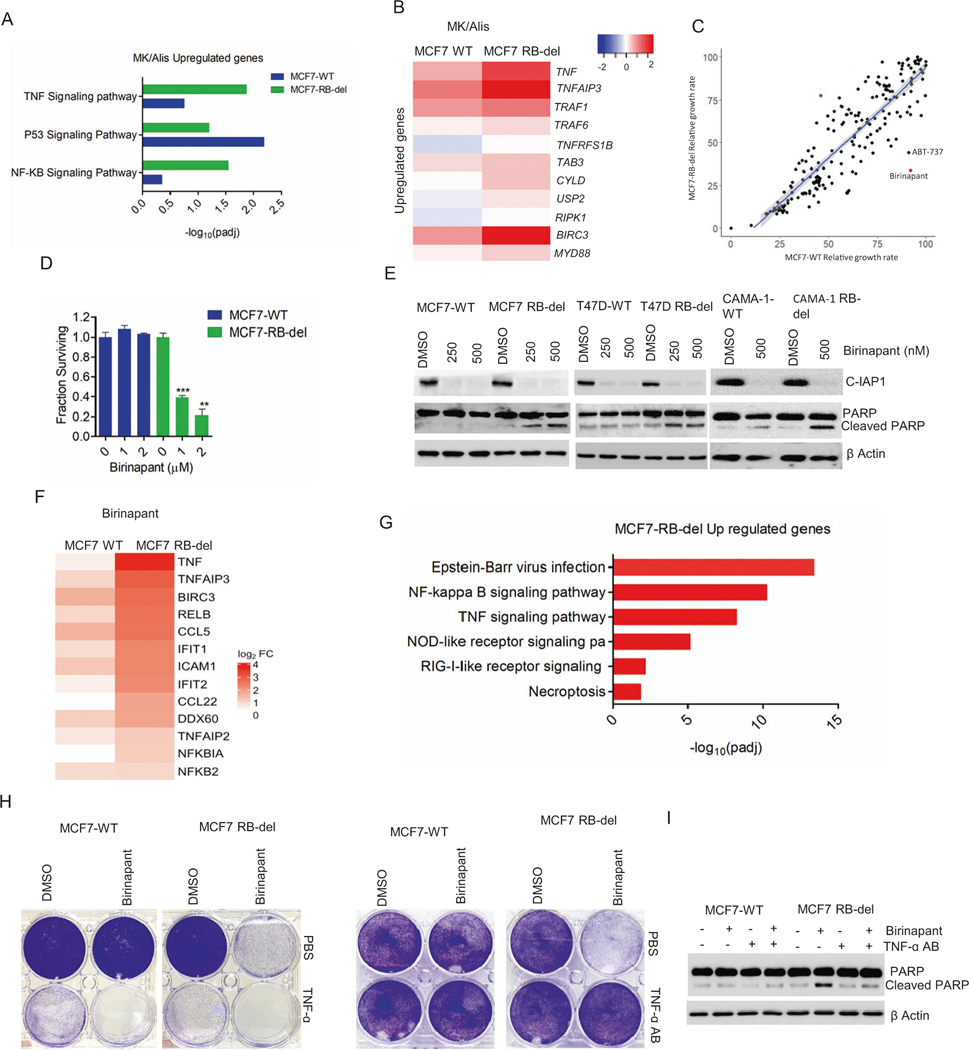
To delineate the mechanism that drives apoptosis in RB-deficient cells in the presence of MK1775 and alisertib, we hypothesized that the replication of 4 N population that selectively occurs in the RB-del cells leads to cell death while the cell cycle arrest in wild-type cells protect them from undergoing apoptosis. However, in the presence of aphidicolin, which inhibited the DNA replication of both 2 N and 4 N population cells in RB-del cells following the treatment with alisertib + MK1775 could not rescue the cells from undergoing apoptosis (Fig. S12). This suggests the involvement of other cellular pathways that causes the RB-deficient cells to activate apoptotic cell death. Further investigation on the RNA sequencing data was carried out to identify genes that were induced in the MCF7 cells following alisertib+MK1775 treatment. GeneOntology analysis revealed high enrichment of genes associated with TNF signaling pathway that were more significantly upregulated in the RB-deleted cells as compared to the wild-type cells in the presence of alisertib+MK1775 (Fig. 4A, Table S3). However, the difference in magnitude between the wild-type and RB-deleted cells was subtle, suggesting that alisertib+MK1775 activated TNF signaling pathway in an RB independent manner (Fig. 4B). Activation of TNF pathway leads either to apoptosis or cell survival depending on the stabilization of death-inducing signaling complex (DISC), which is comprised of RIPK1/FAD88/ Caspase 8 [47]. Since, alisertib+MK1775 induces apoptosis selectively in RB-deleted cells, we hypothesized that RB loss renders the cells more sensitive to the activation of TNF pathway.
Fig. 4. Regulatory function of RB on apoptosis.
A ENRICHR analysis to represent the top gene ontology sets that were significantly upregulated in MCF7-WT and MCF7-RB-del cells treated with MK/Alis based on RNA sequencing analysis. B Heat map depicting the fold change of indicated genes in MCF7-WT and RB-del cells following the treatment with MK1775 and Alisertib. C Scatter plot with X and Y axis representing the relative growth rate of MCF7-WT and RB-deleted cells respectively that were subjected to drug screen analysis at 500 nM. Each dot represents an individual drug from the library. D CTG assay in MCF7-WT and RB-del cells treated with the increasing concentrations of birinapant up to 6 days. Graphs represent mean and SD from triplicates. Experiments were done at two independent experiments (**p < 0.01, ***p < 0.001 as determined by two-way ANOVA). E Western blot analysis on the indicated proteins from the RB-proficient and RB-deficient cells MCF7, T47D, and CAMA-1 cells treated with different concentrations of birinapant for 72 H. F Heat map representing the fold change of indicated genes in MCF7-WT and RB-del cells following the treatment with birinapant (500 nM) up to 48 h. RNA sequencing was done in triplicates. G ENRICHR analysis to represent the top gene ontology sets that were significantly upregulated in MCF7-WT and MCF7-RB-del cells treated with birinapant (500 nM) based on RNA sequencing analysis. H Representative crystal violet images from MCF7-WT and RB-del cells that were treated with birinapant in the absence and presence of exogenous TNF growth factor (5 ng/μl) and TNF-α antibody (100 ng/μl). I Western blot to determine the effect of birinapant (500 nM) on cleaved PARP in MCF7-WT and RB-del cells in the absence and presence of TNF-α antibody.
Recent studies have shown that the TNF pathway-mediated apoptosis could be pharmacologically induced by targeting CIAP, which is involved in the ubiquitination of RIPK1 thereby destabilizing DISC [48]. Based on our primary drug screen a pharmacological inhibitor against CIAP, birinapant, showed marked selectivity that has essentially no activity against RB-proficient cells, while partially inhibiting RB-deficient cells (Fig. 4C, Fig. S13). Further validation by treating MCF7-WT and RB-del cells with different concentrations of birinapant, revealed that the RB-deficient cells were significantly more vulnerable to CIAP inhibition (Fig. 4D). Mechanistically, birinapant induced cell death in all the RB-deficient ER+ models, MCF7, T47D, and CAMA-1, which is mediated through activation of apoptosis based on cleaved PARP (Fig. 4E, Fig. S13). Further investigation on the cellular impact of birinapant was carried out by performing RNA sequencing in MCF7 cells. Based on this analysis, there were more differentially expressed genes in the birinapant-treated MCF7-RB-del cells as compared to the MCF7-WT cells, indicating that RB loss primes the cells for a rapid response (Fig. S13). Interestingly, those genes were significantly upregulated selectively in the RB-deficient cells and were highly enriched for NF-κB and TNF signaling pathways indicating that the activation of these pathways is required for the cellular effect of birinapant (Fig. 4F, G). This phenomenon is consistent with previous studies that have shown that, IAP antagonists require secretion of autocrine TNFs to activate the apoptotic pathway [49]. As a proof of concept, exogenous addition of recombinant-TNF enhanced the toxic effect of birinapant in both RB-proficient and deficient MCF7 cells (Fig. 4H). Conversely, the effect of birinapant could be selectively attenuated in MCF7-RB-del cells by neutralizing the TNF pathway using antibodies further confirming that the activation of this pathway selectively occurs in the RB-deficient models. (Fig. 4H, I).
Combination approaches to enhance RB-selective cell death
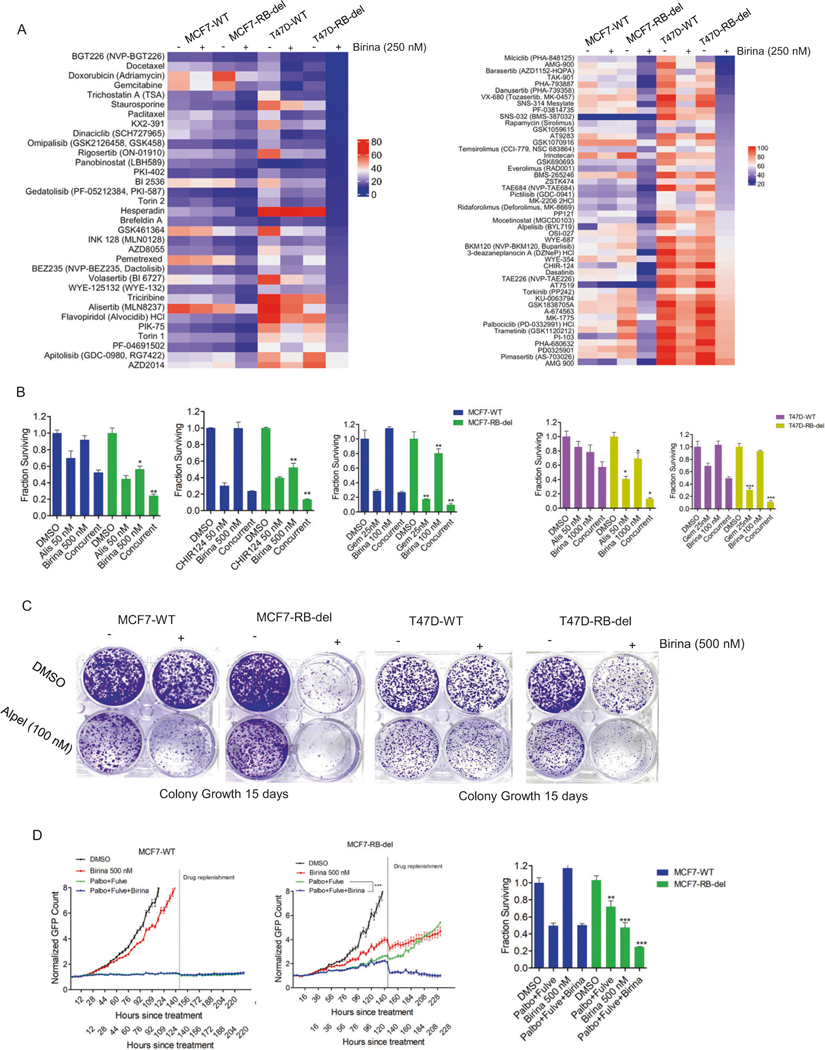
To further enhance the therapeutic efficacy of birinapant, we investigated potential combination approaches that could induce cooperative cell death in the ER+ models. MCF7-WT/RB-del and T47D-WT/RB-del cells were subjected to drug sensitivity analyses following the pretreatment with birinapant (250 nM) for 24 h (Fig. S13). Our drug screen data revealed that chemotherapeutic agents such as, gemcitabine, docetaxel, pemetrexed and AURK inhibitor, alisertib resulted in more enhanced cell killing in combination with birinapant in RB-deficient MCF7 and T47D cells as compared to the wild-type cells (Fig. 5A). Interestingly, several molecular targeted agents and the standard of care drugs for ER+ breast cancer such as PI3K/mTOR inhibitors also induced selective killing in the RB-deficient MCF7 and T47D cells in combination with birinapant (Fig. 5A). Validation experiment confirmed the cooperative cytotoxic effect selectively in RB-deleted MCF7 and T47D cells when birinapant used in combination with AURK and CHK1 inhibitors and gemcitabine (Fig. 5B). Colony formation assay revealed that the residual cell population that escaped the effect of PI3K inhibitor, alpelisib could be selectively killed by birinapant in the RB-deleted MCF7 and T47D cells (Fig. 5C). Similarly, the concurrent administration of birinapant to the standard of therapy, palbociclib+fulvestrant leads to selective cell death in the RB-deficient MCF7 cells (Fig. 5D). Overall these data suggest that limiting anti-apoptotic pathways selectively sensitizes the RB-deficient models to multiple clinical treatment modalities.
Fig. 5. Combination treatment approach using birinapant.
A Heat maps representing the relative cell viability of MCF7-WT, MCF7-RB-del, T47D-WT and T47D-RB-del cells in the presence of drug library (100 nM) following the pretreatment with birinapant (250 nM) from clusters 1 & 2. B Cell viability assays from MCF7 and T47D WT/RB-del that were treated with birinapant in combination with the indicated drugs at the indicated concentrations up to 6 days. Mean and SD were calculated from triplicates. Experiment was done at 2 independent times. (*p < 0.05, **p < 0.01 as determined by two-way ANOVA). C Crystal violet staining from MCF7-WT, MCF7-RB-del, T47D-WT and T47D-RB-del cells that were treated with Alpelisib (100 nM) in combination with DMSO or birinapant (500 nM). Cells were allowed to form colonies up to 15 days. D Live cell imaging to monitor the proliferation of MCF7-WT and RB-del cells that were treated with DMSO, birinapant (500 nM) and palbociclib (50 nM) + fulvestrant (100 nM) ± birinapant. The live cell imaging was terminated, and the cells were subjected to CTG assay to determine the viable cells. Mean and SD were calculated from 6 replicates. (**p < 0.01, ***p < 0.001 as determined by two-way ANOVA).
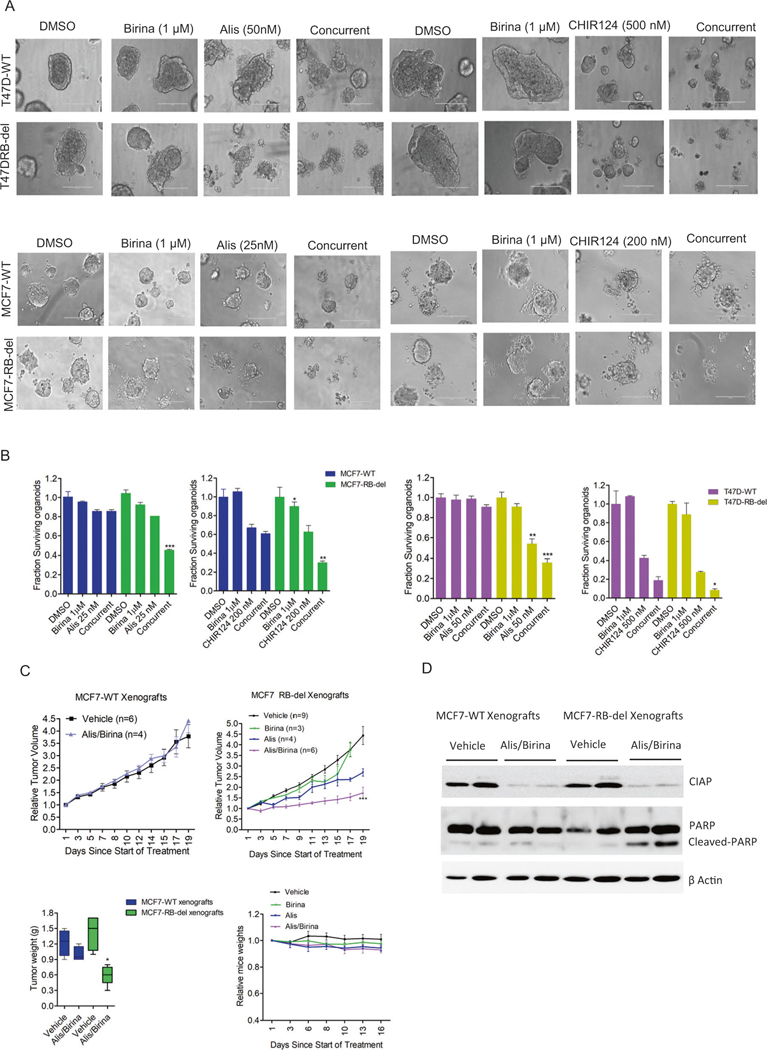
To investigate the selective effect of birinapant against RB-del tumor cells, we tested its efficacy using organoid models that were developed from MCF7-WT/RB-del and T47D-WT/ RB-del cells. Unlike the 2D-cell-culture data, birinapant as single agent had limited effect on the viability of organoids derived from MCF7-WT/RB-del and T47D-WT/ RB-del cells (Fig. 6A). One plausible explanation is that activation of apoptotic pathway may not be sufficient to affect the survival of organoids. However, in combination with other drugs such as alisertib and CHIR124, the impact of the apoptotic signaling enhances the cytotoxic effect that leads to cell death, which selectively occurs in the organoids derived from RB-del MCF7 and T47D cells (Fig. 6B). We examined the effect of alisertib in combination with birinapant in vivo by developing xenograft models derived from the ER+ breast cancer MCF7-WT/RB-del cell (Fig. S11). The treatment with Alisertib in combination with birinapant, did not inhibit the tumor growth of MCF7-WT xenografts whereas it exerted a selective anti-tumor effect against the RB-del xenografts and resulted in a durable disease control as compared to the single agent treatments (Fig. 6C). The combination treatments in mice were well tolerated as no significant weight loss was observed (Fig. 6C). To validate the on-target effect of birinapant in vivo, tumors excised from the vehicle and alisertib + birinapant-treated groups were subjected to western blot analysis. It was evident that the expression of CIAP1, which is the intra-cellular target of birinapant was inhibited in both wild-type and RB-del xenografts (Fig. 6D). However, the RB-del tumor models were more vulnerable to alisertib+birinapant treatment, which resulted in the activation of apoptosis as determined cleaved PARP from the tumor tissues (Fig. 6D). These data provide new avenues for using combinatorial strategies to target RB-deficient tumor models by exploiting drugs with complementary mechanisms of action.
Fig. 6. In vivo efficacy of Alisertib in combination with birinapant.
A Representative images of organoids derived from MCF7-WT, MCF7-RBdel, T47D-WT, and RB-del cells that were treated with birinapant in combination with alisertib and CHIR124 at indicated concentrations for 6 days. B The viability of the organoids were determined by CTG assay. Graphs represent mean and SD from triplicates (*p < 0.05, **p < 0.01, ***p < 0.001 as determined student t-test). C Mice bearing MCF7-WT and RB-del xenografts were randomized for treatment with vehicle, Alisertib (10 mg/kg), birinapant (15 mg/kg) and the combination (Birina/Alis) for 3 weeks and the tumor growth was monitored regularly. Data show mean SEM (***p < 0.001 as determined two-way ANOVA). Box plots represent the tumor weights. Mean and SD are shown (*p < 0.05 as determined by 2-way ANOVA). D Western blot analysis on CIAP1 and cleaved PARP from the MCF7-WT and RB-del tumors at the end of treatment.
DISCUSSION
Genetic aberrations deregulate PI3K/mTOR pathway and cell cycle machinery in ER+ breast cancer models, thereby providing a rational to pharmacologically inhibit these molecular targets as a therapeutic approach [12, 13]. In metastatic setting, the use of PI3K, MTOR, or CDK4/6 inhibitors have significant clinical benefits in combination with endocrine therapy [14, 50]. However, the development of resistance on treatment remains a major challenge. In our current study, drug screening analysis revealed a class of inhibitors that target PI3K and MTOR and possessed antiproliferative effects that is most apparent in T47D cells. We utilized live cell imaging to directly monitor cell division in real time. This approach is not susceptible to confounding effects on tumor metabolism that can influence the veracity of indirect methods (e.g. MTT assays) [51]. We provide mechanistic evidence that the impact of PI3K and mTOR inhibitors on cell cycle depends on the reduction of cyclin D1 levels and subsequent inhibition of CDK2 kinase activity.
CDK4/6 inhibition could help overcome the resistance to PI3K, mTOR inhibitors as indicated by a synergistic effect that completely stopped cell division. Importantly, the synergy depends on RB expression as the RB-deleted isogenic cell lines continued proliferation via active CDK2. While RB loss is a well-known determinant of resistance to CDK4/6 inhibitors, our data suggest that it will also impact the efficacy of the standard of care cytostatic combination treatments involving endocrine therapy and PI3K/mTOR inhibitors. A recent review illustrated the clinical evidence of RB loss mutation, which occurs in approximately 4% of patients with metastatic ER+ breast cancer patients, that would encompass more than 6000 patients annually. Consistent to previous studies, our clinical data further confirmed that RB loss results in short progression-free survival in patients who received CDK4/6 inhibitor in combination with endocrine therapy [34, 52].
Utilizing drug screening approaches and isogenic models we identified different classes of drugs whose sensitivity is increased with the specific deletion of RB. Most of these drugs represent inhibitors of AURK, PLK1 and select chemotherapies that rapidly activate apoptosis in RB-deficient tumor models. To data AURK and PLK1 inhibitor drugs have had a relatively limited clinical success, ostensibly due to the pan-essentiality of AURK and PLK1. In an effort to surmount this challenge, we employed combination strategies involving multiple agents that are selective for RB deficiency. In this context dual targeting of AURK and WEE1 resulted in aberrant mitosis and cell death through mitotic catastrophe in preclinical models [53]. In the current study we demonstrate that the acute response to AURK inhibitor (Alisertib) in combination with a WEE1 inhibitor (MK1775) induced mitotic arrest, whereas prolonged exposure resulted in selective lethality in RB-deficient cell lines. We confirmed the efficacy of alisertib + MK1775 at low doses using xenograft models where RB loss cooperatively delayed tumor progression while the RB-proficient tumors had limited effect. This approach in principle lowers toxic effects and expands treatment possibilities for RB deficiency.
The differential cellular effect rendered by alisertib + MK1775 between RB-proficient and deficient models had a major impact on the activation of apoptosis pathway. RB has been proposed to control a variety of process beyond cell cycle including apoptosis, epigenetic states and tumor metabolism [54]. Hence, we examined the differential response to other classes of drugs that target these pathways. Although the drug library includes a number of inhibitors that impact on chromatin (e.g. entinostat) and metabolism (e.g. CP-613, Phenformin), birinapant, a proapoptotic agent was the only “non-cell cycle” lead to be identified as a highly selective agent against the RB-deficient cell lines. Birinapant is a SMAC mimetic antagonist that targets the IAP family of proteins (cIAP and XIAP) with higher affinity towards cIAP (Kd < 1 nM) as compared to XIAP (Kd= 45 nM) [55]. cIAPS are anti-apoptotic proteins that negatively regulate the TNF-α mediated cell death by destabilizing a pro-death complex or ripoptosome via ubiquitination [47, 56]. Although birinapant degrades the cIAP expression in both RB-proficient and deficient ER+ breast cancer cell lines, the apoptosis is activated selectively in the RB-deleted cells. This suggests that the impact of RB loss primes the cells to undergo apoptosis following the stabilization of ripoptosome. The role of RB and TNF-mediated apoptosis has been investigated in previous studies that RB degradation is an early prerequisite event for the activation of apoptosis suggesting the anti-apoptotic functions of RB [57, 58]. Using organoid models, we showed that birinapant had limited effect as single agent, however, it augments the effect of alisertib selectively in organoids derived from RB-deleted cells. This suggests that limiting the anti-apoptotic pathway using birinapant yields synthetic lethality in combination with drugs that are selective for RB loss. Furthermore, the combination of alisertib and birinapant induced apoptosis in vivo selectively in RB-deficient ER+ breast cancer xenografts, that paved way for introducing novel combination approaches to treat RB-deleted breast cancer models.
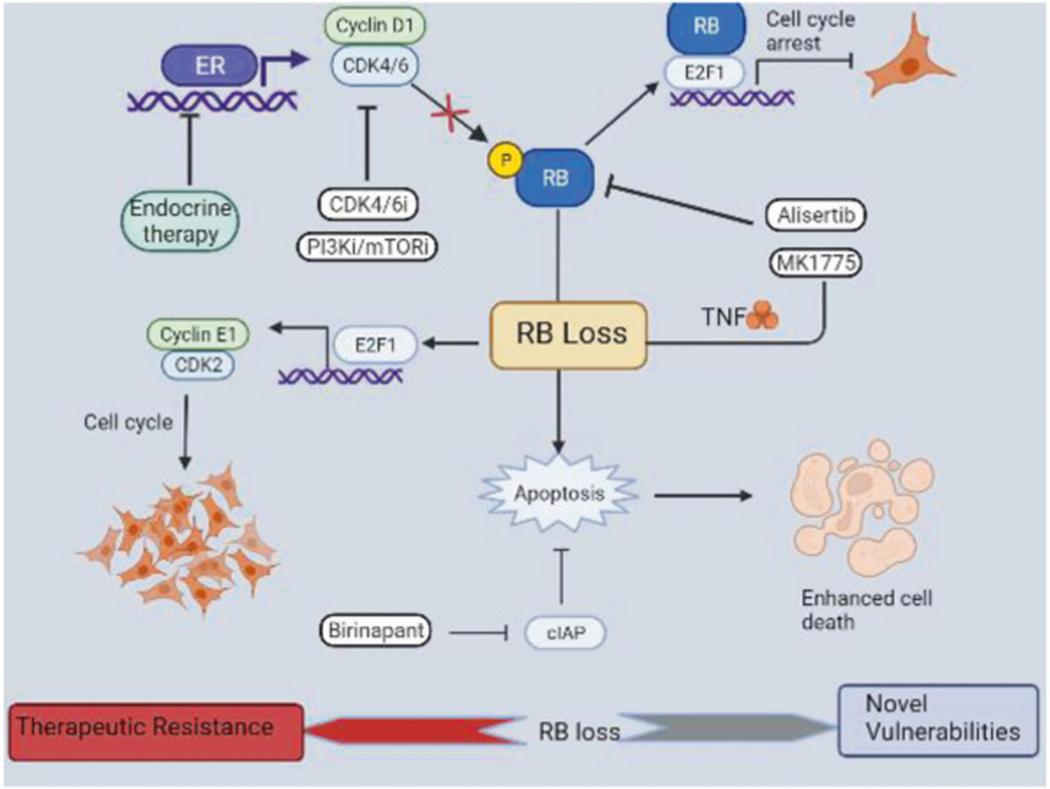
In conclusion, we provide mechanistic evidence to demonstrate that cell cycle aberrations due to RB loss or enhanced cyclin D1/CDK4 could limit the benefit of using CDK4/6 inhibition in combination with targeted therapies or endocrine therapy, which is further supported by a clinical study. Additionally, we indicate that the RB-deletion in ER+ breast cancer models renders them to be more vulnerable to cellular stress induced by the pharmacological agents that impact DNA replication and mitosis and prime the cells to undergo rapid apoptosis (Fig. 7). This phenomenon was further exploited in our study to selectively target the RB-deficient cancer cells by pharmacologically inhibiting an anti-apoptotic factor, CIAP (Fig. 7). Furthermore, our study demonstrates that birinapant could be effectively utilized to selectively kill the RB-deficient ER+ breast cancer models that evade the cytostatic treatment options involving PI3K inhibitors or endocrine therapy. Thus, we implemented a novel therapeutic approach for the ER+ breast cancer models that are resistant to the standard treatment options via RB loss.
Fig. 7. Scheme illustrating the function of RB loss.
Endocrine therapy, CDK4/6, and PI3K/mTOR inhibitors impact cyclin D1/CDK4/6 function thereby activating RB to induce cell cycle arrest. Loss of RB results in sustained E2F activity even in the presence of cytostatic drugs that transactivates cyclin E to drive cell cycle via CDK2 activity. Alisertib in combination with MK1775 induces apoptosis selectively in RB-deficient setting. Birinapant, an inhibitor of IAP, which is an anti-apoptotic protein results in enhanced apoptotic mediated cell death in RB -deficient ER + breast cancer models. Overall, RB loss leads to therapeutic resistance to the standard cytostatic treatment options and becomes more vulnerable to pharmacological agents that induce apoptosis.
METHODS
Cell culture and therapeutic agents
Isogenic breast cancer cell lines, MCF7-WT/ RB-del were maintained in DMEM media containing 10% FBS. T47D cells were grown in RPMI media, supplemented with 10% FBS. CAMA-1 cells were purchased from ATCC and were cultured in EMEM media (Cat # 30–2003) that was supplemented with 10% FBS. All the cells lines were grown at 37 °C and 5% CO2 and were confirmed to be mycoplasma free. Cell line authentication was performed using STR analysis. Palbociclib, Alisertib, and Birinapant were purchased from MedChemExpress (NJ, USA). Mk1775 was purchased from Chemitek (IN, USA). Fulvestrant, Everolimus, and Alpelisib were purchased from SelleckChem. MK1775 was purchased from ChemiTek. All the compounds were dissolved in DMSO to a final concentration of 10 mM. A customized drug library, comprising 311 compounds was purchased from SelleckChem.
Plasmids and infection procedures.
Lentiviral overexpression vectors (pLX304) containing the ORFs clones for CCND1 and CDK4 were purchased from Cancer genetics and Genomics core; Roswell Park Cancer Center. Lentiviral infections were carried out in exponentially growing T47D cells in the presence of Polybrene (Sigma Aldrich). The infected cells were selected using Blasticidine (5 μg/ml) and the protein overexpression was validated using western blotting.
CSII-EF lentiviral vector containing the cDNA for HDHB fused to mCHERRY was a gift from Dr. Steven Pruitt’s laboratory (Roswell Park Cancer Center). Lentiviral infection was performed in exponentially growing H2B-GFP-labeled MCF7 and T47D cells in the presence of Polybrene (Sigma Aldrich). Isogenic MCF7-RB-del and T47D-RB-del cell lines were developed by CRISPR mediated deletion using guide sequences designed to target exon 2 of RB1 as previously described [42].
Drug screen.
Cytation 5 and IncuCyte live cell imaging systems were used to perform drug screen analysis in isogenic MCF7 and T47D cell lines. Cells were seeded in 384-well dish and pretreated with DMSO or palbociclib (50 nM) or birinapant (250 nM) for 24 h. Following 24 h pretreatment, drugs from the library were added at two different doses 100 and 500 nM. Cell growth was monitored for 5 days and the growth rate of the cells in each well was calculated. To define drug efficacy, the growth rate for each compound was normalized to the mean of the DMSO wells within the drug plate. At the end of the drug screen the cell viability was measured by CTG assay. The normalized relative growth rate and cell viability were utilized for further downstream analysis. All the relative growth rates and cell viabilities of the drug library are shown in Table S1.
Knockdown experiments.
Cells were reverse transfected with siRNA using Dharmacon Human On-target plus siRNA: Cyclin D1 (L-003210–00-0005), AURKA (L-003545–01-0005), WEE1 (L-005050–00-0005) and non-targeting siRNA (D-001810–10-05). Transfection was performed using Lipofectamine RNAiMax Transfection Reagent (Invitrogen, 13778150) to a final concentration of 12.5 nM. Following 24 H transfection cells were replenished with media containing DMSO or test compound for another 48 h and 120 h for western blotting and CTG assay, respectively.
CTG assay.
To evaluate cell viability, ER+ breast cancer cells were seeded in 96-well dish (1000 cells/well). Following 24 H the cells were exposed to different concentrations of our experimental drugs as single agents or in combination and exposed up to 6 days. At the end of the experiment, the viability of the cells was measured using the CellTiterGlo reagent from Promega according the manufacturer’s protocol.
Colony formation assay.
MCF7-WT, T47D-WT, MCF7-RB-del, and T47D-RB-del cells were seeded in a six-well dish (5000 cells/well) and were treated with alpelisib (100 nM) in combination with birinapant (500 nM). Colonies were allowed to form for 15 days and the media with drugs were replenished every 5 days. To determine the impact of TNF pathway, MCF7-WT and RB-del cells were seeded in 6 wells dish (10,000 cells/well) and pretreated with PBS, TNF-α (5 ng/ μl) exogenous supplement, and the TNF-α antibody (100 ng/ μl). Following 24 h, the cells were exposed to DMSO and birinapant (500 nM) and the colonies were allowed to form for 15 days. Media was replenished with birinapant, TNF-α antibody and TNF-α for every 5 days.
To investigate the effect of birinapant in combination with palbociclib + fulvestrant, GFP-labeled MCF7-WT and MCF7-RB-del cells were seeded in 96-well dish (1000 cells/well) and treated with DMSO, palbociclib (50 nM) + fulvesetrant (100 nM), birinapant (500 nM) and the combination. The cell growth was monitored for 10 days using live cell imaging, and the drugs were replenished after 6 days. Data were exported to GraphPad Prism for statistical analysis and graph generation. Cells from each condition were seeded in triplicates and the experiment was performed at two independent times.
Organoid cell culture.
The MCF7 and T47D-WT/RB-del cell lines were seeded in 96-well dish (4000 cells/well) that were precoated with 50% Matrigel (Corning; 354234). Cells were allowed to form organoids up to 48 H and were treated with the experimental drugs. The viability of organoids was determined using Cell-Titer Glo 3D cell viability assay kit (Promega). Data were exported to GraphPad Prism for statistical analysis and graph generation. Cells from each condition were seeded in triplicates and the experiment was performed at two independent times.
Mice and xenografts.
NSG mice were maintained at Roswell Park Cancer Center animal care facilities. All animal care, drug treatments, and sacrifice were approved by the Roswell Park Cancer Center Institutional Animal Care and Use Committee (IACUC) in accordance with the NIH guide for the care and use of laboratory animals. Mice were subcutaneously implanted with the early passage MCF7 cells (1 × 107 cells/mouse) were subcutaneously injected into 8–10 weeks old female NSG mice. Mice were supplemented with estrogen pellet (17 mg/mouse) to promote tumor growth. Once the tumor volume reached 150–200 mm3 mice were randomized to different treatment cohorts as follows in a non-blinded manner. Vehicle (n = 9) (30% PEG300 + 5%Tween 80 + ddH2O) was administered orally. For the combination arm Alisertib (n = 8) (10 mg/kg) (30% PEG300 + 5%Tween 80 + ddH2O) was administered orally 6 days a week for 3 weeks. MK1775 (n = 8) (30 mg/kg) was administered orally every other day for 3 weeks. Birinapant (n = 6) (15 mg/kg) (40% PEG300, 5% Tween 80, 45% saline and 10% DMSO) was administered via IP injection for 3 days/week up to 3 weeks. Tumor size was measured every other day using digital calipers and the tumor volume was calculated based on the following equation: (greatest diameter × (shortest diameter2))/2. Mice were sacrificed, and the tumors were harvested at the end of treatment or when the tumor size reached 2000 mm3. Mice that became sick or found dead during the course of treatment were excluded from the analysis.
Patient cohort.
The association between RB loss and therapeutic outcome to CDK4/6 inhibitor was investigated using a total of 71 tumor specimens from 222 patients with metastatic ER+ breast cancer (NCT04526587) who received CDK4/6 inhibitor in combination with endocrine therapy. Patients with recurrent disease underwent either prior chemotherapy or endocrine therapy. Different methods of solid tumor genetic testing were performed, which included Foundation one testing (n = 56), Omniseq testing (n = 13), and tumors from two patients were subjected to both testing methods. Kaplan–Meier curves and associated log-rank tests were created by splitting these 71 patients into 2 groups who harbor wild-type RB (n = 68) and RB-deletion mutation (n = 3). The remaining patients with tumor tissues not subjected to genetic testing were excluded from our analysis. The primary endpoint, progression-free survival, was defined as the time, in months, from the first dose of a CDK4/6 inhibitor in combination with endocrine therapy to either scan- or marker-proven progression, or death while on therapy. Curves were completed using R version 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria) along with the ggplot2 package. The patient study was approved by Roswell Park Cancer Center Institutional review board (IRB). Informed consent was obtained from all the patients.
Gene expression analysis.
RNA was isolated from MCF7-WT and RB-del cell lines that were treated with palbociclib (250 nM) in combination with Everolimus (500 nM), Alisertib (200 nM) in combination with MK1775 (200 nM) and birinapant (500 nM) up to 48 h using RNeasy Plus kit (Quiagen). Extracted RNA was used for 50 bp paired-end RNA sequencing as described in previous studies [59, 60]. The fastq sequences were used as input to run the nf-core/RNA-Seq (version 3.3) pipeline installed on the High-Performance Computing cluster, Center for Computational Research, University at Buffalo. All sequences passed QC and aligned on GRCh37 reference genome using STAR2 with default parameters. Raw read counts were generated using RSEM with default parameters. For differential gene expression analysis, the DESeq2 (version 1.34.0) Bioconductor package was used to remove batch effects, normalize raw read counts and make final differentially expressed gene list. For visualization, the CompexHeatmap (version 2.2.0) Bioconductor package was used to create heat map for genes of interests. The downregulated and upregulated genes were generated based on the log-fold change and Student’s two-sided t test p value for each treatment conditions relative to the control from the normalized reads. Genes that were significantly upregulated and downregulated were used for gene ontology analysis using ENRICHR as described previously [45]. All log-fold change and statistical information is provided in the supplemental data tables S2–S4.
Data deposition.
RNA sequencing data are deposited in GEO: GSE182631 and GSE199919.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all members of the laboratory group and colleagues in the discussion and preparation of the manuscript. We thank Emily Schultz for acquiring the patient information and the genetic characteristics of the tumor tissues. Dr. Steven Pruitt provided the lentiviral vector for CDK2 sensor. Drug screening was performed through Small Molecule Screening facility at RoswellPark Cancer Center. We would like to thank Dr. Sandra Sexton, Facility director of Laboratory Animal Resources at Roswell Park for assisting us with xenografts. The research was supported by a grant to AKW and ESK from National Cancer Institute (NCI).
COMPETING INTERESTS
ESK and AKW have received research funding from Eli Lilly, Novartis and Pfizer over the last 5 years. There is no current research support from these entities and the study was written in the absence of input from any pharmaceutical company.
Footnotes
ADDITIONAL INFORMATION
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41388-022-02362-2.
Reprints and permission information is available at http://www.nature.com/reprints
REFERENCES
- 1.Gong Y, Liu YR, Ji P, Hu X, Shao ZM. Impact of molecular subtypes on metastatic breast cancer patients: a SEER population-based study. Sci Rep. 2017;7:45411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad A. Breast cancer statistics: recent trends. Adv Exp Med Biol. 2019;1152:1–7. [DOI] [PubMed] [Google Scholar]
- 3.Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980–9. [PubMed] [Google Scholar]
- 4.Howell A. The endocrine prevention of breast cancer. Best Pract Res Clin Endocrinol Metab. 2008;22:615–23. [DOI] [PubMed] [Google Scholar]
- 5.Mouridsen H, Gershanovich M, Sun Y, Perez-Carrion R, Boni C, Monnier A, et al. Superior efficacy of letrozole versus tamoxifen as first-line therapy for postmenopausal women with advanced breast cancer: results of a phase III study of the International Letrozole Breast Cancer Group. J Clin Oncol. 2001;19:2596–606. [DOI] [PubMed] [Google Scholar]
- 6.McKeage K, Curran MP, Plosker GL. Fulvestrant: a review of its use in hormone receptor-positive metastatic breast cancer in postmenopausal women with disease progression following antiestrogen therapy. Drugs. 2004;64:633–48. [DOI] [PubMed] [Google Scholar]
- 7.Manna S, Holz MK. Tamoxifen action in ER-negative breast cancer. Sign Transduct Insights. 2016;5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiff R, Massarweh SA, Shou J, Bharwani L, Mohsin SK, Osborne CK. Cross-talk between estrogen receptor and growth factor pathways as a molecular target for overcoming endocrine resistance. Clin Cancer Res. 2004;10:331S–6S. [DOI] [PubMed] [Google Scholar]
- 9.Pu M, Messer K, Davies SR, Vickery TL, Pittman E, Parker BA, et al. Research-based PAM50 signature and long-term breast cancer survival. Breast Cancer Res Treat. 2020;179:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31:2783–90. [DOI] [PubMed] [Google Scholar]
- 11.Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle. 2006;5:2198–202. [DOI] [PubMed] [Google Scholar]
- 12.Mills JN, Rutkovsky AC, Giordano A. Mechanisms of resistance in estrogen receptor positive breast cancer: overcoming resistance to tamoxifen/aromatase inhibitors. Curr Opin Pharmacol. 2018;41:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provenzano A, Kurian S, Abraham J. Overcoming endocrine resistance in breast cancer: role of the PI3K and the mTOR pathways. Exp Rev Anticancer Ther. 2013;13:143–7. [DOI] [PubMed] [Google Scholar]
- 14.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 15.Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, et al. mTORC1 inhibition is required for sensitivity to PI3K p110alpha inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5:196ra99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–65. [DOI] [PubMed] [Google Scholar]
- 17.Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci. 2013;38:12–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thu KL, Soria-Bretones I, Mak TW, Cescon DW. Targeting the cell cycle in breast cancer: towards the next phase. Cell Cycle. 2018;17:1871–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin Cancer Res. 2014;20:3379–83. [DOI] [PubMed] [Google Scholar]
- 21.Turner NC, Slamon DJ, Ro J, Bondarenko I, Im SA, Masuda N, et al. Overall survival with palbociclib and fulvestrant in advanced breast cancer. N Engl J Med. 2018;379:1926–36. [DOI] [PubMed] [Google Scholar]
- 22.Dickler MN, Tolaney SM, Rugo HS, Cortes J, Dieras V, Patt D, et al. MONARCH 1, A Phase II study of abemaciclib, a CDK4 and CDK6 inhibitor, as a single agent, in patients with refractory HR(+)/HER2(−) metastatic breast cancer. Clin Cancer Res. 2017;23:5218–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–32. [DOI] [PubMed] [Google Scholar]
- 24.O’Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8:1390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnedos M, Bayar MA, Cheaib B, Scott V, Bouakka I, Valent A, et al. Modulation of Rb phosphorylation and antiproliferative response to palbociclib: the preoperative-palbociclib (POP) randomized clinical trial. Ann Oncol. 2018;29:1755–62. [DOI] [PubMed] [Google Scholar]
- 27.Condorelli R, Spring L, O’Shaughnessy J, Lacroix L, Bailleux C, Scott V, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29:640–5. [DOI] [PubMed] [Google Scholar]
- 28.Wander SA, Cohen O, Gong X, Johnson GN, Buendia-Buendia JE, Lloyd MR, et al. The genomic landscape of intrinsic and acquired resistance to cyclin-dependent kinase 4/6 inhibitors in patients with hormone receptor positive metastatic breast cancer. Cancer Discov. 2020;10:174–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Razavi P, Li Q, Toy W, Liu B, Ping C, et al. Loss of the FAT1 tumor suppressor promotes resistance to CDK4/6 inhibitors via the hippo pathway. Cancer Cell. 2018;34:893–905e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Presti D, Quaquarini E. The PI3K/AKT/mTOR and CDK4/6 pathways in endocrine resistant HR+/HER2-metastatic breast cancer: biological mechanisms and new treatments. Cancers. 2019;11:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaloglou C, Crafter C, Siersbaek R, Delpuech O, Curwen JO, Carnevalli LS, et al. Combined inhibition of mTOR and CDK4/6 is required for optimal blockade of E2F function and long-term growth inhibition in estrogen receptor-positive breast cancer. Mol Cancer Ther. 2018;17:908–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarducci C, Bonechi M, Benelli M, Biagioni C, Boccalini G, Romagnoli D, et al. Cyclin E1 and Rb modulation as common events at time of resistance to palbociclib in hormone receptor-positive breast cancer. NPJ Breast Cancer. 2018;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asghar US, Kanani R, Roylance R, Mittnacht S. Systematic review of molecular biomarkers predictive of resistance to CDK4/6 inhibition in metastatic breast cancer. JCO Precis Oncol 2022;6:e2100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dean JL, McClendon AK, Knudsen ES. Modification of the DNA damage response by therapeutic CDK4/6 inhibition. J Biol Chem. 2012;287:29075–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oser MG, Fonseca R, Chakraborty AA, Brough R, Spektor A, Jennings RB, et al. Cells Lacking the RB1 tumor suppressor gene are hyperdependent on Aurora B kinase for survival. Cancer Discov. 2018;9:230–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong X, Du J, Parsons SH, Merzoug FF, Webster Y, Iversen PW, et al. Aurora-A kinase inhibition is synthetic lethal with loss of the RB1 tumor suppressor gene. Cancer Discov. 2019;9:48–63. [DOI] [PubMed] [Google Scholar]
- 38.Witkiewicz AK, Chung S, Brough R, Vail P, Franco J, Lord CJ, et al. Targeting the vulnerability of RB tumor suppressor loss in triple-negative breast cancer. Cell Rep. 2018;22:1185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigelt B, Warne PH, Downward J. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 2011;30:3222–33. [DOI] [PubMed] [Google Scholar]
- 40.Gomez Tejeda Zanudo J, Mao P, Alcon C, Kowalski K, Johnson GN, Xu G, et al. Cell line-specific network models of ER(+) breast cancer identify potential PI3Kalpha Inhibitor Resistance Mechanisms and Drug Combinations. Cancer Res. 2021;81:4603–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226–31. [DOI] [PubMed] [Google Scholar]
- 42.Knudsen ES, Nambiar R, Rosario SR, Smiraglia DJ, Goodrich DW, Witkiewicz AK. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun Biol. 2020;3:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature 2019;569:560–4. [DOI] [PubMed] [Google Scholar]
- 44.Kumarasamy V, Vail P, Nambiar R, Witkiewicz AK, Knudsen ES. FunctionaL determinants of cell cycle plasticity and sensitivity to CDK4/6 inhibition. Cancer Res. 2021;81:1347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumarasamy V, Ruiz A, Nambiar R, Witkiewicz AK, Knudsen ES. Chemotherapy impacts on the cellular response to CDK4/6 inhibition: distinct mechanisms of interaction and efficacy in models of pancreatic cancer. Oncogene. 2020;39:1831–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawlinson R, Massey AJ. gammaH2AX and Chk1 phosphorylation as predictive pharmacodynamic biomarkers of Chk1 inhibitor-chemotherapy combination treatments. BMC Cancer. 2014;14:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster JD, Vucic D. The balance of TNF mediated pathways regulates inflammatory cell death signaling in healthy and diseased tissues. Front Cell Dev Biol. 2020;8:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lalaoui N, Merino D, Giner G, Vaillant F, Chau D, Liu L, et al. Targeting triple-negative breast cancers with the Smac-mimetic birinapant. Cell Death Differ. 2020;27:2768–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yardley DA, Noguchi S, Pritchard KI, Burris HA 3rd, Baselga J, Gnant M, et al. Everolimus plus exemestane in postmenopausal patients with HR(+) breast cancer: BOLERO-2 final progression-free survival analysis. Adv Ther. 2013;30:870–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franco J, Balaji U, Freinkman E, Witkiewicz AK, Knudsen ES. Metabolic reprogramming of pancreatic cancer mediated by CDK4/6 inhibition elicits unique vulnerabilities. Cell Rep. 2016;14:979–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knudsen ES, Shapiro GI, Keyomarsi K. Selective CDK4/6 inhibitors: biologic outcomes, determinants of sensitivity, mechanisms of resistance, combinatorial approaches, and pharmacodynamic biomarkers. Am Soc Clin Oncol Educ Book. 2020;40:115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JW, Parameswaran J, Sandoval-Schaefer T, Eoh KJ, Yang DH, Zhu F, et al. Combined Aurora Kinase A (AURKA) and WEE1 inhibition demonstrates synergistic antitumor effect in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2019;25:3430–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knudsen ES, Pruitt SC, Hershberger PA, Witkiewicz AK, Goodrich DW. Cell cycle and beyond: exploiting new RB1 controlled mechanisms for cancer therapy. Trends Cancer. 2019;5:308–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Condon SM, Mitsuuchi Y, Deng Y, LaPorte MG, Rippin SR, Haimowitz T, et al. Birinapant, a smac-mimetic with improved tolerability for the treatment of solid tumors and hematological malignancies. J Med Chem. 2014;57:3666–77. [DOI] [PubMed] [Google Scholar]
- 56.Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43:449–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chau BN, Chen TT, Wan YY, DeGregori J, Wang JY. Tumor necrosis factor alpha-induced apoptosis requires p73 and c-ABL activation downstream of RB degradation. Mol Cell Biol. 2004;24:4438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boutillier AL, Trinh E, Loeffler JP. Caspase-dependent cleavage of the retinoblastoma protein is an early step in neuronal apoptosis. Oncogene. 2000;19:2171–8. [DOI] [PubMed] [Google Scholar]
- 59.Knudsen ES, Balaji U, Mannakee B, Vail P, Eslinger C, Moxom C, et al. Pancreatic cancer cell lines as patient-derived avatars: genetic characterisation and functional utility. Gut. 2018;67:508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knudsen ES, Kumarasamy V, Ruiz A, Sivinski J, Chung S, Grant A, et al. Cell cycle plasticity driven by MTOR signaling: integral resistance to CDK4/6 inhibition in patient-derived models of pancreatic cancer. Oncogene. 2019;38:3355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.