Abstract
In this study, 30 strains of the pathogenic free-living amoeba Naegleria fowleri were investigated by using the randomly amplified polymorphic DNA (RAPD) method. The present study confirmed our previous finding that RAPD variation is not correlated with geographical origin. In particular, Mexican strains belong to the variant previously detected in Asia, Europe, and the United States. In France, surprisingly, strains from Cattenom gave RAPD patterns identical to those of the Japanese strains. In addition, all of these strains, together with an additional French strain from Chooz, exhibited similarities to South Pacific strains. The results also confirmed the presence of numerous variants in Europe, whereas only two variants were detected in the United States. The two variants found in the United States were different from the South Pacific variants. These findings do not support the previous hypothesis concerning the origin and modes of dispersal of N. fowleri.
Molecular markers are of great importance for understanding the genetic structures and the modes of dispersion of various species. This is particularly true in microorganisms for which morphological and biochemical criteria are very limited. For these organisms, randomly amplified polymorphic DNA (RAPD) (28, 29) is one of the most variable markers which allow genome analysis at the intraspecific level. The RAPD method has proven to be a powerful molecular tool for diversity studies and typing, especially with unicellular organisms (1, 10, 11, 17, 24). This method consists of using a single arbitrary short primer that generates polymorphic fragments following PCR amplification. These polymorphic fragments, used as fingerprints, allow discrimination between different strains within species. In addition, only very small quantities of DNA are required, and no prior knowledge of the species genome is necessary. This is very useful for environmental isolates which are difficult to grow under laboratory conditions.
The RAPD method has proven to be suitable for population studies of the free-living amoeba Naegleria fowleri. This thermotolerant species causes fatal primary amoebic meningoencephalitis (PAM) in humans (2). Although it is present throughout the world, particularly in warm-water habitats, its population structure and mode of colonization are unknown. Variability within the species was first detected by restriction fragment length polymorphisms (RFLP) in whole-cell DNA (6, 7, 18) and in mitochondrial or ribosomal DNA plasmids (3, 19). According to De Jonckheere, the pathogenic species N. fowleri could be separated into two major groups, a European group and an oceanic group (6, 7). Isolates from the United States displayed both RFLP types. Other molecular methods, such as interrepeat PCR, revealed very little polymorphism within the species (26), whereas RAPD markers detected a high level of diversity, with several variants present on several continents (20).
In an attempt to detect further variability in N. fowleri and to better understand the dispersal of this organism, additional strains from America, Asia, and Europe were analyzed by using RAPD markers.
MATERIALS AND METHODS
N. fowleri strains.
The origins and the years of isolation of the N. fowleri strains used are shown in Table 1. American strains EPA 988s-GP2, EPA 1020s-GP2, and EPA 1027w-GP2 were obtained from the laboratory of D. John. The French strains were isolated from the nuclear power stations at Chooz, Cattenom, Golfech, and St. Laurent. All of the strains were grown axenically on SCGYEM medium (4).
TABLE 1.
Geographic origins and years of isolation of the N. fowleri strains examined
| Strain | Origin | Year of isolation | Sourcea |
|---|---|---|---|
| NH1 | New Zealand | 1974 | H |
| Morgan | Australia | 1972 | H |
| ORAM | Australia | 1971 | H |
| PA34 | Australia | 1972 | E |
| PA90 | Australia | 1972 | E |
| PA105 | Australia | 1972 | E |
| PA117 | Australia | 1972 | E |
| MW4u | Australia | 1972 | E |
| J16(1)42E | Japan | 1990 | E |
| J26(50)45E | Japan | 1990 | E |
| Na1165b | Cattenom, France | 1988 | E |
| Ch2-1-e13b | Chooz, France | 1997 | E |
| G1-3-f18 | Golfech, France | 1997 | E |
| L2-1-d11 | St. Laurent, France | 1994 | E |
| MCM | England | 1978 | H |
| RPE-1 | Mexico | 1990 | H |
| PLC-2 | Mexico | 1990 | H |
| RTW-4 | Mexico | 1990 | H |
| RC-5 | Mexico | 1990 | H |
| EjI-SON | Mexico | 1990 | E |
| Enterprise | United States | 1976 | E |
| 124 | United States | 1984 | E |
| TY | United States | 1969 | H |
| CJ | United States | 1967 | H |
| WM | United States | 1969 | H |
| SW1 | United States | 1976 | E |
| SW2 | United States | 1976 | E |
| EPA 988s-GP2 | United States | 1994 | E |
| EPA 1020s-GP2 | United States | 1994 | E |
| EPA 1027w-GP2 | United States | 1994 | E |
H, human; E, environmental.
DNA isolation and PCR amplification.
DNA was isolated by phenol-chloroform extraction as described previously (6). Three different Taq DNA polymerases (Appligene, Illkirch, France; Promega, Madison, Wis.; Gibco-BRL, Burlington, Ontario, Canada) were used to test the reproducibility of the RAPD method. The 10-nucleotide primers (Bioprobe Systems, Montreuil, France) used were primers A1 (5′CAGGCCCTTC3′), A15 (5′TTCCGAACCC3′), B10 (5′CTGCTGGGAC3′), B12 (5′CCTTGACGCA3′), and B18 (5′CCACAGCAGT3′). Each amplification reaction mixture (final volume, 25 μl) contained 10 mM Tris-HCl (pH 9), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, each deoxynucleoside triphosphate at a concentration of 100 μM, 4 μM primer, 1 U of Taq DNA polymerase, and 25 to 50 ng of genomic DNA. Amplifications were carried out with two thermal cyclers, a GeneAmp model 2400 thermal cycler and a model 9600 thermal cycler (Perkin-Elmer-Cetus, Norwalk, Conn.), and each amplification reaction consisted of one denaturation step at 94°C for 1 min, followed by 35 cycles consisting of denaturation at 94°C for 30 s, annealing at 36°C for 45 s, and extension at 72°C for 45 s. A final elongation step at 72°C for 5 min was included. Amplification products were visualized in a 1.2% agarose gel or in a 3% NuSieve gel in Tris-borate-EDTA buffer stained with ethidium bromide. A 123-bp DNA ladder (Gibco-BRL) was used as a size marker in the gels.
RESULTS AND DISCUSSION
The efficiency of the RAPD method when it is used with microorganisms has been discussed previously (25, 27). Artifactual variations generated by the RAPD markers are a possible source of error (9). For this reason, it was essential to test the reproducibility of the RAPD method by varying the PCR conditions (namely, by modifying the MgCl2 concentration and the primer/template DNA ratio). In addition, different thermal cyclers with different cycle times, as well as different thermostable DNA polymerases, were used. The amplification protocol described here (see above) is a procedure that is suitable for the three DNA polymerases that we used in this study. As observed previously (20), slight variations were observed when different parameters were varied. Perturbations were observed particularly when the concentration of the primers was modified for a given amount of DNA template (25 ng). When the primer concentration was between 0.4 and 1.0 μM, the number of amplified fragments increased. When the primer concentration was less than 0.4 μM, amplification was weaker and the polymorphic fragments were difficult to visualize. Previously, we detected intraspecific variation in N. fowleri by using a combination of five RAPD discriminating primers (20). The different patterns, reflecting the gain or loss of fragments, made it possible to distinguish several variants distributed throughout the world. Of the five primers previously used with the Biometra thermal cycler (20), only primer B7 gave poor amplification with the Perkin-Elmer thermal cycler. An additional primer, B10, was included in the analyses. The patterns obtained for the 30 strains examined with primers A1, A15, B10, B12, and B18 were consistent with most of the previous results (20). As shown in Table 2, the RAPD markers revealed five major variants distributed on different continents.
TABLE 2.
RAPD patterns for the five variants
| Variant | Strain(s) | Additional amplified fragment generated witha:
|
||||
|---|---|---|---|---|---|---|
| Primer A1 | Primer A15 | Primer B10 | Primer B12 | Primer B18 | ||
| South Pacific | Australian strains Morgan, ORAM, MW4u, PA90, PA105, PA34, and PA117 | (−) | + | − | − | − |
| New Zealand strain NH1 | (−) | + | + | + | − | |
| Chooz | French strain Ch2-1-e13b | (−) | + | + | − | − |
| Cattenom | French strain Na1165b | (+) | + | + | − | + |
| Japanese strains J16(1)42E and J26(50)45E | (+)/(−)b | + | + | − | + | |
| Widespread | French strain G1-3-f18; United States strains 124 and Enterprise; Mexican strains RPE-1, PLC-2, RTW-4, RC-5, and EjI-SON | (−) | + | + | +c | − |
| Euro-American | French strain L2-1-d11; English strain MCM; United States strains CJ, SW1, SW2, TY, WM, EPA 988s-GP2, EPA 1020s-GP2, and EPA 1027w-GP2 | (−) | − | + | + | − |
+, additional amplified fragment present; −, additional amplified fragment absent; (+), strong amplified fragment present; (−), weak amplified fragment present.
Strongly and weakly amplified fragment observed in J16(1)42E and J26(50)45E, respectively.
A specific band in addition to the additional fragment was present.
South Pacific variant.
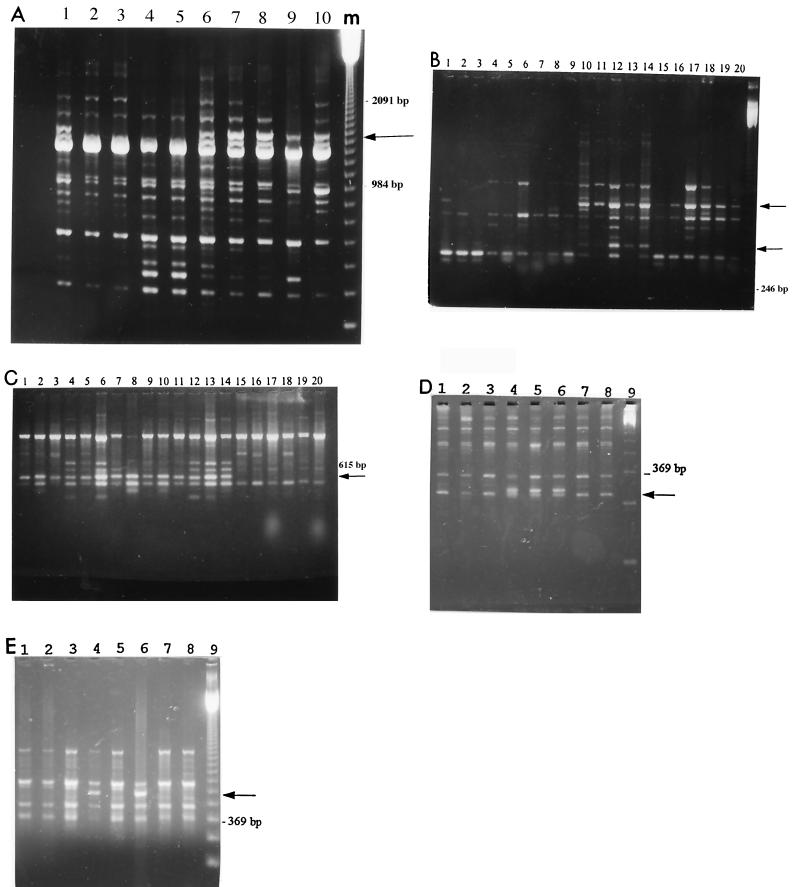
The seven new Australian strains investigated (ORAM, Morgan, PA117, PA105, PA34, PA90, and MW4u) produced RAPD profiles similar to the profiles obtained previously (20). As shown in Fig. 1A and Table 2, the Australian strains were distinguishable from the other strains since they did not generate the additional fragment with primer B10. In addition, as observed previously (20), the New Zealand strains represented here by strain NH1 were different from the Australian strains when the primer B12 profile was examined (Fig. 1B). Thus, the RAPD results support the conclusion from the RFLP and PCR interrepeat analyses that South Pacific strains are different from other strains (3, 6, 7, 18, 26).
FIG. 1.
(A through E) RAPD patterns obtained with the GeneAmp model 2400 thermal cycler and primers B10, B12, A15, B18, and A1, respectively, by using DNAs from various strains of N. fowleri. With these five primers, the three Taq DNA polymerases that we used produced similar discriminating fragments. The Taq DNA polymerase from Gibco-BRL was used for the gels shown in panels A and D, and the Taq DNA polymerase from Appligene was used for the gels shown in panels B, C, and E. The arrows indicate the positions of the additional amplified fragments. (A) All of the strains except the Australians strains produced the 1,580-bp additional fragment generated with primer B10. Lane 1, NH1; lane 2, Morgan; lane 3, ORAM; lane 4, PA90; lane 5, MW4u; lane 6, J26(50)45E; lane 7, Na1165b; lane 8, Ch2-1-e13b; lane 9, RC-5; lane 10, L2-1-d11; lane m, molecular size marker (123-bp DNA ladder). (B and C) Lane 1, NH1; lane 2, Morgan; lane 3, ORAM; lane 4, MW4u; lane 5, PA105; lane 6, Ch2-1-e13b; lane 7, Na1165b; lane 8, J16(1)42E; lane 9, J26(50)45E; lane 10, G1-3-f18; lane 11, RC-5; lane 12, EjI-SON; lane 13, 124; lane 14, Enterprise; lane 15, TY; lane 16, CJ; lane 17, EPA 988s-GP2; lane 18, WM; lane 19, MCM; lane 20, L2-1-d11. (B) When primer B12 was used, in addition to the 930-bp fragment, one specific band at 510 bp differentiated the widespread variant from the other variants. (C) When primer A15 was used, the Euro-American variant was distinguishable from the other variants since it did not produce the 615-bp additional fragment. (D and E) Lane 1, NH1; lane 2, Morgan; lane 3, Ch2-1-e13b; lane 4, J16(1)42E; lane 5, J26(50)45E; lane 6, Na1165b; lane 7, 124; lane 8, EPA 988s-GP2; lane 9, 123-bp DNA ladder. (D) When primer B18 was used, the two Japanese strains and the French strain from Cattenom were the only strains which produced the 310-bp additional fragment. (E) Only one of the two Japanese strains produced the strongly amplified 600-bp fragment obtained with primer A1.
Chooz variant.
Surprisingly, the French Chooz site strain, strain Ch2-1-e13b, produced the same patterns as the South Pacific strains with most of the primers, demonstrating that similar variants were present on different continents (see in particular primers B12 and A15 in Fig. 1B and C and Table 2). The Chooz strain could be differentiated from the Australian strains only by using primer B10 (Fig. 1A).
Cattenom variant.
Another RAPD variant, the variant from Cattenom, France, represented by Na1165b, also exhibited similarities with Australian strains but differed from them when primers B10, B18, and A1 were used (Fig. 1A, D, and E). However, the strongly amplified fragment generated by primer A1 was detected previously in some Australian and New Zealand strains (20). The Cattenom variant, considered to be an Australian RFLP type (21), was detected previously by using the RAPD technique (20). Moreover, the patterns of the two Japanese strains, J16(1)42E and J26(50)45E, were similar to those of the Cattenom variant (Table 2). Indeed, as shown in Fig. 1D, primer B18, although previously thought to give a specific pattern with the Cattenom variant, generated the same small additional fragment with the Japanese strains. Within this geographic group, little variation was observed among the primer A1 profiles, since only strain J16(1)42E produced the strongly amplified fragment (Fig. 1E).
Widespread variant.
The strain from the French Golfech site gave RAPD profiles similar to those of the five Mexican strains (RPE-1, PLC-2, RTW-4, RC-5, and EjI-SON) and the American strains (124 and Enterprise). In particular, these strains generated the same additional fragments with primers B12 and A15 (Fig. 1B and C). Moreover, a characteristic 510-bp fragment amplified by primer B12 differentiated all of the strains belonging to this variant from the other strains (Fig. 1B). Interestingly, this group produced profiles similar to those of strains AR12 (from the United States), NG894 (Hong Kong), and Bugey (France) (20). Thus, American strains which previously had been found to exhibit the Australian RFLP type (6, 7) now appear to represent a distinct RAPD variant. Recently, hybridization with a repetitive clone detected RFLP that clustered strains NG894 and 124 and a Czech strain (16). It is clear now that there is a well-dispersed RAPD variant.
Euro-American variant.
All of the American strains examined except 124 and Enterprise (i.e., strains CJ, SW1, SW2, TY, and WM), as well as the English strain MCM and the French strain from St. Laurent (L2-1-d11), produced profiles corresponding to the profiles detected previously in France and in the United States (i.e., the profiles for the European strain KUL and American strains Lovell, Lee, and HB1). In contrast to the other variants, the Euro-American variant did not produce the additional fragment with primer A15 (Fig. 1C). The same was true for American strains EPA 988s-GP2, EPA 1020s-GP2, and EPA 1027w-GP2, recently isolated by D. John (13). This variant, which includes American and European strains, was identified by most authors when the RFLP techniques were used (3, 6, 7, 15, 18).
N. fowleri is the only pathogenic free-living amoeba which produces 100% mortality in inoculated mice, suggesting that there is no variation in the degree of virulence in this species (13). However, virulence does decrease after axenic cultivation (5, 14). In previous studies (6, 7, 18, 26), no differences in virulence were observed between isolates obtained from cases of PAM and environmental strains. Similarly, as indicated in Tables 1 and 2, three of the five RAPD variants (the South Pacific, Euro-American, and widespread variants) contained at least one isolate obtained from a case of PAM together with environmental isolates. This strongly suggests that the molecular variation detected in N. fowleri is not correlated with the degree of virulence of the pathogenic species.
Diversity observed in the United States and in Europe.
The 10 new strains originating from the United States were not different from the American strains studied previously (20). Consequently, only the Euro-American and widespread RAPD variants have been found in this region. In addition, neither of these two variants was similar to the Australian variant. This is in contrast with the findings of De Jonckheere (6, 7) but supports the recent RFLP results of Kilvington and Beeching (16). The American strains found by De Jonckheere to display the Australian type were classified by Kilvington and Beeching as a new RFLP type of strains from very different geographic areas. Moreover, this new RFLP cluster correlates perfectly with the widespread cluster determined by the RAPD method.
Four different RAPD variants were obtained from the four French sites (Cattenom, Chooz, Golfech, and St. Laurent), confirming once again the high intraspecific diversity observed in France. Furthermore, in France, the distinct RAPD variants were isolated from different nuclear power station sites that are located within a 800-km radius, while the American strains were collected from a much wider geographic area. The detection of only two RAPD variants in the United States could be explained by the predominance of these variants compared with other minor strains.
Dispersion of N. fowleri.
Two single-characteristic RFLP patterns were found in strains from Europe and Oceania, whereas both types were observed in the strains from the United States (6, 7). On the basis of this heterogeneity, De Jonckheere formulated two hypotheses: either N. fowleri originated in the United States, or the two types colonized this country. The Japanese strains analyzed by the RFLP method turned out to be most similar to the strains from Oceania, confirming the hypothesis that N. fowleri strains are correlated with geographical regions. The RAPD results did not corroborate this hypothesis, since greater diversity was found in Europe than in the United States and very similar variants were found on different continents. The Hong Kong strain, NG894, was identical to some European and American strains (20). The results of the recent study of Kilvington and Beeching also supported this finding (16).
Examination of the Cattenom and Chooz sites demonstrated that there are two South Pacific-like variants in Europe. In agreement with our previous hypothesis, we concluded that the Cattenom variant is not a local variant since it has also been detected in England by RFLP analysis (16). The strain from Chooz provides additional evidence that strains related to the South Pacific variant are present in Europe. The presence of South Pacific-like strains in Europe could indicate that one ancestral clone successfully and progressively colonized large geographical areas. The clones derived from the primitive clone could have become established in new regions and evolved independently, generating distinct but related variants. However, this interpretation is not entirely satisfactory since it cannot account for the fact that the Cattenom variant was found to be identical to the Japanese strains. We expected to find some differences between French and Japanese strains. In addition, slight differences were found between the Japanese strains from two geographically close areas. Variation was also found in an RFLP analysis (8). This could indicate that the Japanese strains are sufficiently ancient to have diverged. One possible explanation is that a clone that produces a profile corresponding to the Cattenom profile has recently been introduced into new regions. Although little is known about the mode of dissemination of N. fowleri, some factors could contribute to its proliferation in new areas. Amoeba species have been found in various organisms, particularly birds (12). It is possible that these organisms are involved in the introduction of pathogenic species into distant countries. This hypothesis implies that different variants are present in the same localities. Another possible explanation is that different colonizations occurred in Europe and Oceania at different times. The geographical distribution of the Cattenom variant could be the result of recent colonization. The coexistence of distinct variants in one site has been observed previously (20, 21).
The wide geographic distribution of some variants could suggest that clonal dispersion occurs. This mode of propagation has been observed for several protozoan organisms (23), including other Naegleria species (22). The similarities between the New Zealand and Australia strains and the Chooz and Cattenom strains (classified as South Pacific and South Pacific-like, respectively) could suggest that they all were derived from a unique ancestral group. The slight heterogeneity detected in this cluster, the variation in the South Pacific strains, and the distinction between the two geographically close French variants could indicate that the cluster represents an ancient group. On the other hand, the variants detected in the United States were more homogeneous genetically, and this could suggest that colonization was more recent. In all cases, RAPD data did not allow the origin of N. fowleri dispersion to be determined. In contrast to the hypothesis of De Jonckheere (6, 7), the high level of European diversity could indicate that colonization of the pathogenic species started in Europe. However, this conclusion is premature, since several regions, such as Africa and South America, have not been considered yet. Similarly, the strains from Asia and Oceania analyzed were not sufficiently representative of these regions. Extensive analysis is needed to establish the relatedness between the different variants. The results should allow retracing of the evolutionary history of the pathogenic species N. fowleri and a better understanding of its mode of dispersal throughout the world.
ACKNOWLEDGMENTS
We are grateful to D. John for providing N. fowleri strains. We thank S. Roche and C. Jullien for identification of the environmental strains and D. Tuffery for technical assistance. We also thank J. Briolay and C. Gautier (Centre d’Analyse Moléculaire de la Biodiversité) for laboratory facilities and S. Brown for correcting the manuscript.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter R F. Description of a Naegleria sp. isolated from two cases of primary amoebic meningo-encephalitis, and of the experimental pathological changes induced by it. J Pathol. 1970;100:217–244. doi: 10.1002/path.1711000402. [DOI] [PubMed] [Google Scholar]
- 3.Clark C G, Cross G A M, De Jonckheere J F. Evaluation of evolutionary divergence in the genus Naegleria by analysis of ribosomal DNA plasmid restriction patterns. Mol Biochem Parasitol. 1989;34:281–296. doi: 10.1016/0166-6851(89)90057-1. [DOI] [PubMed] [Google Scholar]
- 4.De Jonckheere J F. Use of an axenic medium for differentiation between pathogenic and nonpathogenic Naegleria fowleri isolates. Appl Environ Microbiol. 1977;33:751–757. doi: 10.1128/aem.33.4.751-757.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Jonckheere J. Differences in virulence of Naegleria fowleri. Pathol Biol. 1979;8:453–458. [PubMed] [Google Scholar]
- 6.De Jonckheere J F. Characterization of Naegleria species by restriction endonuclease digestion of whole-cell DNA. Mol Biochem Parasitol. 1987;24:55–66. doi: 10.1016/0166-6851(87)90115-0. [DOI] [PubMed] [Google Scholar]
- 7.De Jonckheere J F. Geographic origin and spread of pathogenic Naegleria fowleri deduced from restriction enzyme patterns of repeated DNA. Biosystems. 1988;21:269–275. doi: 10.1016/0303-2647(88)90022-6. [DOI] [PubMed] [Google Scholar]
- 8.De Jonckheere J F, Yagita K, Endo T. Restriction-fragment-length polymorphism and variation in electrophoretic karyotype in Naegleria fowleri from Japan. Parasitol Res. 1992;78:475–478. doi: 10.1007/BF00931566. [DOI] [PubMed] [Google Scholar]
- 9.Ellsworth D L, Rittenhouse K D, Honeycutt R L. Artifactual variation in randomly amplified polymorphic DNA banding patterns. BioTechniques. 1993;14:214–217. [PubMed] [Google Scholar]
- 10.Fani R, Damiani G, Di Serio C, Gallori E, Grifoni A, Bazzicalupo M. Use of random amplified polymorphic DNA (RAPD) for generating specific DNA probes for microorganisms. Mol Ecol. 1993;2:243–250. doi: 10.1111/j.1365-294x.1993.tb00014.x. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z G, Johnson A M. Genetic characterization of Toxoplasma gondii strains by random amplified polymorphic DNA polymerase chain reaction. Parasitology. 1995;111:127–132. doi: 10.1017/s0031182000064866. [DOI] [PubMed] [Google Scholar]
- 12.Jadin J B, Willaert E, Hermanne J. Présence d’amibes limax dans l’intestin de l’homme et des animaux. Bull Acad R Sci Outre-Mer. 1973;3:520–526. [Google Scholar]
- 13.John D T, Howard M J. Seasonal distribution of pathogenic free-living amebae in Oklahoma waters. Parasitol Res. 1995;81:193–201. doi: 10.1007/BF00937109. [DOI] [PubMed] [Google Scholar]
- 14.Kadlec V. Different virulence of Naegleria fowleri strains isolated from a swimming pool. Folia Parasitol (Prague) 1981;28:97–103. [PubMed] [Google Scholar]
- 15.Kilvington S, Beeching J. Identification and epidemiological typing of Naegleria fowleri with DNA probes. Appl Environ Microbiol. 1995;61:2071–2078. doi: 10.1128/aem.61.6.2071-2078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilvington S, Beeching J. Detection of a novel restriction fragment length polymorphism type in Naegleria fowleri (free-living amoeba) isolates from electricity power stations in England and France. Eur J Protistol. 1997;33:186–191. [Google Scholar]
- 17.Lehmann P F, Lin D, Lasker B A. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J Clin Microbiol. 1992;30:3249–3254. doi: 10.1128/jcm.30.12.3249-3254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaughlin G L, Brandt F H, Visvesvara G S. Restriction fragment length polymorphisms of the DNA of selected Naegleria and Acanthamoeba amebae. J Clin Microbiol. 1988;26:1655–1658. doi: 10.1128/jcm.26.9.1655-1658.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milligan S M, Band R N. Restriction endonuclease analysis of mitochondrial DNA as an aid in the taxonomy of Naegleria and Vahlkampfia. J Protozool. 1988;35:198–204. doi: 10.1111/j.1550-7408.1988.tb04323.x. [DOI] [PubMed] [Google Scholar]
- 20.Pélandakis M, Kaundun S S, De Jonckheere J F, Pernin P. DNA diversity among the free-living amoeba Naegleria fowleri detected by the RAPD method. FEMS Microbiol Lett. 1997;151:31–39. [Google Scholar]
- 21.Pernin P, De Jonckheere J F. Appearance in Europe of Naegleria fowleri displaying the Australian type of restriction-fragment-length-polymorphism. Parasitol Res. 1992;78:479–481. doi: 10.1007/BF00931567. [DOI] [PubMed] [Google Scholar]
- 22.Pernin P, Cariou M L. Evidence for clonal structure of natural populations of free-living amoebae of the genus Naegleria. Genet Res. 1997;69:173–181. [Google Scholar]
- 23.Tibayrenc M, Kjellberg F, Ayala F J. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tibayrenc M, Neubauer K, Barnabé C, Guerrini F, Skarecky D, Ayala F J. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Belkum A, De Jonckheere J, Quint W G V. Genotyping Naegleria spp. and Naegleria fowleri isolates by interrepeat polymerase chain reaction. J Clin Microbiol. 1992;30:2595–2598. doi: 10.1128/jcm.30.10.2595-2598.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Belkum A. Amplification-based DNA fingerprinting: from artifactual to definitive typing and in between. J Clin Microbiol. 1997;35:3008–3009. doi: 10.1128/jcm.35.11.3008-3009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams J G K, Hanafey M K, Rafalski J A, Tingey S V. Genetic analysis using random amplified polymorphic DNA markers. Methods Enzymol. 1993;218:704–740. doi: 10.1016/0076-6879(93)18053-f. [DOI] [PubMed] [Google Scholar]