Abstract
Vegetative incompatibility in the chestnut blight fungus, Cryphonectria parasitica, in Europe is controlled by six unlinked vic loci, each with two alleles. Four previously identified vic loci (vic1, vic2, vic3, and vic4) were polymorphic in European vegetative compatibility (vc) types. Two new loci, vic6 and vic7, also were identified among European vc types. In one cross, vic genes segregated independently at five loci, and 194 progeny were assigned to 32 vc types; none of these loci were linked. A total of 64 vc types were identified from all crosses. All 64 genotypes possible from six vic loci, each with two alleles (26 = 64), were identified and assigned to vc types. Based on our model, vc types v-c 5 and v-c 10, which had been used in previous genetic studies, differ by only five vic genes. Future studies of vc types in C. parasitica can use knowledge of vic genotypes for analysis of population genetic structure based on vic allele frequencies and to determine the effect of each vic gene on virus transmission between vc types.
Vegetative (or heterokaryon) incompatibility is a self-nonself recognition system in filamentous fungi that regulates the formation of heterokaryons (5, 13, 19) and the transmission of cytoplasmic elements between strains (8, 12, 21). In most filamentous ascomycetes, incompatibility is controlled by allelic interactions; two strains are incompatible when they have different alleles at one or more vegetative incompatibility (vic [or het for heterokaryon incompatibility]) loci (5, 13, 19). Eight to 17 het loci have been found in Neurospora crassa, Aspergillus nidulans, and Podospora anserina (reviewed in references 5, 13, and 19), while segregation of large numbers of vegetative compatibility (vc) types suggests the existence of multiple vic loci in other ascomycetes as well (2, 7, 14, 16, 24). Among ascomycetes, most vic loci have only two alleles, although multiple alleles have been found for some loci in A. nidulans and N. crassa (11, 15).
Vegetative incompatibility has been a valuable phenotype for studying genetic diversity and population biology in fungi (reviewed in references 13 and 19). Although vc type diversity has been determined in numerous populations, more-detailed analyses would be possible if the vic genotypes of vc types were known. However, with the exception of a few vc types used in laboratory genetic studies, there has been no attempt to assign vic genotypes to vc types. If vic genotypes were known for most of the vc types in a population, population genetic analyses that require estimates of allele frequencies would be possible by using vc type data. For example, the multilocus genetic structure of populations could be analyzed to make inferences about recombination (23), or differentiation between populations could be estimated to study gene flow (22). Furthermore, in fungi in which mycoviruses may be a significant factor in population biology, as in the chestnut blight fungus, Cryphonectria parasitica (27, 32), the potential for virus transmission at the population level could be assessed (17, 21), especially if knowledge of the effect of each vic gene on virus transmission were available (16, 17). Without vic genotype data, however, it is difficult to link laboratory findings on virus transmission to field populations.
The simplest populations in which to determine vic genotypes are those in which vc type diversity is relatively low. In C. parasitica, only 31 vc types were found in samples of over 1,000 isolates from Italy and Switzerland, and most subpopulations had 10 or fewer vc types (10). This limited number of vc types could be explained by a minimum of five polymorphic vic loci (assuming two alleles for each locus). Vegetative incompatibility in C. parasitica is postulated to be controlled by allelic interactions at five to seven vic loci (1–3, 16). Anagnostakis (2) clearly identified two vic loci (vic1 and vic2), and Huber (16) recently identified three more (vic3, vic4, and vic5). Our objectives in this study were to determine the number of polymorphic vic loci in C. parasitica populations in Europe, to identify all possible vic genotypes, and to determine the genotypes of vc types used in previous genetic studies.
MATERIALS AND METHODS
The 31 vc types found to date in Europe are referred to as EU-1 to EU-31 (10). Single-conidial field isolates of vc types EU-1 to EU-20 were used as vc testers (9). These tester isolates, and additional single-conidial field isolates, were used as parents in crosses (Table 1); names for field isolates are preceded by two-letter codes (PC, VO, TE, LI, FI, VA, and SA). Isolate JA17, used as a parent in cross MJ1, is a single-conidial field isolate from Japan (26). Field isolate vc testers were later replaced with ascospore isolates of the same vc type; ascospore isolates with novel recombinant vc types were given new EU numbers. Cross numbers (except MJ1) were designated with the letter P (Table 1). Ascospore isolates were designated with the letter P followed by the cross number and the ascospore isolate from that cross (e.g., P1-11 is ascospore isolate 11 from cross P1).
TABLE 1.
Crosses of C. parasitica for studying the genetics of vegetative incompatibility
| Cross no. | Parent 1 vc type | Isolate no. | Parent 2 vc type | Isolate no. | No. of progeny | vic locia |
|---|---|---|---|---|---|---|
| P1 | EU-4 | PC17b | EU-5 | PC7 | 41 | 1, 2, 4 |
| P2 | EU-1 | VO54 | EU-2 | VO56 | 42 | 2 |
| P3 | EU-1 | VO54 | EU-4 | PC6 | 42 | 1,2 |
| P4 | EU-1 | VO54 | EU-5 | PC7 | 42 | 4 |
| P5 | EU-1 | VO54 | EU-3 | VO64 | 42 | 6 |
| P8 | EU-2 | VO1 | EU-3 | VO29 | 40 | 2, 6 |
| P9 | EU-3 | VO64 | EU-4 | PC17 | 41 | 1, 2, 6 |
| P10 | EU-3 | VO29 | EU-5 | PC7 | 39 | 4, 6 |
| P11 | EU-5 | P1-11 | EU-26 | P3-3 | 41 | 1, 4 |
| P12 | EU-3 | P8-7 | EU-26 | P3-3 | 45 | 1, 6 |
| P13 | EU-4 | P1-4 | EU-14 | P9-1 | 39 | 1, 6 |
| P16 | EU-10 | TE56 | EU-12 | SA16 | 44 | 1, 3 |
| P17 | EU-1 | P5-1 | EU-12 | SA31 | 45 | 1, 2, 6, 7 |
| P19 | EU-12 | SA16 | EU-13 | PC83 | 45 | 2, 4 |
| P20 | EU-12 | SA16 | EU-19 | FI38 | 43 | 2, 4, 6 |
| P21 | EU-1 | P5-1 | EU-10 | TE63 | 42 | 2, 3, 6, 7 |
| P24 | EU-1 | P4-4 | EU-13 | SA26 | 45 | 1, 4, 6, 7 |
| P25 | EU-10 | TE63 | EU-13 | PC83 | 41 | 1, 2, 3, 4 |
| P26 | EU-9 | PC39 | EU-13 | PC83 | 44 | 1, 2 |
| P27 | EU-13 | SA25 | EU-19 | VA35 | 12 | 6 |
| P28 | EU-4 | P3-7 | EU-31 | P1-5 | 10 | 2, 4 |
| P29 | EU-21 | P10-18 | EU-30 | P12-39 | 12 | 1, 4 |
| P30 | EU-31 | P11-23 | EU-29 | P13-23 | 42 | 2, 4, 6 |
| P32 | EU-9 | PC39 | EU-19 | VA35 | 41 | 1, 2, 6 |
| P33 | EU-9 | PC39 | EU-10 | TE56 | 19 | 3, 4 |
| P35 | EU-14 | P8-3 | EU-21 | P10-18 | 12 | 2, 4 |
| P36 | EU-2 | P9-11 | EU-17 | P16-6 | 24 | 6, 7 |
| P37 | EU-8 | P17-2 | EU-11 | P17-25 | 22 | 2, 6 |
| P38 | EU-19 | P27-5 | EU-24 | P29-1 | 24 | 6, 7 |
| P40 | EU-17 | P16-6 | EU-42 | P19-4 | 24 | 1, 4 |
| P42 | EU-5 | P1-11 | EU-20 | LI13 | 37 | 1, 2 |
| P45 | EU-25 | P24-9 | EU-45 | P25-13 | 20 | 3, 6 |
| P47 | EU-30 | P9-2 | EU-45 | P25-13 | 20 | 3, 7 |
| P48 | EU-3 | P8-7 | EU-33 | P21-11 | 20 | 3, 7 |
| P49 | EU-10 | P21-9 | EU-14 | P8-3 | 20 | 3, 7 |
| P50 | EU-21 | P10-18 | EU-46 | P25-24 | 20 | 3, 7 |
| P52 | EU-45 | P25-13 | EU-46 | P25-24 | 19 | 1, 4 |
| P54 | EU-10 | P16-2 | EU-22 | P17-4 | 20 | 3, 6 |
| P55 | EU-13 | P27-1 | EU-37 | P25-12 | 20 | 3 |
| P56 | EU-7 | P17-7 | EU-33 | P25-9 | 20 | 3, 6 |
| P57 | EU-34 | P25-27 | EU-27 | P30-9 | 20 | 3, 7 |
| P58 | EU-8 | P17-2 | EU-40 | P16-7 | 40 | 3, 6 |
| P59 | EU-34 | P57-2 | EU-43 | P32-4 | 20 | 3, 6 |
| P60 | EU-47 | P45-5 | EU-48 | P47-14 | 20 | 6, 7 |
| P61 | EU-32 | P49-6 | EU-39 | P54-5 | 20 | 6, 7 |
| P62 | EU-35 | P22-6 | EU-41 | P21-35 | 20 | 6, 7 |
| P64 | EU-36 | P25-6 | EU-39 | P21-18 | 20 | 4, 6 |
| P65 | EU-42 | P19-4 | EU-50 | P57-5 | 39 | 3, 7 |
| P66 | EU-36 | P33-1 | EU-23 | P35-3 | 20 | 3, 7 |
| P67 | EU-6 | P1-6 | EU-36 | P25-6 | 36 | 3, 6, 7 |
| P69 | EU-29 | P9-13 | EU-40 | P58-22 | 20 | 3, 7 |
| P70 | EU-49 | P58-13 | EU-50 | P57-5 | 39 | 4, 6, 7 |
| P72 | EU-13 | P43-3 | EU-31 | P11-23 | 20 | 6, 7 |
| P73 | EU-50 | P65-1 | EU-53 | P59-12 | 30 | 6, 7 |
| P74 | EU-5 | P1-11 | EU-51 | P50-16 | 20 | 3, 6 |
| P76 | EU-19 | P72-6 | EU-37 | P25-12 | 20 | 3, 6 |
| P78 | EU-46 | P50-4 | EU-60 | P74-7 | 20 | 6, 7 |
| P79 | EU-31 | P1-5 | EU-63 | P76-13 | 20 | 3, 7 |
| P80 | EU-50 | P65-1 | EU-51 | P74-10 | 20 | 1, 2 |
| MJ1 | EU-22 | P17-8 | EU-24 | JA17 | 194 | 1, 2, 4, 6, 7 |
vic loci at which segregation occurred in each cross. The nomenclature for these loci is explained in text.
Isolate designations beginning with two-letter codes are field isolates from Italy (9) from the Bergamo (PC), Crevoladossola (VO), Teano (TE), Tonara (SA), Pomino (FI), Ponte in Valtellina (VA), and Pigna (LI) populations. Isolates whose designations begin with the letter P are ascospore progeny isolates; the first number following the P is the cross number, and the second number is the ascospore isolate from that cross (e.g., P1-11 is ascospore isolate 11 from cross P1).
Vegetative incompatibility was assayed as the appearance of dark discoloration and/or barrage formation between colonies on agar medium as described previously (9, 28). Crosses were made on autoclaved chestnut (Castanea sativa or C. dentata) stems embedded in water agar as described previously (1, 20). For most crosses, we picked approximately 20 to 50 random ascospore progeny, although 194 ascospores (ca. 50 from each of four perithecia) were analyzed in cross MJ1.
Our strategy for determining vic genotypes was to start with vc types that had been assigned genotypes at four vic loci. D. Huber gave us isolates representing 13 of 16 possible genotypes determined by vic1, vic2, vic3, and vic4 (16, 17). Six of these isolates were of vc types v-c 5, v-c 8, v-c 16, v-c 39, v-c 56, and v-c 71 which had been analyzed previously (1, 2, 29, 30). Our initial set of crosses was done to determine genotypes at vic1 to vic4 for the three remaining vc types that had not been assigned genotypes. Subsequent crosses were made to analyze the vic genotypes of additional vc types. Based on previous studies of C. parasitica (2, 16), we expected to find 2n vc types in the progeny of each cross, with alleles segregating at n vic loci. Therefore, we used the number of progeny vc types to infer the number of vic genes segregating in each cross. Interpretation of crosses was done sequentially and depended on results from each previous cross.
We also compared isolates of vc types v-c 10, v-c 17, and v-c 40 which had been used in crosses to study vc genetics (1, 2) with vc types from our crosses to determine their vic genotypes. Incompatibility caused by vic5 is weak and cannot be detected on potato dextrose agar (16) or on potato dextrose agar containing bromocresol green (9, 28; unpublished observations); therefore, this locus was not considered in our genetic analyses.
RESULTS
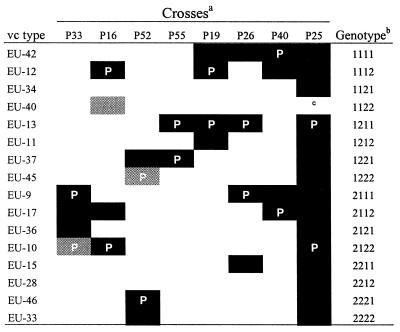
In our initial set of crosses among vc types with genotypes previously defined by vic1 to vic4 (Table 2) we determined the genotypes of the three remaining vc types. In each cross (P33, P16, and P52), three of the four vc types in the progeny had known vic genotypes, making it possible to infer the fourth genotype. Progeny vc types from five additional crosses conformed to those expected from Huber’s model (16). For example, in cross P25, the parents (EU-10 and EU-13) differed at all four vic loci and 15 of the 16 predicted vc types were found in the 30 progeny. Eight of the 31 vc types from Europe were compatible with testers in this set of 16 genotypes.
TABLE 2.
Segregation of vc types defined by vic1 to vic4 in crosses of C. parasitica
Crosses are defined in Table 1. Solid cells indicate vc types that segregated in each cross. Parental vc types are indicated by the letter P. Lightly shaded cells represent the assignment of a genotype to a vc type in a particular cross.
Genotypes for vic1 to vic4 as defined by Huber (16).
EU-40 was expected but was not observed in this cross.
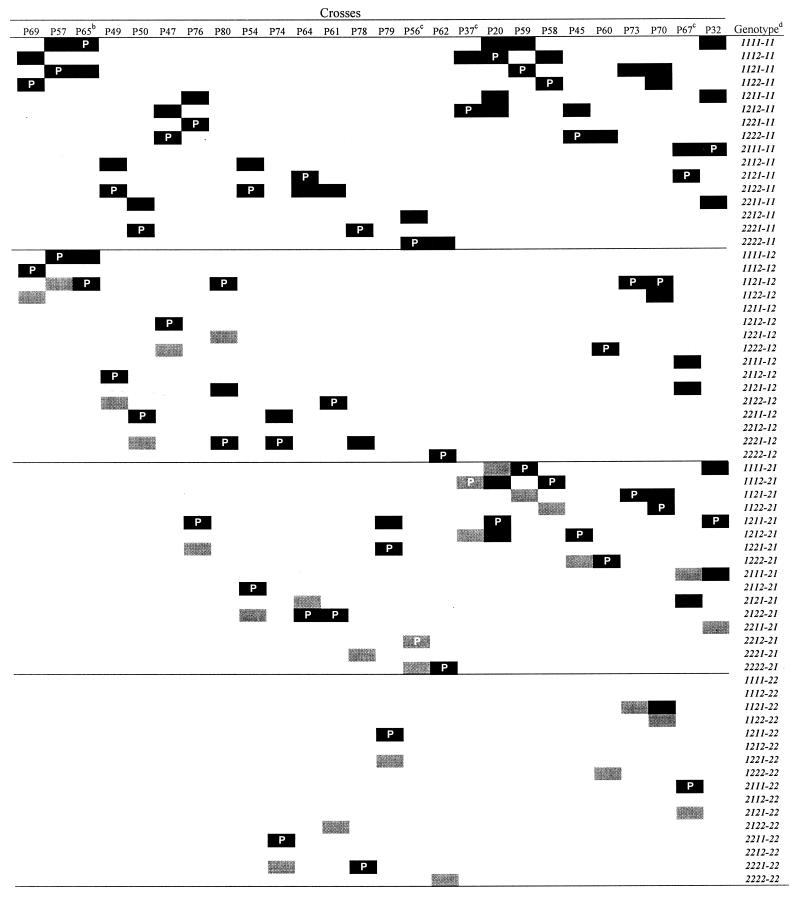
In our second set of crosses, we found 48 additional vc types, for a total of 64, suggesting segregation at six vic loci (Table 3). Crosses are discussed in the order in which they appear in Table 3 to explain how each vic genotype was determined. Crosses with single vic genes segregating were found between EU-1 and EU-2 (cross P2), EU-1 and EU-5 (cross P4), and EU-13 and EU-19 (cross P27).
TABLE 3.
Segregation of vc types in crosses of C. parasitica
Crosses are defined in Table 1. Solid cells indicate vc types that segregated in each cross. Parental vc types are indicated by the letter P. Lightly shaded cells represent the assignment of a genotype(s) to a vc type(s) in a particular cross.
Crosses P38 and P65 confirm segregation results in crosses P72 and P57, respectively.
Crosses discussed in detail in the text but not in order.
Genotypes for vic1 to vic4, vic6, and vic7 are abbreviated with allele numbers only. The first 16 genotypes are the same as those in Table 2.
Progeny from cross MJ1 (EU-22 × EU-24) were of 32 vc types, indicating segregation at five vic loci in this cross. Only eight progeny vc types were among the 16 genotypes defined by vic1 to vic4 (i.e., those shown in Table 2); these eight types all shared allele vic3-1. From this result, we concluded that all of the progeny in this cross had allele vic3-1. We also concluded that alleles were segregating at vic1, vic2, and vic4 and at two new vic loci, which we designated vic6 and vic7. Alleles vic6-1 and vic7-1 were arbitrarily assigned to the 16 genotypes defined by vic1 to vic4.
Three crosses (P24, P17, and P21) were analyzed in which there were 16 vc types in the progeny; we concluded that four vic genes were segregating in each cross. In each of these crosses, 4 of the 16 progeny vc types were in the subset of 16 vc types whose genotypes are defined by vic1 to vic4 (Table 2). We interpreted this to mean that there was segregation at only two of loci vic1 to vic4 and that there was segregation at both vic6 and vic7. Based on the known genotypes of progeny vc types, we inferred that all of the progeny of P24 had alleles vic2-2 and vic3-1 because no segregation was evident at these loci; similarly, all of the progeny of P17 had vic3-1 and vic4-2, and progeny of P21 had vic1-2 and vic4-2. Since EU-1 was a progeny vc type in these three crosses and in cross MJ1, we determined its genotype for vic1 to vic4 (as described above). Because there was segregation at vic6 and vic7 in cross P21, and parent EU-10 has alleles vic6-1 and vic7-1, the other parent, EU-1, had alleles vic6-2 and vic7-2. Therefore, EU-1 has the genotype vic1-2 vic2-2 vic3-1 vic4-2 vic6-2 vic7-2. To simplify the notation, genotypes hereafter are designated simply by their allele numbers for these six loci, e.g., 2212-22 for EU-1; the hyphen indicates that the allele for vic5 is not known. Lightly shaded cells in Table 3 indicate the cross in which a genotype was assigned to a vc type.
In cross P5, EU-1 and EU-3 were found to differ at one vic locus. By the same reasoning as that used for EU-1, EU-3 has genotype 2212--- because it was found in crosses MJ1, P24, P17, and P21; therefore, segregation must have occurred at either vic6 or vic7 in cross P5. Since vic6 and vic7 were not yet defined separately, we arbitrarily assigned alleles vic6-1 and vic7-2 to EU-3. When EU-3 (2212-12) was crossed (in cross P48) with EU-33 (2222-11), two vic genes segregated, at vic3 and vic7, allowing us to assign genotype 2222-12 to recombinant vc type EU-35 because the other recombinant genotype, 2212-11, was already assigned to EU-28. For most of the remaining crosses (not discussed explicitly), we assigned genotypes to vc types in this way; i.e., from crosses in which all but one progeny vc type had genotypes already determined from previous crosses (Table 3). Exceptions to this strategy are described below.
Cross P12.
When EU-3 (2212-12) was crossed with EU-26 (-212---), segregation at vic6 was evident because EU-1 (2212-22), which has vic6-2, was a recombinant vc type. Therefore, vic6-2 was assigned to parent EU-26 and vic6-1 was assigned to the other recombinant type, EU-30. EU-30 could not also have had allele vic7-1 because it is not among the 16 vc types defined by vic1 to vic4 that have both vic6-1 and vic7-1 (Table 2). Therefore, vic7-2 was assigned to both EU-26 and EU-30. With four different progeny vc types in cross P12, there must have been segregation at vic1 in addition to vic6 because vic2-2, vic3-1, vic4-2, and vic7-2 were common to both parents. Therefore, genotypes 1212-22 and 1212-12 were assigned to EU-26 and EU-30, respectively.
Cross P8.
We deduced the genotypes for EU-2 and EU-14 from cross P8 by using a rationale similar to that used for cross P12. From crosses MJ1, P17, and P21, we knew that EU-2 and EU-14 both had alleles vic1-2, vic3-1, and vic4-2. As in P12, segregation at vic6 was evident because EU-1, which has vic6-2, was a recombinant vc type (2212-22), while parent EU-3 (2212-12) had vic6-1. Therefore, vic6-2 was assigned to parent EU-2, while vic6-1 was assigned to the other recombinant type, EU-14. EU-14 could not have had allele vic7-1 because it was not among the 16 vc types defined by vic1 to vic4 that have both vic6-1 and vic7-1 (Table 2). Therefore, vic7-2 was assigned to both EU-2 and EU-14. Since there were four progeny types, alleles must have segregated at vic2 in addition to vic6, and vic2-1 was assigned to both EU-2 and EU-14. Thus, genotypes 2112-22 and 2112-12 were assigned to EU-2 and EU-14, respectively.
Cross P10.
Parent EU-3 (2212-12) and recombinant type EU-1 (2212-22) differ at vic6, demonstrating segregation at this locus. Therefore, parent EU-5 must have had vic6-2, while recombinant EU-21 must have vic6-1. From crosses MJ1 and P24, we knew that parent EU-5 had alleles vic2-2 and vic3-1. After determining the genotype of EU-4 in cross P3, we deduced from cross P1 that EU-5 had vic1-2 because EU-4, the other parent in cross P1, had vic1-1, but there was segregation at vic1 (Table 3). EU-21 could not have had allele vic7-1 because it was not among the 16 vc types defined by vic1 to vic4 that have both vic6-1 and vic7-1 (Table 2). Therefore, vic7-2 was assigned to both EU-5 and EU-21. Since there were four progeny types in cross P10, alleles must have segregated at vic4 in addition to vic6, and vic4-1 was assigned to both EU-5 and EU-21. Thus, genotypes 2211-22 and 2211-12 were assigned to EU-5 and EU-21, respectively.
Cross P56.
Parent EU-33 (2222-11) and recombinant type EU-28 (2212-11) differ at vic3, demonstrating segregation at this locus. Therefore, parent EU-7 had vic3-1, while recombinant EU-41 had vic3-2. One parent (EU-7) and one recombinant type (EU-41) were not among the 16 vc types defined by vic1 to vic4 that have both vic6-1 and vic7-1 (Table 2). Therefore, alleles segregated at either vic6 or vic7 in addition to vic3. However, genotypes 2212-12 and 2222-12 were already defined, leaving genotypes 2212-21 and 2222-21 to be assigned to EU-7 and EU-41, respectively.
Cross P37.
Parent EU-11 (1212-11) and recombinant type EU-12 (1112-11) differ at vic2, demonstrating segregation at this locus. Therefore, the other parent, EU-8, had vic2-1, while recombinant EU-25 had vic2-2. EU-8 and EU-25 were both assigned vic6-2 because 32 vc types had already been assigned genotypes with vic6-1. With segregation at only two loci (vic2 and vic6), there could not have been segregation at any other locus. Therefore, genotypes 1112-21 and 1212-21 were assigned to EU-8 and EU-25, respectively.
Cross P67.
Six of the eight progeny vc types, including both parents (EU-6 and EU-36), had known genotypes; genotypes were not yet known for EU-16 and EU-59. The only progeny genotypes in this cross that were not already assigned to vc types were 2111-21 and 2121-22. Because EU-16 was a progeny vc type in cross MJ1 and EU-59 was not, EU-16 had vic3-1 and was assigned genotype 2111-21, leaving 2121-22 to be assigned to EU-59.
All 64 possible vic genotypes, vc types, and tester isolates are summarized and cross-listed with vc types and testers from previous genetic studies in Table 4. vc types v-c 10, v-c 17, and v-c 40, which were previously used in genetic studies but without known vic genotypes, were compatible with EU-1, EU-26, and EU-5, respectively.
TABLE 4.
Genotypes and tester isolates of 64 C. parasitica vc types in this and previous studies
| Results of:
| ||||||
|---|---|---|---|---|---|---|
| This study
|
Previous studies
|
|||||
| vic geno- typea | EU typeb | Isolate no. | Crossc | vc typed | Isolate no. | ATCC no. |
| 1111-11 | EU-42 | P19-4 | —e | v-c 5 | EP389, 389.7 | 38980 |
| 1112-11 | EU-12 | P16-1 | — | v-c 56 | EP243, F3.2 | |
| 1121-11 | EU-34 | P25-27 | — | N1.9 | ||
| 1122-11 | EU-40 | P16-7 | P16 | |||
| 1211-11 | EU-13 | P1-5 | — | v-c 71 | J2.43 | |
| 1212-11 | EU-11 | P19-2 | — | J2.6 | ||
| 1221-11 | EU-37 | P25-12 | — | M1.6 | ||
| 1222-11 | EU-45 | P25-13 | P52 | |||
| 2111-11 | EU-9 | P26-1 | — | v-c 39 | EP388 | 38979 |
| 2112-11 | EU-17 | P16-6 | — | F3.13 | ||
| 2121-11 | EU-36 | P25-6 | — | K1.43 | ||
| 2122-11 | EU-10 | P16-2 | P33 | |||
| 2211-11 | EU-15 | P26-10 | — | v-c 8 | 22508f, A1.8 | 22508 |
| 2212-11 | EU-28 | P21-16 | — | J2.20, L1.16 | ||
| 2221-11 | EU-46 | P25-24 | — | v-c 16 | EP29, K2.30 | 38754 |
| 2222-11 | EU-33 | P21-11 | — | L1.39 | ||
| 1111-12 | EU-27 | P30-9 | P30 | |||
| 1112-12 | EU-29 | P9-13 | P13 | |||
| 1121-12 | EU-50 | P57-5 | P57 | |||
| 1122-12 | EU-56 | P69-5 | P69 | |||
| 1211-12 | EU-24 | P29-1 | P29 | |||
| 1212-12 | EU-30 | P9-2 | P12 | |||
| 1221-12 | EU-52 | P80-1 | P80 | |||
| 1222-12 | EU-48 | P47-14 | P47 | |||
| 2111-12 | EU-23 | P35-3 | P35 | |||
| 2112-12 | EU-14 | P8-3 | P8 | |||
| 2121-12 | EU-58 | P66-9 | P66 | |||
| 2122-12 | EU-32 | P21-5 | P49 | |||
| 2211-12 | EU-21 | P10-18 | P10 | |||
| 2212-12 | EU-3 | P5-2 | P48 | |||
| 2221-12 | EU-51 | P50-3 | P50 | |||
| 2222-12 | EU-35 | P22-6 | P48 | |||
| 1111-21 | EU-43 | P20-2 | P20 | |||
| 1112-21 | EU-8 | P17-2 | P37 | |||
| 1121-21 | EU-53 | P59-9 | P59 | |||
| 1122-21 | EU-49 | P58-13 | P58 | |||
| 1211-21 | EU-19 | P27-1 | P72 | |||
| 1212-21 | EU-25 | P24-9 | P37 | |||
| 1221-21 | EU-63 | P76-6 | P76 | |||
| 1222-21 | EU-47 | P45-4 | P45 | |||
| 2111-21 | EU-16 | P32-3 | P67 | |||
| 2112-21 | EU-22 | P17-4 | P36 | |||
| 2121-21 | EU-57 | P64-2 | P64 | |||
| 2122-21 | EU-39 | P21-18 | P54 | |||
| 2211-21 | EU-18 | P24-33 | P32 | |||
| 2212-21 | EU-7 | P17-7 | P56 | |||
| 2221-21 | EU-64 | P78-1 | P78 | |||
| 2222-21 | EU-41 | P21-35 | P56 | |||
| 1111-22 | EU-20 | P1-2 | P28 | |||
| 1112-22 | EU-4 | P1-4 | P3 | |||
| 1121-22 | EU-61 | P73-24 | P73 | |||
| 1122-22 | EU-62 | P70-11 | P70 | |||
| 1211-22 | EU-31 | P1-5 | P11 | |||
| 1212-22 | EU-26 | P1-16 | P12 | v-c 17 | EP78 | 38752 |
| 1221-22 | EU-55 | P79-6 | P79 | |||
| 1222-22 | EU-54 | P60-3 | P60 | |||
| 2111-22 | EU-6 | P1-6 | P42 | |||
| 2112-22 | EU-2 | P2-4 | P8 | |||
| 2121-22 | EU-59 | P67-7 | P67 | |||
| 2122-22 | EU-38 | P21-14 | P61 | |||
| 2211-22 | EU-5 | P1-11 | P10 | v-c 40 | EP155 | 38755 |
| 2212-22 | EU-1 | P4-4 | P21 | v-c 10 | EP67 | 38753 |
| 2221-22 | EU-60 | P74-3 | P74 | |||
| 2222-22 | EU-44 | P21-1 | P62 | |||
Genotypes for vc types were based on four vic loci (vic1 to vic4) named by Huber (16), followed by two new loci found in this study (vic6 and vic7). Only the allele numbers for each locus are shown. The hyphen after the allele at the fourth locus is to signify no data for vic5.
Nomenclature for EU types is based on that of Cortesi et al. (10). Isolate numbers refer to tester isolates derived from the crosses listed in Table 1.
Cross number in which a vic genotype was assigned to a vc type.
The v-c nomenclature is based on that of Anagnostakis (1, 3). ATCC (American Type Culture Collection) numbers correspond to isolate numbers beginning with EP; all other isolates were from Huber (16).
These genotypes were determined previously by Huber (16).
No EP number is known for this isolate; Huber (16) used the ATCC number instead.
There was no evidence for linkage among any of the six vic loci (Table 5). Data from multiple crosses with segregation at the same two vic loci were tested for homogeneity (α = 0.10) before pooling (31).
TABLE 5.
Analysis of linkages among six vic loci identified in C. parasitica
| No. of crosses pooleda | nb |
vic locic
|
No. of progeny with vic genotypesd
|
Chi squaree | ||||
|---|---|---|---|---|---|---|---|---|
| i | j | 1, 1 | 1, 2 | 2, 1 | 2, 2 | |||
| 10 | 546 | 1 | 2 | 148 | 135 | 133 | 130 | 1.4 |
| 2 | 85 | 1 | 3 | 26 | 16 | 16 | 27 | 5.2 |
| 8 | 417 | 1 | 4 | 113 | 95 | 105 | 104 | 1.6 |
| 7 | 450 | 1 | 6 | 114 | 115 | 120 | 101 | 1.8 |
| 3 | 284 | 1 | 7 | 70 | 74 | 71 | 69 | 0.2 |
| 2 | 83 | 2 | 3 | 16 | 26 | 22 | 19 | 2.6 |
| 8 | 428 | 2 | 4 | 108 | 117 | 108 | 95 | 2.3 |
| 9 | 470 | 2 | 6 | 121 | 113 | 125 | 111 | 1.1 |
| 3 | 236 | 2 | 7 | 56 | 65 | 62 | 53 | 1.5 |
| 2 | 60 | 3 | 4 | 11 | 15 | 17 | 17 | 1.6 |
| 9 | 196 | 3 | 6 | 46 | 52 | 46 | 52 | 0.7 |
| 11 | 235 | 3 | 7 | 63 | 62 | 51 | 59 | 1.5 |
| 7 | 383 | 4 | 6 | 100 | 97 | 97 | 89 | 0.7 |
| 3 | 233 | 4 | 7 | 64 | 59 | 55 | 55 | 0.9 |
| 14 | 534 | 6 | 7 | 133 | 132 | 137 | 132 | 0.1 |
Number of crosses pooled for each pair of loci. Chi square tests of homogeneity were conducted before pooling (see text).
Total number of progeny from all crosses analyzed for each locus pair.
Pairs of vic loci, i and j, for which linkage analyses were done.
Genotypes were defined as allele 1 or 2 at loci i and j.
Chi square goodness-of-fit statistics (three degrees of freedom) for a genotype ratio of 1:1:1:1.
DISCUSSION
Genetic analyses of vc types in C. parasitica in Europe are consistent with a model of six unlinked vic loci, each with two alleles. Our results confirm that vegetative incompatibility is controlled by allelic interactions. Four of the vic loci we identified in this study were previously described by Huber (16); therefore, we have identified two additional vic loci. We concur with Huber (16) that—except for vic1 and vic2—the tentative genotype assignments made by Anagnostakis (1) and Rizwana and Powell (29, 30) should be disregarded and replaced by Huber’s nomenclature (16). Therefore, we called the two new vic loci found in this study vic6 and vic7. Incompatibility caused by an additional vic locus, vic5, cannot be detected on agar media (16; unpublished results) and was therefore disregarded in this study.
There was no evidence for multiple alleles at any vic locus for the vc types found in the field in Europe (EU-1 to EU-31). Thirty of the 31 European vc types were among the progeny of cross MJ1, in which alleles segregated at each of five vic loci; there was no segregation at vic3 in this cross. EU-10 was the only vc type found in the field in Europe that was not found among the progeny of cross MJ1 (Table 3). EU-10 was shown to have allele vic3-2 (cross P33, Table 2), but all of the other European vc types had allele vic3-1 (Table 2). Therefore, since all 64 vc types were either found in the field or derived from field isolates, only two alleles were found for each vic locus.
Our results (Table 2) are fully in agreement with Huber’s (16) and agree with six of the nine crosses reported by Anagnostakis (1, 2). However, our results disagree with three of Anagnostakis’ crosses. In general, we predict fewer vc types segregating than were observed in some crosses. We speculate that the excess vc types found in previous studies were artifacts caused by vc assays. Recent improvements in vc testing techniques have reduced these ambiguities considerably (9, 28). First, Anagnostakis (1) reported finding only parental vc types in a cross between v-c 5 and v-c 16. In contrast, Rizwana and Powell (29) found 17 nonparental vc types among the progeny of a cross between v-c 5 and v-c 16; however, the ratios of progeny vc types in this cross were highly irregular. According to our interpretation, v-c 5 and v-c 16 are compatible with EU-42 and EU-46, respectively, which differ by three vic genes and should produce progeny in eight vc types; Huber (16) also found that these two vc types differed by vic1, vic2, and vic3. The second cross Anagnostakis (1) reported that differed from our results was between v-c 8 and v-c 17 (EU-15 and EU-26, respectively). Anagnostakis reported finding 22 vc types in the progeny, but we would predict only 16 because the parents differ at four vic loci (Table 4). Finally, the third cross, with which our results are not fully consistent, is between v-c 5 and v-c 10 (2). This cross has often been cited because it was the basis for estimating the minimum number of vic loci controlling vc types in C. parasitica. Anagnostakis (2) estimated segregation at seven vic loci after observing 106 vc types in the progeny of this cross. A lower estimate of five vic loci, however, was based on the observation that approximately 1/25 (11 of 263 and 37 of 973) of the progeny from this same cross were of one parental type. One thirty-second of the progeny would have been expected to be compatible with each parent if alleles had segregated independently at five vic loci (2, 3), whereas 1/128 would have been expected to be compatible if alleles had segregated at seven loci. Our results show that v-c 5 and v-c 10 (compatible with EU-42 and EU-1, respectively; Table 4) differ by only five, not seven, vic genes.
Six vic loci, each with two alleles, may be sufficient to explain much of the diversity of vic genotypes, or vc types, found in populations of C. parasitica in North America. Anagnostakis and Kranz (4) found 48 vc types in a small forest plot in Connecticut; other studies have shown similar levels of vc type diversity in North America (18, 20, 25). Our preliminary studies have shown that most of the 31 vc types found in one population in Maryland (20) are compatible with testers in the 64 genotypes found in this study (unpublished results). More testing is required to determine if additional vic loci are polymorphic in North America or if multiple alleles occur at known loci. In Asia, there are probably additional polymorphic vic loci since 131 vc types were identified among 231 isolates of C. parasitica (33). Furthermore, only two of the 71 vc types found in Japan are compatible with the 64 types from this study (21a). Unfortunately, the effort required to identify additional vic genotypes doubles with every locus that is identified. For example, one additional vic locus in C. parasitica would bring the total number of possible vc types to 128 and two loci would raise it to 256. Alternatively, a third allele at any vic locus would increase the number of possible genotypes by 50%. Multiple alleles at several loci—in relatively high frequencies in populations—would be necessary to explain the diversity of vc types observed in Asia and the lack of compatibility with the 64 vc types we found. However, we found no evidence for multiple alleles in European populations.
Our study has significantly extended the findings of previous genetic studies on vc types in C. parasitica because we have successfully assigned 64 (26) vic genotypes to vc types (Table 4). To our knowledge, such extensive information on vic genotypes is not available for any other fungus. All 31 of the European vc types (EU-1 to EU-31) found in the field to date (10) have been assigned a vic genotype. Thirty of the 32 possible vc types (all except EU-42 and EU-43) defined by five vic loci (vic1, vic2, vic4, vic6, and vic7) have been found in the field. Interestingly, allele vic3-2 was only found in two populations in southern Italy, in vc type EU-10 (9, 10); all of the other vc types found in Europe have vic3-1. Knowledge of vic genotypes will allow us to reanalyze vc type survey data more thoroughly (e.g., references 6, 9, and 10). For example, we will be able to test whether vc types occur at frequencies that would be expected from random mating; this analysis requires estimates of vic allele frequencies along with vc type frequencies (see reference 23). In addition, comparison of the 64 vc types from this study with vc types found in the United States is revealing a high proportion of successful matches (unpublished results), which opens the possibility of reanalyzing vc type population data in the United States based on vic genotypes. Finally, with our knowledge of vic genotypes, we can now examine the effect of each vic allele on virus transmission (17, 21; unpublished results) to make inferences about the potential of virus transmission in populations of C. parasitica.
ACKNOWLEDGMENTS
This research was supported by a NATO Cooperative Research Grant, USDA NRI competitive grant 93-37303-9035, and a fellowship from the National Research Council of Italy, Special Project RAISA, to M.G.M.
We thank David Huber for sharing his laboratory isolates and prepublication drafts; Sandy Anagnostakis for sending us vc testers from her collection; Paola Allegra, Luca Rancati, Sabrina Senzacqua, and Kazue Takeuchi for assisting with vc tests; and Marco Bisiach for helpful discussions.
REFERENCES
- 1.Anagnostakis S L. Cryphonectria parasitica, cause of chestnut blight. Adv Plant Pathol. 1988;6:123–136. [Google Scholar]
- 2.Anagnostakis S L. Genetic analyses of Endothia parasitica: linkage data for four single genes and three vegetative compatibility types. Genetics. 1982;102:25–28. doi: 10.1093/genetics/102.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostakis S L. Vegetative incompatibility in Endothia parasitica. Exp Mycol. 1977;1:306–316. [Google Scholar]
- 4.Anagnostakis S L, Kranz J. Population dynamics of Cryphonectria parasitica in a mixed-hardwood forest in Connecticut. Phytopathology. 1987;77:751–754. [Google Scholar]
- 5.Bégueret J, Turcq B, Clave C. Vegetative incompatibility in filamentous fungi: het genes begin to talk. Trends Genet. 1994;10:441–446. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 6.Bissegger M, Rigling D, Heiniger U. Population structure and disease development of Cryphonectria parasitica in European chestnut forests in the presence of natural hypovirulence. Phytopathology. 1997;87:50–59. doi: 10.1094/PHYTO.1997.87.1.50. [DOI] [PubMed] [Google Scholar]
- 7.Brasier C M. Inter-mycelial recognition systems in Ceratocystis ulmi: their physiological properties and ecological importance. In: Jennings D, Rayner A D M, editors. The ecology and physiology of the fungal mycelium. Cambridge, England: Cambridge University Press; 1984. pp. 451–497. [Google Scholar]
- 8.Caten C E. Vegetative incompatibility and cytoplasmic infection in fungi. J Gen Microbiol. 1972;72:221–229. doi: 10.1099/00221287-72-2-221. [DOI] [PubMed] [Google Scholar]
- 9.Cortesi P, Milgroom M G, Bisiach M. Distribution and diversity of vegetative compatibility types in subpopulations of Cryphonectria parasitica in Italy. Mycol Res. 1996;100:1087–1093. [Google Scholar]
- 10.Cortesi P, Rigling D, Heiniger U. Comparison of vegetative compatibility types in Italian and Swiss subpopulations of Cryphonectria parasitica. Eur J For Pathol. 1998;28:167–176. [Google Scholar]
- 11.Dales R B G, Moorhouse J, Croft J H. Evidence for a multi-allelic heterokaryon incompatibility (het) locus detected by hybridization among three heterokaryon-incompatibility groups of Aspergillus nidulans. Heredity. 1993;70:537–543. doi: 10.1038/hdy.1993.77. [DOI] [PubMed] [Google Scholar]
- 12.Debets F, Yang X, Griffiths A J G. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet. 1994;26:113–119. doi: 10.1007/BF00313797. [DOI] [PubMed] [Google Scholar]
- 13.Glass N L, Kuldau G A. Mating type and vegetative incompatibility in filamentous ascomycetes. Annu Rev Phytopathol. 1992;30:201–224. doi: 10.1146/annurev.py.30.090192.001221. [DOI] [PubMed] [Google Scholar]
- 14.Guerber J C, Sherrill J F, Correll J C. Genetic analysis of sexual and vegetative compatibility in Colletotrichum gloeosporioides. Phytopathology. 1997;87:S36. [Google Scholar]
- 15.Howlett B J, Leslie J F, Perkins D D. Putative multiple alleles at the vegetative (heterokaryon) incompatibility loci het-c and het-8 in Neurospora crassa. Fungal Genet News. 1993;40:40–42. [Google Scholar]
- 16.Huber D H. Ph.D. dissertation. East Lansing: Michigan State University; 1996. [Google Scholar]
- 17.Huber D H, Fulbright D W. Preliminary investigations on the effect of individual vic genes upon the transmission of dsRNA in Cryphonectria parasitica. In: Double M L, MacDonald W L, editors. Proceedings of the International Chestnut Conference. Morgantown: West Virginia University Press; 1994. pp. 15–19. [Google Scholar]
- 18.Kuhlman E G, Bhattacharyya H, Nash B L, Double M L, MacDonald W L. Identifying hypovirulent isolates of Cryphonectria parasitica with broad conversion capacity. Phytopathology. 1984;74:676–682. [Google Scholar]
- 19.Leslie J F. Fungal vegetative compatibility. Annu Rev Phytopathol. 1993;31:127–150. doi: 10.1146/annurev.py.31.090193.001015. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y-C, Cortesi P, Double M L, MacDonald W L, Milgroom M G. Diversity and multilocus genetic structure in populations of Cryphonectria parasitica. Phytopathology. 1996;86:1344–1451. [Google Scholar]
- 21.Liu Y-C, Milgroom M G. Correlation between hypovirus transmission and the number of vegetative incompatibility (vic) genes different among isolates from a natural population of Cryphonectria parasitica. Phytopathology. 1996;86:79–86. [Google Scholar]
- 21a.Liu, Y.-C., and M. G. Milgroom. Unpublished data.
- 22.McDermott J M, McDonald B A. Gene flow in plant pathosystems. Annu Rev Phytopathol. 1993;31:353–373. [Google Scholar]
- 23.Milgroom M G. Recombination and the multilocus structure of fungal populations. Annu Rev Phytopathol. 1996;34:457–477. doi: 10.1146/annurev.phyto.34.1.457. [DOI] [PubMed] [Google Scholar]
- 24.Milgroom M G, Brasier C M. Potential diversity in vegetative incompatibility types of Ophiostoma novo-ulmi in North America. Mycologia. 1997;89:722–726. [Google Scholar]
- 25.Milgroom M G, MacDonald W L, Double M L. Spatial analysis of vegetative compatibility groups in the chestnut blight fungus, Cryphonectria parasitica. Can J Bot. 1991;69:1407–1413. [Google Scholar]
- 26.Milgroom M G, Wang K, Zhou Y, Lipari S E, Kaneko S. Intercontinental population structure of the chestnut blight fungus, Cryphonectria parasitica. Mycologia. 1996;88:179–190. [Google Scholar]
- 27.Nuss D L. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol Rev. 1992;56:561–576. doi: 10.1128/mr.56.4.561-576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell W A. Vegetative incompatibility and mycelial death of Cryphonectria parasitica detected with a pH indicator. Mycologia. 1995;87:738–741. [Google Scholar]
- 29.Rizwana R, Powell W A. Ultraviolet light-induced heterkaryon formation and parasexuality in Cryphonectria parasitica. Exp Mycol. 1995;19:48–60. [Google Scholar]
- 30.Rizwana R, Powell W A. Ultraviolet light-induced instability of vegetative compatibility groups of Cryphonectria parasitica. Phytopathology. 1992;82:1206–1211. [Google Scholar]
- 31.Strickberger M W. Genetics. 2nd ed. New York, N.Y: MacMillan Publishing Co.; 1976. pp. 149–150. [Google Scholar]
- 32.Van Alfen N K, Jaynes R A, Anagnostakis S L, Day P R. Chestnut blight: biological control by transmissible hypovirulence in Endothia parasitica. Science. 1975;189:890–891. doi: 10.1126/science.189.4206.890. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Shao J, Lu J. On vegetative compatibility of Cryphonectria parasitica in Jiangsu and Anhui. J Nanjing Agric Univ. 1991;14:44–48. [Google Scholar]