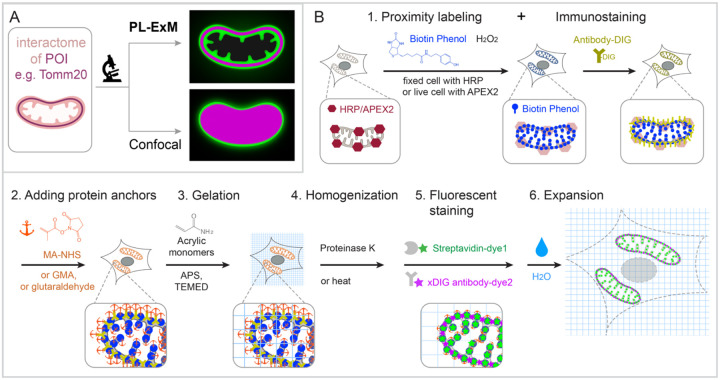
Figure 1. Graphic abstract and workflow of PL-ExM.
In the showcase, Tomm20 is the bait for the PL and the target for the immunostaining. (A) Graphic abstract of PL-ExM method. PL-ExM offers super resolution to visualize small interactome structures that present the ground truth. Diffraction-limited microscopy, such as confocal microscopy, misses structural details in the ground truth. (B) The PL-ExM workflow comprises six steps. 1. Proximity labeling catalyzed by enzymes (HRP, APEX, etc.) and delivered by biotin phenol. Following PL, a protein of interest is labeled with antibodies conjugated with DIG. 2. Adding protein anchors, such as MA-NHS, GMA or glutaraldehyde. 3. Gelation with acrylic and acrylate monomers. 4. Denaturation using proteinase K or heat denaturation. 5. Fluorescent staining: stain the biotin and DIG with fluorescently conjugated streptavidin and anti-DIG antibodies. 6. Expansion: expand hydrogel through immersion in pure water.