Abstract
The production of benzaldehyde from phenylalanine has been studied in various microorganisms, and several metabolic pathways have been proposed in the literature for the formation of this aromatic flavor compound. In this study, we describe benzaldehyde formation from phenylalanine by using a cell extract of Lactobacillus plantarum. Phenylalanine was initially converted to phenylpyruvic acid by an aminotransferase in the cell extract, and the keto acid was further transformed to benzaldehyde. However, control experiments with boiled cell extract revealed that the subsequent conversion of phenylpyruvic acid was a chemical oxidation step. It was observed that several cations could replace the extract in the conversion of phenylpyruvic acid to benzaldehyde. Addition of Cu(II) ions to phenylpyruvic acid resulted not only in the formation of benzaldehyde, but also in the generation of phenylacetic acid, mandelic acid, and phenylglyoxylic acid. These compounds have been considered intermediates in the biological conversion of phenylalanine. The chemical conversion step of phenylpyruvic acid was dependent on temperature, pH, the availability of cations, and the presence of oxygen.
The increasing consumer preference for products of natural origin has directed research towards the exploitation of microbial biosynthetic pathways to produce natural flavors. Natural aromatic compounds, including benzaldehyde and vanillin, represent a very large market in the flavor industry. In quantity, benzaldehyde is, after vanillin, the most important of these compounds (24). The natural benzaldehyde market in 1995 was about 100 metric tons/year and is growing at about 5% per year (6). Natural benzaldehyde is found as a glycoside (amygdalin) in the pits of almonds and cherries and can be released by enzymatic hydrolysis. A drawback is that toxic byproducts, such as hydrocyanic acid, may be formed.
Microbial production of benzaldehyde from phenylalanine offers an attractive alternative way to produce benzaldehyde which can be labeled “natural.” Benzaldehyde formation in medium supplemented with phenylalanine has been reported for cultures of Ischnoderma benzoinum (9, 14), Polyporus tuberaster (13), and Phanerochaete chrysosporium (12). Immobilization of the white-rot fungus Bjerkandera adusta resulted in an increased production of benzaldehyde in a medium containing l-phenylalanine (15). Among bacteria, benzaldehyde formation has been reported for a strain of Pseudomonas putida. In this strain, benzaldehyde was formed as a metabolic intermediate in the mandelic acid pathway during the degradation of mandelic acid (22). Several metabolic pathways have been proposed in the literature for the formation of benzaldehyde from phenylalanine.
Benzaldehyde was found in the volatile fractions of several cheeses and may contribute to the flavor of these products (2, 19). Additionally, benzaldehyde was formed in a complex medium inoculated with several different strains of lactic acid bacteria (11, 21). However, there is no information about the contribution of lactic acid bacteria to the production of benzaldehyde.
In this study, we investigated whether a lactic acid bacterium was able to produce benzaldehyde by using phenylalanine as a substrate. Incubation of a cell extract of Lactobacillus plantarum with phenylalanine revealed that benzaldehyde was indeed formed. More detailed studies of the mechanism involved revealed that benzaldehyde formation was initiated by an aminotransferase, resulting in phenylpyruvic acid. The keto acid formed was subsequently subjected to a chemical reaction, leading to benzaldehyde. The chemical conversion of phenylpyruvic acid was demonstrated under various mild conditions.
MATERIALS AND METHODS
Chemicals.
Phenylalanine, phenylpyruvic acid, α-ketoglutaric acid, pyridoxal 5′-phosphate (PLP), lysozyme, benzaldehyde, phenylglyoxylic acid, and phenylethanol were obtained from Sigma Chemical Co. (St. Louis, Mo.). Phenylacetic acid, trans-cinnamic acid, and mandelic acid were purchased from Acros Chimica (Geel, Belgium), and benzoic acid was obtained from Merck (Darmstadt, Germany).
Strain, growth medium, and preparation of cell extract.
L. plantarum URL-LcL1 (Unilever Research Laboratory, Vlaardingen, The Netherlands) was used in this study. The organism was routinely maintained in 10% sterile litmus milk (Difco) and stored at −80°C until use. L. plantarum was cultured overnight at 30°C in MRS broth (Merck). The cells were harvested by centrifugation (16,000 × g, 15 min, 4°C) and washed twice in 50 mM sodium phosphate buffer (pH 7.0). The cells were resuspended in 50 mM triethanolamine buffer (pH 7.0) containing 20% sucrose and 0.4 mg of lysozyme/ml. After incubation for 30 min at 30°C, the suspension was centrifuged (12,000 × g, 30 min, 4°C) and resuspended in 50 mM potassium phosphate buffer (pH 7.0) containing 0.1 mM PLP. The cells were disrupted by sonication for five periods of 30 s each on ice. Cell debris was removed by centrifugation (20,000 × g, 30 min, 4°C) to give a crude cell extract. The protein concentration in the cell extract was determined by the method of Lowry et al. (18), with bovine serum albumin as the standard.
Phenylalanine catabolism by cell extract.
The cell extract of L. plantarum was incubated with phenylalanine in sterile sealed bottles which were agitated at 37°C. The reaction mixture and the cell extract were sterilized by passing them through a sterile filter (0.2-μm pore size; Schleicher & Schuell, Dassel, Germany). Aliquots of 1 ml were withdrawn at various times to analyze the reaction products by high-performance liquid chromatography (HPLC). The reaction mixture contained 8 mM substrate, 2 ml of cell extract of L. plantarum, and 0.02 mM PLP in 8 ml of 50 mM Tris-HCl (pH 8.0). For incubations with phenylalanine as a substrate, 8 mM α-ketoglutaric acid was added to the reaction mixture. In control samples, the cell extract was boiled for 10 min.
Chemical conversion of phenylpyruvic acid to benzaldehyde.
The reaction mixture for the chemical conversion of phenylpyruvic acid to benzaldehyde contained 8 mM phenylpyruvic acid in 50 mM Tris-HCl (pH 8.0). Sealed bottles were filled with reaction mixture, and CuSO4 · 5H2O was added from a stock solution in 50 mM Tris-HCl (pH 8.0) to a final concentration of 350 μM. Incubations were performed at either 37, 30, or 25°C.
The effect of metal ions on the chemical conversion of phenylpyruvic acid to benzaldehyde was studied with stock solutions of Fe2(SO4)3 · 5H2O, FeSO4 · 7H2O, MnSO4 · H2O, Mn(III) acetate dihydrate, ZnSO4 · 7H2O, MgSO4 · 7H2O, and CuSO4 · 5H2O in Tris-HCl (pH 8.0). The reaction mixtures were incubated at 37°C, and samples were taken after 6 h of incubation.
The effect of pH on the chemical conversion of phenylpyruvic acid to benzaldehyde was studied with 50 mM sodium phosphate buffer (pHs 6.0, 7.0, and 8.0) and 50 mM Tris-HCl buffer (pHs 8.0 and 9.0). All incubations were performed at 37°C in the presence of 350 μM CuSO4 · 5H2O.
HPLC analysis.
Samples of the reaction mixtures were diluted to appropriate concentrations with 50 mM Tris-HCl (pH 8.0). Each diluted sample (900 μl) was mixed with 100 μl of 6 N HCl, and the supernatant fluid obtained after centrifugation for 5 min was analyzed for reaction products. Analyses were performed with a Hewlett-Packard (Waldbronn, Germany) HPLC Chemstation (Pascal series) equipped with an HP 1050 pumping system. Reaction products were separated onto a Chromspher5 C18 column (Chrompack, Bergen op Zoom, The Netherlands) with solvent A (0.115% trifluoroacetic acid) and solvent B (0.1% trifluoroacetic acid, 60% acetonitrile) and detected by an HP 1040 M-series II diode array detector. The products were separated by using the following linear gradient (0.5 ml min−1, 30°C): 25% solvent B at 0 min to 65% solvent B at 21 min followed by equilibration under the initial conditions for 6 min. Detection was by UV at 260 nm for benzaldehyde, 280 nm for phenylpyruvic acid, and 216 nm for phenylacetic acid, mandelic acid, and benzoic acid. Concentrations were calculated from standard curves of the pure compounds.
GC-MS analysis.
For gas chromatography-mass spectrometry (GC-MS) analysis, the reaction mixture was mixed with 10% (vol/vol) 6 N HCl. The acidified reaction mixture was then extracted with ethyl acetate. The extract obtained was used for analysis on an HP5973A quadrupole MS coupled to an HP6980 gas chromatograph equipped with a fused silica capillary column (HP-5MS; 30 m by 0.25 mm inside diameter; film thickness, 0.25 μm). The following operating conditions were used: injector temperature, 220°C; temperature program, 70 to 250°C at 7°C min−1, hold 10 min; injection volume, 1.0 μl; split ratio, 1:50; flow rate of carrier gas (helium), 1.0 ml min−1. Electron impact mass spectra were obtained at 70 eV.
RESULTS
Incubation of cell extract of L. plantarum with phenylalanine.
Incubation of the cell extract of L. plantarum with phenylalanine in the presence of both α-ketoglutaric acid and PLP resulted in the production of two compounds. The HPLC chromatogram showed two UV-absorbing peaks with the retention times of phenylpyruvic acid and benzaldehyde, respectively. The identities of the compounds were confirmed by GC-MS for benzaldehyde and by comparison of the UV spectrum with that of the pure compound for phenylpyruvic acid.
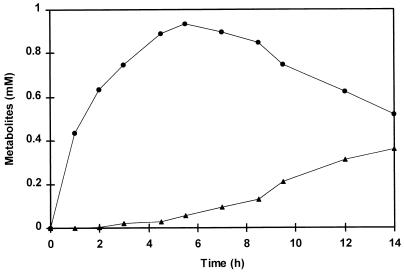
Figure 1 shows the formation of phenylpyruvic acid and benzaldehyde over time. Neither phenylpyruvic acid nor benzaldehyde was formed if α-ketoglutaric acid or PLP was omitted from the reaction mixture or if boiled cell extract was used. Phenylpyruvic acid reached a maximum concentration after 5.5 h, but the benzaldehyde concentration increased continuously up to 0.36 mM after 14 h of incubation.
FIG. 1.
The formation over time of phenylpyruvic acid (•) and benzaldehyde (▴) from phenylalanine during incubation of a cell extract of L. plantarum. Incubations were performed at 37°C in a reaction mixture containing cell extract (8.6 mg of protein), 8 mM phenylalanine, 0.02 mM PLP, and 8 mM α-ketoglutaric acid in 50 mM Tris buffer (pH 8.0). Small amounts of phenylacetic acid, mandelic acid, and phenylglyoxylic acid were detected at concentrations of 0.05, 0.02, and 0.02 mM, respectively, after 14 h of incubation.
Pathway of phenylalanine degradation.
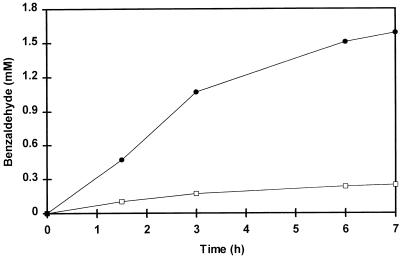
The incubation of cell extract with phenylalanine described above suggested that an aminotransferase is involved in the initial formation of phenylpyruvic acid, which is then converted into benzaldehyde. However, information on the subsequent conversion of phenylpyruvic acid to benzaldehyde was lacking. Therefore, both phenylpyruvic acid and several other compounds (proposed in the literature as intermediates in the degradation pathway of phenylalanine) were tested as the substrate for benzaldehyde production by the cell extract of L. plantarum. Phenylpyruvic acid was converted by the cell extract and resulted in the production of benzaldehyde at a rate six times higher than that with phenylalanine (Fig. 2). Neither phenylethanol, phenylacetic acid, mandelic acid, phenylglyoxylic acid, nor cinnamic acid was a substrate for the formation of benzaldehyde by the cell extract. These results strongly suggested that phenylpyruvic acid is an intermediate in the pathway leading from phenylalanine to benzaldehyde.
FIG. 2.
Benzaldehyde formation by a cell extract of L. plantarum with phenylalanine (□) or phenylpyruvic acid (•) as a substrate. Incubations were performed at 37°C in 50 mM Tris buffer, pH 8.0, containing 8 mM phenylalanine or phenylpyruvic acid, cell extract (11.0 mg of protein), and 0.02 mM PLP. α-Ketoglutaric acid (8 mM) was added to the mixture containing phenylalanine.
Conversion of phenylpyruvic acid.
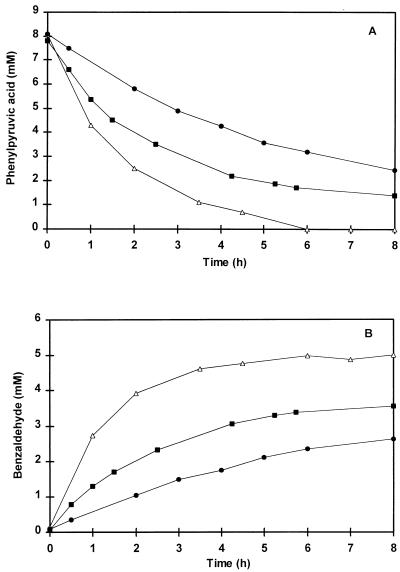
More detailed studies of the conversion of phenylpyruvic acid by the cell extract were performed. Surprisingly, it was observed that the rates of benzaldehyde formation were similar in incubations containing either cell extract or boiled cell extract. However, no benzaldehyde formation from phenylpyruvic acid was observed when the incubation systems containing either normal or boiled extract were flushed with nitrogen gas to create anoxic conditions. These observations suggested that a chemical oxidation reaction rather than an enzymatic step is involved in the conversion of phenylpyruvic acid to benzaldehyde. However, phenylpyruvic acid was not degraded if either boiled or untreated cell extract was omitted from the reaction mixture, suggesting that a component of the cell extract was essential for the conversion of phenylpyruvic acid into benzaldehyde. Several components present in the extract were tested, and it was observed that several cations could replace cell extract in the conversion of phenylpyruvic acid. Initially, the effects of Cu(II) ions on the conversion of phenylpyruvic acid were studied. Figure 3 demonstrates the conversion of phenylpyruvic acid into benzaldehyde in the presence of 350 μM CuSO4 at 37, 30, and 25°C. The conversion of phenylpyruvic acid to benzaldehyde was temperature dependent. Decreasing the incubation temperature to 30 or 25°C reduced the amount of phenylpyruvic acid converted after 8 h of incubation.
FIG. 3.
Chemical conversion of phenylpyruvic acid (A) to benzaldehyde (B) over time. Incubations were performed in a reaction mixture containing 8 mM phenylpyruvic acid in 50 mM Tris buffer, pH 8.0, in the presence of 350 μM CuSO4 at 37 (▵), 30 (■), and 25°C (•). Cell extract was omitted from the reaction mixture.
At 37°C, the conversion of phenylpyruvic acid was completed after 6 h of incubation, yielding 5.0 mM benzaldehyde, which corresponded to 63% conversion to benzaldehyde on a molar basis. Besides benzaldehyde, the substrate was converted to phenylacetic acid (13% [Fig. 4C]) and mandelic acid (5.6%) and phenylglyoxylic acid (5.3%) (results not shown). The identity of these compounds was confirmed by GC-MS (phenylacetic acid) and by UV spectra (mandelic acid and phenylglyoxylic acid). Benzoic acid was only found in trace amounts and could not account for the missing 13% of the phenylpyruvic acid. Addition of CuSO4 to phenylacetic acid, mandelic acid, or phenylglyoxylic acid did not result in either benzaldehyde formation or degradation of these compounds.
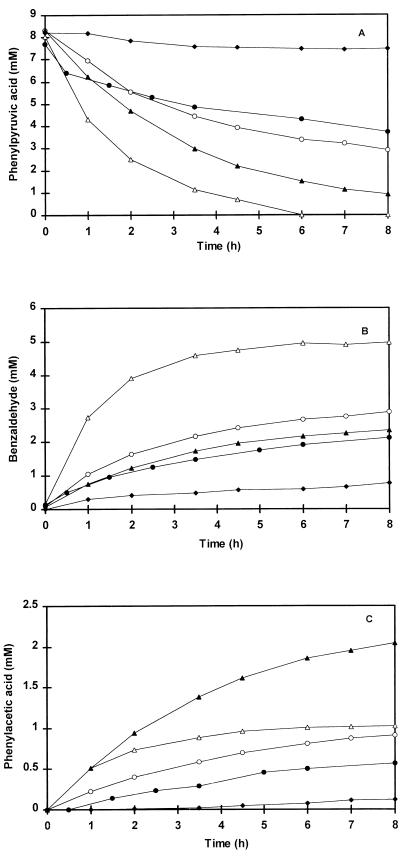
FIG. 4.
Chemical conversion of phenylpyruvic acid (A) and formation of benzaldehyde (B) and phenylacetic acid (C). Incubations were performed at 37°C in the presence of 350 μM CuSO4 in 50 mM Tris buffer, pH 8.0 (▵) and pH 9.0 (▴), or in 50 mM phosphate buffer, pH 8.0 (○), pH 7.0 (•), and pH 6.0 (⧫). Cell extract was omitted from the reaction mixture.
Effect of pH on the chemical formation of benzaldehyde.
Figure 4 shows the effect of pH on the chemical conversion of phenylpyruvic acid to benzaldehyde. At 37°C in the presence of CuSO4, the highest conversion rate was obtained with a Tris buffer of pH 8.0. Either an increase or a decrease in the pH of the reaction mixture resulted in a lower rate of phenylpyruvic acid conversion. After 8 h of incubation, 100 and 89% of the initial 8 mM phenylpyruvic acid was converted in Tris buffer at pHs 8.0 and 9.0, respectively, while percentages of 65, 51, and 9% were obtained in phosphate buffer at pHs 8.0, 7.0, and 6.0, respectively. However, increasing the pH resulted in a higher rate of phenylacetic acid formation.
Effect of metal ions on the chemical formation of benzaldehyde.
Apart from Cu(II) ions, Fe(III), Fe(II), Mn(II), and Mn(III) ions also catalyzed the conversion of phenylpyruvic acid to benzaldehyde at different rates (Table 1). Fe(III) catalyzed the conversion more effectively than Fe(II). Due to its poor solubility, Mn(III) was only tested at 10 μM and showed a slightly lower conversion of the substrate than did Mn(II) at this concentration. Mg(II) and Zn(II) ions had no effect on the conversion of phenylpyruvic acid. The effect of Cu(II) was concentration dependent, and benzaldehyde formation was completely inhibited in the presence of the metal ion chelator EDTA or in the absence of oxygen.
TABLE 1.
Benzaldehyde formation from phenylpyruvic acid in the presence of different metal ions
| Addition to reaction mixturea | Benzaldehyde produced (mM) |
|---|---|
| No addition | <0.001 |
| 350 μM Cu(II) | 4.19 |
| 100 μM Cu(II) | 2.13 |
| 50 μM Cu(II) | 1.03 |
| 100 μM Cu(II) + 200 μM EDTA | <0.001 |
| 100 μM Cu(II) flushed with N2 gas | <0.001 |
| 100 μM Fe(II) | 0.22 |
| 100 μM Fe(III) | 1.88 |
| 100 μM Mn(II) | 1.57 |
| 10 μM Mn(II) | 0.55 |
| 10 μM Mn(III) | 0.38 |
| 100 μM Zn(II) | <0.001 |
| 350 μM Mg(II) | <0.001 |
Each reaction mixture contained 8 mM phenylpyruvic acid in 50 mM Tris buffer, pH 8.0. Incubations were performed at 37°C. In all cases, SO42− was the counterion of the metal ion added.
DISCUSSION
The production of benzaldehyde from phenylalanine has been studied in various microorganisms. Fungi were investigated in most studies, but we have studied the production of benzaldehyde from phenylalanine by lactic acid bacteria because these organisms are important in the generation of flavor in dairy products.
Benzaldehyde was formed from phenylpyruvic acid, which accumulated from phenylalanine. Keto acids can be formed from amino acids by various enzymes, including amino acid oxidases (3, 10, 17), aminotransferases (10, 16), and dehydrogenases (1, 7, 10). In L. plantarum, a transaminase reaction, which depended on α-ketoglutaric acid as an amino group acceptor and on the cofactor PLP, was active. In lactic acid bacteria, very little is known about the degradation of amino acids. Only recently, aminotransferases acting on several aromatic and branched-chain amino acids were purified and characterized from Lactococcus strains (8, 25).
Various research groups have worked on the biotechnological production of benzaldehyde from phenylalanine, and several metabolic pathways for benzaldehyde formation have been postulated. Krings et al. (14) suggested that phenylalanine degradation is initiated by a deamination step leading to phenylpyruvic acid in the fungus I. benzoinum. In the next steps, phenylpyruvic acid was converted by decarboxylation, oxidation, and hydroxylation reactions. Intermediates suggested in this pathway include phenylacetaldehyde, phenylacetic acid, mandelic acid, and phenylglyoxylic acid. Benzaldehyde in turn could either be oxidized to benzoic acid or reduced to benzyl alcohol.
Another pathway proposed for benzaldehyde formation involves the enzyme phenylalanine ammonia-lyase, resulting in cinnamic acid as an intermediate, as described for several fungi (12, 13). Benzaldehyde formation through 2-phenylethanol was suggested by Fabre et al. (9). However, intermediary molecules derived from phenylalanine consumption were not detected.
In our study, none of the intermediates in the conversion of phenylpyruvic acid to benzaldehyde suggested by the above-cited authors was converted by a cell extract of L. plantarum. However, it was demonstrated that phenylpyruvic acid was converted to benzaldehyde in the presence of boiled cell extract, indicating that the conversion of phenylpyruvic acid in the system under study involves a chemical rather than an enzymatic step. Strong support for this conclusion is provided by the observation that the addition of Cu(II), Fe(III), Fe(II), Mn(II), or Mn(III) ions to phenylpyruvic acid catalyzed the formation of benzaldehyde under aerobic conditions in the absence of cell extract. Benzaldehyde formation in the presence of either untreated or boiled cell extract may therefore be due to the introduction of metal ions with the extract.
The results of the present study throw new light on the biological conversion of phenylalanine to benzaldehyde. Incubation of phenylpyruvic acid in the presence of catalyzing metal ions resulted not only in the formation of benzaldehyde but also in the generation of phenylacetic acid, mandelic acid, and phenylglyoxylic acid. These compounds were considered by Krings et al. (14) to be intermediates in the route leading to benzaldehyde. These compounds did not seem to be involved in the chemical pathway leading to benzaldehyde, since no degradation of the separate compounds was observed in the presence of Cu(II). They rather seem to derive from phenylpyruvic acid through a different reaction.
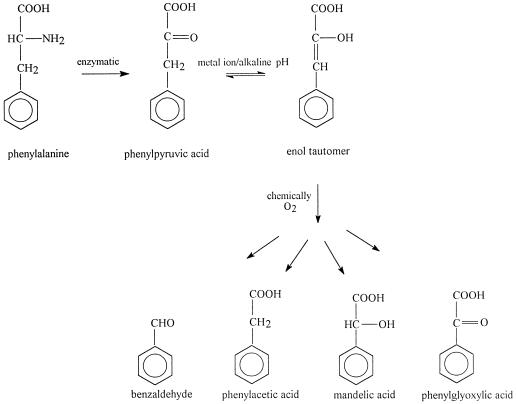
The role of the chemical conversion of phenylpyruvic acid to benzaldehyde in phenylalanine degradation pathways has received no attention in the literature. Nevertheless, oxidation of phenylpyruvic acid to benzaldehyde and oxalic acid had been reported by Pitt (20) as early as 1962. He observed that it was not the keto acid itself but the enol tautomer of the compound that was oxidized. The presence of bivalent metal ions can accelerate this tautomerization reaction (4). Based on the results of the present study, a proposed enzymatic-chemical pathway for benzaldehyde formation from phenylalanine is presented in Fig. 5. Phenylpyruvic acid is formed enzymatically from phenylalanine and is an unstable compound. The presence of catalyzing metal ions or alkaline conditions can enhance the enol tautomer of the keto acid, which is chemically converted to benzaldehyde, phenylacetic acid, mandelic acid, and phenylglyoxylic acid in the presence of oxygen. Benzaldehyde may be further oxidized to benzoic acid, although in our study only trace amounts of this compound were detected. More recently, Casey and Dobb (5) converted microbially produced phenylpyruvic acid to benzaldehyde by heating the fermentation broth containing phenylpyruvic acid to 90°C at pH 9.5. The results of the present study show that in the presence of catalyzing metal ions, the conversion of phenylpyruvic acid to benzaldehyde can occur under far milder conditions.
FIG. 5.
Proposed mechanism for benzaldehyde formation from phenylalanine by both enzymatic and chemical steps.
The phosphate buffer may bind the metal ions in the reaction mixture and thereby reduce their availability. This can cause the difference between benzaldehyde formation at pH 8.0 in phosphate buffer and that in Tris buffer. The reduced conversion rate of phenylpyruvic acid to benzaldehyde at pH 9.0 compared with that at pH 8.0 in Tris buffer may be explained by a dual effect of the pH on the chemical conversion rate. The formation of the enol tautomer is induced by alkaline conditions (20). This effect may be opposed by the reduced solubility of metal ions under alkaline conditions. It can be speculated that the effect of pH on the chemical conversion of phenylpyruvic acid is a combination of these two effects.
Amino acid degradation is believed to be important for flavor development in cheese. Straight-out chemical reactions are not believed to play a major role in the production of cheese flavor but rather seem to be enhanced by enzymes (23). In this study, benzaldehyde formation was initiated by an aminotransferase present in the cell extract of L. plantarum. Aminotransferases from lactic acid bacteria have shown activity under cheese-ripening conditions (8, 25), but the low oxygen concentration, low ripening temperature, and low pH in cheese do not favor the chemical conversion of phenylpyruvic acid to benzaldehyde. However, considering the long time involved in cheese ripening, this mechanism may still make a significant contribution.
ACKNOWLEDGMENT
We thank Henk Swarts for performing the GC-MS analyses.
REFERENCES
- 1.Asano Y, Nakazawa A, Endo K. Novel phenylalanine dehydrogenase from Sporosarcina ureae and Bacillus sphaericus. J Biol Chem. 1987;262:10346–10354. [PubMed] [Google Scholar]
- 2.Bosset J O, Gauch R. Comparison of the volatile flavor compounds of six European ‘AOC’ cheeses by using a new dynamic headspace GC-MS method. Int Dairy J. 1993;3:359–377. [Google Scholar]
- 3.Brearley G M, Price C P, Atkinson T, Hammond P M. Purification and partial characterisation of a broad range l-amino acid oxidase from Bacillus carotarum 2Pfa isolated from soil. Appl Microbiol Biotechnol. 1994;41:670–676. [Google Scholar]
- 4.Bücher T, Kirberger E. Über die enol-keto-tautomerie der p-oxy-phenylbrenztraubensäure. Biochim Biophys Acta. 1952;8:401–406. doi: 10.1016/0006-3002(52)90065-6. [DOI] [PubMed] [Google Scholar]
- 5.Casey J, Dobb R. Microbial routes to aromatic aldehydes. Enzyme Microb Technol. 1992;14:739–747. [Google Scholar]
- 6.Clark G S. An aroma chemical profile. Benzaldehyde Perfum Flavor. 1995;20:53–60. [Google Scholar]
- 7.de Boer L, van Rijssel M, Euverink G J, Dijkhuizen L. Purification, characterization and regulation of a monomeric l-phenylalanine dehydrogenase from the facultative methylotroph Nocardia sp. 239. Arch Microbiol. 1989;153:12–18. [Google Scholar]
- 8.Engels W J M, Alting A C, Arntz M M T G, Gruppen H, Voragen A G J, Visser S. Volatile and non-volatile compounds in ripened cheese: their formation and their contribution to flavor. Ph.D. thesis. Wageningen, The Netherlands: Wageningen Agricultural University; 1997. [Google Scholar]
- 9.Fabre C E, Blanc P J, Goma G. Production of benzaldehyde. Sci Aliments. 1996;16:61–68. [Google Scholar]
- 10.Hemme D, Bouillanne C, Métro F, Desmazeaud M-J. Microbial catabolism of amino acids during cheese ripening. Sci Aliments. 1982;2:113–123. [Google Scholar]
- 11.Imhof R, Glättli H, Bosset J O. Volatile organic compounds produced by thermophilic and mesophilic single strain dairy starter cultures. Lebensm-Wiss Technol. 1995;28:78–86. [Google Scholar]
- 12.Jensen K A, Jr, Evans K M C, Kirk T K, Hammel K E. Biosynthetic pathway for veratryl alcohol in the ligninolytic fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:709–714. doi: 10.1128/aem.60.2.709-714.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawabe T, Morita H. Production of benzaldehyde and benzylalcohol by the mushroom Polyporus tuberaster K2606. J Agric Food Chem. 1994;42:2556–2560. [Google Scholar]
- 14.Krings U, Hinz M, Berger R G. Degradation of [2H]phenylalanine by the basidiomycete Ischnoderma benzoinum. J Biotechnol. 1996;51:123–129. [Google Scholar]
- 15.Lapadatescu C, Ferron G, Vergoignan C, Djian A, Durand A, Bonnarme P. Influence of cell immobilization on the production of benzaldehyde and benzyl alcohol by the white-rot fungi Bjerkandera adusta, Ischnoderma benzoinum and Dichomitus squalens. Appl Microbiol Biotechnol. 1997;47:708–714. [Google Scholar]
- 16.Lee C-W, Lucas S, Desmazeaud M J. Phenylalanine and tyrosine catabolism in some cheese coryneform bacteria. FEMS Microbiol Lett. 1985;26:201–205. [Google Scholar]
- 17.Lee Y-H, Chu W-S. d-Amino acid oxidase from Rhodosporidium toruloides. Lett Appl Microbiol. 1996;23:283–286. [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Molimard P, Spinnler H E. Review: compounds involved in the flavor of surface mold-ripened cheeses: origins and properties. J Dairy Sci. 1996;79:169–184. [Google Scholar]
- 20.Pitt B M. Oxidation of phenylpyruvates to aromatic aldehydes and oxalate. Nature. 1962;196:272–273. doi: 10.1038/196272a0. [DOI] [PubMed] [Google Scholar]
- 21.Tracey R P, Britz T J. Freon 11 extraction of volatile metabolites formed by certain lactic acid bacteria. Appl Environ Microbiol. 1989;55:1617–1623. doi: 10.1128/aem.55.6.1617-1623.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsou A Y, Ransom S C, Gerlt J A. Mandelic acid pathway of Pseudomonas putida: sequence relationships involving mandelic acid racemase, (S)-mandelic acid dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry. 1990;29:9856–9862. doi: 10.1021/bi00494a015. [DOI] [PubMed] [Google Scholar]
- 23.Urbach G. Contribution of lactic acid bacteria to flavour compound formation in dairy products. Int Dairy J. 1995;5:877–903. [Google Scholar]
- 24.Welsh F W, Murray W D, Williams R E. Microbiological production of flavor and fragrance chemicals. Crit Rev Microbiol. 1989;9:105–169. [Google Scholar]
- 25.Yvon M, Thirouin S, Rijnen L, Fromentier D, Gripon J C. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl Environ Microbiol. 1997;63:414–419. doi: 10.1128/aem.63.2.414-419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]