Children with neurofibromatosis type 1 (NF1) are at increased risk of developing autism spectrum disorder (ASD), with approximately 13% of individuals displaying severe-range elevations in quantitative autistic trait (QAT) burden measured using the Social Responsiveness Scale, 2nd Edition (SRS-2).1 While there are no established risk factors for ASD in children with NF1, recent studies have revealed that first-degree family members with NF1 are concordant for QAT severity.1,2 These findings suggest a high degree of mutational specificity for ASD symptomatology in NF1, and raise the intriguing possibility that the germline NF1 gene mutation is one potential risk factor.1,2 In this report, we explore the correlation between the type and location of the NF1 gene mutation and QAT burden in individuals with NF1.
Methods
A retrospective cross-sectional analysis was performed on a previously assembled cohort of individuals with NF1 under an approved Human Studies protocol.2 From this cohort of 117 patients, 63 unrelated individuals had germline NF1 gene mutation and SRS-2 data available.2 Three patients with total NF1 gene deletions were excluded, given this well-established NF1 genotype–phenotype correlation,3 as well as 3 patients with known ASD-associated chromosomal abnormalities identified by clinical chromosomal microarray analysis (CMA). CMA data were not available for the remaining cohort. Data obtained included sex, age, NF1 gene mutation, and SRS-2 total T score; T scores 60–75 are associated with mild to moderate ASD symptomatology, and T scores ≥76 are associated with severe-range ASD traits.2
Categorical variables were analyzed using χ2 tests of independence, and odds ratios (ORs) were computed using logistic regression methods. Continuously distributed traits, adhering to both conventional normality assumptions and homogeneity of variances, were compared using analysis of variance methods.
Results
Of the 57 patients with NF1 mutations and QAT data, there were equal numbers of male (n = 28) and female (n = 29) participants, and the ages ranged from 2.5 to 58 years (median, 13 years; 41 patients <18 years of age). NF1 mutations spanned exon 1 to exon 53,4 with 24 nonsense (42.1%), 18 frameshift (31.6%), 12 splice-site (21.1%), and 3 missense variants (5.3%), representative of the mutational spectrum observed in the NF1 population.5 The mean total T score was 61.2 (SD 15.2): 30 (52.6%) individuals scored ≥60, and 11 (19.3%) scored ≥76. No correlation between sex or NF1 gene mutation type on QAT burden was observed.
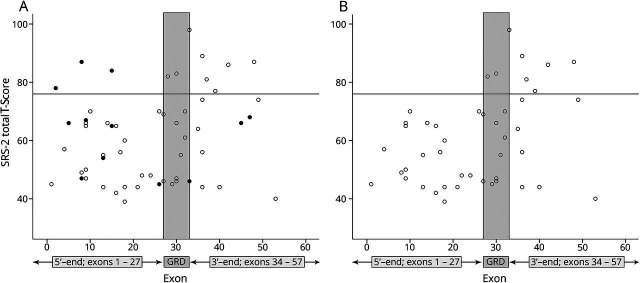
Initial analysis including all mutations (n = 57) revealed a location-dependent association of QAT burden, such that individuals harboring mutations within the 5′-end of the NF1 gene had lower QAT scores relative to those harboring mutations within the 3′-end of the gene (57.4 vs 67.9; p = 0.03; figure, A). Furthermore, 90% of individuals (n = 27) with 5′-end mutations had a total T score of ≤75 compared to 64% of patients (n = 9) with 3′-end mutations (OR 5.0; 95% confidence interval [CI] 0.99–25.21; p = 0.05).
Figure. Scatterplot of Social Responsiveness Scale, 2nd Edition (SRS-2) total T scores vs NF1 mutation location.
Scatterplot of SRS-2 total T scores vs NF1 mutation location for (A) all variants and (B) variants in coding region only. Gray box: GAP-related domain (GRD) spanning exons 27–33. Black dots: splice site mutations. Solid line: total T score cutoff of 76.
Individuals not adhering to this pattern (5′-end mutations and high ASD scores; n = 3) all had splice site mutations predicted to result in in-frame exon skipping. Subsequent analyses, including only those patients harboring mutations within the coding region of the NF1 gene (n = 45), strengthened this genotype–phenotype association (5′-end: 53.8 vs 3′-end: 68.0; p = 0.006; figure, B). As such, 100% of individuals (n = 21) with 5′-end mutations had a total T score of ≤75 compared to 58.3% of patients (n = 7) harboring 3′-end mutations (OR 31.5; 95% CI 1.55–641.05; p = 0.02). Collectively, mutations within the 5′-end of the NF1 gene demonstrated a sensitivity and specificity for detecting normal to moderate QAT burden of 79.4% and 100.0%, respectively. No statistically significant differences were observed using a SRS-2 T score cutoff value of 60; however, more individuals with T scores <60 harbored 5′-end coding variants (66.7% vs 33.3%; p = 0.06).
Discussion
While NF1 is a completely penetrant genetic disorder, QAT burden in NF1 is remarkably variable and is often not clinically evident until later in childhood.1 Coupled with an absence of early prognostic tools, it is difficult to initiate early interventions for these at-risk individuals. Herein, we demonstrate that mutation location within the NF1 gene correlates with QAT severity, such that mutations within the coding region of the 5′-end are associated with significantly higher odds of having lower ASD symptom burden. Taken together with reports demonstrating associations between the location of germline mutations and other NF1 clinical phenotypes (e.g., optic glioma, neurofibromas),6,7 the present findings suggest that the specific germline NF1 mutation is one modifier of QAT severity which, in combination with other to-be-identified risk factors, may allow for early risk stratification prior to the onset of clinically detectable ASD symptomatology. Since this study is limited by an absence of data regarding comorbid behavioral impairments and additional genetic aberrations, future investigations will be required to define the biological mechanisms responsible for these genotype–phenotype correlations.
Author contributions
Stephanie M. Morris, MD: study concept and design, acquisition of data, analysis and interpretation of data, critical revision of manuscript for intellectual content. David H. Gutmann, MD, PhD: study concept and design, acquisition of data, critical revision of manuscript for intellectual content, study supervision.
Study funding
D.H.G. is supported by a Research Program Award from the National Institute of Neurologic Disorders and Stroke (1-R35-NS097211-01). S.M.M. is supported by the Neurologic Sciences Academic Development Award (K12 NS001690).
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Morris SM, Acosta MT, Garg S, et al. Disease burden and symptom structure of autism in neurofibromatosis type 1: a study of the international NF1-ASD consortium team (INFACT). JAMA Psychiatry 2016;73:1276–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Constantino JN, Zhang Y, Holzhauer K, et al. Distribution and within-family specificity of quantitative autistic traits in patients with neurofibromatosis type 1. J Pediatr 2015;167:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehrer-Sawatzki H, Mautner VF, Cooper DN. Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum Genet 2017;136:349–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anastasaki C, Le LQ, Kesterson RA, Gutmann DH. Updated nomenclature for human and mouse neurofibromatosis type 1 genes. Neurol Genet 2017;3:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ars E, Kruyer H, Morell M, et al. Recurrent mutations in the NF1 gene are common among neurofibromatosis type 1 patients. J Med Genet 2003;40:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anastasaki C, Morris SM, Gao F, Gutmann DH. Children with 5′-end NF1 gene mutations are more likely to have glioma. Neurol Genet 2017;3:e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinna V, Lanari V, Daniele P, et al. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur J Hum Genet 2015;23:1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]