An estimated 105 DNA lesions occur daily in the mammalian genome as a consequence of spontaneous decay, replication errors, and cell metabolism, including reactive oxygen species produced by the mitochondria. Oxidative stress is a major mechanism of DNA damage in the nervous system. Damaged DNA must be repaired to allow the proper reading of the genetic code. The response to DNA damage involves DNA damage recognition first, followed by resection of the affected site, DNA processing, filling the gap by action of DNA polymerases, and sealing of the nick by DNA ligases. Severe DNA damage also triggers chromatin remodeling, transient interruption of the cell cycle, and, if left unrepaired, programmed cell death. Whereas disturbances in DNA repair have been primarily linked to carcinogenesis or immunodeficiency, they can also affect development or survival of cells in the nervous system. A prototype neurologic disorder of DNA repair is ataxia telangiectasia (A-T) due to mutation of the A-T mutated (ATM) gene encoding A-T mutated (ATM), a kinase that coordinates responses to double-strand DNA breaks. Ataxia with oculomotor apraxia (AOA) results from mutations of key proteins involved in DNA end-processing and transcription regulation. Many neurodegenerative disorders are associated with inability to repair oxidative base modifications in both nuclear and mitochondrial DNA. Mismatch and base excision–repair are important modifiers in trinucleotide repeat expansion disorders. There are several reviews on the complex mechanisms involved in DNA repair1–7 and the neurologic disorders associated with defective DNA repair pathways.8–16 A comprehensive discussion of these is beyond the scope of this review and only selected topics are discussed here.
Illustrative case
A 15-year-old boy was diagnosed with A-T at 1 year of age after he developed an unsteady gait and conjunctival telangiectasias. At age 2, he developed a tremor of the hands and trunk. By age 10, truncal dystonia and choreoathetotic movements of the upper extremities were prominent. The patient's gait gradually worsened and he was wheelchair dependent at the age of 13. In addition, he was unable to read because of saccadic intrusions, and he complained of chronic fatigability. Neurologic examination revealed dysarthric and slowed speech with oculomotor apraxia and impaired upgaze. The patient had severe truncal and limb ataxia with dysmetria and dystonic posturing of the feet. The upper extremities demonstrated mild choreiform movements. He had a history of recurrent respiratory tract infections. Blood tests showed elevated hypoalbuminemia and elevated α-fetoprotein levels.
Comments
This case illustrates the characteristic neurologic findings of A-T: early onset, progressive cerebellar ataxia with choreoathetosis, oculomotor apraxia, and conjunctival telangiectasias. A-T is a systemic disorder with immunodeficiency and an increased risk for malignancy, particularly lymphoma and leukemia. Patients with A-T are also sensitive to radiation due to the inability to repair double-strand DNA breaks. A-T is an autosomal recessive disorder and there may be substantial variability among affected individuals, while A-T carriers may be asymptomatic or exhibit radiosensitivity and cancer predisposition. The diagnosis is made clinically with genetic testing to identify pathologic mutations in the ATM gene. Neurologic treatment of patients with A-T is symptomatic and supportive, while specific treatment of systemic manifestations targets immunodeficiency and pulmonary disease.17
Basic concepts
Mechanisms of DNA damage in the nervous system
DNA repair is critical during development and in response to damage of the nervous system. Neocortical and cerebellar neurogenesis involve phases of rapid proliferation, differentiation, and migration, which require rapid and efficient mechanisms to repair DNA breaks originating from DNA transcription, replication, or naturally occurring modifications due to oxidation. Oxidative base modifications in nuclear DNA, mitochondrial DNA, or both promote neuronal loss associated with neurodegeneration.11 Single-strand breaks are one of the most common DNA lesions affecting lesions and typically result from oxidative attack by hydroxyl radicals. Neurons are particularly susceptible to DNA breaks given their longevity, high energy demands, high production of oxygen free radicals, and high transcriptional activity; within neurons, mitochondrial DNA is particularly vulnerable given its proximity to the respiratory chain and lack of protective histones. Age-related accumulation of DNA damage and alterations of DNA repair processes are both region- and cell-specific.18 The most susceptible neurons appear to be large neurons such as cerebellar Purkinje cells, neurons of the dorsal root ganglia, and motor neurons. Studies on Atm-deficient mice indicate that the neurons in the cerebellum may be particularly susceptible to oxidative stress and DNA breaks due to low levels of nicotinamide adenine diphosphate, a major cofactor of antioxidant enzymes.19 Single-strand breaks may produce blockade of DNA replication during the S (synthetic) phase of the cell cycle and stall RNA polymerase progression during transcription. DNA double-strand breaks are the most deleterious form of DNA damage because they do not leave an intact complementary DNA that can be used as a template for DNA repair.1,6 If left unrepaired, double-strand breaks can lead to chromosome breaks and translocations that may result in not only developmental deficits and neurodegeneration, but also immunodeficiency, radiosensitivity, and predisposition to cancer.1
Overview of mechanisms of DNA repair
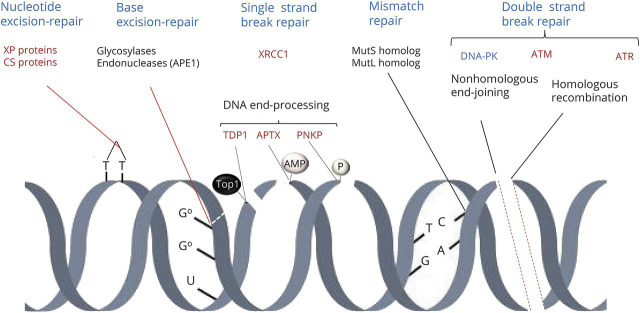
There are several mechanisms of DNA repair1–5 (figure 1). Base excision–repair removes damaged bases, mismatch repair recognizes errors in incorporation of bases to the polynucleotide, nucleotide excision–repair removes bulky DNA adducts, and crosslink repair removes intrastrand crosslinks. Single-strand break repair and double-strand repair also involve chromatin remodeling. Whereas recognition and signaling of DNA lesions varies among the different repair mechanisms, all involve resection of the affected site, DNA processing, filling the gap by action of DNA polymerases, and sealing of the nick by DNA ligases. The DNA repair response involves several protein complexes that act as sensors of DNA damage, signal the site of damage, recruit and regulate mediators of the damage response, or act as final effectors of repair. One of the earliest events of DNA repair is recruitment of poly (adenosine diphosphate [ADP] ribose) polymerase (PARP)1.20 This enzyme binds to and is activated by DNA breaks and subsequently modifies repair proteins, including protein kinases, nucleases, scaffold proteins, and histones, by addition of branches of poly (ADP) ribose, a process called (PAR)ylation. The PARP1 signal is transient; poly (ADP) ribose is rapidly degraded by glycohydrolase, which makes PARP1 again available for other repair reactions.20 Many proteins involved in DNA repair function as scaffolds to recruit other proteins involved in recognition, signal amplification, or execution of DNA repair. Typical examples are X-ray-repair cross-complementing (XRCC) proteins and proliferating cell nuclear antigen 1, which provide a scaffold for several other DNA repair proteins, including DNA polymerases and ligases. DNA polymerases catalyze DNA replication by adding free nucleotides to the 3′ end of the newly forming strand. The main DNA polymerase in the nucleus is polymerase β; polymerases δ/ε contribute to DNA repair; and polymerase γ mediates DNA replication and repair in the mitochondria. DNA ligases catalyze the formation of phosphodiester bonds between 3′ hydroxyl (3′-OH) ends of one nucleotide with the 5′ phosphate (5′P) end of the next. Proteins involved in the DNA damage response are tightly regulated and include posttranslational modifications, such as addition of small ubiquitin-like molecules (SUMOylation), and PARylation.7
Figure 1. Mechanisms of DNA repair.
There are several mechanisms of DNA repair. Base excision–repair removes damaged bases; mismatch repair recognizes errors in incorporation of bases to the polynucleotide; nucleotide excision–repair removes bulky DNA adducts; and crosslink repair removes intrastrand crosslinks. Single-strand break repair and double-strand repair also involve chromatin remodeling. One of the earliest events of DNA repair is recruitment of poly (adenosine diphosphate [ADP] ribose) polymerase (PARP)1 (not shown). Nucleotide excision–repair involves proteins that are affected in xeroderma pigmentosum (XP) and Cockayne syndrome (CS). Base excision–repair involves the sequential action of glycosylases and endonucleases, including apurinic/apyrimidinic site endonuclease 1 (APE1). One important scaffold protein in DNA repair is X-ray-repair cross-complementing protein–1 (XRCC1). DNA end-processing is necessary for the effects of DNA polymerases and ligases during repair mechanisms. This involves tyrosyl-DNA phosphodiesterase 1 (TDP1), polynucleotide 3′kinase phosphatase (PNKP), and aprataxin (APTX). Mismatch repair involves mut short (mutS) and mut long (mutL) homolog proteins. The double-strand DNA damage response involves multiple protein complexes that accumulate locally as foci at the damaged sites. These complexes recruit and regulate repair enzymes, modify the chromatin structure, act as a scaffold for repair and signaling factors, and regulate chromosome mobility, cell cycle, and transcription.1,3 Double-strand break repair occurs through either nonhomologous end-joining or homologous recombination and is controlled by DNA-dependent kinase (DNA-PK), ataxia telangiectasia mutated (ATM), and ataxia telangiectasia Rad3 (ATR) protein. AMP = adenosyl monophosphate; Top1 = topoisomerase 1.
Specific DNA repair pathways
Base excision repair
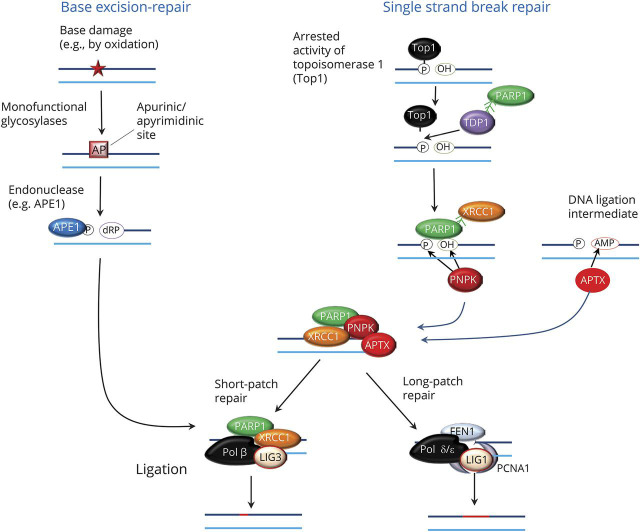
Base excision–repair is the main pathway for removal of oxidative base modifications in nuclear or mitochondrial DNA.21 The first step is the removal of the damaged base from the deoxyribose moiety of the DNA backbone by action of substrate-specific glycosylases. This process generates a site lacking a purine or a pyrimidine (abasic site), which is then cleaved by endonucleases, such as the apurinic/apyrimidinic site endonuclease 1. This creates a nick in the phosphodiester backbone that yields a free 5′P group on the apurinic/apyrimidinic site and a free 3′-OH on the normal nucleotide. DNA end-processing then occurs via mechanisms shared with single-strand repair (see below), allowing filling of the DNA gap via DNA polymerases and ligases (figure 2). Short-patch repair involves the incorporation of a single nucleotide into the DNA gap by action of polymerase β (in the nucleus) or γ (in the mitochondria) followed by sealing of the nick via the XRCC1-ligase 3 complex. Long-patch repair involves the incorporation of 2 to 7 nucleotides into the gap; this is mediated by DNA polymerase δ/ε and is followed by cleavage of the resulting 5′ flap by the flap endonuclease 1 and completed by action of ligase 1 in the presence of proliferating cell nuclear antigen–1 (figure 2).
Figure 2. Mechanisms of based excision–repair and single-strand break repair.
The first step is the removal of the damaged base from the deoxyribose moiety of the DNA by substrate-specific glycosylases. This generates a site lacking a purine or a pyrimidine (AP) site, which is then cleaved by endonucleases, such as the apurinic/apyrimidinic site endonuclease 1 (APE1). Single-strand breaks occur as a consequence of oxidative damage, stalled topoisomerase 1 (Top1) activity, abortive DNA ligase activity resulting in 5′adenosyl monophosphate (AMP), or other mechanisms. DNA end-processing requires this involves tyrosyl-DNA phosphodiesterase 1 (TDP1), polynucleotide 3′kinase phosphatase (PNKP), and aprataxin (APTX). X-ray-repair cross-complementing protein-1 (XRCC1) serves as a scaffold that interacts with poly (adenosine diphosphate [ADP] ribose) polymerase (PARP)1 and with DNA polymerase β (Pol β) and ligase 3 (LIG3) to complete short-patch repair processes. Long-patch repair is mediated by DNA polymerase δ/ε, flap endonuclease 1 (FEN1), and ligase 1 (LIG1) in the presence of proliferating cell nuclear antigen–1 (PCNA1).
Mismatch repair
Mismatch repair recognizes and repairs base–base mismatches that originate from errors or intermediates of DNA replication or homologous recombination.22,23 To prevent genome integrity, this process must occur selectively on the new strand of DNA that contains the wrong nucleotide. Mismatch repair involves a group of proteins homologous to the Mutator short (MutS) and Mutator long (MutL) proteins described in Drosophila; these mutation short homolog and mutation long homolog proteins form different complexes that participate at different stages of the mismatch repair process.22–24
Nucleotide excision–repair
The nucleotide excision–repair system recognizes base lesions that distort the normal helical structure of DNA.25 In nucleotide excision–repair, the damage is excised as a 22–30 base oligonucleotide, producing a nick in a single-stranded DNA strand that is eventually repaired by DNA polymerases followed by DNA ligases. There are 2 nucleotide excision–repair subpathways that differ only at the step of recognition, but utilize a final common machinery that involves several proteins encoded by genes mutated in xeroderma pigmentosum (XP proteins).25 Global genome nucleotide excision–repair removes helix-distorting UV light-provoked lesions and affects all given nucleotides in the genome. This response is initiated by a complex consisting of XPC and the UV excision–repair protein RAD23B, which binds to a DNA damage binding protein complex on the strand opposite to the lesion. Transcription-coupled nucleotide excision–repair involves lesion recognition by Cockayne syndrome (CS) proteins CS2A and CSB, which respond to stalling of RNA polymerase at a lesion on the transcribed strand.26 In both cases, lesion recognition is followed by recruitment of the transcription factor IIH complex that includes the helicases XPB and XPD, which promote opening of the DNA duplex around the lesion. This creates a platform for recruitment of other XP proteins, including exonucleases that target the 3′ and the 5′ ends at the damage. DNA polymerase carries on the gap filling and the nick is sealed via either XRCC1–ligase 3 complex or flap endonuclease 1–ligase 1 complex.25
Single-strand break repair
Single-strand breaks are one of the most common DNA lesions affecting the nervous system and most commonly result from oxidative attack by hydroxyl radicals. They may also occur as a product of other repair pathways, abortive activity of DNA topoisomerase, or erroneous incorporation of ribonucleotides into DNA.13 Single-strand breaks are detected primarily by PARP1, which promotes recruitment and accumulation of XRCC1, a scaffold protein that activates other components of the repair machinery13 (figure 2).
A critical step in single-strand break repair and other repair pathways is DNA end-processing. The 3′ and 5′ termini of damaged DNA strands must be restored to conventional 3′-OH and 5′P moieties in order to fill the gap by action of DNA polymerases followed by ligases. There are 3 main DNA end-processing proteins (figure 2). Polynucleotide 3′kinase phosphatase (PNKP) acts on the damaged 3′P terminus and through by its dual action yields 3′OH and a 5′P ends at the DNA breaks.27 Stalled topoisomerase 1 cleavage complexes, resulting from interruption of the ligation step following initial DNA cleavage by the enzyme, contain 3′P dead ends that are linked to the enzyme. Tyrosyl-DNA phosphodiesterase 1 (TDP1) hydrolyzes the phosphodiester bond between these 3′ ends and the catalytic tyrosyl residue of topoisomerase 1; the 3′ terminus left behind by TDP1 activity is then converted into a 3′OH group by PNKP. Aprataxin (APTX) deadenylates the abnormal 5′AMP termini resulting from abortive DNA ligase activity into normal 5′P ends. Gap filling to complete DNA repair may occur by insertion of either a single nucleotide (short-patch repair) or multiple nucleotides (long-patch repair), as described above (figure 2).
Double-strand break repair
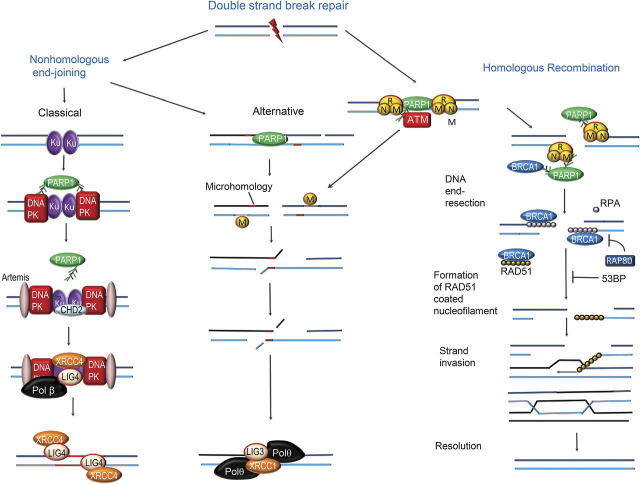
The double-strand DNA damage response involves multiple protein complexes that accumulate locally as foci at the damaged sites. These complexes both recruit and regulate repair enzymes, modify the chromatin structure, act as a scaffold for repair and signaling factors, and regulate chromosome mobility, cell cycle, and transcription.1,3 Double-strand break repair is controlled by 3 master (or apical) phosphoinositide 3′kinase–related kinases: DNA-dependent kinase, ATM, and ataxia telangiectasia Rad3 (ATR) protein.28 PARP1 is an initial sensor of double-strand DNA breaks and activates these kinases as well as other proteins involved in the repair processes.20 DNA repair is tightly coordinated with chromatin remodeling and progression of the cell cycle, in part via activation of effector checkpoint kinases 1 and 2.6 Cells utilize 2 major pathways for repair of double-strand DNA breaks: nonhomologous end-joining and homologous recombination (figure 3). These pathways are complementary and operate under different circumstances.
Figure 3. Overview of mechanisms of double-strand break repair.
Poly (adenosine diphosphate [ADP] ribose) polymerase (PARP)1 is an initial sensor of double-strand DNA breaks. The 2 major pathways for double-strand DNA breaks are nonhomologous end-joining and homologous recombination. The classical nonhomologous end-joining involves an initial recognition step mediated by Ku70/Ku80, followed by recruitment and activation of the DNA-dependent protein kinase (DNA-PK). This is followed by cleavage of the juxtaposed broken DNA ends by the nuclease Artemis. PARP1 recruits the chromodomain-helicase-DNA-binding protein 2 (CHD2). The repair process is completed by DNA polymerase followed by ligation by the X-ray-repair cross-complementing protein-4 (XRCC4)–ligase 4 (LIG4) complex. In an alternative nonhomologous end-joining pathway, after initial processing the DNA ends are sealed by microhomology-mediated base-pairing of DNA single strands, followed by nucleolytic trimming of DNA flaps; DNA double-strand breaks are first recognized by the MRN protein complex, consisting of meiotic recombination 11 (MRE11), DNA repair bridging protein 50 (RAD50), and Nijmegen breakage syndrome–1 (NBS-1). MRE11 elicits DNA end-resection, yielding 3′-overhanging single-strand DNA fragments that bind to replication protein A (RPA); this allows loading of the RAD51 recombinase onto the single-stranded DNA. This process is facilitated by proteins such as breast cancer 1 (BRCA)1 protein and inhibited by p53 binding protein (5eBP) and receptor-associated protein 80 (RAP 80)7 (figure 3). The RAD51-coated single DNA strand forms a nucleoprotein filament that mediates strand invasion into the sister chromatid to search for the homologue template DNA polymerase, DNA ligase 1, and DNA helicase mediates the remaining processes leading to intact repaired DNA molecules.
Nonhomologous end-joining
Nonhomologous end-joining is the predominant double-strand break repair pathway; it mediates the direct ligation of broken DNA ends and operates throughout the cell cycle. This process is potentially mutagenic because deletions or insertions may occur at the sites of repair. The classical nonhomologous end-joining process involves an initial recognition step, during which both ends of the double-strand DNA breaks are first bound by the Ku70/Ku80 heterodimers; this then recruits and activates the catalytic subunit of DNA-dependent protein kinase (figure 3). The juxtaposed broken DNA ends then undergo cleavage by the nuclease Artemis and then DNA end-processing enzymes PNKP and APTX. PARP1 recruits the chromodomain-helicase-DNA-binding protein 2, which triggers chromatin remodeling and regulates the assembly of nonhomologous end-joining complexes. The repair process is completed by DNA polymerase, followed by ligation by the XRCC4–ligase 4 complex. An alternative nonhomologous end-joining process follows initial processing steps that are similar to those involved in homologous repair (see below). After initial processing, the DNA ends are sealed by microhomology-mediated base-pairing of DNA single strands, followed by nucleolytic trimming of DNA flaps, DNA gap filling, and DNA ligation (figure 3).29
Homologous recombination
Homologous recombination requires the presence of a sister chromatid as a template and therefore only occurs during the S (synthetic) and G2 (postmitotic) phases of the cell cycle.30 The double-strand breaks are first recognized by the MRN protein complex, consisting of meiotic recombination 11 (MRE11), DNA repair bridging protein 50 (RAD50), and Nijmegen breakage syndrome–1 (NBS-1)31; this complex is recruited by PARP1 to the site of double-strand damage (figure 3). A major step in homologous recombination is DNA end-resection, which consists of the processing of the broken DNA ends by MRE11, yielding 3′-overhanging single-strand DNA fragments. These fragments bind to replication protein A, which stabilizes the single DNA strands, and this allows loading of the RAD51 recombinase onto the single-stranded DNA. This process is facilitated by proteins such as breast cancer 1 (BRCA1) protein and inhibited by p53 binding protein and receptor-associated protein 807 (figure 3). The RAD51-coated single DNA strand forms a nucleoprotein filament that mediates strand invasion into the sister chromatid to search for the homologue template.30 After the actions of DNA polymerase and DNA ligase 1, DNA helicase mediates the cleavage and resolution of the homologue repair intermediate, to yield intact repaired DNA molecules.
ATM and coordinated response to double-strand break repair
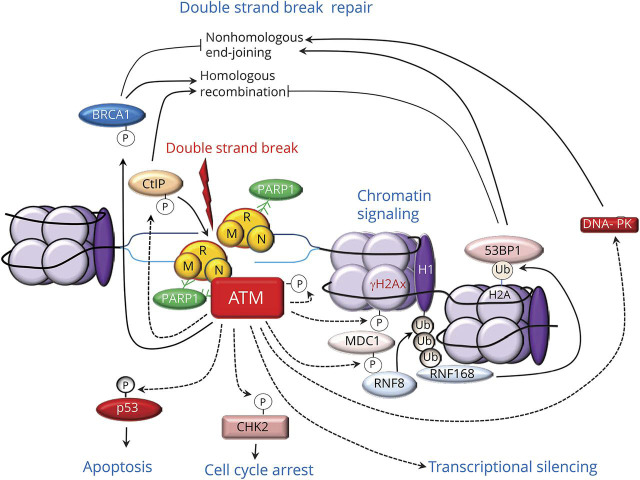
ATM functions as a master regulator of cellular responses to double-strand breaks28,32 (figure 4). Following its recruitment at the damaged sites by interactions with PARP1 and the MRN complex, ATM phosphorylates and activates hundreds of substrate proteins, including other protein kinases, providing a mechanism for signal amplification.28 ATM promotes double-strand break repair both via homologous recombination and nonhomologous end-joining. For example, ATM stimulates DNA end-resection by phosphorylating a protein called C-terminal binding protein–interacting protein, which is a major activator of MRE11.33 ATM also phosphorylates BRCA1, which facilitates DNA end resection and thus homologous recombination.34 ATM also promotes nonhomologous end-joining by activating DNA protein kinase and by facilitating the accumulation of the p53 binding protein at the site of the damaged chromatin.35 ATM triggers chromatin remodeling and signaling. For example, it phosphorylates the histone variant H2AX, yielding γH2AX; this recruits the mediator of DNA damage checkpoint 1 protein, which regulates cell cycle checkpoints and recruits other repair proteins (figure 4). ATM-triggered chromatin signaling involves phosphorylation-ubiquitylation cascades that are sequentially mediated by 2 ubiquitin E3 ligases targeting H1 linker histone and H2A histones. Additional targets of ATM are the effector checkpoint kinases 1 and 2, which spread the repair signals to the nucleus. In addition to regulating DNA repair pathways and chromatin remodeling, ATM regulates mRNA transcription and processing. ATM is recruited and activated at the site of R-loops, which consist of a DNA: RNA hybrid and the associated nontemplate single-stranded DNA; R-loops form at the site of DNA lesions that block transcription. At this level, ATM elicits local inhibition of RNA polymerase–mediated transcription and further processing of the spliceosome.36 Of note, the helicase senataxin (STX) prevents R-loop formation and promotes appropriate termination of transcription.37 Finally, ATM also regulates cell cycle and death pathways by phosphorylating the tumor suppressor transcription factor p53, inhibiting its ubiquitylation and proteasome degradation. Activated p53 binds DNA and activates expression of several genes, including that encoding p21, which binds to cyclin dependent kinase 2, thereby arresting the cell cycle at the G1/S transition. Activated p53 also promotes transcription of mediators of apoptosis (see below).
Figure 4. Multiple functions of ataxia telangiectasia mutated (ATM) protein in the DNA double-strand break response.
After binding to the site of DNA damage via interactions with poly (adenosine diphosphate [ADP] ribose) polymerase (PARP)1 and the MRN protein complex, consisting of meiotic recombination 11 (MRE11), DNA repair bridging protein 50 (RAD50), and Nijmegen breakage syndrome–1 (NBS-1), ATM phosphorylates and activates hundreds of substrate proteins, providing a mechanism for signal amplification. ATM stimulates DNA end-resection by phosphorylating C-terminal binding protein–interacting protein (CtIP), a major activator of MRE11, and breast cancer 1 (BRCA)1 protein. ATM also promotes nonhomologous end-joining by activating DNA protein kinase (DNA-PK) and facilitating the accumulation of the p53 binding protein 1 (53BP1). ATM also phosphorylates the histone variant H2AX, yielding γH2AX; this recruits the mediator of DNA damage checkpoint 1 protein (MDC1) and triggers phosphorylation-ubiquitylation (Ub) cascades that are sequentially mediated by the ring finger (RNF) 8 and RNF168 ubiquitin E3 ligases. Another target of ATM-mediated phosphorylation are the effector checkpoint kinases 2 (CHK2), which elicit cell cycle arrest; and p53, which also triggers apoptosis. ATM is recruited and activated at the site of R-loops and elicits transcriptional silencing.
Whereas ATM and DNA protein kinase are active in both actively replicating and postmitotic cells in response to double-strand breaks, ATR is essential in proliferating cells and is activated by a wide range of genotoxic stresses.28 A key function of ATR kinase is to activate the checkpoint kinase 1, which leads to arrest of cell cycle progression. A unique target activated by ATR is the Fanconi anemia pathway, which promotes repair of DNA interstrand crosslinks.38
Mechanisms of neurodegeneration in DNA repair disorders
Disruption of DNA repair mechanism may have distinct deleterious effects at different stages of neural development. Homologous recombination disruptions predominate at the early stage; nonhomologous end-joining predominates at middle stages; and single-strand break repair and transcription-coupled nucleotide-excision repair in response to oxidative stress predominate at late stages and postnatally. Thus, impairment of homologous recombination typically leads to embryonic lethality; defects of nonhomologous end-joining manifest with microcephaly; disorders affecting single-strand break repair are associated with neurodegeneration.14 Impaired DNA repair may produce programmed cell death via 2 main pathways. One is mediated by p53, which after stabilization and activation by ATM promotes transcription of genes encoding proapoptotic proteins that promote release of cytochrome c from mitochondria. The second results from excessive activation of PARP1, which leads to depletion of cellular nicotinamide adenine dinucleotide (NAD+) and adenosine triphosphate required for synthesis of poly(ADP) ribose. Depletion of NAD+ triggers mitochondrial release of apoptosis inducing factor; this form of PARP1-induced programmed cell is called parnathos.14
Clinical correlations
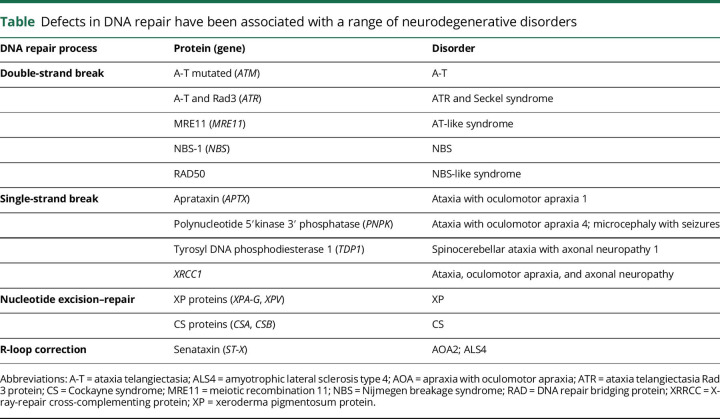
Defects in DNA repair have been associated with a wide range of neurodegenerative disorders, including several hereditary autosomal recessive cerebellar ataxias8 (table). These disorders are clinically and genetically very heterogeneous and are characterized by cerebellar ataxia, frequently associated with peripheral sensorimotor neuropathy. Ataxias associated with single-strand break repair appear to affect only the nervous system, although some may have some laboratory abnormalities. In contrast, defects in double-strand break repair, such as A-T, not only affect the nervous system but also manifest with systemic effects, such as immunodeficiency and malignancy due to genome instability.39 A full discussion of the clinical manifestations of these disorders is beyond the scope of the review and only single points are emphasized here.
Table.
Defects in DNA repair have been associated with a range of neurodegenerative disorders
A-T and other disorders of double-strand break repair
A-T is an autosomal recessive disorder caused by lack or inactivation of the ATM protein kinase. The neurologic and extraneurologic manifestations of this disorder reflect the critical function of ATM as a master regulator of cellular responses to double-strand breaks. As shown in the illustrative case introducing this review, A-T presents with early-onset, progressive cerebellar ataxia with oculomotor apraxia and conjunctival telangiectasias, immunodeficiency with frequent sinopulmonary infections, and susceptibility to ionizing radiation and cancer.17 Mutations affecting the components of the MRN complex also produce neurologic disorders associated with susceptibility to ionizing radiation. MRE11 mutations cause A-T-like disorder (ATLD), an autosomal recessive disorder characterized by progressive cerebellar ataxia and oculomotor apraxia; unlike patients with A-T, patients with ATLD do not have telangiectasias or immune deficiency and tend to have a slower disease course. Nijmegen breakage syndrome, due to mutation of the NBS1 gene,40 and Seckel syndrome linked to mutation of the ATR gene,41 manifest with severe growth retardation, microcephaly, and intellectual disability, sometimes associated with dysmorphic features, immunodeficiency, or radiosensitivity. Mutations of genes encoding proteins involved in DNA repair through nonhomologous end-joining also produce severe immunodeficiency; these include LIG4 mutations impairing function of ligase 4 and XRCC4 mutations affecting XRCC4 encoding a ligase 4 partner.42
AOA and related disorders
AOA is a subgroup of autosomal recessive disorders typically associated with impaired single-strand DNA break repair. Like A-T and ATLD, they are characterized by cerebellar ataxia, oculomotor apraxia, chorea, myoclonus, and severe sensorimotor axonal neuropathy. However, chromosomal instability, immunodeficiency, and sensitivity to ionizing radiation do not occur in AOA. Several genes linked to AOA encode proteins that are involved in DNA end-processing, particularly in response to oxidative stress.43 AOA1 is due to mutations of the APTX gene encoding aprataxin; hypoalbuminemia and hypercholesterolemia may be present in this disorder. AOA4 to the mutation of the PNPK gene encoding PNPK44,45; these mutations may also cause a severe neurodevelopmental disorder characterized by microcephaly, early-onset intractable seizures, and developmental delay.45 More recently, mutations of the XRCC1 gene encoding XRCC1, a molecular scaffold involved in DNA single-strand break repair, were associated with a phenotype of cerebellar ataxia, oculomotor apraxia, and axonal neuropathy resembling AOA4 linked to PNPK mutations.46 As discussed above, PNPK is one of the molecular partners of XRCC1 involved in single-strand break repair; studies in knockout mice indicate that PARP1 hyperactivation is the cause of ataxia in these disorders.46 Spinocerebellar ataxia with axonal neuropathy (SCAN1) is linked to mutations in the TDP1 gene encoding TDP1, which like APTX and PNPK is involved in DNA end-processing. SCAN1 is also characterized by late-childhood-onset slowly progressive cerebellar ataxia, followed by progressive peripheral neuropathy.47 Like in AOA1, hypoalbuminemia and hypercholesterolemia may occur in SCAN1, but oculomotor apraxia is not a typical feature.
Other forms of AOA reflect defects in control of regulation of DNA transcription. The typical example is AOA2, which is linked to mutations of the STX gene encoding senataxin, a DNA helicase that is critical to correct formation of R-loops comprising RNA/DNA hybrids and a displaced single-stranded DNA.37,48 Senataxin mutations have also been linked to amyotrophic lateral sclerosis type 4, emphasizing the importance of this helicase II R-loop-associated neurodegenerative disease.37 A phenotype resembling AOA2, and referred to as AOA3, has been linked to mutations in the PI3KR5 gene encoding phosphatidylinositol 3′kinase regulatory subunit 5, which has a critical role in cerebellar development.49 However, the term AOA3 has also been used for a form of spinocerebellar ataxia associated with deficient DNA damage–induced activation of p53 in response to ATM.50
XP and CS
XP and CS are autosomal recessive disorders due to impaired transcription-coupled nucleotide excision–repair, which is a major mechanism of repair of DNA damage due to UV radiation.51 XP is associated with mutations of genes encoding XPA-G, XPB, XPC, XPD, XPE, XPF, XPG, and XPV, and is characterized by slow-healing sunburns after brief sun exposure and high risk of skin cancer. Approximately one-fourth of all patients with XP have neurologic symptoms, including microcephaly, intellectual disability, hearing loss, ataxia, and neuropathy. CS, due to mutations of the CSA or CSB genes, is a progeria syndrome characterized by photosensitivity, growth failure, intellectual disability, spasticity, ataxia, hearing and vision loss, joint contractures and early mortality, but not cancer. Some patients have a combined XP-CS phenotype.
DNA repair and age-related neurodegenerative disorders
Several studies show age-related accumulation of oxidative DNA damage in the brain. Some areas, such as the substantia nigra pars compacta, are more vulnerable than others to oxidative DNA modification. Aging is also associated with a reduced capacity of DNA repair, including base excision–repair in the nucleus and mitochondria.11 The accumulation of DNA damage depends on the degree of age-dependent cell loss; for example, it is prominent in hippocampal pyramidal and cerebellar granule cells but not in Purkinje cells.18 There is evidence of oxidative damage in nuclear and mitochondrial DNA in several neurodegenerative disorders, including Alzheimer disease,52 Parkinson disease, Huntington disease, and amyotrophic lateral sclerosis, as reflected by accumulation of oxidized bases and PARP1. Defective DNA repair mechanisms may contribute to these findings.11
Studies in mouse models and cell lines show that mismatch repair, transcription-coupled nucleotide excision–repair, and base excision–repair can contribute to repeat instability and somatic expansion of trinucleotide repeats in trinucleotide repeat disorders.16 Repeat expansions trigger transient formation of unusual DNA structures, such as hairpin, loops, and triplet helices that initiate DNA repair mechanisms. Mismatch repair mediators may be activated after recognition of mismatched bases within the expanded triplet repeat and modulate expansion of repeat tracts.16,24,53 Mismatch repair mediators may promote expansion of CAG repeats, like in the case of the huntingtin (HTT) gene. In some cases, these mechanisms may affect the molecular phenotype of disorders such as Friedreich ataxia.54 Variants of genes involved in DNA repair pathways influence the degree of CAG expansion instability and thus age at disease onset in multiple CAG (polyglutamine) repeat diseases.55 Reciprocally, mutant proteins can affect DNA repair mechanisms. For example, mutant HTT accumulates at the site of DNA breaks and binds Ku70, inhibiting DNA repair via nonhomologous end-joining,56 and reduces recruitment of BRCA1 to the nucleus, thereby affecting homologous recombination.57
Perspective
Defects in DNA repair constitute an important disease pathway for neurologic disorders. The increased recognition of specific defects affecting key components of the response to DNA damage provides potential targets for treatment. However, excessive activation of DNA repair pathways may also have a deleterious effect. Therefore, whereas small molecular inhibitors of ATM and other phosphoinositide 3′ kinase–related kinases involved in DNA damage response are a potential approach for anticancer therapy, preventing excessive activation of PARP1, ATM, and other signals triggered by DNA damage may also have potential neuroprotective effects. For example, ATM inhibition had neuroprotective effect in mouse models of Huntington disease58; inhibition of PARP-1 may also be a potential target in Alzheimer disease or Parkinson disease.59
Glossary
- A-T
ataxia telangiectasia
- ADP
adenosine diphosphate
- AOA
apraxia with oculomotor apraxia
- APTX
aprataxin
- ATLD
ataxia-telangiectasia-like disorder
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia Rad 3 protein
- BRCA1
breast cancer 1
- CS
Cockayne syndrome
- MRE11
meiotic recombination 11
- NAD+
nicotinamide adenine dinucleotide
- NBS-1
Nijmegen breakage syndrome–1
- PARP1
polyadenosyl diphosphate (ADP) ribose polymerase 1
- PNKP
polynucleotide 3′kinase phosphatase
- RAD50
DNA repair bridging protein 50
- SCAN1
spinocerebellar ataxia with axonal neuropathy
- TDP1
tyrosyl–DNA phosphodiesterase 1
- XP
xeroderma pigmentosum
- XRRCC
X-ray-repair cross-complementing
Author contributions
E.A. Coon: analysis of data, drafting the manuscript, and revising the manuscript for intellectual content. E.E. Benarroch: design and conceptualization, analysis of data, drafting the manuscript and revising the manuscript for intellectual content.
Study funding
No targeted funding reported.
Disclosure
E.A. Coon reports no disclosures. E.E. Benarroch receives a stipend in his capacity as section editor of Clinical Implications of Neuroscience Research for Neurology®. Go to Neurology.org/N for full disclosures.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010;40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirbu BM, Cortez D. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 2013;5:a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldecott KW. DNA single-strand break repair. Exp Cell Res 2014;329:2–8. [DOI] [PubMed] [Google Scholar]
- 5.Iyama T, Wilson DM III. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair 2013;12:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 2011;25:409–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombardi PM, Matunis MJ, Wolberger C. RAP80, ubiquitin and SUMO in the DNA damage response. J Mol Med 2017;95:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulson HL, Miller VM. Breaks in coordination: DNA repair in inherited ataxia. Neuron 2005;46:845–848. [DOI] [PubMed] [Google Scholar]
- 9.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci 2009;10:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell 2007;130:991–1004. [DOI] [PubMed] [Google Scholar]
- 11.Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience 2007;145:1318–1329. [DOI] [PubMed] [Google Scholar]
- 12.Kulkarni A, Wilson DM III. The involvement of DNA-damage and -repair defects in neurological dysfunction. Am J Hum Genet 2008;82:539–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet 2008;9:619–631. [DOI] [PubMed] [Google Scholar]
- 14.Rulten SL, Caldecott KW. DNA strand break repair and neurodegeneration. DNA Repair 2013;12:558–567. [DOI] [PubMed] [Google Scholar]
- 15.Jiang B, Glover JN, Weinfeld M. Neurological disorders associated with DNA strand-break processing enzymes. Mech Ageing Dev 2017;161:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones L, Houlden H, Tabrizi SJ. DNA repair in the trinucleotide repeat disorders. Lancet Neurol 2017;16:88–96. [DOI] [PubMed] [Google Scholar]
- 17.Rothblum-Oviatt C, Wright J, Lefton-Greif MA, McGrath-Morrow SA, Crawford TO, Lederman HM. Ataxia telangiectasia: a review. Orphanet J Rare Dis 2016;11:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rutten BP, Schmitz C, Gerlach OH, et al. The aging brain: accumulation of DNA damage or neuron loss? Neurobiol Aging 2007;28:91–98. [DOI] [PubMed] [Google Scholar]
- 19.Stern N, Hochman A, Zemach N, et al. Accumulation of DNA damage and reduced levels of nicotine adenine dinucleotide in the brains of Atm-deficient mice. J Biol Chem 2002;277:602–608. [DOI] [PubMed] [Google Scholar]
- 20.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017;18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson DM III, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair 2007;6:544–559. [DOI] [PubMed] [Google Scholar]
- 22.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 2006;7:335–346. [DOI] [PubMed] [Google Scholar]
- 23.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem 2006;281:30305–30309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer RR, Pluciennik A, Napierala M, Wells RD. DNA triplet repeat expansion and mismatch repair. Annu Rev Biochem 2015;84:199–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 2013;5:a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullenders L. DNA damage mediated transcription arrest: step back to go forward. DNA Repair 2015;36:28–35. [DOI] [PubMed] [Google Scholar]
- 27.Shimada M, Dumitrache LC, Russell HR, McKinnon PJ. Polynucleotide kinase-phosphatase enables neurogenesis via multiple DNA repair pathways to maintain genome stability. EMBO J 2015;34:2465–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 2017;66:801–817. [DOI] [PubMed] [Google Scholar]
- 29.Seol JH, Shim EY, Lee SE. Microhomology-mediated end joining: good, bad and ugly. Mutat Res Epub 2017 Jul 16. [DOI] [PMC free article] [PubMed]
- 30.Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol 2015;7:a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 2007;85:509–520. [DOI] [PubMed] [Google Scholar]
- 32.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007;316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Shi LZ, Wong CC, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet 2013;9:e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol 2016;19:1–9. [DOI] [PubMed] [Google Scholar]
- 35.Thorslund T, Ripplinger A, Hoffmann S, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 2015;527:389–393. [DOI] [PubMed] [Google Scholar]
- 36.Tresini M, Warmerdam DO, Kolovos P, et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature 2015;523:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groh M, Albulescu LO, Cristini A, Gromak N. Senataxin: genome guardian at the interface of transcription and neurodegeneration. J Mol Biol 2016;429:3181–3195. [DOI] [PubMed] [Google Scholar]
- 38.Ceccaldi R, Sarangi P, D'Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol 2016;17:337–349. [DOI] [PubMed] [Google Scholar]
- 39.Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair 2004;3:1219–1225. [DOI] [PubMed] [Google Scholar]
- 40.Antoccia A, Kobayashi J, Tauchi H, Matsuura S, Komatsu K. Nijmegen breakage syndrome and functions of the responsible protein, NBS1. Genome Dyn 2006;1:191–205. [DOI] [PubMed] [Google Scholar]
- 41.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 2003;33:497–501. [DOI] [PubMed] [Google Scholar]
- 42.Murray JE, van der Burg M, IJspeert H, et al. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am J Hum Genet 2015;96:412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Ber I, Rivaud-Pechoux S, Brice A, Durr A. Autosomal recessive cerebellar ataxias with oculomotor apraxia [in French]. Rev Neurol 2006;162:177–184. [DOI] [PubMed] [Google Scholar]
- 44.Bras J, Alonso I, Barbot C, et al. Mutations in PNKP cause recessive ataxia with oculomotor apraxia type 4. Am J Hum Genet 2015;96:474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paucar M, Malmgren H, Taylor M, et al. Expanding the ataxia with oculomotor apraxia type 4 phenotype. Neurol Genet 2016;2:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoch NC, Hanzlikova H, Rulten SL, et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 2017;541:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fam HK, Salih MAM, Takashima H, Boerkoel CF. Spinocerebellar ataxia with axonal neuropathy, autosomal recessive. In: Pagon RA, Adam MP, Ardinger HH, et al., eds. GeneReviews. Seattle: University of Washington; 1993. [Google Scholar]
- 48.Suraweera A, Lim Y, Woods R, et al. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum Mol Genet 2009;18:3384–3396. [DOI] [PubMed] [Google Scholar]
- 49.Al Tassan N, Khalil D, Shinwari J, et al. A missense mutation in PIK3R5 gene in a family with ataxia and oculomotor apraxia. Hum Mutat 2012;33:351–354. [DOI] [PubMed] [Google Scholar]
- 50.Gueven N, Chen P, Nakamura J, et al. A subgroup of spinocerebellar ataxias defective in DNA damage responses. Neuroscience 2007;145:1418–1425. [DOI] [PubMed] [Google Scholar]
- 51.Natale V, Raquer H. Xeroderma pigmentosum-Cockayne syndrome complex. Orphanet J Rare Dis 2017;12:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabbita SP, Lovell MA, Markesbery WR. Increased nuclear DNA oxidation in the brain in Alzheimer's disease. J Neurochem 1998;71:2034–2040. [DOI] [PubMed] [Google Scholar]
- 53.Lokanga RA, Zhao XN, Usdin K. The mismatch repair protein MSH2 is rate limiting for repeat expansion in a fragile X premutation mouse model. Hum Mutat 2014;35:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ezzatizadeh V, Sandi C, Sandi M, Anjomani-Virmouni S, Al-Mahdawi S, Pook MA. MutLalpha heterodimers modify the molecular phenotype of Friedreich ataxia. PLoS One 2014;9:e100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bettencourt C, Hensman-Moss D, Flower M, et al. DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann Neurol 2016;79:983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enokido Y, Tamura T, Ito H, et al. Mutant huntingtin impairs Ku70-mediated DNA repair. J Cell Biol 2010;189:425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeon GS, Kim KY, Hwang YJ, et al. Deregulation of BRCA1 leads to impaired spatiotemporal dynamics of gamma-H2AX and DNA damage responses in Huntington's disease. Mol Neurobiol 2012;45:550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu XH, Mattis VB, Wang N, et al. Targeting ATM ameliorates mutant Huntingtin toxicity in cell and animal models of Huntington's disease. Sci Transl Med 2014;6:268ra178. [DOI] [PubMed] [Google Scholar]
- 59.Martire S, Mosca L, d'Erme M. PARP-1 involvement in neurodegeneration: a focus on Alzheimer's and Parkinson's diseases. Mech Ageing Dev 2015;146–148:53–64. [DOI] [PubMed] [Google Scholar]