Abstract
A new dimeric alkaloid plakoramine A [(±)-1] was identified from a marine sponge Plakortis sp. Chiral-phase HPLC separation of (±)-1 led to the purified enantiomers (+)-1 and (−)-1 which both potently inhibited CBL-B E3 ubiquitin ligase activities. The absolute configurations of the enantiomers were determined by quantum chemical calculations. Scrutinization of the purification conditions revealed a previously undescribed, nonenzymatic route to form (±)-1 via photochemical conversion of its naturally occurring monomeric counterpart, plakinidine B (2).
Graphical Abstract
Dimerization is an energetically economical biosynthetic strategy that nature commonly adopts to generate complex natural product architectures.1,2 In 2004, a survey on ca. 3000 articles estimated that about 15–20% of natural products were derivatized by a dimerization process.3 The past decade has seen a significant increase in the study of dimeric natural products mainly arising from their potential therapeutical utility. A quick search of “dimeric natural products” as a keyword in SciFinder returned 849 publications from 2010 to date comparing to the total of only 561 publications in all years before 2010 (Figure S1). Many dimeric natural products deliver a broader spectrum of biological activities than their corresponding monomeric counterparts especially when target proteins rely on the formation of dimeric species for activity.4–6 The ability to simultaneously bind two separate monomers of a dimeric receptor could potentially increase potency and efficacy or lead to the activation of cell-signaling pathways. Thus, dimerization has been increasingly adopted as a useful medicinal chemistry strategy to enhance the therapeutical potential of natural or synthetic molecules.4–7
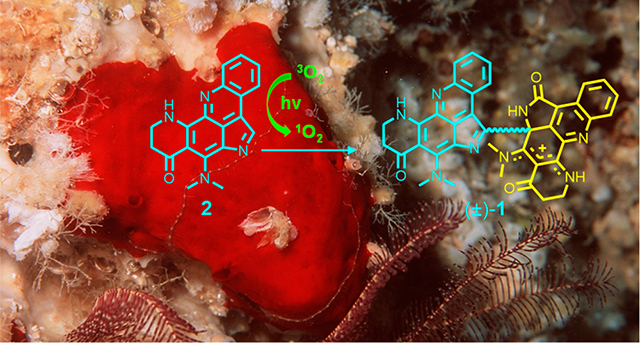
The E3 ubiquitin–protein ligase, Casitas B-lineage lymphoma proto-oncogene-b (CBL-B), plays a crucial role in the regulation of both innate and adaptive immunity.8,9 CBL-B suppresses the antitumor activities of both T cells and NK cells by negatively regulating their cellular signaling pathways.10,11 Genetic ablation of CBL-B in mice demonstrated that the deprivation of CBL-B catalytic activity provided protection against both transplanted and spontaneous tumors.12 Thus, CBL-B represents an attractive target for immunotherapeutic intervention in cancer. A previously established high-throughput assay was used to screen a library of >175,000 natural product fractions for inhibitors of CBL-B catalytic activity.13,14 Bioassay-guided subfractionation of the active fractions of the marine sponge Plakortis sp. revealed a new dimeric alkaloid plakoramine A (1) (Figure 1) that potently inhibited the CBL-B catalytic activity. Plakoramine A (1) represents the first reported dimer of the plakinidine class of alkaloids.15–18 Further mechanistic studies revealed an efficient self-sensitized photodimerization process for the monomeric counterpart plakinidine B (2) (Figure 1). The structure elucidation, CBL-B inhibitory activity, and photochemical synthesis of plakoramine A (1) are described in this report.
Figure 1.
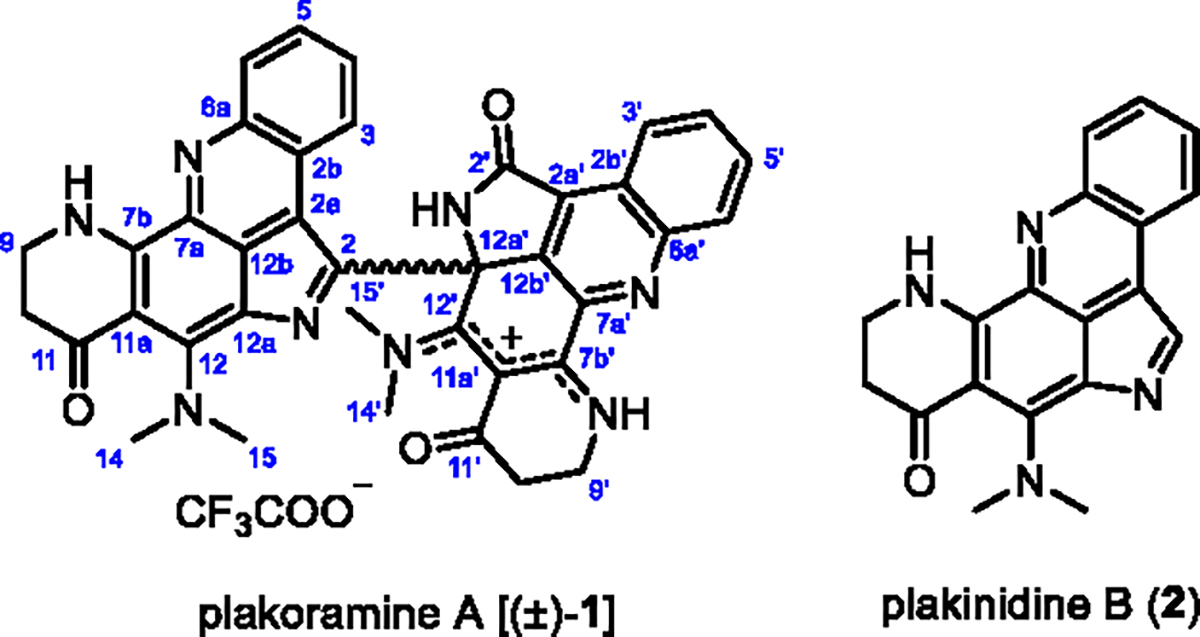
Structures of compounds (±)-1 and 2.
Plakoramine A (1) was isolated as a red solid from the organic extract of a Plakortis sponge which was collected in Tonga. The molecular formula of 1 was determined as C38H31N8O3 given the positively charged ion at m/z 647.2510 (calcd for C38H31N8O3+, m/z 647.2514) detected in its HRESIMS spectrum. The 1H NMR spectroscopic data (Table S1) displayed three NH protons (δH 10.62, 10.37, and 10.29), the coupled spin systems of two ortho-disubstituted benzenes (δH 9.05/8.14/8.20/8.43 and 8.15/7.41/6.74/6.30), four N-methyls (δH 3.95, 3.89, 3.39, and 3.35), and four methylenes (δH 3.74/3.64, 2.52/2.39, 3.75, and 2.56). The 13C NMR data (Table S1) exhibited 31 out of 38 carbons in the downfield region (δC 102–188), indicating a high degree of aromaticity. In comparison with plakinidine B (2), the major metabolite of the producing Plakortis sponge, half of the 1H and 13C NMR resonances (Table S1) of 1 were nearly identical to the 1D NMR data of 217 except for the methine at position 2 (δH 8.76, δC 126.2) which was replaced by a quaternary carbon in 1 (δC 139.5). Further analysis of the 2D NMR [1H–1H COSY, HMBC (pulse sequences optimized for nJCH 8 and 2 Hz), and ROESY] data (Figure 2, Table S1) confirmed that the substructure A of 1 was identical to the structure of 2 while the substituent on C-2 (substructure B) shared a similar conjugated ring system to that of substructure A. Significant changes in substructure B were observed for C-2′ (δC 168.6) and C-12a′ (δC 70.3) which were substituted for an amide carbonyl and a sp3 quaternary carbon, respectively, from two sp2 quaternary carbons in substructure A. Moreover, the sp2 quaternary C-12′ resonance shifted downfield significantly from δC 153.6 to δC 174.8 indicating the presence of an exocyclic C═N double bond. The aforementioned structural modifications in substructure B were supported by the HMBC (pulse sequences optimized for nJCH 8 Hz) correlations from NH-1′ (δH 10.37) to C-2′, C-2a′, C-12a′, and C-12b′ and from CH3-14′ (δH 3.39) to C-12′. The 13′-12′-11a′-7b′-8′ ensemble can be viewed as an N-8′ protonated (δH 10.62) resonance structure with a cation delocalizing between N-13′ and N-8′. The structural assignment was further secured by the long-range HMBC (pulse sequences optimized for nJCH 2 Hz) correlations from NH-1′ to C-2b′, C-7a′, and CH3-15′, from CH3-15′ (δH 3.95) to C-12a′ and C-12b′, and from CH3-14′ to C-11′ and C-11a′. Finally, the structural elucidation of 1 was completed by connecting substructures A and B through a single bond between C-2 and C-12a′, which was supported by the long-range HMBC correlations from NH-1′ and CH3-15′ to C-2.
Figure 2.
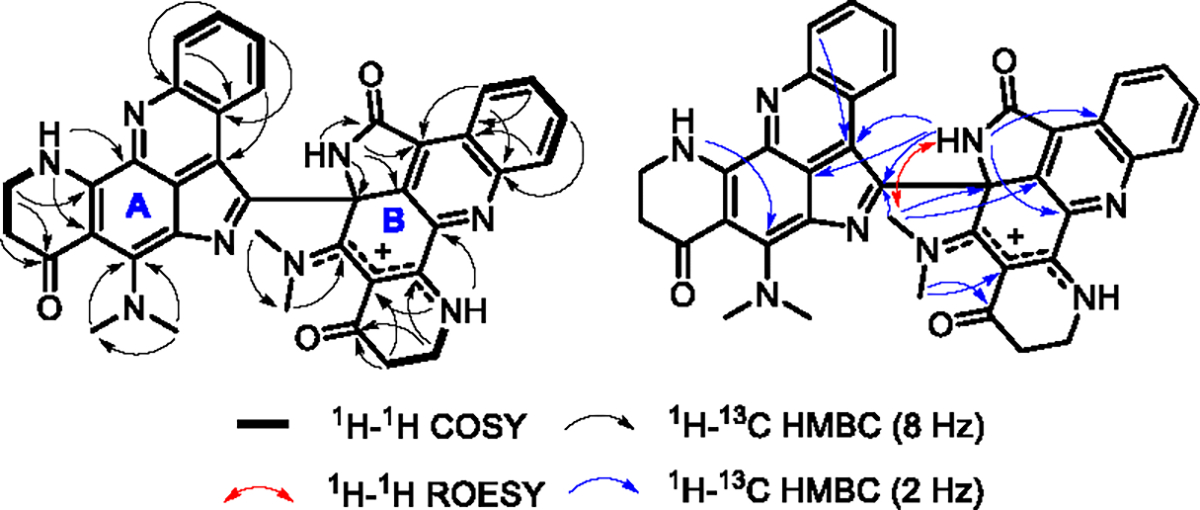
Selected 2D NMR correlations of compound (±)-1.
To determine the absolute configuration of C-12′, the chiroptical properties of 1 were measured with optical polarimetry and electronic circular dichroism (ECD) spectroscopy. Compound 1 showed no optical rotation ([α]D = 0) or Cotton effect in its ECD spectrum (Figure S2) indicating it as a racemic mixture [(±)-1]. Chiral-phase HPLC separation of (±)-1 (Figure S3) on a Lux 5 μm i-Amylose-3 column led to the optically pure enantiomers (+)-1 ([α]20D 340) and (−)-1 ([α]20D –335). The absolute configurations of the purified enantiomers were determined by comparison of their experimental ECD spectra with the computationally calculated spectrum of the 12a′S isomer (Figure 3). A conformational search was carried out using the GMMX methodology with an energy cutoff of 3 kcal/mol. Lowest energy conformers were optimized with Gaussian ‘16 using the B3LYP/DGDZVP method with the COSMO solvation model. Following TDDFT calculation at the same levels on six optimized conformers led to the simulated ECD spectrum of the 12a′S isomer. The ECD spectrum of (+)-1 displayed a strong negative Cotton effect at ca. 263 nm and an intense positive Cotton effect at ca. 352 nm which were consistent with the calculated ECD data for the 12a′S isomer (Figure 3). The ECD spectrum of (−)-1 displayed Cotton effects of equal magnitude but opposite sign to those of the 12a′S isomer. Thus, the absolute configurations of (+)-1 and (−)-1 were determined as 12a′S and 12a′R, respectively (Figure 3).
Figure 3.
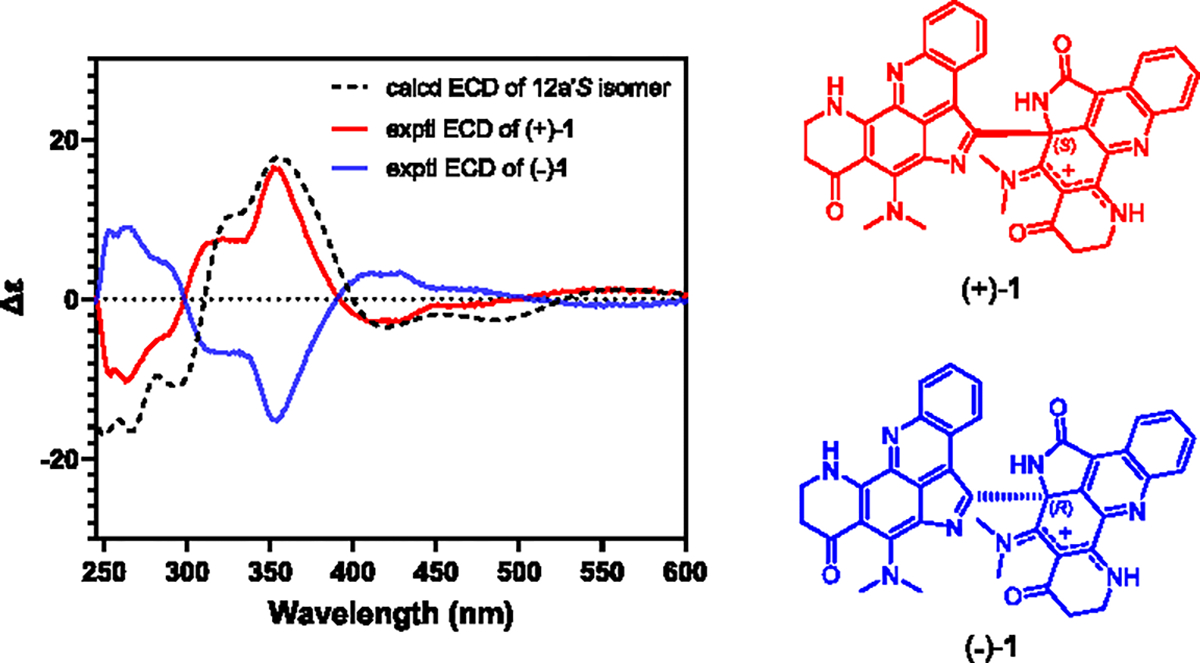
Comparison of the experimental ECD spectra of (+)-1 and (−)-1 with the calculated ECD spectrum of the 12a′S isomer of (±)-1.
The activities of (+)-1 and (−)-1 against the E3 ubiquitin-protein ligase CBL-B were evaluated in a biochemical assay as previously described.13 Both (+)-1 and (−)-1 inhibited CBL-B activity with IC50 values of 7.5 and 9.7 μM, respectively (Figure 4). In contrast, the monomeric precursor 2 showed ~20 fold less potency (IC50 167 μM) against CBL-B than its dimeric counterparts. It is also noteworthy that the monomer 2 showed potent antiproliferative activities (average GI50 0.18 μM) (Figures S5 and S6) in the NCI-60 cell lines screen,19 while neither (+)-1 or (−)-1 affected cell growth or viabilities even at high micromolar concentrations (average GI50s > 30 μM, Figures S7 and S8). It is known that CBL-B dimerization is required for its ubiquitin ligase activities.20 The activation of CBL-B relies on the two critical interaction surfaces of the dimerized CBL-B ubiquitin-associated (UBA) domain which is activated by ubiquitin binding.20 Thus, it will be interesting to further study the mechanism of plakoramine A [(±)-1] by investigating the importance of its dimeric architecture for its interaction with CBL-B and regulation of CBL-B activities.
Figure 4.
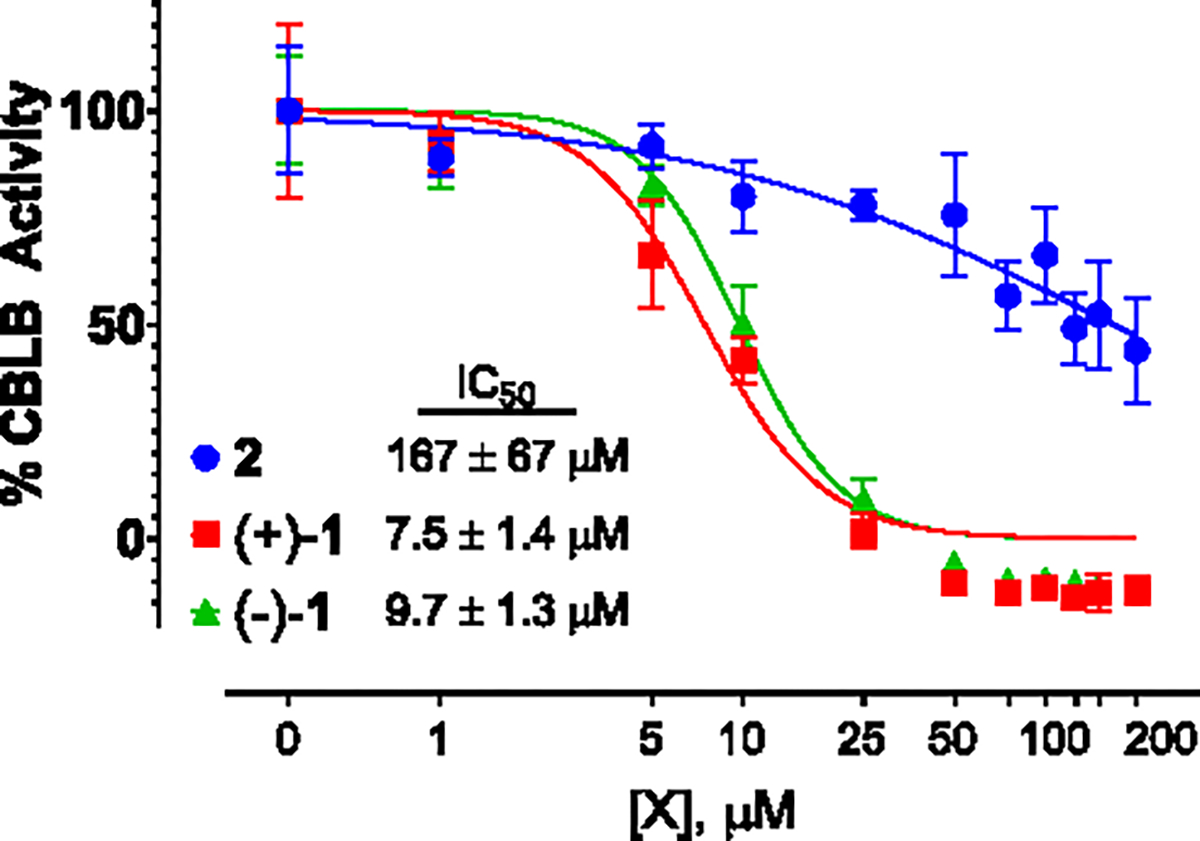
Dose–response curves of 2, (+)-1, and (−)-1 in the CBL-B biochemical assay.
In light of the racemic nature of plakoramine A [(±)-1], it is reasonable to hypothesize that the dimeric structure of (±)-1 was derived from a nonenzymatic process. It was found that a trace amount of (±)-1 was constantly detected in preparative HPLC fractions that contained mainly 2, which provided the starting conditions for testing our hypothesis. First, a pure sample of 2 was stirred in a common HPLC solvent system [MeCN/H2O (1:1)] at room temperature in the dark overnight, but the starting material was unchanged (Figure S9B). Further attempts to scrutinize the reaction conditions revealed that (±)-1 was readily formed through the spontaneous dimerization of 2 in MeCN/H2O (1:1) under both UV (Figure S9C) and white light (Figure 5A) irradiations. In contrast, the reaction of 2 in anhydrous MeCN did not provide readily detectable (±)-1 with either UV or white light irradiations (Figure S9D) suggesting both light and solvent have a dramatic effect on conversion efficiency. Manipulation of the irradiation parameters (i.e., light source, irradiation distance, and exposure period) afforded the optimized yield of (±)-1 at ~30% with white light irradiation of 2 in MeCN/H2O (1:1) for 4 h using a 15 W LED (Figure 5A). Interestingly, reaction of 2 in N2-sparged solvents led to a significant decrease of the yield of (±)-1 from ~30% to only ~9% (Figure 5A). In total, the data suggested that the dimerization of 2 was likely driven by a photooxidative process that was catalyzed by certain reactive oxygen species (ROS) generated through the photoactivation of molecular oxygen in the aqueous environment.
Figure 5.
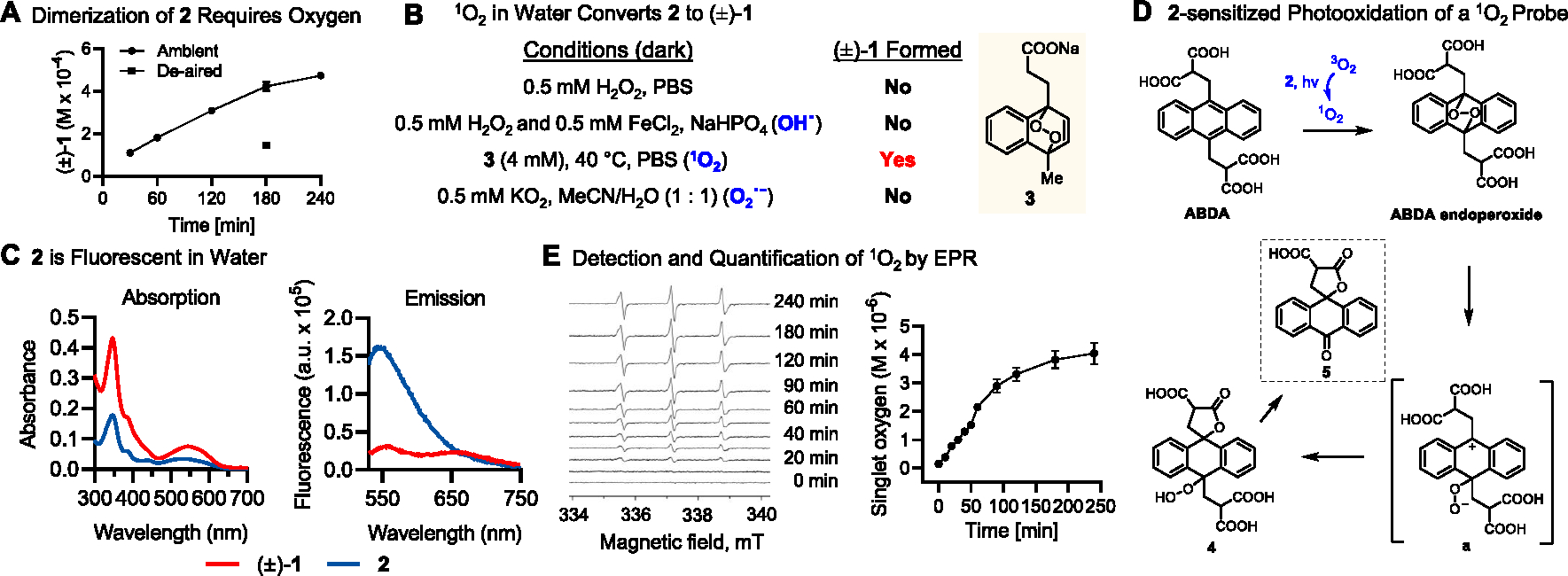
Mechanistic studies of the photochemical dimerization of 2. (A) Time-dependent production of (±)-1 was monitored by HPLC-PDA following irradiation of 2 (1 mg/mL) in 1:1 MeCN/H2O with white light (15 W, LED) for 4 h under ambient and deaired conditions. (B) Evaluating the dimerization of 2 to (±)-1 with independently generated ROS under dark conditions. (C) UV–vis absorption and fluorescence emission (520 nm excitation) spectra of 2 and (±)-1. (D) Plakinidine B (2)-sensitized photooxidation of the 1O2 probe ABDA. (E) Detection and quantification of 1O2 by electron paramagnetic resonance (EPR) using spin traps and the 4-Oxo-TEMP redox probe. Time-dependent production of 1O2 was monitored following irradiation of 2 (1 mg/mL) in 1:1 MeCN/H2O with white light for 4 h under ambient conditions. All quantitative values were obtained from three independent experiments and graphed as mean ± SEM.
To address the responsible ROS, the reactivity of 2 was tested under dark conditions with hydrogen peroxide (H2O2) and chemically generated singlet oxygen (1O2), hydroxyl radical (OH•), and superoxide anion (O2•−). The results clearly showed that (±)-1 was efficiently and exclusively formed in the presence of thermally generated singlet oxygen (1O2) (Figures 5B and S10). The highly reactive singlet oxygen (1O2) is normally produced by irradiation of molecule oxygen (3O2) in the presence of a photosensitizer, such as rose bengal, methylene blue, or porphyrins.21 The Abs/Em spectra (Figure 5C) indicated 2 was moderately fluorescent and probably functioned as a photosensitizer. The hypothesis was tested by irradiating 2 in MeCN/H2O (1:1) with the presence of the singlet oxygen probe 9,10-anthracenediyl-bis(methylene)-dimalonic acid (ABDA) which can be specifically oxidized to its endoperoxide analog by singlet oxygen (1O2).22 After 2 h irradiation with white light, ABDA was completely consumed and partially converted to two γ-lactone derivatives 4 and 5 (Figure 5D and S11). It is reasonable to hypothesize that 4 and 5 were formed through a similar anthracene endoperoxide degradation pathway as previously described23 with the participation of a key zwitterion intermediate a (Figure S12). Finally, the 2-sensitized photosynthesis of singlet oxygen (1O2) was confirmed by indirect detection and quantification of 1O2 by electron paramagnetic resonance (EPR) using a 1O2-specific spin trap 2,2,6,6-tetramethyl-4-piperidone (4-Oxo-TEMP).24 Singlet oxygen (1O2) was steadily generated in a time-dependent manner with continuous white light irradiation. The production of 1O2 reached its near-to-maximal yield after around 4 h (Figure 5E) showing good agreement with the kinetics of (±)-1 formation (Figure 5A).
Based on the aforementioned evidence, a plausible mechanism was proposed for the photochemical dimerization of 2 (Scheme 1). First, aqueous conditions may facilitate formation of the zwitterionic resonance form of 2. Upon light irradiation, the triplet excited state of 2 sensitizes molecular oxygen (3O2) to form the reactive singlet oxygen (1O2) which directly attacks the pyrrole moiety of the zwitterion form via [4 + 2] cycloaddition to yield the endoperoxide intermediate a.25–27 Well-precedented ring opening of the endoperoxide reveals an electrophilic iminium ion b, which is then subject to nucleophilic attack by C-2 of another molecule of 2 to form the C-12a′/C-2 bridge. Final hydroperoxide decomposition forms the γ-lactam product (±)-1.
Scheme 1.
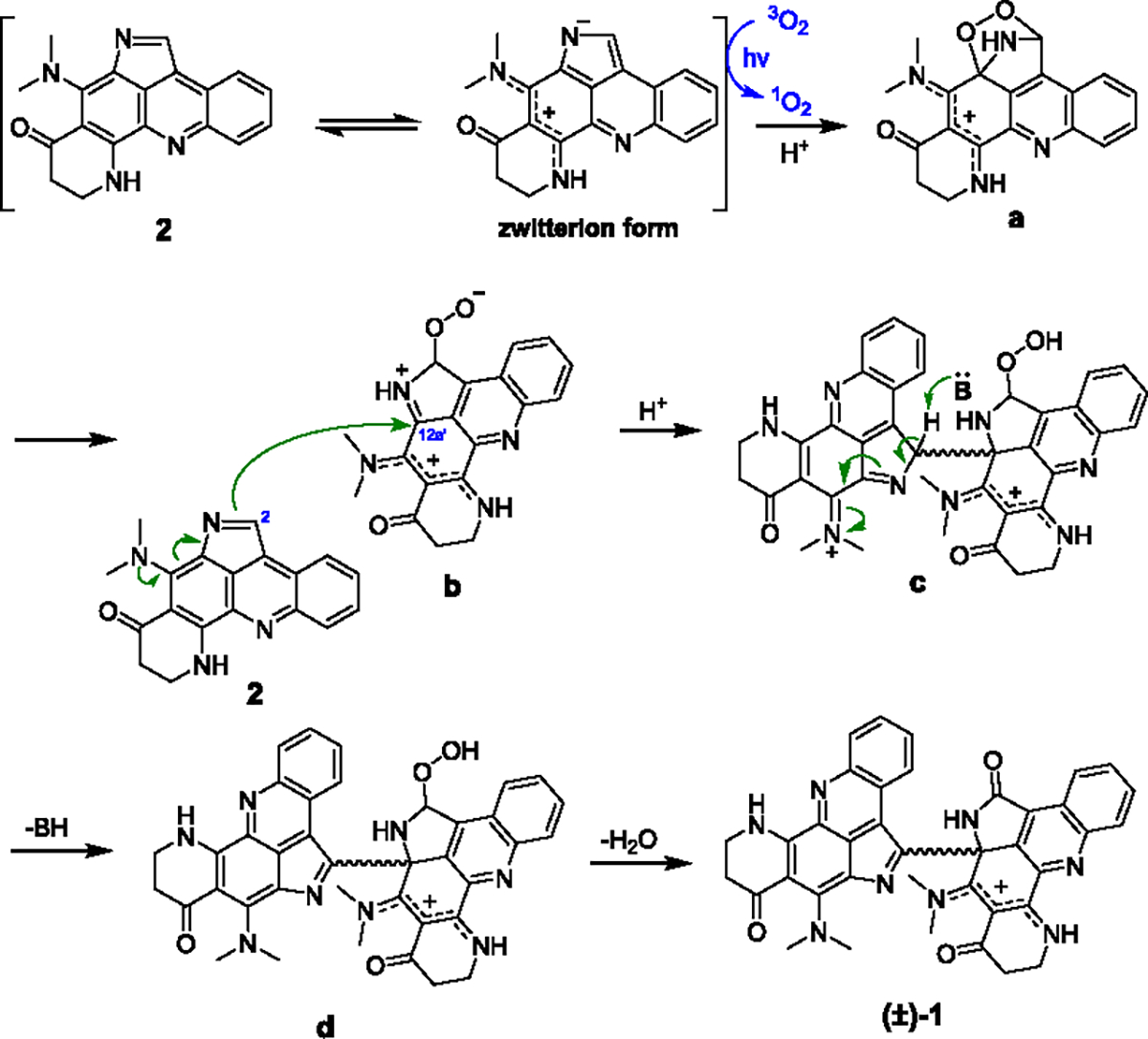
Proposed Mechanism of the Photochemical Dimerization of 2
Plakoramine A [(±)-1] represents the first example of a novel class of heterodimeric alkaloids that provides a promising pharmacophore for chemical intervention of the anticancer target E3 ubiquitin-protein ligase CBL-B. The naturally occurring, monomeric counterpart plakinidine B (2) also stands for a new functional fluorophore with potential utility as both a photosensitizer and a photochemically triggered electrophilic agent.
Supplementary Material
Acknowledgments
We thank the Natural Products Support Group (NCI at Frederick) for the extract preparation and Emily Smith (Basic Science Program, Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research) for CBL-B assay support. We also thank Lauren Procter, Morgan Pagonis, and Jane Jones of the FNLCR Protein Expression Laboratory for their recombinant protein production efforts.
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.orglett.2c03922
Contributor Information
Quan T. Khong, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States
Donghao Li, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 20850, United States.
Brice A. P. Wilson, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States
Kalina Ranguelova, Bruker BioSpin Corp, Billerica, Massachusetts 01821, United States.
Masoumeh Dalilian, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States; Basic Science Program, Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research, Frederick, Maryland 21702-1201, United States.
Emily A. Smith, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States Basic Science Program, Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research, Frederick, Maryland 21702-1201, United States.
Antony Wamiru, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States; Basic Science Program, Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research, Frederick, Maryland 21702-1201, United States.
Ekaterina I. Goncharova, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States Advanced Biomedical Computational Science, Frederick National Laboratory for Cancer Research, Frederick, Maryland 21702-1201, United States.
Tanja Grkovic, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States; Natural Products Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Frederick, Maryland 21701-1201, United States.
Donna Voeller, Women’s Malignancies Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892-1578, United States.
Stanley Lipkowitz, Women’s Malignancies Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892-1578, United States.
Martin J. Schnermann, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 20850, United States
Barry R. O’Keefe, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States Natural Products Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Frederick, Maryland 21701-1201, United States.
Lin Du, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States.
REFERENCES
- (1).Snyder SA Organic chemistry: symmetrizing the unsymmetrical. Nature 2010, 465, 560–561. [DOI] [PubMed] [Google Scholar]
- (2).Liu J; Liu A; Hu Y Enzymatic dimerization in the biosynthetic pathway of microbial natural products. Nat. Prod. Rep. 2021, 38, 1469–1505. [DOI] [PubMed] [Google Scholar]
- (3).Voloshchuk T; Farina NS; Wauchope OR; Kiprowska M; Haberfield P; Greer A Molecular bilateral symmetry of natural products: prediction of selectivity of dimeric molecules by density functional theory and semiempirical calculations. J. Nat. Prod. 2004, 67, 1141–1146. [DOI] [PubMed] [Google Scholar]
- (4).Berube G Natural and synthetic biologically active dimeric molecules: anticancer agents, anti-HIV agents, steroid derivatives and opioid antagonists. Curr. Med. Chem. 2006, 13, 131–154. [DOI] [PubMed] [Google Scholar]
- (5).Paquin A; Reyes-Moreno C; Berube G Recent advances in the use of the dimerization strategy as a means to increase the biological potential of natural or synthetic molecules. Molecules 2021, 26, 2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hadden MK; Blagg BS Dimeric approaches to anti-cancer chemotherapeutics. Anticancer Agents Med. Chem. 2008, 8, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sun J; Yang H; Tang W Recent advances in total syntheses of complex dimeric natural products. Chem. Soc. Rev. 2021, 50, 2320–2336. [DOI] [PubMed] [Google Scholar]
- (8).Bhoj VG; Chen ZJ Ubiquitylation in innate and adaptive immunity. Nature 2009, 458, 430–437. [DOI] [PubMed] [Google Scholar]
- (9).Liu YC Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 2004, 22, 81–127. [DOI] [PubMed] [Google Scholar]
- (10).Liyasova MS; Ma K; Lipkowitz S Molecular pathways: Cbl proteins in tumorigenesis and antitumor immunity-opportunities for cancer treatment. Clin. Cancer Res. 2015, 21, 1789–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Paolino M; Choidas A; Wallner S; Pranjic B; Uribesalgo I; Loeser S; Jamieson AM; Langdon WY; Ikeda F; Fededa JP; Cronin SJ; Nitsch R; Schultz-Fademrecht C; Eickhoff J; Menninger S; Unger A; Torka R; Gruber T; Hinterleitner R; Baier G; Wolf D; Ullrich A; Klebl BM; Penninger JM The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 2014, 507, 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chiang JY; Jang IK; Hodes R; Gu H Ablation of Cbl-b provides protection against transplanted and spontaneous tumors. J. Clin. Invest. 2007, 117, 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wilson BAP; Voeller D; Smith EA; Wamiru A; Goncharova EI; Liu G; Lipkowitz S; O’Keefe BR In Vitro Ubiquitination Platform Identifies Methyl Ellipticiniums as Ubiquitin Ligase Inhibitors. SLAS Discovery 2021, 26, 870–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wilson BAP; Thornburg CC; Henrich CJ; Grkovic T; O’Keefe BR Creating and screening natural product libraries. Nat. Prod. Rep. 2020, 37, 893–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ford PW; Davidson BS Plakinidine D, a new pyrroloacridine alkaloid from the ascidian Didemnum rubeum. J. Nat. Prod. 1997, 60, 1051–1053. [DOI] [PubMed] [Google Scholar]
- (16).Smith CJ; Venables DA; Hopmann C; Salomon CE; Jompa J; Tahir A; Faulkner DJ; Ireland CM Plakinidine D, a new pyrroloacridine alkaloid from two ascidians of the genus Didemnum. J. Nat. Prod. 1997, 60, 1048–1050. [DOI] [PubMed] [Google Scholar]
- (17).West RR; Mayne CL; Ireland CM; Brinen LS; Clardy J Plakinidines: Cytotoxic alkaloid pigments from the Fijian sponge Plakortis sp. Tetrahedron Lett. 1990, 31, 3271–3274. [Google Scholar]
- (18).Inman WD; Oneilljohnson M; Crews P Novel Marine Sponge Alkaloids. 1. Plakinidine A and B, anthelmintic active alkaloids from a Plakortis sponge. J. Am. Chem. Soc. 1990, 112, 1–4. [Google Scholar]
- (19).Shoemaker RH The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [DOI] [PubMed] [Google Scholar]
- (20).Peschard P; Kozlov G; Lin T; Mirza IA; Berghuis AM; Lipkowitz S; Park M; Gehring K Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell 2007, 27, 474–485. [DOI] [PubMed] [Google Scholar]
- (21).Greer A Christopher Foote’s discovery of the role of singlet oxygen [1O2 (1Δg)] in photosensitized oxidation reactions. Acc. Chem. Res. 2006, 39, 797–804. [DOI] [PubMed] [Google Scholar]
- (22).Entradas T; Waldron S; Volk M The detection sensitivity of commonly used singlet oxygen probes in aqueous environments. J. Photochem. Photobiol., B 2020, 204, 111787. [DOI] [PubMed] [Google Scholar]
- (23).Mao X; Zhang J; Wang X; Zhang H; Wei P; Sung HHY; Williams ID; Feng X; Ni XL; Redshaw C; Elsegood MRJ; Lam JWY; Tang BZ An air-stable organic radical from a controllable photoinduced domino reaction of a hexa-aryl substituted anthracene. J. Org. Chem. 2021, 86, 7359–7369. [DOI] [PubMed] [Google Scholar]
- (24).Lin Y; Yang Q; Geng F; Feng H; Chen M; Hu B Suppressing singlet oxygen formation during the charge process of Li-O2 batteries with a Co3O4 solid catalyst revealed by operando electron paramagnetic resonance. J. Phys. Chem. Lett 2021, 12, 10346–10352. [DOI] [PubMed] [Google Scholar]
- (25).Lightner DA; Bisacchi GS; Norris RD On the mechanism of the sensitized photooxygenation of pyrroles. J. Am. Chem. Soc. 1976, 98, 802–807. [Google Scholar]
- (26).Alberti MN; Vougioukalakis GC; Orfanopoulos M Photosensitized oxidations of substituted pyrroles: unanticipated radical-derived oxygenated products. J. Org. Chem. 2009, 74, 7274–7282. [DOI] [PubMed] [Google Scholar]
- (27).Howard JK; Rihak KJ; Bissember AC; Smith JA The oxidation of pyrrole. Chem.—Asian J 2016, 11, 155–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


