Abstract
PURPOSE
Osteosarcoma research advancement requires enhanced data integration across different modalities and sources. Current osteosarcoma research, encompassing clinical, genomic, protein, and tissue imaging data, is hindered by the siloed landscape of data generation and storage.
MATERIALS AND METHODS
Clinical, molecular profiling, and tissue imaging data for 573 patients with pediatric osteosarcoma were collected from four public and institutional sources. A common data model incorporating standardized terminology was created to facilitate the transformation, integration, and load of source data into a relational database. On the basis of this database, a data commons accompanied by a user-friendly web portal was developed, enabling various data exploration and analytics functions.
RESULTS
The Osteosarcoma Explorer (OSE) was released to the public in 2021. Leveraging a comprehensive and harmonized data set on the backend, the OSE offers a wide range of functions, including Cohort Discovery, Patient Dashboard, Image Visualization, and Online Analysis. Since its initial release, the OSE has experienced an increasing utilization by the osteosarcoma research community and provided solid, continuous user support. To our knowledge, the OSE is the largest (N = 573) and most comprehensive research data commons for pediatric osteosarcoma, a rare disease. This project demonstrates an effective framework for data integration and data commons development that can be readily applied to other projects sharing similar goals.
CONCLUSION
The OSE offers an online exploration and analysis platform for integrated clinical, molecular profiling, and tissue imaging data of osteosarcoma. Its underlying data model, database, and web framework support continuous expansion onto new data modalities and sources.
Osteosarcoma Explorer: a research data commons with clinical, genomic, protein, and image data.
INTRODUCTION
Some recent osteosarcoma research works have been empowered by rapid and significant advances in biomedical technologies, such as next-generation sequencing, digital pathology, and electronic health records (EHRs).1-7 These technological developments have led to the generation of massive amounts of data over the past decade. Data sets of large volumes and diverse types further benefit research, especially when incorporating advanced analysis methods and AI technologies.2,6,8 However, the often-poor accessibility and interoperability of these novel data sets are limiting their value in research. Technical and organizational barriers exist in almost every step of using these data: from data generation to standardization, storage, and sharing, which results in siloed data resources across health care facilities, industrial vendors, academic institutions, and other research organizations.
CONTEXT
Key Objective
To develop a large and comprehensive data commons that integrates clinical, molecular profiling, and tissue imaging data for osteosarcoma research.
Knowledge Generated
To address the siloed landscape of osteosarcoma research data, we have developed and publicly released the Osteosarcoma Explorer (OSE), which captures data for 573 patients with osteosarcoma integrated from various sources and modalities. Providing a user-friendly web interface for data exploration and analysis, the OSE has experienced an increasing utilization by the osteosarcoma research community.
Relevance (F. Lin)
-
The OSE project offers a comprehensive platform for gaining insight into the biology of pediatric osteosarcoma. This resource is achieved through integrated exploration of clinical data, molecular profiling, and tissue imaging data. The interface provides an accessible gateway to the data for a broad range of researchers at various levels, allowing empirical analysis to be done to gain correlative insights rapidly. This tool is directly relevant to translational research into this rare disease.*
*Relevance section written by JCO CCI Associate Editor Frank Lin, PhD, MBChB, FRACP, FAIDH.
The challenges of constrained data availability and interoperability are amplified for rare diseases such as pediatric osteosarcoma. Although osteosarcoma is the most common type of bone cancer in children and adolescents, the total number of patients is much smaller than more common cancers in children such as leukemia and central nervous system tumors.9,10 In the United States, the annual incidence rate of osteosarcoma in the 0-19 years age group is estimated to be 4.7 per million person-years, with approximately 800 new cases occurring every year.10 Unlike other pediatric cancers, no substantive improvements in patient outcome have been seen for over 30 years. There is an urgent need for more data, in terms of both volume and type, to improve our understanding of osteosarcoma biology and develop new therapeutics with better efficacy and fewer side effects.
Data commons represents an emerging solution to overcome the siloed data landscape in biomedical research.11,12 A well-designed data commons should incorporate harmonized data sets (generated from various sources and technology platforms), meaningful data standards and terminology, and a user-friendly web interface for data visualization and analytics.13 There are currently several large-scale data commons for cancer research, such as cBioPortal and Genomic Data Commons.13-15 Although these general purpose data commons projects have collected different types of data for various cancer types, research on each type of cancer usually demands a unique set of data elements and modalities depending on disease characteristics and management strategies. In addition, it is often challenging to integrate multimodal data (eg, clinical, genomics, proteomics, and tissue imaging) in a meaningful way, limiting the comprehensive characterization of a selected cohort. A recent effort initiated by the Children's Oncology Group (COG) and the QuadW Foundation focused on developing osteosarcoma data resources on the basis of biospecimens and clinical annotations obtained through COG study protocols. As an initial proof of concept, the High Dimensional Data (HDD) platform was developed by the QuadW-COG Childhood Sarcoma Biostatistics and Annotation Office (CSBOA).16 The HDD project established a workflow whereby projects using COG osteosarcoma biospecimens can share data through the platform. Our study builds upon this previous effort by integrating multisource, multimodal osteosarcoma data into a data commons.
The goal of this study is to develop an osteosarcoma-focused data commons, known as the Osteosarcoma Explorer (OSE), which integrates publicly available and institutional osteosarcoma data of various data types and provides a user-friendly web interface for data visualization and analysis. The OSE web portal is now accessible to the public.17 By providing public access to this resource, we hope to enhance osteosarcoma awareness within both research and health care communities.
MATERIALS AND METHODS
Data Collection and Integration
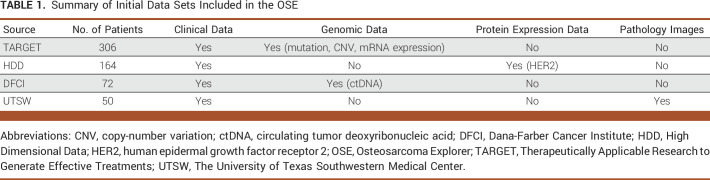
The COG and institutional review boards at the University of Texas Southwestern Medical Center (UTSW) and the Dana-Farber Cancer Institute (DFCI) have approved this study and the use of deidentified patient information in this study. Deidentified clinical and research data from patients with pediatric osteosarcoma were collected from four sources, summarized in Table 1. The Therapeutically Applicable Research to Generate Effective Treatments (TARGET) data set (306 patients) includes clinical annotations, gene expression, and copy-number variation (CNV) data collected through the TARGET Osteosarcoma project.18 The HDD data set (164 patients) includes clinical annotation and human epidermal growth factor receptor 2 (HER2) protein expression established through the CSBOA.16 The DFCI data set (72 patients) includes clinical annotations and circulating tumor DNA (ctDNA) status for patients enrolled on the COG osteosarcoma biology study AOST06B1.19 The UTSW data set (50 patients) includes clinical annotations and hematoxylin and eosin (H&E)–stained pathology images for patients treated at UTSW/Children's Medical Center Dallas.2 Patients were matched using the Unique Specimen Identifier (USI; assigned through COG studies) where applicable. For privacy protection, no germline data were included.
TABLE 1.
Summary of Initial Data Sets Included in the OSE
To integrate data from different sources, we designed a concept map (Fig 1) that captures typical clinical workflow in osteosarcoma patient care, including initial diagnosis, treatment, biospecimen collection and characterization (imaging, molecular profiling, etc), follow-up, and continuing evaluation of disease status. On the basis of the concept map, a common data model was constructed by matching and consolidating variables from different sources. A standardized terminology system was developed by harmonizing existing values from all data sources and mapping onto the concept unique identifiers (CUIs) in the National Cancer Institute (NCI) Metathesaurus database.20 The clinical data were then transformed onto this common data model, resulting in an integrated, standardized data set.
FIG 1.
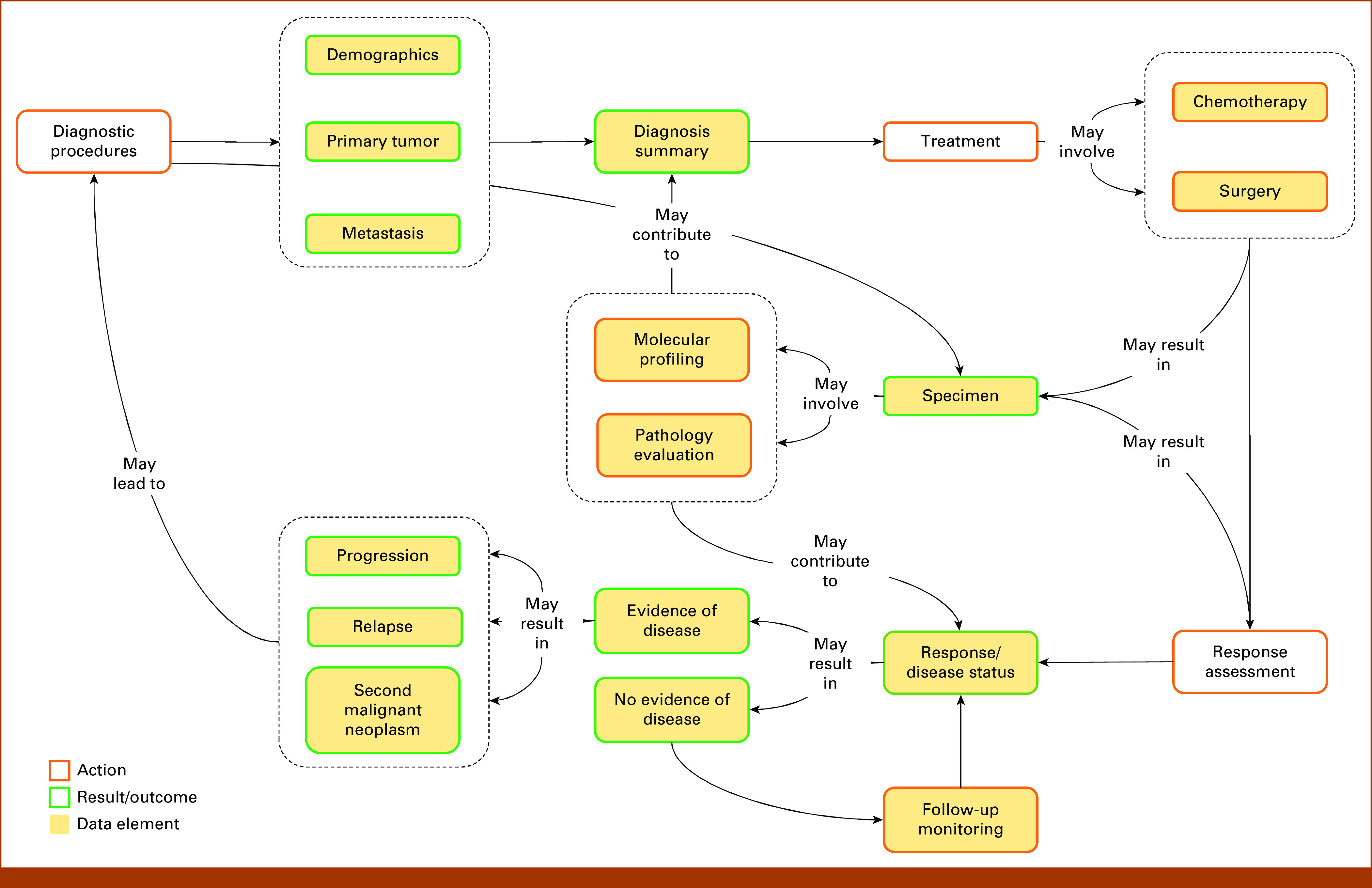
Concept map of the general clinical practice workflow for patients with osteosarcoma. Care actions are denoted by red-edged boxes. Observational results and patient outcomes are denoted by green-edged boxes. Yellow-filled boxes show from which element data have been captured. The design of the concept map was guided by pediatric oncologists with osteosarcoma expertise (L.F.C. and L.J.K.).
Gene expression (89 patients) and CNV (88 patients) data from patient tumor samples were downloaded from TARGET with the corresponding clinical annotations (306 patients).18 After data processing (Data Supplement), the gene expression data were normalized and categorized into high and low groups on the basis of median gene expression level in the cohort for each given gene, and the CNV data were categorized into three groups: copy-number gain, copy-number loss, and diploid. The ctDNA data were acquired from a previous study, where ultra-low-pass whole-genome sequencing was used to detect copy-number alterations in blood samples of patients with osteosarcoma (72 patients; source: DFCI).19 Patients were categorized by ctDNA positivity into positive and negative groups. The HER2 protein expression data (164 patients; source: HDD) were categorized into positive and negative groups via a semiquantitative approach on the basis of the protein expression levels determined through immunohistochemistry staining using CB11 antibody, where positive indicates that 51%-100% of cells were stained.16,21,22 Raw, deidentified H&E images (50 patients; 712 images; source: UTSW) were converted to portable network graphics (PNG) files at 10,000 × 10,000 pixel resolution using OpenSlide.23 The PNG files were then converted to tiled image pyramids in deep zoom image (DZI) format using deepzoom.py24 for database import and online visualization using OpenSeadragon.25
Database and Web Portal Development
On the basis of the OSE common data model, a relational database was developed using MySQL. To build the data-binding system, all codes were compacted into a single, reusable package. Database structure was described by JavaScript object notation (JSON) files and organized by table, column, and value. On the front end, data visualization was built using Chart.js and D3.js. We developed interactive charts for cohort summary information, a table- and graph-formatted individual patient dashboard, and survival analysis on the basis of user-selected grouping variables. Data filtering was bound with database query information for real-time updating. Through this two-way data binding system, the total number, summary charts, and table of the selected cohort are updated in real time when any combination of data filters is selected by users. We further implemented a local instance of the cBioPortal platform using the integrated OSE data set.13,14
RESULTS
Concept Map
The concept map outlines the general clinical practice workflow for osteosarcoma care (Fig 1). Episodes in this workflow are categorized as care actions (red-edged boxes) or observational results and patient outcomes (green-edged boxes). Initially, patients suspected of having osteosarcoma undergo the standard diagnostic procedures, which yield information about demographics, characteristics of the primary tumor(s), and metastasis. These together form a diagnosis summary, which determines first-line treatments. Two major types of first-line treatments (chemotherapy and surgery) are captured. Next, response assessment procedure is conducted to evaluate treatment response and disease status. If no evidence of disease is found, the routine follow-up procedure is to be applied. Any evidence of disease (eg, progression, relapse, or second malignant neoplasm) may trigger a new cycle of care. Importantly, several procedures may generate biospecimens. Each sample collection activates a separate subcycle that may involve pathology evaluation and molecular profiling. These biospecimen-derived data may further inform initial diagnosis and assessment of treatment response to yield more comprehensive diagnosis and prognosis profiles.
Data Model and Data Integration
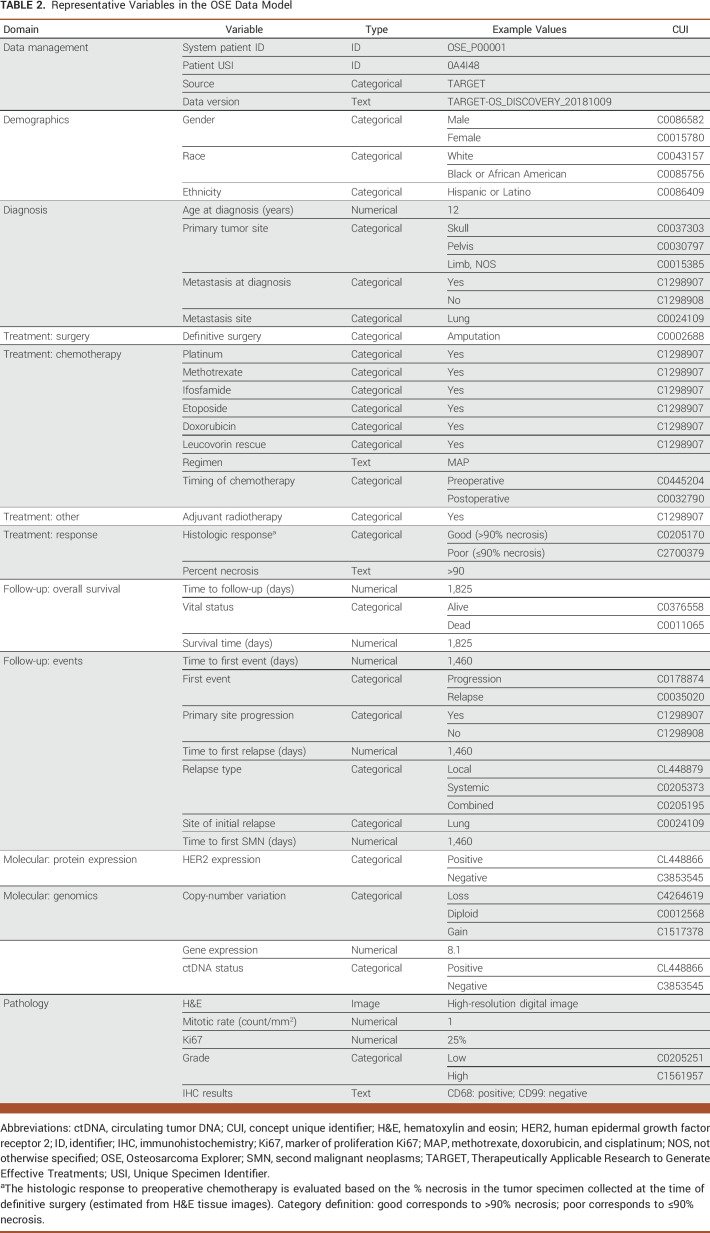
The OSE data model captures all key episodes in the concept map (yellow-filled boxes, Fig 1). Representative variables are shown in Table 2 (see the Data Supplement for specifications). The standardized terminology system is connected to the NCI Metathesaurus through CUIs to facilitate data interoperability and future updates. In total, five molecular profiling data tables are modeled into OSE, including gene mutation landscape, CNV, mRNA expression, ctDNA status, and HER2 protein expression (Data Supplement).
TABLE 2.
Representative Variables in the OSE Data Model
After identifier (ID) matching through USI, 573 unique patients were included, while overlapped patients were found between TARGET and HDD (n = 7), and between TARGET and DFCI (n = 6; Table 1). For source data version control, a data version ID is attached to each data record. The finalized, integrated OSE data set, including a harmonized clinical data table, five molecular profiling data tables, and 712 H&E tissue images, was loaded into the OSE database.
Online Data Exploration and Analysis
The OSE web portal provides an online graphical user interface for the exploration, visualization, and analysis of the integrated OSE dataset. In the Cohort Discovery module (Fig 2), major variables describing demographic, clinical, molecular, and imaging data are provided for patient selection and data query. Data variables are organized in a hierarchical structure reflecting the concept map (Fig 2A). When any set of variables is applied as filter, the pie charts (Fig 2B) and patient list (Fig 2C) are updated in real time to reflect the composition of the selected cohort. By clicking a patient ID hyperlink (Fig 2C), the Patient Dashboard module is opened, presenting a one-stop-shop view of all available data elements for the selected patient which originally were scattered at different sources (Fig 3A). The Patient Dashboard shows an individual patient's demographic and clinical data, HER2 protein expression and mRNA expression, CNV landscape, and a table of genes with somatic mutation linked to CNV status and mRNA expression (Fig 3A; Data Supplement). High-resolution whole-slide tissue images can be visualized in the Image Viewer module (Fig 3B). Users can open any image directly from the Cohort Discovery module. Actions such as zoom-in, zoom-out, and drag are smoothly realized using the mouse cursor. Figure 3C shows a clear zoomed-in view of an image.
FIG 2.
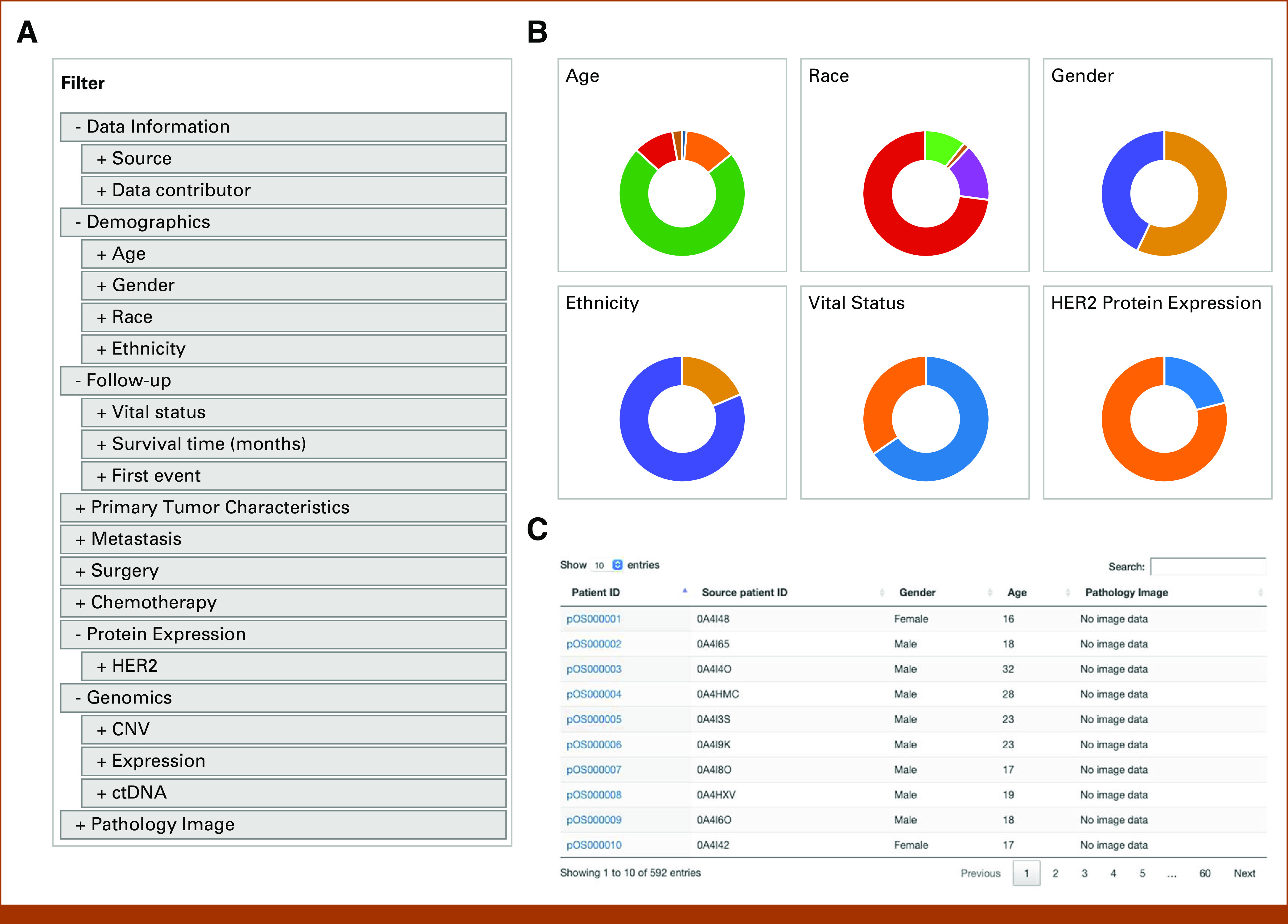
Cohort Discovery module on the OSE. (A) The list of variable filters is provided on the left side of the web page and is organized in a hierarchical structure. (B) Cohort summary panel on the right side of the web page displays six distribution charts for the selected cohort. (C) The patient table at the lower portion of the web page lists the patients in the selected cohort and provides the entry to the Patient Dashboard of each listed patient. The table is sortable by each column and capable of whole-table search. When any filter(s) is selected, the cohort summary panel and patient table are updated in real time. CNV, copy-number variation; ctDNA, circulating tumor DNA; HER2, human epidermal growth factor receptor 2; ID, identifier; OSE, Osteosarcoma Explorer.
FIG 3.
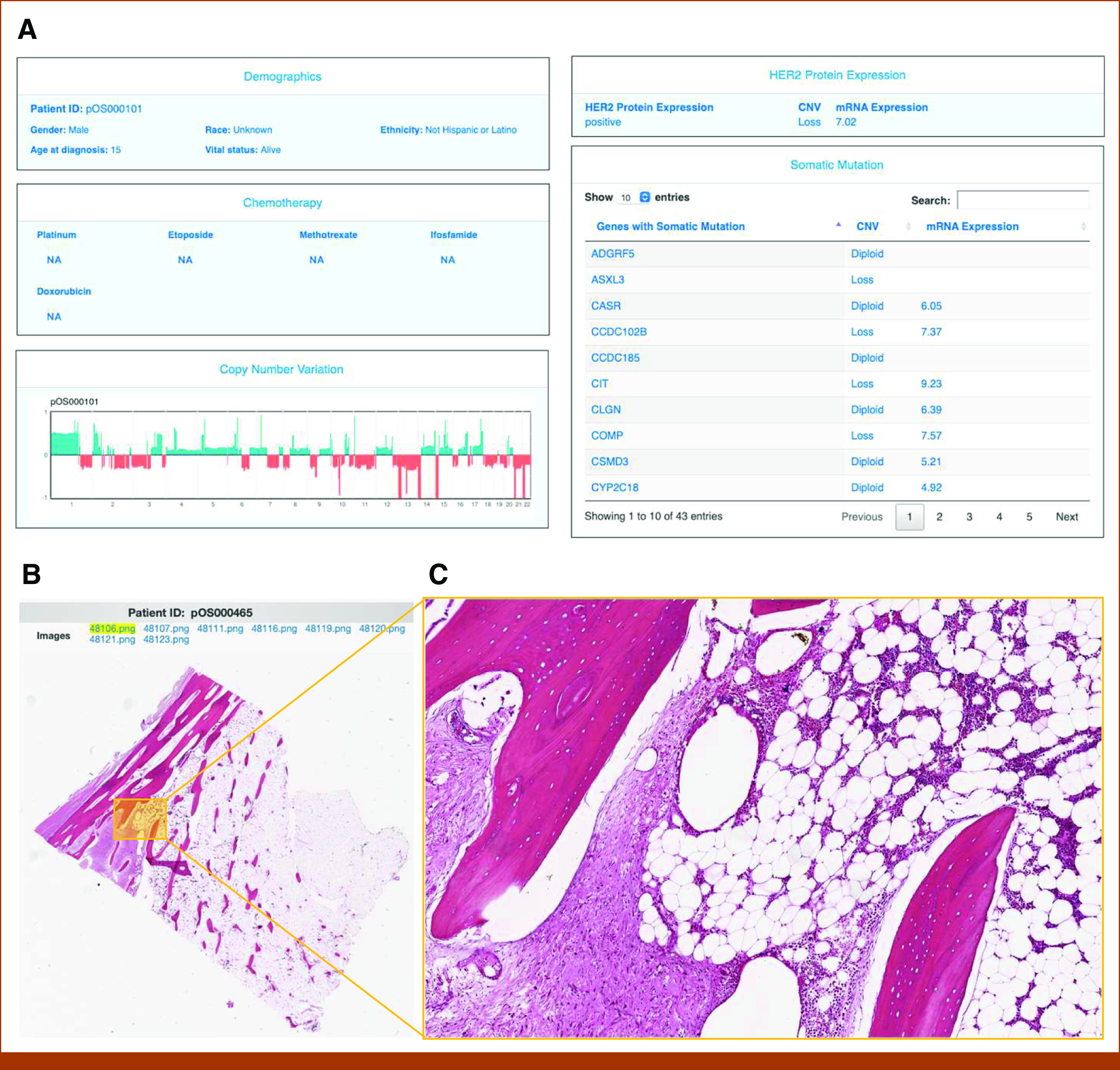
Patient Dashboard module and Image Viewer module on the OSE. (A) Individual patient-level data is displayed by sections, including demographics, chemotherapy, HER2 protein expression, an overview graph of CNV, and a table of CNV and mRNA expression status for genes with somatic mutation. This example is obtained from patient pOS000101. Access to this module is limited to registered users only. CNV graph: x-axis shows the position of a genomic region; y-axis shows CNV signatures of the region; green indicates copy-number gain; red indicates copy-number loss. The gene mutation table can be sorted and searched by users. (B) High-resolution, whole-slide tissue images are displayed for each patient. In this example, eight H&E images are available for the selected patient. (C) In-browser zoom-in action can smoothly focus on regions of interest with cell-level details. This example is obtained from patient pOS000465. Access to this module is limited to registered users only. CNV, copy-number variation; H&E, hematoxylin and eosin; HER2, human epidermal growth factor receptor 2; ID, identifier; NA, not applicable; OSE, Osteosarcoma Explorer.
The Online Analysis module (Fig 4) allows users to perform various types of data analysis using the integrated OSE data set. To conduct survival analysis, users can group the patients on the basis of a clinical, gene expression, CNV, ctDNA, or HER2 protein expression feature (Fig 4A). Both overall survival and event-free survival data are available for the analysis. For example, Figure 4B shows the Kaplan-Meier curve of overall survival on the basis of ctDNA status. The local, customized cBioPortal instance is integrated with OSE portal (Fig 4C) to provide analysis functions using the OSE genomic data. Users can perform association analysis and create custom plots for data visualization. Gene mutation and CNV landscape are illustrated on the front page of the OSE-cBioPortal instance (Fig 4C).
FIG 4.
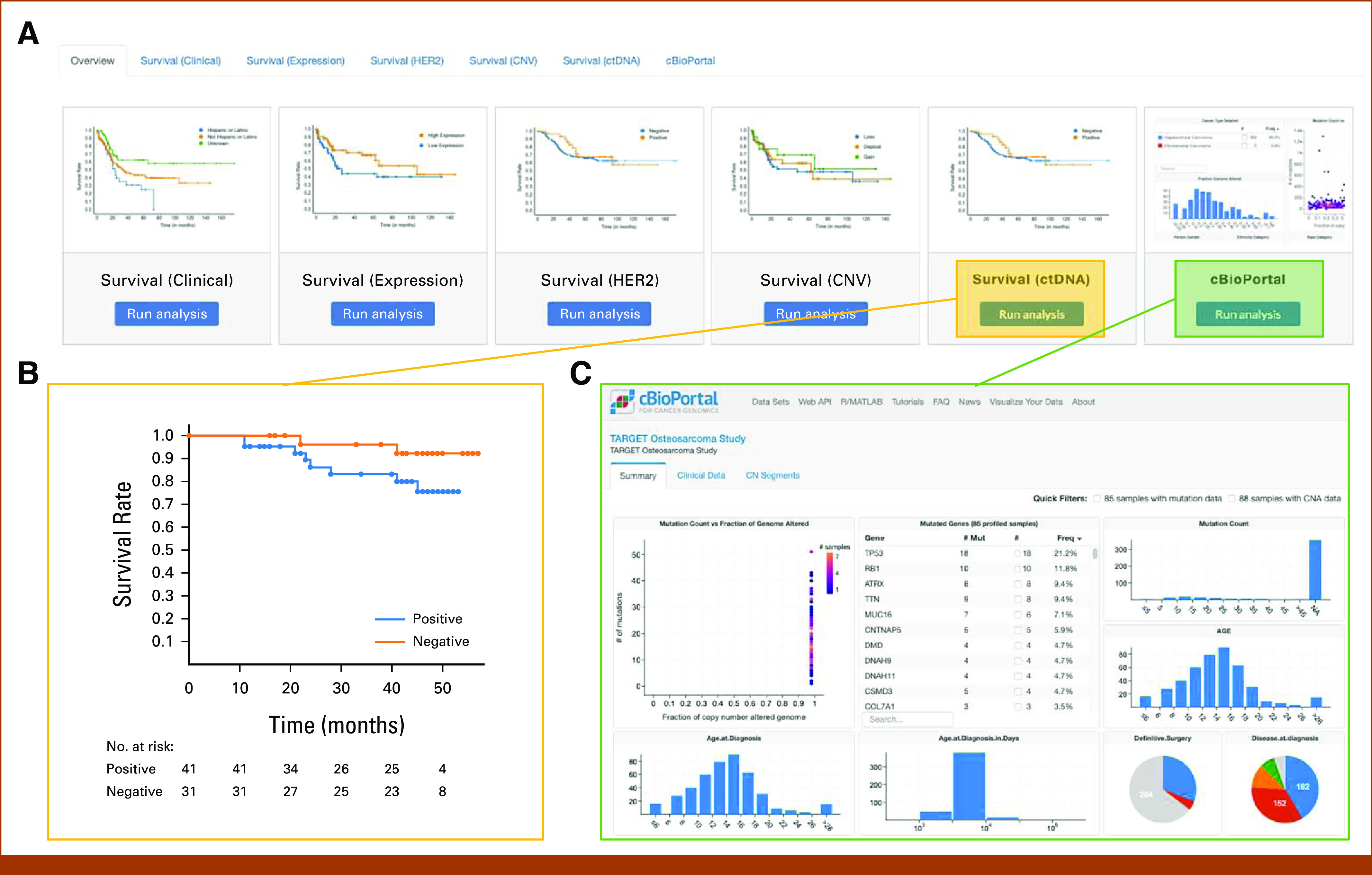
Online Analysis module on the OSE. (A) Various analysis tasks are provided in the module, including overall survival and event-free survival analysis based on clinical, gene expression, HER2 protein expression, CNV, ctDNA status, and analysis tools on cBioPortal. (B) An example Kaplan-Meier curve for overall survival based on ctDNA status. Users can easily switch to event-free survival using the provided dropdown list and the analysis plot will be refreshed in real time. To caution users against p-hacking, an alert message is provided with the survival analysis tool. (C) The analysis tools provided through a local instance of cBioPortal platform. CNV, copy-number variation; ctDNA, circulating tumor DNA; HER2, human epidermal growth factor receptor 2; OSE, Osteosarcoma Explorer; TARGET, Therapeutically Applicable Research to Generate Effective Treatments.
Public Release, Data Security, and User Support
The OSE web portal was released to the public in November 2021.17 To ensure data security, only deidentified data are stored in the OSE database. General users can view group-level summary charts in the Cohort Discovery module without registration or login. Access to individual-level data, images, and analysis is available to registered users only. Registration is currently open to research, educational, and government institutions by restricting email domains to .edu and .gov. From its public release in November 2021 to March 2023, the OSE has received an average of 132 site visits per month and accumulated 66 registered users from various countries. User support provided by the OSE team includes general technical support, user-specific consultations covering database design and data analysis, and a data repository service for data sharing needs. For instance, users can request an entry to upload and share deidentified osteosarcoma data used in a published work.
DISCUSSION
Data resources that meet the Findable, Accessible, Interoperable, and Reusable (FAIR) standard15 are in high demand for biomedical research, especially for rare diseases such as pediatric osteosarcoma. Data commons has become the main solution to providing such resources. Pediatric osteosarcoma research data have been hosted in different data repositories maintained by organizations such as COG, research and health care institutions, and individual research laboratories. The OSE project serves as a model for integrating clinical, genomic, protein, and tissue image data from various sources following the FAIR standard. The successfully developed OSE data model, web portal, and the experiences in community engagement and user support presented here can benefit the data integration and international data sharing efforts for other rare diseases and facilitate clinical and translational research.
A key contribution of the OSE project is an established workflow for integrating data from scattered sources into a comprehensive and harmonized data set. In this workflow, data source tracking plays a central role. The OSE centralized ID system identifies unique patients using USI, which allows various data records for a given patient to be linked and consolidated. Data acquisition for OSE is designed as a continuous process. For example, the TARGET Osteosarcoma project has published clinical annotation data in different versions. The OSE assigns unique data version IDs (eg, TARGET-OS_DISCOVERY_20181009) to each individual data record. By default, the OSE uses data elements (eg, survival times) from the latest version to overwrite the previous version on a patient-by-patient basis while keeping all historical versions in the backend, ensuring data accuracy and traceability.
Another key aspect of a successful data integration is the data model design. In the OSE data model, major clinical episodes in the concept map are constructed as distinct data domains, which is reflected in both the back-end database and the front-end web interface. Mapping of the OSE terminology system onto NCI Metathesaurus further ensures clear variable definitions, feasibility of data exchange with other data models/standards, and capacity for further expansion of the OSE data model and terminology system when new data sources become available.26 Importantly, the established data integration workflow for OSE can be readily applied to other projects of similar purposes.
The OSE web portal is the main interface for researchers to access this data resource. The Cohort Discovery module greatly simplifies the cohort formation process by enabling data query across different sources and data types. In the Patient Dashboard module, users have an integrated view of clinical characteristics and molecular profiling data features for a given patient, which were originally scattered across different sources. Similarly, the Pathology Image module offers high-resolution images and related clinical variables in one place, enhancing the data visualization capabilities. By providing a user-friendly web portal, OSE transforms the traditional way of accessing a research data set through interpersonal communications with the data provider or curator, which often limits the volume and efficiency of user support. Instead, OSE users can fulfill most of the common requests, such as data exploration, cohort formation, visualization, and analysis, directly by themselves through the web portal. The steadily increasing user access and interests demonstrate OSE's usability and usefulness in osteosarcoma research. This user interaction framework can be applied to other informatics projects of similar scope.
The current, initial version of OSE serves as a solid foundation for future endeavors to enhance the data commons for osteosarcoma research. Limited availability and interoperability of source data remains a significant challenge for rare cancers such as osteosarcoma. For example, the clinical annotation data acquired from the TARGET Osteosarcoma project cover basic data elements such as individual chemotherapy drug names but lack more granular elements such as timing of chemotherapy and regimen information. To overcome this challenge, EHRs can potentially be leveraged. By successfully integrating the UTSW EHR-based data into OSE, we demonstrate the feasibility of incorporating essential data elements directly from institutional EHRs. We plan to integrate more EHR-based osteosarcoma data by engaging other health care and research institutions.
Solid digital pathology components, in the form of both images and structured pathologic features, are crucial for osteosarcoma research and care. The current version of OSE supports high-resolution tissue images and the associated features such as grade and histologic response to preoperative chemotherapy. More comprehensive pathologic features, such as mitotic rate and immunochemistry markers, are typically available in the pathology reports. A major challenge to fully utilizing these reports is the complex and intensive natural language processing required for extracting structured data elements from free texts. The recent advancements in large language models (eg, GPT-4 and Med-PaLM) show unprecedented promises for effectively and efficiently processing medical notes.27 We are in the process of applying the large language model technology to processing and curating pathology notes, which can potentially populate the next version of OSE with more abundant pathologic features.
Another prospective component of the OSE is data from patient-derived xenograft (PDX), which is an important resource for translational research. A key challenge for integrating and managing PDX data is a well-designed data model that captures and simplifies the multilevel lineage relations between patient-level and sample-level data. In the future, we plan to incorporate the PDX data component into OSE by leveraging an in-house, web-based biospecimen management tool that features a proved data model for handling patient-sample lineage. Building upon this, we further plan to integrate sample and molecular profiling data for PDX generated at various sources.
In conclusion, we developed the OSE data commons, which integrates osteosarcoma clinical, molecular profiling, and tissue imaging data from multiple sources. Data of heterogenous structure and terminology were transformed onto a common data model. A relational database and a user-friendly web portal were built to provide functionality to query, explore, and analyze data online. Since its public release, the OSE has continuously supported the osteosarcoma research community. Its successful initial deployment has laid a strong foundation for expanding the project to incorporate new data sources and a broader range of data types.
ACKNOWLEDGMENT
The authors thank the COG and QuadW Foundation for providing the HDD data set and the support for the development of the OSE as a means to improve outcomes for patients with osteosarcoma.
Donghan M. Yang
Employment: Novartis (immediate family member)
Stock and Other Ownership Interests: Novartis (immediate family member)
Dinesh Rakheja
Consulting or Advisory Role: ClearNano Inc (immediate family member)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/591421
Tammy Lo
Employment: Johnson and Johnson
Donald A. Barkauskas
Employment: Genentech
Patents, Royalties, Other Intellectual Property: US patent based on PhD research in glioblastoma
David S. Shulman
Consulting or Advisory Role: Boehringer Ingelheim
Katherine Janeway
Honoraria: Foundation Medicine, Takeda
Consulting or Advisory Role: Bayer, Ipsen, Illumina, Bayer
Travel, Accommodations, Expenses: Bayer
Chand Khanna
Leadership: VuJaDe Lifesciences
Stock and Other Ownership Interests: VuJaDe Lifesciences
Richard Gorlick
Research Funding: Eisai (Inst)
Christopher Menzies
Employment: Children's Health
Leadership: Children's Health
Guanghua Xiao
Consulting or Advisory Role: Adjuvant Genomics Inc
Laura J. Klesse
Uncompensated Relationships: Alexion Pharmaceuticals
Brian D. Crompton
Employment: Acceleron Pharma, Generate Biomedicines
Leadership: New Age Industries
Stock and Other Ownership Interests: Acceleron Pharma
Consulting or Advisory Role: PetDx, Animal Cancer Foundation, AstraZeneca
Research Funding: Gradalis
No other potential conflicts of interest were reported.
Yang Xie
Consulting or Advisory Role: Adjuvant Genomics Inc
PRIOR PRESENTATION
Presented in part at the Connective Tissue Oncology Society (CTOS) 2021 Annual Meeting, virtual, November 2021.
SUPPORT
Supported by the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30CA142543 (Y.X. and D.M.Y.), R35GM136375 (Y.X.), U01AI156189 (Y.X.), R01GM140012 (G.X.), R01GM141519 (G.X.), R01DE030656 (G.X.), U01CA249245 (G.X.), and the Cancer Prevention and Research Institute of Texas RP180805 (Y.X.). This work is also supported by NCTN Operations Center Grant U10CA180886, Human Specimen Banking Grant U24CA196173, NCTN Statistics and Data Center Grant U10CA180899 of the Children's Oncology Group (COG) from the National Cancer Institute of the National Institutes of Health. Additional support for research is provided by a grant from the WWWW (QuadW) Foundation, Inc (www.QuadW.org) to the COG.
D.M.Y. and Q.Z. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Donghan M. Yang, Qinbo Zhou, Lauren Furman Cline, Katherine Janeway, Richard Gorlick, Stephen X. Skapek, Brian D. Crompton, Yang Xie
Financial support: Yang Xie
Administrative support: Yang Xie
Provision of study materials or patients: Patrick J. Leavey, Dinesh Rakheja, David S. Shulman, Chand Khanna, Richard Gorlick, Christopher Menzies, Lin Xu, Brian D. Crompton, Yang Xie
Collection and assembly of data: Donghan M. Yang, Qinbo Zhou, Lauren Furman Cline, Xian Cheng, Danni Luo, Hongyin Lai, Yueqi Li, Kevin W. Jin, Bo Yao, Patrick J. Leavey, Dinesh Rakheja, Tammy Lo, David Hall, Donald A. Barkauskas, David S. Shulman, Chand Khanna, Richard Gorlick, Christopher Menzies, Lin Xu, Laura J. Klesse, Brian D. Crompton, Yang Xie
Data analysis and interpretation: Donghan M. Yang, Qinbo Zhou, Lauren Furman Cline, Danni Luo, Hongyin Lai, Yueqi Li, Patrick J. Leavey, Dinesh Rakheja, David Hall, David S. Shulman, Xiaowei Zhan, Guanghua Xiao, Stephen X. Skapek, Lin Xu, Laura J. Klesse, Yang Xie
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Donghan M. Yang
Employment: Novartis (immediate family member)
Stock and Other Ownership Interests: Novartis (immediate family member)
Dinesh Rakheja
Consulting or Advisory Role: ClearNano Inc (immediate family member)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/591421
Tammy Lo
Employment: Johnson and Johnson
Donald A. Barkauskas
Employment: Genentech
Patents, Royalties, Other Intellectual Property: US patent based on PhD research in glioblastoma
David S. Shulman
Consulting or Advisory Role: Boehringer Ingelheim
Katherine Janeway
Honoraria: Foundation Medicine, Takeda
Consulting or Advisory Role: Bayer, Ipsen, Illumina, Bayer
Travel, Accommodations, Expenses: Bayer
Chand Khanna
Leadership: VuJaDe Lifesciences
Stock and Other Ownership Interests: VuJaDe Lifesciences
Richard Gorlick
Research Funding: Eisai (Inst)
Christopher Menzies
Employment: Children's Health
Leadership: Children's Health
Guanghua Xiao
Consulting or Advisory Role: Adjuvant Genomics Inc
Laura J. Klesse
Uncompensated Relationships: Alexion Pharmaceuticals
Brian D. Crompton
Employment: Acceleron Pharma, Generate Biomedicines
Leadership: New Age Industries
Stock and Other Ownership Interests: Acceleron Pharma
Consulting or Advisory Role: PetDx, Animal Cancer Foundation, AstraZeneca
Research Funding: Gradalis
No other potential conflicts of interest were reported.
Yang Xie
Consulting or Advisory Role: Adjuvant Genomics Inc
REFERENCES
- 1.Zhao J Dean DC Hornicek FJ, et al. : Emerging next-generation sequencing-based discoveries for targeted osteosarcoma therapy. Cancer Lett 474:158-167, 2020 [DOI] [PubMed] [Google Scholar]
- 2.Arunachalam HB Mishra R Daescu O, et al. : Viable and necrotic tumor assessment from whole slide images of osteosarcoma using machine-learning and deep-learning models. PLoS One 14:e0210706, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ligon JA Choi W Cojocaru G, et al. : Pathways of immune exclusion in metastatic osteosarcoma are associated with inferior patient outcomes. J Immunother Cancer 9:e001772, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paludo J Fritchie K Haddox CL, et al. : Extraskeletal osteosarcoma: Outcomes and the role of chemotherapy. Am J Clin Oncol 41:832-837, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misaghi A Goldin A Awad M, et al. : Osteosarcoma: A comprehensive review. SICOT J 4:12, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J Pan H Li M, et al. : Predictive model for the 5-year survival status of osteosarcoma patients based on the SEER database and XGBoost algorithm. Sci Rep 11:5542, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X Wu Q Gong X, et al. : Osteosarcoma: A review of current and future therapeutic approaches. Biomed Eng Online 20:24, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anisuzzaman D Barzekar H Tong L, et al. : A deep learning study on osteosarcoma detection from histological images. Biomed Signal Process Control 69:102931, 2021 [Google Scholar]
- 9.Mirabello L, Troisi RJ, Savage SA: Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer 115:1531-1543, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linabery AM, Ross JA: Trends in childhood cancer incidence in the U.S. (1992-2004). Cancer 112:416-432, 2008 [DOI] [PubMed] [Google Scholar]
- 11.Grossman RL Heath A Murphy M, et al. : A case for data commons: Toward data science as a service. Comput Sci Eng 18:10-20, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen MA Ferretti V Grossman RL, et al. : The NCI Genomic Data Commons as an engine for precision medicine. Blood 130:453-459, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J Aksoy BA Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E Gao J Dogrusoz U, et al. : The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401-404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heath AP Ferretti V Agrawal S, et al. : The NCI genomic data commons. Nat Genet 53:257-262, 2021 [DOI] [PubMed] [Google Scholar]
- 16.Glover J Man TK Barkauskas DA, et al. : Osteosarcoma enters a post genomic era with in silico opportunities: Generation of the High Dimensional Database for facilitating sarcoma biology research: A report from the Children's Oncology Group and the QuadW Foundation. PLoS One 12:e0181204, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osteosarcoma Explorer. https://datacommons.swmed.edu/cce/ose
- 18.TARGET Osteosarcoma project. https://ocg.cancer.gov/programs/target/projects/osteosarcoma
- 19.Shulman DS Klega K Imamovic-Tuco A, et al. : Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children's Oncology Group. Br J Cancer 119:615-621, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.NCI Metathesaurus (NCIm). https://ncimetathesaurus.nci.nih.gov/ncimbrowser/
- 21.Ebb D Meyers P Grier H, et al. : Phase II trial of trastuzumab in combination with cytotoxic chemotherapy for treatment of metastatic osteosarcoma with human epidermal growth factor receptor 2 overexpression: A report from the Children's Oncology Group. J Clin Oncol 30:2545-2551, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorlick S Barkauskas DA Krailo M, et al. : HER-2 expression is not prognostic in osteosarcoma; a Children's Oncology Group prospective biology study. Pediatr Blood Cancer 61:1558-1564, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.OpenSlide. https://openslide.org/api/python/
- 24.deepzoom.py. https://github.com/openzoom/deepzoom.py
- 25.OpenSeadragon. https://openseadragon.github.io/
- 26.Ci B Yang DM Krailo M, et al. : Development of a data model and data commons for germ cell tumors. JCO Clin Cancer Inform 4:555-566, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Will ChatGPT transform healthcare? Nat Med 29:505-506, 2023 [DOI] [PubMed] [Google Scholar]