Abstract
This study aimed to explore the mode of action of Yiqiyangyinquyu prescription (YP) against Sjögren’s syndrome (SS) by combining network pharmacology with molecular docking techniques. YP’s active components and target proteins were identified using the BATMAN-traditional Chinese medicine database. Concurrently, targets associated with SS were extracted from databases, including Genecards, Online Mendelian Inheritance in Man, and Therapeutic Target Database. The standard targets were then imported into the STRING database to construct a protein-protein interaction network. We then conducted gene ontology and Kyoto encyclopedia of genes and genomes enrichment analyses, which were succeeded by molecular docking studies to validate core active components and key targets. Finally, in vitro experiments and molecular dynamics simulation were conducted to substantiate the therapeutic efficacy of YP in treating SS. A total of 206 intersection targets and 46 active compounds were identified. Gene ontology analysis unveiled that YP targets were primarily enriched in cellular responses to chemical stress, inflammation, and cell proliferation. Key enriched signaling pathways encompassed the interleukin 17, hypoxia-inducible factor-1, tumor necrosis factor (TNF-α), and advanced glycation end products-receptor for AGEs (AGE-RAGE) signaling pathways. Molecular docking results demonstrated high-affinity between neotanshinone C, tanshiquinone B, miltionone I, TNF-α, interleukin 1 beta (IL-1β), and interleukin 6 (IL-6). Noteworthy, TNF-α, considered the most important gene in YP against SS, binds to YP most stably, which was further validated by molecular dynamics simulation. In vitro experiments confirmed YP’s capacity to reduce TNF-α, IL-1β, and IL-6 expression, effectively alleviating SS-related inflammation. YP demonstrated a significant anti-inflammatory effect by suppressing inflammatory cytokines (TNF-α, IL-6, and IL-1β), providing experimental evidence for its clinical application in treating SS.
Keywords: experimental validation, molecular docking, network pharmacology, Sjögren’s syndrome, Yiqiyangyinquyu Prescription
1. Introduction
Sjögren’s syndrome (SS) is a chronic autoimmune disease primarily characterized by dryness in the mouth and eyes, resulting from secretory dysfunction of exocrine glands, particularly the lacrimal and salivary glands, due to local inflammation.[1] The disorder primarily targets middle-aged women, presenting a female-to-male prevalence ratio of about 9:1 to 20:1.[2] The complex pathogenesis of SS entails a multitude of immune cells, inflammatory factors, and signaling pathways. Nonetheless, applying systemic immunosuppression in SS has demonstrated limited efficacy when juxtaposed with its effectiveness in addressing other autoimmune diseases. Current treatment strategies rely on empirical approaches centered on symptom management. Notably, in the realm of SS, biological agents like anakinra, tocilizumab, and rituximab have fallen short of attaining primary outcomes in randomized, double-blind controlled trials.[3] As a result, traditional Chinese medicine (TCM) interventions are receiving heightened scrutiny and interest.
TCM primarily considers the etiology and pathogenesis of SS as complex, with Qi deficiency, Yin deficiency, and blood stasis, with dryness-heat manifesting as the symptomatic expression. In adherence to this principle, the Yiqiyangyinquyu prescription (YP) was formulated for the clinical treatment of SS using selected herbs: Ophiopogon japonicus, Rehmannia glutinosa, Scrophularia ningpoensis Hemsl, Astragalus mongholicus Bunge, Salvia miltiorrhiza Bunge, Leonurus japonicus Houtt, Dendrobium nobile Lindl, and Paeonia lactiflora Pall. Previous research conducted by our team has provided evidence that YP effectively ameliorates dry mouth and dry eye symptoms while lowering the expression levels of serum immunoglobulin G, interleukin 10, B cell-activating factor, and Toll-like receptors 9 in SS patients.[4] YP plays a pivotal role in modulating both innate and adaptive immunity, elevating the quality of life for patients, diminishing the disease activity index, reducing reliance on glucocorticoids, and enhancing the immune microenvironment and exocrine function in patients.[5–8] Additionally, utilizing an SS-like spontaneous model in NOD mice, our research unveiled that YP can modulate the Toll-like receptor-interferon (IFN)-B-cell stimulating factor pathway in mice’s peripheral blood and submandibular glands. Moreover, it improves the blood flow microenvironment and alleviates injury in the submandibular glands of NOD mice.[9] In summary, the therapeutic potential of YP in the context of SS demands additional exploration and study.
Network pharmacology, a multidisciplinary approach that incorporates elements of chemoinformatics, bioinformatics, network biology, and pharmacology, aligns seamlessly with the holistic principles of TCM. It effectively aids in unraveling the mechanisms of drug action.[10] In this study, we investigated the mechanism that underlies the effects of YP on SS, utilizing a combination of network pharmacology and molecular docking methodologies. Furthermore, we assessed the influence of YP on inflammatory responses through in vitro experiments. Our discoveries establish a solid groundwork for the clinical utilization and development of YP and present fresh research angles for the progressive evolution of Chinese medicine.
2. Materials and methods
2.1. Identification of active compounds and targets of YP
The constituent drugs of YP, which include O japonicus, R glutinosa, S ningpoensis Hemsl, A mongholicus Bunge, S miltiorrhiza Bunge, L japonicus Houtt, D nobile Lindl, and P lactiflora Pall, were input into BATMAN-TCM database (http://bionet.ncpsb.org.cn/batman-tcm/index.php/) to acquire potential active compounds and their corresponding targets.[11] Utilizing a prediction score exceeding 20 and a P value <.05 as selection benchmarks, 1560 key targets associated with 115 active compounds were discerned.
2.2. Selection of SS-related targets
Targets relevant to SS were sourced from the GeneCards database (https://www.genecards.org/) and the Online Mendelian Inheritance in Man (OMIM, https://www.genecards.org/).[12] Furthermore, targets associated with SS were also retrieved from the Therapeutic Target Database (TTD) (http://db.idrblab.net/ttd/)[13] using the keyword “Sjögren’s syndrome.”
2.3. Construction of drug-disease target network and Venn analysis
The targets associated with “Sjögren’s syndrome” and the ingredients of YP were submitted to obtain the intersection targets of “SS-YP” using Venn online tool, available at (https://bioinfogp.cnb.csic.es/tools/venny/).[14]
2.4. Construction of protein-protein interaction (PPI) network and screening of core proteins
The intersection targets of SS and YP were entered into the STRING platform, accessible at (https://cn.string-db.org/).[15] These intersection targets were subjected to PPI network analysis using the STRING platform. The generated PPI network was transferred into Cytoscape 3.9.0 for in-depth network analysis to pinpoint key proteins within the intersection of drug and disease targets.
2.5. Pathway enrichment analysis
The essential proteins derived from the SS-YP intersection were subjected to gene ontology (GO) functional analysis and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis using the R programming language. GO analysis spanned 3 categories: molecular function (MF), biological process (BP), and cellular component (CC). These results were analyzed using Cytoscape 3.9.0, allowing for organizing GO functions and pathways based on rankings of network node degree values.
2.6. Molecular docking analysis and visualization
Core target proteins and their associated active chemical components in the treatment of SS were identified based on network node degree values. The AutoDock Vina software was employed to verify molecular docking interactions between the core target proteins and chemical components. Preparing macromolecular receptor files involved extracting the 3D protein structure from the Protein Data Bank (PDB) at (https://www.rcsb.org/) and configuring it using Pymol.[16] This process encompassed several steps, such as eliminating solvents and organics, importing hydrogenation into Autodock, specifying the receptor, and saving the structure as a PDBQT protein receptor file. The preparation of drug-receptor files involved obtaining the 2-dimensional molecular formula structure by downloading it from PubChem (https://pubchem.ncbi.nlm.nih.gov/)[17] Furthermore, this involved converting the 2-dimensional molecular formula structure into 3D structures with minimized energy using ChemOffice 20.0, typically achieved through the MM2 (Molecular Mechanics 2) method. The acquired 3D structures were subsequently adjusted within the AutoDock tool by incorporating hydrogen atoms and protonation. Nonpolar hydrogen atoms were added to the respective carbon atoms, and the rotation and torsion settings were configured automatically within the software. The structures were saved as PDBQT ligand files, and molecular docking procedures were executed using Vina software. Docking affinity was assessed and scored according to the computed docking binding free energy.[18] Pymol was employed for visualizing the 3D docking model, focusing on the one with the lowest binding affinity. Binding energies <0 were indicative of spontaneous ligand-receptor binding.
2.7. Molecular dynamics (MD) simulation
The MD simulations were carried out by GROMACS 2020.3 software. The amber99sb-ildn force field and the general Amber force field were used to generate the parameter and topology of proteins and ligands, respectively. The simulation box size was optimized with the distance between each atom of the protein and the box >1.0 nm. Then, fill the box with water molecules based on a density of 1. To make the simulation system electrically neutral, the water molecules were replaced with Cl- and Na + ions. Following the steepest descent method, energy optimization of 5.0 × 104 steps was performed to minimize the energy consumption of the entire system, and finally to reduce the unreasonable contact or atom overlap in the entire system. After energy minimization, first-phase equilibration was performed with the NVT ensemble at 300 K for 100 ps to stabilize the temperature of the system. Second-phase equilibration was simulated with the NPT ensemble at 1 bar and 100 ps. The primary objective of the simulation is to optimize the interaction between the target protein and the solvent and ions so that the simulation system is fully pre-equilibrated. All MD simulations were performed for 100 ns under an isothermal and isostatic ensemble with a temperature of 300 K and a pressure of 1 atmosphere. The temperature and pressure were controlled by the V-rescale and Parrinello-Rahman methods, respectively, and the temperature and pressure coupling constants were 0.1 and 0.5 ps, respectively. Lennard-Jones function was used to calculate the Van der Waals force, and the nonbond truncation distance was set to 1.4 nm. The bond length of all atoms was constrained by the LINCS algorithm. The long-range electrostatic interaction was calculated by the Particle Mesh-Ewald method with the Fourier spacing of 0.16 nm.
2.8. Preparation of YP freeze-dried powder
The herbal formulation YP, consisting of 8 components, was obtained from the pharmacy of The Second Affiliated Hospital of Zhejiang Chinese Medical University. The composition included O japonicus (15 g), R glutinosa (24 g), S ningpoensis Hemsl (15 g), A mongholicus Bunge (24 g), S miltiorrhiza Bunge (30 g), L japonicus Houtt (15 g), D nobile Lindl (12 g), and P lactiflora Pall (12 g). The YP mixture was condensed to a volume of 100 mL and then transferred into a dedicated freeze-drying container. Subsequently, the mixture was rapidly frozen to liquid nitrogen and dried at–50˚C in a freeze dryer.
After 48 hours, the lyophilized powder was removed, crushed, and stored under cryogenic drying conditions.
2.9. Cell culture
Human salivary gland (HSG) cells, sourced from the Shanghai Tongpai Institute of Biotechnology in Shanghai, China, were cultivated in Roswell Park Memorial Institute (RPMI/1640) medium. The medium was fortified with 10% fetal bovine serum, 100 μg/mL of streptomycin, and 100 U/mL of penicillin. The cells were kept under culture conditions at 37˚C with a 5% CO2 environment.
2.10. RNA isolation and reverse transcription
The cells were seeded into 6-well culture plates with cell slides. Once the cells reached 80% growth density, they were divided into various treatment groups, which included control, LPS (lipopolysaccharide) at a concentration of 10 μg/mL, LPS (10 μg/mL) + Low-YP at 200 μg/mL, LPS (10 μg/mL) + Middle-YP at 400 μg/mL, and LPS (10 μg/mL) + High-YP at 800 μg/mL. In each specified group, 5.5 × 10^5 cells were collected, and the total RNA was isolated using Trizol Reagent from Takara in Kusatsu, Shiga, Japan. The isolated RNA was then subjected to reverse transcription to generate complementary DNA. This process was carried out using the ReverTra Ace qPCR RT Kit from Toyobo in Osaka, Japan, within a T100 Thermal Cycler from Bio-Rad, CA, USA.
Following the reverse transcription, complementary DNA was subjected to reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. This analysis was performed using the UltraSYBR Mixture from Cwbiotech in Taizhou, Jiangsu, China, and it took place in a Light Cycler 96 by Roche, located in Basel, Switzerland. Primer sequences were designed utilizing an online resource available at (https://pga.mgh.harvard.edu/primerbank/). These primer sequences are detailed in Table 1.
Table 1.
Primer sequences used for quantitative real-time PCR.
| Sequence (5′–3′) | |
|---|---|
| TNF-α-F | CCTCTCTCTAATCAGCCCTCTG |
| TNF-α-R | GAGGACCTGGGAGTAGATGAG |
| IL-6-F | ACTCACCTCTTCAGAACGAATTG |
| IL-6-R | CCATCTTTGGAAGGTTCAGGTTG |
| IL-1β-F | ATGATGGCTTATTACAGTGGCAA |
| IL-1β-R | GTCGGAGATTCGTAGCTGGA |
| GAPDH-F | TCAAGGCTGAGAACGGGAAG |
| GAPDH-R | GACTCCACGACGTACTCAGC |
IL-1β = interleukin 1 beta, IL-6 = interleukin 6, TNF-α = tumor necrosis factor.
2.11. Immunofluorescence detection of tumor necrosis factor (TNF-α), IL6, and interleukin 1 beta (IL-1β) expression
The cells were placed into 24-well plates with cell slides. When they reached approximately 70% density, they were assigned to the 5 treatment groups mentioned earlier. Every well was subjected to 3 rounds of washing with PBS (Phosphate-Buffered Saline), lasting 5 minutes. Subsequently, 500 μL of a 40 g/L paraformaldehyde solution was used to fix the cells for 30 minutes. The cells were later blocked with a solution containing 3% fetal bovine serum and 0.5% Triton X-100 at room temperature for 2 hours. After blocking, the cells were incubated with antibodies specific to TNF-α, IL-1β, and interleukin 6 (IL-6) (obtained from Huabio in Hangzhou, China). This incubation was performed at a 1:200 dilution overnight at 4˚C, followed by goat anti-rabbit IgG secondary antibodies (diluted at 1:500 from Huabio in Hangzhou, China) at room temperature for 2 hours. Following an 8-min incubation with Hoechst (diluted at 1:10,000) in a dark environment, the cells were visualized and captured using a fluorescence microscope.
2.12. Statistical analysis
All statistical analyses were conducted using GraphPad Prism 9, a software tool developed by GraphPad Software in La Jolla, CA, United States. Multiple group differences were evaluated using analysis of variance. The analysis regarded a P value of <.05 as statistically significant.
3. Results
3.1. Identification of active ingredients and targets in YP
By considering both prediction scores and P values, 115 unique and effective compounds from the YP formula were identified following the screening process that removed duplicates. Among these compounds, there were 18 effective compounds from P lactiflora Pall, 39 from S miltiorrhiza Bunge, 2 from R glutinosa, 23 from A mongholicus Bunge, 17 from O japonicus, 5 from D nobile Lindl, 6 from S ningpoensis Hemsl, and 9 from L japonicus Houtt. In aggregate, these compounds were linked to 2961 target proteins, with specific numbers as follows: 142 targets for P lactiflora Pall, 770 for S miltiorrhiza Bunge, 205 for R glutinosa, 424 for A mongholicus Bunge, 193 for O japonicus, 35 for D nobile Lindl, 502 for S ningpoensis Hemsl, and 690 for L japonicus Houtt.
3.2. Identification of common targets between disease and drug
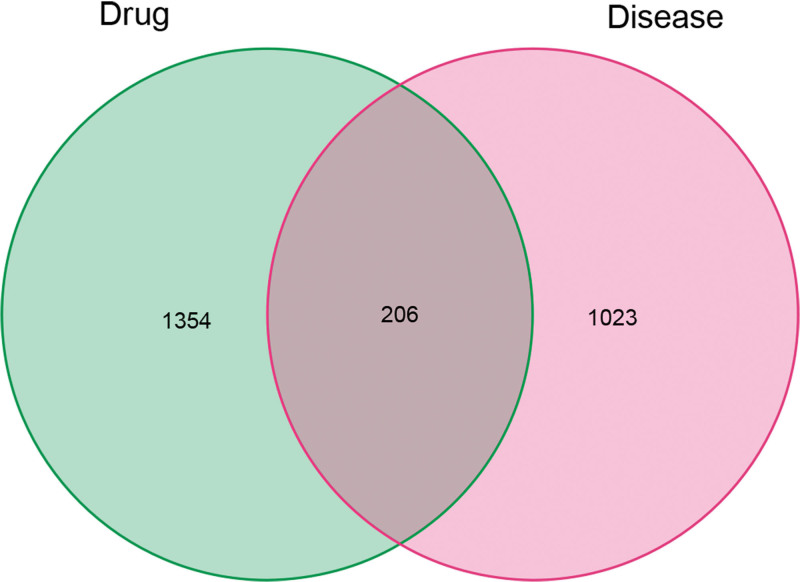
A total of 1229 disease-related targets were retrieved from the GeneCards, OMIM, and TTD databases. The corresponding targets were analyzed using an online Venn diagram tool to identify the common targets between YP and SS. This enabled the determination of intersecting targets between YP and SS (YP-SS). From this analysis, 206 overlapping targets were identified (Fig. 2).
Figure 2.
Venn diagram summarizing the intersection targets of the SS and YP. SS = Sjögren’s syndrome, YP = Yiqiyangyinquyu prescription.
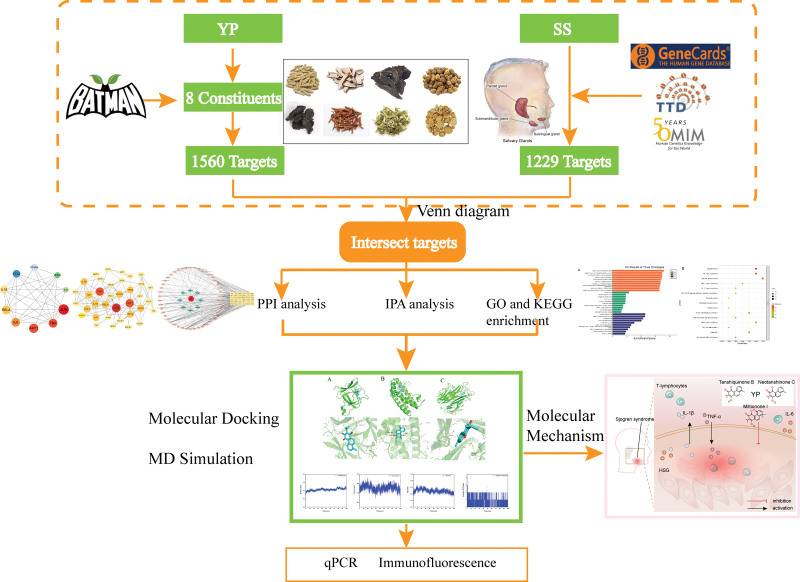
Figure 1.
The flow chart of the study.
3.3. PPI network of intersecting targets
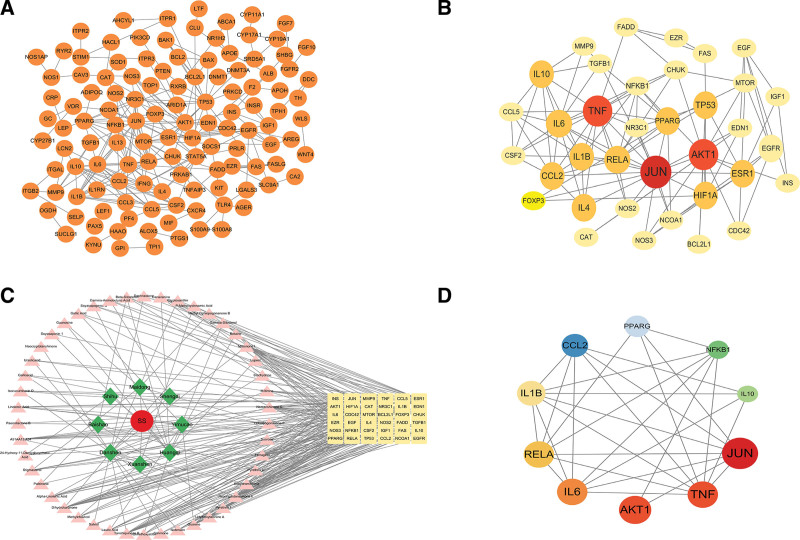
The identified intersecting targets were imported into the STRING database, where a confidence threshold of 0.9 was applied, and any disconnected nodes in the network were concealed. Through this procedure, a PPI network graph for the 206 core proteins was created (Fig. 3A). The PPI results were then integrated into Cytoscape for topological analysis, encompassing a comprehensive network of 206 nodes and 395 edges. Through CytoNCA calculations, both the ranking of core targets and the PPI network diagram for the YP and SS (YP-SS) were derived (Fig. 3B and C).
Figure 3.
PPI Network of YP-SS: The interactive PPI network was constructed using data from the STRING database, with a minimum required interaction score set at 0.9. It includes a total of 206 nodes and 395 edges. Each node corresponds to relevant targets in this network, while the edges represent protein-protein associations. These associations encompass various types, such as known interactions (indicated by azure for curated databases and purple for experimentally determined interactions), predicted interactions (highlighted in green for gene neighborhood, red for gene fusions, and blue for gene co-occurrence), as well as other interactions (shown in light green for text mining, black for co-expression, and light blue for protein homology). (B) The core PPI network of core proteins is derived from (A); it consists of 36 nodes and 114 edges. (C) The C-D-T network for YP against SS consists of 91 nodes and 262 edges. In this network, red circles represent diseases, blue diamonds represent drugs, pink triangles indicate active drug ingredients, and yellow round rectangles signify target genes. (D) The top ten genes were selected based on degree value. PPI = protein-protein interaction, YP-SS = Yiqiyangyinquyu prescription-Sjögren’s syndrome.
3.4. Cytoscape network analysis of drug-disease interactions
Based on SS-YP intersection, 46 core target chemical components were identified through a comprehensive calculation process. These include 1 in R glutinosa, 6 in P lactiflora Pall, 18 in S miltiorrhiza Bunge, 10 in A mongholicus Bunge, 4 in O japonicus, 1 in D nobile Lindl, 3 in S ningpoensis Hemsl, and 6 in L japonicus Houtt. Additionally, 1 component was found in both S miltiorrhiza Bunge and A mongholicus Bunge, 1 in P lactiflora Pall and A mongholicus Bunge, and 1 in A mongholicus Bunge and O japonicus. The top 10 active compounds are displayed in Table 2, and the top ten core genes are presented in Table 3 and Figure 3D.
Table 2.
Top ten active compounds of YP.
| Compound name | Degree | Betweenness centrality | Closeness centrality | Compound source |
|---|---|---|---|---|
| Neotanshinone C | 19 | 537.7209275 | 0.4285714 | Salvia miltiorrhiza Bunge |
| Miltionone I | 19 | 537.7209275 | 0.4285714 | S miltiorrhiza Bunge |
| Tanshiquinone B | 19 | 537.7209275 | 0.4285714 | S miltiorrhiza Bunge |
| Neocryptotanshinone Ii | 17 | 431.6127373 | 0.4205607 | S miltiorrhiza Bunge |
| Dihydrokaranone | 14 | 668.5626116 | 0.4205607 | S miltiorrhiza Bunge |
| Guanine | 12 | 485.4388433 | 0.4017857 | Leonurus japonicus Houtt |
| Ferruginol | 10 | 168.2455603 | 0.3913043 | S miltiorrhiza Bunge |
| Linolenic Acid | 7 | 340.7252137 | 0.387931 | L japonicus Houtt |
| Alpha-Linolenic Acid | 7 | 340.7252137 | 0.387931 | L japonicus Houtt |
| Canavanine | 6 | 273.2912794 | 0.3383459 | Astragalus mongholicus Bunge |
YP = Yiqiyangyinquyu prescription.
Table 3.
Top ten genes information in PPI network.
| Name | Degree | Betweenness | Closeness |
|---|---|---|---|
| JUN | 17 | 318.3767399 | 0.636363636 |
| TNF | 15 | 183.7220113 | 0.573770492 |
| AKT1 | 14 | 249.0402653 | 0.573770492 |
| IL-6 | 12 | 69.82714508 | 0.514705882 |
| RELA | 11 | 52.8468254 | 0.538461538 |
| IL-1β | 10 | 25.54900655 | 0.5 |
| CCL2 | 9 | 19.87120657 | 0.492957746 |
| PPARG | 8 | 54.59042069 | 0.514705882 |
| NFKB1 | 8 | 32.9013653 | 0.52238806 |
| IL-10 | 8 | 14.3469697 | 0.406976744 |
AKT1 = serine/threonine kinase 1, AP-1 = transcription factor subunit, CCL2 = C-C motif chemokine ligand 2, IL-10 = interleukin 1, IL-6 = interleukin 6, JUN = Jun proto-oncogene, NFKB1 = nuclear factor kappa B subunit 1, NF-κB = nuclear factor κB, IL-1β = interleukin 1 beta, PPARG = peroxisome proliferator-activated receptor gamma, PPI = protein-protein interaction, RELA = RELA proto-oncogene, TNF-α = tumor necrosis factor α.
3.5. Enrichment analysis: elucidating the functional pathways and potential anti-SS gene associations of YP
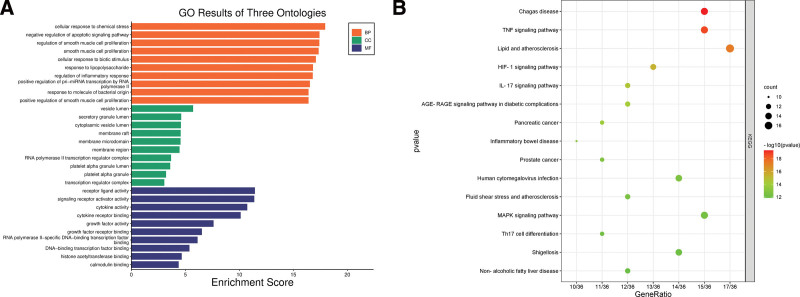
We carried out an extensive GO analysis on the 36 intersecting targets of YP, which is proposed for the treatment of SS. This analysis was performed using the R programming language. The analysis identified 3361 GO entries, which were subsequently categorized into 3 distinct groups: BPs, CCs, and MFs. In the BPs category, 3005 entries were identified. Notably, these entries encompassed cellular responses to chemical stress, negative regulation of apoptotic signaling pathways, regulation of inflammatory responses, cell proliferation, and responses to LPSs. Within the CCs category, 142 entries were primarily linked to vesicle lumen, membrane rafts, granular lumen, and membrane microdomains. The MFs category encompassed 214 entries, with a strong presence of functions related to cytokine activity, receptor-ligand activity, DNA transcription factor binding, and actin binding. Following this, we conducted a KEGG functional enrichment analysis, identifying 191 pathways. Upon visually inspecting the top 15 ranked entries, it became evident that the primary enrichment of central targets was in the TNF signaling pathway (Fig. 4). This discovery suggests that the TNF signaling pathway might play a pivotal role in the potential therapeutic effects of YP in SS treatment.
Figure 4.
GO and KEGG enrichment analysis plots. (A) The bar diagram of GO enrichment analysis of YP-SS genes, including the top 10 significant enrichment terms of biological process (BP), cellular component (CC), and molecular function (MF). (B) The bubble plot diagram of KEGG enrichment pathways (top 10). In these plots, the size of a dot corresponds to the number of genes annotated within the respective entry. A larger dot represents a higher number of genes. Additionally, the dot’s color, with a redder shade, indicates a lower q value, signifying a more significant pathway enrichment. GO = gene ontology, KEGG = Kyoto encyclopedia of genes and genomes, YP-SS = Yiqiyangyinquyu prescription-Sjögren’s syndrome.
3.6. Molecular docking analysis
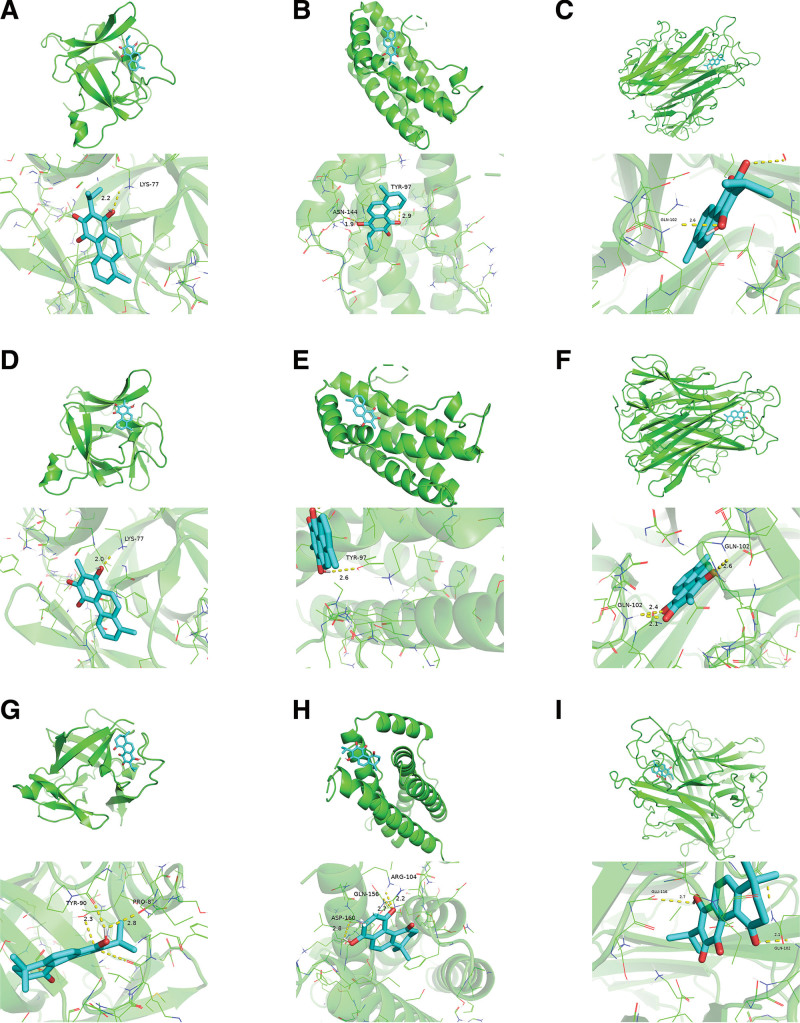
Guided by the insights from network pharmacology, the top 3 compounds—tanshiquinone B, neotanshinone C, and miltionone I—were chosen for molecular docking with the target proteins TNF-α (PDB ID: 1A8M), IL-1β (PDB ID: 1I1B), and IL-6 (PDB ID: 1ALU).
Molecular docking outcomes revealed that tanshiquinone B established a hydrogen bond with IL-1β at LYS-77 (Fig. 5A) and hydrogen bonds with IL-6 at ASN-144 and TYR-97 (Fig. 5B). Furthermore, tanshiquinone B formed a hydrogen bond with TNF-α at GLN-102 (Fig. 5C).
Figure 5.
Molecular docking results of main chemical components of YP. YP = Yiqiyangyinquyu prescription.
Neotanshinone C formed a single hydrogen bond with IL-1β at LYS-77 (Fig. 5D), 1 hydrogen bond with IL-6 at TYR-97 (Fig. 5E), and 3 hydrogen bonds with TNF-α at GLN-102 (Fig. 5F).
Miltionone I formed hydrogen bonds with IL-1β at TYR-90 and PRO-87 (Fig. 5G), as well as with IL-6 at ASP-160, GLN-156, and ARG-104 (Fig. 5H). Furthermore, miltionone I established hydrogen bonds with TNF-α at GLU-116 and GLN-102 (Fig. 5I).
A lower binding energy in molecular docking signifies a stronger affinity between the receptor and ligand, leading to a more stable conformation. Typically, binding energies below–5 kcal/mol indicate favorable binding activity between the ligand and receptor.[19] The molecular docking results revealed that all binding energies were below–5 kcal/mol, indicating a high affinity of the compound for the respective protein. Notably, neotanshinone C displayed the highest binding affinity to TNF-α, with a binding energy of–8.6 kcal/mol. In contrast, its weakest binding was observed with IL-6, where the binding energy was–6.9 kcal/mol. Further detailed information is available in Table 4. In light of these discoveries, we propose that all these compounds play a pivotal role in treating SS.
Table 4.
Component—target docking binding energy.
| Target | Target (PDB ID) | Compound | Affinity (kcal/mol) |
|---|---|---|---|
| IL-6 | 1ALU | Neotanshinone C | −6.9 |
| Tanshiquinone B | −7.4 | ||
| Miltionone I | −7.1 | ||
| TNF-α | 1A8M | Neotanshinone C | −8.6 |
| Tanshiquinone B | −7.8 | ||
| Miltionone I | −7.5 | ||
| IL-1β | 1I1B | Neotanshinone C | −7.4 |
| Tanshiquinone B | −7.6 | ||
| Miltionone I | −7.6 |
IL-1β = interleukin 1 beta, IL-6 = interleukin 6, TNF-α = tumor necrosis factor.
3.7. MD simulation
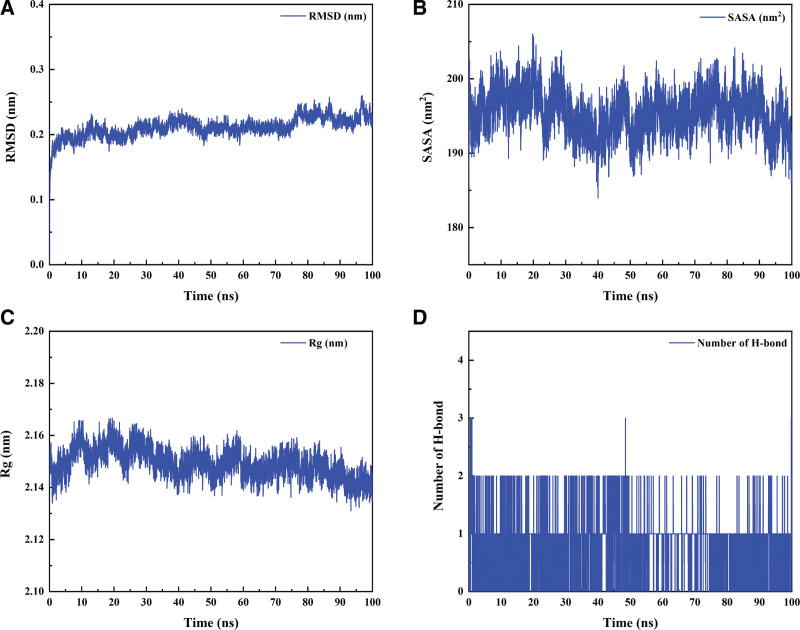
MD simulations have been widely employed to assess the structural features of protein-ligand systems and to study the binding stability between proteins and molecules. In this study, we selected TNF-α with neotanshinone C, which had the lowest binding energy in molecular docking, for further analysis of binding stability. Analysis of the MD simulation results showed that the protein ligand reached equilibrium after 5 ns, indicating that the whole simulation process was stable and reliable (Fig. 6A). The solvent-accessible surface areas (SASA) were calculated using Van der Waals energy and solvent molecules; consequently, the changes in SASA could predict the changes in protein structure. The SASA values of proteins in protein-ligand complexes exhibited no significant changes during all complex simulations, indicating that the protein ligands were tightly bound (Fig. 6B). The radius of gyration (Rg) was used to demonstrate the tightness of the protein structure during the simulations, and Figure 6C shows that the Rg values showed no significant changes. To explore the interaction between proteins and ligands, we performed a hydrogen bonding analysis of the protein-ligands. As indicated in Figure 6D, the average distribution of the number of hydrogen bonds between the proteins and small molecules was 0.844, suggesting the existence of hydrogen-bonding interactions between the proteins and the small molecules. Finally, we determined the binding energies of all protein-ligand complexes at equilibrium using the gmx_mmpbsa (https://jerkwin.github.io/gmxtools/) script. The binding energies of proteins and ligands are listed in Table 5. In the protein-ligand complex system, the free energy of protein-small molecule binding is −71.112 kJ/mol, indicating that protein-small molecule binding is stable and that the main interaction energy is Van der Waals interaction. Overall, MD simulation results are consistent with molecular docking results.
Fig.6.
Root-mean-square deviation (A), the solvent-accessible surface areas (B), radius of gyration (C) and the number of hydrogen bonds (D) over time during protein-ligand complex simulations.
Table 5.
Protein ligand MM-PBSA analysis.
| Energy | Complex |
|---|---|
| Van der Waals energy (KJ/mol) | −156.194 |
| Electrostatic energy (kJ/mol) | −56.908 |
| Polar solvation energy (KJ/mol) | 158.597 |
| Nonpolar solvation energy (KJ/mol) | −16.608 |
| Total binding energy (KJ/mol) | −71.112 |
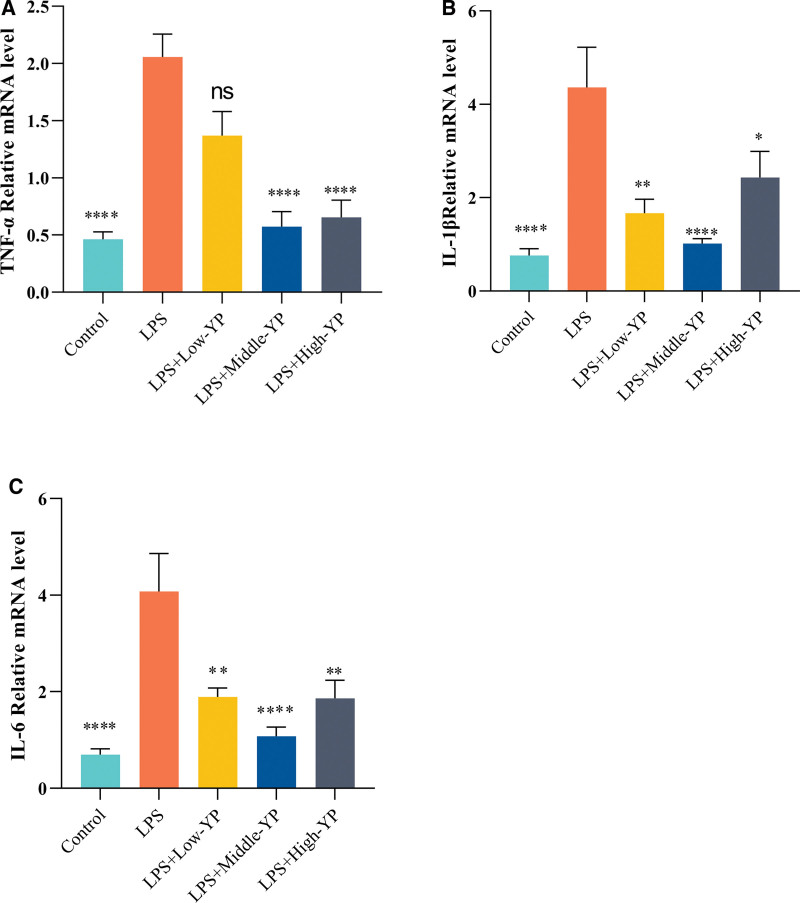
3.8. To verify the levels of TNF-α, IL-1β, and IL-6 mRNA levels in HSG cells
To examine the effect of YP on HSG cells, we assessed the expression levels of TNF-α, IL-1β, and IL-6 in HSG cells using RT-qPCR. TNF-α, IL-1β, and IL-6 expression in HSG cells exhibited an increase following LPS induction (Fig. 7). Upon the inhibition of TNF-α, IL-1β, and IL-6 expression in HSG cells after YP intervention, it was observed that the medium concentration appeared to have a more favorable impact among the 3 groups. This result suggested that YP can effectively inhibit LPS-induced high expression of inflammatory factors in HSG cells.
Figure 7.
TNF-α, IL-1β, and IL-6 expression levels in different subgroups of YP concentration. (A) Following YP treatment, the level of TNF-α was lower than that in the model group. This effect was most significant at medium doses (B). Following YP treatment, the level of IL-1β was lower than that in the model group. This effect was most prominent at medium doses (C). After YP treatment, the level of IL-6 was lower than that in the model group, and this effect was most notable at medium doses. P < .0001; ****, P < .0001; ***, P < .0005; **, P < .005; *, P < .05. IL-1β = interleukin 1 beta, IL-6 = interleukin 6, TNF-α = tumor necrosis factor, YP = Yiqiyangyinquyu prescription.
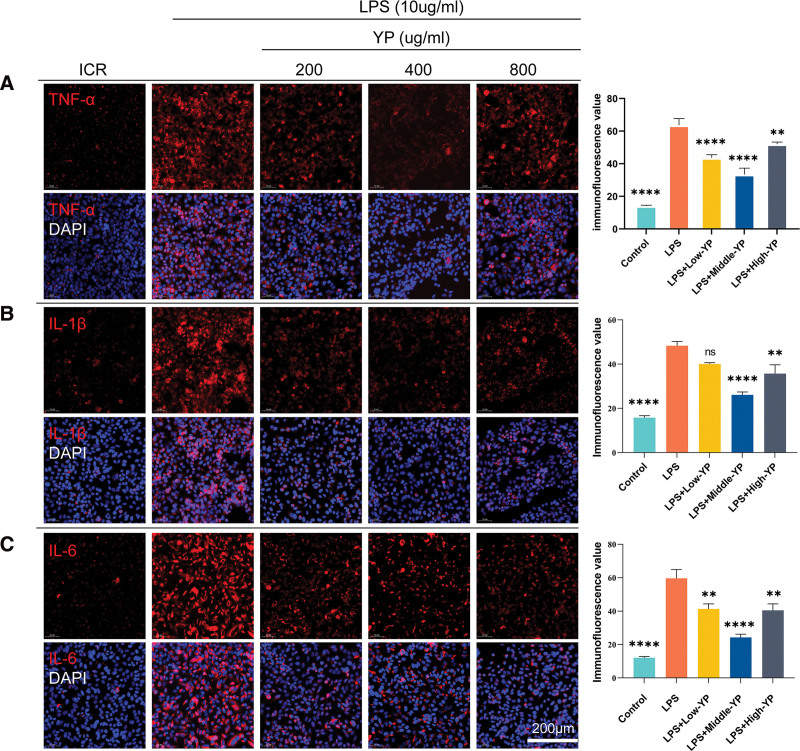
3.9. YP inhibits LPS-induced inflammatory expression in HSG cells
Given that YP significantly suppressed the expression of inflammatory factors in RT-qPCR induced by LPS. Immunofluorescence assays yielded consistent results (Fig. 8). Following 24 hours of LPS stimulation, the expression of TNF-α (Fig. 8A), IL-1β (Fig. 8B), and IL-6 (Fig. 8C) was notably elevated in the model group. In contrast, the administration of YP in the LPS + Middle-YP group exhibited a significantly more effective inhibitory effect than the other 2 drug concentrations. We also observed IL-1β expression in the LPS + Low-YP group; although there was a trend toward inhibition, there was no significant difference between the 2 groups compared to the LPS group (Fig. 8B).
Figure 8.
YP suppresses LPS-induced inflammatory expression in HSG cells. HSG cells were pretreated with LPS (10 µg/mL) or not for 2 h and subsequently incubated with varying concentrations of YP for 24 h. Immunofluorescence was used to evaluate the expression of TNF-α, IL-1β, and IL-6. The cells were subjected to immunostaining using CY3-labeled antibodies specific to TNF-α, IL-1β, and IL-6. Simultaneously, the cell nuclei were stained blue with DAPI. (A) The levels of TNF-α were reduced after YP treatment compared to the model group, and moderate doses had a significant inhibitory effect on the release of inflammatory factors (B). The levels of IL-1β were decreased after YP treatment in comparison to the model group. Moderate doses of YP notably suppressed the release of inflammatory factors, as observed in 3 representative independent experiments. (C) The levels of IL-6 were diminished after YP treatment compared to the model group, and moderate doses significantly inhibited the release of inflammatory factors. ****, P < .0001; ***, P < .0005; **, P < .005; *, P < .05; ns, no statistically significant difference (P ≥ .05). HSG = Human salivary gland, IL-1β = interleukin 1 beta, IL-6 = interleukin 6, LPS = lipopolysaccharide, TNF-α = tumor necrosis factor, YP = Yiqiyangyinquyu prescription.
4. Discussion
SS ranks as the second most common autoimmune disease, following rheumatoid arthritis (RA), and impacts about 1% of the population.[20] Recent research has highlighted the crucial role of salivary gland epithelial cells (SGECs) in SS as significant mediators of immune response initiation and propagation. When stimulated by antigens, SGECs release proinflammatory factors like IFN, TNF-α, IL-6, and interleukin 17 (IL-17). This, in turn, amplifies subsequent inflammation. Additionally, IFN-γ prompts SGECs to discharge alarmins into the extracellular environment, perpetuating a harmful auto-inflammatory cycle contributing to local and systemic damage.[21,22]
Current clinical treatments for SS predominantly center around alleviating symptoms and managing organ damage resulting from immune responses. These approaches involve using artificial tears, artificial saliva, and eye lubricants to ease dry eyes and mouth. Immunosuppressive or immunomodulatory therapies, including medications such as hydroxychloroquine, methotrexate, sulfasalazine, and leflunomide, are commonly employed. Furthermore, biological agents such as rituximab and belimumab have shown clinical effectiveness.[20,23] However, the aforementioned pharmacological treatments have their limitations. They can lead to adverse effects, including but not limited to anaphylaxis (characterized by urticaria and angioedema), excessive sweating, retinopathy, and acute and chronic myocardial adverse reactions.[24–26] Recently, there has been growing interest in TCM interventions. These approaches are noted for their reduced side effects and reliable efficacy. Wei[27] demonstrated TCM has demonstrated its potential to effectively treat SS by upregulating aquaporins, inhibiting apoptosis, and suppressing the abnormal activation of B and T lymphocytes.
Extensive clinical practice has demonstrated that YP effectively alleviates patients’ dry mouth and dry eye symptoms, providing a novel therapeutic approach for managing SS. The mode of action of YP is multifaceted, involving various components and targets. Contemporary pharmacological research suggests that P lactiflora Pall, one of the constituents of YP, exhibits anti-inflammatory, analgesic, and immunomodulatory properties. It is widely used to treat autoimmune diseases, including RA, systemic lupus erythematosus, SS, and psoriasis.[28] On the other hand, S miltiorrhiza Bunge can reduce the production of reactive oxygen species and effectively protect against endothelial dysfunction. When combined with A mongholicus Bunge, S miltiorrhiza Bunge can activate the PI3K/AKT signaling pathway and inhibit the TLR4/nuclear factor κB signaling pathway, resulting in significant anti-inflammatory and antioxidant effects.[29,30] In this study, we identified 46 chemical constituents from the BATMAN-TCM database, including compounds such as neotanshinone C, tanshiquinone B, miltionone I, neocryptotanshinone II, and dihydrokaranone. It has been observed that the main chemical components can interact with other active compounds on multiple targets within the network.[31] Within the predicted active ingredients, neotanshinone C and miltionone I may influence BPs related to inflammatory responses and chemokine metabolism.[32] Neocryptotanshinone II, a diterpenoid derived from S miltiorrhiza Bunge, is known for its potent anti-inflammatory properties. It targets explicitly crucial molecules such as TNF-α and IL-6.[33]
We collected 206 shared targets for SS and YP from the GeneCard, OMIM, and TTD databases. The construction of a PPI network indicated that TNF-α, IL-1β, and IL-6 may be the core targets for addressing inflammation. Recent studies have shown that inflammatory factors like TNF-α, IL-1β, and IL-6 are significantly more expressed in SS patients’ tears than in healthy individuals.[34–37] TNF-α, a classic inflammatory factor, predominantly signals through 2 receptors, TNF-R1 and TNF-R2, and regulates inflammation, antitumor responses, and immune system homeostasis.[38] Antagonists of TNF-α have demonstrated significant effectiveness in the treatment of autoinflammatory diseases.[39]
Additionally, TNF-α can activate various inflammatory pathways and stimulate the secretion of IL-6.[36] IL-6, a pivotal proinflammatory factor, plays a significant role in developing chronic inflammatory and autoimmune diseases and regulates the balance between Th17 and Treg cells.[40] Elevated levels of IL-6 can lead to damage in SS monocytes and enhance the activation of the epithelial-mesenchymal transition program in SGECs, thereby worsening the progression of the disease.[41,42] IL-1β, a significant inflammatory mediator in the IL-1 family, can be triggered by the NLRP3 inflammasome and plays a vital role in the host’s defense against infections and injuries.[37,43] Moreover, IL-1β and TNF-α can increase the expression of IL-33, another member of the IL-1 family. This contributes to the initiation of an inflammatory cycle and plays a role in the pathogenesis of SS through the activation of the IL-33/ST2 pathway.[44]
Based on the hypothesis and the integration of GO and KEGG enrichment analysis, it can be postulated that the mechanism of YP in SS primarily involves signaling pathways like TNF, IL-17, hypoxia-inducible factor-1 (HIF-1), AGE-RAGE, and others, with critical targets such as TNF-α, IL-1β, IL-6, and processes including apoptosis, proliferation, inflammatory response, and LPS response. TNF and IL-17 are indeed prototypical inflammatory pathways that have been implicated in the pathogenesis of various autoinflammatory diseases, including psoriasis, SS, inflammatory bowel disease, ankylosing spondylitis, RA, and systemic lupus erythematosus. These pathways play a significant role in the inflammatory processes and immune dysregulation associated with these conditions.[45–47] Indeed, IL-17 has been shown to impair salivary gland function by affecting calcium movement.[48] It has been linked to promoting salivary gland inflammation and cell death in conditions like SS,[49] which can activate nuclear factor κB. This transcription factor plays a leading role in regulating the expression of various proinflammatory cytokines (TNF-α, IL-1β, IL-6, and C-C motif chemokine ligand 2).[42]
The AGE-RAGE pathway indeed plays a significant role in inflammation and autoimmune diseases. AGEs can interact with the RAGE to trigger a cascade of events leading to the upregulation of adhesion molecules, chemokines, proinflammatory cytokines, and other inflammatory factors. This perpetuates a proinflammatory cycle associated with chronic inflammation in autoimmune diseases, further contributing to tissue damage and disease progressionc.[50,51]
HIF-1 signaling pathway is key in regulating various immune components and inflammation. It modulates the function of T cells, neutrophils, dendritic cells, and other immune cells within the immune system.[52] One of its essential functions is its involvement in promoting the development of Th17 cells, a subset of T helper cells known for their proinflammatory properties.[53] Additionally, HIF-1 plays a critical role in processes such as B cell activation, macrophage polarization, and oxidative stress.[54–57] Based on these findings, we speculate that YP may exert its mechanism of action by mediating the inflammatory response.
In our study, we conducted molecular docking analysis to explore the potential molecular mechanisms of YP in treating SS. We focused on 3 critical targets: TNF-α, IL-1β, and IL-6, which are involved in the inflammatory processes of the disease. We also analyzed the interactions of 3 active YP components, neotanshinone C, tanshiquinone B, and miltionone I, with these targets. The results suggest that these components may play a role in mitigating inflammation in SS, which is a key aspect of the disease’s pathogenesis. Our findings demonstrated effective binding between the 3 compounds and the target proteins, with neotanshinone C showing the most stable interaction, particularly with TNF-α. This suggests a potential mechanism by which YP may exert its therapeutic effects in treating SS by modulating the activity of these inflammatory proteins. Following in vitro experiments, we validated the efficacy of YP in significantly reducing the elevated levels of TNF-α, IL-1β, and IL-6 in HSG cells through various methods, including RT-qPCR and immunofluorescence assays. This supports the potential anti-inflammatory action of YP in the context of SS. This study has certain limitations, such as the need for scientific validation of the accuracy and currency of database data, the limited presence of neotanshinone C, tanshiquinone B, and miltionone I in previous studies, and the requirement for pharmacodynamic experiments and molecular biology investigations to substantiate the research findings.
5. Conclusion
This study utilized network pharmacology, molecular docking techniques, and in vitro experimental validation to preliminarily investigate YP’s primary components and potential mechanisms in treating SS. The findings suggest that neotanshinone C, tanshiquinone B, and miltionone I may be the key active constituents of YP. Concurrently, the network’s TNF-α, IL-6, and IL-1β could serve as potential therapeutic targets. These targets may exert their effects through the IL-17 signaling pathway, TNF signaling pathway, HIF-1 signaling pathway, and AGE-RAGE signaling pathway.
Author contributions
Conceptualization: Xin-Chang Wang.
Data curation: Tao Hong, Ding-Qi Lu.
Formal analysis: Ya-Ting Ren.
Investigation: Xin-Yi Yao.
Methodology: Ya-Ting Ren, Xin-Chang Wang.
Project administration: Yi-Han Wang.
Resources: Wu Chen, Yi-Han Wang.
Software: Wu Chen, Kai-Yuan Zhang.
Supervision: Xin-Chang Wang.
Validation: Ding-Qi Lu.
Writing – original draft: Tao Hong.
Abbreviations:
- AGE
- advanced glycation end products
- HIF-1
- hypoxia-inducible factor-1
- HSG
- human salivary gland
- IFN
- interferon
- IL-17
- interleukin 17
- IL-6
- interleukin 6
- LPS
- lipopolysaccharide
- MD
- molecular dynamics
- OMIM
- Online Mendelian Inheritance in Man
- RA
- rheumatoid arthritis
- RAGE
- receptor for AGEs
- RT-qPCR
- reverse transcription-quantitative polymerase chain reaction
- SASA
- solvent-accessible surface areas
- SGECs
- salivary gland epithelial cells
- SS
- Sjögren’s syndrome
- TNF-α
- tumor necrosis factor
- YP
- Yiqiyangyinquyu prescription
This research was funded by the Research Fund of Zhejiang Chinese Medical University, grant number 2022YKJ05; the National Natural Science Foundation of China, grant number 82074341.
The authors have no conflicts of interest to disclose.
This study was approved by an ethics review board, and the requirement to obtain informed written consent was waived.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Hong T, Chen W, Ren Y-T, Wang Y-H, Lu D-Q, Zhang K-Y, Yao X-Y, Wang X-C. Network pharmacology identifies the inhibitory effect of Yiqiyangyinquyu prescription on salivary gland inflammation in Sjögren’s syndrome. Medicine 2023;102:47(e36144).
Contributor Information
Tao Hong, Email: 799815826@qq.com.
Wu Chen, Email: chenwzm1007@163.com.
Ya-Ting Ren, Email: renjunor@163.com.
Yi-Han Wang, Email: ossani@126.com.
Ding-Qi Lu, Email: ludingqi745@163.com.
Kai-Yuan Zhang, Email: 202111120611045@zcmu.edu.cn.
Xin-Yi Yao, Email: xy18868113715@163.com.
References
- [1].Jonsson R, Brokstad KA, Jonsson MV, et al. Current concepts on Sjögren’s syndrome – classification criteria and biomarkers. Eur J Oral Sci. 2018;126 (Suppl 1):37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stefanski AL, Tomiak C, Pleyer U, et al. The diagnosis and treatment of Sjögren’s syndrome. Dtsch Arztebl Int. 2017;114:354–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mavragani CP, Moutsopoulos HM. Sjögren’s syndrome: old and new therapeutic targets. J Autoimmun. 2020;110:102364. [DOI] [PubMed] [Google Scholar]
- [4].Wang X, Tang X, Huang S, et al. Effect of Yiqi Yangyin Quyu method on serum IgG and BAFF in patients with Sjogren’s syndrome. J Zhejiang Chin Med Univ. 2013;37:223–5 + 33. [Google Scholar]
- [5].Wang X, Cao L, Fan Y. Discussion of the etiology and pathogenesis of traditional Chinese medicine of Sjogren’s syndrome. J Zhejiang Chin Med Univ. 2011;35:643–4. [Google Scholar]
- [6].Li Z, Wang X, Sun H, et al. Diagnosis and treatment of 120 cases of Sjogren’s syndrome by Tonifying Qi, nourishing Yin, and resolving blood stasis method. J Shandong Univ Tradit Chin Med. 2017;41:231–4. [Google Scholar]
- [7].Ling C, Qing Y, Sun H, et al. Effects of Yiqi Yangyin Quyu formula on TLR-IFN-BAFF signaling pathway in patients with primary Sjogren syndrome. China J Tradit Chin Med Pharm. 2018;33:3706–9. [Google Scholar]
- [8].Wang X, Xie Z, Wen C, et al. Influence of medicine of notifying Qi to nourish Yin and remove stasis on sexual hormone of dry syndrome. J Zhejiang Chin Med Univ. 2009;33:48–9 + 52. [Google Scholar]
- [9].Ling C, Huang S, Qing Y, et al. Effect of Chinese herbs for Qi benefiting, Yin nourishing, stasis removing on TLR-IFN-BAFF signaling pathway in submaxillary gland of NOD mice with Sj(o)gren’s syndrome. Chin J Integr Tradit West Med. 2017;37:1487–90. [Google Scholar]
- [10].Zhang L, Han L, Wang X, et al. Exploring the mechanisms underlying the therapeutic effect of Salvia miltiorrhiza in diabetic nephropathy using network pharmacology and molecular docking. Biosci Rep. 2021;41:BSR20203520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci Rep. 2016;6:21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bai LL, Chen H, Zhou P, et al. Identification of tumor necrosis factor-alpha (TNF-α) inhibitor in rheumatoid arthritis using network pharmacology and molecular docking. Front Pharmacol. 2021;12:690118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48:D1031–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jia A, Xu L, Wang Y. Venn diagrams in bioinformatics. Brief Bioinform. 2021;22:bbab108. [DOI] [PubMed] [Google Scholar]
- [15].Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Burley SK, Berman HM, Kleywegt GJ, et al. Protein Data Bank (PDB): the single global macromolecular structure archive. Methods Mol Biol. 2017;1607:627–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim S, Chen J, Cheng T, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu J, Liu J, Tong X, et al. Network pharmacology prediction and molecular docking-based strategy to discover the potential pharmacological mechanism of Huai Hua San against ulcerative colitis. Drug Des Devel Ther. 2021;15:3255–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li X, Wei S, Niu S, et al. Network pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Huanglian Jiedu decoction against sepsis. Comput Biol Med. 2022;144:105389. [DOI] [PubMed] [Google Scholar]
- [20].Vivino FB, Bunya VY, Massaro-Giordano G, et al. Sjogren’s syndrome: an update on disease pathogenesis, clinical manifestations, and treatment. Clin Immunol. 2019;203:81–121. [DOI] [PubMed] [Google Scholar]
- [21].Sarrand J, Baglione L, Parisis D, et al. The involvement of alarmins in the pathogenesis of Sjögren’s syndrome. Int J Mol Sci. 2022;23:5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tian Y, Yang H, Liu N, et al. Advances in pathogenesis of Sjögren’s syndrome. J Immunol Res. 2021;2021:5928232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fox RI, Fox CM, Gottenberg JE, et al. Treatment of Sjögren’s syndrome: current therapy and future directions. Rheumatology (Oxford). 2021;60:2066–74. [DOI] [PubMed] [Google Scholar]
- [24].Karmacharya P, Poudel DR, Pathak R, et al. Rituximab-induced serum sickness: a systematic review. Semin Arthritis Rheum. 2015;45:334–40. [DOI] [PubMed] [Google Scholar]
- [25].Noaiseh G, Baker JF, Vivino FB. Comparison of the discontinuation rates and side-effect profiles of pilocarpine and cevimeline for xerostomia in primary Sjögren’s syndrome. Clin Exp Rheumatol. 2014;32:575–7. [PubMed] [Google Scholar]
- [26].Nirk EL, Reggiori F, Mauthe M. Hydroxychloroquine in rheumatic autoimmune disorders and beyond. EMBO Mol Med. 2020;12:e12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wei SJ, He QM, Zhang Q, et al. Traditional Chinese medicine is a useful and promising alternative strategy for treatment of Sjogren’s syndrome: a review. J Integr Med. 2021;19:191–202. [DOI] [PubMed] [Google Scholar]
- [28].Zhang L, Wei W. Anti-inflammatory and immunoregulatory effects of paeoniflorin and total glucosides of paeony. Pharmacol Ther. 2020;207:107452. [DOI] [PubMed] [Google Scholar]
- [29].Wang N, Zhang X, Ma Z, et al. Combination of tanshinone IIA and astragaloside IV attenuate atherosclerotic plaque vulnerability in ApoE(-/-) mice by activating PI3K/AKT signaling and suppressing TRL4/NF-κB signaling. Biomed Pharmacother. 2020;123:109729. [DOI] [PubMed] [Google Scholar]
- [30].Cheng YC, Hung IL, Liao YN, et al. Salvia miltiorrhiza protects endothelial dysfunction against mitochondrial oxidative stress. Life (Basel). 2021;11:1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li S, Wang N, Hong M, et al. Hepatoprotective effects of a functional formula of three chinese medicinal herbs: experimental evidence and network pharmacology-based identification of mechanism of action and potential bioactive components. Molecules. 2018;23:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zeng P, Liu W, Zhang S, et al. Integrated network pharmacology and mice model to investigate Qing Zao Fang for treating Sjögren’s syndrome. Evid Based Complement Alternat Med. 2022;2022:3690016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang F, Liu Y, Zheng S, et al. Pharmacological network reveals the active mechanism of qi-replenishing, spleen-strengthening, phlegm-dispelling, and blood-nourishing Fufang on coronary heart disease. Evid Based Complement Alternat Med. 2020;2020:1062325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao H, Li Q, Ye M, et al. Tear luminex analysis in dry eye patients. Med Sci Monit. 2018;24:7595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee SY, Han SJ, Nam SM, et al. Analysis of tear cytokines and clinical correlations in Sjögren syndrome dry eye patients and non-Sjögren syndrome dry eye patients. Am J Ophthalmol. 2013;156:247–253.e1. [DOI] [PubMed] [Google Scholar]
- [36].Wang T, He C. Proinflammatory cytokines: the link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 2018;44:38–50. [DOI] [PubMed] [Google Scholar]
- [37].Kim SK, Choe JY, Lee GH. Enhanced expression of NLRP3 inflammasome-related inflammation in peripheral blood mononuclear cells in Sjögren’s syndrome. Clin Chim Acta. 2017;474:147–54. [DOI] [PubMed] [Google Scholar]
- [38].Mehta AK, Gracias DT, Croft M. TNF activity and T cells. Cytokine. 2018;101:14–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. 2019;20:6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hirano T. IL-6 in inflammation, autoimmunity and cancer. Int Immunol. 2021;33:127–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yoshimoto K, Tanaka M, Kojima M, et al. Regulatory mechanisms for the production of BAFF and IL-6 are impaired in monocytes of patients of primary Sjögren’s syndrome. Arthritis Res Ther. 2011;13:R170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sisto M, Tamma R, Ribatti D, et al. IL-6 contributes to the TGF-β1-mediated epithelial to mesenchymal transition in human salivary gland epithelial cells. Arch Immunol Ther Exp (Warsz). 2020;68:27. [DOI] [PubMed] [Google Scholar]
- [43].Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011;22:189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Soyfoo MS, Nicaise C. Pathophysiologic role of Interleukin-33/ST2 in Sjögren’s syndrome. Autoimmun Rev. 2021;20:102756. [DOI] [PubMed] [Google Scholar]
- [45].Schinocca C, Rizzo C, Fasano S, et al. Role of the IL-23/IL-17 pathway in rheumatic diseases: an overview. Front Immunol. 2021;12:637829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149–60. [DOI] [PubMed] [Google Scholar]
- [47].Kamiya K, Kishimoto M, Sugai J, et al. Risk factors for the development of psoriasis. Int J Mol Sci. 2019;20:4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Xiao F, Du W, Zhu X, et al. IL-17 drives salivary gland dysfunction via inhibiting TRPC1-mediated calcium movement in Sjögren’s syndrome. Clin Transl Immunology. 2021;10:e1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hwang SH, Woo JS, Moon J, et al. IL-17 and CCR9(+)α4β7(-) Th17 cells promote salivary gland inflammation, dysfunction, and cell death in Sjögren’s syndrome. Front Immunol. 2021;12:721453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Du C, Whiddett RO, Buckle I, et al. Advanced glycation end products and inflammation in type 1 diabetes development. Cells. 2022;11:3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yu W, Tao M, Zhao Y, et al. 4’-Methoxyresveratrol alleviated AGE-induced inflammation via RAGE-mediated NF-κB and NLRP3 inflammasome pathway. Molecules. 2018;23:1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mcgettrick AF, O’Neill LAJ. The role of HIF in immunity and inflammation. Cell Metab. 2020;32:524–36. [DOI] [PubMed] [Google Scholar]
- [53].Dang EV, Barbi J, Yang HY, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dasgupta S, Dasgupta S, Bandyopadhyay M. Regulatory B cells in infection, inflammation, and autoimmunity. Cell Immunol. 2020;352:104076. [DOI] [PubMed] [Google Scholar]
- [55].Islam SMT, Won J, Khan M, et al. Hypoxia-inducible factor-1 drives divergent immunomodulatory functions in the pathogenesis of autoimmune diseases. Immunology. 2021;164:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Liu R, Li X, Ma H, et al. Spermidine endows macrophages anti-inflammatory properties by inducing mitochondrial superoxide-dependent AMPK activation, Hif-1α upregulation and autophagy. Free Radic Biol Med. 2020;161:339–50. [DOI] [PubMed] [Google Scholar]
- [57].Quijano C, Trujillo M, Castro L, et al. Interplay between oxidant species and energy metabolism. Redox Biol. 2016;8:28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]