Abstract
PURPOSE
Triple-negative breast cancer (TNBC) is a heterogeneous disease. We previously showed that homologous recombination deficiency (HRD) and the DNA damage immune response (DDIR) signature are prognostic in TNBC. We hypothesized that these biomarkers reflect related but not completely interdependent biological processes, that their combined use would be prognostic, and that simultaneous assessment of the immunologic microenvironment and susceptibility to DNA damaging therapies might be able to identify subgroups with distinct therapeutic vulnerabilities.
METHODS
We analyzed the dual DDIR/HRD classification in 341 patients with TNBC treated with adjuvant anthracycline-based chemotherapy on the SWOG S9313 trial and corroborated our findings in The Cancer Genome Atlas breast cancer data set.
RESULTS
DDIR/HRD classification is highly prognostic in TNBC and identifies biologically and immunologically distinct subgroups. Immune-enriched DDIR+/HRD+ TNBCs have the most favorable prognosis, and DDIR+/HRD– and DDIR–/HRD+ TNBCs have favorable intermediate prognosis, despite the latter being immune-depleted. DDIR–/HRD– TNBCs have the worst prognosis and represent an internally heterogeneous group of immune-depleted chemoresistant tumors.
CONCLUSION
Our findings propose DDIR/HRD classification as a potentially clinically relevant approach to categorize tumors on the basis of therapeutic vulnerabilities.
Dual classification of #TNBC by HRD and DDIR is prognostic and may identify specific therapeutic vulnerabilities.
INTRODUCTION
Triple-negative breast cancer (TNBC) accounts for 10%-15% of all breast cancers in the United States and is characterized by the lack of expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2).1-4 At the molecular level, TNBC is a heterogeneous disease.
CONTEXT
Key Objective
Triple-negative breast cancer (TNBC) is a heterogeneous disease. Multiple prognostic biomarkers have been identified and several classification systems have been described, although none of these are able to predict response to discrete therapeutic agents. This study examines the association between homologous recombination deficiency (HRD) and the DNA damage immune response (DDIR) signature, and dual classification of TNBCs by DDIR and HRD as prognostic and potentially predictive biomarkers.
Knowledge Generated
We show that HRD is positively associated with the DDIR gene expression signature and that dual classification of TNBCs by DDIR and HRD define favorable and unfavorable prognostic groups with differential chemosensitivity and different immune microenvironment features that may reflect differential susceptibility to or benefit from immunotherapy.
Relevance
Dual classification of TNBC by DDIR and HRD may be useful in individualizing systemic therapy based on therapeutic vulnerabilities.
Genetic or epigenetic inactivation of the homologous recombination (HR)/Fanconi anemia (FA) DNA repair pathway is observed in more than half of TNBCs. Inactivation of this pathway results in defective repair of DNA double-strand breaks (DSBs) and accumulation of stable genomic scars from low-fidelity repair of DSBs by nonhomologous end joining.5-7 The homologous recombination deficiency (HRD) phenotype predicts for hypersensitivity to DNA-damaging chemotherapy, poly(ADP) ribose polymerase (PARP) inhibitors, and ionizing radiation.8 We and others have shown that HRD is prognostic in patients with TNBC treated with DNA-damaging chemotherapy.9,10
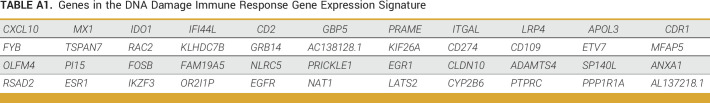
Defective DSB repair in HRD+ cells results in the formation of cytosolic micronuclei. When these micronuclei rupture, double-stranded DNA (dsDNA) activates cyclic GMP-AMP synthase (cGAS), resulting in the synthesis of 2′3′-cGAMP, activation of stimulator of interferon genes (STING), and induction of a type I interferon response.11 The 44-gene DDIR gene expression signature (Appendix Table A1) reflects activation of the cGAS-STING pathway,12 and we and others have previously shown that DDIR is prognostic in patients with TNBC treated with chemotherapy.13,14 DDIR has also been shown to be associated with upregulation of immune checkpoint blockade (ICB) therapy targets including PD-L1,14 suggesting that it may be a useful marker of susceptibility to immunotherapy.
Neoadjuvant chemoimmunotherapy is the current standard of care for stage II-III TNBC.15,16 Predictive biomarkers that align biologically with therapeutic vulnerabilities could enable individualized treatment approaches in TNBC. We hypothesized that HRD would be associated with DDIR and that combined use of these biomarkers could enable identification of immune-enriched and immune-depleted prognostic groups that could be further differentiated based on susceptibility to DNA-damaging chemotherapy. To test this, we determined the HRD status using the genomic instability (GI) score (Myriad Genetics, Salt Lake City, UT) and DDIR status (Almac Diagnostic Services, Northern Ireland) in a cohort of 341 early-stage TNBC cases treated with uniform adjuvant doxorubicin/cyclophosphamide (AC) on the SWOG S9313 protocol and correlated HRD and DDIR status with stromal tumor-infiltrating lymphocyte (sTIL) infiltration, leukocyte type(s), and survival outcomes. We also corroborated our findings in patients with TNBC within The Cancer Genome Atlas (TCGA) data set.
METHODS
Details of molecular and statistical analysis are provided in the Data Supplement (Supplemental Methods).
S9313 TNBC Patients
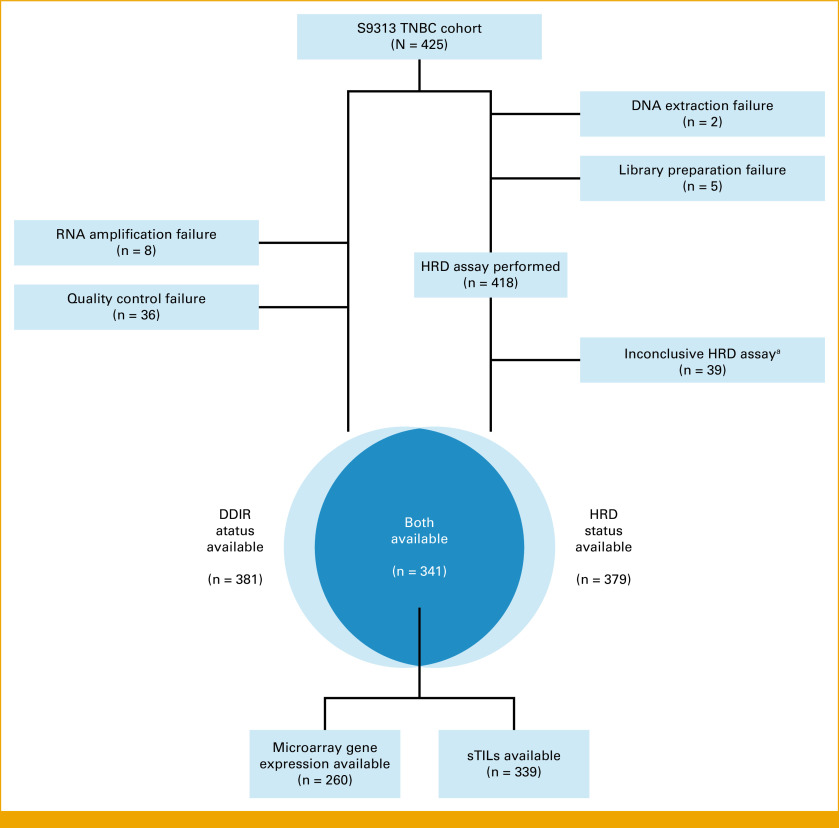
Patient selection, signature performance, and data analysis are reported according to Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria.17 The S9313 TNBC study cohort has been described previously,9,13 additional details are provided in the Data Supplement (Supplemental Methods), and the final subset of patients used in this study is described in Appendix Figure A1.
TCGA TNBC Patients
We selected a cohort of 192 TNBC samples that have been previously described.18 Molecular data for these tumors were downloaded from cBioportal19 and analyzed as described in the Data Supplement (Supplemental Methods).
RESULTS
Identification of the Study Population
We previously evaluated DDIR and HRD in the SWOG S9313 adjuvant chemotherapy trial.9,13 Among the 425 patients with centrally confirmed TNBC in S9313, we were able to determine the DDIR status for 381/425 (89.6%) patients and the HRD status for 379/425 (89.2%) patients (Appendix Fig A1). Both biomarkers were available for the 341/425 (80.2%) patients who comprise the final analysis cohort for this study. There is no difference in disease-free survival (DFS) or overall survival (OS) in patients in whom DDIR and HRD status are known and unknown.9,13 Findings were corroborated in a cohort of patients with TNBC within the TCGA data set.
Patient Demographics
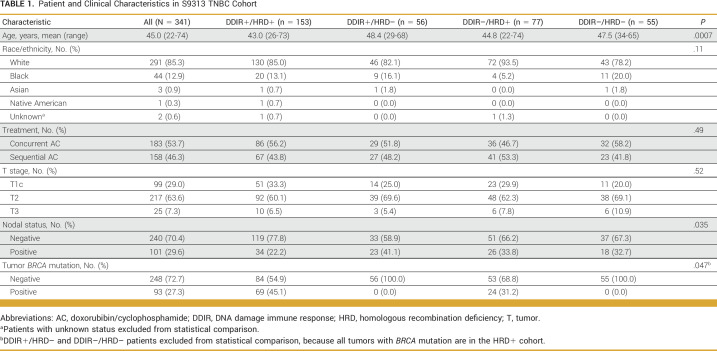
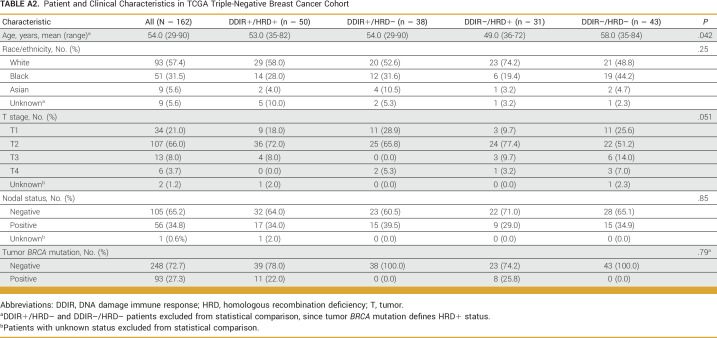
Demographic and clinical characteristics of the 341 patients with TNBC in the S9313 cohort are shown in Table 1. At a median follow-up of 12.6 years, there have been 133 DFS and 103 OS events. Median follow-up for the TCGA cohort was 24.6 months. Appendix Table A2 provides demographic characteristics of the TCGA cohort (N = 162). Hazard ratios (HRs) with 95% CIs and P values are descriptive and do not account for multiple comparisons.
TABLE 1.
Patient and Clinical Characteristics in S9313 TNBC Cohort
HRD Is Associated With DDIR
HRD+ TNBCs exhibited significantly higher DDIR scores (Mann-Whitney P = .003; odds ratio [OR], 3.26 [95% CI, 1.42 to 7.49]; P = .005) and were more likely to be DDIR+ by dichotomous classification (P = .006; Fig 1A). Among patients with TNBC in the TCGA data set, there was a suggestion of association between HRD+ status and DDIR (Fig 1B). Similarly, we observed that TNBCs with tumor BRCA mutations also exhibited higher continuous DDIR scores (Mann-Whitney P = .003; OR, 3.24 [95% CI, 1.48 to 7.09]; P = .003) and were more likely to be DDIR+ by dichotomous classification (P = .003; Fig 1C). Using an established threshold for DDIR status in the S9313 data set, and a gene expression–derived dichotomization threshold for DDIR in the TCGA data set (Appendix Fig A2), we partitioned all tumors into four groups on the basis of dual DDIR and HRD status (Figs 1D and 1E).
FIG 1.
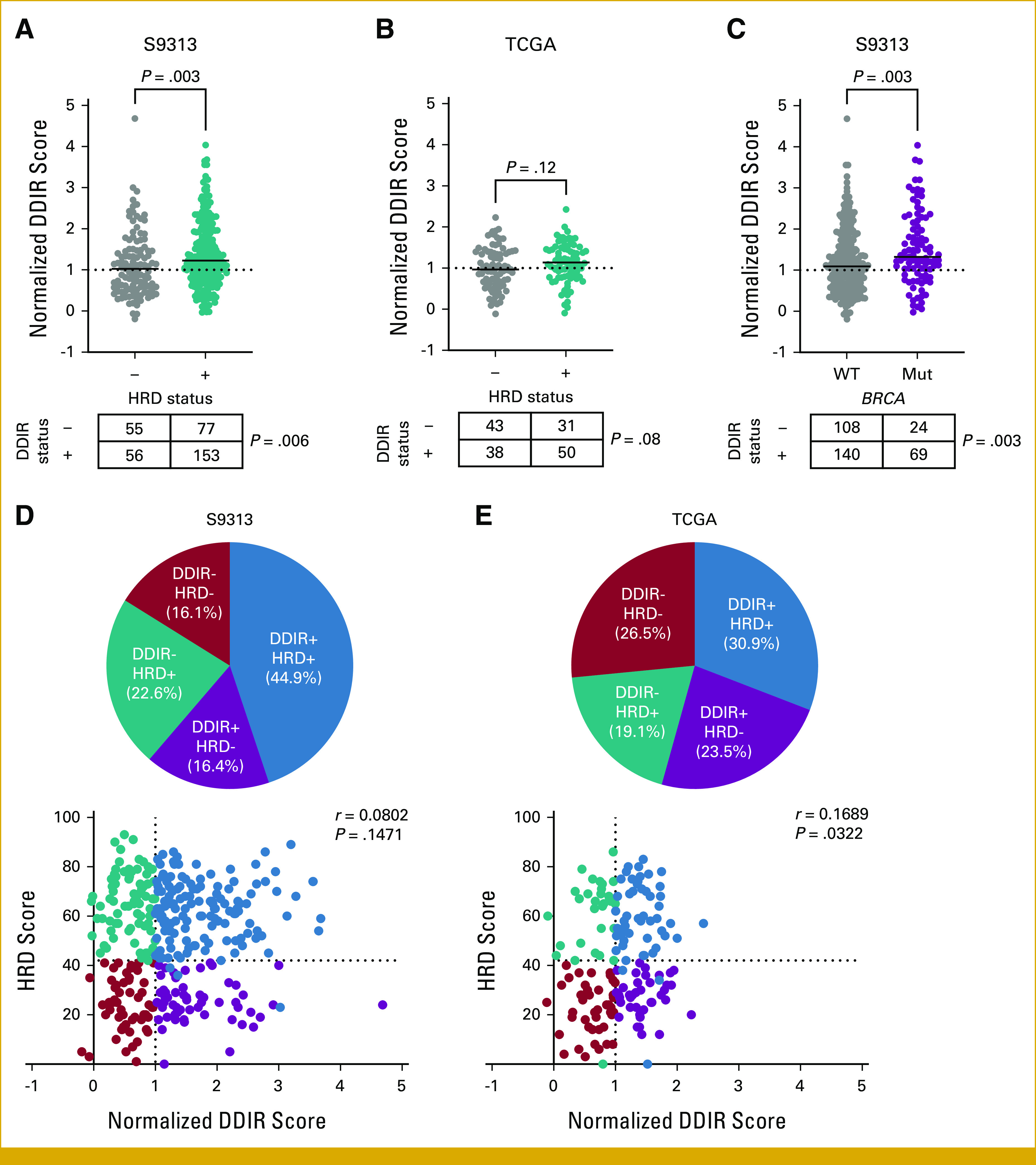
(A) Continuous and categorical comparison of threshold-normalized DDIR score and status by HRD status within the S9313 TNBC cohort. For continuous comparison, P is Mann-Whitney; for categorical comparison, P is chi-square. (B) Continuous and categorial comparison of threshold normalized DDIR score and status by HRD status within the TCGA cohort. For continuous comparison P is Mann-Whitney; for categorical comparison P is chi-square. (C) Continuous and categorial comparison of threshold normalized DDIR score and status by tumor BRCA mutation status within the S9313 TNBC cohort. For continuous comparison P is Mann-Whitney; for categorical comparison P is chi-square. (D and E) Distribution of DDIR/HRD classes and threshold normalized DDIR and HRD scores within the (D) S9313 and (E) TCGA TNBC cohorts. r is Spearman's coefficient. DDIR, DNA damage immune response; HRD, homologous recombination deficiency; TCGA, The Cancer Genome Atlas; TNBC, triple-negative breast cancer.
Dual Classification by DDIR/HRD Is Prognostic
In the S9313 cohort, 5-year DFS was 80.9% (DDIR+/HRD+), 74.7% (DDIR+/HRD–), 74.0% (DDIR–/HRD+), and 56.4% (DDIR–/HRD–). Adjusting for tumor size, positive nodes, age, and treatment arm, the DFS HRs relative to the DDIR–/HRD– group are as follows: DDIR+/HRD+ (HR, 0.36 [95% CI, 0.20 to 0.63]); DDIR+/HRD– (HR, 0.46 [95% CI, 0.23 to 0.89]); and DDIR–/HRD+ (HR, 0.46 [95% CI, 0.25 to 0.83]). In a multivariable model of both biomarkers adjusting for tumor size, positive nodes, age, and treatment arm, the HR for DDIR+ versus DDIR– is 0.62 (95% CI, 0.41 to 0.95; P = .028) and for HRD+ versus HRD– is 0.58 (95% CI, 0.37 to 0.91; P = .017), suggesting that each biomarker provides independent prognostic information. Five-year OS was 87.5% (DDIR+/HRD+), 85.5% (DDIR+/HRD–), 83.1% (DDIR–/HRD+), and 69.1% (DDIR–/HRD–; Figs 2A and 2B). Adjusting for the same variables, the OS HRs relative to the DDIR–/HRD– group are as follows: DDIR+/HRD+ (HR, 0.41 [95% CI, 0.21 to 0.82]); DDIR+/HRD– (HR, 0.36 [95% CI, 0.15 to 0.85]); and DDIR–/HRD+ (HR, 0.47 [95% CI, 0.23 to 0.98]). In a multivariable model of both biomarkers adjusting for the same variables, the HRs for DDIR+ versus DDIR– is 0.60 (95% CI, 0.35 to 1.01; P = .055) and for HRD+ versus HRD– is 0.69 (95% CI, 0.40 to 1.19; P = .18). These findings were corroborated in the TCGA TNBC data set, where DDIR+ and DDIR–/HRD+ subgroups had better DFS and OS compared with the DDIR–/HRD– subgroup (Figs 2C and 2D).
FIG 2.
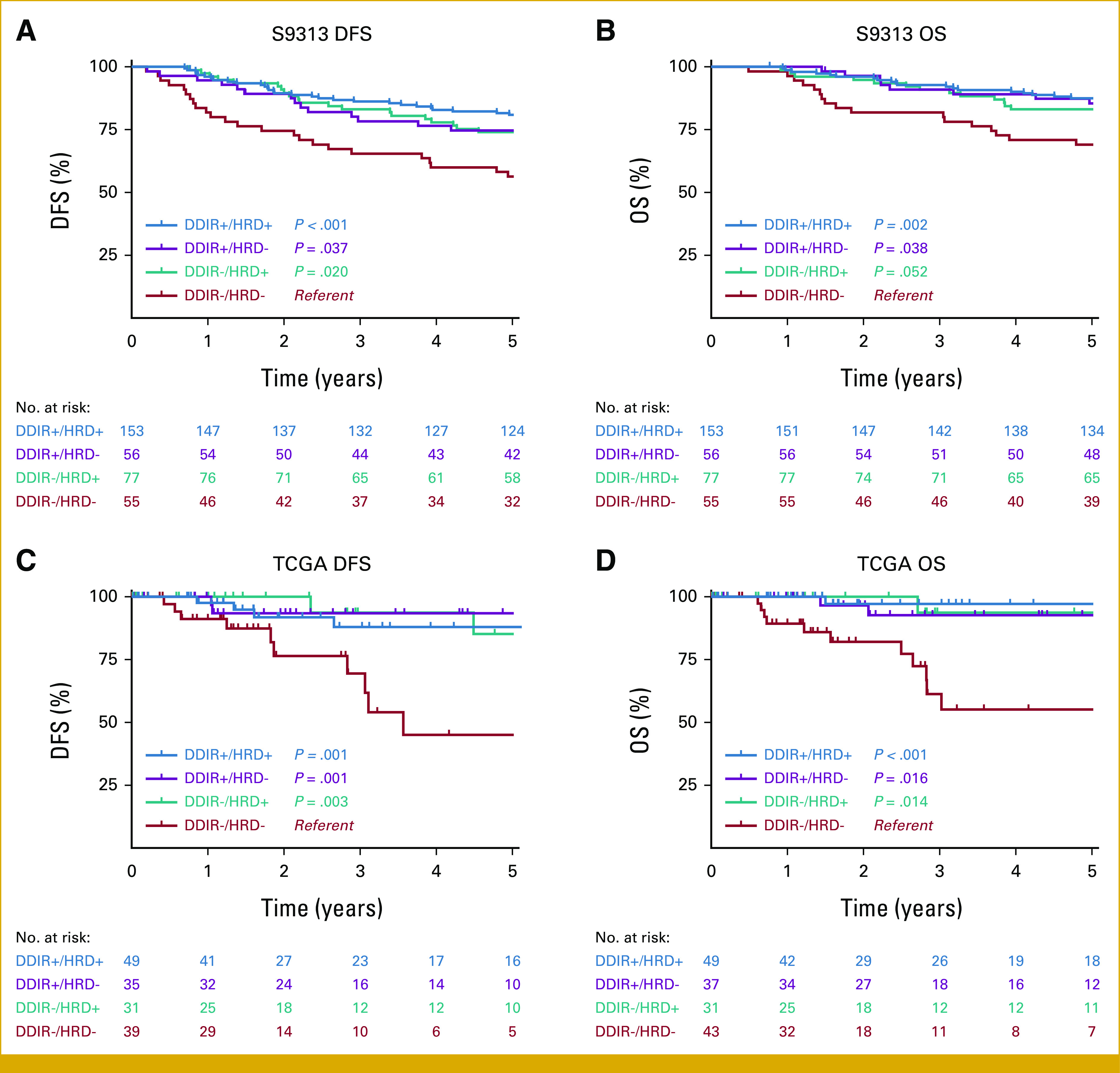
(A) DFS and (B) OS by DDIR/HRD class in the S9313 TNBC cohort. (C) DFS and (D) OS by DDIR/HRD class in the TCGA cohort. P value is log-rank, with DDIR–/HRD– class as referent. DDIR, DNA damage immune response; DFS, disease-free survival; HRD, homologous recombination deficiency; OS, overall survival; TCGA, The Cancer Genome Atlas; TNBC, triple-negative breast cancer.
DDIR/HRD Classes Are Biologically and Immunologically Distinct
We evaluated the distribution of TNBC molecular subtypes and the immunomodulatory (IM) gene expression signature20 in the context of DDIR/HRD dual classification. The distribution of molecular subtypes was highly skewed among the DDIR/HRD classes in both the S9313 (Fig 3A) and TCGA (Fig 3B) data sets (P < .001). In both data sets, we observed enrichment of basal-like subtypes and the IM signature among DDIR+ tumors, regardless of HRD status. By contrast, there was virtual absence of the IM signature among DDIR– tumors, regardless of HRD status, and enrichment of the mesenchymal subtype among DDIR–/HRD+ tumors. There was no clear over-representation of a subtype in the poor-prognosis DDIR–/HRD– tumors.
FIG 3.
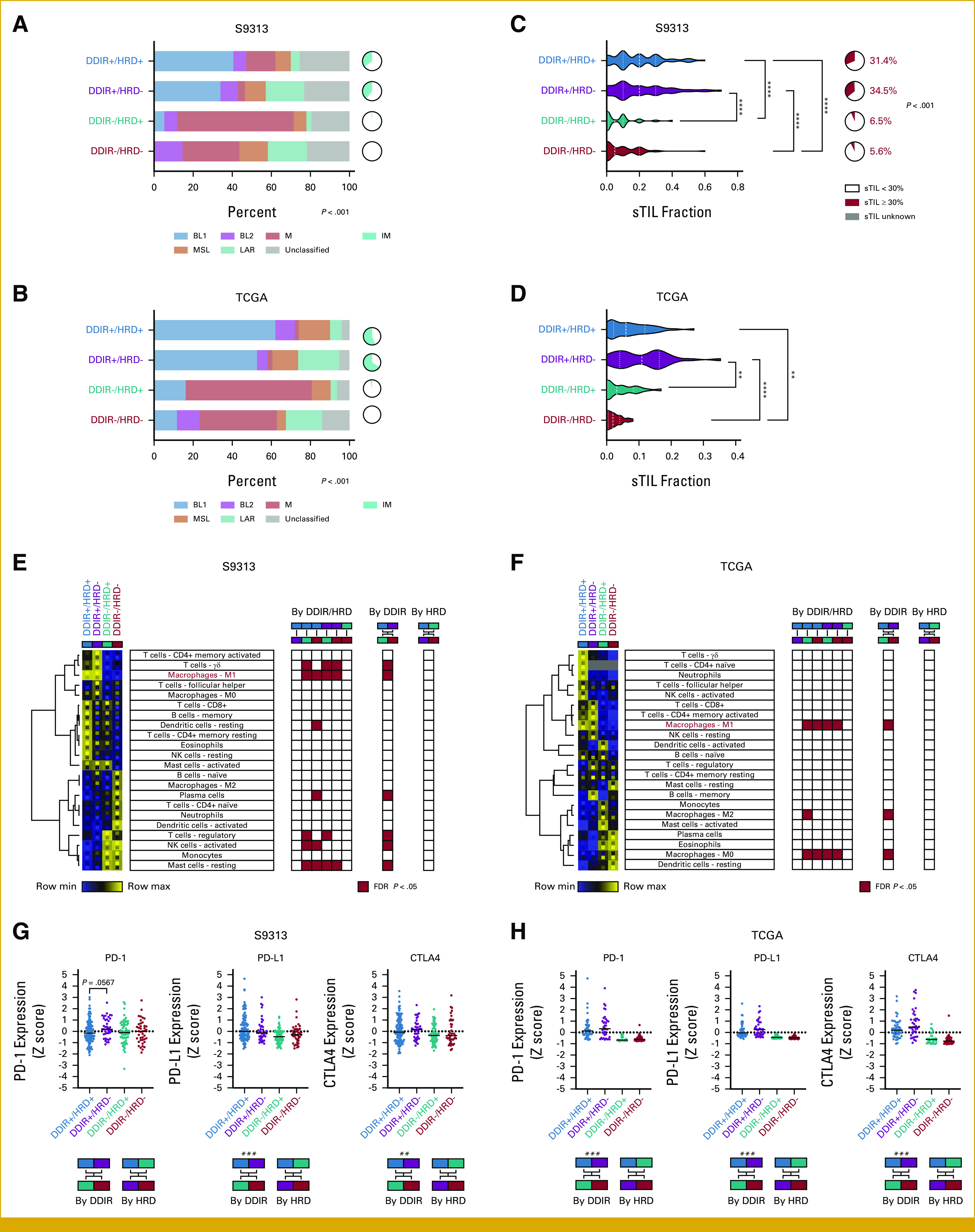
(A and B) TNBC molecular subtype and immunomodulatory status in the (A) S9313 and (B) TCGA TNBC cohorts. (C) Distribution of sTIL frequency and fraction of tumors with ≥30 sTILs within the S9313 TNBC cohort. For continuous comparison, P is Kruskal-Wallis test with Dunn's multiple comparison correction. For comparison by ≥30% threshold, P is Fisher-Freeman-Halton test. (D) sTIL frequency in the TCGA TNBC cohort. P is Kruskal-Wallis test with Dunn's multiple comparison correction. *<0.05, **<0.01, ***<0.001, ****<0.0001. (E and F) Relative CIBERSORTx leukocyte fractions within the (E) S9313 and (F) TCGA TNBC cohorts. Heatmap reflects mean leukocyte fraction within DDIR/HRD class with upper and lower inset boxes reflecting mean + SE and mean - SE, respectively. Red boxes in grids denote significant differences for each comparison (FDR <0.05). (G and H) Expression of PD-1, PD-L1, and CTLA4 by DDIR/HRD class in the (G) S9313 and (H) TCGA TNBC cohorts. P is Kruskal-Wallis test with Dunn's multiple comparison correction. Binary comparisons by DDIR or HRD status alone are Mann-Whitney, with *<0.05, **<0.01, ***<0.001, ****<0.0001. DDIR, DNA damage immune response; FDR, false discovery rate; HRD, homologous recombination deficiency; sTIL, stromal tumor infiltrating lymphocyte; TCGA, The Cancer Genome Atlas; TNBC, triple-negative breast cancer.
The IM gene expression signature reflects enrichment of sTILs.20 As expected, we observed robust sTIL infiltration in DDIR+ cancers, regardless of HRD status (median 20% sTILs in both DDIR+/HRD+ and DDIR+/HRD– classes), and paucity of sTILs in DDIR– cancers (median 5% sTILs in both DDIR–/HRD+ and DDIR–/HRD– classes; Fig 3C). Using a previously reported 30% cutoff,21,22 DDIR+ tumors in the S9313 data set were significantly more likely to have high sTILs than DDIR– tumors (P < .001). These findings were confirmed in the TCGA TNBC data set where similar enrichment of sTILs in DDIR+ tumors and paucity of sTILs in DDIR– tumors was noted, although the absolute values differed between studies, likely because of differences in sTIL quantification methodology (Fig 3D).
Relative leukocyte subtype frequencies were computed for each sample using CIBERSORTx digital cytometry,23 and then, cluster analysis was performed to identify patterns of immune cell infiltration within each DDIR/HRD class. In the S9313 data set, we observed a significant enrichment of γδ T cells, M1 macrophages, and resting dendritic cells, and significant depletion of plasma cells, regulatory T cells (Tregs), activated natural killer (NK) cells, and resting mast cells among the DDIR/HRD classes (Fig 3E). Comparison on the basis of DDIR status alone and HRD status alone showed that these cell populations differed only based on DDIR status (Fig 3E). CIBERSORTx analysis on the TCGA TNBC data set identified DDIR status–dependent differences in M0, M1, and M2 macrophage populations (Fig 3F). The only immune cell population that demonstrated a congruent significant difference in both the S9313 and TCGA data sets was enrichment of M1 macrophages in DDIR+ tumors.
Because DDIR status affects expression of ICB target genes,14 we evaluated the expression of PD-1, PD-L1, and CTLA4 on the basis of DDIR/HRD class. Unsurprisingly, we saw upregulation of PD-L1 in DDIR+ cancers since expression of its gene (CD274) is one component of the DDIR score. Interestingly, there was a trend toward upregulation of PD-1 in DDIR+/HRD– tumors compared with DDIR+/HRD+ tumors in the S9313 data set (P = .0567; Fig 3G) and this was numerically reflected in the TCGA data set as well, although it did not meet statistical significance. Analysis on the basis of DDIR and HRD status alone showed significant upregulation of PD-L1 and CTLA4 among DDIR+ compared with DDIR– cancers (Fig 3G). We performed similar analyses in the TCGA TNBC cohort, where highly significant differences in PD-1, PD-L1, and CTLA4 gene expression between DDIR+ and DDIR– cancers were observed (Fig 3H).
Evaluation of Other Gene Expression Signatures and Cancer Hallmarks by DDIR/HRD Classes
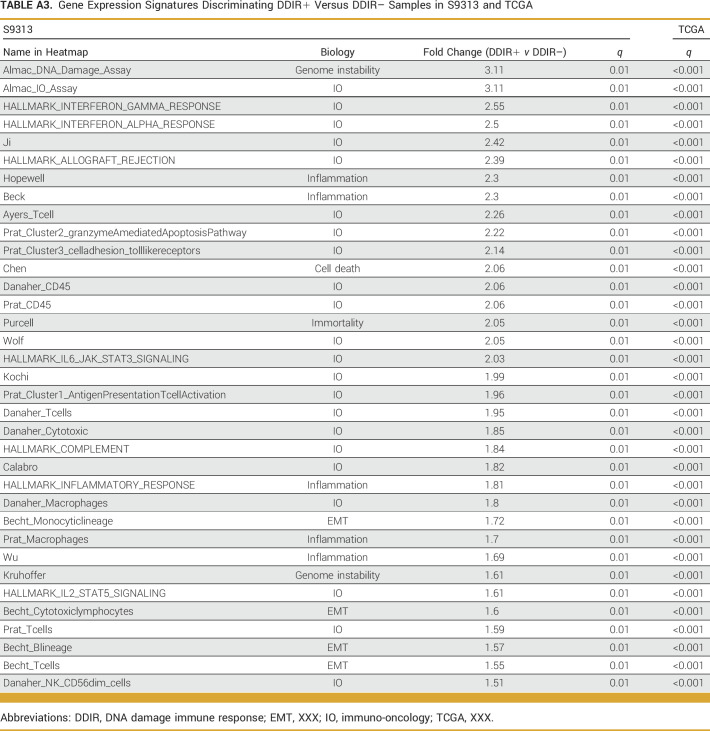
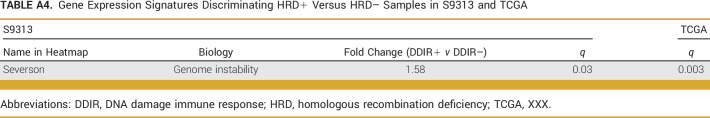
claraT analysis (Almac Diagnostic Services) was used to compare known gene expression signatures and cancer hallmarks between DDIR/HRD classes. Since our data were from bulk tissue gene expression, it was necessary to account for the contribution of immune and stromal cell populations in our comparisons. Using a fold-change (FC) cutoff of 1.5 with a false discovery rate (FDR) of <0.05, we identified 35 gene expression signatures that were significantly enriched in DDIR+ compared with DDIR– cancers (Appendix Fig A3A, Appendix Table A3). As expected, the DDIR signature (Almac_DNA_Damage_Assay and Almac_IO_Assay) was the top discriminator since group assignments were made a priori on the basis of this signature. Among the other 33 signatures that were significant, the majority were classified as immuno-oncology or inflammatory signatures. These signatures were validated in the TCGA TNBC cohort by performing k-means clustering and evaluating the distribution of the DDIR/HRD classes within the two resulting clusters. This resulted in robust discrimination of DDIR+ and DDIR– cancers (Appendix Fig A3B, Appendix Table A3) within the TCGA cohort. An analysis to identify signatures that were significantly enriched or depleted in HRD+ compared with HRD– cancers was also performed using the same FC and FDR cutoffs. The only signature that emerged as significant was a signature of BRCAness (Appendix Fig A3C, Appendix Table A4).24 We confirmed that this signature was also differentially expressed in the TCGA TNBC cohort on the basis of HRD status (Appendix Fig A3D, Appendix Table A4).
The Poor-Prognosis DDIR–/HRD– Class Is Heterogeneous and Contains a Subset of Tumors With a 3p21 Expression Profile With an Exceptionally Poor Prognosis
We interrogated the TCGA data set to try to understand unique biological features in poor-prognosis DDIR–/HRD– TNBCs. For all features, we compared DDIR–/HRD– TNBCs with DDIR–/HRD+ TNBCs to minimize the influence of interference from the immune-enriched tumor microenvironment in DDIR+ TNBCs. There were no significantly different mutational alterations or gene methylation patterns in DDIR–/HRD– compared with DDIR–/HRD+ TNBCs. We next compared DDIR–/HRD– and DDIR–/HRD+ TNBCs using gene set enrichment analysis (GSEA; Fig 4A). The Chr3p21 positional gene set emerged with a highly significant normalized enrichment score in DDIR–/HRD– compared with DDIR–/HRD+ TNBCs (NES = 2.17, FDR q = 0.012; Fig 4B). Analysis of copy-number alterations at this locus showed that an increased rate of loss at Chr3p21 was observed in DDIR–/HRD+ TNBCs (Fig 4C), suggesting that retention of one or more genes at this locus in DDIR–/HRD– tumors is driving aggressive behavior. We identified 42 of the 200 genes in the Chr3p21 positional gene set that were able to discriminate DDIR–/HRD– from DDIR–/HRD+ TNBCs with FDR < 0.05 (Fig 4D). Using hierarchical clustering within the DDIR/HRD classes, it was noted that there was clearly a subset of DDIR–/HRD– tumors with dramatically enriched expression of these 42 genes (Fig 4D). Outcomes among the DDIR–/HRD– tumors with enriched expression of this Chr3p21 cluster were exceptionally poor (Fig 4E). To understand if the Chr3p21 expression cluster represented a known biological phenotype, we examined the distribution of TNBC molecular subtypes among the two Chr3p21 expression patterns. No significant difference in molecular subtype were found between the two Chr3p21 patterns (P = .296). Only six of the 42 genes were represented in the array expression data in the S9313 data set, limiting our ability to validate this cluster among that cohort.
FIG 4.
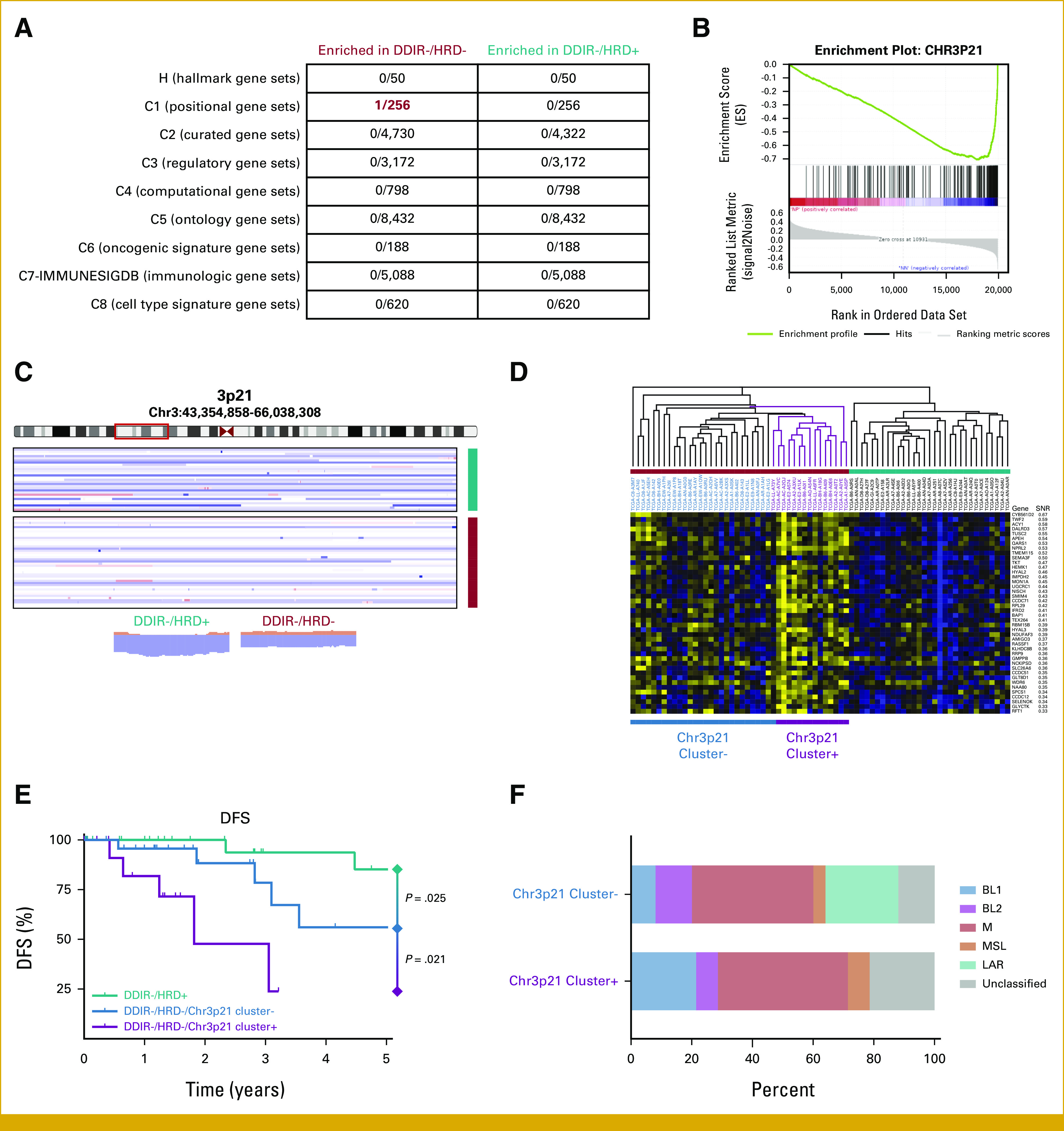
(A) Gene set enrichment analysis. Numerator in each box denotes significant (FDR <0.05) gene sets within the entire set of genes analyzed (denominator). (B) Enrichment plot for Chr3p21 positional gene set between DDIR–/HRD– and DDIR–/HRD+ tumors. (C) Copy number plot for Chr3p21 locus. Plots at bottom show summary copy-number changes among DDIR–/HRD– and DDIR–/HRD+ classes. (D) Expression profile of Chr3p21 genes within DDIR–/HRD– and DDIR–/HRD+ tumors, with identification of Chr3p21 cluster+ tumors within DDIR–/HRD– class. (E) DFS among DDIR–/HRD– tumors with and without the Chr3p21 expression cluster signature and DDIR–/HRD+ tumors. P is log-rank. (F) TNBC molecular subtype by Chr3p21 cluster expression within DDIR–/HRD– tumors. DDIR, DNA damage immune response; DFS, disease-free survival; FDR, false discovery rate; HRD, homologous recombination deficiency; TNBC, triple-negative breast cancer.
DISCUSSION
We show that combined use of DDIR and HRD enables classification of both the immunologic state and intrinsic chemosensitivity of TNBCs, and that combinatorial use of these two biomarkers is prognostic.
It has long been known that breast cancers arising in women with BRCA1 mutations were associated with robust mononuclear immune cell infiltration.26-28 Recently, the link between HRD and augmented immune response has been attributed to cGAS-mediated STING-dependent induction of type I interferon expression in response to cytosolic dsDNA.11,12,29 Although there was clearly an association between HRD and DDIR, these biomarkers were not completely overlapping. Indeed, approximately 40% of tumors in both the S9313 and TCGA data sets demonstrated this lack of overlap, where tumors were positive for HRD alone or DDIR alone. This suggests that there are HRD-independent mechanisms of STING activation, and there are HRD+ tumors that, despite potentially immunogenic genomic instability, ultimately fail to elicit a DNA-dependent immune response.
Integrating ICB therapy with cytotoxic chemotherapy has resulted in substantial clinical benefit in patients with both localized and metastatic TNBC.16,30-34 Interestingly, we observed a trend toward higher PD-1 expression in DDIR+/HRD– compared with DDIR+/HRD+ TNBCs. Although expression of PD-1/PD-L1 is not currently used to select patients with early-stage TNBC who will benefit from ICB therapy, low PD-1 expression might suggest that the antitumor immune response is already invigorated and that the benefit of anti–PD-1/PD-L1 therapy in DDIR+/HRD+ patients may be less pronounced.
We found that DDIR+ TNBCs have a favorable prognosis, with the best outcomes noted in tumors that are also positive for HRD. The robust infiltration of sTILs with low PD-1 expression (a marker of lymphocyte exhaustion)35 in the DDIR+/HRD+ tumors suggests that the immune microenvironment is permissive and that the infiltrated lymphocytes are not anergic. As such, we speculate that these tumors may be exquisitely sensitive to DNA-damaging therapy and may not receive substantial incremental benefit from immunotherapy (Fig 5A).
FIG 5.
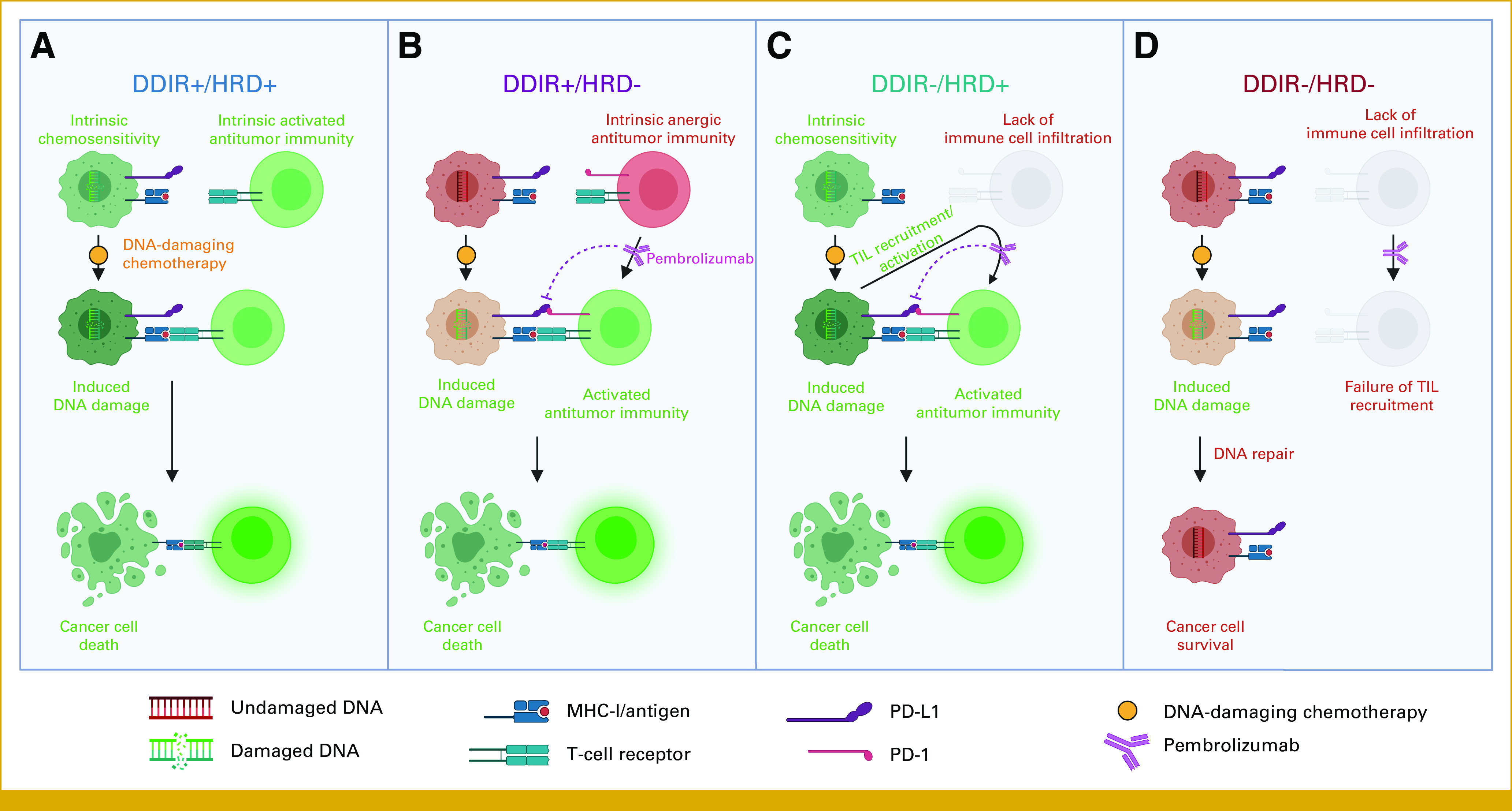
DDIR/HRD provides a therapeutic vulnerability–based classification scheme in TNBC. DDIR, DNA damage immune response; HRD, homologous recombination deficiency; TNBC, triple-negative breast cancer. Figure created on BioRender.com.25
Although sTILs are enriched in DDIR+/HRD– tumors to a similar degree to DDIR+/HRD+ tumors, our finding that PD-1 expression is increased in DDIR+/HRD– compared with DDIR+/HRD+ tumors suggests that DDIR+/HRD– tumors have developed an anergic or exhausted tumor microenvironment. In this tumor-immune microenvironment, DNA-damaging chemotherapy can result in augmented innate and adaptive immune responses,36-41 and anti–PD-1 immunotherapy can potentially reverse anergy in the infiltrated antitumor immune population (Fig 5B).
Existence of a DDIR–/HRD+ subgroup suggests that there are instances where loss of HR function does not result in constitutive activation of the cGAS-STING pathway. The mechanism by which HRD-associated micronucleus-induced inflammatory signaling is suppressed in these tumors is unknown. Regardless, in DDIR–/HRD+ tumors, the presence of HRD will result in hypersensitivity to DNA-damaging chemotherapy, and studies suggest that TNBCs with BRCA mutations (the most robust cause of HRD in TNBC) are more likely to develop vigorous postneoadjuvant chemotherapy immune infiltrates compared with tumors without underlying BRCA mutations.42 Previous in vitro work has shown that loss of DSB repair pathway function is associated with significant upregulation of PD-L1 after genotoxic stress in cancer cells.43 Taken together, these data suggest that for the DDIR–/HRD+ subgroup, combination chemoimmunotherapy will exploit both the intrinsic chemosensitivity of these tumors and the tendency to use PD-L1 to suppress lymphocytes that are recruited after sustaining DNA damage from chemotherapy (Fig 5C). Although prognosis of DDIR+/HRD– and DDIR–/HRD+ tumors was equivalent to DDIR+/HRD+ tumors, we suggest that potential differences in therapeutic vulnerabilities and the immune microenvironment between these favorable prognosis subgroups support classifying them as separate entities.
The outcome for the DDIR–/HRD– subgroup, which has neither intrinsic chemosensitivity nor an active tumor immune microenvironment, was poor (Fig 5D). Using gene set enrichment analysis, we were able to identify differential expression of genes at the Chr3p21 locus among a subset of DDIR–/HRD– tumors and found that tumors with robust expression from this locus had exceptionally poor prognosis. Alterations at the 3p21 locus are common in a variety of epithelial cancers, including breast cancer, and this genomic region contains several putative tumor suppressor genes.44,45 The significance of the Chr3p21 expression cluster should be validated in independent data sets, and further work should be performed to understand which gene(s) at this locus are driving poor prognosis.
By interrogating the molecular data across subtypes and treatment arms of I-SPY2, Wolf et al46 showed that immune status defined by a composite dendritic cell and STAT1 signature was predictive for response to pembrolizumab, and DNA repair deficiency (DRD) defined using a multigene signature termed VCpred_TN was predictive for response to veliparib plus carboplatin. As in our study, they also found that patients with TNBC who were neither immune-enriched nor positive for DRD had low pathologic response rates to all treatments. The I-SPY2 Immune/DRD classification and our DDIR/HRD scheme are likely to result in similar biological partitioning. The DDIR assay (claraT, Almac Diagnostic Services) has already been validated in numerous clinical cohorts,13,14,47 and the HRD test we used (myChoice CDx, Myriad Genetics) is FDA approved and in routine clinical use. As such, there is a pathway toward clinical implementation of DDIR/HRD.
Major strengths of our study include the prospective nature of the S9313 study with known long-term outcomes and central assessment of TNBC status. There are certain limitations as well, including that the chemotherapy in S9313 was devoid of taxanes and immunotherapy, which are standard-of-care contemporary systemic treatments for TNBC. It will be of critical importance to test this molecular classification scheme in patients treated with modern chemoimmunotherapy. Furthermore, details on the nature of DFS events (distant, locoregional, contralateral breast cancer, and so forth) from S9313 are not readily available; therefore, we were unable to examine other end points such as distant DFS or invasive DFS. We have validated our findings in patients with TNBC within the TCGA data set, although details of systemic therapy in that cohort are relatively incomplete.
In summary, we demonstrate that immune activation and DNA damage repair defects identify related but therapeutically distinct vulnerabilities in TNBC and illuminate DDIR/HRD as a clinically relevant combination to classify this heterogeneous group of tumors. Future studies should explore the predictive utility of this classification scheme with respect to discrete systemic therapy agents.
APPENDIX
FIG A1.
REMARK diagram for S9313 TNBC cohort selection. aN = 6 were HRD-negative by GI score analysis but BRCA mutation could not be determined; N = 33 were wild-type BRCA but GI score analysis failed. TNBC, triple-negative breast cancer.
FIG A2.
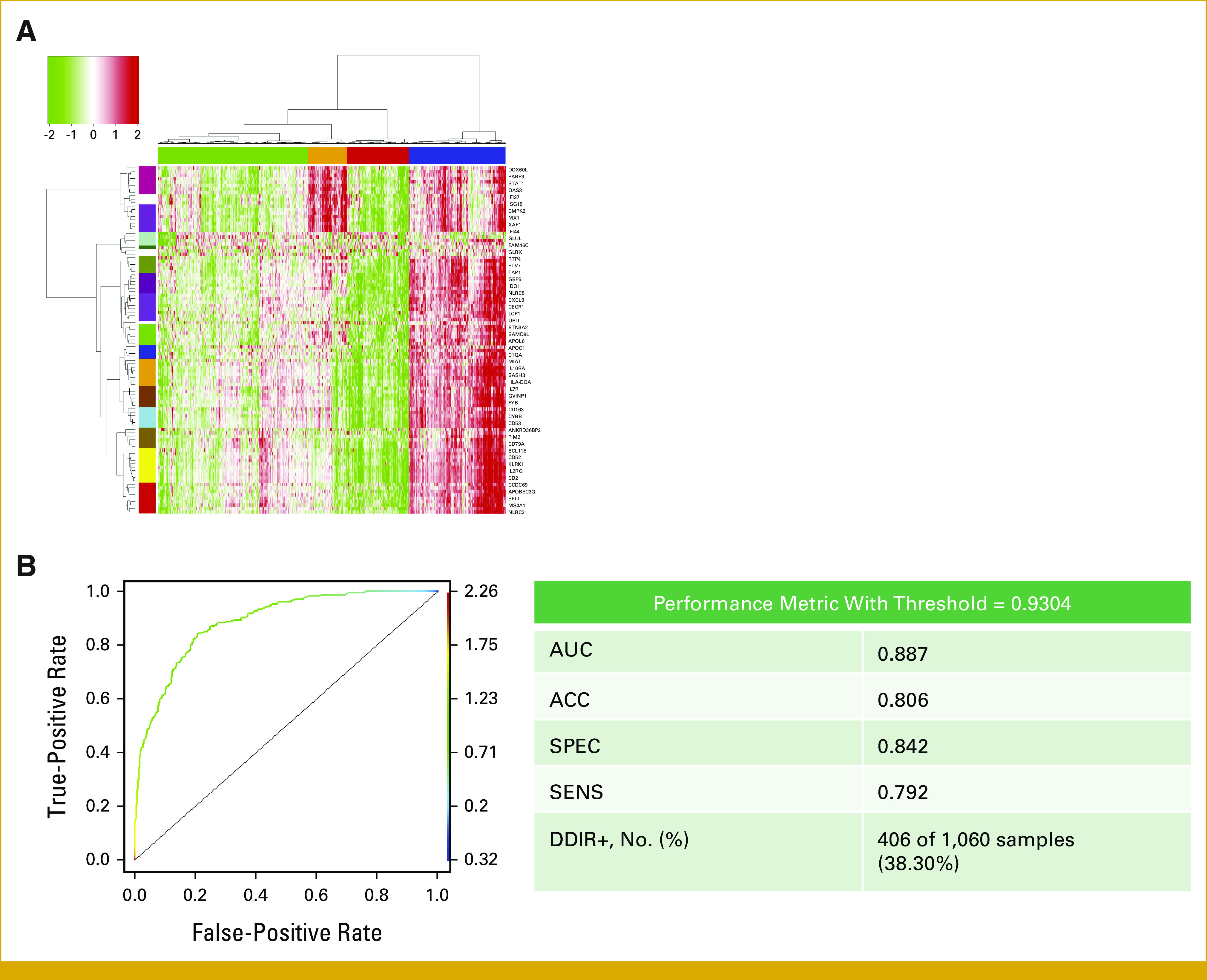
(A) Cluster and (B) ROC analysis to define optimal DDIR threshold in the TCGA cohort. ACC, accuracy; DDIR, DNA damage immune response; ROC, receiver operating characteristic; SENS, sensitivity; SPEC, specificity; TCGA, The Cancer Genome Atlas.
FIG A3.
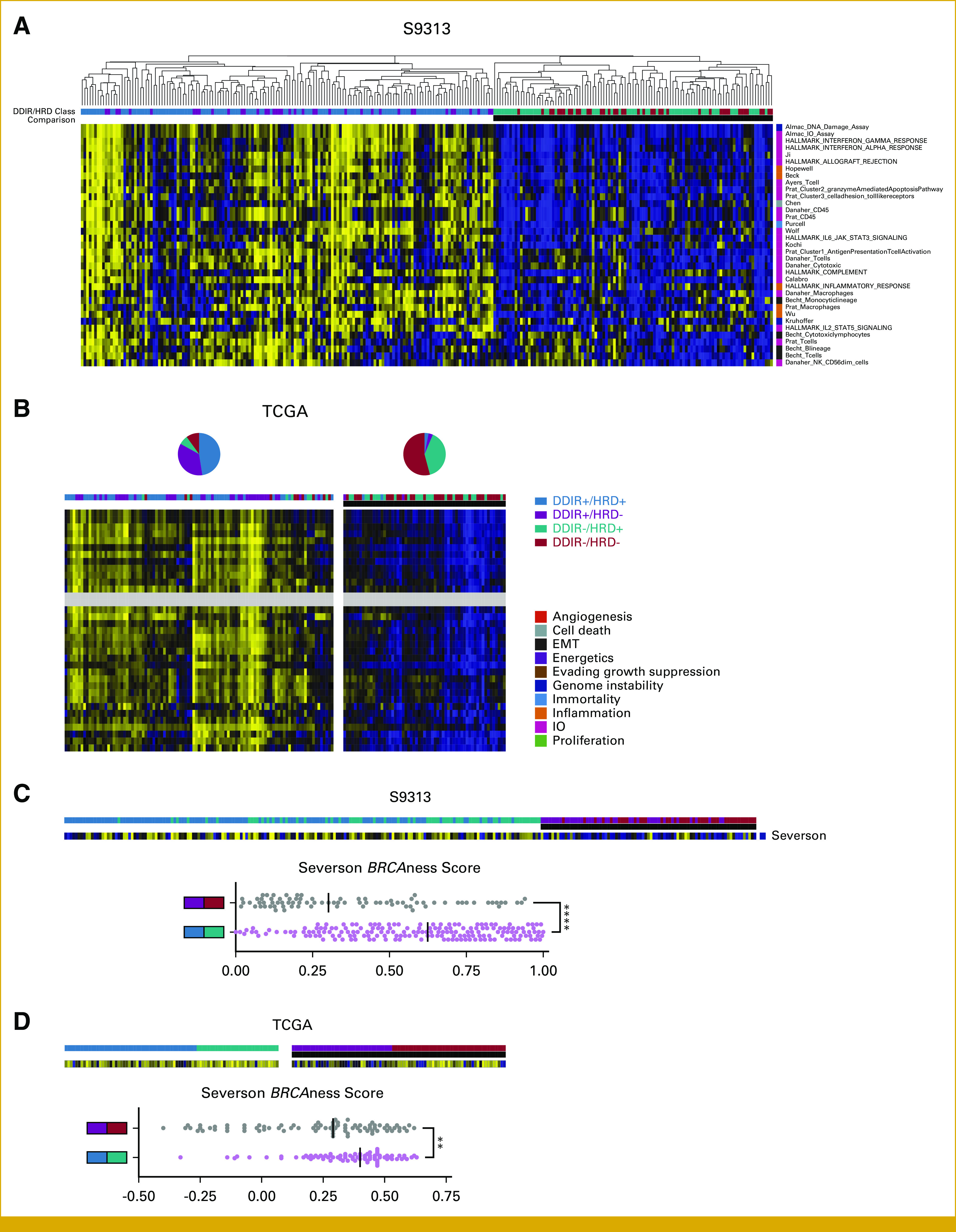
(A) Gene expression signatures that differ between DDIR+ and DDIR– tumors within the S9313 cohort. (B) Analysis of discriminating gene expression signatures by DDIR status in the S9313 cohort by k-means clustering (k = 2) within the TCGA cohort. Pie charts denote distribution of DDIR/HRD classes within the two clusters. (C) Gene expression signature that differs between HRD+ and HRD– tumors within the S9313 cohort, and (D) analysis of discriminating gene expression signature by HRD status in the S9313 within the TCGA cohort. DDIR, DNA damage immune response; EMT, epithelial-mesenchymal transition; HRD, homologous recombination deficiency; IO, immuno-oncology; TCGA, The Cancer Genome Atlas.
TABLE A1.
Genes in the DNA Damage Immune Response Gene Expression Signature
TABLE A2.
Patient and Clinical Characteristics in TCGA Triple-Negative Breast Cancer Cohort
TABLE A3.
Gene Expression Signatures Discriminating DDIR+ Versus DDIR– Samples in S9313 and TCGA
TABLE A4.
Gene Expression Signatures Discriminating HRD+ Versus HRD– Samples in S9313 and TCGA
Shane R. Stecklein
Research Funding: Natera
William Barlow
Research Funding: Merck (Inst), AstraZeneca (Inst)
Lajos Pusztai
Honoraria: bioTheranostics, natera, OncoCyte, Athenex
Consulting or Advisory Role: H3 Biomedicine, Merck, Novartis, Seagen, Syndax, AstraZeneca, Roche/Genentech, Bristol Myers Squibb, Clovis Oncology, Immunomedics, Eisai, Almac Diagnostics, Pfizer
Research Funding: Merck (Inst), Genentech (Inst), Seagen (Inst), AstraZeneca (Inst), Bristol Myers Squibb (Inst), Pfizer (Inst)
Travel, Accommodations, Expenses: AstraZeneca
Uncompensated Relationships: NanoString Technologies, Foundation Medicine
Open Payments Link: https://openpaymentsdata.cms.gov/physician/110878
Kirsten Timms
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Inventor on numerous patents
Travel, Accommodations, Expenses: Myriad Genetics
Richard Kennedy
Employment: Almac Diagnostics
Leadership: Almac Diagnostics
Patents, Royalties, Other Intellectual Property: Gene Signature for Immune Therapies in Cancer Publication number: 20190316203 Almac Diagnostics (Inst), Molecular Diagnostic Test for Cancer Publication number: 20140031260 (Inst)
Gemma E. Logan
Employment: Almac Diagnostics
Rob Seitz
Employment: OncoCyte
Leadership: OncoCyte
Stock and Other Ownership Interests: OncoCyte
Consulting or Advisory Role: Thermo Fisher Scientific, Biogeneration Ventures, Biogeneration Ventures, OncoCyte
Patents, Royalties, Other Intellectual Property: Patent with technologies licensed to and in partnership with Oncocyte, Inc
Sunil Badve
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: Personalis, Dako/Agilent Technologies, Ventana Medical Systems, Bristol Myers Squibb Foundation
Consulting or Advisory Role: PAIGE.AI, Ventana Medical Systems, Agilent, BMS
Speakers' Bureau: Genomic Health, Targos GmbH, Agilent
Research Funding: Dako/Agilent Technologies (Inst), Lilly (Inst), Bristol Myers Squibb Foundation (Inst)
Patents, Royalties, Other Intellectual Property: EarlyR—signature for ER+ breast cancer (Inst), E-Score for predicting recurrence of DCIS (Inst)
Yesim Gökmen-Polar
Other Relationship: Elsevier, Elsevier
Hannah Linden
Leadership: Evolent, Progyny
Stock and Other Ownership Interests: Evolent
Consulting or Advisory Role: Sanofi, Pfizer, GE Healthcare, Novartis, Gilead Sciences
Research Funding: Sanofi/Aventis (Inst), Zymeworks (Inst), Zionexa (Inst), Tolmar (Inst), Pfizer (Inst), Zeno Pharmaceuticals (Inst), Veru
Debu Tripathy
Consulting or Advisory Role: Novartis, Pfizer, GlaxoSmithKline, Genomic Health, AstraZeneca, OncoPep, Sermonix Pharmaceuticals, Personalis, Ambrx, Roche, Roche, Gilead Sciences, Gilead Sciences, Gilead Sciences, Gilead Sciences, Gilead Sciences
Research Funding: Novartis (Inst), Polyphor (Inst), Pfizer (Inst), Ambrx (Inst)
Travel, Accommodations, Expenses: Novartis, AstraZeneca
Gabriel N. Hortobagyi
Consulting or Advisory Role: Novartis, Seagan, Blueprint Medicines, AstraZeneca
Research Funding: Novartis (Inst)
Andrew K. Godwin
Employment: University of Kansas Health System
Stock and Other Ownership Interests: Clara Biotech, Exokeryx
Honoraria: Sinochips Kansas
Consulting or Advisory Role: Biovica
Research Funding: BioFluidica (Inst), VITRAC Therapeutics (Inst), Clara Biotech (Inst), Predicine (Inst)
Patents, Royalties, Other Intellectual Property: Provisional Patent application entitled—“WJMSC-Derived Small Extracellular Vesicle (sEVs) Enhance T Cell Suppression Through Checkpoint PD-L1”—Dr Joseph McGuirk (applicant) (Inst), Provisional—US Patent number 63/496,270 entitled “Treatment of Neoplasm by Inhibiting KIF15” (Inst), Provisional—US Patent number 63/391,657 entitled “Extracellular Vesicle Proteomic Biomarker Panel for Ovarian Cancer Screening and the Early Detection of Disease” (Inst)
Alastair Thompson
Employment: Lilly
Stock and Other Ownership Interests: Lilly
Travel, Accommodations, Expenses: Endomagnetics
Daniel F. Hayes
Stock and Other Ownership Interests: InBiomotion, Cellworks, Xilis
Honoraria: Tempus
Consulting or Advisory Role: Cepheid, Freenome, Epic Sciences, Cellworks, BioVica, Oncocyte, Turnstone Bio, Guardant Health, L-Nutra, Macrogenics, Tempus, xilis, Exact Sciences, Centrix, Arvinis, bioTheranostics
Research Funding: AstraZeneca (Inst), Pfizer (Inst), Menarini Silicon Biosystems (Inst), Cepheid/Danaher (Inst), Angle (Inst)
Patents, Royalties, Other Intellectual Property: Royalties from licensed technology, Diagnosis and Treatment of Breast Cancer. Patent No. US 8,790,878 B2. Date of Patent: July 29, 2014. Applicant Proprietor: University of Michigan. Dr Daniel F. Hayes is designated as inventor/co-inventor, Circulating Tumor Cell Capturing Techniques and Devices. Patent No.: US 8,951,484 B2. Date of Patent: February 10, 2015. Applicant Proprietor: University of Michigan. Dr Daniel F. Hayes is designated as inventor/co-inventor, Title: A method for predicting progression free and overall survival at each follow-up timepoint during therapy of metastatic breast cancer patients using circulating tumor cells. Patent no. 05725638.0-1223-US2005008602
Other Relationship: UpToDate, Cancer Expert Now
Priyanka Sharma
Stock and Other Ownership Interests: Amgen, Johnson & Johnson/Janssen, Roche/Genentech, Gilead Sciences, Sanofi, Pfizer
Consulting or Advisory Role: Novartis, Merck, AstraZeneca, Pfizer, Gilead Sciences, GlaxoSmithKline, Sanofi, Boston Scientific, Cipla, Salient Pharmaceuticals
Research Funding: Novartis (Inst), Bristol Myers Squibb (Inst), Merck (Inst), Novartis (Inst), Gilead Sciences (Inst)
Patents, Royalties, Other Intellectual Property: UpToDate
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at the San Antonio Breast Cancer Symposium San Antonio, TX, December 8-11, 2020.
AUTHOR CONTRIBUTIONS
Conception and design: Shane R. Stecklein, Rob Seitz, Sunil Badve, Yesim Gökmen-Polar, Peggy Porter, Alastair Thompson, Daniel F. Hayes, Priyanka Sharma
Financial support: Sunil Badve, Peggy Porter, Priyanka Sharma
Administrative support: Shane R. Stecklein, Lajos Pusztai, Gemma E. Logan, Sunil Badve, Peggy Porter, Gabriel N. Hortobagyi, Priyanka Sharma
Provision of study materials or patients: Sunil Badve, Yesim Gökmen-Polar, Hannah Linden, Alastair Thompson, Daniel F. Hayes
Collection and assembly of data: Shane R. Stecklein, William Barlow, Kirsten Timms, Gemma E. Logan, Rob Seitz, Sunil Badve, Yesim Gökmen-Polar, Peggy Porter, Gabriel N. Hortobagyi, Alastair Thompson, Daniel F. Hayes
Data analysis and interpretation: Shane R. Stecklein, William Barlow, Lajos Pusztai, Richard Kennedy, Rob Seitz, Sunil Badve, Yesim Gökmen-Polar, Hannah Linden, Debu Tripathy, Andrew K. Godwin, Alastair Thompson, Priyanka Sharma
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.23.00197.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Shane R. Stecklein
Research Funding: Natera
William Barlow
Research Funding: Merck (Inst), AstraZeneca (Inst)
Lajos Pusztai
Honoraria: bioTheranostics, natera, OncoCyte, Athenex
Consulting or Advisory Role: H3 Biomedicine, Merck, Novartis, Seagen, Syndax, AstraZeneca, Roche/Genentech, Bristol Myers Squibb, Clovis Oncology, Immunomedics, Eisai, Almac Diagnostics, Pfizer
Research Funding: Merck (Inst), Genentech (Inst), Seagen (Inst), AstraZeneca (Inst), Bristol Myers Squibb (Inst), Pfizer (Inst)
Travel, Accommodations, Expenses: AstraZeneca
Uncompensated Relationships: NanoString Technologies, Foundation Medicine
Open Payments Link: https://openpaymentsdata.cms.gov/physician/110878
Kirsten Timms
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Inventor on numerous patents
Travel, Accommodations, Expenses: Myriad Genetics
Richard Kennedy
Employment: Almac Diagnostics
Leadership: Almac Diagnostics
Patents, Royalties, Other Intellectual Property: Gene Signature for Immune Therapies in Cancer Publication number: 20190316203 Almac Diagnostics (Inst), Molecular Diagnostic Test for Cancer Publication number: 20140031260 (Inst)
Gemma E. Logan
Employment: Almac Diagnostics
Rob Seitz
Employment: OncoCyte
Leadership: OncoCyte
Stock and Other Ownership Interests: OncoCyte
Consulting or Advisory Role: Thermo Fisher Scientific, Biogeneration Ventures, Biogeneration Ventures, OncoCyte
Patents, Royalties, Other Intellectual Property: Patent with technologies licensed to and in partnership with Oncocyte, Inc
Sunil Badve
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: Personalis, Dako/Agilent Technologies, Ventana Medical Systems, Bristol Myers Squibb Foundation
Consulting or Advisory Role: PAIGE.AI, Ventana Medical Systems, Agilent, BMS
Speakers' Bureau: Genomic Health, Targos GmbH, Agilent
Research Funding: Dako/Agilent Technologies (Inst), Lilly (Inst), Bristol Myers Squibb Foundation (Inst)
Patents, Royalties, Other Intellectual Property: EarlyR—signature for ER+ breast cancer (Inst), E-Score for predicting recurrence of DCIS (Inst)
Yesim Gökmen-Polar
Other Relationship: Elsevier, Elsevier
Hannah Linden
Leadership: Evolent, Progyny
Stock and Other Ownership Interests: Evolent
Consulting or Advisory Role: Sanofi, Pfizer, GE Healthcare, Novartis, Gilead Sciences
Research Funding: Sanofi/Aventis (Inst), Zymeworks (Inst), Zionexa (Inst), Tolmar (Inst), Pfizer (Inst), Zeno Pharmaceuticals (Inst), Veru
Debu Tripathy
Consulting or Advisory Role: Novartis, Pfizer, GlaxoSmithKline, Genomic Health, AstraZeneca, OncoPep, Sermonix Pharmaceuticals, Personalis, Ambrx, Roche, Roche, Gilead Sciences, Gilead Sciences, Gilead Sciences, Gilead Sciences, Gilead Sciences
Research Funding: Novartis (Inst), Polyphor (Inst), Pfizer (Inst), Ambrx (Inst)
Travel, Accommodations, Expenses: Novartis, AstraZeneca
Gabriel N. Hortobagyi
Consulting or Advisory Role: Novartis, Seagan, Blueprint Medicines, AstraZeneca
Research Funding: Novartis (Inst)
Andrew K. Godwin
Employment: University of Kansas Health System
Stock and Other Ownership Interests: Clara Biotech, Exokeryx
Honoraria: Sinochips Kansas
Consulting or Advisory Role: Biovica
Research Funding: BioFluidica (Inst), VITRAC Therapeutics (Inst), Clara Biotech (Inst), Predicine (Inst)
Patents, Royalties, Other Intellectual Property: Provisional Patent application entitled—“WJMSC-Derived Small Extracellular Vesicle (sEVs) Enhance T Cell Suppression Through Checkpoint PD-L1”—Dr Joseph McGuirk (applicant) (Inst), Provisional—US Patent number 63/496,270 entitled “Treatment of Neoplasm by Inhibiting KIF15” (Inst), Provisional—US Patent number 63/391,657 entitled “Extracellular Vesicle Proteomic Biomarker Panel for Ovarian Cancer Screening and the Early Detection of Disease” (Inst)
Alastair Thompson
Employment: Lilly
Stock and Other Ownership Interests: Lilly
Travel, Accommodations, Expenses: Endomagnetics
Daniel F. Hayes
Stock and Other Ownership Interests: InBiomotion, Cellworks, Xilis
Honoraria: Tempus
Consulting or Advisory Role: Cepheid, Freenome, Epic Sciences, Cellworks, BioVica, Oncocyte, Turnstone Bio, Guardant Health, L-Nutra, Macrogenics, Tempus, xilis, Exact Sciences, Centrix, Arvinis, bioTheranostics
Research Funding: AstraZeneca (Inst), Pfizer (Inst), Menarini Silicon Biosystems (Inst), Cepheid/Danaher (Inst), Angle (Inst)
Patents, Royalties, Other Intellectual Property: Royalties from licensed technology, Diagnosis and Treatment of Breast Cancer. Patent No. US 8,790,878 B2. Date of Patent: July 29, 2014. Applicant Proprietor: University of Michigan. Dr Daniel F. Hayes is designated as inventor/co-inventor, Circulating Tumor Cell Capturing Techniques and Devices. Patent No.: US 8,951,484 B2. Date of Patent: February 10, 2015. Applicant Proprietor: University of Michigan. Dr Daniel F. Hayes is designated as inventor/co-inventor, Title: A method for predicting progression free and overall survival at each follow-up timepoint during therapy of metastatic breast cancer patients using circulating tumor cells. Patent no. 05725638.0-1223-US2005008602
Other Relationship: UpToDate, Cancer Expert Now
Priyanka Sharma
Stock and Other Ownership Interests: Amgen, Johnson & Johnson/Janssen, Roche/Genentech, Gilead Sciences, Sanofi, Pfizer
Consulting or Advisory Role: Novartis, Merck, AstraZeneca, Pfizer, Gilead Sciences, GlaxoSmithKline, Sanofi, Boston Scientific, Cipla, Salient Pharmaceuticals
Research Funding: Novartis (Inst), Bristol Myers Squibb (Inst), Merck (Inst), Novartis (Inst), Gilead Sciences (Inst)
Patents, Royalties, Other Intellectual Property: UpToDate
No other potential conflicts of interest were reported.
REFERENCES
- 1.Bauer KR Brown M Cress RD, et al. : Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California Cancer Registry. Cancer 109:1721-1728, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Hammond ME Hayes DF Wolff AC, et al. : American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. JCO Oncol Pract 6:195-197, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohler BA Sherman RL Howlader N, et al. : Annual report to the nation on the status of cancer, 1975-2011, featuring Incidence of breast cancer subtypes by race/ethnicity, poverty, and state. J Natl Cancer Inst 107:djv048, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolff AC Hammond MEH Hicks DG, et al. : Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical Practice guideline update. J Clin Oncol 31:3997-4013, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Abkevich V Timms KM Hennessy BT, et al. : Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 107:1776-1782, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birkbak NJ Wang ZC Kim JY, et al. : Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2:366-375, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popova T Manié E Rieunier G, et al. : Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 72:5454-5462, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Stecklein SR, Jensen RA: Identifying and exploiting defects in the Fanconi anemia/BRCA pathway in oncology. Transl Res 160:178-197, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Sharma P Barlow W Godwin A, et al. : Impact of homologous recombination deficiency biomarkers on outcomes in patients with triple-negative breast cancer treated with adjuvant doxorubicin and cyclophosphamide (SWOG S9313). Ann Oncol 29:654-660, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Telli ML Metzger O Timms K, et al. : Evaluation of homologous recombination deficiency (HRD) status with pathological response to carboplatin +/- veliparib in BrighTNess, a randomized phase 3 study in early stage TNBC. J Clin Oncol 36, 2018. (suppl 15; abstr 519) [Google Scholar]
- 11.Mackenzie KJ Carroll P Martin CA, et al. : cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548:461-465, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkes EE Walker SM Taggart LE, et al. : Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J Natl Cancer Inst 109:djw199, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma P Barlow WE Godwin AK, et al. : Validation of the DNA damage immune response signature in patients with triple-negative breast cancer from the SWOG 9313c trial. J Clin Oncol 37:3484-3492, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkes EE Savage KI Lioe T, et al. : Activation of a cGAS-STING-mediated immune response predicts response to neoadjuvant chemotherapy in early breast cancer. Br J Cancer 126:247-258, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid P Cortes J Dent R, et al. : Event-free survival with pembrolizumab in early triple-negative breast cancer. N Engl J Med 386:556-567, 2022 [DOI] [PubMed] [Google Scholar]
- 16.Schmid P Cortes J Pusztai L, et al. : Pembrolizumab for early triple-negative breast cancer. N Engl J Med 382:810-821, 2020 [DOI] [PubMed] [Google Scholar]
- 17.McShane LM Altman DG Sauerbrei W, et al. : Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 97:1180-1184, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Lehmann BD Colaprico A Silva TC, et al. : Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat Commun 12:6276, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami, et al. : The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery 2:401, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehmann BD Jovanović B Chen X, et al. : Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One 11:e0157368, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth S Huo L Rauch GM, et al. : Delineating longitudinal patterns of response to neoadjuvant systemic therapy (NAST) in triple-negative breast cancer (TNBC): Profiling results from a randomized, TNBC enrolling trial to confirm molecular profiling improves survival (ARTEMIS; NCT02276443). J Clin Oncol 37, 2019. (suppl 15; abstr 586) [Google Scholar]
- 22.Sharma P Kimler BF O'Dea A, et al. : Randomized phase II trial of anthracycline-free and anthracycline-containing neoadjuvant carboplatin chemotherapy regimens in stage I-III triple-negative breast cancer (NeoSTOP). Clin Cancer Res 27:975-982, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM Steen CB Liu CL, et al. : Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37:773-782, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severson TM Wolf DM Yau C, et al. : The BRCA1ness signature is associated significantly with response to PARP inhibitor treatment versus control in the I-SPY 2 randomized neoadjuvant setting. Breast Cancer Res 19:99, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.BioRender: http://www.biorender.com
- 26.Tan PH Ellis I Allison K, et al. : The 2019 World Health Organization classification of tumours of the breast. Histopathology 77:181-185, 2020 [DOI] [PubMed] [Google Scholar]
- 27.Lakhani SR Jacquemier J Sloane JP, et al. : Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst 90:1138-1145, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Marcus JN Watson P Page DL, et al. : Hereditary breast cancer: Pathobiology, prognosis, and BRCA1 and BRCA2 gene linkage. Cancer 77:697-709, 1996 [DOI] [PubMed] [Google Scholar]
- 29.Chopra N Tovey H Pearson A, et al. : Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat Commun 11:2662, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortes J Cescon DW Rugo HS, et al. : Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 396:1817-1828, 2020 [DOI] [PubMed] [Google Scholar]
- 31.Loibl S Schneeweiss A Huober JB, et al. : Durvalumab improves long-term outcome in TNBC: Results from the phase II randomized GeparNUEVO study investigating neodjuvant durvalumab in addition to an anthracycline/taxane based neoadjuvant chemotherapy in early triple-negative breast cancer (TNBC). J Clin Oncol 39, 2021. (suppl 15; abstr 506) [Google Scholar]
- 32.Mittendorf EA Zhang H Barrios CH, et al. : Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): A randomised, double-blind, phase 3 trial. Lancet 396:1090-1100, 2020 [DOI] [PubMed] [Google Scholar]
- 33.Schmid P Cortes J Dent R, et al. : VP7-2021: KEYNOTE-522: Phase III study of neoadjuvant pembrolizumab + chemotherapy vs. placebo + chemotherapy, followed by adjuvant pembrolizumab vs. placebo for early-stage TNBC. Ann Oncol 32:1198-1200, 2021 [Google Scholar]
- 34.Schmid P Rugo HS Adams S, et al. : Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 21:44-59, 2020 [DOI] [PubMed] [Google Scholar]
- 35.Jiang Y, Li Y, Zhu B: T-cell exhaustion in the tumor microenvironment. Cell Death Dis 6:e1792, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galluzzi L Buqué A Kepp O, et al. : Immunological effects of Conventional chemotherapy and targeted anticancer agents. Cancer Cell 28:690-714, 2015 [DOI] [PubMed] [Google Scholar]
- 37.Kepp O Galluzzi L Martins I, et al. : Molecular determinants of immunogenic cell death elicited by anticancer chemotherapy. Cancer Metastasis Rev 30:61-69, 2011 [DOI] [PubMed] [Google Scholar]
- 38.Martins I Wang Y Michaud M, et al. : Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ 21:79-91, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obeid M Tesniere A Ghiringhelli F, et al. : Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med 13:54-61, 2007 [DOI] [PubMed] [Google Scholar]
- 40.Vincent J Mignot G Chalmin F, et al. : 5-Fluorouracil selectively kills tumor-associated myeloid-derived suppressor cells resulting in enhanced T cell-dependent antitumor immunity. Cancer Res 70:3052-3061, 2010 [DOI] [PubMed] [Google Scholar]
- 41.Yamazaki T Hannani D Poirier-Colame V, et al. : Defective immunogenic cell death of HMGB1-deficient tumors: Compensatory therapy with TLR4 agonists. Cell Death Differ 21:69-78, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grandal B Evrevin C Laas E, et al. : Impact of BRCA mutation status on tumor infiltrating lymphocytes (TILs), response to treatment, and prognosis in breast cancer patients treated with neoadjuvant chemotherapy. Cancers (Basel) 12:3681, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato H Niimi A Yasuhara T, et al. : DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun 8:1751, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hesson LB, Cooper WN, Latif F: Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene 26:7283-7301, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Kok K, Naylor SL, Buys CH: Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res 71:27-92, 1997 [DOI] [PubMed] [Google Scholar]
- 46.Wolf DM Yau C Wulfkuhle J, et al. : Redefining breast cancer subtypes to guide treatment prioritization and maximize response: Predictive biomarkers across 10 cancer therapies. Cancer Cell 40:609-623 e6, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulligan JM Hill LA Deharo S, et al. : Identification and validation of an anthracycline/cyclophosphamide-based chemotherapy response assay in breast cancer. J Natl Cancer Inst 106:djt335, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.23.00197.