Abstract
Fluorescent in situ hybridization (FISH) is now a widely used method for identification of bacteria at the single-cell level. With gram-positive bacteria, the thick peptidoglycan layer of a cell wall presents a barrier for entry of horseradish peroxidase (HRP)-labeled probes. Therefore, such probes do not give any signal in FISH unless cells are first treated with enzymes which hydrolyze the peptidoglycan. We explored this feature of FISH to detect cells which have undergone permeabilization due to expression of autolytic enzymes. Our results indicate that FISH performed with HRP-labeled probes provides a sensitive method to estimate the states of cell walls of individual gram-positive bacteria.
Fluorescent in situ hybridization (FISH) is a recognized tool for phylogenetic identification of bacteria at the level of individual cells. This method is based on hybridization of fixed cells with oligonucleotide probes, generally targeted to rRNA (1–3). The use of horseradish peroxidase (HRP)-labeled probes, followed by treatment with fluorescent tyramide, was recently reported to increase the sensitivity of FISH by 10- to 20-fold, compared to that of FISH with probes labeled directly with fluorochrome (14). With either type of probe, cells are specifically colored according to their phylogenetic affiliation and can be visualized by epifluorescence microscopy.
For the probe to enter a bacterium, paraformaldehyde-fixed cells are permeabilized with cell wall-degrading enzymes like lysozyme (14). This is particularly important for gram-positive bacteria because of their thick cell walls (16). The permeabilization of fixed bacterial cells is especially important when large probes labeled with high-molecular-mass enzymes such as HRP are used (the molecular mass of HRP is ∼44 kDa, compared to ∼330 Da for fluorescein; these two molecules are joined to a 20-nucleotide oligonucleotide of ∼6 kDa). The requirement for permeabilization is considered a disadvantage where characterization of complex bacterial communities by FISH is concerned, because of difficulties in finding permeabilization conditions which are effective for different types of bacteria (14).
In this work, we exploit the nonpermeability of cells to large probes to estimate the state of the bacterial cell wall. We reasoned that expression of intracellular, peptidoglycan-hydrolyzing enzymes like autolysins or phage-encoded lysins would permeabilize the cell wall and make cells detectable by FISH. We performed experiments on gram-positive bacteria, since gram-negative Escherichia coli cells gave a strong FISH signal with HRP-labeled probes even without permeabilization (not shown).
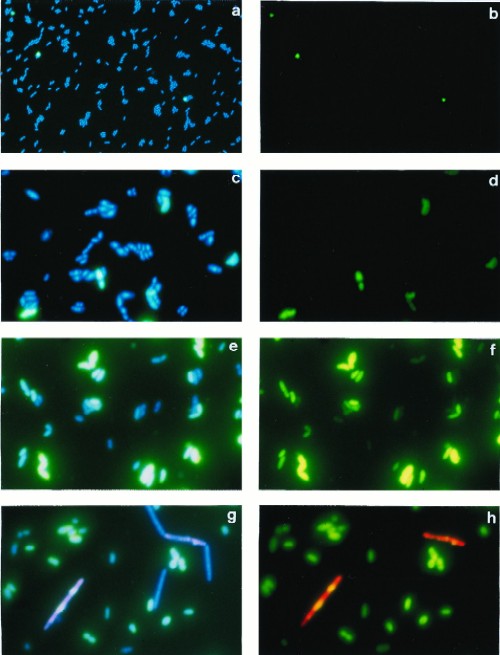
To test out our approach, we examined the effect of phage infection on the ability of the gram-positive bacterium Lactococcus lactis to hybridize to a general HRP-labeled 16S RNA-targeted probe (EUB338) (3, 12). L. lactis subsp. lactis IL1403 (7) was infected with virulent bacteriophage bIL66 (10) as follows. An overnight culture of IL1403, grown at 30°C in M17 medium supplemented with 0.5% (wt/vol) glucose (18), was diluted 100-fold in the same medium and grown to an optical density (OD) at 600 nm of 0.1, CaCl2 (10 mM) was added, and cultures were infected with bacteriophage at a multiplicity of infection of 10. Cell samples were fixed with paraformaldehyde, hybridized with the probe, and colored as described previously (14) by addition of a fluorophore-tyramide substrate (NEN Life Science Products, Boston, Mass.). To visualize the entire cell population, bacteria were colored with 4′,6-diamidino-2-phenylindole (DAPI; 2.5 μg/ml; Sigma, St. Louis, Mo.) and added directly to glycerol–phosphate-buffered saline mountant (Citifluor Ltd., Canterbury, United Kingdom). Probes were purchased from Cybergene (Saint Malo, France). Photographs of cells were taken with an epifluorescence microscope equipped with a three-band filter set for emission light (Nikon, Tokyo, Japan) and Sensia 400 film (Fuji, Tokyo, Japan). Filters appropriate for excitation light (i.e., green, red, or blue) were used to visualize cells which were colored by a particular fluorophore. Hybridized cells were counted on images captured with the image analysis system Visiolab 1000 (Biocom, Les Ulis, France). Total cell counts were determined from images taken without a filter for excitation light; this allows cells colored with DAPI, fluorescein, and rhodamine to be counted simultaneously. The percentage of hybridized cells (seen as green in Fig. 1b, d, and f) among the total number of cells (seen as blue and turquoise in Fig. 1a, c, and e) is referred to hereafter as the hybridization frequency (HF).
FIG. 1.
Epifluorescence micrographs of L. lactis IL1403 before infection with bacteriophage bIL66 (40× lens objective) (a and b), 30 min after infection (100× lens objective) (c and d), and 40 min after infection (100× lens objective) (e and f) and of an artificial mixture of fixed cultures of IL1403 and Lactobacillus helveticus CNRZ493, infected with their specific phages (100× lens objective) (g and h). Micrographs were taken without a filter for excitation light (a, c, e, and g), with a filter for fluorescein (b, d, and f), and with double exposure and filters for fluorescein and rhodamine (h).
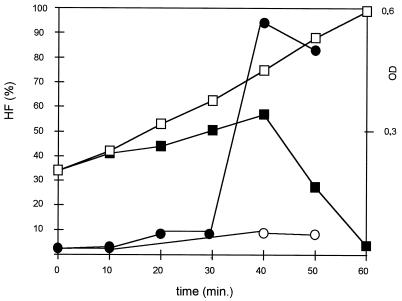
We observed that in an exponentially growing culture of IL1403 only a few cells hybridized, giving an HF of only about 4% (Fig. 2, time point 0; Fig. 1a and b). After infection and at 10 min before the start of culture lysis, as judged by a drop in OD, HF increased sharply to 90% (Fig. 1e and f and 2). We observed similar kinetics of HFs after infection of L. lactis subsp. cremoris MG1363 (9) with phage c2 (11) and after infection of Lactobacillus helveticus CNRZ493 (15) with phage hv (reference 15 and data not shown). In the latter experiments, growth conditions were the same as for IL1403, except that MRS medium (Difco, Detroit, Mich.) was used for Lactobacillus helveticus. As with IL1403, the HF was low prior to and in the early stages of infection and was greatest just before lysis. We consider that the appearance of a FISH signal in phage-infected bacteria is due to permeabilization of the cell walls by phage lysins.
FIG. 2.
Kinetics of the HF of strain IL1403 after infection with bacteriophage bIL66. Open squares, ODs of noninfected culture; filled squares, ODs of infected culture; open circles, HFs of noninfected culture; filled circles, HFs of infected culture. Values are representative of results of five independent measurements.
To further confirm that a hybridization signal with HRP-labeled probes reflects increased permeability of the cell wall, we examined hybridization of untreated cells with the same oligonucleotide probe, but which was labeled with fluorescein (Eurobio, Les Ulis, France), making it a smaller molecule. In contrast to the larger probe, which did not hybridize to cells with intact cell walls, hybridization with the smaller probe gave a weak and uneven signal. These results suggest that small but not large molecules are able to penetrate untreated cells. In contrast, when cells were treated with egg white lysozyme (10 mg/ml, 10 min at 37°C; Serva, Heidelberg, Germany), strong uniform signals were achieved with both HRP-labeled and fluorescein-labeled probes (data not shown).
Lysozyme attacks the glycosidic bond of peptidoglycan (6, 20). Some phage lysins also attack this bond (e.g., the lysin of the prolate-headed phage c2 [8, 10]), while others are of the amidase type and affect peptide bonds (produced by a group of small isometric phages, exemplified by bIL66 [5, 10, 20]). Both types of lysins appear to increase cell permeability. Our results indicate that the increase in the HF before lysis of phage-infected cells is due to permeabilization of the cell wall by phage lysins. The phage-encoded lysins destroy the host cell wall at the end of an infection cycle (8); at this point, just prior to cell lysis, we observed a strong increase in the HF (Fig. 2).
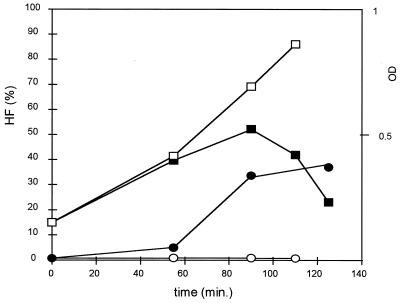
We used another approach to examine FISH signals as a function of cell permeability. The HF of L. lactis subsp. cremoris NCDO 712 was monitored upon induction of its prophage φT712 (9) by mitomycin C (MC). For this, MC (0.5 μg/ml; Boehringer Mannheim GmbH, Mannheim, Germany) was added to exponentially growing NCDO 712 cells. The HF of strain MG1363, a prophage-cured derivative of NCDO 712 (9), was low (about 0.5%) and was unaffected by MC addition (Fig. 3). Similarly, the HF of NCDO 712 without MC addition did not change during the growth period. In contrast, addition of MC resulted in an increase in the HF prior to lysis of the culture.
FIG. 3.
Kinetics of the HF of strain NCDO 712 after induction of prophage with MC. Open squares, ODs of noninduced culture; filled squares, ODs of induced culture; open circles, HFs of noninduced culture; filled circles, HFs of induced culture. Values are representative of results of five independent measurements.
Finally, we asked whether our approach could be used with mixed cultures to identify strains bearing activated prophage. To test whether HFs can also be determined in a mixed bacterial population, we examined an artificial mixture of fixed L. lactis IL1403 and Lactobacillus helveticus CNRZ493 cells, both infected with phages. The mixture was hybridized first with the general EUB-HRP probe with the fluorescein-tyramide substrate. Cells were then heated to 80°C for 20 min to inactivate the HRP and hybridized a second time with probe Lbh1-HRP, which is specific to Lactobacillus helveticus (17). After the second hybridization, cells were colored with rhodamine-tyramide. The hybridized L. lactis cells (green) could be easily distinguished from Lactobacillus helveticus (red or yellow) and from nonhybridized (DAPI-colored blue) cells (Fig. 1g and h). These results confirm that the states of cell walls of different bacteria can be studied with a complex culture.
The above-reported results show that gram-positive bacteria which are permeabilized through the actions of phage-encoded lysins (amidases of bIL66 or lysozyme of c2) can be specifically detected with HRP-labeled probes and by FISH. We tested whether the same approach can be used to examine the actions of chromosome-encoded autolysins, like N-acetylmuramidase, the major autolysin of L. lactis, which is encoded by the acmA gene (4). Strain MG1363, carrying intact acmA, was grown in the buffered medium which maintains a neutral pH (this condition favors expression of the autolytic activity of L. lactis [4, 13]). The HF of MG1363 in these conditions reached ∼87% after 72 h of growth. The HF of the isogenic acmA mutant strain remained low (<1%) during the same period of growth. These results suggest that the high HF of MG1363 seen under these conditions reflects expression of the acmA gene product.
The application of FISH without an additional permeabilization step constitutes a novel method to monitor the state of the bacterial cell wall, as well as expression of lytic enzymes, at the level of individual cells. The percentage of hybridized cells, or HF, is a potentially valuable measurement to estimate the level of autolysis in a culture.
In this work, the actions of two different phage-encoded lysins and a chromosomally encoded autolysin of L. lactis were monitored by this method. We expect that this approach will allow us to screen strains with increased or decreased spontaneous autolysis. The level of spontaneous HF can be an indication of the presence or absence of prophages or may reflect activities of genes encoding autolytic enzymes. It will also be useful in detecting cell wall damage due to other autolytic enzymes and will allow us to determine environmental conditions which lead to cell wall damage and eventual cell death. This method may have particular use in defining conservation conditions for dairy starter. Since induction of prophages may result from activation of global regulatory networks, like the SOS response network (19), the proposed method should also enable us to monitor the induction of such networks at the level of individual cells. We suggest that other in situ coloration techniques which require diffusion into the cells of large-molecular-weight molecules, such as enzymes, antibodies, and streptavidin, can also be used for this purpose. The particular advantage of FISH with HRP-labeled probes, besides its high sensitivity, is that it enables us to color the cells specifically, according to the nucleotide sequence of the probe. Thus, FISH can be used to estimate the state of the cell wall of a particular bacterial species in mixed populations, such as starter cultures for the dairy industry and dairy products. Our technique may be of particular use in examining cell lysis in opaque medium, where simple OD measurements cannot be performed.
Acknowledgments
We thank A. Gruss for numerous improvements to the manuscript and M.-C. Chopin, P. Duwat, S. D. Ehrlich, A.-M. Le Coq, O. Matte-Tailliez, G. Le Bourhis, P. Quénée, and W. Schönhuber for stimulating discussions and helpful readings of the manuscript.
This work was partially supported by the Bureau des Ressources Génétiques, Paris, France.
REFERENCES
- 1.Amann R I. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, section 3.3.6. In: Akkerman A D C, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publisher; 1995. pp. 1–15. [Google Scholar]
- 2.Amann R I. Fluorescently labeled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol Ecol. 1995;4:543–554. [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema D, Haandrilman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandry P S, Moore S C, Boyce J D, Davidson B E, Hiller A J. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol. 1997;26:49–64. doi: 10.1046/j.1365-2958.1997.5491926.x. [DOI] [PubMed] [Google Scholar]
- 6.Chapot-Chartier M-P. Autolysins of lactic acid bacteria. Lait. 1996;76:91–109. [Google Scholar]
- 7.Chopin M-C, Moillo-Batt A, Rouault A. Plasmid-mediated UV protection in Streptococcus lactis. FEMS Microbiol Lett. 1985;26:243–245. [Google Scholar]
- 8.Fastrez J. Phage lysozymes. In: Jollès P, editor. Lysozymes: model enzymes in biochemistry and biology. Basel, Switzerland: Birkhäuser Verlag; 1996. pp. 35–64. [Google Scholar]
- 9.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis A W, Fitzgerald G F, Mata M, Mercenier A, Neve H, Powell I B, Ronda C, Saxelin M, Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32:2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- 11.Lubbers M W, Waterfield N R, Beresford T P J, Le Page R W F, Jarvis A W. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of structural genes. Appl Environ Microbiol. 1995;61:4348–4356. doi: 10.1128/aem.61.12.4348-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rallu, F., and E. Maguin. 1998. Unpublished data.
- 14.Schönhuber W, Fuchs B, Juretschko S, Amann R. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3268–3273. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Séchaud L, Russeau M, Fayard B, Callegari M L, Quénée P, Accolas J-P. Comparative study of 35 bacteriophages of Lactobacillus helveticus: morphology and host range. Appl Environ Microbiol. 1992;58:1001–1018. doi: 10.1128/aem.58.3.1011-1018.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shockman G D, Barett J F. Structure, function and assembly of cell walls of Gram-positive bacteria. Annu Rev Microbiol. 1983;37:501–527. doi: 10.1146/annurev.mi.37.100183.002441. [DOI] [PubMed] [Google Scholar]
- 17.Tailliez, P., and O. Matte-Tailliez. 1997. Unpublished data.
- 18.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Environ Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker G. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1400–1416. [Google Scholar]
- 20.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]