Abstract
Background
The T2-FLAIR mismatch sign is defined by signal loss of the T2-weighted hyperintense area with Fluid-Attenuated Inversion Recovery (FLAIR) on magnetic resonance imaging, causing a hypointense region on FLAIR. It is a highly specific diagnostic marker for IDH-mutant astrocytoma and is postulated to be caused by intercellular microcystic change in the tumor tissue. However, not all IDH-mutant astrocytomas show this mismatch sign and some show the phenomenon in only part of the lesion. The aim of the study is to determine whether the T2-FLAIR mismatch phenomenon has any prognostic value beyond initial noninvasive molecular diagnosis.
Methods
Patients initially diagnosed with histologically lower-grade (2 or 3) IDH-mutant astrocytoma and with at least 2 surgical resections were included in the GLASS-NL cohort. T2-FLAIR mismatch was determined, and the growth pattern of the recurrent tumor immediately before the second resection was annotated as invasive or expansive. The relation between the T2-FLAIR mismatch sign and tumor grade, microcystic change, overall survival (OS), and other clinical parameters was investigated both at first and second resection.
Results
The T2-FLAIR mismatch sign was significantly related to Grade 2 (80% vs 51%), longer post-resection median OS (8.3 vs 5.2 years), expansive growth, and lower age at second resection. At first resection, no relation was found between the mismatch sign and OS. Microcystic change was associated with areas of T2-FLAIR mismatch.
Conclusions
T2-FLAIR mismatch in IDH-mutant astrocytomas is correlated with microcystic change in the tumor tissue, favorable prognosis, and Grade 2 tumors at the time of second resection.
Keywords: astrocytoma, glioma, magnetic resonance imaging
Key Points.
T2-FLAIR mismatch is correlated with micocystic change.
T2-FLAIR mismatch is associated with better survival and a higher probability of Grade 2 at recurrence.
T2-FLAIR mismatch at initial diagnosis is often retained at recurrence.
Importance of the Study.
Astrocytomas, IDH-mutant that start as lower-grade tumors are known to undergo malignant transformation sooner or later, leading to a rapid deterioration of the patient and resistance to therapy. The T2-FLAIR mismatch sign is a known diagnostic marker that is highly specific of this type of glioma, but its clinical relevance beyond initial diagnosis is unknown. In this study, we find that the T2-FLAIR mismatch sign is related to a low-grade and longer median survival at second resection, indicating that it could be used as a prognostic marker at the time of tumor recurrence. Even in the absence of contrast enhancement, which is an established indicator of low grade, we find that the T2-FLAIR mismatch sign can provide additional evidence of a better prognosis and low grade.
Glioma Longitudinal AnalySiS (GLASS)-NL is a multicenter consortium in the Netherlands (NL) and part of the international GLASS initiative.1 GLASS-NL focusses on changes underlying malignant progression in astrocytomas, IDH-mutant (henceforth “astrocytomas”) through the analysis of molecular characteristics of repeated resections (Vallentgoed et al., unpublished manuscript) and longitudinal magnetic resonance imaging (MRI). A highly specific imaging feature to this type of glioma is the presence of a near-complete mismatch between the signal on T2-weighted (T2w) and T2-weighted Fluid-Attenuated Inversion Recovery (FLAIR) MRI, also called the T2-FLAIR mismatch sign. In this study, we investigate the longitudinal characteristics of this mismatch phenomenon and its relation to tumor malignancy grade and patient prognosis.
The signal on FLAIR is similar to T2w within brain tissue, but the free fluid signal is dark FLAIR whereas on T2w it is hyperintense. A nonenhancing lesion is generally hyperintense on both sequences, but in low-grade, glioma it has been noted that in areas where T2w shows a distinct high signal, the FLAIR signal may sometimes be relatively hypointense. The T2-FLAIR mismatch sign describes the phenomenon where nearly the entire lesion shows this signal intensity mismatch except for a bright outer rim. It was first reported by Patel et al.2 as an imaging marker for noninvasive diagnosis of astrocytoma, and subsequently validated in multiple studies3,4 to be highly specific (approximately 99%) for these tumors within the group of adult-type diffuse low-grade gliomas (LGGs). However, the sensitivity of the T2-FLAIR mismatch sign was found to be low, as it is described to be present in approximately half of all astrocytomas at the initial diagnosis. A clear relationship was reported between the T2-FLAIR mismatch sign and the presence of microcysts in histological slides of the tumors.2,5,6 Previous studies reported no significant difference in outcome or clinical parameters between cases with and without the mismatch sign.3,5,6 However, these analyses were performed on studies with small sample sizes (n < 50), and only MRI information at initial diagnosis was used. To date, no longitudinal study has been performed on the T2-FLAIR mismatch sign.
So far, it remains unclear whether the T2-FLAIR mismatch sign has clinical relevance beyond an MRI-based (ie, noninvasive) indication of astrocytoma in adults. Astrocytomas that start as lower-grade tumors are known to sooner or later undergo malignant transformation, so a noninvasive marker for tumor grade can potentially inform treatment decisions for recurrent disease. The GLASS-NL cohort provides a unique opportunity to relate T2-FLAIR mismatch to tumor grade at progression in a clinically well-defined cohort. Furthermore, the longitudinal nature of the data acquisition allows us to assess the growth pattern of the recurrent tumor and the change in T2-FLAIR mismatch over time.
The aim of this work is to validate the correlation between the T2-FLAIR mismatch phenomenon and microcystic change in the GLASS-NL-cohort and to investigate the clinical relevance of the mismatch phenomenon during the course of tumor evolution. Specifically, we aim to investigate whether T2-FLAIR mismatch is related to tumor grade and overall survival (OS) and whether it has added prognostic value when considering the presence of contrast enhancement and tumor grade. It is also possible that a part of the lesion shows a T2-FLAIR mismatch, but the mismatch is not “near-complete” as is required for the T2-FLAIR mismatch sign. In this study, we consider also these cases as a T2-FLAIR mismatch area, distinct from the mismatch sign, as this distinction may be relevant for the relation with histopathology and clinical parameters.
Methods
Patient Inclusion
The GLASS-NL cohort is a retrospective multicenter cohort from the Netherlands. Patients were included from Amsterdam UMC, Erasmus MC, and UMC Utrecht according to the following inclusion criteria:
Patient was first diagnosed as an adult (>18 years old);
The initial histological diagnosis was lower-grade (Grade II or III) astrocytoma, IDH-mutant according to the CNS WHO-2016 classification7;
Patient underwent surgical resection at least twice, with a second surgery performed after progression and with at least 6 months’ time difference;
Both surgeries yielded tumor tissue sufficient for molecular diagnosis;
For this study, samples from the GLASS-NL cohort were selected where preoperative MR imaging was available, meeting the following requirements:
The following sequences were available: (1) T2-weighted, (2) T2-weighted FLAIR, (3) T1-weighted, and (4) T1-weighted after administration of a gadolinium-based contrast agent (postcontrast T1-weighted).
The image quality was sufficient to delineate the lesion.
The latest available preoperative scan meeting these criteria was used for image analysis and annotation. This study was approved by the ethical review board of Amsterdam UMC (VUMC 2019.085). The study was performed in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Clinical Characteristics
The location of the lesion and presence in or near eloquent areas according to Sawaya et al.8 were annotated by a radiologist (A.L.) for each scan, both at first and second resection. Clinical characteristics and treatment history were retrieved from electronic health records. OS and progression-free survival (PFS) were measured from both the date of first (OS-R1, PFS-R1) and second resection (OS-R2, PFS-R2) till the time of death or progression respectively, or censored at last follow-up date. As clinical practice has changed over time, with an initial period of watchful waiting being more common in earlier samples, the time of resection was preferred over the time of diagnosis as a reference date for survival analysis. Tumor grade was re-evaluated according to the WHO-2021 classification.9
Volume Measurement and Annotation of Enhancement
The tumor was delineated automatically using HD-GLIO10,11 and corrected semiautomatically using ITK-Snap.12 The resulting two-class segmentation was used to compute the contrast-enhancing tumor (CET) volume and whole tumor (WT) volume (T2-weighted abnormalities + CET). If parts of the abnormalities on T2-weighted imaging were clearly attributable to treatment effects, they were excluded from the volume.
The presence and thickness of the contrast-enhancing margin were annotated by a radiologist (A.L.) according to the VASARI features.13 If there was a disconcordance between the annotation of contrast enhancement and the presence of contrast-enhancing volume in the segmentation in recurrent lesions, which may be a result of postsurgery changes, a board-certified and expert neuroradiologist (M.S.) was consulted to decide whether the lesion was enhancing or nonenhancing.
T2-FLAIR Mismatch Annotation
A distinction was made between the presence of a T2-FLAIR mismatch area and the T2-FLAIR mismatch sign. A lesion was annotated as having a mismatched area if an area was found with the following characteristics:
The T2w sequence is homogeneously hyperintense in this area;
The FLAIR sequence is clearly hypointense in this area in comparison to FLAIR-hyperintense areas elsewhere in the same lesion (eg, a hyperintense rim);
The T2-FLAIR mismatched area is not contrast-enhancing, necrotic, or a cyst, although these aspects may be present near or within the mismatched area.
In order to be classified as having the mismatch sign, the following criteria were used:
In initial lesions: almost the entire lesion shows T2-FLAIR mismatch area, except for a thin hyperintense rim;
In recurrent lesions: the lesion identified as a recurrent tumor almost entirely shows T2-FLAIR mismatch, with the presence of a hyperintense rim at the interface of the recurrent lesion and healthy-appearing brain.
This is according to the recommendations by Jain et al.14 for initial lesions. However, the criteria for recurrent lesions were adapted as preexisting and potentially treatment-induced abnormalities can be hyperintense on the T2-weighted FLAIR scan and thereby result in an incomplete T2-FLAIR mismatch phenomenon. Both the T2-FLAIR mismatch area and the T2-FLAIR mismatch sign were annotated for all included preoperative scans as (YES/NO). Note that a mismatch area is a prerequisite for the mismatch sign, so all cases with a mismatch sign necessarily also show a mismatch area. If the first rater (K.A.v.G.) was not sure whether a T2-FLAIR mismatch (sign or area) was present, a board-certified and expert neuroradiologist (M.S.) was consulted to reach a decision in consensus.
Growth Pattern Annotation
The growth pattern was annotated for the recurrent lesions, by the same rater as the T2-FLAIR mismatch (K.A.v.G.), by comparing the preoperative scan to a prior reference scan, selected to show the most recent visible growth of the tumor. The following categories were used:
Mostly invasive: the recurrent lesion mostly infiltrates formerly healthy appearing tissue;
Mostly expansive: the recurrent lesion barely seems to invade surrounding preexistent brain tissue, but rather shows expansive growth, thereby variably displacing/compressing the surrounding brain tissue;
Mixed: both patterns of growth are present and neither is clearly dominant;
Not sure: the growth pattern cannot be distinguished, for example, because there is not enough growth visible or the scan quality is insufficient.
Not available: there is no imaging available.
Only the category “Not available” was considered missing values and excluded from the statistical analysis.
Histopathology
Histological slides of the tumors were stained with hematoxylin-eosin (H&E) and digitized using a whole slide scanner (Hamamatsu NanoZoomer 2.0 HT). The presence/absence of microcysts was assessed on the section originally used for histopathological diagnosis in all samples where sufficient preoperative imaging was available for the assessment of T2-FLAIR mismatch sign, by a board-certified, expert-neuropathologist (J.M.K.) who was blinded to MRI data. If only a small part of the samples showed microcysts, or if the microcystic change was incipient but visible, it was still annotated as present. If the scanned samples did not have sufficient quality to identify microcysts, the sample was annotated as NOT SURE and excluded from the analysis.
Statistical Analysis
Statistical analysis was performed using Python (v3.8.13) and the packages SciPy (v1.8.0) and lifelines (v0.27.4). The group of tumors with T2-FLAIR mismatch sign and those with T2-FLAIR mismatch area, the latter by definition also including tumors with the T2-FLAIR mismatch sign, were compared to the group of tumors without T2-FLAIR mismatch sign/area. The analysis was performed at first and second resections, including for each resection all patients with available imaging data at that time. Additionally, the clinical characteristics of the entire cohort (including those without imaging data) were reported at time of first and second resections. Fisher’s exact test was used to test differences in categorical variables. The difference in continuous variables (volume and age) between groups was tested using the Mann–Whitney-U test. A threshold of P < .05 was used for significance. In case of missing values in the clinical parameters, the number of patients missing the parameter was reported and they were excluded from statistical testing for that specific parameter. The first and second resections were analyzed separately as availability of imaging was different at both time points.
Survival analysis was performed by visualizing the Kaplan–Meier estimates for groups and comparing these using the log-rank test. The groups were defined based on the presence of T2-FLAIR mismatch area and sign, tumor grade according to the WHO-2021 classification,9 and the presence of microcysts.
As the analysis at the second resection may include cases with clear malignant transformation, a stratified analysis of clinical characteristics and OS-R2 was performed in the subset of nonenhancing samples at second resection. Additionally, a stratified analysis of OS was performed in cases of CNS WHO-2021 Grade 2.
To assess whether the longitudinal changes in T2-FLAIR mismatch affected survival, another comparison was made for OS-R2 for T2-FLAIR mismatch area and sign in longitudinal categories, creating 4 groups for each: patients where the mismatch sign/area was present in both first and second resection (“preserved”), those where it was present at second resection but not at first (“gained”), those where it was present at first resection but not at second (“lost”) and those where it was never present (“never”).
Results
Patient Inclusion and Characteristics
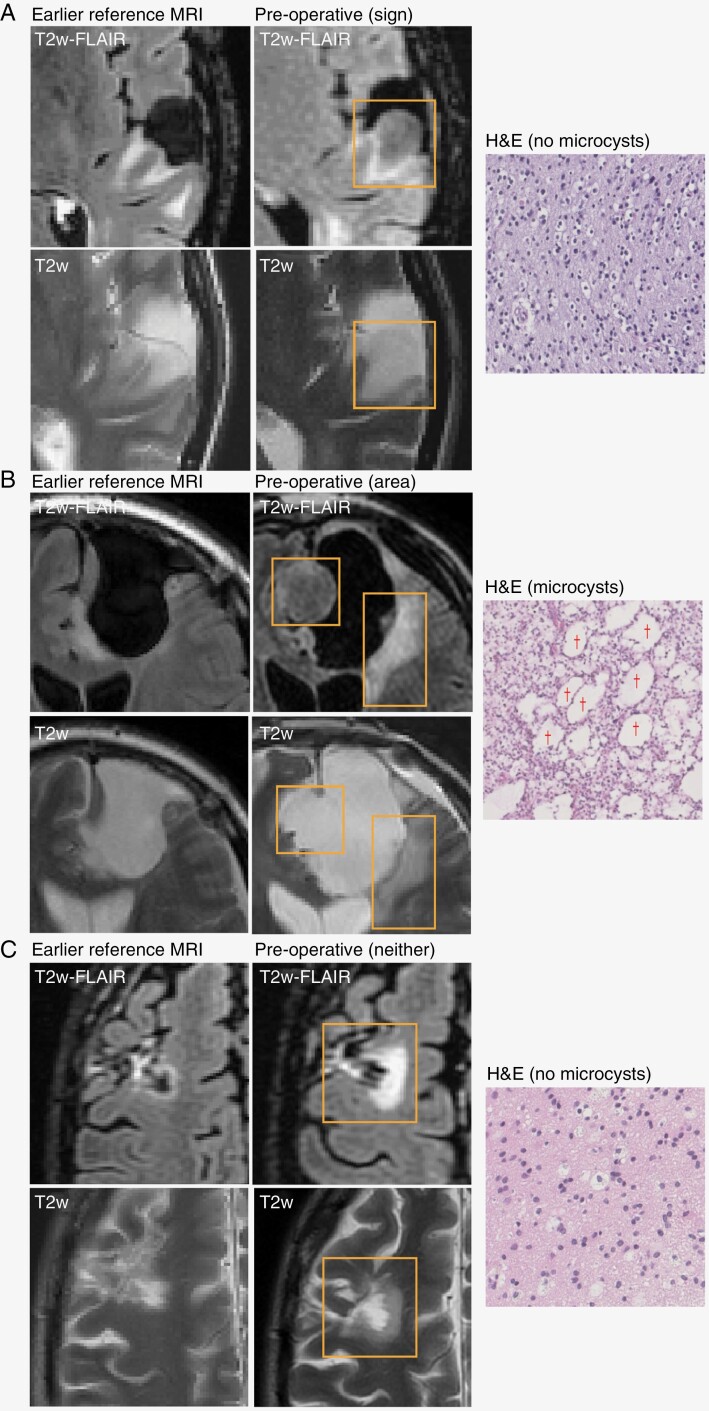
A total of 101 patients were included in GLASS-NL, with a total of 224 tissue samples. Although there were tissue samples of at least 2 resections for each patient, for some patients the tissue of the first or second resection was missing. A total of 98 samples were included at first resection and 97 at second resection. For 32 and 4 patients, a sample could be included from a third and fourth resection, respectively. Figure 1 shows examples of the T2-FLAIR mismatch sign and area at second resection, including the corresponding H&E slides and growth pattern.
Figure 1.
Examples of FLAIR and T2w MRI and H&E slides for 3 cases. The last preoperative MRI is shown with an earlier reference MRI used to assess the growth pattern. Recurrent lesions are outlined with a rectangle. Examples of microcysts are indicated by a “†” symbol. (A) Recurrent tumor with mismatch sign; preoperative MRI shows pre-existing treatment effect, but recurrent growth is mismatched with hyperintense rim; growth pattern is mostly expansive; H&E does not show microcysts. (B) Recurrent tumor that is partly mismatched, so this case shows a T2-FLAIR mismatch area but no mismatch sign; growth pattern is mixed; H&E shows large microcysts. Note that the source of the histopathology (mismatch area or not) is unknown. (C) Recurrent tumor without T2-FLAIR mismatch; growth pattern is mostly invasive; H&E shows no microcysts.
Clinical Characteristics at First Resection
At first resection, 45 samples were included. A complete overview of patient characteristics at first resection, stratified by the presence/absence of the T2-FLAIR mismatch sign is shown in Supplementary Table 1. Supplementary Table 2 contains the same characteristics stratified by T2-FLAIR mismatch area. The mean age at diagnosis was 32 years, median OS was 9.6 years (95% CI: 8.8–10.7) and median PFS was 2.8 years (95% CI: 2.6–3.3). The T2-FLAIR mismatch sign was found in 13 patients (29%) and a T2-FLAIR mismatch area was found in an additional 26 patients (58%). The remaining 6 patients (13%) showed no T2-FLAIR mismatch at all. There was no significant difference between patients with and without the T2-FLAIR mismatch sign for any of the clinical parameters at first resection. Patients with a T2-FLAIR mismatch area received chemotherapy less often than those without either a T2-FLAIR mismatch area or sign (1/39 vs 3/6 patients, 3% vs 50%) and were less often diagnosed with Grade 4 (1/39 vs 3/6 patients, 3% vs 50%, reclassified according to WHO-2021 criteria).
Clinical Characteristics at Second Resection and Beyond
At second resection, 76 samples could be included. Table 1 shows an overview of patient characteristics at second resection, stratified by the presence/absence of the T2-FLAIR mismatch sign. Supplementary Table 3 shows the same overview stratified by T2-FLAIR mismatch area. Median OS-R2 was 5.8 years (95% CI: 4.2–7.4) and median PFS-R2 was 2.5 years (95% CI: 1.6–3.5).
Table 1.
Patient Characteristics and Annotation Results at Second Resection, Stratified by the Presence of T2-FLAIR Mismatch Sign
| T2-FLAIR mismatch sign | Absent (n = 51) | Present (n = 25) | P-value |
|---|---|---|---|
| Female sex (%) | 17 (33%) | 15 (60%) | 0.05 |
| Age at diagnosis (y) median (range) | 34.0 (18.0–70.0) | 28.0 (19.0–53.0) | 0.03 |
| Median time since diagnosis in y (range) | 4.5 (1.2–23.5) | 3.9 (1.0–14.9) | 0.3 |
| Median overall post-resection survival (OS-R2) in y (95% CI) | 5.2 (2.8–5.9) | 8.3 (6.4—N/A) | 0.001 |
| Median progression-free post-resection survival (PFS-R2) in y (95% CI) | 1.6 (1.3–2.8) | 4.1 (2.5—N/A) | 0.01 |
| Median time to second resection in y (range) | 3.6 (1.0–17.5) | 3.6 (0.9–13.9) | 0.8 |
| CNS WHO-2021 grade | |||
| Grade 2 | 26 (51%) | 20 (80%) | 0.02 |
| Grade 3 | 7 (14%) | 2 (8%) | 0.71 |
| Grade 4 | 18 (35%) | 3 (12%) | 0.05 |
| KPS before surgery | |||
| 100 | 21 (41%) | 14 (56%) | 0.33 |
| 90 | 19 (37%) | 8 (32%) | 0.8 |
| <90 | 11 (22%) | 3 (12%) | 0.37 |
| Location | |||
| Side of lesion center | |||
| Left | 24 (47%) | 13 (52%) | 0.81 |
| Right | 27 (53%) | 12 (48%) | 0.81 |
| Location in or near eloquent regions (Sawaya et al.) | |||
| Eloquent (III) | 36 (71%) | 9 (36%) | 0.006 |
| Near-eloquent (II) | 8 (16%) | 7 (28%) | 0.23 |
| Noneloquent (I) | 7 (14%) | 9 (36%) | 0.04 |
| Tumor site (multiple sites possible) | |||
| Frontal lobe | 39 (76%) | 22 (88%) | 0.36 |
| Temporal lobe | 23 (45%) | 6 (24%) | 0.09 |
| Insula | 21 (41%) | 5 (20%) | 0.08 |
| Corpus callosum | 7 (14%) | 0 (0%) | 0.09 |
| Parietal lobe | 15 (29%) | 3 (12%) | 0.15 |
| Occipital lobe | 3 (6%) | 0 (0%) | 0.55 |
| Brainstem | 1 (2%) | 0 (0%) | 1 |
| Basal ganglia | 1 (2%) | 0 (0%) | 1 |
| Thalamus | 1 (2%) | 0 (0%) | 1 |
| Treatment | |||
| Extent of resection | |||
| Partial resection | 36 (71%) | 13 (52%) | 0.13 |
| Complete resection | 15 (29%) | 12 (48%) | 0.13 |
| Radiotherapy | 15 (29%) | 12 (48%) | 0.13 |
| Chemotherapy | 18 (35%) | 8 (32%) | 1.00 |
| Prior treatment | |||
| Radiotherapy | 21 (41%) | 5 (20%) | 0.08 |
| Number of radiotherapy treatments | |||
| 1 | 20 (39%) | 5 (20%) | 0.12 |
| 2 | 1 (2%) | 0 (0%) | 1.00 |
| Chemotherapy | 11 (22%) | 1 (4%) | 0.09 |
| Number of chemotherapy treatments | |||
| 1 | 7 (14%) | 1 (4%) | 0.26 |
| 2 | 2 (4%) | 0 (0%) | 1.00 |
| Biopsy | 6 (12%) | 1 (4%) | 0.41 |
| Radiological features | |||
| Median contrast-enhancing volume in mL (range) | 0.1 (0.0–63.8) | 0.0 (0.0–0.3) | <0.001 |
| Median whole tumor volume in mL (range) | 22.5 (1.7–149.3) | 12.1 (1.3–29.0) | 0.01 |
| Thickness of enhancing margin | |||
| Not applicable | 25 (49%) | 20 (80%) | 0.01 |
| Thin (<3 mm) | 11 (22%) | 4 (16%) | 0.76 |
| Thick/nodular (≥3 mm) | 10 (20%) | 1 (4%) | 0.09 |
| Solid | 5 (10%) | 0 (0%) | 0.16 |
| Growth pattern | |||
| Mostly invasive | 13 (27%) | 4 (18%) | 0.55 |
| Mostly expansive | 2 (4%) | 5 (23%) | 0.03 |
| Not sure | 18 (38%) | 7 (32%) | 0.79 |
| Mixed | 15 (31%) | 6 (27%) | 0.79 |
| Not available | 3 | 3 | 1 |
| Mismatch sign at first resection | |||
| Yes | 4 (14%) | 7 (50%) | 0.02 |
| No | 24 (86%) | 7 (50%) | 0.02 |
| Not available | 23 | 11 | 1 |
Note: First column (all) includes patients where no sufficient imaging was available. P-value compares columns absent and present. Missing values are reported “as Not available,” but not included in the computation of percentages and P-values.
The T2-FLAIR mismatch sign was found in 25 patients (33%), and an additional 11 patients (14%) showed a T2-FLAIR mismatch area but not the sign. The remaining 40 patients (53%) did not show any T2-FLAIR mismatch.
Both the presence of the T2-FLAIR mismatch area and sign were related to a lower mean age at initial diagnosis and a lower probability of the lesion being in an eloquent area. The T2-FLAIR mismatch area and sign were related to a lower contrast-enhancing volume and a higher probability of being nonenhancing and of an expansive growth pattern. The median WT volume was significantly lower in patients with the T2-FLAIR mismatch sign (12.1 vs 22.5 mL, P = .01), but not in patients with the T2-FLAIR mismatch area (16.9 vs 20.4 mL, P = .29).
Longitudinal T2-FLAIR Characteristics
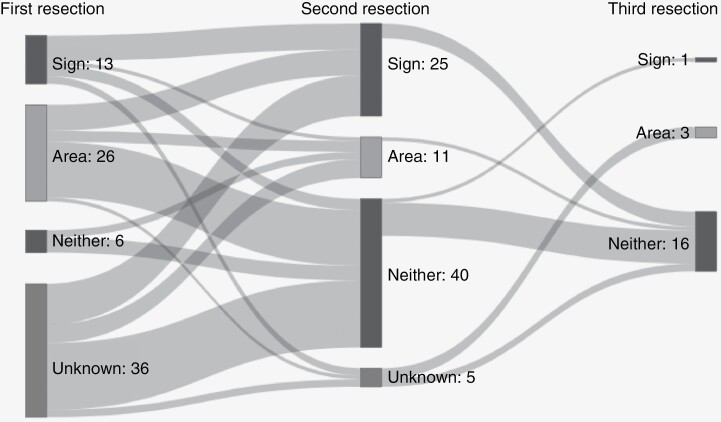
The prevalence of T2-FLAIR mismatch area decreased with second resection (87% vs 47% without mismatch, P < .001), but the prevalence of the T2-FLAIR mismatch sign did not change significantly between first and second resection (29% vs 33%, P = .69). There was a significant positive relation between the presence of T2-FLAIR mismatch sign at first and second resection (P = .02), although 4 patients lost the T2-FLAIR mismatch sign and 7 patients gained the T2-FLAIR mismatch sign at second resection. At the third resection, 20 samples could be matched with a preoperative scan and annotated, of which one showed T2-FLAIR mismatch sign. At the fourth resection, with 2 matching scans available, none of the scans showed the T2-FLAIR mismatch phenomenon anymore.
See Sankey diagram in Figure 2 for visualization of the number of cases showing T2-FLAIR mismatch sign and/or area in the sequential resection specimens.
Figure 2.
Sankey diagram of T2-FLAIR mismatch sign and area over repeated resections (left: first, middle: second, and right: third). Area indicates that there was an area of T2-FLAIR mismatch, but the lesion did not meet the criteria for the mismatch sign. Resections of the same patients are connected by gray bands.
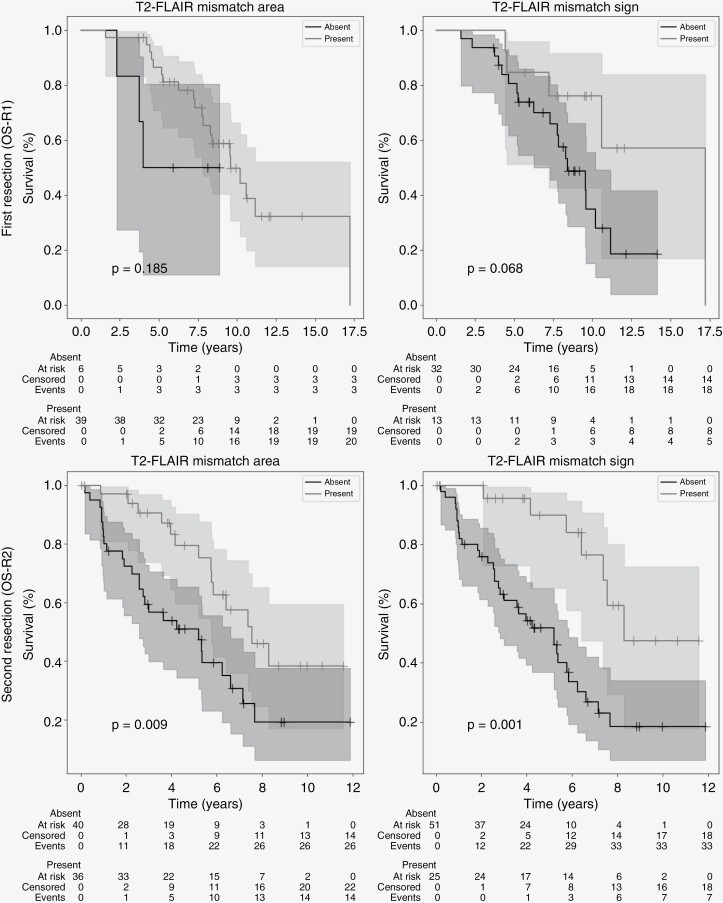
Survival Analysis
Figure 3 shows the Kaplan–Meier OS curves for presence/absence of both the T2-FLAIR mismatch area and sign at first (OS-R1) and second resection (OS-R2). There was no significant difference in OS-R1 in patients with or without a T2-FLAIR mismatch area (P = .19) or sign (P = .07). The tumor grade at first resection, according to WHO-2021 guidelines, was also not a significant prognostic factor in this cohort (P = .12 CNS WHO Grade 2 vs 3 or 4, P = .73 Grade 2 or 3 vs 4), see Supplementary Figure 5. The median PFS-R1 was significantly higher in patients with than in patients without the T2-FLAIR mismatch sign (3.5 vs 2.2 years, P = .002). When looking at the T2-FLAIR mismatch area, the difference in PFS-R1 was not significant (2.8 vs 2.8 years, P = .51).
Figure 3.
Kaplan–Meier curves for OS-R2 of mismatch area (left) and mismatch sign (right) at first (top) and second (bottom) resection. Starting date for the analysis is the date of first and second resection, respectively, and T2-FLAIR mismatch area/sign was annotated on the last available MRI before resection. Censored patients indicated by a “+” at date of last follow-up. Shaded areas indicate 95% confidence intervals.
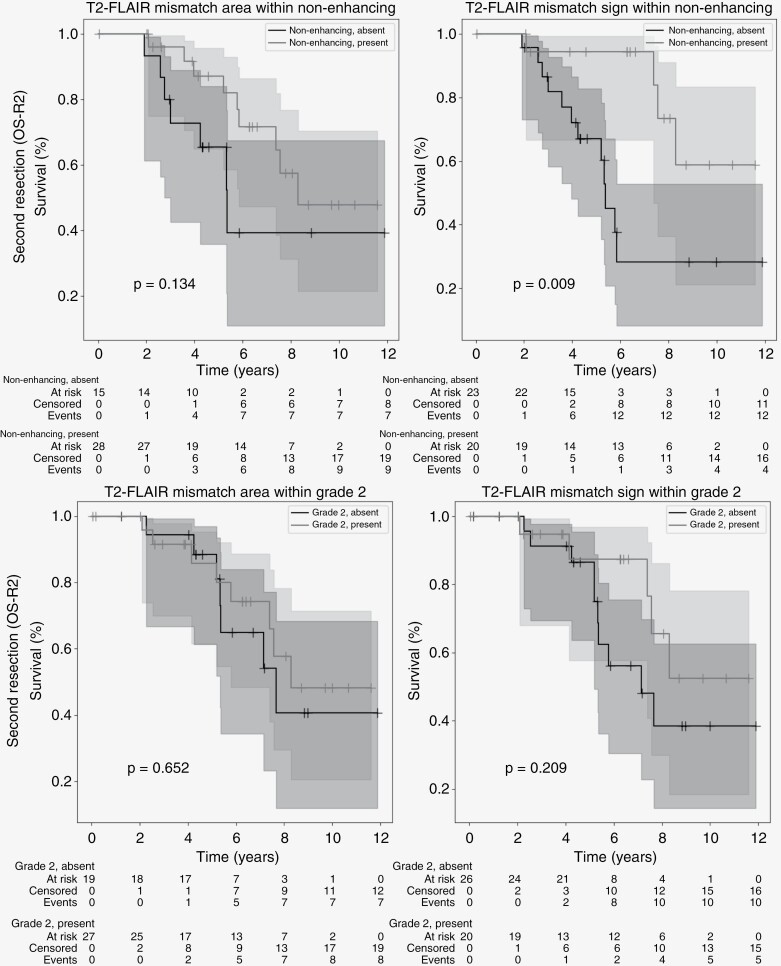
At second resection, both the presence of T2-FLAIR mismatch area and sign were related to longer OS-R2 (P = .001 sign, P = .009 area; Figure 3) and PFS-R2 (P = .01, P = .07 area). Tumor grade in general was also a prognostic factor at second resection (P < .001 Grade 2 vs 3 or 4, P < .001 Grade 2 or 3 vs 4). When considering only cases of CNS WHO Grade 2, there was no significant difference in OS-R2 between patients with or without the T2-FLAIR mismatch sign at second resection (P = .21). When considering only nonenhancing lesions, the presence of the T2-FLAIR mismatch sign was still a strongly significant prognostic factor (P = .009). Figure 4 contains the Kaplan–Meier curves for these stratified analyses. No significant difference in OS was found between patients with and without microcysts at first resection (OS-R1, P = .12) or second resection (OS-R2, P = .11), see Supplementary Figure 6.
Figure 4.
Kaplan–Meier curves of mismatch area (left) and sign (right) combined with other indicators of good prognosis at second resection. Top: Nonenhancing lesions. Bottom: WHO-2021 Grade 2. Starting date of the analysis is date of second resection. T2-FLAIR mismatch area/sign and enhancement were annotated on the last available MRI before resection. Censored patients indicated by a “+” at date of last follow-up. Shaded areas indicate 95% confidence intervals.
For the longitudinal categories of T2-FLAIR mismatch sign/area, there was a significant difference in OS-R2 for patients where the T2-FLAIR mismatch area was preserved versus gained (P = .004). For the T2-FLAIR mismatch sign, there was no significant difference between preserved or gained. Supplementary Figure 7 contains the Kaplan–Meier curves for all 4 categories.
Analysis in Nonenhancing Recurrent Lesions
When considering only the nonenhancing lesions at second resection 20 out of 43 patients (47%) showed the T2-FLAIR mismatch sign. The group with the T2-FLAIR mismatch sign had a longer OS-R2 (P = .009) after resection, but there was no significant difference in PFS-R2 (P = .13), tumor grade (P = .18), age at diagnosis (P = .90) or tumor volume (P = .53). The complete overview of clinical characteristics for nonenhancing lesions at second resection is shown in Table 2.
Table 2.
Patient Characteristics and Annotation Results at Second Resection for Nonenhancing Lesions only, Stratified According to the Presence of T2-FLAIR Mismatch Sign
| T2-FLAIR mismatch sign | Absent (n = 23) | Present (n = 20) | P-value |
|---|---|---|---|
| Female sex (%) | 9 (39%) | 12 (60%) | 0.23 |
| Median age at diagnosis in y (range) | 27.0 (18.0–54.0) | 28.0 (19.0–41.0) | 0.90 |
| Median time since diagnosis in y (range) | 3.6 (1.2–18.7) | 3.8 (1.3–14.9) | 0.80 |
| Median overall post-resection survival (OS-R2) in y (95% CI) | 5.4 (4.0—N/A) | N/A (7.4—N/A) | 0.009 |
| Median progression-free post-resection survival (PFS-R2) in y (95% CI) | 2.7 (1.4–7.7) | 5.2 (1.8—N/A) | 0.13 |
| CNS WHO-2021 grade | |||
| Grade 2 | 15 (65%) | 17 (85%) | 0.18 |
| Grade 3 | 4 (17%) | 2 (10%) | 0.67 |
| Grade 4 | 4 (17%) | 1 (5%) | 0.35 |
| KPS before resection | |||
| 100 | 14 (61%) | 11 (55%) | 0.76 |
| 90 | 8 (35%) | 6 (30%) | 1.00 |
| <90 | 1 (4%) | 3 (15%) | 0.32 |
| Location | |||
| Side of lesion center | |||
| Left | 13 (57%) | 12 (60%) | 1.00 |
| Right | 10 (43%) | 8 (40%) | 1.00 |
| Center | 0 (0%) | 0 (0%) | 1.00 |
| Location in or near eloquent regions (Sawaya et al.) | |||
| Eloquent (III) | 14 (61%) | 7 (35%) | 0.13 |
| Near-eloquent (II) | 5 (22%) | 6 (30%) | 0.73 |
| Noneloquent (I) | 4 (17%) | 7 (35%) | 0.29 |
| Tumor site (multiple sites possible) | |||
| Frontal lobe | 16 (70%) | 18 (90%) | 0.14 |
| Temporal lobe | 11 (48%) | 5 (25%) | 0.21 |
| Insula | 7 (30%) | 4 (20%) | 0.50 |
| Corpus callosum | 5 (22%) | 0 (0%) | 0.05 |
| Parietal lobe | 4 (17%) | 1 (5%) | 0.35 |
| Occipital lobe | 1 (4%) | 0 (0%) | 1.00 |
| Treatment | |||
| Extent of resection | |||
| Partial resection | 18 (78%) | 10 (50%) | 0.06 |
| Complete resection | 5 (22%) | 10 (50%) | 0.06 |
| Radiotherapy | 9 (39%) | 9 (45%) | 0.76 |
| Chemotherapy | 5 (22%) | 6 (30%) | 0.73 |
| Prior treatment | |||
| Radiotherapy | 3 (13%) | 2 (10%) | 1.00 |
| Chemotherapy | 0 (0%) | 0 (0%) | 1.00 |
| Biopsy | 1 (4%) | 1 (5%) | 1.00 |
| Radiological features | |||
| Median contrast-enhancing volume in mL (range) | 0.0 (0.0–0.1) | 0.0 (0.0–0.1) | 0.79 |
| Median whole tumor volume in mL (range) | 11.5 (1.7–73.1) | 11.4 (1.3–29.0) | 0.53 |
| Thickness of enhancing margin | |||
| Not applicable | 21 (91%) | 18 (90%) | 1.00 |
| Thin (<3 mm) | 2 (9%) | 2 (10%) | 1.00 |
| Growth pattern | |||
| Mostly invasive | 7 (32%) | 3 (17%) | 0.46 |
| Mostly expansive | 0 (0%) | 4 (22%) | 0.03 |
| Mixed | 8 (36%) | 5 (28%) | 0.74 |
| Not sure | 7 (32%) | 6 (33%) | 1.00 |
| Not availablea | 1 | 2 | 0.59 |
Notes: Missing values are reported as “Not available,” but not included in the computation of percentages and P-values. *The growth pattern could not be assessed if no reference scan was available prior to progression (see Section 2.5).
aThe growth pattern could not be assessed if no reference scan was available prior to progression (see section 2.5).
When we compared the lack of contrast enhancement to the T2-FLAIR mismatch sign as a marker for Grade 2, we found that the positive predictive value (PPV) was higher (80%) for the T2-FLAIR mismatch sign than the absence of contrast enhancement (74%), although the sensitivity was lower (43% vs 70%). When we combine the 2 markers, considering lesions that are nonenhancing and show the T2-FLAIR mismatch sign, the PPV for Grade 2 increases to 85%, while the sensitivity is 39%. The confusion matrices for these 3 imaging markers are shown in Supplementary Table 4.
Microcysts
When combining all samples, including third and fourth resections, 88 out of 137 samples (64%) contained microcysts and the relation with T2-FLAIR mismatch area was significant (P = .03). However, there was no significant relation with the T2-FLAIR mismatch sign (P = .16).
At first resection, 42 samples were evaluated of which 23 (55%) were found to have microcysts. Two samples had to be excluded due to insufficient quality. In the samples with microcysts, all MRI scans also showed a T2-FLAIR mismatch area. Only 6 (26%) of the samples with microcysts had the T2-FLAIR mismatch sign on MRI. In the samples without microcysts, 13 MRI scans (68%) still showed a T2-FLAIR mismatch area and 5 (26%) showed the T2-FLAIR mismatch sign. There was a significant correlation between the presence of microcysts and T2-FLAIR mismatch area (P = .005), but there was no significant relation with the T2-FLAIR mismatch sign (P = .99).
At the second resection, all 76 samples could be annotated and 52 (68%) contained microcysts. In the samples with microcysts, 29 (56%) also showed a T2-FLAIR mismatch area and 21 (40%) also showed the T2-FLAIR mismatch sign on MRI. There was a significant difference in the presence of microcysts for both the T2-FLAIR mismatch area (P = .03) and sign (P = .04). Supplementary Table 5 shows the confusion matrix for microcysts and T2-FLAIR mismatch sign/area at first and second resection.
Discussion
In this study, the presence of T2-FLAIR mismatch area and the sign was analyzed in the GLASS-NL cohort, a longitudinal study of astrocytomas, IDH-mutant. In general, we find that the T2-FLAIR mismatch sign is related to a higher probability of Grade 2 and a better prognosis at recurrence. Considering the risk of malignant progression in this patient group, the presence of the T2-FLAIR mismatch sign is a potential clinically relevant marker for low-grade recurrence that is highly specific but not sensitive. Comparing to the presence/absence of contrast enhancement, which is a well-established indicator of malignant progression, in astrocytoma the T2-FLAIR mismatch sign can be considered an additional strong indicator of low-grade recurrence and good prognosis.
The interpretation of OS in this cohort is difficult, as it is prone to survivorship bias and confounding factors. Patients included in this cohort survived at least up to 6 months after the first resection and were eligible for a second resection as per the inclusion criteria, indicating a relatively good condition and a tumor at a location accessible for surgery. This may also contribute to our finding that tumor grade at first resection was not a prognostic marker for OS in this cohort.
Although OS is generally measured from the date of diagnosis, the time between diagnosis and the first or second resection may vary due to changing treatment standards. Therefore, the dates of resection were used as a starting point for survival analysis. Considering that the patients became eligible for inclusion in the GLASS-NL study only at the time of the second resection, we took this date as a starting point for the analysis as well. However, in interpreting the results at second resection we must be careful to consider the potential confounding effect of treatment decisions. It is possible that a longer survival is caused by a difference in intervention rather than overall prognosis, especially when considering radiological parameters. These lesions more often showed an expansive growth pattern and tended to appear more well-delineated, which could make them more likely to be considered for resection and amenable to gross total resection. However, there was no significant difference in the time between diagnosis and first or second resection for patients with or without the T2-FLAIR mismatch sign.
The T2-FLAIR mismatch sign at second resection was also correlated to age, location, WT volume, and contrast-enhancing tumor volume, which are potential confounding variables for OS. However, this analysis includes enhancing lesions that show clear malignant progression (such as contrast enhancement). When considering only nonenhancing lesions, the T2-FLAIR mismatch sign was still strongly correlated with OS while the correlations with age, volume, and location were no longer found.
Although the GLASS-NL cohort is unique in its availability of tissue at least two-time points in the disease process, it is not suitable to draw firm conclusions about prognosis. It is possible that the findings at recurrence also extend to the initial presentation, but the homogeneity of the cohort and low availability of MRI at first resection in our study likely cause insufficient power to distinguish any correlation between the presence of the T2-FLAIR mismatch sign and tumor grade or OS. When considering the T2-FLAIR mismatch sign and area at both resections combined, we found evidence of an improved prognosis for those who showed a mismatched area already at first resection, versus those who gained it at second resection. If the mismatch sign is a representation of some biological property that reduces malignancy, then it is unlikely that this property will be gained over time. This suggests that a mismatch sign gained at recurrence is less reliable as a marker for low grade. For future research, it would be worthwhile to study the relation between T2-FLAIR mismatch at initial presentation and recurrence in a large cohort of astrocytomas without selection for treatment or initial histological grade.
The distribution of the T2-FLAIR mismatch area and sign was different at first and second resections. At the first resection, the presence of a T2-FLAIR mismatch area (but no sign) was common, while the absence of any T2-FLAIR mismatch was rare. At the time of second resection, the absence of a T2-FLAIR mismatch area was more common, but a T2-FLAIR mismatch area (without sign) was rare and the T2-FLAIR mismatch sign was found at approximately the same rate as in the first resection. This is an indication that the criteria for the T2-FLAIR mismatch sign as further refined by Jain et al.14 was strictly interpreted at initial diagnosis, while consideration of treatment-related changes caused the T2-FLAIR mismatch area (without sign) to be annotated less often. In general, the potential ambiguity in the definition of the T2-FLAIR mismatch sign at recurrence is a limitation for its use in clinical practice and interpretation in research. Recent studies also show that the detection of the mismatch phenomenon may be highly dependent on acquisition parameters of the FLAIR scan,15 which could cause cases of T2-FLAIR mismatch sign or area to remain undetected.
From this study, it is unclear how the cases with a T2-FLAIR mismatch area, but without meeting all criteria for the T2-FLAIR mismatch sign at recurrence should be interpreted. In future research, it would be worthwhile to measure the mismatch sign as a continuous variable and investigate the percentage of T2-FLAIR mismatch in the lesion as a prognostic marker. However, this would not be feasible for clinical practice without a robust automated volume measurement. In general, the T2-FLAIR mismatch sign shows a stronger relation to prognosis and grade than a T2-FLAIR mismatch area, suggesting that a strict interpretation of the marker and adherence to the criteria should be preferred while taking into account potential treatment effects in recurrent lesions.
The presence of microcysts as observed by histopathological analysis was significantly correlated with the presence of a T2-FLAIR mismatch area, which supports the hypothesis that the mismatch phenomenon is a direct result of microcystic change. A limitation of this analysis is that the exact location of acquisition of the histopathology slides was unknown, making it impossible to correlate the T2-FLAIR mismatch area with microcystic change. Previous studies have shown a correlation between the T2-FLAIR mismatch sign and microcystic change,2,5,6 but did not include recurrent samples or a distinction between T2-FLAIR mismatch area sign. Small areas of T2-FLAIR mismatch that do not meet the criteria for the T2-FLAIR mismatch sign are not specific to astrocytoma, and neither is microcystic change, so this correlation would likely be found in other LGGs such as oligodendroglioma, IDH-mutant and 1p19q-codeleted. The longitudinal analysis shows a correlation between the T2-FLAIR mismatch sign at first and second resection, suggesting that there could be a distinct property that causes the T2-FLAIR mismatch sign in a subgroup of astrocytomas. Further research would be needed to find an underlying cause or property that explains the presence of the T2-FLAIR mismatch sign.
To conclude, in the GLASS-NL cohort, we found that the T2-FLAIR mismatch sign is related to a low grade and better prognosis at recurrence and that it has prognostic relevance in addition to the absence of contrast enhancement. However, it is possible that the treatment regimen affected the results in this retrospective study, as the appearance of lesions with the T2-FLAIR mismatch sign may have affected the diagnosis of progression and the decision to undergo a second resection. Due to the design of the cohort, we cannot draw firm conclusions concerning the prognostic value of the T2-FLAIR mismatch sign at the initial presentation. We conclude that the T2-FLAIR mismatch sign is a potential additional marker for favorable prognosis in recurrent astrocytoma, IDH-mutant that should be investigated further.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (https://academic.oup.com/neuro-oncology).
Contributor Information
Karin A van Garderen, Department of Radiology & Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands; Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands; Medical Delta, Delft, The Netherlands.
Wies R Vallentgoed, Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands; Department of Neurology, Erasmus MC, Rotterdam, The Netherlands.
Anna Lavrova, Department of Radiology & Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands.
Johanna M Niers, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; Cancer Center Amsterdam, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Wendy W J de Leng, Department of Pathology, UMC Utrecht, Utrecht, The Netherlands.
Youri Hoogstrate, Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands; Department of Neurology, Erasmus MC, Rotterdam, The Netherlands.
Iris de Heer, Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands; Department of Neurology, Erasmus MC, Rotterdam, The Netherlands.
Bauke Ylstra, Cancer Center Amsterdam, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; Department of Pathology, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Erik van Dijk, Cancer Center Amsterdam, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; Department of Pathology, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Stefan Klein, Department of Radiology & Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands.
Kaspar Draaisma, Department of Neurosurgery, UMC Utrecht, The Netherlands.
Pierre A J T Robe, Department of Neurosurgery, UMC Utrecht, The Netherlands.
Roel G W Verhaak, Jackson Laboratory for Genomic Medicine, Farmington, CT, USA; Department of Neurosurgery, Amsterdam UMC, Amsterdam, The Netherlands.
Bart A Westerman, Department of Neurosurgery, Amsterdam UMC, Amsterdam, The Netherlands.
Pim J French, Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands; Department of Neurology, Erasmus MC, Rotterdam, The Netherlands.
Martin J van den Bent, Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands; Department of Neurology, Erasmus MC, Rotterdam, The Netherlands.
Mathilde C M Kouwenhoven, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; Cancer Center Amsterdam, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Johan M Kros, Department of Pathology, Erasmus MC, Rotterdam, The Netherlands.
Pieter Wesseling, Department of Pathology, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Marion Smits, Department of Radiology & Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands; Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands; Medical Delta, Delft, The Netherlands.
Funding
Dutch Cancer Society (KWF 11026, GLASS-NL); Medical Delta (to K.A.v.G.).
Conflict of Interest
M.S. reports receiving speaker fees (paid to the institution) from GE Health care and AuntMinnie.com, and consulting fees (paid to the institution) from Bracco SpA. The other authors declare no conflicts of interest.
Authorship Statement
Concept and design: K.A.v.G., W.R.V., P.J.F, M.J.v.d.B., M.C.M.K., J.M.K., P.W., M.S. Data curation and patient inclusion: K.A.v.G., W.R.V., J.M.N., W.W.J.d.L., I.d.H., K.D., P.A.J.T.R, M.C.M.K., J.M.K. Annotation of imaging and histopathology: K.A.v.G., A.L., M.S., J.M.K. Drafting of the manuscript and statistical analysis: K.A.v.G. Supervision: P.J.F., M.J.v.d.B., M.S., S.K. Obtained funding: P.W., M.J.v.d.B., M.C.M.K., P.F., B.A.W., M.S., R.G.W.V., J.M.K. Critical revision of the manuscript: all authors. Unpublished manuscripts cited: Vallentgoed et al.
Data Availability
Additional raw data will be made available upon reasonable request.
References
- 1. Aldape K, Amin SB, Ashley DM, et al. Glioma through the looking GLASS: molecular evolution of diffuse gliomas and the Glioma Longitudinal Analysis Consortium. Neuro-Oncol. 2018;20(7):873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patel SH, Poisson LM, Brat DJ, et al. T2–FLAIR mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: a TCGA/TCIA project. Clin Cancer Res. 2017;23(20):6078–6085. [DOI] [PubMed] [Google Scholar]
- 3. Corell A, Ferreyra Vega S, Hoefling N, et al. The clinical significance of the T2-FLAIR mismatch sign in grade II and III gliomas: a population-based study. BMC Cancer. 2020;20(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Broen MPG, Smits M, Wijnenga MMJ, et al. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro Oncol. 2018;20(10):1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deguchi S, Oishi T, Mitsuya K, et al. Clinicopathological analysis of T2-FLAIR mismatch sign in lower-grade gliomas. Sci Rep. 2020;10(1):111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamashita S, Takeshima H, Kadota Y, et al. T2-fluid-attenuated inversion recovery mismatch sign in lower grade gliomas: correlation with pathological and molecular findings. Brain Tumor Pathol. 2022;39(2):88–98. [DOI] [PubMed] [Google Scholar]
- 7. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 8. Sawaya R, Hammoud M, Schoppa D, et al. Neurosurgical outcomes in a modern series of 400 craniotomies for treatment of parenchymal tumors. Neurosurgery. 1998;42(5):1044–55; discussion 1055. [DOI] [PubMed] [Google Scholar]
- 9. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kickingereder P, Isensee F, Tursunova I, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20(5):728–740. [DOI] [PubMed] [Google Scholar]
- 11. Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH.. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203–211. [DOI] [PubMed] [Google Scholar]
- 12. Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 13. Nicolasjilwan M, Hu Y, Yan C, et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol. 2015;42(4):212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jain R, Johnson DR, Patel SH, et al. “Real world” use of a highly reliable imaging sign: “T2-FLAIR mismatch” for identification of IDH mutant astrocytomas. Neuro Oncol. 2020;22(7):936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kinoshita M, Arita H, Takahashi M, et al. Impact of inversion time for FLAIR acquisition on the T2-FLAIR mismatch detectability for IDH-mutant, non-CODEL astrocytomas. Front Oncol. 2021;10:3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional raw data will be made available upon reasonable request.