Abstract
Glioma is the most common malignant intracranial tumor, accounting for 80 % of all malignant brain tumors. Growing evidence suggests that lncRNAs are involved in the growth, angiogenesis, metastasis, and therapeutic resistance in a variety of tumors, including glioma. In this study, lncBIRC3-OT (NONHSAT159592.1), which is highly expressed in glioma, was screened by RNA-seq method and verified by quantitative reverse transcription polymerase chain reaction. Subsequently, we knocked down the endogenous expression of lncBIRC3-OT in U87 and U251 cells and found that down-regulated lncBIRC3-OT inhibited cell proliferation, colony formation, migration, and invasion. Mechanically, lncBIRC3-OT could guide RELA protein to the stanniocalcin-1 (STC1) promoter, initiate STC1 transcription, and ultimately promote the progression of glioma. Together, these findings suggest that lncBIRC3-OT is an important regulator promoting glioma progression, and may be a promising therapeutic target for glioma.
Keywords: Glioma, Long non-coding RNA, LncBIRC3-OT, RELA, Stanniocalcin-1
1. Background
Glioma is the most common malignant intracranial tumor, accounting for 80 % of all malignant brain tumors [1]. In recent years, advances in new technologies and research have greatly improved the clinical diagnosis, prognosis assessment, and treatment of glioma [2]. However, the prognosis of glioma patients remains unfavorable, with overall survival of only 14–17 months [3]. Therefore, there is an urgent need to elucidate and decipher the biological mechanisms of glioma occurrence and development, and to develop effective therapeutic strategies to overcome this deadly disease.
Long non-coding RNAs (LncRNAs) are a class of non-coding transcripts with more than 200 nucleotides [4]. LncRNAs are involved in transcriptional, post-transcriptional, and epigenetic regulation, thereby participating in various physiological and pathological processes [5,6]. There is growing evidence that lncRNAs are involved in tumor growth, angiogenesis, metastasis, and drug resistance in gliomas [[7], [8], [9]]. For example, lncRNA XLOC013218 confers temozolomide resistance by activating PI3K/AKT signaling in glioblastoma [10]. LncRNA LINREP promotes glioma progression by protecting polypyrimidine tract-binding protein 1 from ubiquitin-proteasome degradation [11]. LncRNA-FAM66C modulates the tumor microenvironment and hypoxia-related pathways in glioblastoma [12]. Overall, lncRNAs have been recognized as important epigenetic regulators of gliomas.
In this study, we identified lncBIRC3-OT (NONHSAT159592.1) as a conserved and significantly upregulated lncRNA in gliomas via RNA sequencing and qRT-PCR. Next, we demonstrate that lncBIRC3-OT knockdown inhibited the proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) process in U87 and U251 cells. Regarding the mechanism, lncBIRC3-OT recruits RELA to the STC1 promoter, and then initiates STC1 transcription, thereby enhancing the malignant behaviors of glioma cells. Thus, lncBIRC3-OT may be a promising therapeutic target for glioma.
2. Materials and methods
2.1. Cells and cell culture
Human glioma cell lines (U87 and U251) were purchased from the American Type Culture Collection (Rockville, Maryland, USA) and cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco, Carlsbad, USA) supplemented with 10 % fetal bovine serum (Gibco, Carlsbad, USA).
2.2. Clinical specimens
The 22 glioma tissues and 15 normal brain tissues used in this study were collected from the Characteristic Medical Center of the Chinese People's Armed Police Force with written informed consent. This study was approved by the Ethical Review Committees of the Characteristic Medical Center of the Chinese People's Armed Police Force (No. 2022-0002.1). Written informed consent has been obtained from the participant or guardian.
2.3. Database
The mRNA expression and survival analysis of STC1 in glioblastoma multiform (GBM) were analyzed by the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn) [13]. The JASPAR database was used to predict the potential transcription factor binding site of transcription factor RELA on the STC1 promoter [14].
2.4. Cell transfection
RELA overexpressed plasmids (pcDNA3.1-RELA), STC1 overexpressed plasmids (pcDNA3.1-STC1), lncBIRC3-OT shRNAs (sh-lncBIRC3-OT, 5′-AAGAAAGAACATGTAAAGTGTGT-3′) and RELA siRNA (si-RELA, 5′-CTCTTCTCAAGTGCCTTAATAGT-3′) were provided by GenePharma (Shanghai, China). Transfection of pcDNA3.1-RELA, pcDNA3.1-STC1, and sh-lncBIRC3-OT was performed by Lipofectamine 2000 (Thermo Fisher, Waltham, MA, USA). Transfection of si-RELA was conducted by Lipofectamine RNAiMAX (Thermo Fisher, Waltham, MA, USA).
2.5. Quantitative reverse transcription polymerase chain reaction (QRT–PCR)
The expression levels of lncBIRC3-OT and STC1 mRNA were examined by qRT-PCR. TRIzol (Invitrogen, USA) was applied for RNA extraction. SYBR Green (Takara, Japan) was used to prepare the reaction mix. QRT-PCR was performed in the ABI 7500 Real-Time PCR System. The relative expression of indicated genes was calculated using the 2−ΔΔCt method, normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primer sequences for qRT-PCR are listed in Table 1.
Table 1.
The primer sequences for RT-PCR.
| Genes | Primer sequences |
|---|---|
| NONHSAT183359.1 | Forward: 5′-AGCCTCAGTAGCCG-3′ Reverse: 5′-TCCCGTCACTTTCAG-3′ |
| STC1 | Forward: 5′-AGCTGCCCAATCACTTCTCC-3′ Reverse: 5′-CTCATTGGTGCGTCTCCTGT-3′ |
| RELA | Forward: 5′- ATTTCCGCCTCTGGCGAATG-3′ Reverse: 5′- TGATCTCCACATAGGGGCCA-3′ |
| GAPDH | Forward: 5′-GACAGTCAGCCGCATCTTCT-3′ Reverse: 5′-GCGCCCAATACGACCAAATC-3′ |
2.6. CCK-8 assay
1 × 103 cells were seeded into a 96-well plate. After 24, 48, or 72 h of incubation, 10 μL CCK-8 solution was added to each well for staining. After incubation for 1 h, the absorbance was detected at 450 nm using an enzyme immunoassay analyzer.
2.7. Colony formation assay
Cells (1 × 103/well) were implanted into 12-well plates. After two weeks, cells were stained with 0.1 % crystal violet (Solarbio, Beijing, China) for 10 min. Finally, cell clones were observed under a microscope and photographed.
2.8. Cell invasion assay
Transwell chambers (Corning, NY, USA) were pretreated with Matrigel (BD Bioscience, Bedford, USA) and were used for cell invasion assays. Cells (1 × 105 cells) in 200 μL of FBS-free DMEM were added to the upper chamber, and 600 μL of complete medium to the lower chamber. After 48 h of culture, cells were fixed with 4 % paraformaldehyde for 30 min. After staining with 1 % crystal violet solution for 15 min, the cells that invaded the lower chamber were photographed and counted under a microscope.
2.9. Western blot
Protein was isolated with Lysis Buffer (Sangon, Shanghai, China). BCA Protein Quantitation Kit (Sangon, Shanghai, China) was used for protein quantification. Then, the protein samples were separated by SDS-PAGE, transferred to nitrocellulose membranes, and blocked with 5 % BSA. The membranes were incubated with primary antibodies overnight at 4 °C and then incubated with secondary antibodies for 1 h. The membranes were treated with ECL luminescence reagent (Sangon, Shanghai, China). The protein bands were then exposed and scanned. Image J software was used to analyze the grayscale values of protein bands. The primary antibodies used in this study are listed as follows: E-cadherin (1:5000, #60335-1-lg), N-cadherin (1:2000, #66219-1-lg), Vimentin (1:2000, #60330-1-lg), RELA (1:1000, #66535-1-lg), STC1 (1:1000, #20621-1-AP) and GAPDH (1:10000, #60004-1-lg) were obtained from Proteintech (Wuhan, China).
2.10. RNA pull-down assay
RNA pull-down assay was performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (ThermoFisher, Shanghai, China) as previously reported [15]. The biotin-labeled lncBIRC3-OT probes (Zoonbio, Nanjing, China) were incubated with streptavidin magnetic beads (ThermoFisher, Shanghai, China) for 30 min at room temperature to generate probe-bound beads. Cell lysates were mixed with probe-bound beads. After 1 h of incubation, the RNA-binding protein complexes were collected, washed, and eluted. The proteins were separated and subjected to SDS-PAGE. Specific bands were excised and analyzed by mass spectrometry (MS) or Western blot.
2.11. RNA immunoprecipitation (RIP) assay
EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Merck, Beijing, China) was used for RIP assay as previously reported [16]. Cells were harvested and lysed with polysome lysis buffer. Then, cell lysates were incubated with 5 μg RELA or IgG antibody (Proteintech, Wuhan, China) overnight at 4 °C, and then incubated with protein A/G beads for 1 h at 4 °C. After centrifugation at 2500 rpm for 30 s, bead precipitates were resuspended in 500 μL RIP buffer, washed, and then incubated with TRIzol RNA extraction reagent to isolate coprecipitated RNAs. Subsequently, the purified RNA samples were determined by qRT-PCR to assess the enrichment of lncBIRC3-OT to RELA.
2.12. Chromatin immunoprecipitation (ChIP)
To verify whether RELA binds to the STC1 promoter, a ChIP assay was conducted using the SimpleChIP® Plus Enzymatic Chromatin IP Kit (CST, Massachusetts, USA) [17]. Briefly, the cells were treated with 1 % formaldehyde for 10 min to produce the protein-DNA cross-linked complexes. The complex was then lysed and digested by micrococcal nucleases to form chromatin fragments of 200–800 bp. Approximately 5 % of the sample was saved and served as the input control before adding beads. The prepared fragments were incubated with the RELA antibodies bound to magnetic beads (IgG was the negative control). The magnetic bead-antibody-protein-DNA complexes were fully washed, and the protein-DNA complexes were purified and eluted. Protease K was used to unlink the DNA-protein crosslink. Finally, the obtained DNA fragments were analyzed by qRT-PCR and the fold changes of % input were used for analysis.
2.13. Data analysis
SPSS 22.0 software (SPSS Inc., USA) was used for all statistical analysis. Data were shown as Mean ± standard deviation (SD). Student's t-test or analysis of variance (ANOVA) was used to compare two or more groups. P < 0.05 was considered significant.
3. Results
3.1. LncBIRC3-OT expression is upregulated in gliomas
In previous study [18], we identified 243 up-regulated and 1163 down-regulated lncRNAs (p < 0.05 and |log2FC| > 2) in GBM tumor tissues relative to peritumor tissues by RNA sequencing (Fig. 1A). We focused on an uncharacterized lncRNA, NONHSAT159592.1, which is located on human chromosome 11 and overlaps with exons 8–10 of baculoviral IAP repeat containing 3 (BIRC3) gene (Fig. 1B). Therefore, we termed it lncRNA BIRC3 overlapping gene (lncBIRC3-OT). LncBIRC3-OT consists of three exons spanning 1041 bp and is a modestly conserved gene (Fig. 1B). The expression of lncBIRC3-OT was further examined in 22 gliomas and 15 normal brain tissues. lncBIRC3-OT was found to be significantly highly expressed in glioma samples (Fig. 1C). Further analysis shows no correlation of lncBIRC3-OT expression with patient sex, WHO grade, MGMT, GFAP, and Ki-67 expression (Table 2). In addition, we validated that lncBIRC3-OT was significantly upregulated in several glioma cells compared to NHA cells, with U87 and U251 cells exhibiting the highest expression of lncBIRC3-OT (Fig. 1D).
Fig. 1.
LncBIRC3-OT is highly expressed in glioma. (A) Differentially expressed lncRNAs in primary glioma tumors compared with peritumor tissues. (B) The UCSC database showed the genomic locus of lncBIRC3-OT on chromosome 11 and its conversation using Phylop software. (C) LncBIRC3-OT expression was higher in glioma tumor tissues (n = 22) than in normal brain tissues (n = 15). Data were normalized to endogenous GAPDH expression. (D) LncBIRC3-OT was detected in glioma cell lines and NHA cell lines. **P < 0.01, ***P < 0.001.
Table 2.
The relationship between lncBIRC3-OT expression and clinical characteristics.
| Characteristics | LncBIRC3-OT Expression |
P value | |
|---|---|---|---|
| Low (n = 11) | High (n = 11) | ||
| Age (Range) | 14–37 | ||
| Sex | |||
| Female | 2 | 2 | 1.000 |
| Male | 9 | 9 | |
| WHO Grade | 0.193 | ||
| III | 6 | 3 | |
| IV | 5 | 8 | |
| MGMT | 0.500 | ||
| Negative | 7 | 6 | |
| Positvie | 4 | 5 | |
| GFAP | 0.107 | ||
| Negative | 3 | 0 | |
| Positvie | 8 | 11 | |
| Ki-67 | 0.762 | ||
| ≤5 % | 1 | 1 | |
| >5 % | 10 | 10 | |
3.2. LncBIRC3-OT is required for malignant behaviors of glioma cells
To determine the biological role of lncBIRC3-OT in the malignant behaviors of glioma cells, we designed a series of shRNAs targeting lncBIRC3-OT to knockdown lncBIRC3-OT expression in U87 and U251 cells. As shown in Figs. S1A–B, sh#3 and sh#6 could significantly down-regulated lncBIRC3-OT expression but had no or little effect on BIRC3 expression. Moreover, the expression of lncBIRC3-OT in sh#3-transfected cells was lower than that in #6-transfected cells, so sh#3 (named sh-lncBIRC3-OT) was screened for subsequent experiments (Fig. 2A). Moreover, transfection of sh-lncBIRC3-OT decreased the expression of lncBIRC3-OT in both cytoplasm and nucleus (Fig. S1C). Next, CCK-8 assay showed that lncBIRC3-OT depletion significantly inhibited cell viability compared to negative control (NC) cells (Fig. 2B). Furthermore, lncBIRC3-OT knockdown remarkably inhibited the ability of a single cell to form a colony in U87 and U251 cells (Fig. 2C). Additionally, wound healing assay indicated that lncBIRC3-OT depletion reduced the migration of U87 and U251 cells (Fig. 2D). Furthermore, transwell assay showed that the number of lncBIRC3-OT-deficient cells passing through matrix gel was significantly reduced in U87 and U251 cells (Fig. 2E). Notably, lncBIRC3-OT depletion decreased the expression of N-cadherin and Vimentin compared to NC group. In contrast, lncBIRC3-OT knockdown increased the expression of epithelial marker E-cadherin (Fig. 2F). These findings indicated that lncBIRC3-OT is required for malignant behaviors of glioma cells and knockdown of lncBIRC3-OT inhibits proliferation, migration, invasion, and EMT.
Fig. 2.
LncBIRC3-OT is required for malignant behaviours of glioma cells. (A) LncBIRC3-OT was silenced in U87 and U251 cells by shRNAs. (B) Cell viability was detected in lncBIRC3-OT-depleted cells by CCK-8 assay. (C) LncBIRC3-OT depletion causes a diminished colony-forming capacity in U87 and U251 cells. (D) Cell migration was detected in lncBIRC3-OT-depleted cells by wound-healing assays. (E) Cell invasion was analyzed in lncBIRC3-OT-silenced U87 and U251 cells by Transwell invasion assay. (F) N-cadherin, Vimentin, and E-cadherin were analyzed in lncBIRC3-OT-depleted cells by Western blot analysis. Uncropped images of blots are shown in Supplementary material. Data were shown as mean ± SD from three independent experiments. **P < 0.01, ***P < 0.001 vs. the NC groups.
3.3. LncBIRC3-OT physically interacts with RELA
A large number of studies have shown that lncRNA-binding proteins are closely related to various physiological and pathological processes [19]. We speculated that lncBIRC3-OT may promote tumor progression of glioma by binding proteins to regulate downstream gene expression. To figure out the mechanism of lncBIRC3-OT in glioma, we explored the potential interacting proteins of lncBIRC3-OT through RNA pull-down assay and mass spectrometry -based quantitative proteomics. Silver-staining results showed that a specific band was observed in the immunoprecipitation complex enriched with lncBIRC3-OT probe (Fig. 3A). Further LS-MS analysis revealed that it was probably RELA (Fig. 3B). Western blot assay showed that RELA was enriched in the lncBIRC3-OT-pulldown complex (Fig. 3C). To further support this view, RELA antibody was used for RIP experiments, and the results showed that lncBIRC3-OT expression in the RELA antibody-enriched complex was significantly higher than that in the lgG group, confirming the interaction of lncBIRC3-OT with RELA (Fig. 3D). Moreover, immunofluorescence assay revealed that lncBIRC3-OT was colocalized with RELA in the nuclei of U251 cells (Fig. 3E). These data indicate that lncBIRC3-OT interacts with RELA in glioma cells.
Fig. 3.
LncBIRC3-OT associates with RELA. (A) Silver staining of proteins retrieved by lncBIRC3-OT probe (Sense). Scramble probe (Antisense) was used as a negative control. Whole cell lysates (Input) was used as a positive control. (B) RELA-specific peptides were identified with peptide mass fingerprinting using mass spectrometry analysis. (C) Western blot was used to measure RELA expression. Uncropped images of blots are shown in Supplementary material. (D) RIP assay was conducted to examine the interaction of lncBIRC3-OT with RELA. Uncropped images of blots are shown in Supplementary material. (E) LncBIRC3-OT was visualized by RNA-FISH, and immunofluorescence staining of RELA in U251 cells was performed.
3.4. LncBIRC3-OT functioned with RELA to promote STC1 expression
Previous studies showed that RELA is a transcription factor to facilitates STC1 transcription in esophageal squamous cell carcinoma [20]. STC1 has been identified as a crucial oncogene in glioma [21]. The expression of STC1 mRNA was upregulated in glioma (Fig. 4A), and high levels of STC1 predicted poor prognosis via the GEPIA database (Fig. 4B). JARPAR database showed the binding motif of RELA to STC1 promoter (Fig. 4C). To determine whether RELA regulates STC1 expression in glioma, ChIP experiment was performed. As shown in Fig. 4D, PCR amplification was performed on P1–P4 fragments without binding sites and P5 fragments with binding sites. It was found that the P5 fragment on the STC1 promoter region was significantly enriched in the immunoprecipitants of anti-RELA antibody. However, other fragments did not show enrichment. Furthermore, we performed a ChIP assay under the lncBIRC3-OT knockdown condition. The results showed that lncBIRC3-OT knockdown reduced the enrichment of the P5 fragment (Fig. 4E). Moreover, the endogenous expression of RELA was knocked down in U87 and U251 cells (Fig. 4F). We found that RELA deletion reduced the expression of STC1 mRNA in U87 and U251 cells (Fig. 4G). In addition, overexpression of RELA upregulated the expression of STC1 mRNA in glioma cells, whereas knockdown of lncBIRC3-OT abolished the promotion (Fig. 4H), indicating that lncBIRC3-OT is essential for RELA-mediated STC1 transcription in glioma tissues.
Fig. 4.
LncBIRC3-OT recruits RELA to initiate STC1 expression. (A) STC1 was overexpressed in glioma samples compared to control samples. (B) The survival analysis of STC1 in glioma. (C) JASPAR database showed the binding sites of RELA on STC1 promoter. (D) ChIP-PCR assays indicated that RELA is bound to the STC1 promoter in U87 and U251 cells. TSS, transcriptional start site. ***P < 0.001 vs. lgG group. (E) ChIP assay under lncBIRC3-OT knockdown condition was performed. ***P < 0.001 vs. NC group. (F) The mRNA levels of RELA in RELA-deletion glioma cells. (G) The mRNA levels of STC1 in RELA-deletion glioma cells. (H) U87 and U251 cells were co-transfected with sh-lncBIRC3-OT and pcDNA3.1-RELA, and the mRNA levels of STC1 were examined by RT-qPCR methods. Data were shown as mean ± SD from three independent experiments. *P < 0.05, ***P < 0.001.
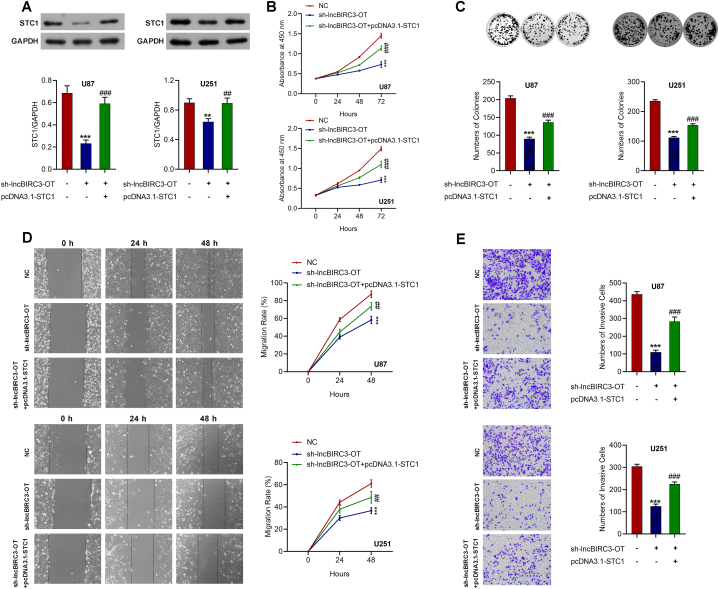
3.5. Overexpression of STC1 reverses the proliferation, migration, and invasion induced by lncBIRC3-OT deletion
To explore the role of STC1 in lncBIRC3-OT-mediated malignant phenotype in glioma cells, STC1-expressing plasmids were constructed and confirmed to upregulate the expression of STC1 in U87 and U251 cells (Fig. S2). Then, STC1-expressing plasmids were transfected into lncBIRC3-OT-deleted glioma cells (Fig. 5A). It was found that knockdown of lncBIRC3-OT suppressed malignant biological properties of U87 and U251 cells. In contrast, overexpression of STC1 in lncBIRC3-OT-deleted glioma cells reversed the proliferation (Fig. 5B), colony formation (Fig. 5C), migration (Fig. 5D), and invasion (Fig. 5E). Thus, our results demonstrated that lncBIRC3-OT promotes glioma progression by facilitating RELA-mediated STC1 transcription.
Fig. 5.
Restoration of STC1 rescues lncBIRC3-OT deletion-mediated suppression of glioma cell proliferation, migration, and invasion. (A) The expression of STC1 was upregulated by transient transfection in lncBIRC3-OT-silencing cells, which was detected by Western blot. Uncropped images of blots are shown in Supplementary material. (B) CCK8 assay and (C) colony-forming assay demonstrated that STC1 overexpression reversed the proliferation inhibited by lncBIRC3-OT knockdown in U87 and U251 cells. (D) Wound healing and (E) Transwell invasion assays showed that STC1 reversed migration and invasion induced by the knockdown of lncBIRC3-OT U87 and U251 cells. Data were shown as mean ± SD from three independent experiments. ***P < 0.001 vs. the NC groups; ##P < 0.01, ###P < 0.001 vs. the sh-lncBIRC3-OT groups.
4. Discussion
Here, we identified a novel glioma-associated lncRNA termed LncBIRC3-OT, which is a crucial epigenetic regulator of glioma cell proliferation and invasion. We found that lncBIRC3-OT is one of the most upregulated lncRNAs in glioma and is critical for glioma cell proliferation and invasion, as the knockdown of lncBIRC3-OT impairs cell proliferation, migration, and invasion. Importantly, lncBIRC3-OT guides RELA to STC1 promoter, and enhances STC1 transcription, which in turn promotes glioma progression. Take together, our study identifies lncBIRC3-OT as a tumor-promoting lncRNA and reveals its role and mechanism in the malignant behaviors of gliomas. These data indicate that lncBIRC3-OT may be a promising target for the treatment of gliomas.
Glioma is a highly aggressive tumor and is the most lethal primary brain tumor in adults. Only 5 % of glioma patients survive more than 5 years after diagnosis [22]. In recent years, the discovery of ncRNAs has added a new dimension to the diagnosis and treatment of glioma [23]. Among them, lncRNAs account for about 80–90 % of ncRNAs. LncRNAs are usually expressed in specific tissues or developmental stages [24]. Thus, they are considered as promising diagnostic and prognostic biomarkers for gliomas [25]. In recent years, increasing evidence revealed that dysregulation of several lncRNAs is closely associated with glioma development, progression, and treatment resistance [[26], [27], [28]]. For example, lncRNA LINREP linc-RoR has been identified as a pro-oncogenic molecule and promotes glioma progression [11,29]. Moreover, several lncRNAs including LINC00520, XLOC013218, and PDIA3P1, contribute to the acquisition of temozolomide resistance [10,30,31]. In the present study, we found that lncBIRC3-OT was significantly highly expressed in glioma tissues and cells. Loss-of-function experiments revealed that knockdown of lncBIRC3-OT suppressed the malignant phenotype of glioma cells, as evidenced by impaired cell proliferation, migration, and invasive capacity, as well as the EMT process. These findings highlight the important role of lncBIRC3-OT in glioma proliferation and progression.
Due to their structural diversity, lncRNAs can interact with miRNAs, RNAs, or proteins to regulate downstream genes and signaling pathways [32]. Here, we confirmed that lncBIRC3-OT had a high affinity for RELA by RNA pull-down assay followed by LC-MS and Western blot. RELA (also known as p65), an important member of the NF-κB pathway that drives tumor progression in many cancers, including gliomas [33]. Pharmacological or biological inhibition of RELA suppresses glioma tumorigenesis and progression [33,34]. Furthermore, a recent report showed that downregulation of phosphorylated RELA enhanced temozolomide sensitization in gliomas [35]. And, knockout of RELA sensitized GSCs to CAR-mediated antitumor activity [36].
RELA acts as a transcription factor that enhances the expression of multiple proliferation- or migration-related gene in tumors [37,38]. For example, RELA activates lncRNA NEAT1, which promotes the malignant behavior of pancreatic ductal adenocarcinoma cells [39]. RELA drives the expression of CXCL13 and CXCR5, which promotes breast cancer progression [40]. In addition, RELA modulates the expression of crucial immunosuppressive molecules in tumors [41]. A previous study showed that RELA facilitates STC1 transcription and promotes esophageal squamous cell carcinoma metastasis [20]. STC1 has been identified as a prognostic biomarker in gliomas and participates in glioma cell migration and invasion [42]. Suppression of STC1 restrains the malignant behaviors of glioma cells [43]. We demonstrated that RELA could bind the STC1 promoter to drive its transcription. Functional assays demonstrated that overexpression of STC1 restored lncBIRC3-OT depletion-mediated inhibition of malignant behaviors in glioma cells. Therefore, we concluded that lncBIRC3-OT promotes glioma progression by recruiting RELA to the STC1 promoter to facilitate STC1 transcription.
5. Conclusion
This study elucidates that lncBIRC3-OT promotes glioma cell proliferation, migration, and invasion by guiding RELA protein to the STC1 promoter and promoting the transcription of STC1. This study suggests that lncBIRC3-OT may be a potential target for the treatment of gliomas.
Ethical approval
The study was approved by Committees for Ethical Review in the Characteristic Medical Center of Chinese People's Armed Police Force (No. 2022-0002.1) and written informed consent was obtained from all participants.
Data availability statement
Data will be made available on request.
Funding
This work was supported by the National Natural Science Foundation of China, China (No. 81871086, toHQ.L.).
CRediT authorship contribution statement
Renjie Wang: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Validation, Visualization, Writing – original draft. Qi Li: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Writing – original draft. Xiaolei Chu: Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – review & editing. Nan Li: Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. Haiqian Liang: Conceptualization, Data curation, Methodology, Project administration, Supervision, Writing – review & editing. Feng He: Conceptualization, Data curation, Formal analysis, Methodology, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21777.
Contributor Information
Haiqian Liang, Email: lianghaiqian711@163.com.
Feng He, Email: heaven@tju.edu.cn.
Appendix A. Supplementary data
The following are the supplementary data to this article.
figs1.
figs2.
figs3.
References
- 1.Montella L., et al. Epigenetic alterations in glioblastomas: diagnostic, prognostic and therapeutic relevance. 2023;153(3):476–488. doi: 10.1002/ijc.34381. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong T.S., et al. Glioma patient-reported outcome assessment in clinical care and research: a Response Assessment in Neuro-Oncology collaborative report. Lancet Oncol. 2020;21(2):e97–e103. doi: 10.1016/S1470-2045(19)30796-X. [DOI] [PubMed] [Google Scholar]
- 3.Kumari S., et al. Multiple therapeutic approaches of glioblastoma multiforme: from terminal to therapy. Biochim. Biophys. Acta Rev. Canc. 2023;1878(4) doi: 10.1016/j.bbcan.2023.188913. [DOI] [PubMed] [Google Scholar]
- 4.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L., Xu X., Su X. Modifications of noncoding RNAs in cancer and their therapeutic implications. Cell. Signal. 2023;108 doi: 10.1016/j.cellsig.2023.110726. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad M., et al. Involvement of lncRNAs in cancer cells migration, invasion and metastasis: cytoskeleton and ECM crosstalk. 2023;42(1):173. doi: 10.1186/s13046-023-02741-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mousavi S.M., et al. Non-coding RNAs and glioblastoma: insight into their roles in metastasis. Mol Ther Oncolytics. 2022;24:262–287. doi: 10.1016/j.omto.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balandeh E., et al. Roles of non-coding RNAs and angiogenesis in glioblastoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.716462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin J., et al. Roles of long noncoding RNAs in conferring glioma progression and treatment. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.688027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J., et al. lncRNA XLOC013218 promotes cell proliferation and TMZ resistance by targeting the PIK3R2-mediated PI3K/AKT pathway in glioma. 2022;113(8):2681–2692. doi: 10.1111/cas.15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji X., et al. 2022. N(6)-Methyladenosine-modified lncRNA LINREP Promotes Glioblastoma Progression by Recruiting the PTBP1/HuR Complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D., et al. LncRNA-FAM66C was identified as a key regulator for modulating tumor microenvironment and hypoxia-related pathways in glioblastoma. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.898270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Z., et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro-Mondragon J.A., et al. Jaspar 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50(D1):D165–d173. doi: 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y.P., Liu Y. Induction of cancer cell stemness in glioma through glycolysis and the long noncoding RNA HULC-activated FOXM1/AGR2/HIF-1α axis. 2022;102(7):691–701. doi: 10.1038/s41374-021-00664-9. [DOI] [PubMed] [Google Scholar]
- 16.Bian E., et al. Super-enhancer-associated TMEM44-AS1 aggravated glioma progression by forming a positive feedback loop with Myc. J. Exp. Clin. Cancer Res. 2021;40(1):337. doi: 10.1186/s13046-021-02129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou C., et al. Circular RNA mitochondrial translation optimization 1 homologue (CircMTO1) induced by zinc finger protein 460 (ZNF460) promotes oral squamous cell carcinoma progression through the microRNA miR-320a/alpha thalassemia/mental retardation, X-linked (ATRX) axis. Bioengineered. 2021;12(2):9585–9597. doi: 10.1080/21655979.2021.1997699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R., et al. 2023. Sequencing and Bioinformatics Analysis of lncRNA/circRNA-miRNA-mRNA in Glioblastoma Multiforme. [DOI] [PubMed] [Google Scholar]
- 19.Herman A.B., Tsitsipatis D., Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. 2022;82(12):2252–2266. doi: 10.1016/j.molcel.2022.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., et al. MLL2 promotes cancer cell lymph node metastasis by interacting with RelA and facilitating STC1 transcription. Cell. Signal. 2020;65 doi: 10.1016/j.cellsig.2019.109457. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Y., Wang Q. STC1 regulates glioblastoma migration and invasion via the TGF-β/SMAD4 signaling pathway. Mol. Med. Rep. 2019;20(4):3055–3064. doi: 10.3892/mmr.2019.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander B.M., Cloughesy T.F. Adult glioblastoma. J. Clin. Oncol. 2017;35(21):2402–2409. doi: 10.1200/JCO.2017.73.0119. [DOI] [PubMed] [Google Scholar]
- 23.Goenka A., Tiek D.M. The role of non-coding RNAs in glioma. 2022;10(8) doi: 10.3390/biomedicines10082031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bridges M.C., Daulagala A.C., Kourtidis A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021;220(2) doi: 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Z., Liu C., Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer. 2018;17(1):61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T., et al. A positive feedback loop of lncRNA-RMRP/ZNRF3 axis and Wnt/β-catenin signaling regulates the progression and temozolomide resistance in glioma. Cell Death Dis. 2021;12(11):952. doi: 10.1038/s41419-021-04245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu M., et al. LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene. 2020;39(45):6879–6892. doi: 10.1038/s41388-020-01466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N., et al. Machine learning-based identification of tumor-infiltrating immune cell-associated lncRNAs for improving outcomes and immunotherapy responses in patients with low-grade glioma. Theranostics. 2022;12(13):5931–5948. doi: 10.7150/thno.74281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovalenko T.F., et al. Functions of long non-coding RNA ROR in patient-derived glioblastoma cells. Biochimie. 2022;200:131–139. doi: 10.1016/j.biochi.2022.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Yuan S., et al. STAT3-mediated upregulation of LINC00520 contributed to temozolomide chemoresistance in glioblastoma by interacting with RNA-binding protein LIN28B. 2022;22(1):248. doi: 10.1186/s12935-022-02659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Z., et al. PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPβ degradation to facilitate proneural-to-mesenchymal transition. Cancer Cell Int. 2022;41(1):223. doi: 10.1186/s13046-022-02431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu K., et al. Evolving insights into the biological function and clinical significance of long noncoding RNA in glioblastoma. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.846864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J.K., et al. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J. Neuro Oncol. 2017;131(2):255–265. doi: 10.1007/s11060-016-2308-5. [DOI] [PubMed] [Google Scholar]
- 34.Zhen Z.G., et al. Linarin suppresses glioma through inhibition of NF-κB/p65 and up-regulating p53 expression in vitro and in vivo. Biomed. Pharmacother. 2017;95:363–374. doi: 10.1016/j.biopha.2017.08.023. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., et al. Overexpressed GNA13 induces temozolomide sensitization via down-regulating MGMT and p-RELA in glioma. Am J Transl Res. 2021;13(10):11413–11426. [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D., et al. CRISPR screening of CAR T cells and cancer stem cells reveals critical dependencies for cell-based therapies. Cancer Discov. 2021;11(5):1192–1211. doi: 10.1158/2159-8290.CD-20-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capece D., et al. Cancer secretome and inflammation: the bright and the dark sides of NF-κB. Semin. Cell Dev. Biol. 2018;78:51–61. doi: 10.1016/j.semcdb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Nakazato A., et al. RELA is required for CD271 expression and stem-like characteristics in hypopharyngeal cancer. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-22736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Z., et al. RELA/NEAT1/miR-302a-3p/RELA feedback loop modulates pancreatic ductal adenocarcinoma cell proliferation and migration. J. Cell. Physiol. 2019;234(4):3583–3597. doi: 10.1002/jcp.27039. [DOI] [PubMed] [Google Scholar]
- 40.Biswas S., et al. RelA driven co-expression of CXCL13 and CXCR5 is governed by a multifaceted transcriptional program regulating breast cancer progression. Biochim. Biophys. Acta, Mol. Basis Dis. 2019;1865(2):502–511. doi: 10.1016/j.bbadis.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Zhang M., et al. CECR2 drives breast cancer metastasis by promoting NF-κB signaling and macrophage-mediated immune suppression. Sci. Transl. Med. 2022;14(630) doi: 10.1126/scitranslmed.abf5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo W., et al. Stanniocalcin 1 is a prognostic biomarker in glioma. Oncol. Lett. 2020;20(3):2248–2256. doi: 10.3892/ol.2020.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakata J., et al. MicroRNA regulating stanniocalcin-1 is a metastasis and dissemination promoting factor in glioblastoma. J. Neuro Oncol. 2019;142(2):241–251. doi: 10.1007/s11060-019-03113-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.