Abstract
Machine learning and deep learning are two subsets of artificial intelligence that involve teaching computers to learn and make decisions from any sort of data. Most recent developments in artificial intelligence are coming from deep learning, which has proven revolutionary in almost all fields, from computer vision to health sciences. The effects of deep learning in medicine have changed the conventional ways of clinical application significantly. Although some sub-fields of medicine, such as pediatrics, have been relatively slow in receiving the critical benefits of deep learning, related research in pediatrics has started to accumulate to a significant level, too. Hence, in this paper, we review recently developed machine learning and deep learning-based solutions for neonatology applications. We systematically evaluate the roles of both classical machine learning and deep learning in neonatology applications, define the methodologies, including algorithmic developments, and describe the remaining challenges in the assessment of neonatal diseases by using PRISMA 2020 guidelines. To date, the primary areas of focus in neonatology regarding AI applications have included survival analysis, neuroimaging, analysis of vital parameters and biosignals, and retinopathy of prematurity diagnosis. We have categorically summarized 106 research articles from 1996 to 2022 and discussed their pros and cons, respectively. In this systematic review, we aimed to further enhance the comprehensiveness of the study. We also discuss possible directions for new AI models and the future of neonatology with the rising power of AI, suggesting roadmaps for the integration of AI into neonatal intensive care units.
Subject terms: Translational research, Paediatric research
Introduction
The AI tsunami fueled by advances in artificial intelligence (AI) is constantly changing almost all fields, including healthcare; it is challenging to track the changes originated by AI as there is not a single day that AI is not applied to anything new. While AI affects daily life enormously, many clinicians may not be aware of how much of the work done with AI technologies may be put into effect in today’s healthcare system. In this review, we fill this gap, particularly for physicians in a relatively underexplored area of AI: neonatology. The origins of AI, specifically machine learning (ML), can be tracked all the way back to the 1950s, when Alan Turing invented the so-called “learning machine” as well as military applications of basic AI1. During his time, computers were huge, and the cost of increased storage space was astronomical. As a result, their capabilities, although substantial for their day, were restricted. Over the decades, incremental advancements in theory and technological advances steadily increased the power and versatility of ML2.
How do machine learning (ML) and deep learning (DL) work? ML falls under the category of AI2. ML’s capacity to deal with data brought it to the attention of computer scientists. ML algorithms and models can learn from data, analyze, evaluate, and make predictions or decisions based on learning and data characteristics. DL is a subset of ML. Different from this larger class of ML definitions, the underlying concept of DL is inspired by the functioning of the human brain, particularly the neural networks responsible for processing and interpreting information. DL mimics this operation by utilizing artificial neurons in a computer neural network. In simple terms, DL finds weights for each artificial neuron that connects to each other from one layer to another layer. Once the number of layers is high (i.e., deep), more complex relationships between input and output can be modeled3–5. This enables the network to acquire more intricate representations of the data as it learns. The utilization of a hierarchical approach enables DL models to autonomously extract features from the data, as opposed to depending on human-engineered features as is customary in conventional ML3. DL is a highly specialized form of ML that is ideally modified for tasks involving unstructured data, where the features in the data may be learnable, and exploration of non-linear associations in the data can be possible6–8.
The main difference between ML and DL lies in the complexity of the models and the size of the datasets they can handle. ML algorithms can be effective for a wide range of tasks and can be relatively simple to train and deploy6,7,9–11. DL algorithms, on the other hand, require much larger datasets and more complex models but can achieve exceptional performance on tasks that involve high-dimensional, complex data7. DL can automatically identify which aspects are significant, unlike classical ML, which requires pre-defined elements of interest to analyze the data and infer a decision10. Each neuron in DL architectures (i.e., artificial neural networks (ANN)) has non-linear activation function(s) that help it learn complex features representative of the provided data samples9.
ML algorithms, hence, DL, can be categorized as either supervised, unsupervised, or reinforcement learning based on the input-output relationship. For example, if output labels (outcome) are fully available, the algorithm is called “supervised,” while unsupervised algorithms explore the data without their reference standards/outcomes/labels in the output3,12. In terms of applications, both DL and ML are typically used for tasks such as classification, regression, and clustering6,9,10,13–15. DL methods’ success depends on the availability of large-scale data, new optimization algorithms, and the availability of GPUs6,10. These algorithms are designed to autonomously learn and develop as they gain experience, like humans3. As a result of DL’s powerful representation of the data, it is considered today’s most improved ML method, providing drastic changes in all fields of medicine and technology, and it is the driving force behind virtually all progress in AI today5 (Fig. 1).
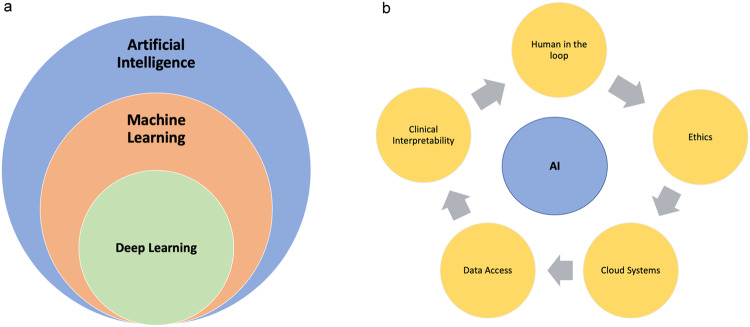
Fig. 1. Exploring AI Hierarchy and Challenges in Healthcare.
a Hierarchical diagram of AI. How do machine learning (ML) and deep learning (DL) work? ML falls under the category of AI. DL is a subset of ML. b Ongoing hurdles of AI when applied to healthcare applications. Key concerns related to AI and each concern affects the outcome of AI in Neonatology including; (1) challenges with clinical interpretability; (2) knowledge gaps in decision-making mechanisms, with the latter requiring human-in-the-loop systems (3) ethical considerations; (4) the lack of data and annotations, and (5) the absence of Cloud systems allowing for secure data sharing and data privacy.
There are three major problem types in DL in medical imaging: image segmentation, object detection (i.e., an object can be an organ or any other anatomical or pathological entity), and image classification (e.g., diagnosis, prognosis, therapy response assessment)3. Several DL algorithms are frequently employed in medical research; briefly, those approaches belong to the following family of algorithms:
Convolutional Neural Networks (CNNs) are predominantly employed for tasks related to computer vision and signal processing. CNNs can handle tasks requiring spatial relationships where the columns and rows are fixed, such as imaging data. CNN architecture encompasses a sequence of phases (layers) that facilitate the acquisition of hierarchical features. Initial phases (layers) extract more local features such as corners, edges, and lines, later phases (layers) extract more global features. Features are propagated from one layer to another layer, and feature representation becomes richer this way. During feature propagation from one layer to another layer, the features are added certain nonlinearities and regularizations to make the functional modeling of input-output more generalizable. Once features become extremely large, there are operations within the network architecture to reduce the feature size without losing much information, called pooling operations. The auto-generated and propagated features are then utilized at the end of the network architecture for prediction purposes (segmentation, detection, or classification)3,16.
Recurrent Neural Networks (RNNs) are designed to facilitate the retention of sequential data, namely text, speech, and time-series data such as clinical data or electronic health records (EHRs). They can capture temporal relationships between data components, which can be helpful for predicting disease progression or treatment outcomes11,17,18. RNNs use similar architecture components that CNNs have. Long Short-Term Memory (LSTM) models are types of RNNs and are commonly used to overcome their shortcomings because they can learn long-term dependencies in data better than conventional RNN architectures. They are utilized in some classification tasks, including audio17,19. LSTM utilizes a gated memory cell in the network architecture to store information from the past; hence, the memory cell can store information for a long period of time, even if the information is not immediately relevant to the current task. This allows LSTMs to learn patterns in data that would be difficult for other types of neural networks to learn.
Generative adversarial networks (GANs) are a class of DL models that can be used to generate new data that is like existing data. In healthcare, GANs have been used to generate synthetic medical images. There are two CNNs (generator and discriminator); the first CNN is called the generator, and its primary goal is to make synthetic images that mimic actual images. The second CNN is called the discriminator, and its main objective is to identify between artificially generated images and real images20. The generator and discriminator are trained jointly in a process called adversarial training, where the generator tries to create data that is so realistic that the discriminator cannot distinguish it from real data. GANs are used to generate a variety of different types of data, including images, videos, and text. GANs are used to enhance image quality, signal reconstruction, and other tasks such as classification and segmentation too20–22.
Transfer learning (TL) is a concept derived from cognitive science that states that information is transferred across related activities to improve performance on a new task. It is generally known that people can accomplish similar tasks by building on prior knowledge23. TL has been implemented to minimize the need for annotation by transferring DL models with knowledge from a previous task and then fine-tuning them in the current task24. The majority of medical image classification techniques employ TL from pretrained models, such as ImageNet, which has been demonstrated to be inefficient due to the ImageNet consisting of natural images25. The approaches that utilized ImageNet pre-trained images in CNNs revealed that fine-tuning more layers provided increased accuracy26. The initial layers of ImageNet-pretrained networks, which detect low-level image characteristics, including corners and borders, may not be efficient for medical images25,26.
New and more advanced DL algorithms are developed almost daily. Such methods could be employed for the analysis of imaging and non-imaging data in order to enhance performance and reliability. These methods include Capsule Networks, Attention Mechanisms, and Graph Neural Networks (GNNs)27–30. Briefly, these are:
Capsule Networks are a relatively new form of DL architecture that aim to address some of the shortcomings of CNNs: pooling operations (reducing the data size) and a lack of hierarchical relations between objects and their parts in the data. Capsules can capture spatial relationships between features and are more capable of handling rotations and deformations of image objects thanks to their vectorial representations in neuronal space. Capsule Networks have shown potential in image classification tasks and could have applications in medical imaging analysis27. However, its implementation and computational time are two hurdles that restrict its widespread use.
Attention Mechanisms, represented by Transformers, have contributed to the development of computer vision and language processing. Unlike CNNs or RNNs, transformers allow direct interaction between every pair of components within a sequence, making them particularly effective at capturing long-term relationships29,30. More specifically, a self-attention mechanism in Transformers is an important piece of the DL model as it can dynamically focus on different parts of the input data sequence when producing an output, providing better context understanding than CNN based systems.
Graph Neural Networks (GNNs) are a form of data structure that describes a collection of objects (nodes) and their relationships (edges). There are three forms of tasks, including node-level, edge-level, and graph level31. Graphs may be used to denote a wide range of systems, including molecular interaction networks, and bioinformatics31–33. GNNs have demonstrated potential in both imaging and non-imaging data analysis28,34.
Physics-driven systems are needed in imaging field. Several studies have demonstrated the effectiveness of DL methods in the medical imaging field35–39. As the field of DL continues to evolve, it is likely that new methods and architectures will emerge to address the unique challenges and constraints of various types of data. One of the most common problems faced with DL-based MRI construction35. Specific algorithms for this problem can be essentially categorized into two groups: data driven and physics driven algorithms. In purely data-driven approaches, a mapping is learned between the aliased image and the image without artifacts39. Acquiring fully sampled (artifact-free) datasets is impractical in many clinical imaging studies when organs are in motion, such as the heart, and lung. Recently developed models can employ these under sampled MRI acquisitions as input and generate output images consistent with fully-sampled (artifact free) acquisitions37–39.
What is the Hybrid Intelligence? A highly desirable way of incorporating advances in AI is to let AI and human intellect work together to solve issues, and this is referred to as “hybrid intelligence“40 (e.g., one may call this “mixed intelligence” or “human-in-the-loop AI systems”). This phenomenon involves the development of AI systems that serve to supplement and amplify human decision-making processes, as opposed to completely replacing them3. The concept involves integrating the respective competencies of artificial intelligence and human beings in order to attain superior outcomes that would otherwise be unachievable41. AI algorithms possess the ability to process extensive amounts of data, recognize patterns, and generate predictions rapidly and precisely. Meanwhile, humans can contribute their expertise, understanding, and intuition to the discussion to offer context, analyze outcomes, and render decisions42. The hybrid intelligence strategy can help decision-makers in a variety of fields make decisions that are more precise, effective, and efficient by combining these qualities3,4,43,44. Human in the loop and hybrid intelligence systems are promising for time-consuming tasks in healthcare and neonatology.
Where do we stand currently? AI in medicine has been employed for over a decade, and it has often been considered that clinical implementation is not completely adapted to daily practice in most of the clinical field5,45,46. In recent years, increasingly complex computer algorithms and updated hardware technologies for processing and storing enormous datasets have contributed to this achievement6,7,46,47. It has only been within the last decade that these systems have begun to display their full potential6,9. The field of AI research appears to have been taken up with differing degrees of enthusiasm across disciplines. When analyzing the thirty years of research into AI, DL, and ML conducted by several medical subfields between the years 1988 and 2018, one-third of publications in DL yielded to radiology, and most of them are within the imaging sciences (radiology, pathology, and cell imaging)48. Software systems work by utilizing biomedical images with predictive/diagnostic/prognostic features and integrating clinical or pre-clinical data. These systems are designed with ML algorithms46. Such breakthrough methods in DL are nowadays extensively applied in pathology, dermatology, ophthalmology, neurology, and psychiatry6,47,49. AI has its own difficulties with the increasing utilization of healthcare (Fig. 1b).
What are the needs in clinics? Clinicians are concerned about the healthcare system’s integration with AI: there is an exponential need for diagnostic testing, early detection, and alarm tools to provide diagnosis and novel treatments without invasive tests and procedures50. Clinicians have higher expectations of AI in their daily practices than before. AI is expected to decrease the need for multiple diagnostic invasive tests and increase diagnostic accuracy with less invasive (or non-invasive) tests. Such AI systems can easily recognize imaging patterns on test images (i.e., unseen or not utilized efficiently in daily routines), allowing them to detect and diagnose various diseases. These methods could improve detection and diagnosis in different fields of medicine.
The overall goal of this systematic review is to explain AI’s potential use and benefits in the field of neonatology. We intend to enlighten the potential role of AI in the future in neonatal care. We postulate that AI would be best used as a hybrid intelligence (i.e., human-in-the-loop or mixed intelligence) to make neonatal care more feasible, increase the accuracy of diagnosis, and predict the outcome and diseases in advance. The rest of the paper is organized as follows: In results, we explain the published AI applications in neonatology along with AI evaluation metrics to fully understand their efficacy in neonatology and provide a comprehensive overview of DL applications in neonatology. In discussion, we examine the difficulties of AI utilization in neonatology and future research discussions. In the methods section, we outline the systematic review procedures, including the examination of existing literature and the development of our search strategy.
We review the past, current, and future of AI-based diagnostic and monitoring tools that might aid neonatologists’ patient management and follow-up. We discuss several AI designs for electronic health records, image, and signal processing, analyze the merits and limits of newly created decision support systems, and illuminate future views clinicians and neonatologists might use in their normal diagnostic activities. AI has made significant breakthroughs to solve issues with conventional imaging approaches by identifying clinical variables and imaging aspects not easily visible to human eyes. Improved diagnostic skills could prevent missed diagnoses and aid in diagnostic decision-making. The overview of our study is structured as illustrated in Fig. 2. Briefly, our objectives in this systematic review are:
to explain the various AI models and evaluation metrics thoroughly explained and describe the principal features of the AI models,
to categorize neonatology-related AI applications into macro-domains, to explain their sub-domains and the important elements of the applicable AI models,
to examine the state-of-the-art in studies, particularly from the past several years, with an emphasis on the use of ML in encompassing all neonatology,
to present a comprehensive overview and classification of DL applications utilized and in neonatology,
to analyze and debate the current and open difficulties associated with AI in neonatology, as well as future research directions, to offer the clinician a comprehensive perspective of the actual situation.
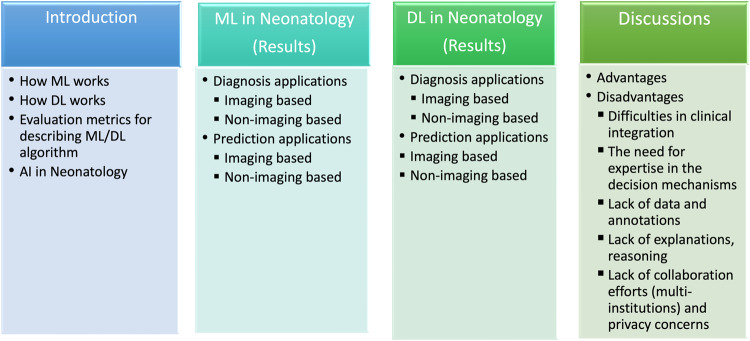
Fig. 2. An overview of the structure of this paper.
It is provided an overview of our paper’s structure and objectives: 1. Explaining AI Models and Evaluation Metrics: 2. Evaluating ML applied studies in Neonatology 3. Evaluating DL applied studies in Neonatology 4. Analyzing Challenges and Future Directions.
AI covers a broad concept for the application of computing algorithms that can categorize, predict, or generate valuable conclusions from enormous datasets46. Algorithms such as Naive Bayes, Genetic Algorithms, Fuzzy Logic, Clustering, Neural Networks (NN), Support Vector Machines (SVM), Decision Trees, and Random Forests (RF) have been used for more than three decades for detection, diagnosis, classification, and risk assessment in medicine as ML methods9,10. Conventional ML approaches for image classification involve using hand-engineered features, which are visual descriptions and annotations learned from radiologists, that are encoded into algorithms.
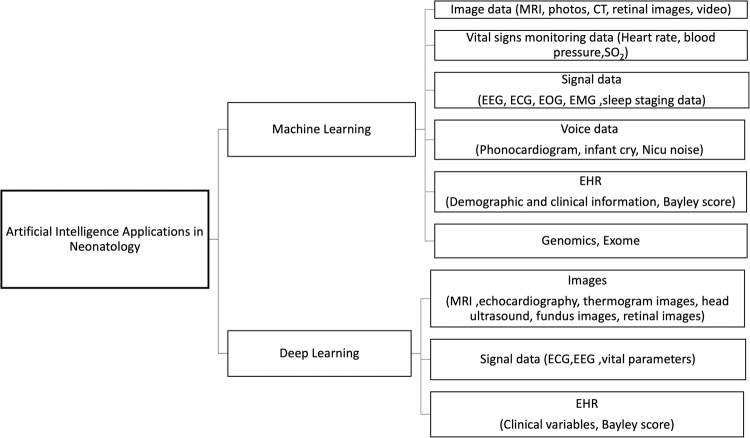
Images, signals, genetic expressions, EHR, and vital signs are examples of the various unstructured data sources that comprise medical data (Fig. 3). Due to the complexity of their structures, DL frameworks may take advantage of this heterogeneity by attaining high abstraction levels in data analysis.
Fig. 3. An overview of AI applications in neonatology.
Unstructured data such as medical images, vital signals, genetic expressions, EHRs, and signal data contribute to the wide variety of medical information. Analyzing and interpreting different data streams in neonatology requires a comprehensive strategy because each has unique characteristics and complications.
While ML requires manual/hand-crafted selection of information from incoming data and related transformation procedures, DL performs these tasks more efficiently and with higher efficacy9,10,46. DL is able to discover these components by analyzing a large number of samples with a high degree of automation7. The literature on these ML approaches is extensive before the development of DL5,7,45.
It is essential for clinicians to understand how the suggested ML model should enhance patient care. Since it is impossible for a single metric to capture all the desirable attributes of a model, it is customarily necessary to describe the performance of a model using several different metrics. Unfortunately, many end-users do not have an easy time comprehending these measurements. In addition, it might be difficult to objectively compare models from different research models, and there is currently no method or tool available that can compare models based on the same performance measures51. In this part, the common ML and DL evaluation metrics are explained so neonatologists could adapt them into their research and understand of upcoming articles and research design51,52.
AI is commonly utilized everywhere, from daily life to high-risk applications in medicine. Although slower compared to other fields, numerous studies began to appear in the literature investigating the use of AI in neonatology. These studies have used various imaging modalities, electronic health records, and ML algorithms, some of which have barely gone through the clinical workflow. Though there is no systematic review and future discussions in particular in this field53–55. Many studies were dedicated to introducing these systems into neonatology. However, the success of these studies has been limited. Lately, research in this field has been moving in a more favorable direction due to exciting new advances in DL. Metrics for evaluations in those studies were the standard metrics such as sensitivity (true-positive rate), specificity (true-negative rate), false-positive rate, false-negative rate, receiver operating characteristics (ROC), area under the ROC curves (AUC), and accuracy (Table 1).
Table 1.
Evaluation metrics in artificial intelligence.
| Term | Definition |
|---|---|
| True Positive (TP) | The number of positive samples that have been correctly identified. |
| True Negative (TN) | The number of samples that were accurately identified as negative. |
| False Positive (FP) | The number of samples that were incorrectly identified as positive. |
| False Negative (FN) | The number of samples that were incorrectly identified as negative. |
| Accuracy (ACC) |
The proportion of correctly identified samples to the total sample count in the assessment dataset. The accuracy is limited to the range [0, 1], where 1 represents properly predicting all positive and negative samples and 0 represents successfully predicting none of the positive or negative samples. |
| Recall (REC) | The sensitivity or True Positive Rate (TPR) is the proportion of correctly categorized positive samples to all samples allocated to the positive class. It is computed as the ratio of correctly classified positive samples to all samples assigned to the positive class. |
| Specificity (SPEC) | The negative class form of recall (sensitivity) and reflects the proportion of properly categorized negative samples. |
| Precision (PREC) | The ratio of correctly classified samples to all samples assigned to the class. |
| Positive Predictive Value (PPV) | The proportion of correctly classified positive samples to all positive samples. |
| Negative Predictive Value (NPV) | The ratio of samples accurately identified as negative to all samples classified as negative. |
| F1 score (F1) | The harmonic mean of precision and recall, which eliminates excessive levels of either. |
| Cross Validation | A validation technique often employed during the training phase of modeling, without no duplication among validation components. |
| AUROC (Area under ROC curve - AUC) | A function of the effect of various sensitivities (true-positive rate) on false-positive rate. It is limited to the range [0, 1], where 1 represents properly predicting all cases of all and 0 represents predicting the none of cases. |
| ROC | By displaying the effect of variable levels of sensitivity on specificity, it is possible to create a curve that illustrates the performance of a particular predictive algorithm, allowing readers to easily capture the algorithm’s value. |
| Overfitting | Modeling failure indicating extensive training and poor performance on tests. |
| Underfitting | Modeling failure indicating inadequate training and inadequate test performance. |
| Dice Similarity Coefficient | Used for image analysis. It is limited to the range [0, 1], where 1 represents properly segmenting of all images and 0 represents successfully segmenting none of images. |
Results
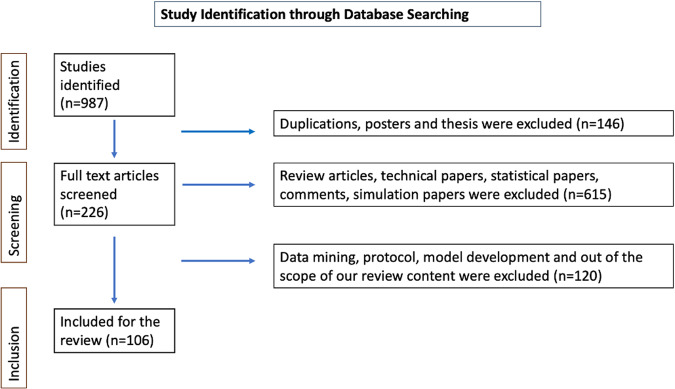
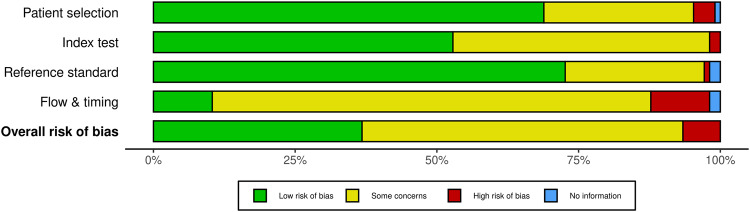
This systematic review was guided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) protocol56. The search was completed on 11st of July 2022. The initial search yielded many articles (approximately 9000), and we utilized a systematic approach to identify and select relevant articles based on their alignment with the research focus, study design, and relevance to the topic. We checked the article abstracts, and we identified 987 studies. Our search yielded 106 research articles between 1996 and 2022 (Fig. 4). Risk of bias summary analysis was done by the QUADAS-2 tool (Figs. 5 and 6)57–59.
Fig. 4. Identification of studies through database searches.
Initial research conducted on 11th of July 2022, yielded 9000 articles, of which 987 article abstracts were screened. Of those, 106 research articles published between 1996 and 2022 were eligible for inclusion in this systematic review. The PRISMA flow diagram illustrates the study selection process in more detail.
Fig. 5. Bias summary of all research according to the QUADAS-2.
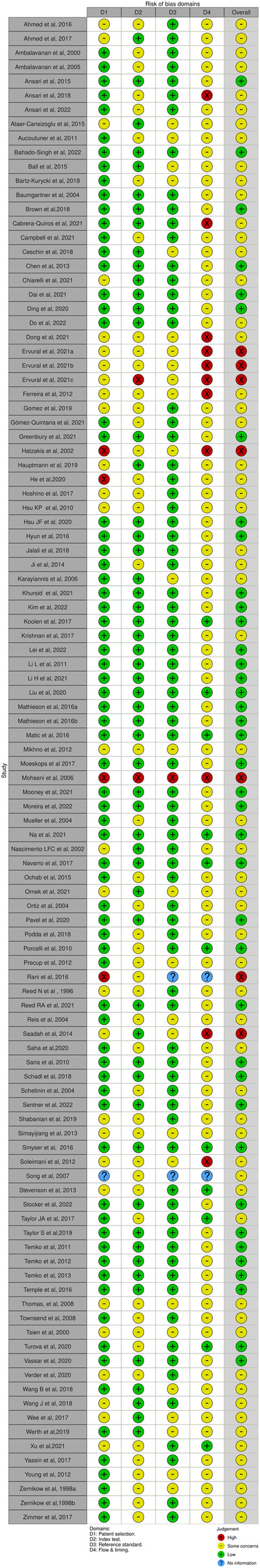
Risk of bias summary analysis was done by the QUADAS-2 tool.
Fig. 6. Bias summary of all studies according to the QUADAS-2.
Risk of bias summary analysis was done by the QUADAS-2 tool.
Our findings are summarized in two groups of tables: Tables 2–5 summarize the AI methods from the pre-deep learning era (“Pre-DL Era”) in neonatal intensive care units according to the type of data and applications. Tables 6, 7, on the other hand, include studies from the DL Era. Applications include classification (i.e., prediction and diagnosis), detection (i.e., localization), and segmentation (i.e., pixel level classification in medical images).
Table 2.
ML based (non-DL) studies in neonatology using imaging data for diagnosis.
| Study | Approach | Purpose | Dataset | Type of data | Performance | Pros(+) |
|---|---|---|---|---|---|---|
| Cons(-) | ||||||
| Hoshino et al., 2017194 | CLAFIC, logistic regression analysis |
To determine optimal color parameters predicting Biliary atresia (BA)stools |
50 neonates | 30 BA and 34 non-BA images | 100% (AUC) |
+ Effective and convenient modality for early detection of BA, and potentially for other related diseases |
| - Small sample size | ||||||
| Dong et al., 2021195 | Level Set algorithm |
To evaluate the postoperative enteral nutrition of neonatal high intestinal obstruction and analyze the clinical treatment effect of high intestinal obstruction |
60 neonates | CT images | 84.7% (accuracy) | + Segmentation algorithm can accurately segment the CT image, so that the disease location and its contour can be displayed more clearly. |
|
- EHR (not included AI analysis) - Small sample size - Retrospective design | ||||||
| Ball et al., 201590 | Random Forest (RF) | To compare whole-brain functional connectivity in preterm newborns at term-equivalent age with healthy term-born neonates in order to determine if preterm birth leads in particular changes to functional connectivity by term-equivalent age. | 105 preterm infants and 26 term controls |
Both resting state functional MRI and T2-weighted Brain MRI |
80% (accuracy) |
+ Prospective + Connectivity differences between term and preterm brain |
| - Not well-established model | ||||||
| Smyser et al., 201688 |
Support vector machine (SVM)-multivariate pattern analysis (MVPA) |
To compare resting state-activity of preterm-born infants (Scanned at term equivalent postmenstrual age) to term infants |
50 preterm infants (born at 23–29 weeks of gestation and without moderate–severe brain injury) 50 term-born control infants studied |
Functional MRI data + Clinical variables |
84% (accuracy) |
+ Prospective + GA at birth was used as an indicator of the degree of disruption of brain development + Optimal methods for rs-fMRI data acquisition and preprocessing for this population have not yet been rigorously defined |
| - Small sample size | ||||||
| Zimmer et al., 201793 | NAF: Neighborhood approximation forest classifier of forests | To reduce the complexity of heterogeneous data population, manifold learning techniques are applied, which find a low-dimensional representation of the data. |
111 infants (NC, 70 subjects), affected by IUGR (27 subjects) or VM (14 subjects). |
3 T brain MRI |
80% (accuracy) |
+ Combining multiple distances related to the condition improves the overall characterization and classification of the three clinical groups (Normal, IUGR, Ventriculomegaly) |
|
- The lack of neonatal data due to challenges during acquisition and data accessibility - Small sample size | ||||||
| Krishnan et al., 2017100 |
Unsupervised machine learning: Sparse Reduced Rank Regression (sRRR) |
Variability in the Peroxisome Proliferator Activated Receptor (PPAR) pathway would be related to brain development |
272 infants born at less than 33 wk gestational age (GA) |
Diffusion MR Imaging Diffusion Tractography Genome wide Genotyping |
63% (AUC) | + Inhibited brain development found in individuals exposed to the stress of a preterm extrauterine world is controlled by genetic variables, and PPARG signaling plays a previously unknown cerebral function |
|
- Further work is required to characterize the exact relationship between PPARG and preterm brain development, notably to determine whether the effect is brain specific or systemic | ||||||
| Chiarelli et al., 202191 | Multivariate statistical analysis |
To better understand the effect of prematurity on brain structure and function, |
88 newborns |
3 Tesla BOLD and anatomical brain MRI Few clinical variables |
The multivariate analysis using motion information could not significantly infer GA at birth |
+ Prematurity was associated with bidirectional alterations of functional connectivity and regional volume |
|
- Retrospective design - Small sample size | ||||||
| Song et al., 200794 | Fuzzy nonlinear support vector machines (SVM). |
Neonatal brain tissue segmentation in clinical magnetic resonance (MR) images |
10 term neonates | Brain MRI T1 and T2 weighted |
70%–80% (dice score-gray matter) 65%–80% (dice score-white matter) |
+ Nonparametric modeling adapts to the spatial variability in the intensity statistics that arises from variations in brain structure and image inhomogeneity + Produces reasonable segmentations even in the absence of atlas prior |
| - Small sample size | ||||||
| Taylor et al., 2017137 | Machine Learning |
Technology that uses a smartphone application has the potential to be a useful methodology for effectively screening newborns for jaundice |
530 newborns |
Paired BiliCam images total serum bilirubin (TSB) levels |
High-risk zone TSB level was 95% for BiliCam and 92% for TcB (P = 0.30); for identifying newborns with a TSB level of ≥17.0, AUCs were 99% and 95%, respectively (P =0.09). |
+ Inexpensive technology that uses commodity smartphones could be used to effectively screen newborns for jaundice + Multicenter data + Prospective design |
| - Method and algorithm name were not explained | ||||||
| Ataer-Cansizoglu et al., 2015134 |
Gaussian Mixture Models i-ROP |
To develop novel computer based image analysis system for grading plus diseases in ROP | 77 wide-angle retinal images |
95% (accuracy) |
+ Arterial and venous tortuosity (combined), and a large circular cropped image (with radius 6 times the disc diameter), provided the highest diagnostic accuracy + Comparable to the performance of the 3 individual experts (96%, 94%, 92%), and significantly higher than the mean performance of 31 nonexperts (81%) |
|
|
- Used manually segmented images with a tracing algorithm to avoid the possible noise and bias that might come from an automated segmentation algorithm - Low clinical applicability | ||||||
| Rani et al., 2016133 | Back Propagation Neural Networks | To classify ROP |
64 RGB images of these stages have been taken, captured by RetCam with 120 degrees field of view and size of 640 × 480 pixels. |
90.6% (accuracy) |
- No clinical information - Required better segmentation - Clinical adaptation |
|
| Karayiannis et al., 2006101 | Artificial Neural Networks (ANN) |
To aim at the development of a seizure-detection system by training neural networks with quantitative motion information extracted from short video segments of neonatal seizures of the myoclonic and focal clonic types and random infant movements |
54 patients |
240 video segments (Each of the training and testing sets contained 120 video segments (40 segments of myoclonic seizures, 40 segments of focal clonic seizures, and 40 segments of random movements |
96.8% (sensitivity) 97.8% (specificity) |
+ Video analysis |
|
- Not be capable of detecting neonatal seizures with subtle clinical manifestations (Subclinical seizures) or neonatal seizures with no clinical manifestations (electrical-only seizures - Not include EEG analysis - Small sample size - No additional clinical information |
Table 5.
ML based (non-DL) studies in neonatology using non-imaging data for prediction.
| Reference | Approach | Purpose | Dataset | Type of data | Performance | Pros(+) |
|---|---|---|---|---|---|---|
| Cons(-) | ||||||
| Soleimani et al., 2012141 |
Multilayer perceptron (MLP) (ANN) |
Predict developmental disorder | 6150 infants’ |
Infant Neurological International Battery (INFANIB) and prenatal factors |
79% (AUC) | + Neural network ability includes quantitative and qualitative data |
|
- Relying on preexisting data - Missing important topics - Small sample size | ||||||
| Zernikow et al., 199868 | ANN | To predict the individual neonatal mortality risk | 890 preterm neonates | Clinical records | 95% (AUC) | + ANN predict mortality accurately |
| - Its high rate of prediction failure | ||||||
| Ji et al., 2014139 | Generalized linear mixed-effects models |
To develop the NEC diagnostic and prognostic models |
520 infants | Clinical variables | 84%–85% (AUC) | + Prediction of NEC and risk stratification. |
| - Non-image data | ||||||
| Young et al., 2012203 | Multilayer perceptron (MLP) ANN |
To forecasting the sound loads in NICUs |
72 individual data | Voice record- | + Prediction of noise levels | |
| - Limited only to time and noise level | ||||||
| Nascimento LFC et al., 200264 | A fuzzy linguistic model |
To estimate the possibility of neonatal mortality. |
58 neonatal deaths in 1351 records. | EHR |
It depends on the GA, APGAR score and BW 90% (accuracy) |
+ Not to compare this model with other predictive models because the fuzzy model does not use blood analyses and current models such as PRISM, SNAP or CRIB do not use the fuzzy variables |
| - No change over the time | ||||||
| Reis et al., 2004204 | Fuzzy composition | Determine if more intensive neonatal resuscitation procedures will be required during labor and delivery |
Nine neonatologists facing which a degree of association with the risk of occurrence of perinatal asphyxia |
61 antenatal and intrapartum clinical situations | 93% (AUC) |
+ Maternal medical, obstetric and neonatal characteristics to the clinical conditions of the newborn, providing a risk measurement of need of advanced neonatal resuscitation measures |
|
- Implement a supplemental system to help health care workers in making perinatal care decisions. - Eighteen of the factors studied were not tested by experimental analysis, for which testing in a multicenter study or over a very long period of time in a prospective study would be probably needed | ||||||
| - No image | ||||||
| Jalali et al., 2018147 | SVM | To predict the development of PVL by analyzing vital sign and laboratory data received from neonates shortly following heart surgery | 71 neonates(including HLHS and TGA) |
Physiological and clinical data Up to 12 h after cardiac surgery |
88% (AUC) | + Might be used as an early prediction tool |
|
- Retrospective observational study - Other variables did not collected which precipitated the PVL | ||||||
| Ambalavanan et al., 2000140 | ANN | To predict adverse neurodevelopmental outcome in ELBW |
218 neonates 144 for training 74 for test set |
Clinical variables and Bayley scores at 18 months | 62% (Major handicapped-AUC) | + Neural network is more sensitive detection individual mortality |
|
- Short follow-up - Underperformance of neural network | ||||||
| Saria et al., 2010 146 |
Bayesian modeling paradigm Leave one out algorithm |
To develop morbidity prediction tool | To identify infants who are at risk of short- and long-term morbidity in advance | Electronically collected physiological data from the first 3 hours of life in preterm newborns (<34 weeks gestation, birth weight <2000 gram) of 138 infants | 91.9% (AUC-predicting high morbidity) | + Physiological variables, notably short-term variability in respiratory and heart rates, contributed more to morbidity prediction than invasive laboratory tests. |
| Saadah et al., 2014205 | ANN |
To identify subgroups of premature infants who may benefit from palivizumab prophylaxis during nosocomial outbreaks of respiratory syncytial virus (RSV) infection |
176 infants 31 (17.6%) received palivizumab during the outbreaks |
EHR |
In male infants whose birth weight was less than 0.7 kg and who had hemodynamically significant congenital heart disease. |
- Retrospective analysis using an AI model - No external validation - Low generalizability - Small sample size |
| Mikhno et al., 2012128 | Logistic Regression Analysis |
Developed a prediction algorithm to distinguish patients whose extubation attempt was successful from those that had EF |
179 neonates |
EHR 57 candidate features Retrospective data from the MIMIC-II database |
87.1% (AUC) |
+ A new model for EF prediction developed with logistic regression, and six variables were discovered through ML techniques |
|
- 2 hour prior extubation took into consideration - Longer duration should be encountered | ||||||
| Gomez et al., 201974 |
AdaBoost Bagged Classification Trees (BCT) Random Forest(RF) Logistic Regression (LR) SVM |
To predict sepsis in term neonates within 48 hours of life monitoring heart rate variability(HRV) and EHR |
79 newborns 15 were diagnosed with sepsis |
4 EHR variables and HRV variables. HRV variables were analyzed with the ML methods |
94.3% (AUC) AdaBoost 88.8% (AUC) Bagged Classification Trees Lowest AUC 64% (k-NN) |
+ Noninvasive methods for sepsis prediction |
|
- Small sample size - Need an extra software for HRV analysis - Not included EHR into ML analysis - No Adequate Clinical Information | ||||||
| Verder et al., 2020125 | Support vector machine (SVM) |
To develop a fast bedside test for prediction and early targeted intervention of bronchopulmonary dysplasia (BPD) to improve the outcome |
61 very preterm infants were included in the study |
Spectral pattern analysis of gastric aspirate combined with specific clinical data points |
Sensitivity: 88% Specificity: 91% |
+ Multicenter non-interventional diagnostic cohort study + Early prediction and targeted intervention of BPD have the potential to improve the outcome + First algorithm developed by AI to predict BPD after shortly birth with high sensitivity and specificity. |
| - Small sample size | ||||||
| Ochab et al., 2015126 | SVM and logistic regression | To predict BPD in LBW infant | 109 neonates | EHR (14 risk factors) | 83.2% (accuracy) | + Decision support system |
|
- Small sample size - Few clinical variables - Low accuracy with SVM - A single-center design leads to missing data and unavoidable biases in identifying and recruiting participants | ||||||
| Townsend et al., 200862 | ANN | To predict events in the NICU |
Data collected by the CNN between January 1996 and October 1997 contains data from 17 NICUs |
27 clinical variables | 85% (AUC) |
+ Modeling life-threatening complications will be combined with a case-presentation tool to provide physicians with a patient’s estimated risk for several important outcomes |
|
+ Annotations would be created prospectively with adequate details for understanding any surrounding clinical conditions occurring during alarms | ||||||
|
- The methodology employed for data annotation - Retrospective design - Not confirmed with real clinical situations - Data may not capture short-lived artifacts and thus these models would not be effectively designed to detect such artifacts in a prospective setting | ||||||
| Ambalavanan et al., 200563 | ANN and logistic regression | To predict death of ELBW infant | 8608 ELBW infants | 28 clinical variables |
84% (AUC) 85% (AUC) |
+ The difficulties of predicting death should be acknowledged in discussions with families and caregivers about decisions regarding initiation or continuation of care |
|
- Chorioamnionitis, timing of prenatal steroid therapy, fetal biophysical profile, and resuscitation variables such as parental or physician wishes regarding resuscitation) could not be evaluated because they were not part of the data collected. | ||||||
| Bahado-Singh et al., 2022200 |
Random forest (RF), support vector machine (SVM), linear discriminant analysis (LDA), prediction analysis for microarrays (PAM), and generalized linear model (GLM) |
Prediction of coarctation in neonates | Genome-wide DNA methylation analysis of newborn blood DNA |
24 patients 16 controls |
97% (80%–100%) (AUC) |
+ AI in epigenomics + Accurate prediction of CoA |
|
- Small dataset - Not included other CHD | ||||||
| Bartz-Kurycki et al., 2018142 |
Random forest classification (RFC), and a hybrid model (combination of clinical knowledge and significant variables from RF) |
To predict neonatal surgical site infections (SSI) |
16,842 neonates | EHR | 68% (AUC) |
+ Large dataset + Important neonatal outcome |
|
-Retrospective study -Bias in missing data | ||||||
| Do et al., 202265 | Artificial neural network (ANN), random forest (RF), and support vector machine (SVM) | To predict mortality of very low birth weight infants (VLBWI) | 7472 VLBWI data from Korean neonatal network | EHR |
84.5% (81.5%–87.5%) (ANN-AUC) 82.6% (79.5%–85.8%) (RF-AUC) 63.1% (57.8%–68.3%). SVM-AUC |
+VLBWI mortality prediction using ML methods would produce the same prediction rate as the standard statistical LR approach and may be appropriate for predicting mortality studies utilizing ML confront a high risk of selection bias. |
| - Low prediction rate with ML | ||||||
| Podda et al., 201866 | ANN |
Development of the Preterm Infants Survival Assessment (PISA) predictor |
Between 2008 and 2014, 23747 neonates (<30 weeks gestational age or <1501 g birth weight were recruited Italian Neonatal Network | 12 easily collected perinatal variables |
91.3% (AUC) 77.9% (AUC) 82.8% (AUC) 88.6% (AUC) |
+ NN had a slightly better discrimination than logistic regression |
|
- Like all other model-based methods, is still too imprecise to be used for predicting an individual infant’s outcome - Retrospective design - Lack of variables | ||||||
| Turova et al., 202085 | Random Forest | To predict intraventricular hemorrhage in 23–30 weeks of GA infants | 229 infants |
Clinical variables and cerebral blood flow (extracted from mathematical calculation) were used 10 fold validation |
86%–93% (AUC) Vary on the extracted features in and feature weight in the model |
+ Good accuracy |
|
- Retrospective - Gender distribution was not standardized between the groups - Not corresponding lab value according to the IVH time | ||||||
| Cabrera-Quiros et al., 2021145 |
Logistic regressor, naive Bayes, and nearest mean classifier |
Prediction of late-onset sepsis (starting after the third day of life) in preterm babies based on various patient monitoring data 24 hours before onset |
32 premature infants with sepsis and 32 age-matched control patients |
Heart rate variability, respiration, and body motion, differences between late-onset sepsis and Control group were visible up to 5 hours preceding the cultures, resuscitation, and antibiotics started here (CRASH) point |
Combination of all features showed a mean accuracy 79% and mean precision rate 82% 3 hours before the onset of sepsis Naive Bayes accuracy: 71% Nearest Mean: 70% |
+ Monitoring of vital parameters could be predicted late onset sepsis up to 5 hours. |
|
- Small sample size - Retrospective - Gestational age, postnatal age, sepsis and culture | ||||||
| Reed et al., 2021143 |
Comparison least absolute shrinkage and selection operator (LASSO) and random forest (RF) to expert-opinion driven logistic regression modeling |
Prediction of 30-day unplanned rehospitalization of preterm babies |
5567 live-born babies and 3841 were included to the study Data derived exclusively from The population-based prospective cohort study of French preterm babies, EPIPAGE 2. |
The logistic regression model comprised 10 predictors, selected by expert clinicians, while the LASSO and random forest included 75 predictors |
65% (AUC) RF 59% (AUC) LASSO 57% (AUC) LR |
+ The first comparison of different modeling methods for predicting early rehospitalization + Large cohort with data variation |
|
- No accurate evaluation of rehospitalization causes - Data collection after discharge based on survey filled by mothers - 9% of babies were rehospitalized | ||||||
| Khursid et al., 202170 |
K-nearest neighbor, random forest, artificial neural network, stacking neural network ensemble |
To predict, on days 1, 7, and 14 of admission to neonatal intensive care, the composite outcome of BPD/death prior to discharge. |
<33 weeks GA cohort (n = 9006) And < 29 weeks GA were included |
For each set of models (Days 1, 7, 14), stratified random sampling. 80% of used were training. 20% of used were test set. 10-fold cross validation for test dataset |
81%–86% (AUC) for, 33 weeks 70–79% (AUC) for, 29 weeks |
+ Large dataset |
|
- Not having good performance scores - No data sharing - Not included important predictors (FiO2 and presence of PDA before 7th days) | ||||||
| Moreira et al., 202272 | Logistic regression and Random Forest | To develop an early prediction model of neonatal death on extremely low gestational age(ELGA) infants |
< 28 weeks Swedish Neonatal Quality Registry 2011- May 2021 3752 live born ELGA infants |
Birthweight, Apgar score at 5 min, gestational age were selected as features and new model (BAG) designed to predict mortality |
76.9%(AUC) Validation cohort 68.9% (AUC) |
+ Model development cohort and validation cohort included + BAG model had better AUC than individual birthweight and gestational age model. + Code is available + Online calculator is available |
|
- BAG model does not include clinical variables and clinical practice. Birthweight and gestational age could not be changed. Only Apgar scores could be changed. | ||||||
| Hsu et al., 202071 |
RF KNN ANN XGBoost Elastic-net |
To predict mortality of neonates when they were on mechanical intubation |
1734 neonates 70% training 30% test |
Mortality scores Patient demographics Lab results Blood gas analysis Respirator parameters Cardiac inotrop agents from onset of respiratory failure to 48 hours |
93.9% (AUC) RF has achieved the highest prediction of mortality |
+ Employed several ML and statistics + Explained the feature analysis and importance into analysis |
|
- Two center study - Algorithmic bias - Inability to real time prediction | ||||||
| Stocker et al., 202275 | RF | To predict blood culture test positivity according to the all variables, all variables without biomarkers, only biomarkers, only risk factors, and only clinical signs |
1710 neonates from 17 centers Secondary analysis of NeoPInS data |
Biomarkers(4 variables) Risk factors (4 variables) Clinical signs(6 variables) Other variables(14) All variables (28) They included to RF analysis to predict culture positive early onset sepsis |
Only biomarkers 73.3% (AUC) All variables 83.4% (AUC) Biomarkers are the most important contributor |
+ CRP and WBC are the most important variables in the model + Decrease the overtreatment + Multi-center data |
|
- Overfitting of the model due to the discrepancy with currently known clinical practice - Seemed not evaluated the clinical signs and risk factors which are really important in daily practice | ||||||
| Temple et al., 2016229 | supervised ML and NLP |
To identify patients that will be medically ready for discharge in the subsequent 2–10 days. |
4693 patients (103,206 patient-days178 |
NLP using a bag of words (BOW) surgical diagnoses, pulmonary hypertension, retinopathy of prematurity, and psychosocial issues |
63.3% (AUC) 67.7% (AUC) 75.2% (AUC) 83.7% (AUC) |
+ Could potentially avoid over 900 (0.9%) hospital days |
Table 6.
DL-based studies in neonatology using imaging and non-imaging data for diagnosis.
| Study | Approach | Purpose | Dataset | Type of data (image/non-image) | Performance | Pros(+) |
|---|---|---|---|---|---|---|
| Cons(-) | ||||||
| Hauptmann et al., 2019187 | 3D (2D plus time) CNN architecture |
Ability of CNNs to reconstruct highly accelerated radial real‐time data in patients with congenital heart disease |
250 CHD patients. | Cardiovascular MRI with cine images | +Potential use of a CNN for reconstruction real time radial data | |
| Lei et al., 2022158 | MobileNet-V2 CNN | Detect PDA with AI |
300 patients 461 echocardiograms |
Echocardiography | 88% (AUC) |
+Diagnosis of PDA with AI - Does not detect the position of PDA |
| Ornek et al., 2021189 |
VGG16 (CNN) |
To focus on dedicated regions to monitor the neonates and decides the health status of the neonates (healthy/unhealthy) |
38 neonates | 3800 Neonatal thermograms | 95% (accuracy) | +Known with this study how VGG16 decides on neonatal thermograms |
| -Without clinical explanation | ||||||
| Ervural et al., 2021190 | Data Augmentation and CNN | Detect health status of neonates | 44 neonates |
880 images Neonatal thermograms |
62.2% to 94.5% (accuracy) | +Significant results with data augmentation |
|
-Less clinically applicable -Small dataset | ||||||
| Ervural et al., 2021191 | Deep siamese neural network(D-SNN) | Prediagnosis to experts in disease detection in neonates | 67 neonates, |
1340 images Neonatal thermograms |
99.4% (infection diseases accuracy in 96.4% (oesophageal atresia accuracy), 97.4% (in intestinal atresia-accuracy, 94.02% (necrotising enterocolitis accuracy) |
+D-SNN is effective in the classification of neonatal diseases with limited data |
| -Small sample size | ||||||
| Ceschin et al., 2018188 | 3D CNNs |
Automated classification of brain dysmaturation from neonatal MRI in CHD |
90 term-born neonates with congenital heart disease and 40 term-born healthy controls |
3 T brain MRI | 98.5% (accuracy) |
+ 3D CNN on small sample size, showing excellent performance using cross-validation for assessment of subcortical neonatal brain dysmaturity + Cerebellar dysplasia in CHD patients |
| - Small sample size | ||||||
| Ding et al., 2020169 | HyperDense-Net and LiviaNET | Neonatal brain segmentation |
40 neonates 24 for training 16 for experiment |
3T Brain MRI T1 and T2 |
94% 95%/ 92% (Dice Score) 90%/90%/88% (Dice Score) |
+Both neural networks can segment neonatal brains, achieving previously reported performance |
| - Small sample size | ||||||
| Liu et al., 202099 |
Graph Convolutional Network (GCN) |
Brain age prediction from MRI | 137 preterm |
1.5-Tesla MRI + Bayley-III Scales of Toddler Development at 3 years |
Show the GCN’s superior prediction accuracy compared to state-of-the-art methods |
+ The first study that uses GCN on brain surface meshes to predict neonatal brain age, to predict individual brain age by incorporating GCN-based DL with surface morphological features |
| -No clinical information | ||||||
| Hyun et al., 2016155 |
NLP and CNN AlexNet and VGG16 |
To achieve neonatal brain ultrasound scans in classifying and/or annotating neonatal using combination of NLP and CNN |
2372 de identified NS report |
11,205 NS head Images |
87% (AUC) |
+ Automated labeling |
| - No clinical variable | ||||||
| Kim et al., 2022157 |
CNN(VGG16) Transfer learning |
To assesses whether a convolutional neural network (CNN) can be trained via transfer learning to accurately diagnose germinal matrix hemorrhage on head ultrasound |
400 head ultrasounds (200 with GMH, 200 without hemorrhage) |
92% (AUC) |
+ First study to evaluate GMH with grade and saliency map + Not confirmed with MRI or labeling by radiologists |
|
|
- Small sample size which limited the training, validation and testing of CNN algorithm | ||||||
| Li et al., 2021159 | ResU-Net |
Diffuse white matter abnormality (DWMA) on VPI’s MR images at term-equivalent age |
98 VPI 28 VPI |
3 Tesla Brain MRI T1 and T2 weighted |
87.7% (Dice Score) 92.3% (accuracy) |
+Developed to segment diffuse white matter abnormality on T2-weighted brain MR images of very preterm infants |
| + 3D ResU-Net model achieved better DWMA segmentation performance than multiple peer deep learning models. | ||||||
|
- Small sample size - Limited clinical information | ||||||
| Greenbury et al., 2021170 |
Agnostic, unsupervised ML Dirichlet Process Gaussian Mixture Model (DPGMM) |
To acquire understanding into nutritional practice, a crucial component of neonatal intensive care |
n = 45,679) over a six-year period UK National Neonatal Research Database (NNRD) |
EHR clustering on time analysis on daily nutritional intakes for extremely preterm infants born <32 weeks gestation |
+Identifying relationships between nutritional practice and exploring associations between nutritional practices and outcomes using two outcomes: discharge weight and BPD +Large national multi center dataset |
|
| - Strong likelihood of multiple interactions between nutritional components could be utilized in records | ||||||
| Ervural et al., 2021192 |
CNN Data augmentation |
To detect respiratory abnormalities of neonates by AI using limited thermal image |
34 neonates 680 images 2060 thermal images (11 testing) 23 training) |
Thermal camera image |
85% (accuracy) |
+ CNN model and data enhancement methods were used to determine respiratory system anomalies in neonates. |
|
-Small sample size -There is no follow-up and no clinical information | ||||||
| Wang et al., 2018174 | DCNN | To classify automatically and grade a retinal hemorrhage |
3770 newborns with retinal hemorrhage of different severity (grade 1, 2 and 3) and normal controls from a large cross-sectional investigation in China. |
48,996 digital fundus images |
97.85% to 99.96% (accuracy) 98.9%–100% AUC) |
+The first study to show that a DCNN can detect and grade neonatal retinal hemorrhage at high performance levels |
| Brown et al., 2018171 | DCNN |
To develop and test an algorithm based on DL to automatically diagnose plus disease from retinal photographs |
5511 retinal photographs (trained) independent set of 100 images |
Retinal images |
94% (AUC) 98% (AUC) |
+ Outperforming 6 of 8 ROP expert + Completely automated algorithm detected plus disease in ROP with the same or greater accuracy as human doctors + Disease detection, monitoring, and prognosis in ROP-prone neonates |
|
-No clinical information and no clinical variables | ||||||
| Wang et al., 2018179 |
DNN (Id-Net Gr-Net) |
To automatically develop identification and grading system from retinal fundus images for ROP |
349 cases for identification 222 cases for grading |
Retinal fundus images |
Id-Net: 96.64% (sensitivity) 99.33% (specificity) 99.49% (AUC) Gr-Net: 88.46% (sensitivity) 92.31% (specificity) 95.08% (AUC) |
+ Large dataset including training, testing and, comparison with human experts. + Good example of human in the loop models + Code is available |
|
- No clinical grading included - Dataset is not available | ||||||
| Taylor et al., 2019172 |
DCNN Quantitative score |
To describe a quantitative ROP severity score derived using a DL algorithm designed to evaluate plus disease and to assess its utility for objectively monitoring ROP progression |
Retinal images | 871 premature infants | + ROP vascular severity score is related to disease category at a specific period and clinical course of ROP in preterm | |
|
-Retrospective cohort study -No follow-up for patients -Low generalizability | ||||||
| Campbell et al., 2021173 |
DL(U-Net) Tensor Flow ROP Severity Score(1-9) |
Evaluate the effectiveness of artificial intelligence (AI)-based screening in an Indian ROP telemedicine program and whether differences in ROP severity between neonatal care units (NCUs) identified by using AI are related to differences in oxygen-titrating capability |
4175 unique images from 1253 eye examinations retinopathy of Prematurity Eradication Save Our Sight ROP telemedicine program |
363 infants from 32 NCUs | 98% (AUC) |
+ Integration of AI into ROP screening programs may lead to improved access to care for secondary prevention of ROP and may facilitate assessment of disease epidemiology and NCU resources |
| Xu et al., 2021193 |
-Wireless sensors -Pediatric focused algorithm -ML and data analytics -cloud based dashboards |
To enhance monitoring with wireless sensors |
By the middle of 2021, there were 15,000 pregnant women and up to 500 newborns. 1000 neonates |
+ Future predictive algorithms of clinical outcomes for neonates +As small as 4.4 cm 2.4 cm and as thin as 1 mm in totally wirelessly powered versions, these devices provide continuous monitoring in this sensitive group |
||
| Werth et al., 2019186 |
Sequential CNN ResNet |
Automated sleep state requirement without EEG monitoring | 34 stable preterm infants |
Vital signs were recorded ECG R peaks were analyzed |
Kappa of 0.43 ± 0.08 Kappa of 0.44 ± 0.01 Kappa of 0.33 ± 0.04 |
+ Non-invasive sleep monitoring from ECG signals |
|
- Retrospective study - Video were not used in analysis | ||||||
| Ansari et al., 2022185 | A Deep Shared Multi-Scale Inception Network | Automated sleep detection with limited EEG Channels | 26 preterm infants | 96 longitudinal EEG recordings |
Kappa 0.77 ± 0.01 (with 8-channel EEG) and 0.75 ± 0.01 (with a single bipolar channel EEG |
+ The first study using Inception-based networks for EEG analysis that utilizes filter sharing to improve efficiency and trainability. + Even a single EEG channel making it more practical |
|
- Small sample size - Retrospective - No clinical information | ||||||
| Ansari et al., 2018184 | CNN |
To discriminate quiet sleep from nonquiet sleep in preterm infants (without human labeling and annotation) |
26 preterm infants |
54 EEG recordings for training 43 EEG recording for the test (at 9 and 24 months corrected age, a normal neurodevelopmental outcome score (Bayley Scales of Infant Development-II, mental and motor score >85)) |
92% (AUC) 98% (AUC) |
+ CNN is a viable and rapid method for classifying neonatal sleep phases in preterm babies + Clinical information |
|
- Retrospective - The paucity of EEG recordings below 30 weeks and beyond 38 weeks postmenstrual age - Lack of interpretability of the features | ||||||
| Moeskops et al., 2017199 |
CNN for MRI segmentation230 SVM for neurocognitive outcome prediction |
To predict cognitive and motor outcome at 2–3 years of preterm infants from MRI at 30th and 40th weeks of PMA |
30 weeks (n = 86) 40 weeks (n = 153) |
3 T Brain MRI at 30th and 40th weeks of PMA BSID-III at average age of 29 months (26–35) |
Cognitive Outcome (BSID<85) 78% (AUC) 30 weeks of PMA 70% (AUC) 40 weeks of PMA Motor Outcome BSID<85 80% (AUC) 30 weeks of PMA 71% (AUC) 40 weeks of PMA |
+ Brain MRI can predict cognitive and motor outcome + Segmentations, quantitative descriptors, classification were performed and + Volumes, measures of cortical morphology were included as a predictor |
|
- Small sample size -Retrospective design |
Table 7.
DL-based studies in neonatology using imaging and non-imaging for prediction.
| Study | Approach | Purpose | Dataset | #Non-Image data | #-Image data | AUC/accuracy | Pros(+) |
|---|---|---|---|---|---|---|---|
| Cons(-) | |||||||
| Saha et al., 2020176 | CNN |
To predict abnormal motor outcome at 2 years from early brain diffusion magnetic resonance imaging (MRI) acquired between 29 and 35 weeks postmenstrual age (PMA) |
77 very preterm infants (born <31 weeks gestational age (GA)) |
At 2 years CA, infants were assessed using the Neuro-Sensory Motor Developmental Assessment (NSMDA) |
3 T brain diffusion MRI |
72% (AUC) |
+ Neuromotor outcome can be predicted directly from very early brain diffusion MRI (scanned at ~30 weeks PMA), without the requirement of constructing brain connectivity networks, manual scoring, or pre-defined feature extraction + Cerebellum and occipital and frontal lobes were related motor outcome |
| -Small sample size | |||||||
| Shabanian et al., 2019175 | Based on MRIs, the 3D CNN algorithm can promptly and accurately diagnose neurodevelopmental age |
Neurodevelopmental age estimation |
112 individuals | 1.5T MRI from NIMH Data Achieve |
95% (accuracy) 98.4% (accuracy) |
+ 3D CNNs can be used to accurately estimate neurodevelopmental age in infants based on brain MRIs |
|
|
- Restricted clinical information - No clinical variable - Small sample size which limited the training, validation and testing of CNN algorithm | |||||||
| He et al., 2020177 | Supervised and unsupervised learning | In terms of predicting abnormal neurodevelopmental outcomes in extremely preterm newborns, multi-stage DTL (deep transfer learning) outperforms single-stage DTL. |
33 preterm infants Retrained in 291 neonates |
Bayley Scales of Infant and Toddler Development III at 2 years corrected age |
3 Tesla Brain MRI T1 and T2 weighted |
86% (cognitive deficit-AUC) 66% (language deficit-AUC) 84% (motor deficit-AUC) |
+ Risk stratification at term-equivalent age for early detection of long-term neurodevelopmental abnormalities and directed earlier therapies to enhance clinical outcomes in extremely preterm infants |
| - The investigation of the brain’s functional connectome was based on an anatomical/structural atlas as opposed to a functional brain parcellated atlas. |
ML applications in neonatal mortality
Neonatal mortality is a major factor in child mortality. Neonatal fatalities account for 47 percent of all mortality in children under the age of five, according to the World Health Organization60. It is, therefore, a priority to minimize worldwide infant mortality by 203061.
ML investigated infant mortality, its reasons, and its mortality prediction62–68. In a recent review, 1.26 million infants born from 22 weeks to 40 weeks of gestational age were enrolled67. Predictions were made as early as 5 min of life and as late as 7 days. An average of four models per investigation were neural networks, random forests, and logistic regression (58.3%)67. Two studies (18.2%) completed external validation, although five (45.5%) published calibration plots67. Eight studies reported AUC, and five supplied sensitivity and specificity67. The AUC was 58.3–97.0%67. Sensitivities averaged 63 to 80%, and specificities 78 to 98%67. Linear regression analysis was the best overall model despite having 17 features67. This analysis highlighted the most prevalent AI neonatal mortality measures and predictions. Despite the advancement in neonatal care, it is crucial that preterm infants remain highly susceptible to mortality due to immaturity of organ systems and increased susceptibility to early and late sepsis69. Addressing these permanent risks necessitates the utilization of ML to predict mortality63–66,68,70. Early studies employed ANN and fuzzy linguistic models and achieved an AUC of 85–95% and accuracy of 90%62,68. New studies in a large preterm populations and extremely low birthweight infants found an AUC of 68.9–93.3%65,71. There are some shortcomings in these studies; for example, none of them used vital parameters to represent dynamic changes, and hence, there was no improvement in clinical practice in neonatology. Unsurprisingly, gestational age, birthweight, and APGAR scores were shown as the most important variables in the models64,72. Future research is suggested to focus on external evaluation, calibration, and implementation of healthcare applications67.
Neonatal sepsis, which includes both early onset sepsis and late onset sepsis, is a significant factor contributing to neonatal mortality and morbidity73. Neonatal sepsis diagnosis and antibiotic initiation present considerable obstacles in the field of neonatal care, underscoring the importance of implementing comprehensive interventions to alleviate their profound negative consequences. The studies have predicted early sepsis from heart rate variability with an accuracy of 64–94%74. Another secondary analysis of multicenter data revealed that clinical biomarkers weighed the ML decision by integrating all clinical and lab variables and achieved an AUC of 73–83%75.
ML applications in neurodevelopmental outcome
Recent advancements in neonatal healthcare have resulted in a decrease in the incidence of severe prenatal brain injury and an increase in the survival rates of preterm babies76. However, even though routine radiological imaging does not reveal any signs of brain damage, this population is nonetheless at significant risk of having a negative outcome in terms of neurodevelopment77–80. It is essential to discover early indicators of abnormalities in brain development that might serve as a guide for the treatment of preterm children at a greater risk of having negative neurodevelopmental consequences81,82.
The most common reason for neurodevelopmental impairment is intraventricular hemorrhage (IVH) in preterm infants83. Two studies predicted IVH in preterm infants. Both studies have not deployed the ultrasound images in their analysis, they only predicted IVH according to the clinical variables84,85.
Morphological studies have demonstrated that preterm birth is linked to smaller brain volume, cortical folding, axonal integrity, and microstructural connectivity86,87. Studies concentrating on functional markers of brain maturation, such as those derived from resting-state functional connectivity (rsFC) analyses of blood-oxygen-level dependent (BOLD) fluctuations, have revealed further impacts of prematurity on the developing connectome, ranging from decreased network-specific connectivity82,88,89. Many studies investigated brain connectivity in preterm infants88,90–92 and brain structural analysis in neonates93 and neonatal brain segmentation94 with the help of ML methods. Similarly, one of the most important outcomes of neurodevelopment at 2-year-old-age is neurocognitive evaluations. The studies evaluated the morphological changes in the brain in relation to neurocognitive outcome95–97 and brain age prediction98,99. It has been found that near-term regional white matter (WM) microstructure on diffusion tensor imaging (DTI) predicted neurodevelopment in preterm infants using exhaustive feature selection with cross-validation96 and multivariate models of near-term structural MRI and WM microstructure on DTI might help identify preterm infants at risk for language impairment and guide early intervention95,97 (Table 4). One of the studies that evaluated the effects of PPAR gene activity on brain development with ML methods100 revealed a strong association between abnormal brain connectivity and implicating PPAR gene signaling in abnormal white matter development. Inhibited brain growth in individuals exposed to early extrauterine stress is controlled by genetic variables, and PPARG signaling has a formerly unknown role in cerebral development100 (Table 2).
Table 4.
ML based (non-DL) studies in neonatology using imaging data for prediction.
| Study | Approach | Purpose | Dataset | Type of data | Performance | Pros(+) |
|---|---|---|---|---|---|---|
| Cons(-) | ||||||
| Vassar et al., 202095 |
Multivariate models with leave-one-out cross-validation and exhaustive feature selection |
Very premature infants' structural brain MRI and white matter microstructure as evaluated by diffusion tensor imaging (DTI) in the near term and their impact on early language development | 102 infants |
Brain MRI and DTI + (Bayley Scales of Infant- Toddler Development-III at 18 to 22 months) |
50.2% (language composite score -AUC) 61.7% (expressive language subscore-AUC) 32.2% (receptive language subscore-AUC) |
+ Preterm babies at risk for language impairment may be identified using multivariate models of near-term structural MRI and white matter microstructure on DTI, allowing for early intervention |
|
- Demographic data is not included - Cross validation? - Small sample size | ||||||
| Schadl et al., 201896 |
-Linear models with exhaustive feature selection and leave-one-out cross-validation |
To predict neurodevelopment in preterm children in near term MRI and DTI |
66 preterm infants |
Brain MRI and DTI 51 WM regions (48 bilateral regions, 3 regions of corpus callosum) Bayley Scales of Infant-Toddler Development, 3rd-edition (BSID-III) at 18–22 months. |
100% (AUC, cognitive impairment) 91% (AUC, motor impairment |
- Using structural brain MRI findings of WMA score, lower accuracy - Small cohort - DTI has better implementation and interpretation |
| Wee et al., 201797 | SVM and canonical correlation analysis (CCA) |
To examine heterogeneity of neonatal brain network and its prediction to child behaviors at 24 and 48 months of age |
120 neonates |
1.5-Tesla DW MRI Scans Diffusion tensor imaging (DTI) tractography + Child Behavior Checklist (CBCL) at 24 and 48 months of age. |
89.4% (accuracy) |
+ Neural organization established during fetal development could predict individual differences in early childhood behavioral and emotional problems |
| - Small sample size |
Alternative to morphological studies, neuromonitorization is shown to be an important tool for which ML methods have been frequently employed, for example, in automatic seizure detection from video EEG101–103 and EEG biosignals in infants and neonates with HIE104–108. The detection of artifacts109,110, sleep states102, rhythmic patterns111, burst suppression in extremely preterm infants112,113 from EEG records were studied with ML methods. EEG records are often used for HIE grading114 too. It has been shown in those studies that EEG recordings of different neonate datasets found an AUC of 89% to 96%104,105,115, accuracy 78–87%114,116 regarding seizure detection with different ML methods (Table 3).
Table 3.
ML based (non-DL) studies in neonatology using non-imaging data for diagnosis.
| Study | Approach | Purpose | Dataset | Type of data | Performance | Pros(+) |
|---|---|---|---|---|---|---|
| Cons(-) | ||||||
| Reed et al., 1996135 | Recognition-based reasoning | Diagnosis of congenital heart defects | 53 patients |
Patient history, physical exam, blood tests, cardiac auscultation, X-ray, and EKG data |
+ Useful in multiple defects | |
| - Small sample size-Not real AI - implementation | ||||||
| Aucouturier et al., 2011148 |
Hidden Markov model architecture (SVM, GMM) |
To identify expiratory and inspiration phases from the audio recording of human baby cries |
14 infants, spanning four vocalization contexts in their first 12 months | Voice record- |
86%–95% (accuracy) |
+ Quantify expiration duration, count the crying rate, and other time-related characteristics of baby crying for screening, diagnosis, and research purposes over large populations of infants + Preliminary result |
|
- More data needed - No clinical explanation - Small sample size - Required preprocessing | ||||||
| Cano Ortiz et al., 2004149 |
Artificial neural networks (ANN) |
To detect CNS diseases in infant cry |
35 neonates, nineteen healthy cases and sixteen sick neonates |
Voice record (187 patterns) |
85% (accuracy) |
+ Preliminary result |
| - More data needed for correct classification for | ||||||
| Hsu et al., 2010151 |
Support Vector Machine (SVM) Service-Oriented Architecture (SOA) |
To diagnose Methylmalonic Acidemia (MMA) |
360 newborn samples |
Metabolic substances data collected from tandem mass spectrometry (MS/MS) |
96.8% (accuracy) |
+Better sensitivity than classical screening methods |
|
-Small sample size - SVM pilot stage education not integrated | ||||||
| Baumgartner et al., 2004152 |
Logistic regression analysis (LRA) Support vector machines (SVM) Artificial neural networks (ANN) Decision trees (DT) k-nearest neighbor classifier (k-NN) |
Focusing on phenylketonuria (PKU), medium chain acyl-CoA dehydrogenase deficiency (MCADD |
During the Bavarian newborn screening program all newborns |
Metabolic substances data collected from tandem mass spectrometry (MS/MS) |
99.5% (accuracy) |
+ ML techniques, LRA (as discussed above), SVM and ANN, delivered results of high predictive power when running on full as well as on reduced feature dimensionality. |
|
- Lacking direct interpretation of the knowledge representation | ||||||
| Chen et al., 2013153 | Support vector machine (SVM) |
To diagnose phenylketonuria (PKU), hypermethioninemia, and 3-methylcrotonyl-CoA-carboxylase (3-MCC) deficiency |
347,312 infants (220 metabolic disease suspect) |
Newborn dried blood samples |
99.9% (accuracy) 99.9% (accuracy) 99.9% (accuracy) |
+ Reduced false positive cases |
|
- The feature selection strategies did not include the total features for establishing either the manifested features or total combinations | ||||||
| Temko et al., 2011105 |
Support Vector Machine (SVM) classifier leave-one-out (LOO) cross-validation method. |
To measure system performance for the task of neonatal seizure detection using EEG |
17 newborns system is validated on a large clinical dataset of 267 h All seizures were annotated independently by 2 experienced neonatal electroencephalographers using video EEG |
EEG data | 89% (AUC) |
+ SVM-based seizure detection system can greatly assist clinical staff, in a neonatal intensive care unit, to interpret the EEG. |
|
- No clinical variable - Datasets for neonatal seizure detection are quite difficult to obtain and never too large | ||||||
| Temko et al., 2012104 | SVM |
To use recent advances in the clinical understanding of the temporal evolution of seizure burden in neonates with hypoxic ischemic encephalopathy to improve the performance of automated detection algorithms. |
17 HIE patients | 816.7 hours EEG recordings of infants with HIE |
96.7% (AUC) |
+ Improved seizure detection |
| Temko et al., 2013115 |
Support Vector Machine (SVM) classifier leave-one-out (LOO) cross-validation method |
Robustness of Temko 2011105 |
Trained in 38 term neonates Tested in 51 neonates |
Trained in 479 hours EEG recording Tested in 2540 hours |
96.1% (AUC) Correct detection of seizure burden 70% |
- Small sample size - No clinical information |
| Stevenson et al., 2013116 | Multiclass linear classifier | Automatically grading one hour EEG epoch | 54 full term neonates | One-hour-long EEG recordings | 77.8% (accuracy) |
+ Involvement of clinical expert + Method explained in a detailed way |
| - Retrospective design | ||||||
| Ahmed et al., 2016114 |
-Gaussian mixture model. -Universal Background Model (UBM) -SVM |
An automated system for grading hypoxic–ischemic encephalopathy (HIE) severity using EEG is presented |
54 full term neonates (same dataset as Stevenson et al., 2013) |
One-hour-long EEG recordings | 87% (accuracy) |
+ Provide significant assistance to healthcare professionals in assessing the severity of HIE + Some brief temporal activities (spikes, sharp waves and certain spatial characteristics such as asynchrony and asymmetry) which are not detected by system |
| - Retrospective design | ||||||
| Mathieson et al., 2016103 |
Robusted Support Vector Machine (SVM) classifier leave-one-out (LOO) cross-validation method115 |
Validation of Temko 2013115 |
70 babies from 2 centers 35 Seizure 35 Non Seizure |
Seizure detection Algorithm thresholds is clinically acceptable range Detection rates 52.5%–75% |
+ Clinical information and Cohen score were added + First Multicenter study |
|
| - Retrospective design | ||||||
| Mathieson et al., 2016198 |
Support Vector Machine (SVM) classifier leave-one-out (LOO) cross-validation method.105 |
Analysis of Seizure detection Algorithm and characterization of false negative seizures |
20 babies (10 seizure -10 non seizure) (20 of 70 babies)103 |
Seizure detections were evaluated the sensitivity threshold |
+ Clinical information and Cohen score were added + Seizure features were analyzed |
|
| - Retrospective design | ||||||
| Yassin et al., 2017150 | Locally linear embedding (LLE) |
Explore autoencoders to perform diagnosis of infant asphyxia from infant cry |
One-second segmentation was then performed producing 600 segmented signals, from which 284 were normal cries while 316 were asphyxiated cries |
100% (accuracy) | + 600 MFCC features of normal and non-asphyxiated newborns | |
| - No clinical information | ||||||
| Li et al., 2011136 | Fuzzy backpropagation neural networks | To establish an early diagnostic system for hypoxic ischemic encephalopathy (HIE) in newborns |
140 cases (90 patients and 50 control) |
The medical records of newborns with HIE |
The correct recognition rate was 100% for the training samples, and the correct recognition rate was 95% for the test samples, indicating a misdiagnosis rate of 5%. |
+ High accuracy in the early diagnosis of HIE |
| - Small sample size | ||||||
| Zernikow et al., 199884 | ANN |
To detect early and accurately the occurrence of severe IVH in an individual patient |
890 preterm neonates (50%, 50%) Validation and training |
EHR |
93.5% (AUC) |
+ Observational study + Skipped variables during training of ANN |
| - No image | ||||||
| Ferreira et al., 2012138 | Decision trees and neural networks | Employing data analysis methods to the problem of identifying neonatal jaundice | 227 healthy newborns | 70 variables were collected and analyzed |
89% (accuracy) 84% (AUC) |
+ Predicting subsequent hyperbilirubinemia with high accuracy + Data mining has the potential to assist in clinical decision - making, thus contributing to a more accurate diagnosis of neonatal jaundice |
| - Not included all factors contributing to hyperbilirubinemia | ||||||
| Porcelli et al., 2010228 |
Artificial neural network (ANN) |
To compare the accuracy of birth weight–based weight curves with weight curves created from individual patient records |
92 ELBW infants | Postnatal EHR |
The neural network maintained the highest accuracy during the first postnatal month compared with the static and multiple regression methods |
+ ANN-generated weight curves more closely approximated ELBW infant weight curves, and, using the present electronic health record systems, may produce weight curves better reflective of the patient’s status |
| Mueller et al., 2004130 | Artificial neural network (ANN) and a multivariate logistic regression model (MLR). | To compare extubation failure in NICU |
183 infants (training (130)/validation(53)) |
EHR, 51 potentially predictive variables for extubation decisions | 87% (AUC) |
+ Identification of numerous variables considered relevant for the decision whether to extubate a mechanically ventilated premature infant with respiratory distress syndrome |
|
- Small sample size - 2-hour prior extubation took into consideration - Longer duration should be encountered | ||||||
| Precup et al., 2012129 | Support Vector Machines (SVM) |
To determine the optimal time for extubation that will minimize the duration of MV and maximize the chances of success |
56 infants; 44 successfully extubated and 12 required re-intubation |
Respiratory and ECG signals 3000 samples of the AUC features for each baby |
83.2% (failure class-accuracy) 73.6% (success class-accuracy) |
+ Prospective |
|
- Small sample size - Overfitting | ||||||
| Hatzakis et al., 2002131 | Fuzzy Logic Controller |
To develop modularized components for weaning newborns with lung disease |
10 infants with severe cyanotic congenital heart disease following surgical procedures requiring intra-operative cardiac bypass support |
Through respiratory frequency (RR); tidal volume (VT); minute ventilation (VE); gas diffusion (PaO2, PaCO2, P(A-a)02 and pH); muscle effort parameters of oxygen saturation (SaO2) and heart rate (HR) |
-No evaluation metrics | + More intelligent systems |
|
- Surrogate markers relevant to virus, drug, host, and mechanical ventilation interactions will have to be considered - Retrospective | ||||||
| Dai et al., 2021127 | ML |
To determine the significance of genetic variables in BPD risk prediction early and accurately |
131 BPD infants and 114 infants without BPD | Clinical Exome sequencing(Thirty and 21 genes were included in BPD–RGS and sBPD) |
90.7% (sBPD-AUC) 91.5% (BPD-AUC) |
+ Conducted a case–control analysis based on a prospective preterm cohort + Genetic information contributes to susceptibility to BPD + Data available |
|
- A single-center design leads to missing data and unavoidable biases in identifying and recruiting participants | ||||||
| Tsien et al., 2000144 |
C4.5 Decision tree system (artifact annotation by experts) |
To detect artifact pattern across multiple physiologic data signals | Data from bedside monitors in the neonatal ICU | 200 h of four-signal data (ECG,HR,BP,CO2) |
99.9% (O2-AUC) 93.3% (CO2-AUC) 89.4% (BP-AUC) 92.8% (HR-AUC) |
- Annotations would be created prospectively with adequate details for understanding any surrounding clinical conditions occurring during alarms |
|
- The methodology employed for data annotation - Retrospective design - Not confirmed with real clinical situations data may not - Data may not capture short lived artifacts and thus these models wouldnot be effectively designed to detect such artifacts in a prospective settings | ||||||
| Koolen et al., 2017102 | SVM | To develop an automated neonatal sleep state classification approach based on EEG that can be employed over a wide age range |
231 EEG recordings from 67 infants between 24 and 45 weeks of postmenstrual age. Ten-minute epochs of 8 channel polysomnography (N = 323) from active and quiet sleep were used as a training dataset. |
A set of 57 EEG features |
85% (accuracy) |
+ A robust EEG-based sleep state classifier was developed + The visualization of sleep state in preterm infants which can assist clinical management in the neonatal intensive care unit + Clinical variables |
|
- No integration of physiological variables - Need of longer records | ||||||
| Mohseni et al., 2006111 | Artificial neural network (ANN) | To detect EEG rhythmic pattern detection | 4 infants | 2-hour EEG record |
72.4% (sensitivity) 93.2% (specificity) |
+ Uses very short (0.4 second) segment of the data in compared to the other methods (10 seconds), + Detect seizure sooner and more accurately |
|
- Small sample size - No clinical information | ||||||
| Simayijiang et al., 2013112 | Random Forest (RF) |
To analyze the features of EEG activity bursts for predicting outcome in extremely preterm infants. |
14 extremely preterm infants Eight infants had good outcome and six had poor outcome, defined as neurodevelopmental impairment according to psychological testing and neurological examination at two years age |
One-channel EEG recordings during the first three postnatal days of 14 extremely preterm infants |
71.4% (accuracy) |
+ Each burst six features were extracted and random forest techniques |
| - Small sample size | ||||||
| Ansari et al., 2015109 | SVM | To reduce EEG artifacts in NICU |
17 neonates (for training) 18 neonates for testing |
27 hours recording EEG polygraphy (ECG, EMG, EOG, abdominal respiratory movement signal | False alarm rate drops 42% | + Reduced false alarm rate |
|
- Small sample size - Not fully online | ||||||
| Matic et al., 2016106 |
Least-squares support vector machine (LS-SVM) classifiers low-amplitude temporal profile (LTP). |
To develop an automated algorithm to quantify background electroencephalography (EEG) dynamics in term neonates with hypoxic ischemic encephalopathy |
53 neonates |
The recordings were started 2–48 (median 19) hours postpartum, using a set of 17 EEG electrodes, whereas in some patients, a reduced set of 13 electrodes was used |
91% (AUC) 94% (AUC) 94% (AUC) 97% (AUC) |
+The first study that used an automated method to study EEGs over long monitoring hours and to accurately detect milder EEG discontinuities + Necessary to perform further multicenter validation studies with even larger datasets and characterizing patterns of brain injury on MRI and clinical outcome |
|
- The number of misclassifications was rather high as compared to the EEG expert | ||||||
| Navarro et al., 2017113 | kNN, SVM and LR | To detect EEG burst in preterm infants |
Trained 14 very preterm infants Testing in 21 infants |
EEG recording |
84% (accuracy) |
+ New functionality to current bedside monitors, + Integrating wearable devices or EEG portable headsets) to follow-up maturation in preterm infants after hospital discharge |
| Ahmed et al., 2017107 |
Gaussian dynamic time warping SVM Fusion |
To improve the detection of short seizure events | 17 neonates |
EEG recording (261 h of EEG) |
71.9% (AUC) 69.8% (AUC) 75.2% (AUC) |
+ Achieving a 12% improvement in the detection of short seizure events over the static RBF kernel based system |
|
- Better post processing methods - Small sample size | ||||||
| Thomas, et al., 2008108 |
Basic Gradient Descent (BGD) Least Mean Squares (LMS) Newton Least Mean Squares (NLMS) |
To alert NICU staff ongoing seizures and detect neonatal seizures | 17 full term neonates | EEG recording |
77% (Global classifier-AUC) 80% (BGD-AUC) 79% (LMS-AUC) 80% (NLMS-AUC) |
+ The adapted classifiers outperform the global classifier in both sensitivity and specificity leading to a large increase in accuracy |
|
- Local training data is not representative of the patient’s entire EEG record | ||||||
| Schetinin et al., 2004110 |
Artificial Neural Networks (ANN) (GMDH: Group Method of Data Handling) (DT: Decision Tree) FNN: Feedforward Neural Network PNN: Polynomial Neural Network (Combined (PNN&DT) |
To detect artifacts in clinical EEG of sleeping newborns | 42 neonates |
40 EEG records 20 records containing 17,094 segments were randomly selected for training 20 records containing 21,250 segments were used for testing |
69.8% (DT-accuracy) 70.7% (FNN-accuracy) 73.2% (GMDH- accuracy) 73.2% (PNN-accuracy) 73.5% (PNN&DT) |
+ Keep the classification error done |
| - Not included other signal data (EMG, EOG) | ||||||
| Na et al., 2021123 | Multiple Logistic Regression |
Compare the performance of AI analysis with that of conventional analysis to identify risk factors associated with symptomatic PDA (sPDA) in very low birth weight infants |
10,390 Very low birth weight infant | 47 perinatal risk factors |
77% (75%–79%) (accuracy) 82% (80%–84%) (AUC) |
+ First to use AI to predict sPDA and sPDA therapy and to analyze the main risk factors for sPDA using large-scale cohort data comprising only electronic records |
|
- Low accuracy - Non-image dataset | ||||||
| Gómez-Quintana et al., 2021124 | XGBoost |
Developing an objective clinical decision support tool based on ML to facilitate differentiation of sounds with signatures of Patent Ductus Arteriosus (PDA)/CHDs, in clinical settings |
265 infants | Phonocardiogram | 88% (AUC) | + PDA diagnosis with phonocardiogram |
| - Worst performance in early days of life which is more important for diagnosis | ||||||
| - Low prediction rate with ML. | ||||||
| Sentner et al., 2022201 | Logistic regression, decision tree, and random forest | To develop an automated algorithm based on routinely measured vital parameters to classify sleep-wake states of preterm infants in real-time at the bedside. |
37 infants (PMA: 31.1 ± 1.5 weeks 9 infants (PMA 30.9 ± 1.3) validation |
Sleep-wake state observations were obtained in 1-minute epochs using a behavioral scale developed in-house while vital signs (HR, RR, SO2 were recorded simultaneously) |
80% (AUC) 77% (AUC) |
+ Real-time sleep staging algorithm was developed for the first time for preterm infants + Adapt bedside clinical work based on infants‟ sleep-wake states, potentially promoting the early brain development and well-being of preterm infants without EEG signals, noninvasive tool + Observational study |
|
- Small sample size - No additional clinical information | ||||||
| Pavel et al., 2020197 |
ANSeR Software System SVM GMM Universal Background Model (UBM), |
To detect neonatal seizure with algorithm |
128 neonates in algorithm group 130 neonates in non-algorithm group |
2–100 hours EEG recording for each neonate |
Specificity Sensitivity False Alarm Rate were calculated. AUC and accuracy were not calculated. Seizures detected by algorithm. No difference between the algorithm and non-algorithm group specificity, sensitivity |
+ The first randomized, multicenter clinical investigation to assess the clinical impact of a machine-learning algorithm in real time on neonatal seizure recognition in a clinical setting |
| - The authors mentioned the algorithm103,105,115 but not defined detailed way | ||||||
| Mooney et al., 2021196 | Random Forest | Secondary analysis of Validation of Biomarkers in HIE (BiHiVE study) |
53000 birth screened 409 infants were included 129 infants with HIE |
154 clinical variables Blood gas analysis APGAR |
Three model were used for analysis Best evaluation metrics Accuracy: 94% Specificity: 92% Sensitivity: 100% |
+ Classification with ML + Secondary analysis of prior prospective trial |
| - Not a prospective design |
ML applications in predictions of prematurity complications (BPD, PDA, and ROP)
Another important cause of mortality and morbidity in the NICU is PDA (Patent Ductus Arteriosus). The ductus arteriosus is typically present during the fetal stage, when the circulation in the lungs and body is regularly supplied by the mother; in newborns, the ductus arteriosus closes functionally by 72 h of age117. 20–50% of infants with a gestational age (GA) 32 weeks have the ductus arteriosus on day 3 of life118, while up to 60% of neonates with a GA 29 weeks have the ductus arteriosus. The presence of PDA in preterm neonates is associated with higher mortality and morbidity, and physicians should evaluate if PDA closure might enhance the likelihood of survival vs. the burden of adverse effects119–122.
ML methods were utilized on PDA detection from EHR123 and auscultation records124 such that 47 perinatal factors were analyzed with 5 different ML methods in 10390 very low birth weight infants’ predicted PDA with an accuracy of 76%123 and 250 auscultation records were analyzed with XGBoost and found to have an accuracy of 74%124 (Table 3).
Bronchopulmonary dysplasia (BPD) is a leading cause of infant death and morbidity in preterm births. While various biomarkers have been linked to the development of respiratory distress syndrome (RDS), no clinically relevant prognostic tests are available for BPD at birth125. There are ML studies aiming to predict BPD from birth70,126, gastric aspirate content125 and genetic data127 and it has been shown that BPD could be predicted with an accuracy of up to 86% in the best-case scenario70 (Table 5), analysis of responsible genes with ML could predict BPD development with an AUC of 90%127 (Table 3) and combination of gastric aspirate after birth and clinical information analysis with SVM predicted BPD development with a sensitivity of 88%125 (Table 5).
In relation to published studies in BPD with ML-based predictions, long-term invasive ventilation is considered one of the most important risk factors for BPD, nosocomial infections, and increased hospital stay. There are ML-based studies aiming to predict extubation failure128–130 and optimum weaning time131 using long-term invasive ventilation information. It has been shown in those studies that predicted extubation failure with an accuracy of 83.2% to 87%128–130 (Tables 2 and 3).
Retinopathy of prematurity (ROP) is another area of interest in the application of machine learning in neonatology132. ROP is a serious complication of prematurity that affects the blood vessels in the retina and is a leading cause of childhood blindness in high and middle-income countries, including the United States, among very low-birthweight (1500 g), very preterm (28–32 weeks), and extremely preterm infants (less than 28 weeks)132. Due to a shortage of ophthalmologists available to treat ROP patients, there has been increased interest in the use of telemedicine and artificial intelligence as solutions for diagnosing ROP132. Some ML methods, such as Gaussian mixture models, were employed to diagnose and classify ROP from retinal fundus images in studies132,133,134, and it has been reported that the i-ROP134 system classified pre-plus and plus disease with 95% accuracy. This was close to the performance of the three individual experts (96%, 94%, and 92%, respectively), and much higher than the mean performance of 31 nonexperts (81%)134 (Table 2).
Other ML applications in neonatal diseases
EHR and medical records were featured in ML algorithms for the diagnosis of congenital heart defects135, HIE (Hypoxic Ischemic Encephalopathy)136, IVH (Intraventricular Hemorrhage)84,85, neonatal jaundice137,138, prediction of NEC (Necrotizing Enterocolitis)139, prediction of neurodevelopmental outcome in ELBW (extremely low birth weight) infants65,140,141, prediction of neonatal surgical site infections142, and prediction of rehospitalization143 (Table 5).
Electronically captured physiologic data are evaluated as signal data, and they were analyzed with ML to detect artifact patterns144, late onset sepsis145, and predict infant morbidity146. Electronically captured vital parameters (respiratory rate, heart rate) of 138 infants (≤34 weeks’ gestation, birth weight ≤2000 gram) in the first 3 h of life predicted an accuracy of overall morbidity and an AUC of 91%146 (Table 5).
In addition to physiologic data, clinical data up to 12 h after cardiac surgery in HLHS (hypoplastic left heart syndrome) and TGA (transposition of great arteries) infants were analyzed to predict PVL (periventricular leukomalacia) occurrence after surgery147. The F-score results for infants with HLHS and those without HLHS were 88% and 100%, respectively147 (Table 5). Voice records were used to diagnose respiratory phases in infant cry148, to classify neonatal diseases in infant cry149, and to evaluate asphyxia from infant cry voice records150. Voice records of 35 infants were analyzed with ANN, and accuracy was found 85%149. Cry records of 14 infants in their 1st year of life were analyzed with SVM and GMM, and phases of respiration and crying rate were quantified with an accuracy of 86%148 (Table 3).
SVM was the most commonly used method in the diagnosis of metabolic disorders of newborns, including MMA (methylmalonic acidemia)151, PKU (phenylketonuria)152,153, MCADD (medium-chain acyl CoA dehydrogenase deficiency)152. During the Bavarian newborn screening program, dried blood samples were analyzed with ML and increased the positive predictive value for PKU (71.9% versus 16,2) and for MCADD (88.4% versus 54.6%)152 (Table 3).
Neonatology with deep learning
The main uses of DL in clinical image analysis are categorized into three categories: classification, detection, and segmentation. Classification involves identifying a specific feature in an image, detection involves locating multiple features within an image; and segmentation involves dividing an image into multiple parts7,9,154–160.
Neuroradiological evaluation with AI in neonatology
Neonatal neuroimaging can establish early indicators of neurodevelopmental abnormality to provide early intervention during a time of maximal neuroplasticity and fast cognitive and motor development79,96. DL methods can assist in an earlier diagnosis than clinical signs would indicate.
The imaging of an infant’s brain using MRI can be challenging due to lower tissue contrast, substantial tissue inhomogeneities, regionally heterogeneous image appearance, immense age-related intensity variations, and severe partial volume impact due to the smaller brain size. Since most of the existing tools were created for adult brain MRI data, infant-specific computational neuroanatomy tools are recently being developed. A typical pipeline for early prediction of neurodevelopmental disorders from infant structural MRI (sMRI) is made up of three basic phases. (1) Image preprocessing, tissue segmentation, regional labeling, and extraction of image-based characteristics (2) Surface reconstruction, surface correspondence, surface parcellation, and extraction of surface-based features (3) Feature preprocessing, feature extraction, AI model training, and prediction of unseen subjects161. The segmentation of a newborn brain is difficult due to the decreased SNR (signal to noise ratio) resulting from the shorter scanning duration enforced by predicted motion restrictions and the diminutive size of the neonatal brain. In addition, the cerebrospinal fluid (CSF)-gray matter border has an intensity profile comparable to that of the mostly unmyelinated white matter (WM), resulting in significant partial volume effects. In addition, the high variability resulting from the fast growth of the brain and the continuing myelination of WM imposes additional constraints on the creation of effective segmentation techniques. Several non-DL-based approaches for properly segmenting newborn brains have been presented over the years. These methods may be broadly classified as parametric162–164, classification165, multi-atlas fusion166,167, and deformable models168,169. The Dice Similarity Coefficient metric is used for image segmentation evaluation; the higher the dice, the higher the segmentation accuracy10 (Table 1).
In the NeoBrainS12 2012 MICCAI Grand-Challenge (https://neobrains12.isi.uu.nl), T1W and T2W images were presented with manually segmented structures to assess strategies for segmenting neonatal tissue162. Most methods were found to be accurate, but classification-based approaches were particularly precise and sensitive. However, segmentation of myelinated vs. unmyelinated WM remains a difficulty since the majority of approaches162 failed to consistently obtain reliable results.
Future research in neonatal brain segmentation will involve a more thorough neural segmentation network. Current studies are intended to highlight efficient networks capable of producing accurate and dependable segmentations while comparing them to existing conventional computer vision techniques. In the perspective of comparing previous efforts on newborn brain segmentation, the small sample size of high-quality labeled data must also be recognized as a significant restriction169. The field of artificial intelligence in neonatology has progressed slowly due to a shortage of open-source algorithms and the availability of datasets.
Future research should also focus on improving the accuracy of DL for diagnosing germinal matrix hemorrhage and figuring out how DL can help a radiologist’s workflow by comparing how well sonographers identify studies that look suspicious. More studies could also look at how well DL works for accurately grading germinal matrix hemorrhages and maybe even small hemorrhages that a radiologist can see on an MRI but not on a head ultrasound. This could be useful in improving the diagnostic capabilities of head ultrasound in various clinical scenarios157.
Evaluation of prematurity complications with DL in neonatology
In the above discussion, we have addressed the primary applications of DL in relation to disease prediction. These include DL for analyzing conditions such as PDA (patent ductus arteriosus)158, IVH (intraventricular ventricular hemorrhage)155,157, BPD (bronchopulmonary dysplasia)170, ROP (retinopathy of prematurity)171–173, retinal hemorrhage174 diagnosis. This also includes DL applications for analyzing MR images159,175 and combined with EHR data176,177 for predicting neurocognitive outcome and mortality. Additionally, DL has potential applications in treatment planning and discharge from the NICU178, including customized medicine and follow-up6,67,125 (Tables 6 and 7).
Digital imaging and analysis with AI are promising and cost-effective tools for detecting infants with severe ROP who may need therapy132,171,172,179. Despite limitations such as image quality, interpretation variability, equipment costs, and compatibility issues with EHR systems, AI has been shown to be effective in detecting ROP180. Studies comparing BIO (Binocular Indirect Ophthalmoscope) to telemedicine have shown that both methods have equivalent sensitivity for identifying zone disease, plus disease, and ROP. However, BIO was found to be slightly better at identifying zone III and stage 3 ROP181,182. DL algorithms were applied to 5511 retinal images, achieving an AUC of 94% (diagnosis of normal) and 98% (diagnosis of plus disease), outperforming 6 out of 8 ROP experts171. In another study, DL was used to quantify the clinical progression of ROP by assigning ROP vascular severity scores172. A consecutive study with a large dataset showed in 4175 retinal images from 32 NICUs, resulting in an AUC of 98% for detecting therapy required ROP with DL173. The use of AI in ROP screening programs may increase access to care for secondary prevention of ROP and enable the evaluation of disease epidemiology173 (Table 6).
Signal detection for sleep protection in the NICU is another ongoing discussion. DL has been used to analyze infant EEGs and identify sleep states. Interruptions of sleep states have been linked to problems in neuronal development183. Automated sleep state detection from EEG records184,185 and from ECG monitoring parameters186 were demonstrated with DL. The underperformance of the all-state classification (kappa score 0.33 to 0.44) was likely owing to the difficulties in differentiating small changes between states and a lack of enough training data for minority classes186 (Table 6).
DL has been found to be effective in real-time evaluation of cardiac MRI for congenital heart disease187. Studies have shown that DL can accurately calculate ventricular volumes from images rebuilt using residual UNet, which are not statistically different from the gold standard, cardiac MRI. This technology has the potential to be particularly beneficial for infants and critically ill individuals who are unable to hold their breath during the imaging process187 (Table 6).
DL-based 3D CNN algorithms have been used to demonstrate the automated classification of brain dysmaturation from neonatal brain MRI188. In a study, brain MRIs of 90 term neonates with congenital heart diseases and 40 term healthy controls were analyzed using this method, which achieved an accuracy of 98%. This technique could be useful in detecting brain dysmaturation in neonates with congenital heart diseases188 (Table 6).
DL algorithms have been used to classify neonatal diseases from thermal images189–192. These studies analyzed neonatal thermograms to determine the health status of infants and achieved good AUC scores189–192. However, these studies didn’t include any clinical information (Table 6).
Two large scale studies showed breakthrough results regarding the effect of nutrition practices in NICU170 and wireless sensors in NICU193. A nutrition study revealed that nutrition practices were associated with discharge weight and BPD170. This exemplifies how unbiased ML techniques may be used to effectively bring about clinical practice changes170. Novel, wireless sensors can improve monitoring, prevent iatrogenic injuries, and encourage family-centered care193. Early validation results show performance equal to standard-of-care monitoring systems in high-income nations. Furthermore, the use of reusable sensors and compatibility with low-cost mobile phones may reduce monitoring.
Discussion
The studies in neonatology with AI were categorized according to the following criteria.
-
(i)
The studies were performed with ML or DL,
-
(ii)
imaging data or non-imaging data were used,
-
(iii)
according to the aim of the study: diagnosis or other predictions.
Most of the studies in neonatology were performed with ML methods in the pre-DL era. We have listed 12 studies with ML and imaging data for diagnosis. There are 33 studies that used non-imaging data for diagnosis purposes. Imaging data studies cover BA diagnosis from stool color194, postoperative enteral nutrition of neonatal high intestinal obstruction195, functional brain connectivity in preterm infants82,90,91,94,100, ROP diagnosis133,134, neonatal seizure detection from video records101, newborn jaundice screening137. Non-imaging studies for diagnosis include the diagnosis of congenital heart defects135, baby cry analysis148–150, inborn metabolic disorder diagnosis and screening151–153, HIE grading104,106,114,136,196, EEG analysis102,104,106,107,110–113,115,184,197,198, PDA diagnosis123,124, vital sign analysis and artifact detection144, extubation and weaning analysis129–131,144, BPD diagnosis127. ML studies with imaging data for prediction are focused on neurodevelopmental outcome prognosis from brain MRIs95–97,127,164,199. ML-based non-imaging data for prediction encompassed mortality risk63–65,68, NEC prognosis139, morbidity66,146, BPD125,126.
When it comes to DL applications, there has been less research conducted compared to ML applications. The focus of DL with imaging and non-imaging data focused on brain segmentation159,169,175,177,188, IVH diagnosis157, EEG analysis184,185, neurocognitive outcome176, PDA and ROP diagnosis171–173. Upcoming articles and research will surely be from the DL field, though.
It is worth noting that there have also been several articles and studies published on the topic of the application of AI in neonatology. However, the majority of these studies do not contain enough details, are difficult to evaluate side-by-side, and do not give the clinician a thorough picture of the applications of AI in the general healthcare system66,67,93,95–97,99,125–127,140,142,147,169,174,177,185,188,200–205.
There are several limitations in the application of AI in neonatology, including a lack of prospective design, a lack of clinical integration, a small sample size, and single center evaluations. DL has shown promise in bioscience and biosignals, extracting information from clinical images, and combining unstructured and structured data in EHR. However, there are some issues that limit the success of DL in medicine, which can be grouped into six categories. In the following paragraphs, we’ll examine the key concerns related to DL, which have been divided into six components:
Difficulties in clinical integration, including the selection and validation of models;
the need for expertise in decision mechanisms, including the requirement for human involvement in the process;
lack of data and annotations, including the quality and nature of medical data; distribution of data in the input database; and lack of open-source algorithms and reproducibility;
lack of explanations and reasoning, including the lack of explainable AI to address the “black-box” problem;
lack of collaboration efforts across multi-institutions; and
Difficulties in clinical integration
Despite the accuracy that AI has reached in healthcare in recent years, there are several restrictions that make it difficult to translate into treatment pathways. First, physicians’ suspicion of AI-based systems stems from the lack of qualified randomized clinical trials, particularly in the field of pediatrics, showing the reliability and/or improved effectiveness of AI systems compared to traditional systems in diagnosing neonatal diseases and suggesting appropriate therapies. The studies’ pros and cons are discussed in tables and relevant sections. Studies are mainly focused on imaging-based or signal-based studies in terms of one variable or disease. Neonatologists and pediatricians need evidence-based proven algorithm studies. There are only six prospective clinical trials in neonatology with AI197,207–211. The one is detecting neonatal seizures with conventional EEG in the NICU which is supported by the European Union Cost Program in 8 European NICU197. Neonates with a corrected gestational age between 36 and 44 weeks who had seizures or were at high risk of having seizures and needed EEG monitoring were given conventional EEG with ANSeR (Algorithm for Neonatal Seizure Recognition) coupled with an EEG monitor that displayed a seizure probability trend in real time (algorithm group) or continuous EEG monitoring alone (non-algorithm group)197. The algorithm is not available, and the code is not shared. Another one is a study showing the physiologic effects of music in premature infants208. Even so, it could not be founded on any AI analysis in this study. The third study, “Rebooting Infant Pain Assessment: Using Machine Learning to Exponentially Improve Neonatal Intensive Care Unit Practice (BabyAI),” is newly posted and recruiting209. The fourth study, “Using sensor-fusion and machine learning algorithms to assess acute pain in non-verbal infants: a study protocol,” aims to collect data from 15 subjects: preterm infants, term infants within the first month of age in NICU admission and their follow-up data at 3rd and 6th months of age. They record pain signals using facial electromyography(EMG), ECG, electrodermal activity, oxygen saturation, and EEG in real time, and they will analyze the data with ML methods to evaluate pain in neonates. The data is in iPAS (NCT03330496) and is updated as recruitment completed210. However, no result has been submitted. The fifth study, “Prediction of Extubation Readiness in Extreme Preterm Infants by the Automated Analysis of Cardiorespiratory Behavior: APEX study”211 records revealed that the recruitment was completed in 266 infants. Still, no results have been released yet (NCT01909947). To sum up, there is only one prospective multicenter randomized AI study that has been published with its results.
There is an unmet need to plan clinically integrated prospective and real-time data collection studies in neonatology. The clinical situation of infants changed rapidly, and real-time designed studies would be significant by analyzing multimodal data and including imaging and non-imaging components.
The need for expertise in the decision mechanisms
In terms of neonatologists determining whether to implement a system’s recommendation, it may be required for that system to present supporting evidence95,96,125,202. Many suggested AI solutions in the medical field are not expected to be an alternative to the doctor’s decision or expertise but rather to serve as helpful assistance. When it comes to struggling neonatal survival without sequela, AI may be a game changer in neonatology. The broad range of neonatal diseases and different clinical presentations of neonates according to gestational age and postnatal age make accurate diagnosis even harder for neonatologists. AI would be effective for early disease detection and would assist clinicians in responding promptly and fostering therapy outcomes.
Neonatology has multidisciplinary collaborations in the management of patients, and AI has the potential to achieve levels of efficacy that were previously unimaginable in neonatology if more resources and support from physicians were allocated to it. Neonatology collaborates and closely works with other specialties of pediatrics, including perinatology, pediatric surgery, radiology, pediatric cardiology, pediatric neurology, pediatric infectious disease, neurosurgery, cardiovascular surgery, and other subspecialties of pediatrics. Those multidisciplinary workflows require patient follow-up and family involvement. AI-based predictive analysis tools might address potential risks and neurologic problems in the future. AI supported monitoring systems could analyze real time data from monitors and detect changes simultaneously. These tools could be helpful not only for routine NICU care but also for “family centered care”212,213 implications. Although neonatologists could be at the center of decision making and giving information to parents, AI could be actively used in NICUs. Hybrid intelligence would provide a follow-up platform for abrupt and subtle clinical changes in infants’ clinical situations.
Given that many medical professionals have a limited understanding of DL, it may be difficult to establish contact and communication between data scientists and medical specialists. Many medical professionals, including pediatricians and neonatologists in our instance, are unfamiliar with AI and its applications due to a lack of exposure to the field as an end user. However, the authors also acknowledge the increasing efforts in building bridges among many scientists and institutions, with conferences, workshops, and courses, that clinicians have successfully started to lead AI efforts, even with software coding schools by clinicians214–218.
Neonatal critical conditions will be monitored by the human in the loop systems in the near future, and AI empowered risk classification systems may help clinicians prioritize critical care and allocate supplies precisely. Hence, AI could not replace neonatologists, but there would be a clinical decision support system in the critical and calls for prompt response environment of NICU.
Lack of imaging data and annotations and reproducibility problems
There is a rising interest in building deep learning approaches to predict neurological abnormalities using connectome data; however, their usage in preterm populations has been limited81,88–91. Similar to most DL applications, the training of such models often requires the use of big datasets11; however, large neuroimaging datasets are either not accessible or difficult and expensive to acquire, especially in the pediatric world. Since the success of DL methods currently relies on well-labeled data and high-capacity models requiring several iterative updates across many labeled examples and obtaining millions of labeled examples, is an extreme challenge, there is not enough jump in the neonatal AI applications.
As a side note, accurate labeling always requires physician effort and time, which overcomplicates the current challenges. Unfortunately, there is no established collaboration between physicians and data scientists at a large scale that can ease some of the challenges (data gathering/sharing and labeling). Nonetheless, once these problems are addressed, DL can be used in prevention and diagnosis programs for optimal results, radically transforming clinical practice. In the following, we envision the potential of DL to transform other imaging modalities in the context of neonatology and child health.
The requirement for a massive volume of data is a significant barrier, as mentioned earlier. The quantity of data needed by an AI or ML system can grow in proportion to the sophistication of its underlying architecture; deep neural networks (DNN), for example, have particularly high volume of data needs. It’s not enough that the needed data just be sufficient; they also need to be of good quality in terms of data cleaning and data variability (both ANN and DNN tend to avoid overfitting data if the variability is high). It may be difficult to collect a substantial amount of clean, verified, and varied data for several uses in neonatology. For this reason, there is a data repository shared with neonatal researchers, including EHR202 and clinical variables. Some approaches for addressing the lack of labeled, annotated, verified, and clean datasets include: (1) building and training a model with a very shallow network (only a few thousand parameters) and (2) data augmentation. Data augmentation techniques are not helpful in the medical imaging field or medical setting219.
In the field of neonatal imaging, high-quality labeling and medical imaging data are exceedingly uncommon. One of the other comparable available neonatal datasets the authors are aware of has just ten individuals166,220,221. This pattern holds even in more recent research, as detailed by the majority of studies involving little more than 20 individuals167. Regardless of sample size and technology, it is crucial to be able to generalize to new data in the field of image segmentation, especially considering the wide range of MRI contrasts and variations between scanners and sequences between institutions. Moreover, it is generally known that models based on DL have weak generalization skills on unseen data. This is especially crucial for the future translation of research into reality since (1) there is a shift between images obtained in various situations, and (2) the model must be retrained as these images become accessible. Adopting a strategy of continuous learning is the most practical way to handle this challenge. This method involves progressively retraining deep models while preventing any virtual memory loss on previously viewed data sets that may not be available during retraining. This field of endeavor will advance169.
Most of the studies did not release their algorithms as open source to the libraries. Even though algorithms are available, it should be known whether separate training and testing datasets exist. There is a strong expectation that studies should have clarified which validation method has been chosen. In terms of comparing algorithm success, reproducibility is a crucial point. Methodological bias is another issue with this system. Research is frequently based on databases and guidelines from other nations that may or may not have patient populations similar to ours96. A database that only contains data that is applicable to the specific problem that must be solved; however, obtaining the relevant information may be difficult due to the number of databases.
Lack of explanations and reasoning
The trustworthiness of algorithms is another obstacle222. The most widely used deep learning models use a black-box methodology, in which the model simply receives input and outputs a prediction without explaining its thought process. In high-stakes medical settings, this can be dangerous. Some models, on the other hand, incorporate human judgment (human-in-the-loop) or provide interpretability maps or explainability layers to illuminate the decision-making process. Especially in the field of neonatology, where AI is expected to have a significant impact, this trustworthiness is essential for its widespread adoption.
Lack of collaboration efforts (multi-institutions) and privacy concerns
New collaborations have been forged because of this information; early detection and treatment of diseases that affect children, who make up a large portion of the world’s population, will change treatment and follow-up status. Monitoring systems and knowing mortality and treatment activity with multi-site data will help. Considering the necessity for consent to the processing of personal health data by AI systems as an example of a subject related to the protection of privacy and security96. Efforts involving multiple institutions can facilitate training, but there are privacy concerns associated with the cross-site sharing of imaging data. Federated learning (FL) was introduced recently to address privacy concerns by facilitating distributed training without the transfer of imaging data223. Existing FL techniques utilize conditional reconstruction models to map from under sampled to fully-sampled acquisitions using explicit knowledge of the accelerated imaging operator223. Nevertheless, the data from various institutions is typically heterogeneous, which may diminish the efficacy of models trained using federated learning. SplitAVG is proposed as a novel heterogeneity-aware FL method to surmount the performance declines in federated learning caused by data heterogeneity224.
AI ethics
While AI has great promise for enhancing healthcare, it also presents significant ethical concerns. Ethical concerns in health AI include informed consent, bias, safety, transparency, patient privacy, and allocation, and their solutions are complicated to negotiate225. In neonatology, crucial decision-making is frequently accompanied by a complicated and challenging ethical component. Interdisciplinary approaches are required for progress226. The border of viability, life sustaining treatments227 and the different regulations worldwide made AI utilization in neonatology more complicated. How an ethics framework is implemented in an AI in neonatology has not been reported yet, and there is a need for transparency for trustworthy AI.
The applications of AI in real-world contexts have the potential to result in a few potential benefits, including increased speed of execution; potential reduction in costs, both direct and indirect; improved diagnostic accuracy; increased healthcare delivery efficiency (“algorithms work without a break”); and the potential of supplying access to clinical information even to persons who would not normally be able to utilize healthcare due to geographic or economic constraints4.
To achieve an accurate diagnosis, it is planned to limit the number of extra invasive procedures. New DL technologies and easy-to-implement platforms will enable regular and complete follow-up of health data for patients unable to access their records owing to a physician shortage, hence reducing health costs.
The future of neonatal intensive care units and healthcare will likely be profoundly impacted by AI. This article’s objective is to provide neonatologists in the AI era with a reference guide to the information they might require. We defined AI, its levels, its techniques, and the distinctions between the approaches used in the medical field, and we examined the possible advantages, pitfalls, and challenges of AI. While also attempting to present a picture of its potential future implementation in standard neonatal practice. AI and pediatrics require clinicians’ support, and due to the fact that AI researchers with clinicians need to work together and cooperatively. As a result, AI in neonatal care is highly demanded, and there is a fundamental need for a human (pediatrician) to be involved in the AI-backed up applications, in contrast to systems that are more technically advanced and involve fewer healthcare professionals.
Methods
Literature review and search strategy
We used PubMed™, IEEEXplore™, Google Scholar™, and ScienceDirect™ to search for publications relating to AI, ML, and DL applications towards neonatology. We have done a varying combination of the keywords (i.e., one from technical keywords and one from clinical keywords) for the search. Clinical keywords were “infant,” “neonate,” “prematurity,” “preterm infant,” “hypoxic ischemic encephalopathy,” “neonatology,” “intraventricular hemorrhage,” “infant brain segmentation,” “NICU mortality,” “infant morbidity,” “ bronchopulmonary dysplasia,” “retinopathy of prematurity.” The inclusion criteria were (i) publication date between 1996–2022 and, (ii) being an artificial intelligence in neonatology study, (iii) written in English, (iv) published in a scholarly peer-reviewed journal, and (v) conducted an assessment of AI applications in neonatology objectively. Technical keywords were AI, DL, ML, and CNN. Review papers, commentaries, letters to the editor and papers with only technical improvement without any clinical background, animal studies, and papers that used statistical models like linear regression, studies written in any language other than English, dissertation thesis, posters, biomarker prediction studies, simulation-based studies, studies with infants are older than 28 days of life, perinatal death, and obstetric care studies were excluded. The preliminary investigation yielded a substantial collection of articles, amounting to approximately 9000 in total. Through a meticulous examination of the abstracts of the papers, a subset of 987 research was found (Fig. 4). Ultimately, 106 studies were selected for inclusion in our systematic review (Supplementary file). The evaluation encompassed diverse aspects, including sample size, methodology, data type, evaluation metrics, advantages, and limitations of the studies (Tables 2–7).
Supplementary information
Acknowledgements
This work is partially supported by the NIH NCI funding: R01-CA246704 and R01-CA240639. Dr. E Keles is working as a senior clinical research associate in the Machine and Hybrid Intelligence Lab at the Northwestern University Feinberg School of Medicine, Department of Radiology. Dr. U. Bagci is director of the Machine and Hybrid Intelligence Lab and Associate Professor at the Department of Radiology, Northwestern University, Feinberg School of Medicine.
Author contributions
Both authors contributed to the review design, data collection, interpretation of the data, analysis of data and drafting the report.
Data availability
Dr. E. Keles and Dr. U. Bagci have full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All study materials are available from the corresponding author upon reasonable request.
Competing interests
Dr. E. Keles has no COI. Dr. U. Bagci discloses Ther-AI LLC.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-023-00941-5.
References
- 1.Turing, A.M. & Haugeland, J. In The Turing Test: Verbal Behavior as the Hallmark of Intelligence, 29–56 (1950).
- 2.Padula WV, et al. Machine learning methods in health economics and outcomes research—the PALISADE checklist: a good practices report of an ISPOR task force. Value Health. 2022;25:1063–1080. doi: 10.1016/j.jval.2022.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Bagci, U., Irmakci, I., Demir, U. & Keles, E. in AI in Clinical Medicine: A Practical Guide for Healthcare Professionals 56–65 (2023).
- 4.Burt JR, et al. Deep learning beyond cats and dogs: recent advances in diagnosing breast cancer with deep neural networks. Br. J. Radio. 2018;91:20170545. doi: 10.1259/bjr.20170545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piccialli F, Somma VD, Giampaolo F, Cuomo S, Fortino G. A survey on deep learning in medicine: Why, how and when? Inf. Fusion. 2021;66:111–137. doi: 10.1016/j.inffus.2020.09.006. [DOI] [Google Scholar]
- 6.Rubinger L, Gazendam A, Ekhtiari S, Bhandari M. Machine learning and artificial intelligence in research and healthcare. Injury. 2023;54:S69–S73. doi: 10.1016/j.injury.2022.01.046. [DOI] [PubMed] [Google Scholar]
- 7.Sarker IHDeep. Learning: a comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput. Sci. 2021;2:420. doi: 10.1007/s42979-021-00815-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savadjiev P, et al. Demystification of AI-driven medical image interpretation: past, present and future. Eur. Radio. 2019;29:1616–1624. doi: 10.1007/s00330-018-5674-x. [DOI] [PubMed] [Google Scholar]
- 9.Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018;319:1317–1318. doi: 10.1001/jama.2017.18391. [DOI] [PubMed] [Google Scholar]
- 10.Janiesch C, Zschech P, Heinrich K. Machine learning and deep learning. Electron. Mark. 2021;31:685–695. doi: 10.1007/s12525-021-00475-2. [DOI] [Google Scholar]
- 11.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 12.Wiens J, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat. Med. 2019;25:1337–1340. doi: 10.1038/s41591-019-0548-6. [DOI] [PubMed] [Google Scholar]
- 13.Chen PC, Liu Y, Peng L. How to develop machine learning models for healthcare. Nat. Mater. 2019;18:410–414. doi: 10.1038/s41563-019-0345-0. [DOI] [PubMed] [Google Scholar]
- 14.Futoma J, Simons M, Panch T, Doshi-Velez F, Celi LA. The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Health. 2020;2:e489–e492. doi: 10.1016/S2589-7500(20)30186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakaura T, Higaki T, Awai K, Ikeda O, Yamashita Y. A primer for understanding radiology articles about machine learning and deep learning. Diagn. Inter. Imaging. 2020;101:765–770. doi: 10.1016/j.diii.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Mortazi, A. & Bagci, U. Automatically designing CNN architectures for medical image segmentation. in Machine Learning in Medical Imaging: 9th International Workshop, MLMI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16, 2018, Proceedings 9 98–106 (Springer, 2018).
- 17.Perna, D. & Tagarelli, A. Deep auscultation: predicting respiratory anomalies and diseases via recurrent neural networks. in 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS) 50–55 (2019).
- 18.Murabito, F. et al. Deep recurrent-convolutional model for automated segmentation of craniomaxillofacial CT scans. in 2020 25th International Conference on Pattern Recognition (ICPR) 9062-9067 (IEEE, 2021).
- 19.Aytekin, I. et al. COVID-19 detection from respiratory sounds with hierarchical spectrogram transformers. arXiv https://arxiv.org/abs/2207.09529 (2022). [DOI] [PMC free article] [PubMed]
- 20.Ker J, Wang L, Rao J, Lim T. Deep learning applications in medical image analysis. IEEE Access. 2018;6:9375–9389. doi: 10.1109/ACCESS.2017.2788044. [DOI] [Google Scholar]
- 21.Demir, U. et al. Transformer Based Generative Adversarial Network for Liver Segmentation. in Image Analysis and Processing. ICIAP 2022 Workshops: ICIAP International Workshops, Lecce, Italy, May 23–27, 2022, Revised Selected Papers, Part II 340-347 (Springer, 2022). [DOI] [PMC free article] [PubMed]
- 22.Irmakci, I., Unel, Z. E., Ikizler-Cinbis, N. & Bagci, U. Multi-contrast MRI segmentation trained on synthetic images. in 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) 5030–5034 (IEEE, 2022). [DOI] [PMC free article] [PubMed]
- 23.Kim HE, et al. Transfer learning for medical image classification: a literature review. BMC Med. Imaging. 2022;22:69. doi: 10.1186/s12880-022-00793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang F, et al. A comprehensive survey on transfer learning. Proc. IEEE. 2020;109:43–76. doi: 10.1109/JPROC.2020.3004555. [DOI] [Google Scholar]
- 25.Valverde JM, et al. Transfer learning in magnetic resonance brain imaging: a systematic review. J. Imaging. 2021;7:66. doi: 10.3390/jimaging7040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swati ZNK, et al. Content-based brain tumor retrieval for MR images using transfer learning. IEEE Access. 2019;7:17809–17822. doi: 10.1109/ACCESS.2019.2892455. [DOI] [Google Scholar]
- 27.LaLonde R, Xu Z, Irmakci I, Jain S, Bagci U. Capsules for biomedical image segmentation. Med. image Anal. 2021;68:101889. doi: 10.1016/j.media.2020.101889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X-M, Liang L, Liu L, Tang M-J. Graph neural networks and their current applications in bioinformatics. Front. Genet. 2021;12:690049. doi: 10.3389/fgene.2021.690049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Z, Qu A, He X. Contour-aware semantic segmentation network with spatial attention mechanism for medical image. Vis. Comput. 2022;38:749–762. doi: 10.1007/s00371-021-02075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonçalves, T., Rio-Torto, I., Teixeira, L. F. & Cardoso, J. S. A survey on attention mechanisms for medical applications: are we moving towards better algorithms? IEEE Access (2022).
- 31.Zhou J, et al. Graph neural networks: a review of methods and applications. AI Open. 2020;1:57–81. doi: 10.1016/j.aiopen.2021.01.001. [DOI] [Google Scholar]
- 32.Fout, A., Byrd, J., Shariat, B. & Ben-Hur, A. Protein interface prediction using graph convolutional networks. in Advances in Neural Information Processing Systems 30 (2017).
- 33.Khalil, E., Dai, H., Zhang, Y., Dilkina, B. & Song, L. Learning combinatorial optimization algorithms over graphs. in Advances in Neural Information Processing Systems 30 (2017).
- 34.Gaggion N, Mansilla L, Mosquera C, Milone DH, Ferrante E. Improving anatomical plausibility in medical image segmentation via hybrid graph neural networks: applications to chest X-ray analysis. IEEE Trans. Med. Imaging. 2023;42:546–556. doi: 10.1109/TMI.2022.3224660. [DOI] [PubMed] [Google Scholar]
- 35.Liang D, Cheng J, Ke Z, Ying L. Deep magnetic resonance image reconstruction: inverse problems meet neural networks. IEEE Signal Process Mag. 2020;37:141–151. doi: 10.1109/MSP.2019.2950557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dar SUH, Özbey M, Çatlı AB, Çukur T. A transfer-learning approach for accelerated MRI using deep neural networks. Magn. Reson Med. 2020;84:663–685. doi: 10.1002/mrm.28148. [DOI] [PubMed] [Google Scholar]
- 37.Güngör A, et al. Adaptive diffusion priors for accelerated MRI reconstruction. Med. Image Anal. 2023;88:102872. doi: 10.1016/j.media.2023.102872. [DOI] [PubMed] [Google Scholar]
- 38.Monga V, Li Y, Eldar YC. Algorithm unrolling: Interpretable, efficient deep learning for signal and image processing. IEEE Signal Process. Mag. 2021;38:18–44. doi: 10.1109/MSP.2020.3016905. [DOI] [Google Scholar]
- 39.Yaman B, et al. Self-supervised learning of physics-guided reconstruction neural networks without fully sampled reference data. Magn. Reson. Med. 2020;84:3172–3191. doi: 10.1002/mrm.28378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akata Z, et al. A research agenda for hybrid intelligence: augmenting human intellect with collaborative, adaptive, responsible, and explainable artificial intelligence. Computer. 2020;53:18–28. doi: 10.1109/MC.2020.2996587. [DOI] [Google Scholar]
- 41.RaviPrakash, H. & Anwar, S. M. In AI in Clinical Medicine: A Practical Guide for Healthcare Professionals 94–103 (2023).
- 42.Keles E, Irmakci I, Bagci U. Musculoskeletal MR image segmentation with artificial intelligence. Adv. Clin. Radiol. 2022;4:179–188. doi: 10.1016/j.yacr.2022.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hussein, S., Cao, K., Song, Q. & Bagci, U. Risk stratification of lung nodules using 3D CNN-based multi-task learning. in International Conference on Information Processing in Medical Imaging 249–260 (Springer, 2017).
- 44.Hussein S, Kandel P, Bolan CW, Wallace MB, Bagci U. Lung and pancreatic tumor characterization in the deep learning era: novel supervised and unsupervised learning approaches. IEEE Trans. Med. imaging. 2019;38:1777–1787. doi: 10.1109/TMI.2019.2894349. [DOI] [PubMed] [Google Scholar]
- 45.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 46.Esteva A, et al. A guide to deep learning in healthcare. Nat. Med. 2019;25:24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 47.Sujith AVLN, Sajja GS, Mahalakshmi V, Nuhmani S, Prasanalakshmi B. Systematic review of smart health monitoring using deep learning and Artificial intelligence. Neuroscience Informatics. 2022;2:100028. doi: 10.1016/j.neuri.2021.100028. [DOI] [Google Scholar]
- 48.Stewart JE, Rybicki FJ, Dwivedi G. Medical specialties involved in artificial intelligence research: is there a leader. Tasman Med. J. 2020;2:20–27. [Google Scholar]
- 49.Mesko B, Gorog M. A short guide for medical professionals in the era of artificial intelligence. NPJ Digit Med. 2020;3:126. doi: 10.1038/s41746-020-00333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat. Med. 2022;28:31–38. doi: 10.1038/s41591-021-01614-0. [DOI] [PubMed] [Google Scholar]
- 51.Hicks SA, et al. On evaluation metrics for medical applications of artificial intelligence. Sci. Rep. 2022;12:5979. doi: 10.1038/s41598-022-09954-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier-Hein, L. et al. Metrics reloaded: Pitfalls and recommendations for image analysis validation. Preprint https://arxiv.org/abs/2206.01653 (2022). [DOI] [PMC free article] [PubMed]
- 53.McAdams RM, et al. Predicting clinical outcomes using artificial intelligence and machine learning in neonatal intensive care units: a systematic review. J. Perinatol. 2022;42:1561–1575. doi: 10.1038/s41372-022-01392-8. [DOI] [PubMed] [Google Scholar]
- 54.Kwok TNC, et al. Application and potential of artificial intelligence in neonatal medicine. Semin. Fetal Neonatal Med. 2022;27:101346. doi: 10.1016/j.siny.2022.101346. [DOI] [PubMed] [Google Scholar]
- 55.Jeong, H. & Kamaleswaran, R. Pivotal challenges in artificial intelligence and machine learning applications for neonatal care. In Seminars in Fetal and Neonatal Medicine Vol. 27, 101393 (Elsevier, 2022) [DOI] [PubMed]
- 56.Page MJ, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGuinness LA, Higgins JP. Risk-of-bias VISualization (robvis): an R package and Shiny web app for visualizing risk-of-bias assessments. Res. Synth. Methods. 2021;12:55–61. doi: 10.1002/jrsm.1411. [DOI] [PubMed] [Google Scholar]
- 58.Sounderajah V, et al. A quality assessment tool for artificial intelligence-centered diagnostic test accuracy studies: QUADAS-AI. Nat. Med. 2021;27:1663–1665. doi: 10.1038/s41591-021-01517-0. [DOI] [PubMed] [Google Scholar]
- 59.Yang B, et al. QUADAS-C: a tool for assessing risk of bias in comparative diagnostic accuracy studies. Ann. Intern Med. 2021;174:1592–1599. doi: 10.7326/M21-2234. [DOI] [PubMed] [Google Scholar]
- 60.SDG Target 3.2: End Preventable Deaths of Newborns and Children under 5 Years of Age in 2021 (https://www.who.int/data/gho/data/themes/theme-details/GHO/child-health) (2022).
- 61.United Nations General Assembly. Resolution adopted by the General Assembly on 25 September 2015. 70/1. Transforming our world: the 2030 agenda for sustainable development New York, NY (https://sdgs.un.org/goals) (2015).
- 62.Townsend, D. & Frize, M. Complimentary artificial neural network approaches for prediction of events in the neonatal intensive care unit. In 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society 4605-4608 (IEEE, 2008). [DOI] [PubMed]
- 63.Ambalavanan N, et al. Prediction of death for extremely low birth weight neonates. Pediatrics. 2005;116:1367–1373. doi: 10.1542/peds.2004-2099. [DOI] [PubMed] [Google Scholar]
- 64.Nascimento LFC, Ortega NRS. Fuzzy linguistic model for evaluating the risk of neonatal death. Rev. Saúde. Pública. 2002;36:686–692. doi: 10.1590/S0034-89102002000700005. [DOI] [PubMed] [Google Scholar]
- 65.Do HJ, Moon KM, Jin HS. Machine learning models for predicting mortality in 7472 very low birth weight infants using data from a nationwide neonatal network. Diagnostics. 2022;12:625. doi: 10.3390/diagnostics12030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Podda M, et al. A machine learning approach to estimating preterm infants survival: development of the Preterm Infants Survival Assessment (PISA) predictor. Sci. Rep. 2018;8:13743. doi: 10.1038/s41598-018-31920-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mangold C, et al. Machine learning models for predicting neonatal mortality: a systematic review. Neonatology. 2021;118:394–405. doi: 10.1159/000516891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zernikow B, et al. Artificial neural network for risk assessment in preterm neonates. Arch. Dis. Child.-Fetal Neonatal Ed. 1998;79:F129–F134. doi: 10.1136/fn.79.2.F129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pearlman SA. Advancements in neonatology through quality improvement. J. Perinatol. 2022;42:1277–1282. doi: 10.1038/s41372-022-01383-9. [DOI] [PubMed] [Google Scholar]
- 70.Khurshid F, et al. Comparison of multivariable logistic regression and machine learning models for predicting bronchopulmonary dysplasia or death in very preterm infants. Front Pediatr. 2021;9:759776. doi: 10.3389/fped.2021.759776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu JF, et al. Machine learning algorithms to predict mortality of neonates on mechanical intubation for respiratory failure. Biomedicines. 2021;9:1377. doi: 10.3390/biomedicines9101377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreira A, et al. Development and validation of a mortality prediction model in extremely low gestational age neonates. Neonatology. 2022;119:418–427. doi: 10.1159/000524729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shane AL, Sánchez PJ, Stoll BJ. Neonatal sepsis. lancet. 2017;390:1770–1780. doi: 10.1016/S0140-6736(17)31002-4. [DOI] [PubMed] [Google Scholar]
- 74.Gomez, R., Garcia, N., Collantes, G., Ponce, F. & Redon, P. Development of a non-invasive procedure to early detect neonatal sepsis using HRV monitoring and machine learning algorithms. in 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS) 132–137 (2019).
- 75.Stocker M, et al. Machine learning used to compare the diagnostic accuracy of risk factors, clinical signs and biomarkers and to develop a new prediction model for neonatal early-onset sepsis. Pediatr. Infect. Dis. J. 2022;41:248–254. doi: 10.1097/INF.0000000000003344. [DOI] [PubMed] [Google Scholar]
- 76.Manuck TA, et al. Preterm neonatal morbidity and mortality by gestational age: a contemporary cohort. Am. J. Obstet. Gynecol. 2016;215:103.e101–103.e114. doi: 10.1016/j.ajog.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Volpe JJ. Brain injury in premature infants: a complex amalgam of destructive and developmental disturbances. Lancet Neurol. 2009;8:110–124. doi: 10.1016/S1474-4422(08)70294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson S, et al. Neurodevelopmental disability through 11 years of age in children born before 26 weeks of gestation. Pediatrics. 2009;124:e249–e257. doi: 10.1542/peds.2008-3743. [DOI] [PubMed] [Google Scholar]
- 79.Ment LR, Hirtz D, Hüppi PS. Imaging biomarkers of outcome in the developing preterm brain. Lancet Neurol. 2009;8:1042–1055. doi: 10.1016/S1474-4422(09)70257-1. [DOI] [PubMed] [Google Scholar]
- 80.Ophelders D, et al. Preterm brain injury, antenatal triggers, and therapeutics: timing is key. Cells. 2020;9:1871. doi: 10.3390/cells9081871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rogers CE, Lean RE, Wheelock MD, Smyser CD. Aberrant structural and functional connectivity and neurodevelopmental impairment in preterm children. J. Neurodev. Disord. 2018;10:1–13. doi: 10.1186/s11689-018-9253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smyser CD, et al. Resting-state network complexity and magnitude are reduced in prematurely born infants. Cereb. Cortex. 2016;26:322–333. doi: 10.1093/cercor/bhu251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vohr BR. Neurodevelopmental outcomes of premature infants with intraventricular hemorrhage across a lifespan. Semin. Perinatol. 2022;46:151594. doi: 10.1016/j.semperi.2022.151594. [DOI] [PubMed] [Google Scholar]
- 84.Zernikow B, et al. Artificial neural network for predicting intracranial haemorrhage in preterm neonates. Acta Paediatr. 1998;87:969–975. doi: 10.1111/j.1651-2227.1998.tb01768.x. [DOI] [PubMed] [Google Scholar]
- 85.Turova V, et al. Machine learning models for identifying preterm infants at risk of cerebral hemorrhage. PLoS ONE. 2020;15:e0227419. doi: 10.1371/journal.pone.0227419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keunen K, Counsell SJ, Benders MJ. The emergence of functional architecture during early brain development. Neuroimage. 2017;160:2–14. doi: 10.1016/j.neuroimage.2017.01.047. [DOI] [PubMed] [Google Scholar]
- 87.Sripada K, et al. Trajectories of brain development in school-age children born preterm with very low birth weight. Sci. Rep. 2018;8:15553. doi: 10.1038/s41598-018-33530-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smyser CD, et al. Prediction of brain maturity in infants using machine-learning algorithms. Neuroimage. 2016;136:1–9. doi: 10.1016/j.neuroimage.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao W, Lin W, Grewen K, Gilmore JH. Functional connectivity of the infant human brain: plastic and modifiable. Neuroscientist. 2017;23:169–184. doi: 10.1177/1073858416635986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ball G, et al. Machine-learning to characterise neonatal functional connectivity in the preterm brain. Neuroimage. 2016;124:267–275. doi: 10.1016/j.neuroimage.2015.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiarelli AM, Sestieri C, Navarra R, Wise RG, Caulo M. Distinct effects of prematurity on MRI metrics of brain functional connectivity, activity, and structure: Univariate and multivariate analyses. Hum. Brain Mapp. 2021;42:3593–3607. doi: 10.1002/hbm.25456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shang J, et al. A machine learning investigation of volumetric and functional MRI abnormalities in adults born preterm. Hum. Brain Mapp. 2019;40:4239–4252. doi: 10.1002/hbm.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zimmer VA, et al. Learning and combining image neighborhoods using random forests for neonatal brain disease classification. Med. Image Anal. 2017;42:189–199. doi: 10.1016/j.media.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 94.Song, Z., Awate, S. P., Licht, D. J. & Gee, J. C. Clinical neonatal brain MRI segmentation using adaptive nonparametric data models and intensity-based Markov priors. In International Conference on Medical Image Computing and Computer-assisted Intervention 883–890 (Springer, 2007). [DOI] [PubMed]
- 95.Vassar R, et al. Neonatal brain microstructure and machine-learning-based prediction of early language development in children born very preterm. Pediatr. Neurol. 2020;108:86–92. doi: 10.1016/j.pediatrneurol.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 96.Schadl K, et al. Prediction of cognitive and motor development in preterm children using exhaustive feature selection and cross-validation of near-term white matter microstructure. Neuroimage Clin. 2018;17:667–679. doi: 10.1016/j.nicl.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wee CY, et al. Neonatal neural networks predict children behavioral profiles later in life. Hum. Brain Mapp. 2017;38:1362–1373. doi: 10.1002/hbm.23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Y, et al. Brain connectivity based graph convolutional networks and its application to infant age prediction. IEEE Trans. Med Imaging. 2022;41:2764–2776. doi: 10.1109/TMI.2022.3171778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu, M. et al. Deep learning of cortical surface features using graph-convolution predicts neonatal brain age and neurodevelopmental outcome. in 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) 1335–1338 (IEEE, 2020).
- 100.Krishnan ML, et al. Machine learning shows association between genetic variability in PPARG and cerebral connectivity in preterm infants. Proc. Natl Acad. Sci. USA. 2017;114:13744–13749. doi: 10.1073/pnas.1704907114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karayiannis NB, et al. Automated detection of videotaped neonatal seizures of epileptic origin. Epilepsia. 2006;47:966–980. doi: 10.1111/j.1528-1167.2006.00571.x. [DOI] [PubMed] [Google Scholar]
- 102.Koolen N, et al. Automated classification of neonatal sleep states using EEG. Clin. Neurophysiol. 2017;128:1100–1108. doi: 10.1016/j.clinph.2017.02.025. [DOI] [PubMed] [Google Scholar]
- 103.Mathieson SR, et al. Validation of an automated seizure detection algorithm for term neonates. Clin. Neurophysiol. 2016;127:156–168. doi: 10.1016/j.clinph.2015.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Temko A, Lightbody G, Thomas EM, Boylan GB, Marnane W. Instantaneous measure of EEG channel importance for improved patient-adaptive neonatal seizure detection. IEEE Trans. Biomed. Eng. 2012;59:717–727. doi: 10.1109/TBME.2011.2178411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Temko A, Thomas E, Marnane W, Lightbody G, Boylan GB. Performance assessment for EEG-based neonatal seizure detectors. Clin. Neurophysiol. 2011;122:474–482. doi: 10.1016/j.clinph.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matic V, et al. Improving reliability of monitoring background EEG dynamics in asphyxiated infants. IEEE Trans. Biomed. Eng. 2016;63:973–983. doi: 10.1109/TBME.2015.2477946. [DOI] [PubMed] [Google Scholar]
- 107.Ahmed R, Temko A, Marnane WP, Boylan G, Lightbody G. Exploring temporal information in neonatal seizures using a dynamic time warping based SVM kernel. Comput Biol. Med. 2017;82:100–110. doi: 10.1016/j.compbiomed.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 108.Thomas, E., Greene, B., Lightbody, G., Marnane, W. & Boylan, G. Seizure detection in neonates: improved classification through supervised adaptation. in 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society 903-906 (IEEE, 2008). [DOI] [PubMed]
- 109.Ansari, A. H. et al. Improvement of an automated neonatal seizure detector using a post-processing technique. In 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 5859–5862 (IEEE, 2015). [DOI] [PubMed]
- 110.Schetinin V, Schult J. The combined technique for detection of artifacts in clinical electroencephalograms of sleeping newborns. IEEE Trans. Inf. Technol. Biomed. 2004;8:28–35. doi: 10.1109/TITB.2004.824735. [DOI] [PubMed] [Google Scholar]
- 111.Mohseni, H.R., Mirghasemi, H., Shamsollahi, M.B. & Zamani, M.R. Detection of rhythmic discharges in newborn EEG signals. in 2006 International Conference of the IEEE Engineering in Medicine and Biology Society 6577–6580 (IEEE, 2006). [DOI] [PubMed]
- 112.Simayijiang, Z., Backman, S., Ulén, J., Wikström, S. & Åström, K. Exploratory study of EEG burst characteristics in preterm infants. in 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 4295–4298 (IEEE, 2013). [DOI] [PubMed]
- 113.Navarro X, et al. Multi-feature classifiers for burst detection in single EEG channels from preterm infants. J. Neural Eng. 2017;14:046015. doi: 10.1088/1741-2552/aa714a. [DOI] [PubMed] [Google Scholar]
- 114.Ahmed R, Temko A, Marnane W, Lightbody G, Boylan G. Grading hypoxic-ischemic encephalopathy severity in neonatal EEG using GMM supervectors and the support vector machine. Clin. Neurophysiol. 2016;127:297–309. doi: 10.1016/j.clinph.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 115.Temko A, Boylan G, Marnane W, Lightbody G. Robust neonatal EEG seizure detection through adaptive background modeling. Int. J. neural Syst. 2013;23:1350018. doi: 10.1142/S0129065713500184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stevenson N, et al. An automated system for grading EEG abnormality in term neonates with hypoxic-ischaemic encephalopathy. Ann. Biomed. Eng. 2013;41:775–785. doi: 10.1007/s10439-012-0710-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Clyman RI. Mechanisms regulating the ductus arteriosus. Biol. Neonate. 2006;89:330–335. doi: 10.1159/000092870. [DOI] [PubMed] [Google Scholar]
- 118.Sellmer A, et al. Morbidity and mortality in preterm neonates with patent ductus arteriosus on day 3. Arch. Dis. Child Fetal Neonatal Ed. 2013;98:F505–510. doi: 10.1136/archdischild-2013-303816. [DOI] [PubMed] [Google Scholar]
- 119.El-Khuffash A, Rios DR, McNamara PJ. Toward a rational approach to patent ductus arteriosus trials: selecting the population of interest. J. Pediatr. 2021;233:11–13. doi: 10.1016/j.jpeds.2021.01.012. [DOI] [PubMed] [Google Scholar]
- 120.de Waal K, Phad N, Stubbs M, Chen Y, Kluckow M. A randomized placebo-controlled pilot trial of early targeted nonsteroidal anti-inflammatory drugs in preterm infants with a patent ductus arteriosus. J. Pediatr. 2021;228:82–86.e82. doi: 10.1016/j.jpeds.2020.08.062. [DOI] [PubMed] [Google Scholar]
- 121.El-Khuffash A, et al. A pilot randomized controlled trial of early targeted patent ductus arteriosus treatment using a risk based severity score (The PDA RCT) J. Pediatr. 2021;229:127–133. doi: 10.1016/j.jpeds.2020.10.024. [DOI] [PubMed] [Google Scholar]
- 122.Sung SI, Lee MH, Ahn SY, Chang YS, Park WS. Effect of nonintervention vs oral ibuprofen in patent ductus arteriosus in preterm infants: a randomized clinical trial. JAMA Pediatr. 2020;174:755–763. doi: 10.1001/jamapediatrics.2020.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Na JY, et al. Artificial intelligence model comparison for risk factor analysis of patent ductus arteriosus in nationwide very low birth weight infants cohort. Sci. Rep. 2021;11:22353. doi: 10.1038/s41598-021-01640-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gomez-Quintana S, et al. A framework for AI-assisted detection of patent ductus arteriosus from neonatal phonocardiogram. Healthcare. 2021;9:169. doi: 10.3390/healthcare9020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Verder H, et al. Bronchopulmonary dysplasia predicted at birth by artificial intelligence. Acta Paediatr. 2021;110:503–509. doi: 10.1111/apa.15438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ochab M, Wajs W. Expert system supporting an early prediction of the bronchopulmonary dysplasia. Comput Biol. Med. 2016;69:236–244. doi: 10.1016/j.compbiomed.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 127.Dai D, et al. Bronchopulmonary dysplasia predicted by developing a machine learning model of genetic and clinical information. Front Genet. 2021;12:689071. doi: 10.3389/fgene.2021.689071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mikhno, A. & Ennett, C.bM. Prediction of extubation failure for neonates with respiratory distress syndrome using the MIMIC-II clinical database. in 2012 Annual international conference of the IEEE Engineering in Medicine and Biology Society 5094–5097 (IEEE, 2012). [DOI] [PubMed]
- 129.Precup, D. et al. Prediction of extubation readiness in extreme preterm infants based on measures of cardiorespiratory variability. in 2012 Annual international conference of the IEEE Engineering in Medicine and Biology Society 5630–5633 (IEEE, 2012). [DOI] [PubMed]
- 130.Mueller M, et al. Predicting extubation outcome in preterm newborns: a comparison of neural networks with clinical expertise and statistical modeling. Pediatr. Res. 2004;56:11–18. doi: 10.1203/01.PDR.0000129658.55746.3C. [DOI] [PubMed] [Google Scholar]
- 131.Hatzakis, G. E. & Davis, G. M. Fuzzy logic controller for weaning neonates from mechanical ventilation. in Proceedings of the AMIA Symposium 315 (American Medical Informatics Association, 2002). [PMC free article] [PubMed]
- 132.Barrero-Castillero A, Corwin BK, VanderVeen DK, Wang JC. Workforce shortage for retinopathy of prematurity care and emerging role of telehealth and artificial intelligence. Pediatr. Clin. North Am. 2020;67:725–733. doi: 10.1016/j.pcl.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 133.Rani P, Rajkumar ER. Classification of retinopathy of prematurity using back propagation neural network. Int. J. Biomed. Eng. Technol. 2016;22:338–348. doi: 10.1504/IJBET.2016.081221. [DOI] [Google Scholar]
- 134.Ataer-Cansizoglu E, et al. Computer-based image analysis for plus disease diagnosis in retinopathy of prematurity: performance of the “i-ROP” system and image features associated with expert diagnosis. Transl. Vis. Sci. Technol. 2015;4:5. doi: 10.1167/tvst.4.6.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reed NE, Gini M, Johnson PE, Moller JH. Diagnosing congenital heart defects using the Fallot computational model. Artif. Intell. Med. 1997;10:25–40. doi: 10.1016/S0933-3657(97)00382-5. [DOI] [PubMed] [Google Scholar]
- 136.Li L, et al. The use of fuzzy backpropagation neural networks for the early diagnosis of hypoxic ischemic encephalopathy in newborns. J. Biomed. Biotechnol. 2011;2011:349490. doi: 10.1155/2011/349490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taylor JA, et al. Use of a Smartphone App to Assess Neonatal Jaundice. Pediatrics. 2017;140:e20170312. doi: 10.1542/peds.2017-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ferreira D, Oliveira A, Freitas A. Applying data mining techniques to improve diagnosis in neonatal jaundice. BMC Med. Inform. Decis. Mak. 2012;12:1–6. doi: 10.1186/1472-6947-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ji J, et al. A data-driven algorithm integrating clinical and laboratory features for the diagnosis and prognosis of necrotizing enterocolitis. PLoS ONE. 2014;9:e89860. doi: 10.1371/journal.pone.0089860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ambalavanan N, et al. Prediction of neurologic morbidity in extremely low birth weight infants. J. Perinatol. 2000;20:496–503. doi: 10.1038/sj.jp.7200419. [DOI] [PubMed] [Google Scholar]
- 141.Soleimani F, Teymouri R, Biglarian A. Predicting developmental disorder in infants using an artificial neural network. Acta Med. Iran. 2013;51:347–352. [PubMed] [Google Scholar]
- 142.Bartz-Kurycki MA, et al. Enhanced neonatal surgical site infection prediction model utilizing statistically and clinically significant variables in combination with a machine learning algorithm. Am. J. Surg. 2018;216:764–777. doi: 10.1016/j.amjsurg.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 143.Reed RA, et al. Machine-learning vs. expert-opinion driven logistic regression modelling for predicting 30-day unplanned rehospitalisation in preterm babies: a prospective, population-based study (EPIPAGE 2) Front Pediatr. 2020;8:585868. doi: 10.3389/fped.2020.585868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tsien CL, Kohane IS, McIntosh N. Multiple signal integration by decision tree induction to detect artifacts in the neonatal intensive care unit. Artif. Intell. Med. 2000;19:189–202. doi: 10.1016/S0933-3657(00)00045-2. [DOI] [PubMed] [Google Scholar]
- 145.Cabrera-Quiros L, et al. Prediction of late-onset sepsis in preterm infants using monitoring signals and machine learning. Crit. Care Explor. 2021;3:e0302. doi: 10.1097/CCE.0000000000000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Saria S, Rajani AK, Gould J, Koller D, Penn AA. Integration of early physiological responses predicts later illness severity in preterm infants. Sci. Transl. Med. 2010;2:48ra65–48ra65. doi: 10.1126/scitranslmed.3001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jalali A, Simpao AF, Galvez JA, Licht DJ, Nataraj C. Prediction of periventricular leukomalacia in neonates after cardiac surgery using machine learning algorithms. J. Med. Syst. 2018;42:177. doi: 10.1007/s10916-018-1029-z. [DOI] [PubMed] [Google Scholar]
- 148.Aucouturier JJ, Nonaka Y, Katahira K, Okanoya K. Segmentation of expiratory and inspiratory sounds in baby cry audio recordings using hidden Markov models. J. Acoust. Soc. Am. 2011;130:2969–2977. doi: 10.1121/1.3641377. [DOI] [PubMed] [Google Scholar]
- 149.Cano Ortiz, S. D., Escobedo Beceiro, D. I. & Ekkel, T. A radial basis function network oriented for infant cry classification. in Iberoamerican Congress on Pattern Recognition 374–380 (Springer, 2004).
- 150.Yassin I, et al. Infant asphyxia detection using autoencoders trained on locally linear embedded-reduced Mel Frequency Cepstrum Coefficient (MFCC) features. J. Fundam. Appl. Sci. 2017;9:716–729. doi: 10.4314/jfas.v9i3s.56. [DOI] [Google Scholar]
- 151.Hsu KP, et al. A newborn screening system based on service-oriented architecture embedded support vector machine. J. Med. Syst. 2010;34:899–907. doi: 10.1007/s10916-009-9305-6. [DOI] [PubMed] [Google Scholar]
- 152.Baumgartner C, et al. Supervised machine learning techniques for the classification of metabolic disorders in newborns. Bioinformatics. 2004;20:2985–2996. doi: 10.1093/bioinformatics/bth343. [DOI] [PubMed] [Google Scholar]
- 153.Chen WH, et al. Web-based newborn screening system for metabolic diseases: machine learning versus clinicians. J. Med. Internet Res. 2013;15:e98. doi: 10.2196/jmir.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang W, et al. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hyun, D. & Brickson, L. Classification of Neonatal Brain Ultrasound Scans Using Deep Convolutional Neural Networks. (Stanford CS229, 2016).
- 156.Kelly C, et al. Investigating brain structural maturation in children and adolescents born very preterm using the brain age framework. Neuroimage. 2022;247:118828. doi: 10.1016/j.neuroimage.2021.118828. [DOI] [PubMed] [Google Scholar]
- 157.Kim KY, Nowrangi R, McGehee A, Joshi N, Acharya PT. Assessment of germinal matrix hemorrhage on head ultrasound with deep learning algorithms. Pediatr. Radio. 2022;52:533–538. doi: 10.1007/s00247-021-05239-w. [DOI] [PubMed] [Google Scholar]
- 158.Lei H, Ashrafi A, Chang P, Chang A, Lai W. Patent ductus arteriosus (PDA) detection in echocardiograms using deep learning. Intelligence-Based Med. 2022;6:100054. doi: 10.1016/j.ibmed.2022.100054. [DOI] [Google Scholar]
- 159.Li H, et al. Automatic segmentation of diffuse white matter abnormality on T2-weighted brain MR images using deep learning in very preterm infants. Radio. Artif. Intell. 2021;3:e200166. doi: 10.1148/ryai.2021200166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ding W, Abdel-Basset M, Hawash H, Pedrycz W. Multimodal infant brain segmentation by fuzzy-informed deep learning. IEEE Trans. Fuzzy Syst. 2022;30:1088–1101. doi: 10.1109/TFUZZ.2021.3052461. [DOI] [Google Scholar]
- 161.Mostapha M, Styner M. Role of deep learning in infant brain MRI analysis. Magn. Reson Imaging. 2019;64:171–189. doi: 10.1016/j.mri.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Makropoulos A, et al. Automatic tissue and structural segmentation of neonatal brain MRI using expectation-maximization. MICCAI Gd. Chall. Neonatal Brain Segment. 2012;2012:9–15. [Google Scholar]
- 163.Beare RJ, et al. Neonatal brain tissue classification with morphological adaptation and unified segmentation. Front. Neuroinform. 2016;10:12. doi: 10.3389/fninf.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu M, et al. Patch-based augmentation of Expectation–Maximization for brain MRI tissue segmentation at arbitrary age after premature birth. NeuroImage. 2016;127:387–408. doi: 10.1016/j.neuroimage.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moeskops P, et al. Automatic segmentation of MR brain images of preterm infants using supervised classification. NeuroImage. 2015;118:628–641. doi: 10.1016/j.neuroimage.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 166.Weisenfeld NI, Warfield SK. Automatic segmentation of newborn brain MRI. NeuroImage. 2009;47:564–572. doi: 10.1016/j.neuroimage.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim, H., Lepage, C., Evans, A. C., Barkovich, A. J. & Xu, D. NEOCIVET: Extraction of cortical surface and analysis of neonatal gyrification using a modified CIVET pipeline. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (eds. Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 571–579 (Springer International Publishing, 2015).
- 168.Wang L, et al. 4D Multi-modality tissue segmentation of serial infant images. PLoS ONE. 2012;7:e44596. doi: 10.1371/journal.pone.0044596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ding Y, et al. Using deep convolutional neural networks for neonatal brain image segmentation. Front Neurosci. 2020;14:207. doi: 10.3389/fnins.2020.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Greenbury SF, et al. Identification of variation in nutritional practice in neonatal units in England and association with clinical outcomes using agnostic machine learning. Sci. Rep. 2021;11:7178. doi: 10.1038/s41598-021-85878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brown JM, et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018;136:803–810. doi: 10.1001/jamaophthalmol.2018.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Taylor S, et al. Monitoring disease progression with a quantitative severity scale for retinopathy of prematurity using deep learning. JAMA Ophthalmol. 2019;137:1022–1028. doi: 10.1001/jamaophthalmol.2019.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Campbell JP, et al. Applications of artificial intelligence for retinopathy of prematurity screening. Pediatrics. 2021;147:e2020016618. doi: 10.1542/peds.2020-016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang B, et al. Application of a deep convolutional neural network in the diagnosis of neonatal ocular fundus hemorrhage. Biosci. Rep. 2018;38:BSR20180497. doi: 10.1042/BSR20180497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shabanian, M., Eckstein, E. C., Chen, H. & DeVincenzo, J. P. Classification of neurodevelopmental age in normal infants using 3D-CNN based on brain MRI. in 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2373–2378 (IEEE, 2019).
- 176.Saha S, et al. Predicting motor outcome in preterm infants from very early brain diffusion MRI using a deep learning convolutional neural network (CNN) model. Neuroimage. 2020;215:116807. doi: 10.1016/j.neuroimage.2020.116807. [DOI] [PubMed] [Google Scholar]
- 177.He L, et al. A multi-task, multi-stage deep transfer learning model for early prediction of neurodevelopment in very preterm infants. Sci. Rep. 2020;10:15072. doi: 10.1038/s41598-020-71914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Temple MW, Lehmann CU, Fabbri D. Predicting discharge dates from the NICU using progress note data. Pediatrics. 2015;136:e395–405. doi: 10.1542/peds.2015-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang J, et al. Automated retinopathy of prematurity screening using deep neural networks. EBioMedicine. 2018;35:361–368. doi: 10.1016/j.ebiom.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Valikodath N, Cole E, Chiang MF, Campbell JP, Chan RVP. Imaging in retinopathy of prematurity. Asia Pac. J. Ophthalmol. 2019;8:178–186. doi: 10.22608/APO.201963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Biten H, et al. Diagnostic accuracy of ophthalmoscopy vs telemedicine in examinations for retinopathy of prematurity. JAMA Ophthalmol. 2018;136:498–504. doi: 10.1001/jamaophthalmol.2018.0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chiang MF, et al. Detection of clinically significant retinopathy of prematurity using wide-angle digital retinal photography: a report by the american academy of ophthalmology. Ophthalmology. 2012;119:1272–1280. doi: 10.1016/j.ophtha.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ednick M, et al. A review of the effects of sleep during the first year of life on cognitive, psychomotor, and temperament development. Sleep. 2009;32:1449–1458. doi: 10.1093/sleep/32.11.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ansari AH, et al. Quiet sleep detection in preterm infants using deep convolutional neural networks. J. Neural Eng. 2018;15:066006. doi: 10.1088/1741-2552/aadc1f. [DOI] [PubMed] [Google Scholar]
- 185.Ansari AH, et al. A deep shared multi-scale inception network enables accurate neonatal quiet sleep detection with limited EEG. Channels IEEE J. Biomed. Health Inf. 2022;26:1023–1033. doi: 10.1109/JBHI.2021.3101117. [DOI] [PubMed] [Google Scholar]
- 186.Werth J, Radha M, Andriessen P, Aarts RM, Long X. Deep learning approach for ECG-based automatic sleep state classification in preterm infants. Biomed. Signal Process. Control. 2020;56:101663. doi: 10.1016/j.bspc.2019.101663. [DOI] [Google Scholar]
- 187.Hauptmann A, Arridge S, Lucka F, Muthurangu V, Steeden JA. Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning-proof of concept in congenital heart disease. Magn. Reson Med. 2019;81:1143–1156. doi: 10.1002/mrm.27480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ceschin R, et al. A computational framework for the detection of subcortical brain dysmaturation in neonatal MRI using 3D Convolutional Neural Networks. Neuroimage. 2018;178:183–197. doi: 10.1016/j.neuroimage.2018.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ornek AH, Ceylan M. Explainable artificial intelligence (XAI): classification of medical thermal images of neonates using class activation maps. Trait. Signal. 2021;38:1271–1279. doi: 10.18280/ts.380502. [DOI] [Google Scholar]
- 190.Ervural S, Ceylan M. Classification of neonatal diseases with limited thermal Image data. Multimed. Tools Appl. 2021;81:9247–9275. doi: 10.1007/s11042-021-11391-0. [DOI] [Google Scholar]
- 191.Ervural S, Ceylan M. Thermogram classification using deep siamese network for neonatal disease detection with limited data. Quant. InfraRed Thermogr. J. 2022;19:312–330. doi: 10.1080/17686733.2021.2010379. [DOI] [Google Scholar]
- 192.Ervural S, Ceylan M. Convolutional neural networks-based approach to detect neonatal respiratory system anomalies with limited thermal image. Trait. Signal. 2021;38:437–442. doi: 10.18280/ts.380222. [DOI] [Google Scholar]
- 193.Xu S, et al. Wireless skin sensors for physiological monitoring of infants in low-income and middle-income countries. Lancet Digit. Health. 2021;3:e266–e273. doi: 10.1016/S2589-7500(21)00001-7. [DOI] [PubMed] [Google Scholar]
- 194.Hoshino E, et al. An iPhone application using a novel stool color detection algorithm for biliary atresia screening. Pediatr. Surg. Int. 2017;33:1115–1121. doi: 10.1007/s00383-017-4146-8. [DOI] [PubMed] [Google Scholar]
- 195.Dong Y, et al. Artificial intelligence algorithm-based computed tomography images in the evaluation of the curative effect of enteral nutrition after neonatal high intestinal obstruction operation. J. Health. Eng. 2021;2021:7096286. doi: 10.1155/2021/7096286. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 196.Mooney C, et al. Predictive modelling of hypoxic ischaemic encephalopathy risk following perinatal asphyxia. Heliyon. 2021;7:e07411. doi: 10.1016/j.heliyon.2021.e07411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pavel AM, et al. A machine-learning algorithm for neonatal seizure recognition: a multicentre, randomised, controlled trial. Lancet Child Adolesc. Health. 2020;4:740–749. doi: 10.1016/S2352-4642(20)30239-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mathieson S, et al. In-depth performance analysis of an EEG based neonatal seizure detection algorithm. Clin. Neurophysiol. 2016;127:2246–2256. doi: 10.1016/j.clinph.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Moeskops P, et al. Prediction of cognitive and motor outcome of preterm infants based on automatic quantitative descriptors from neonatal MR brain images. Sci. Rep. 2017;7:2163. doi: 10.1038/s41598-017-02307-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bahado-Singh RO, et al. Precision cardiovascular medicine: artificial intelligence and epigenetics for the pathogenesis and prediction of coarctation in neonates. J. Matern Fetal Neonatal Med. 2022;35:457–464. doi: 10.1080/14767058.2020.1722995. [DOI] [PubMed] [Google Scholar]
- 201.Sentner, T. et al. The Sleep Well Baby project: an automated real-time sleep-wake state prediction algorithm in preterm infants. Sleep45, zsac143 (2022). [DOI] [PMC free article] [PubMed]
- 202.Sirota M, et al. Enabling precision medicine in neonatology, an integrated repository for preterm birth research. Sci. Data. 2018;5:180219. doi: 10.1038/sdata.2018.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Young J, Macke CJ, Tsoukalas LH. Short-term acoustic forecasting via artificial neural networks for neonatal intensive care units. J. Acoust. Soc. Am. 2012;132:3234–3239. doi: 10.1121/1.4754556. [DOI] [PubMed] [Google Scholar]
- 204.Reis M, Ortega N, Silveira PSP. Fuzzy expert system in the prediction of neonatal resuscitation. Braz. J. Med. Biol. Res. 2004;37:755–764. doi: 10.1590/S0100-879X2004000500018. [DOI] [PubMed] [Google Scholar]
- 205.Saadah LM, et al. Palivizumab prophylaxis during nosocomial outbreaks of respiratory syncytial virus in a neonatal intensive care unit: predicting effectiveness with an artificial neural network model. Pharmacotherapy. 2014;34:251–259. doi: 10.1002/phar.1333. [DOI] [PubMed] [Google Scholar]
- 206.Kakarmath S, et al. Best practices for authors of healthcare-related artificial intelligence manuscripts. NPJ Digit Med. 2020;3:134. doi: 10.1038/s41746-020-00336-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Plana D, et al. Randomized clinical trials of machine learning interventions in health care: a systematic review. JAMA Netw. Open. 2022;5:e2233946–e2233946. doi: 10.1001/jamanetworkopen.2022.33946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Caparros-Gonzalez RA, de la Torre-Luque A, Diaz-Piedra C, Vico FJ, Buela-Casal G. Listening to relaxing music improves physiological responses in premature infants: a randomized controlled trial. Adv. Neonatal Care. 2018;18:58–69. doi: 10.1097/ANC.0000000000000448. [DOI] [PubMed] [Google Scholar]
- 209.Pillai Riddell, R. & Fabrizi, L Rebooting Infant Pain Assessment: Using Machine Learning to Exponentially Improve Neonatal Intensive Care Unit Practice (BabyAI) ClinicalTrials.gov Identifier: NCT05579496. https://clinicaltrials.gov/study/NCT05579496?id=NCT05579496%20&rank=1#more-information, https://www.yorku.ca/lamarsh/rebooting-infant-pain-assessment-using-machine-learning-to-exponentially-improveneonatal-intensive-care-unit-practice (2022).
- 210.Roue JM, Morag I, Haddad WM, Gholami B, Anand KJS. Using sensor-fusion and machine-learning algorithms to assess acute pain in non-verbal infants: a study protocol. BMJ Open. 2021;11:e039292. doi: 10.1136/bmjopen-2020-039292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Shalish W, et al. Prediction of Extubation readiness in extremely preterm infants by the automated analysis of cardiorespiratory behavior: study protocol. BMC Pediatr. 2017;17:167. doi: 10.1186/s12887-017-0911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Janvier, A., et al. The ethics of family integrated care in the NICU: Improving care for families without causing harm. Seminars in Perinatology46, 151528 (2022). [DOI] [PubMed]
- 213.Waddington C, van Veenendaal NR, O’Brien K, Patel N. Family integrated care: Supporting parents as primary caregivers in the neonatal intensive care unit. Pediatr. Investig. 2021;5:148–154. doi: 10.1002/ped4.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Morton CE, Smith SF, Lwin T, George M, Williams M. Computer programming: should medical students be learning it? JMIR Med. Educ. 2019;5:e11940. doi: 10.2196/11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. Multimodal biomedical AI. Nat. Med. 2022;28:1773–1784. doi: 10.1038/s41591-022-01981-2. [DOI] [PubMed] [Google Scholar]
- 216.Ahuja AS. The impact of artificial intelligence in medicine on the future role of the physician. PeerJ. 2019;7:e7702. doi: 10.7717/peerj.7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Han E-R, et al. Medical education trends for future physicians in the era of advanced technology and artificial intelligence: an integrative review. BMC Med. Educ. 2019;19:460. doi: 10.1186/s12909-019-1891-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lozano PM, et al. Training the next generation of learning health system scientists. Learn. Health Syst. 2022;6:e10342. doi: 10.1002/lrh2.10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kawahara J, et al. BrainNetCNN: convolutional neural networks for brain networks; towards predicting neurodevelopment. NeuroImage. 2017;146:1038–1049. doi: 10.1016/j.neuroimage.2016.09.046. [DOI] [PubMed] [Google Scholar]
- 220.Alexander B, et al. A new neonatal cortical and subcortical brain atlas: the Melbourne Children’s Regional Infant Brain (M-CRIB) atlas. NeuroImage. 2017;147:841–851. doi: 10.1016/j.neuroimage.2016.09.068. [DOI] [PubMed] [Google Scholar]
- 221.Prastawa M, Gilmore JH, Lin W, Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med. Image Anal. 2005;9:457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 222.Cutillo CM, et al. Machine intelligence in healthcare—perspectives on trustworthiness, explainability, usability, and transparency. npj Digit. Med. 2020;3:47. doi: 10.1038/s41746-020-0254-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Elmas G, et al. Federated learning of generative image priors for MRI reconstruction. IEEE Trans. Med. Imaging. 2022;42:1996–2009. doi: 10.1109/TMI.2022.3220757. [DOI] [PubMed] [Google Scholar]
- 224.Zhang M, Qu L, Singh P, Kalpathy-Cramer J, Rubin DL. SplitAVG: a heterogeneity-aware federated deep learning method for medical imaging. IEEE J. Biomed. Health Inf. 2022;26:4635–4644. doi: 10.1109/JBHI.2022.3185956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Katznelson G, Gerke S. The need for health AI ethics in medical school education. Adv. Health Sci. Educ. 2021;26:1447–1458. doi: 10.1007/s10459-021-10040-3. [DOI] [PubMed] [Google Scholar]
- 226.Mercurio MR, Cummings CL. Critical decision-making in neonatology and pediatrics: the I–P–O framework. J. Perinatol. 2021;41:173–178. doi: 10.1038/s41372-020-00841-6. [DOI] [PubMed] [Google Scholar]
- 227.Lin M, Vitcov GG, Cummings CL. Moral equivalence theory in neonatology. Semin. Perinatol. 2022;46:151525. doi: 10.1016/j.semperi.2021.151525. [DOI] [PubMed] [Google Scholar]
- 228.Porcelli PJ, Rosenbloom ST. Comparison of new modeling methods for postnatal weight in ELBW infants using prenatal and postnatal data. J. Pediatr. Gastroenterol. Nutr. 2014;59:e2–8. doi: 10.1097/MPG.0000000000000342. [DOI] [PubMed] [Google Scholar]
- 229.Temple MW, Lehmann CU, Fabbri D. Natural language processing for cohort discovery in a discharge prediction model for the neonatal ICU. Appl Clin. Inf. 2016;7:101–115. doi: 10.4338/ACI-2015-09-RA-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Moeskops P, et al. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans. Med. Imaging. 2016;35:1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Dr. E. Keles and Dr. U. Bagci have full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All study materials are available from the corresponding author upon reasonable request.