Abstract
These days carbon dots have been developed for multiple biomedical applications. In the current study, the transfection potential of synthesized carbon dots from single biopolymers such as chitosan, PEI-2kDa, and PEI-25kDa (CS-CDs, PEI2-CDs, and PEI25-CDs) and by combining two biopolymers (CP2-CDs and CP25-CDs) through a bottom-up approach have been investigated. The characterization studies revealed successful synthesis of fluorescent, positively charged carbon dots <20 nm in size. Synthesized carbon dots formed a stable complex with plasmid DNA (EGFP-N1) and miRNA-153 that protected DNA/miRNA from serum-induced degradation. In-vitro cytotoxicity analysis revealed minimal cytotoxicity in cancer cell lines (A549 and MDA-MB-231). In-vitro transfection of EGFP-N1 plasmid DNA with PEI2-CDs, PEI25-CDs and CP25-CDs demonstrated that these CDs could strongly transfect A549 and MDA-MB-231 cells. The highest EGFP-N1 plasmid transfection efficiency was observed with PEI2-CDs at a weight ratio of 32:1. PEI25-CDs polyplex showed maximum transfection at a weight ratio of 8:1 in A549 at a weight ratio of 16:1 in MDA-MB-231 cells. CP25-CDs exhibited the highest transfection at a weight ratio of 16:1 in both cell lines. The in-vitro transfection of target miRNA, i.e., miR-153 in A549 and MDA-MB-231 cells with PEI2-CDs, PEI25-CDs, and CP25-CDs suggested successful transfer of miR-153 into cells which induced significant cell death in both cell lines. Importantly, CS-CDs and CP2-CDs could be tolerated by cells up to 200 μg/mL concentration, while PEI2-CDs, PEI25-CDs, and CP25-CDs showed non-cytotoxic behavior at low concentrations (25 μg/mL). Together, these results suggest that a combination of carbon dots synthesized from chitosan and PEI (CP25-CDs) could be a novel vector for transfection nucleic acids that can be utilized in cancer therapy.
Keywords: Carbon dots, Micro RNA, Transfection, Chitosan, PEI, Cancer, Cell line
Abbreviation
- CDs
Carbon dots
- DNA
Deoxyribonucleic acid
- RNA
Ribonucleic acid
- PEI
Polyethylenimine
- CP25-CDs
Chitosan-PEI 25 kDa carbon dots
- CP2-CDs
Chitosan-PEI 2 kDa carbon dots
- CS-CDs
Chitosan carbon dots
- PEI2-CDs
PEI 2 kDa carbon dots
- PEI25-CDs
PEI 25 kDa carbon dots
- PBA
4-carboxyphenylboronic acid
- miRNA
MicroRNA
- FBS
Fetal Bovine Serum
- DMEM
Dulbecco's Modified Eagle's Medium
- RPMI-1640
Roswell Park Memorial Institute-1640 medium
- DMSO
Dimethyl sulphoxide
- FTIR
Fourier transform infrared
- XRD
X-Ray diffraction
- HR-TEM
High Resolution-Transmission electron microscopy
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide
- fc-rPEICdots
Folate-conjugated reducible PEI passivated carbon dots
1. Introduction
The union of nanotechnology and gene therapy has led to the emergence of nanomedicine, where a nanomaterial is used for gene delivery and gene therapy. The major challenge in this area is designing a multifunctional nanocarrier that combines therapeutic and diagnosis capabilities. Many kinds of nanomaterial, such as gold, silica, iron oxide, carbon nanotubes, semiconductor quantum dots, graphene, and nanodiamonds, have been explored as non-viral vectors for nucleic acid delivery [1]. Recently, fluorescent carbon dots (CDs) have drawn massive attention in nanomaterial fields.
Carbon dots (CDs) are carbon-based nanomaterials that became popular due to their special physiochemical, luminescent, and electronic properties [2]. They also possess other advantageous characteristics such as simple method of synthesis, low cost, wavelength-dependent luminescence emission, small size, resistance towards photobleaching, bioconjugation, nontoxicity, and biocompatibility. The surface of CDs possess various functional groups like amino, carboxyl, hydroxyl, epoxy, and carbonyl which provide hydrophobicity and allow modifications on the surface of CDs with numerous polymeric, organic, and biological species. Therefore, CDs have been proven as a potential fluorescence probe in bioimaging, optical sensing, photocatalysis, electrocatalysis, and drug delivery [[2], [3], [4], [5], [6], [7], [8], [9], [10], [11]]. In recent years CDs have been extensively used as potential nanomaterials in bioimaging, sensing, energy conversion, catalysis, and nanomedicine [12,13]. Due to biocompatibility, physicochemical properties, and a huge variation in surface functionalization groups, the CDs have been utilized in gene transfection application, and their fluorescence properties are useful in monitoring the dissociation and association of plasmid DNA in real-time. However, transfection of siRNA/miRNA across the cell membrane is a crucial task, and to overcome this issue researchers have synthesized CDs. In the past few years, CDs have been widely utilized to transfer small interfering RNA (siRNA) in vitro and in vivo. Previously, Carbon dots were synthesized using citric acid and tryptophane, and the surface of CDs was functionalized with PEI to deliver survivin siRNA into MGC803 (human gastric cancer cell line) [14]. Various researchers have utilized CDs for DNA, siRNA, and miRNA transfection resulting in efficient gene delivery and gene knockdown in vitro and in vivo [15,16].
The surface passivated CDs have been utilized for various applications such as broad excitation spectra, photoluminescence, and also possess excellent biocompatibility compared to other semiconductor quantum dots. Generally, CDs can be synthesized by two methods: top-up method and bottom-up method. The Top-up method comprises laser ablation and electrochemical oxidation, whereas the bottom-up approach includes combustion, thermal carbonization, acid dehydration, and ultrasonic treatment [[17], [18], [19]]. The bottom-up approach is more conducive to controlling CDs physical and chemical characteristics, therefore primarily utilized for fabricating distinct types of carbon dots [20].
In recent years, researchers are showing great interest in the fabrication of targeted fluorescence CDs using organic molecules, which is simple, eco-friendly, and economical. Chitosan (N-deacetylated derivative of chitin) is a naturally occurring mucopolysaccharide with biocompatible and nontoxic properties and contains amino and hydroxyl functional groups in large amounts [21]. Chitosan is cationic in nature and thus binds with DNA/RNA, protect them from degradation and serves as a gene delivery vector [22]. Remarkably, polyethylenimine (PEI) is also cationic in nature, containing high amount of amino groups and thus, it is more often used as gene delivery vehicle. PEI have rare proton sponge mechanism which is favorable for endosomal escape in cells [23]. PEI of large molecular weight is stable but toxic in nature, whereas PEI of low molecular weight induces less toxicity [24]. PEI and chitosan both are accepted carbon sources for the fabrication of carbon dots and have already been applied for the synthesis and surface functionalization of carbon dots using various carbonization methods [20,25].
Herein, hybrid fluorescence carbon dots based on chitosan-PEI 25 kDa (CP25-CDs) and chitosan-PEI 2 kDa (CP2-CDs) were synthesized by bottom-up method for targeted delivery of plasmid DNA and miRNA in cancer cells. PBA was utilized in synthesized CDs that will help the CDs to target the salicylic acid overexpressed cancer cells. The CDs were fabricated using one step synthesis method under open atmosphere. Both hybrid CDs exhibited plasmid binding abilities but only CP25-CDs accomplished transfection of EGFP-N1 plasmid DNA in to A549 and MDA-MB-231 cells. CP25-CDs successfully transfected miR-153 in to A549 and MDA-MB-231 cells that leads to apoptosis induced cell death.
2. Materials and methods
2.1. Materials and chemicals
Chitosan, high molecular weight (310,000–375,000 Da) > 75 % deacetylated obtained from crustacean shells, poly (ethyleneimine) solution 2 kDa, polyethyleneimine, branched 25 kDa, 4-carboxyphenylboronic acid (PBA) and miRNA mimic (unscrambled miRNA, negative control) and has-miR-153 were purchased from Merck (India). Fetal Bovine Serum (FBS) heat inactivated (origin, South American), Citric acid anhydrous (cell culture tested), Dulbecco's Modified Eagle's Medium (DMEM), Roswell Park Memorial Institute-1640 medium (RPMI-1640), sodium bicarbonate (cell culture tested) and antibiotic solution 100X (10,000U penicillin and 10 mg streptomycin/mL) were procured from Hi-media (India). Trypsin EDTA solution 1X, dimethyl sulphoxide (DMSO), agarose (low EEO), luria bertani broth (LB Broth), agar type 1 and DEPC-treated water were also procured from Hi-media (India). Lipofectamine2000 reagent was supplied by Invitrogen (India) and heparin sodium was supplied by Biological E. Limited (India). The EGFP-N1 plasmids DNA was isolated from Escherichia coli with QIAprep DNA isolation Kit Qiagen (Germany).
2.2. Synthesis of chitosan and PEI CDs
Fluorescent CDs passivated with chitosan (CS-CDs), PEI 2 kDa (PEI2-CDs), PEI 25 kDa (PEI25-CDs) and hybrid of chitosan-PEI 2 kDa (CP2-CDs) and chitosan-PEI 25 kDa (CP25-CDs) were synthesized by one step thermal decomposition method. Briefly, citric acid (200 mg), chitosan/PEI 2kDa/PEI 25 kDa (500 mg), PBA (300 mg), diluted HCl (2 mL, 0.3 N), were mixed in double distilled water (30 mL) and heated for 2 h at 300 °C. Similarly, for the synthesis of hybrid CDs (CP2 and CP25-CDs) citric acid (200 mg), chitosan (250 mg), PEI-2kDa/PEI-25kDa (250 mg), PBA (300 mg), and diluted HCL (2 mL, 0.3 N) were dissolved in double distilled water (30 mL) and heated for 2 h while stirring at 300 °C. The color-changed products aqueous suspension were further dialysed for 24 h with a 3500 Da MWCO membrane and the resulting solution was further lyophilized by using an Allied Frost lyophilizer to obtain CDs in powder form [26].
2.3. Characterizations of CDs
UV–Vis spectroscopy and excitation-dependent fluorescence emission spectrum of carbon dots were analyzed using Thermo Fischer Scientific (Varioskan LUX 3020–561) spectrometer. The Fourier transform infrared spectroscopy were conducted on Shimadzu (Model QATR-S) at a wavelength of 4000–400 cm−1. X-Ray diffraction pattern were further recorded on X-Ray Diffractometer (Bruker) for 2θ range of 5°- 90° with a scanning step of 0.02° and a scanning rate of 1°/min. Zeta potential was measured by Malvern Particle Size and Zeta Potential Analyser. The morphology and size of carbon dots were examined by High Resolution-Transmission electron microscopy (HR-TEM) (JEOL, JEM 2100 plus).
2.4. Preparation of CDs:plasmid DNA polyplexes
EGFP-N1plamsid DNA was isolated from E. coli and used to prepare different polyplexes. The EGFP-N1 plasmid was mixed with CDs at different weight ratio CS-CDs:EGFP-N1 (5:1, 10:1, 25:1, 50:1, 100:1, 200:1), PEI2-CDs:EGFP-N1 (1:1, 2:1, 4:1, 8:1, 16:1, 24:1, 32:1), PEI25-CDs:EGFP-N1 (0.5:1, 1:1, 2:1, 3:1, 4:1, 5:1), CP2-CDs:EGFP-N1 (1:1, 2:1, 4:1, 8:1, 16:1, 24:1, 32:1), and CP25-CDs:EGFP-N1 (0.5:1, 1:1, 2:1, 3:1, 4:1, 5:1). The various combination mixtures were incubated for 30 min at room temperature to allow complex formation completely. Thereafter, polyplex mixtures were mixed with loading dye, loaded on 1 % agarose gel containing ethidium bromide (0.5 μg/mL) and horizontal electrophoresis experiment was carried out in 1X TAE buffer for 30 min at a constant voltage of 100 V. Then the plasmid DNA bands were visualized through gel documentation system.
2.5. Decomplexation of polyplexes
To achieve the decomplexation, the polyplexes were prepared and then treated with 20 % heparin for 30 min at room temperature. Then the reaction mixture was loaded on 1 % agarose gel and run for 30 min at 100 V.
2.6. Serum stability of polyplexes
Serum stability assay were used to evaluate the capability of various polyplexes to protect nucleic acid from serum degradation. Briefly, first polyplexes were prepared at various weight ratios and incubated with 50 % FBS for 24 h. Then 20 % heparin was added and incubated for 15 min, followed by centrifugation at 3380 g for 10 min. Thereafter, the supernatant was collected and loaded on 1 % agarose gel, followed by electrophoresis in 1X TAE buffer for 30 min.
2.7. Mammalian cell viability assay
Lung adenocarcinoma (A549), and human mammary carcinoma (MDA-MB-231) cells were procured from the National Centre for Cell Sciences, Pune (India). The cells were maintained by standard protocols in humidified CO2 (5 %) incubator. The cytotoxicity effect of carbon dots (CS-CDs, PEI2-CDs, PEI25-CDs, CP2-CDs, and CP25-CDs) on A549 and MDA-MB-231 cell lines was assessed using MTT assay. A549 cells (1 × 104 cells/mL) and MDA-MB-231 cells (2 × 104 cells/mL) were seeded in a 96-well plate and after 12 h the cells were treated with CDs (5–200 μg/mL). After 48 h, CDs containing media was replaced with fresh media (100 μL) containing 0.5 mg/mL MTT. The MTT assay was performed by measuring the absorbance at 590 nm wavelength, and the percentage of cell viability was calculated as:
| Cell viability (%) = Absorbance Sample/ Absorbance Control × 100 |
2.8. In-vitro transfection of EGFP-N1
In-vitro transfection of A549 and MDA-MB-231 cells with EGFP-N1 plasmid using CS-CDs, PEI2-CDs, PEI25-CDs, CP2-CDs and CP25-CDs was analyzed. The cells (2 × 105 cells) were seeded in a 12-well plate and incubated for 24 h. After 24 h, medium was aspirated from each well and fresh medium containing polyplexes (CDs:EGFP-N1 plasmid) at different weight ratios were added to each well (n = 3). Transfection was carried out for 24 h and then polyplexes that were not internalized were aspirated and replaced with fresh medium, followed by incubation for next 24 h in a CO2 humified atmosphere. Thereafter, transfection efficiency was measured through flow cytometer (BD Accuri C6 Flow cytometer). Transfected cells were harvested using trypsin and subjected to flow cytometer. Data were analyzed using the FCS express flow cytometry software.
2.9. Binding of CDs with microRNA (miR-153)
miRNA binding affinity of CDs were determined via binding assay. Briefly, miR-153 was mixed with carbon dots (PEI2-CDs, PEI25-CDs, and CP25-CDs) at different weight ratios: PEI2-CDs:miR-153 (4:1, 8:1, 16:1, 32:1), PEI25-CDs:mir-153 (2:1, 4:1, 8:1, 16:1, 32:1), and CP25-CDs:miR-153 (4:1, 8:1, 16:1, 32:1), and incubated at room temperature for 40 min. Subsequently, the mixture was loaded on 2 % agarose gel containing ethidium bromide (0.5 μg/mL) and electrophoresis was carried out in 1X TAE buffer for 30 min at 100 V. Then bands of miR-153 were visualized through gel documentation system.
2.10. In-vitro transfection of miR-153
Further to analyze if the polyplexes can be utilized for gene silencing. A549 cells (1 × 104 cells/well) and MDA-MB-231 cells (2 × 104 cells/well) were seeded in a 96-well microtiter plate and incubated overnight. Next day medium from each well replaced with medium containing polyplexes with miR-153 at various weight ratios. After 24 h, transfection media was aspirated and replaced with fresh media, and transfected cells were incubated further for 48 h in 5 % CO2 and 37 °C. Afterward, cell cytotoxicity assay was performed on A549 and MDA-MB-231 cells, as mentioned earlier, and the percentage of cell viability was calculated as described above.
2.11. Statistical analyses
All the experiments were done in triplicate and one-way analysis of variance (ANOVA) was employed for statistical analysis. Data have shown as mean ± SEM and p value of <0.05 considered as significance difference between groups.
3. Results
3.1. Characterization of CDs
The average yield of synthesized carbon obtained from chitosan (CS-CDs) recorded with the highest yield i. e 54 % ± 3.14, while the PEI25-CDs and combination carbon dots CP25-CDs provided 48.2 % ± 2.32 and 48.7 % ± 2.65 yield respectively. The CP2-CDs produced 44.1 % ± 2.29 yield, whereas the minimum yield was obtained from PEI2-CDs approximately 26.9 % ± 1.55. The physicochemical properties of synthesized carbon dots (CS-CDs, PEI2-CDs, PEI25-CDs, CP2-CDs, and CP25-CDs) were analyzed and CS-CDs exhibited a broad range UV spectrum at 283 nm (Fig. 1A), while PEI2-CDs and PEI25-CDs exhibited an absorbance peak at 281 nm and a wide band centered at 356 nm and 353 respectively (Fig. 1A). However, the UV–Vis spectroscopy data of CP2-CDs and CP25-CDs revealed absorption maxima at 280 nm (Fig. 1A). The absorption peak around 280 nm corresponds to the π-π* transition of the sp2 domains from the carbon core, while a peak between 300 and 400 nm could be attributed to n–π* transitions of the C O group. According to previous studies on chitosan and PEI carbon dots, chitosan carbon dots exhibit a strong UV–Vis absorption peak at 261 nm which corresponds to the π-π* transition [21], and PEI carbon dots display a sharp absorption peak at 272 nm and a broad absorption peak at 356 nm [27].
Fig. 1.
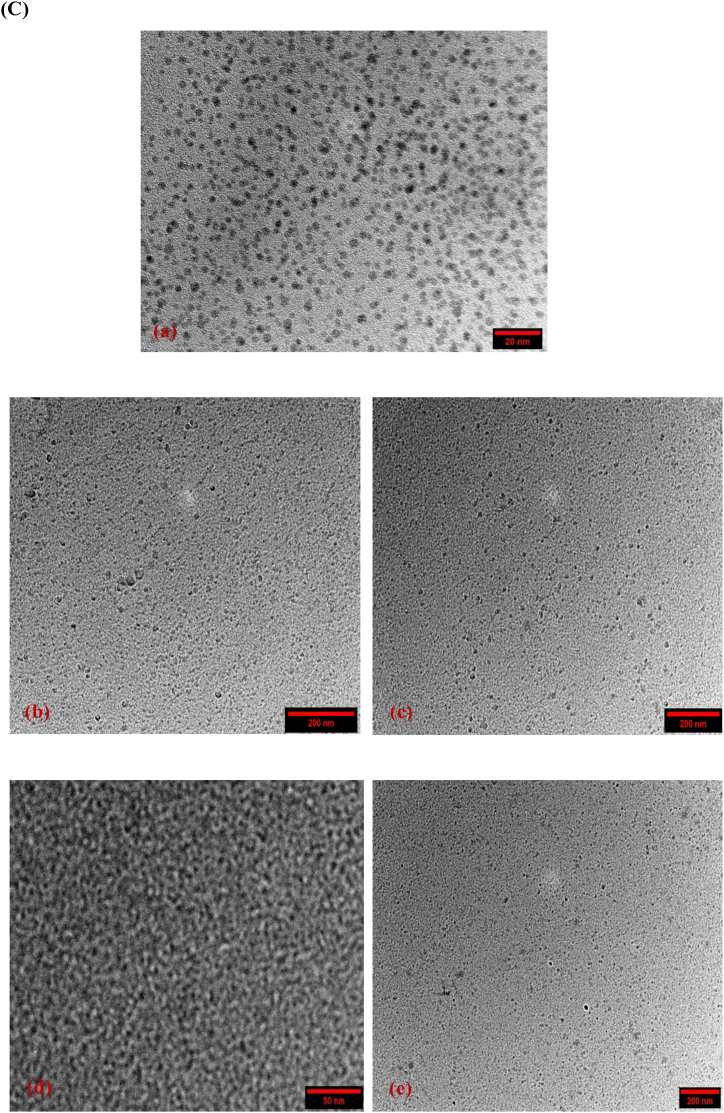
(A) UV absorbance spectra of synthesized carbon dots (B) XRD patterns for synthesized carbon dots and, (C) High resolution TEM images of carbon dots. (a) CS-CDs, (b) PEI2-CDs, (c) PEI25-CDs, (d) CP2-CDs and, (e) CP25-CDs.
The photoluminescence emission spectra of CS-CDs, CP2-CDs, and CP25-CDs exhibited the highest fluorescence emission peak at 320 nm when excited at wavelength of 270 nm (Figs. S1A and S1D and, S1E). The emission peak varied from 270 nm to 410 nm as the excitation wavelengths were increased from 250 nm to 390 nm. When the excitation wavelength was increased from 250 nm to 270 nm, the emission peak became stronger and showed redshift, but when the excitation wavelength was increased 270–390 nm, the emission peak exhibited redshift with constantly decreasing intensity. PEI2-CDs and PEI25-CDs were observed with strong fluorescence emission peak at 457 nm and 455 nm, at an excitation wavelength of 370 nm and did not exhibit a great shift as observed for CS-CDs, CP2-CDs, and CP25-CDs (Figs. S1B and S1C).
The functional groups on CDs were analyzed through FTIR spectra. CS-CDs, PEI2-CDs, PEI25-CDs, CP2-CDs, and CS25-CDs showed similar peaks of citric acid, PBA, raw chitosan, PEI-2kDa, and PEI-25 kDa (Fig. S2A) such as O–H stretching, C O stretching, C–OH stretching, B–O bond, carboxylic acid stretching vibration, stretching and bending of N–H groups, stretching and bending bands of the C–H groups, stretching of the C–N groups, and represented the C C, C N, and C C–O groups (Fig. S2B) which indicates the successful synthesis of these materials (Table 1). Sharp peaks from 1700 to 1725 cm−1 represented the carbonyl stretching frequency. After the formation of carbon dots, decreased intensity of peaks represented to the C C, C N, and C C–O groups were also observed (Fig. S2B). Other sharp peaks from 1617 to 1638 cm−1 indicated the carbonyl stretching frequency of amide.
Table 1.
FTIR peaks and functional groups present on precousor and synthesized carbon dots.
| Sr. No | Name of Compound | Functional Group | Frequency |
|---|---|---|---|
| 1. | Citric Acid | A. O–H stretching vibration | 3283 & 3493 |
| B. C O stretching vibration | 1741 | ||
| C. C–OH stretching vibration | 1138 | ||
| D. CH2 rocking | 769 | ||
| 2. | PBA | A. O–H stretching vibration | 3139 |
| B. B–O stretching vibration | 1322 | ||
| C. C O stretching vibration | 1677 | ||
| D. CH2 bending vibration | 1262, 1188 | ||
| 3. | Raw Chitosan | A. O–H & N–H stretching vibration | 3327 |
| B. C–H stretching vibration | 2875 | ||
| C. NH2 bending vibration | 1648 & 1568 | ||
| D. C C, C N and C C–O stretching | 1375–1148 | ||
| E. CH2 bending vibrations | 1024–900 | ||
| 4. | Raw PEI 2kd | A. N–H stretching vibration | 3300 |
| B. C–H stretching vibration | 2842 & 2946 | ||
| C. NH2 bending vibration | 1603 | ||
| D. CH2 bending vibration | 1464 | ||
| E. C–N stretching vibration | 1107 | ||
| 5. | Raw PEI 25 kd | A. N–H stretching vibration | 3294 |
| B. C–H stretching vibration | 2811 & 2935 | ||
| C. NH2 bending vibration | 1594 | ||
| D. CH2 bending vibration | 1454 | ||
| E. C–N stretching vibration | 1107 | ||
| 6. | CS-CDs | A. O–H and N–H stretching vibration | 3378 |
| B. C O stretching vibration | 1727 | ||
| C. C C stretching vibration | 1638, 1071 | ||
| D. NH2 bending vibration | 1506 | ||
| E. C–O stretching vibration | 1071 | ||
| F. CH2 bending vibration | 1014 | ||
| 7. | PEI2-CDs | A. O–H and N–H stretching vibration | 3335 |
| B. C–H stretching vibration | 2921 | ||
| C. C O stretching vibration | 1700 | ||
| D. C C stretching vibration | 1642 | ||
| E. CH2 bending vibration | 1430 | ||
| F. C–O stretching vibration | 1071 | ||
| 8. | PEI25-CDs | A. O–H and N–H stretching vibration | 3348 |
| B. C–H stretching vibration | 2969 | ||
| C. C O stretching vibration | 1705 | ||
| D. C C stretching vibration | 1637 | ||
| E. CH2 bending vibration | 1456 | ||
| F. C–O stretching vibration | 1055 | ||
| 9 | CP2-CDs | A. O–H and N–H stretching vibration | 3348 |
| B. C–H stretching vibration | 2969 | ||
| C. C O stretching vibration | 1714 | ||
| D. C C stretching vibration | 1617, 1064 | ||
| E. NH2 bending vibration | 1507 | ||
| F. C–N stretching vibration | 1194 | ||
| G. C–O stretching vibration | 1064 | ||
| 10. | CP25-CDs | A. O–H and N–H stretching vibration | 3365 |
| B. C–H stretching vibration | 2929 | ||
| C. C O stretching vibration | 1727 | ||
| D. C C stretching vibration | 1617, 1067 | ||
| E. NH2 bending vibration | 1511 | ||
| F. C–N stretching vibration | 1182 | ||
| G. C–O stretching vibration | 1067 |
The XRD pattern of CS-CDs exhibits small peaks at 12° which corresponds to chitosan but did not exhibit broad peak of graphene (Fig. 1B) while the XRD pattern of PEI2-CDs, PEI25-CDs, CP2-CDs, and CP25-CDs exhibited a broad peak that was centered at about 25°, which was consistent with the (002) crystal planes of graphene (Fig. 1B). The broadening of the peak near 25° confirms the carbonization of the material, which was attributed to highly disordered carbon atoms and confirms the formation of amorphous carbon dots.
The average zeta potential value of PEI25-CDs was +62 ± 1 mV (the maximum positive charge), whereas the minimum positive charge was carried by CS-CDs (+22 ± 0.2 mV). The zeta potential recorded for PEI2-CDs was +25 ± 1 mV, CP2-CDs was +41 ± 1.7 mV, and CP25-CDs was +32 ± 0.3 mV (Fig. S3).
The HR-TEM micrographs showed that carbon dots were spherical and approximately 10 nm in size. The average size recorded was 6 ± 0.2 nm for CS-CDs (Fig. 1C (a)), 9 ± 0.5 nm for PEI2-CDs (Fig. 1C (b)), 10 ± 0.6 nm for PEI25-CDs (Fig. 1C (c)), 6 ± 0.2 nm for CP2-CDs (Fig. 1C (d)), and 5 ± 0.4 nm for CP25-CDs (Fig. 1C (e)). The HR-TEM data revealed that the average diameter of synthesized material was less than 20 nm.
3.2. Binding affinity of CDs:plasmid DNA polyplexes
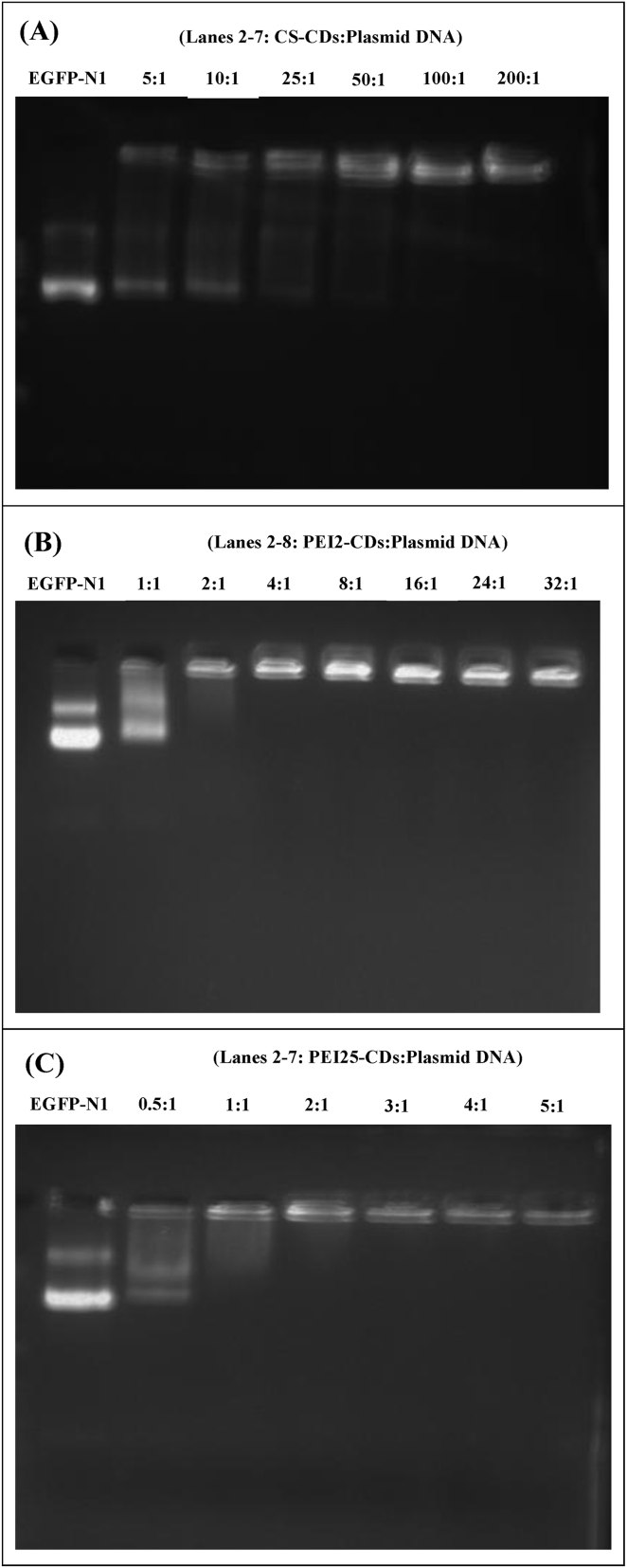
The ability of carbon dots to bind with plasmid DNA was determined through a binding assay. The gradual decrease observed in the intensities of migrating DNA bands on agarose gel, and visible lack of migration of DNA bands on the gel was considered as the appropriate binding ratio for vector and plasmid DNA [24]. The CS-CDs offered the binding with EGFP-N1 plasmid at a weight ratio of 100:1 (Fig. 2A), PEI2 and CP2 polyplexes showed the binding at a ratio of 4:1 (Fig. 2B and D), while PEI25 and CP25 polyplexes exhibited the binding affinity at weight ratio 2:1 (Figs. 2C) and 3:1 subsequently (Fig. 2E).
Fig. 2.
Gel electrophoresis images showing binding affinity of EGFP-N1 plasmid with carbon dots at different weight ratio (w/w). (A) CS-CDs with plasmid, (B) PEI2-CDs with plasmid (C) PEI25-CDs with plasmid, (D) CP2-CDs with plasmid and, (E) CP25-CDs with plasmid.
3.3. Decomplexation of polyplexes
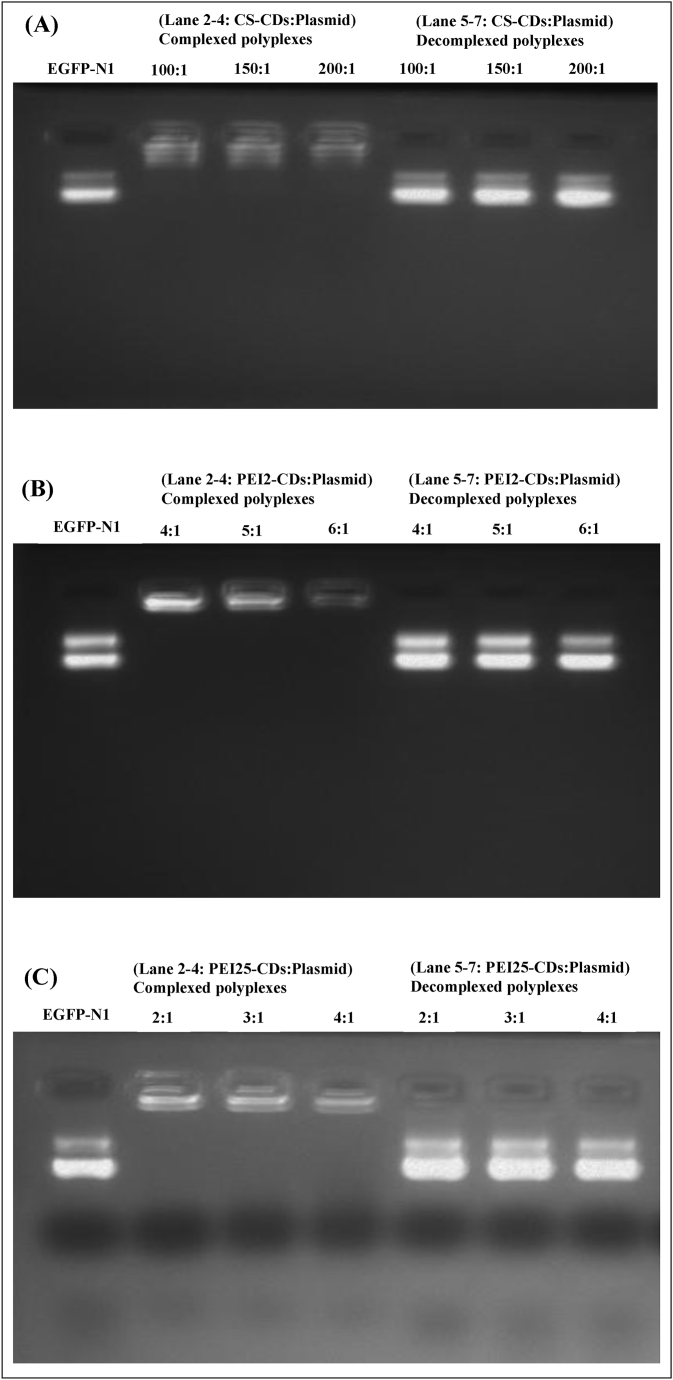
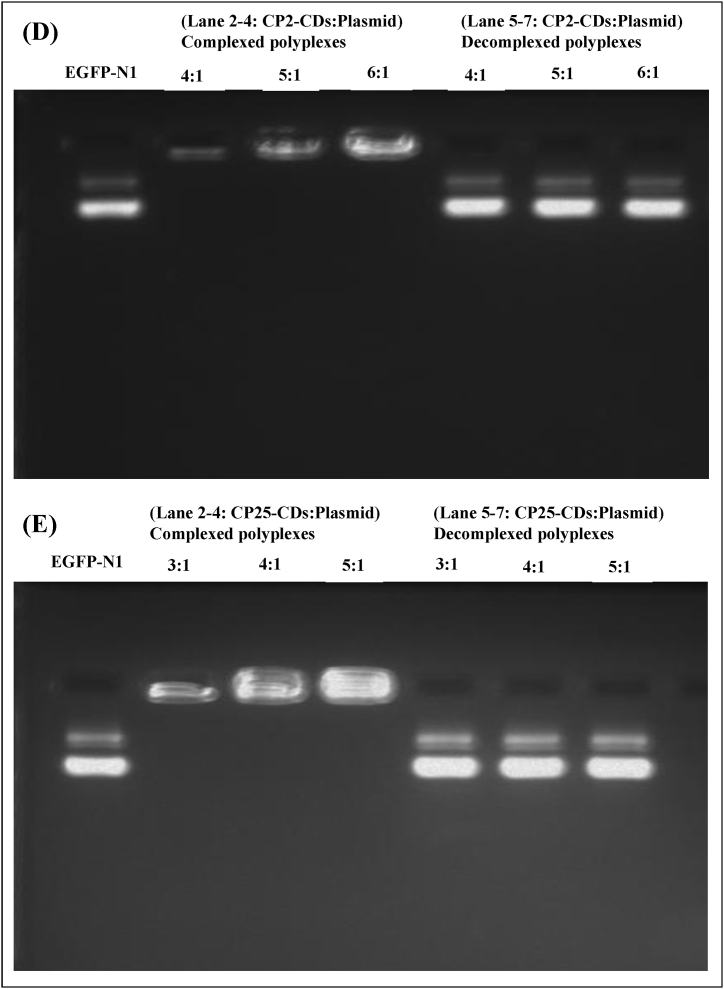
To access the release of EGFP-N1 plasmid from vector: EGFP-N1 plasmid complex, decomplexation assays were performed on all the polyplexes that displayed binding affinity at various weight ratios. To achieve this, polyplexes were incubated with heparin, and decomplexation was confirmed through gel electrophoresis. The gel images revealed that dissociated EGFP-N1 plasmid moved from the well and its band appeared on the gel which conformed decomplexation of plasmid DNA from carbon dots (Fig. 3).
Fig. 3.
Heparin decomplexation assay showing complexation and decomplexation of complexes of various weight ratio with 20 % heparin. (A) CS-CDs-plasmid, (B) PEI2-CDs-plasmid, (C) PEI25-CDs-plasmid, (D) CP2-CDs-plasmid and, (E) CP25-CDs-plasmid. Lane 1: EGFP-N1 plasmid alone, Lane 2–4: CDs:plasmid DNA complex, Lane 5–7: CDs:plasmid DNA complex treated with heparin.
3.4. Serum stability of polyplexes
The presence of serum hinders the nucleic acid transfection inside the cell and degrades DNA. Therefore, successful transfection of nucleic acid requires its protection from serum degradation. The gel electrophoresis images suggested that plasmid DNA that was not complexed with any vector gets degraded in the presence of serum (Fig. 4: lane 2), but the plasmid DNA released from the polyplexes are not showing any degradation (Fig. 4: lane 3–6).
Fig. 4.
Serum stability assay showing degradation of plasmid without CDs, and stability of plasmid DNA in the complexed state. (a) CS-CDs-plasmid, (b) PEI2-CDs-plasmid, (c) PEI25-CDs-plasmid, (d) CP2-CDs-plasmid (e) CP25-CDs-plasmid. Lane1: EGFP-N1 plasmid alone, Lane 2: plasmid treated with FBS, Lane 3–6: CDs:plasmid complex treated with FBS.
The intensity of plasmid DNA bands released from polyplexes (Fig. S4) appeared more (1 %–20 % more) or similar to control, while the EGFP-N1 plasmid treated with serum exhibited very minimum intensity (−50 % to −100 % less) (Fig. S4). The synthesized CDs could bind with plasmid DNA at a different weight ratio (w/w) (Table 2). Further, binding with CDs prevented the degradation of plasmid DNA in the presence of serum at similar weight ratios where they exhibited binding affinity (Table 2).
Table 2.
Binding, decomplexation and stability ratios of polyplexes.
| Sr. No. | Name of Polyplex | Binding affinity weight ratio (w/w) | Decomplexation weight ratio (w/w) | Serum stability weight ratio (w/w) |
|---|---|---|---|---|
| 1 | CS-CDs:EGFP-N1 | 100:1 | 100:1 | 100:1 |
| 2 | PEI2-CDs:EGFP-N1 | 4:1 | 4:1 | 4:1 |
| 3 | PEI25-CDs:EGFP-N1 | 2:1 | 2:1 | 2:1 |
| 4 | CP2-CDs:EGFP-N1 | 4:1 | 4:1 | 4:1 |
| 5 | CP25-CDs:EGFP-N1 | 3:1 | 3:1 | 4:1 |
3.5. Cytotoxicity evaluation of carbon dots
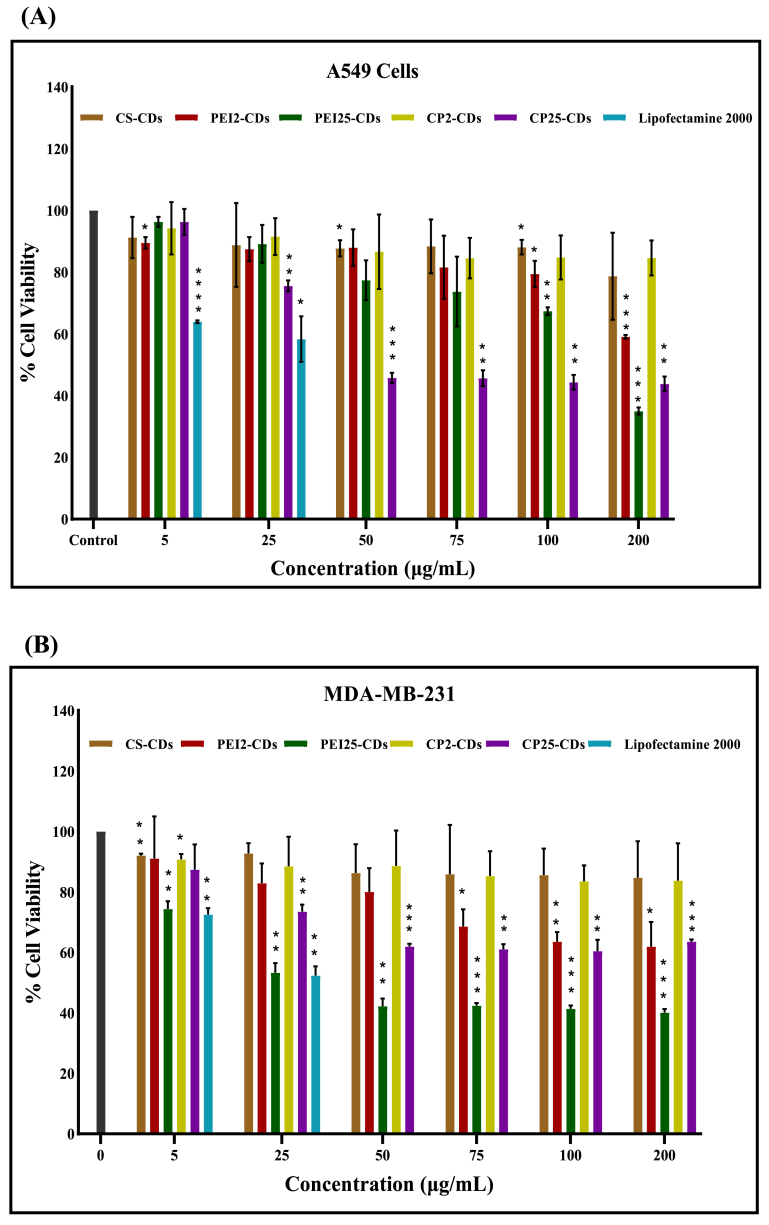
With integration of high transfection ability carbon dots also required less cytotoxicity. Thus, cytotoxicity evaluations were determined on A549 and MDA-MB-231 cells. It was observed from MTT data that CS and CP2 vectors/CDs exhibited superior cytocompatibility in A549 (79 % and 85 % respectively) and MDA-MB-231 cells (nearly 85 %) at concentrations (200 μg/mL); whereas PEI2-CDs displayed 79 % cell viability in A549 cells at 100 μg/mL (Fig. 5A) and 80 % in MDA-MB-231 cells at 50 μg/mL concentration (Fig. 5B). Further we observed toxicity in both cells treated with PEI25-CDs and CP25-CDs at concentrations (200 μg/mL). But at low concentration of PEI25-CDs (50 μg/mL) and CP25-CDs (25 μg/mL), 77 % and 76 % cell viability were detected in A549 cells (Fig. 5A). On the other hand, for MDA-MB-231 cells CP25-CDs exhibited 75 % cell viability at 25 μg/mL and PEI25-CDs exhibited 75 % cell viability at 5 μg/mL concentration (Fig. 5B). PEI25-CDs exhibited slight toxicity towards MDA-MB-231 cells but less than lipofectamine2000. In A549 cells Lipofectamine2000 exhibited 64 % cell viability at 5 μg/mL and 58 % at 25 μg/mL (Fig. 5A), whereas in MDA-MB-231 cells 73 % cell viability was observed at 5 μg/mL and 52 % at 25 μg/mL (Fig. 5B).
Fig. 5.
Cytotoxicity testing of lipofectamine2000 and carbon dots (CS-CDs, PEI2-CDs, PEI25-CDs, CP2-CDs, CP25-CDs) at different concentration (5 μg/mL to 200 μg/mL) against (A) A549 cells and (B) MDA-MB-231 cells. The data expressed as the mean ± SEM where p value < 0.05 represented by *, p value < 0.01 represented **, p value < 0.001 represented *** and, p value < 0.0001 represented with ****.
3.6. In vitro transfection of EGFP-N1 plasmid
In order to evaluate the transfection efficiency of carbon dots, A549 and MDA-MB-231 cells were transfected with EGFP-N1 plasmid via carbon dots at various ratios (w/w). The transfecting agent lipofectamine2000 was used as a control. The flow cytometry results revealed that A549 cells transfected with lipofectamine2000 at weight ratio 2:1 (vector:EGFP-N1 plasmid) showed approximately 47 % transfection (Fig. 6A and Fig. S5) whereas, MDA-MB-231 cells showed 23 % transfection at similar ratio (Fig. 6B and Fig. S7). On the other hand, PEI2-CDs, PEI25-CDs, and CP25-CDs also own the transfection ability and exhibited high transfection efficiency than lipofectamine2000.
Fig. 6.
Percentage of EGFP-N1 transfection with lipofectamine2000 and carbon dots at various weight ratios (vector:EGFP-N1) in (A) A549 and (B) MDA-MB-231 cells. The data are expressed as the mean ± SEM and p value < 0.001 represented by ***, p value < 0.0001 represented with ****.
PEI2-CDs acquired 53 % transfection in A549 cells and 13 % in MDA-MB-231 cells at a weight ratio of 32:1 (Table 3). Maximum 41 % transfection was observed in A549 cells when transfected with PEI25-CDs at a weight ratio of 8:1 and 74 % in MDA-MB-231 cells at 16:1 wt ratio (Table 3) (Fig. 6A and B, Fig. S6 and Fig. S8). CP25-CDs exhibited 74 % transfection in A549 cells at a weight ratio of 16:1 (Fig. 6A and Fig. S6) and it also offered good transfection efficiency (89 %) at similar weight ratio in MDA-MB-231 cells (Table 3) (Fig. 6B and Fig. S8). CS-CDs and CP2-CDs both failed to transfect EGFP-N1 plasmid DNA in A549 and MDA-MB-231 cells even at higher concentration (Fig. S5 and Fig. S7). Thus, only PEI2-CDs, PEI25-CDs, and CP25-CDs acquire the transfection ability, and CP25-CDs exhibited the highest transfection efficiency amongst all of them.
Table 3.
Percentage cell viability and transfection in A549 and MDA-MB-231 cells.
| Name |
% Cell viability (Vector alone) |
% Cell transfection (Vector:EGFP-N1) |
||||
|---|---|---|---|---|---|---|
| Conc. (μg/mL) | A549 | MDA-MB-231 | Weight ratio (w/w) (conc. Of vector (μg/mL)) | A549 | MDA-MB-231 | |
| CP25-CDs | 5 μg/mL | 96 ± 2 | 87 ± 5 | 4:1 (4 μg/mL) | 18 ± 0.2 | 11 ± 1 |
| 25 μg/mL | 76 ± 1 | 74 ± 1 | 8:1 (8 μg/mL) | 28 ± 0.4 | 59 ± 3 | |
| 50 μg/mL | 46 ± 1 | 62 ± 0.5 | 16:1 (16 μg/mL) | 74 ± 4.6 | 89 ± 2 | |
| PEI25-CDs | 5 μg/mL | 96 ± 1 | 74 ± 1 | 2:1 (2 μg/mL) | 29 ± 0.3 | 2 ± 0.2 |
| 25 μg/mL | 89 ± 3.5 | 53 ± 2 | 4:1 (4 μg/mL) | 39 ± 0.7 | 7 ± 0.4 | |
| 50 μg/mL | 77 ± 3.7 | 42 ± 1 | 8:1 (8 μg/mL) | 42 ± 0.3 | 33 ± 0.6 | |
| 16:1 (16 μg/mL) | 32 ± 1 | 74 ± 2 | ||||
| PEI2-CDs | 5 μg/mL | 90 ± 1 | 91 ± 8 | 4:1 (4 μg/mL) | 24 ± 0.4 | 6 ± 0.7 |
| 25 μg/mL | 87 ± 2 | 83 ± 4 | 8:1 (8 μg/mL) | 42 ± 1 | 10 ± 0.7 | |
| 50 μg/mL | 88 ± 3 | 80 ± 4 | 16:1 (16 μg/mL) | 41 ± 1 | 10 ± 0.5 | |
| 32:1 (32 μg/mL) | 53 ± 1 | 13 ± 0.4 | ||||
3.7. Evaluation of miR-153 binding affinity with CDs
miRNA-based gene therapy for cancer treatment requires efficient binding between vector and miRNA. The miR-153 was bound with PEI2-CDs, PEI25-CDs, and CP25-CDs at different vector:miRNA weight ratios. The gel images results displayed that PEI2-CDs and PEI25-CDs bound with miR-153 at weight ratio 16:1 (Fig. 7A and B) while the CP25-CDs bound with miR-153 at ratio 8:1 (Fig. 7C). CP25-CDs, PEI2-CDs, and PEI25-CDs offered the binding with miR-153 therefore, it was further utilized for miR-153 transfection.
Fig. 7.
Gel electrophoresis images showing binding affinity of miRNA with carbon dots at various weight ratio (w/w). (A) PEI2-CDs with miR-153, (B) PEI25-CDs with miR-153 (C) CP25-CDs with miR-153.
3.8. In-vitro evaluation miR-153 transfection
To evaluate the transfection of miRNA, miR-153 was transfected in A549 and MDA-MB 231 cells via CDs (PEI2-CDs, PEI25-CDs and CP25-CDs) at various weight ratios. Unscrambled miRNA was used as negative control. The MTT data revealed that cells transfected with mir-153 alone exhibited 85–90 % cell viability but when transfected with CDs (PEI2-CDs, PEI25-CDs and CP25-CDs) a gradual death of the cells was observed. The cells transfected with unscrambled miRNA alone and with carbon dots exhibited more than 80 % cell viability (Fig. 8A and B, Fig. S9 and Fig. S10). CP25-CDs:miR-153 complex exhibited a gradual decrease in cell viability as the vector:miRNA weight ratios were increased and minimum cell viability (12 % in A549 and 18 % in MDA-MB-231) was observed at a weight ratio of 80:1 (Fig. 8A and B, Fig. S9 and Fig. S10). PEI25-CDs:miR-153 complex displayed 34 % cell viability in A549 and 29 % in MDA-MB-231 cells at a ratio of 80:1, while 19 % cell viability in A549 and 16 % in MDA-MB-231 cells at a ratio of 160:1 (Fig. 8A and B, Fig. S9 and Fig. S10). The PEI2-CDs:miR-153 complex was observed with 47 % (A549) and 54 % (MDA-MB-231) cell viability at a weight ratio of 80:1 whereas, 45 % (A549) and 32 % (MDA-MB-231) cell viability were observed at a ratio of 160:1 (Fig. 8A and B, Fig. S9 and Fig. S10). From these observations, it is concluded that miRNA-153 was also successfully delivered into the cells by synthesized carbon dots (CP25-CDs, PEI25-CDs, and PEI2-CDs). The observed toxicity is due to the transfection of miR-153.
Fig. 8.
Percentage of cells viability after transfection of control miRNA and miR-153 with and without carbon dots (CP25-CDs, PEI25-CDs and PEI2-CDs) at various weight ratio (vector:miRNA) in (A) A549 cells and (B) MDA-MB-231 cells. The data are expressed as the mean ± SEM where p value < 0.05 represented by *, p value < 0.01 represented **, p value < 0.001 represented *** and, p value < 0.0001 represented with ****.
4. Discussion
The main limitation of gene therapy is the lack of suitable gene carrier that exhibits cytocompatibility properties and high gene transfection ability. Although various delivery systems such as cationic lipids and polymers, cell-penetrating peptides, organic and inorganic nanoparticles, and many others have been investigated, still there is no effective gene carrier available. Based on the recent advancement in gene delivery applications by environment-friendly and economically convenient carbon dots, organic fluorescence CDs were fabricated using chitosan and PEI. Synthesized carbon dots, CS-CDs, PEI2-CDs, PEI25-CDs, CP2-CDs, and CP25-CDs exhibited physicochemical properties similar to previously synthesized material from the same precursor's chitosan and PEI [27,28]. CS-CDs, CP2-CDs, and CP25-CDs exhibited excitation-dependent emission behavior, while PEI2-CDs and PEI25-CDs displayed excitation-independent emission behavior.
Sharp peaks of carbonyl stretching were observed in the FT-IR spectrum of carbon dots that might have appeared due to the carbonization process. Another sharp peaks which indicated the carbonyl stretching frequency of amide are due to the formation of amide linkage on the surface of synthesized carbon dots after carbonization [29]. Other physicochemical properties of synthesized carbon dots such as spherical shape, minimum size (<10 nm), and amorphous and cationic nature confirmed that these carbon dots can be utilized as a vector to transfer desired nucleic acids into the cells. Further, the ability of these carbon dots to complex, release and protect plasmid DNA was studied and it is observed that a 100:1 wt ratio of CS-CDs:plasmid DNA, 4:1 wt ratio of PEI2-CDs:plasmid DNA, CP2-CDs:plasmid DNA, and CP25:plasmid DNA and 2:1 wt ratio of PEI25:plasmid DNA is sufficient to complex and protect plasmid DNA (Table 2). The positive charge present on the surface of synthesized carbon dots enables them for condensing DNA through electrostatic interactions. The negatively charged plasmid DNA is shielded by the positively charged carbon dots, hence does not migrate towards the negative electrode, and remains in the sample loading pocket in gel electrophoresis. Further addition of heparin provides more negative charge to the complex which releases plasmid DNA from the complex. The binding of plasmid DNA with CDs prevents the degradation of plasmid DNA in the presence of serum at similar weight ratios where they exhibit binding affinity so that synthesized carbon dots as vectors protects the nucleic acids from serum degradation at the time of transfection.
From cytotoxicity studies of synthesized carbon dots on A549 and MDA-MB-231 cells, it was found that CS-CDs and CP2-CDs displayed maximum cytocompatibility at 200 μg/mL concentration in both cells thus this concentration is safe for transfection. PEI2-CDs exhibit cytocompatibility at 100 μg/mL whereas, PEI25-CDs and CP25-CDs can be used as gene carriers at low concentrations (25 μg/mL).
Transfection studies in A549 and MDA-MB-231 cells revealed that commercial transfection agent lipofectamine2000 is more toxic and less efficient than synthesized carbon dots (CP25-CDs). PEI2-CDs are less toxic at higher concentration in comparison to lipofectamine2000 and also shows higher transfection in A549 cells than lipofectamine2000 however, the transfection efficiency of PEI2-CDs was less than lipofectamine2000 in MDA-MB-231 cells. PEI25-CDs displayed higher transfection efficiency in A549 and MDA-MB-231 cells than lipofectamine2000 but also shows slight toxicity towards MDA-MB-231 cells at a concentration where higher transfection was achieved. In previous studies, branched PEI passivated carbon dots have been reported to significantly enhance luciferase gene expression (104 times) in comparison to linear PEI passivated carbon dots [30]. A previous report indicated that despite less toxicity, PEI-2kDa exhibited less transfection efficiency, whereas PEI-25kDa exhibited more toxicity and good transfection efficiency [31]. Although the hybrid CDs (CP25-CDs) achieved maximum transfection in both A549 and MDA-MB-231 cells and also exhibits good cytocompatibility at the same concentration (Table 3). In further studies on miRNA binding, the binding affinity of CDs with miRNA-153 was achieved. Various studies support the data that miR-153 is downregulated in breast cancer cells (MDA-MB-231) [32,33] and lung cancer cells, and tissues (A549, H460, H157) [34,35]. MiR-153 can also be used as a potential diagnostic biomarker to diagnose distinct types of cancer [36]. Therefore, the ability of CDs to transfect miRNA was examined in A549 and MDA-MB-231 cells. The obtained data suggested that PEI2-CDs, PEI25-CDs, and CP25-CDs transfected miR-153 successfully into A549 and MDA-MB-231 cells which achieved persistent cell death. As it is already discussed that hybrid CDs (CP25-CDs) achieved the highest transfection in both cell lines and here also the maximum cell death in both cells when miR-153 was transfected with CP25-CDs followed by PEI25-CDs and PEI2-CDs was observed. These data suggested that PEI2-CDs, PEI25-CDs, and CP25-CDs are capable to transfect plasmid DNA as well as miRNA into mammalian cells and also confirms the hypothesis that hybrid CDs (CP25-CDs) are more efficient to transfect nucleic acid into the cancer cells. However future studies should focus on establishing the in vivo anti-cancer efficacy of developed carbon dots as well as the underlying mechanism of gene silencing by miR-153 in transfected cells.
5. Conclusion
In the present study, five different carbon dots: CS-CDs, PEI2-CDs, PEI25-CDs, CP2-CDs and CP25-CDs were successfully constructed. Further studies revealed that these CDs played crucial role and exhibit photoluminescence properties, DNA and miRNA binding efficiency and serum stability for successful transfection. The CS-CDs and CP2-CDs did not exhibit any cytotoxicity towards A549 and MDA-MB-231 cells even at higher concentrations whereas PEI2-CDs showed approximately 40 % cytotoxicity, while PEI25-CDs and CP25-CDs exhibited robust cytotoxic response (65 % and 56 % in A549 and 60 % and 37 % in MDA-MB-231 cells) at 200 μg/mL concentrations but displayed low cytotoxicity (25 %) at low concentration. Further, transfection studies revealed that CS-CDs and CP2-CDs were not competent to transfect A549 and MDA-MB-231 cells whereas, PEI2-CDs, PEI25-CDs, and CP25-CDs could successfully transfect both cell types and also exhibited higher transfection efficiency than lipofectamnine2000. Transfection of miR-153 using PEI2-CDs, PEI25-CDs, and CP25-CDs at various weight ratios demonstrated strong cytotoxic response suggesting that miR-153 was successfully transfected in both cell types that ultimately resulted in death of cancer cells. Out of three CDs those exhibited transfection efficiency, the combination CDs i. e CP25-CDs exhibited the maximum transfection of plasmid DNA as well as miR-153. So, this study demonstrated that the synthesized carbon dots PEI2-CDs, PEI25-CDs, and CP25-CDs exhibited potent nucleic acid transfection ability that can be used as a vector to transfect human cells with long plasmid DNA as well as with short miRNA/siRNA which can be utilized in gene therapy.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Saloni Thakur: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. Reena V. Saini: Conceptualization, Data curation, Investigation, Methodology, Supervision. Neelam Thakur: Investigation, Methodology. Rohit Sharma: Investigation, Methodology, Project administration, Validation. Joydeep Das: Conceptualization, Data curation, Investigation, Resources, Supervision, Writing – original draft, Writing – review & editing. Petr Slama: Funding acquisition, Investigation, Resources, Writing – original draft, Writing – review & editing. Hardeep Singh Tuli: Formal analysis, Supervision, Writing – original draft, Writing – review & editing. Shafiul Haque: Funding acquisition, Project administration, Supervision, Writing – original draft. Hatoon A. Niyazi: Funding acquisition, Methodology, Supervision, Visualization, Writing – review & editing. Mohammed Moulay: Funding acquisition, Investigation, Methodology, Validation, Writing – review & editing. Steve Harakeh: Formal analysis, Funding acquisition, Software, Writing – review & editing. Adesh K. Saini: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research work was funded by the Institutional Fund projects under grant no. (IFPIP:1800-141-1443). Therefore, the authors gratefully acknowledge technical and financial support from the Ministry of Education and King Abdulaziz University, Deanship of Scientific Research, Jeddah, Saudi Arabia. Authors are also thankful to Central Research Cell, MM(DU) for support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21824.
Contributor Information
Rohit Sharma, Email: rohit.sharma@shooliniuniversity.com.
Joydeep Das, Email: jdchem83@gmail.com.
Adesh K. Saini, Email: sainiade@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Thakur S., Saini R.V., Singh P., RaizadA P., Thakur V.K., Saini A.K. Nanoparticles as an emerging tool to alter the gene expression: preparation and conjugation methods. Mater. Today Chem. 2020;17:1–16. doi: 10.1016/j.mtchem.2020.100295. [DOI] [Google Scholar]
- 2.Yuan F., Li S., Fan Z., Meng X., Fan L., Yang S. Shining carbon dots: synthesis and biomedical and optoelectronic applications. Nano Today. 2016;11(5):565–586. doi: 10.1016/j.nantod.2016.08.006. [DOI] [Google Scholar]
- 3.Mehta V.N., Jha S., Kailasa S.K. One-pot green synthesis of carbon dots by using Saccharum officinarum juice for fluorescent imaging of bacteria (Escherichia coli) and yeast (Saccharomyces cerevisiae) cells. Mater. Sci. Eng. C. 2014;38:20–27. doi: 10.1016/j.msec.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 4.Mehta V.N., Jha S., Singhal R.K., Kailasa S.K. Preparation of multicolor emitting carbon dots for HeLa cell imaging. New J. Chem. 2014;38(12):6152–6160. doi: 10.1039/C4NJ00840E. [DOI] [Google Scholar]
- 5.Mehta V.N., Jha S., Basu H., Singhal R.K., Kailasa S.K. One-step hydrothermal approach to fabricate carbon dots from apple juice for imaging of mycobacterium and fungal cells. Sensor. Actuator. B Chem. 2015;213:434–443. doi: 10.1016/j.snb.2015.02.104. [DOI] [Google Scholar]
- 6.Kasibabu B.S., D'souza S.L., Jha S., Singhal R.K., Basu H., Kailasa S.K. One-step synthesis of fluorescent carbon dots for imaging bacterial and fungal cells. Anal. Methods. 2015;7(6):2373–2378. doi: 10.1039/C4AY02737J. [DOI] [Google Scholar]
- 7.Kasibabu B.S., D’souza S.L., Jha S., Kailasa S.K. Imaging of bacterial and fungal cells using fluorescent carbon dots prepared from carica papaya juice. J. Fluoresc. 2015;25(4):803–810. doi: 10.1007/s10895-015-1595-0. [DOI] [PubMed] [Google Scholar]
- 8.Bhamore J.R., Jha S., Singhal R.K., Kailasa S.K. Synthesis of water dispersible fluorescent carbon nanocrystals from Syzygium cumini fruits for the detection of Fe3+ ion in water and biological samples and imaging of Fusarium avenaceum cells. J. Fluoresc. 2017;27(1):125–134. doi: 10.1007/s10895-016-1940-y. [DOI] [PubMed] [Google Scholar]
- 9.D'souza S.L., Deshmukh B., Bhamore J.R., Rawat K.A., Lenka N., Kailasa S.K. Synthesis of fluorescent nitrogen-doped carbon dots from dried shrimps for cell imaging and boldine drug delivery system. RSC Adv. 2016;6(15):12169–12179. doi: 10.1039/C5RA24621K. [DOI] [Google Scholar]
- 10.D'souza S.L., Deshmukh B., Rawat K.A., Bhamore J.R., Lenka N., Kailasa S.K. Fluorescent carbon dots derived from vancomycin for flutamide drug delivery and cell imaging. New J. Chem. 2016;40(8):7075–7083. doi: 10.1039/C6NJ00358C. [DOI] [Google Scholar]
- 11.Mehta V.N., S Chettiar S., Bhamore J.R., Kailasa S.K., Patel R.M. Green synthetic approach for synthesis of fluorescent carbon dots for lisinopril drug delivery system and their confirmations in the cells. J. Fluoresc. 2017;27(1):111–124. doi: 10.1007/s10895-016-1939-4. [DOI] [PubMed] [Google Scholar]
- 12.Shen L.M., Liu J. New development in carbon quantum dots technical applications. Talanta. 2016;156:245–256. doi: 10.1016/j.talanta.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J., Yu S.H. Carbon dots: large-scale synthesis, sensing and bioimaging. Mater. Today. 2016;19(7):382–393. doi: 10.1016/j.mattod.2015.11.008. [DOI] [Google Scholar]
- 14.Wang Q., Zhang C., Shen G., Liu H., Fu H., Cui D. Fluorescent carbon dots as an efficient siRNA nanocarrier for its interference therapy in gastric cancer cells. J. Nanobiotechnol. 2014;12(1):1–2. doi: 10.1186/s12951-014-0058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S., Choi Y., Park G., Won C., Park Y.J., Lee Y., Kim B.S., Min D.H. Highly efficient gene silencing and bioimaging based on fluorescent carbon dots in vitro and in vivo. Nano Res. 2017;10(2):503–519. doi: 10.1007/s12274-016-1309-1. [DOI] [Google Scholar]
- 16.Yang X., Wang Y., Shen X., Su C., Yang J., Piao M., Jia F., Gao G., Zhang L., Lin Q. One-step synthesis of photoluminescent carbon dots with excitation-independent emission for selective bioimaging and gene delivery. J. Colloid Interface Sci. 2017;492:1–7. doi: 10.1016/j.jcis.2016.12.057. [DOI] [PubMed] [Google Scholar]
- 17.Chan K.K., Yap S.H.K., Yong K.T. Biogreen synthesis of carbon dots for biotechnology and nanomedicine applications. Nano-Micro Lett. 2018;10(4):1–46. doi: 10.1007/s40820-018-0223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludmerczki R., Mura S., Carbonaro C.M., Mandity I.M., Carraro M., Senes N., Garroni S., Granozzi G., Calvillo L., Marras S., Malfatti L. Carbon dots from citric acid and its intermediates formed by thermal decomposition. Chem.--Eur. J. 2019;25(51):11963–11974. doi: 10.1002/chem.201902497. [DOI] [PubMed] [Google Scholar]
- 19.Khairol Anuar N.K., Tan H.L., Lim Y.P., So’aib M.S., Abu Bakar N.F. A review on multifunctional carbon-dots synthesized from biomass waste: design/fabrication, characterization and applications. Front. Energy Res. 2021;9:1–22. doi: 10.3389/fenrg.2021.626549. [DOI] [Google Scholar]
- 20.Zhang M., Zhao X., Fang Z., Niu Y., Lou J., Wu Y., Zou S., Xia S., Sun M., Du F. Fabrication of HA/PEI-functionalized carbon dots for tumor targeting, intracellular imaging and gene delivery. RSC Adv. 2017;7(6):3369–3375. doi: 10.1039/C6RA26048A. [DOI] [Google Scholar]
- 21.Liu X., Pang J., Xu F., Zhang X. Simple approach to synthesize amino-functionalized carbon dots by carbonization of chitosan. Sci. Rep. 2016;6(1):1–8. doi: 10.1038/srep31100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karimi M., Avci P., Mobasseri R., Hamblin M.R., Naderi-Manesh H. The novel albumin–chitosan core–shell nanoparticles for gene delivery: preparation, optimization and cell uptake investigation. J. Nanoparticle Res. 2013;15(5):1–14. doi: 10.1007/s11051-013-1651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu D., Yan H., Zhou Z., Tang J., Liu X., Hartmann R., Parak W.J., Feliu N., Shen Y. Detailed investigation on how the protein corona modulates the physicochemical properties and gene delivery of polyethylenimine (PEI) polyplexes. Biomater. Sci. 2018;6(7):1800–1817. doi: 10.1039/C8BM00128F. [DOI] [PubMed] [Google Scholar]
- 24.Liu C., Zhang P., Zhai X., Tian F., Li W., Yang J., Liu Y., Wang H., Wang W., Liu W. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials. 2012;33(13):3604–3613. doi: 10.1016/j.biomaterials.2012.01.052. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Li R., Yang B. Carbon dots: a new type of carbon-based nanomaterial with wide applications. ACS Cent. Sci. 2020;6(12):2179–2195. doi: 10.1021/acscentsci.0c01306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur N., Sharma V., Singh T.A., Pabbathi A., Das J. Fabrication of novel carbon dots/cerium oxide nanocomposites for highly sensitive electrochemical detection of doxorubicin. Diam. Relat. Mater. 2022;125:1–12. [Google Scholar]
- 27.Yang X., Wang Y., Shen X., Su C., Yang J., Piao M., Jia F., Gao G., Zhang L., Lin Q. One-step synthesis of photoluminescent carbon dots with excitation-independent emission for selective bioimaging and gene delivery. J. Colloid Interface Sci. 2017;492:1–7. doi: 10.1016/j.jcis.2016.12.057. [DOI] [PubMed] [Google Scholar]
- 28.A Mathew S., Praveena P., Dhanavel S., Manikandan R., Senthilkumar S., Stephen A. Luminescent chitosan/carbon dots as an effective nano-drug carrier for neurodegenerative diseases. RSC Adv. 2020;10(41):24386–24396. doi: 10.1039/D0RA04599C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandra S., Laha D., Pramanik A., Ray Chowdhuri A., Karmakar P., Sahu S.K. Synthesis of highly fluorescent nitrogen and phosphorus doped carbon dots for the detection of Fe3+ ions in cancer cells. Luminescence. 2016;31(1):81–87. doi: 10.1002/bio.2927. [DOI] [PubMed] [Google Scholar]
- 30.Dou Q., Fang X., Jiang S., Chee P.L., Lee T.C., Loh X.J. Multi-functional fluorescent carbon dots with antibacterial and gene delivery properties. RSC Adv. 2015;5(58):46817–46822. doi: 10.1039/C5RA07968C. [DOI] [Google Scholar]
- 31.Yu M., Lei B., Gao C., Yan J., Ma P.X. Optimizing surface-engineered ultra-small gold nanoparticles for highly efficient miRNA delivery to enhance osteogenic differentiation of bone mesenchymal stromal cells. Nano Res. 2017;10(1):49–63. doi: 10.1007/s12274-016-1265-9. [DOI] [Google Scholar]
- 32.Wang J., Liang S., Duan X. Molecular mechanism of miR‐153 inhibiting migration, invasion and epithelial‐mesenchymal transition of breast cancer by regulating transforming growth factor beta (TGF‐β) signaling pathway. J. Cell. Biochem. 2019;120(6):9539–9546. doi: 10.1002/jcb.28230. [DOI] [PubMed] [Google Scholar]
- 33.Zuo Z., Ye F., Liu Z., Huang J., Gong Y. MicroRNA-153 inhibits cell proliferation, migration, invasion and epithelial-mesenchymal transition in breast cancer via direct targeting of RUNX2. Exp. Ther. Med. 2019;17(6):4693–4702. doi: 10.3892/etm.2019.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan N., Shen L., Wang J., He D., Duan C. MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochem. Biophys. Res. Commun. 2015;456(1):385–391. doi: 10.1016/j.bbrc.2014.11.093. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y., Du W., Wang Y., Xu C., Wang J., Zhang Y., Wang H., Ju J., Zhao L., Wang Z., Lu Y. Suppression of AKT expression by miR‐153 produced anti‐tumor activity in lung cancer. Int. J. Cancer. 2015;136(6):1333–1340. doi: 10.1002/ijc.29103. [DOI] [PubMed] [Google Scholar]
- 36.Thakur S., Saini A.K., Das J., Saini V., Balhara P., Nanda J.S., Saini R.V. miR-153 as biomarker for cancer—functional role as tumor suppressor. Biocell. 2022;46(1):13–26. doi: 10.32604/biocell.2022.016953. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.