Abstract
The AHNAK family currently consists of two members, namely AHNAK and AHNAK2, both of which have a molecular weight exceeding 600 kDa. Homologous sequences account for approximately 90% of their composition, indicating a certain degree of similarity in terms of molecular structure and biological functions. AHNAK family members are involved in the regulation of various biological functions, such as calcium channel modulation and membrane repair. Furthermore, with advancements in biological and bioinformatics technologies, research on the relationship between the AHNAK family and tumors has rapidly increased in recent years, and its regulatory role in tumor progression has gradually been discovered. This article briefly describes the physiological functions of the AHNAK family, and reviews and analyzes the expression and molecular regulatory mechanisms of the AHNAK family in malignant tumors using Pubmed and TCGA databases. In summary, AHNAK participates in various physiological and pathological processes in the human body. In multiple types of cancers, abnormal expression of AHNAK and AHNAK2 is associated with prognosis, and they play a key regulatory role in tumor progression by activating signaling pathways such as ERK, MAPK, Wnt, and MEK, as well as promoting epithelial-mesenchymal transition.
Keywords: AHNAK, AHNAK2, biological function, malignant tumors, prognosis
1. Introduction
The AHNAK family currently contains two members, AHNAK (AHNAK1) and AHNAK2, which share approximately 90% homologous sequences with some similarities in molecular structure and biological function (1).
AHNAK, or AHNAK Nucleoprotein, is also known as Desmoyokin. In 1989, Hieda et al. isolated and identified a new desmosomal plaque protein, Desmoyokin, at the periphery of the desmoplasmic plaques in the bovine oral stratified squamous epithelium (2). In 1992, Shtivelman et al. found a giant protein of round about 700 kDa in size when screening for genes that might be reduced or absent in neuroblastoma cells, and then named it AHNAK (which means giant in Hebrew) (3). Subsequent studies by Hashimoto et al. confirmed that AHNAK and Desmoyokin refer to the same protein (4). AHNAK was initially thought to be encoded by a 17-kb intronless gene on human chromosome 11q12 (5). However, it has been recently reported that the gene contains a giant exon flanked by introns and multiple small exons encoding a giant protein (~700 kDa) and a small protein (17 kDa), and that the commonly studied AHNAK is classified as a giant protein of 700 kDa (6). AHNAK consists of three structural domains: an amino terminal containing 251 amino acids and PDZ domain (N-terminal), a central repeating units (CRUs) containing multiple repeating units of 4,392 amino acids (about 128 residues per repeating unit), and a carboxyl terminal (C-terminal) containing 1002 amino acids, where there are more interaction and regulatory sites (7). AHNAK can be localized in the nucleus, cytoplasm, cell membrane, lysosomes, mitochondria and other structures. Its localization is variable, and the changes may be involved in regulating different functions. As report goes, the cellular localization of AHNAK is regulated by the C-terminal (8).
AHNAK2, or AHNAK Nucleoprotein 2, is also known as C14orf78. It is the second member of the AHNAK family and was originally found in mouse heart extracts (9). AHNAK2 is a giant protein with a molecular weight of over 600 kDa with its coding gene located on human chromosome 14q32, possessing an open reading frame of 15 kb (9). Slightly different from AHNAK, AHNKA2 has at least seven exons, six of which are relatively small together with a large exon of approximately 17,559 bp (9). AHNAK2 contains a total of 5,795 amino acids and consists of a three-part structure: a short non-repetitive N-terminal containing the PDZ structural domain, a central structural domain consisting of 24 repeat units (each containing 165 amino acids) forming the central structural domain, and a C-terminus with a molecular weight size of approximately 100 kDa (9, 10).
This review will focus on the biological functions of AHNAK family members with an emphasis on their role in the development of malignant tumors.
2. Functional introduction to the AHNAK family
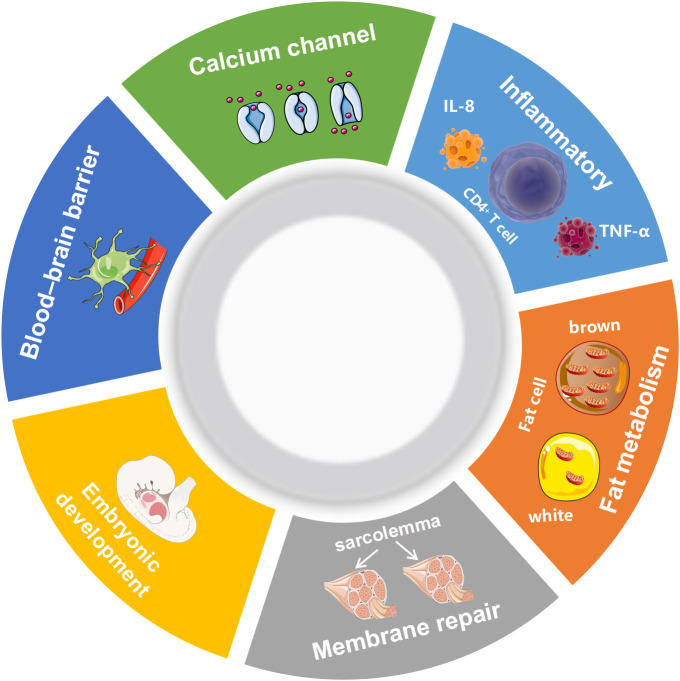
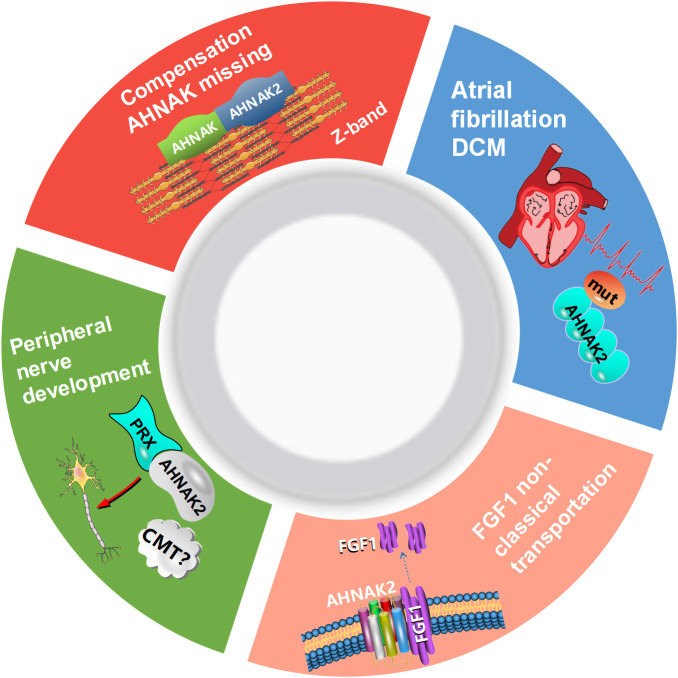
AHNAK can be involved in a wide range of biological processes ( Figure 1 ). It has been recently reported that AHNAK plays a critical role in the regulation of calcium channels, blood-brain barrier formation, embryonic development, lipid metabolism, membrane repair, inflammatory responses and other processes (11). However, compared to AHNAK, the function of AHNAK2 is still relatively poorly studied and its biological function is not well defined. AHNAK2 may play a significant part in sarcolemma assembly, peripheral nervous system development, and cardiac calcium channel regulation ( Figure 2 ). In the following sections, we will briefly review the biological function of AHNAK family.
Figure 1.
Biological processes regulated by AHNAK. TNF-α, Tumor necrosis factor; IL-8, Interleukin 8.
Figure 2.
Biological processes regulated by AHNAK2. CMT, Charcot-Marie-Tooth; DCM, dilated cardiomyopathy; PRX, periaxin; FGF1, fibroblast growth factor.
2.1. Calcium channel regulation
Several studies have confirmed that AHNAK plays an important role in the regulation of calcium channel, in particular L-type voltage gated calcium channels (LVGCC) (1, 12). In cardiomyocytes, AHNAK co-localizes with the β2 subunit of cardiomyocyte L-type calcium channels (Cavβ2) in the myocardium and inhibits LGVCC activity by binding to Cavβ2 through the C-terminal, which will be released for normal function and increased inward calcium flux upon β-adrenergic activation and subsequent protein kinase A (PKA) phosphorylation, thereby regulating cardiomyocyte contraction (13, 14). AHNAK2 and AHNAK can be localized together in the sarcolemmas and Z-bands of mouse cardiomyocytes. It showed no obvious abnormalities to knockdown the AHNAK in mouse models, which suggested that AHNAK2 may compensate for the absence of AHNAK (9). ANRIL is an lncRNA gene whose transcript target is down-regulated in patients with Coronary Artery Disease (CAD) and is involved in the initiation process of CAD. In contrast, decreased expression of AHNAK2 was detected after ANRIL knockdown (15). In patients with atrial fibrillation (AF) and dilated cardiomyopathy, AHNAK2 was one of the three most frequently mutated genes, with respective mutation rates of 52% and 51% (16, 17). These results suggest that AHNAK2 may participate in the regulation of heart disease, although the specific molecular mechanism is not clear. AHNAK is an important player in the regulation of cardiac L-type calcium channels. As its homologue, does AHNAK2 influence the development of cardiac disease by regulating L-type calcium channels? Further studies are needed to clarify its role.
In skeletal muscle, AHNAK can provide mechanical stability during muscle contraction and co-localizes with the β1 subunit of skeletal LVGCC at the T-tubule to regulate skeletal muscle contraction in a manner similar to Cavβ2 regulation (18). In neurons, AHNAK can participate in the regulation of Cav1.2 and Cav1.3 channels, enhancing neuronal excitability and increasing excitatory neurotransmitter release. Besides, in inhibitory synapses, AHNAK can interact with Cavβ4 and participate in its regulation (in a manner similar to Cavβ2 regulation), reducing inhibitory neurotransmitter release and thus ameliorating depressive behavior (19, 20). Additionally, in the presence of arachidonic acid, the center recall unit (CRU) region of AHNAK activates phospholipase C and produces inositol triphosphate and diacylglycerol, which in turn regulate intracellular calcium flux (21). The complete actin cytoskeleton plays a critical part in the regulation of calcium channel currents. AHNAK can interact with G-actin and F-actin, and can participate in cytoskeletal regulation, consequently affecting calcium channel function (22).
2.2. Blood-brain barrier formation
The blood-brain barrier forms the basis for brain homeostasis and low permeability (23). It has been shown that AHNAK is expressed on the plasma membrane of endothelial cells that form the blood-brain barrier but is not found in capillary endothelial cells that undergo molecular exchange between blood and extracellular fluid. Co-culture of astrocytes with brain capillary endothelial cells resulted in the relocalization of AHNAK from the cytoplasm to the plasma membrane when endothelial cells acquired blood-brain barrier properties (24). In the case of spinal cord injury, increased expression of AHNAK was detected in cells with barrier properties and might be involved in the formation of a barrier around the injury parts (25).
2.3. Embryonic development
M. Downs et al. detected AHNAK expression in mouse trophoblast ectoderm derivatives, in the urinary bladder attached to the chorion, and at the tip of the neural plate during neural tube formation (26). A study proved that AHNAK is involved in the formation of germinal bodies and the initial differentiation of pluripotent stem cells by inhibiting c-Myc (27). Impaired migration of enteric neural crest cells during embryonic development leads to a lack of ganglion cells in the distal part of the digestive tract, resulting in congenital megacolonization. It has been reported in studies that there is significant upregulation of AHNAK protein expression and inhibition of cell migration and proliferation in stenotic intestinal tissues from patients with congenital megacolon (28).
2.4. Muscle membrane repair
Dysferlin is a membrane protein involved in skeletal muscle membrane repair, with which AHNAK can co-localize and interact to participate in the sarcolemma repair process (29). During this process, S100A10, annexinA2 and AHNAK can bind to form a complex that promotes faster recruitment of AHNAK proteins to the plasma membrane, then inducing more efficient membrane repair (30).
2.5. Lipid metabolism
Studies show that AHNAK deficiency in mice on a high-fat diet leads to increased insulin sensitivity and that AHNAK plays an important role in β-adrenergic signaling to regulate white adipose tissue browning, catabolism and thermogenesis (31). In hepatocytes, AHNAK deficiency induced upregulation of fibroblast growth factor 21 (FGF21) expression and attenuated fatty liver formation in mice fed a high-fat diet (32). Jong et al. put forward that AHNAK can participate in the regulation of adipocyte differentiation via members of the bone morphogenetic protein family, such as human hone morphogenetic protein 2 (BMP2) and human hone morphogenetic protein 4 (BMP4) (33, 34). These findings confirm that AHNAK plays a critical part in fat metabolism and may have implications for the treatment of obesity or related metabolic diseases.
2.6. Inflammatory response
Studies have found that AHNAK (5758-5775 polypeptide segment) can activate mast cells to release IL-8 and TNF-α, mediated by suppression of tumorigenicity 2 (ST2), and participate in the development and progression of psoriasis (35). In the single nucleated cells and membrane tissues peripheral blood from patients with habitual abortion, the mRNA and protein levels of AHNAK are elevated. And AHNAK with increased CD4+ T cell expression may be involved in the immune dysregulation of RPL in habitual abortion by elevating IL-6 production (36).
2.7. Other functions
In peripheral nerves, AHNAK is of much importance in the formation and maintenance of myelin sheaths (37). Periaxin (PRX) is a protein abundant in the peripheral nervous system and has an important role in the cytoskeleton and myelin formation (38). Both PRX and AHNAK2 have PDZ structures and share more than 50% of the homologous sequences in this region. One report points out that AHNAK2 and PRX can form homodimers, suggesting that AHNAK2 may be involved in regulating cytoskeleton and peripheral nerve development, although further research are required to confirm (39). Charcot-Marie-Tooth (CMT) disease is an inherited peripheral neuropathy affecting motor and sensory neurons. A study using linkage analysis and whole exome sequencing revealed significantly lower mRNA and protein levels of AHNAK2 in fibroblasts from patients with AR-CMT (autosomal recessive CMT), suggesting that AHNAK2 may be involved in the pathogenesis of autosomal recessive CMT in Malaysia (40). These results suggest that the AHNAK family may be involved in the regulation of peripheral nervous system development.
According to research findings, AHNAK/Annexin A2 complex influences the cortical organization of the astral microtubule anchoring complex, and thereby mitotic spindle positioning in human cells (41). One report showed that AHNAK knockout mice had shortened femur and tibia with significantly decreased skeletal strength, as well as morphological abnormalities in the coccyx with age (42). In addition, as a substrate of calpain-3, AHNAK may be closely associated with calpain-specific limb-girdle muscular dystrophy 2A (43). It is suggested that AHNAK may play an important role in skeletons and skeletal muscles development and metabolism. Cleavage of autoantigens by granzyme B and caspase 3 is commonly seen in systemic autoimmune diseases. AHNAK can be cleaved by the both enzymes and identified as a systemic lupus erythematosus (SLE) autoantigen, promising a new target for therapy (44). Although further studies are needed to confirm, AHNAK2 has been identified as a susceptibility gene for SLE by exon sequencing of large samples (45). It has been reported that AHNAK2 and fibroblast growth factor 1 (FGF1) are co-localized near the cell membrane, which are important components in the regulation of non-classical transport of FGF1. It is a non-classically released growth factor that plays an important role in regulating cell growth, tumor invasion, angiogenesis and participates in the regulation of MAPK-ERK and other signaling pathways (46). The role of AHNAKs in malignant tumors has received much attention and will be discussed in detail below.
3. Relationship between AHNAK family and tumor development
3.1. Lung cancer
3.1.1. Lung adenocarcinoma
A study on Lung adenocarcinoma found that UBE3C enhances A549 cell stemness by ubiquitinating the degradation of AHNAK, which in turn disrupts the AHNAK-P53 complex, and that patients with high AHNAK expression have longer survival (47). Another study seemed to confirm the above results, stating that AHNAK expression was decreased in human lung adenocarcinoma, that AHANK-/- mice showed increased lung volume, alveolar wall thickening, and type II alveolar epithelial cell proliferation, and that approximately 20% of aged AHNAK-/- mice developed lung tumors and were more susceptible to lung cancer under urethane induction (48). The above studies seem to indicate a possible oncogenic role of AHNAK in NSCLC. However, it has also been reported that AHNAK expression was elevated during TGF-β induced epithelial mesenchymal transition in human lung adenocarcinoma A549 cells, suggesting that AHNAK may promote invasion and metastasis of Lung adenocarcinoma cells (49).
While the role of AHNAK in lung adenocarcinoma remains controversial, several studies have now confirmed the role of AHNAK2 as a pro-cancer factor in lung cancer. It was reported that AHNAK2 expression was upregulated in lung adenocarcinoma and correlated with poor prognosis, which may be an independent factor in determining prognosis. In addition, the migration of lung adenocarcinoma A549 cells was inhibited by AHNAK2 knockdown (50). Liu et al. reported that downregulation of AHNAK2 inhibited TGF-β1-induced cell migration, invasion, and EMT, and decreased Smad3 signaling activation. Inclusion of Smad3 phosphorylation inhibitors in lung adenocarcinoma cells did not affect the regulation of cell migration, invasion and EMT by TGF-β1 with or without knockdown of AHNAK2, suggesting that AHNAK2 promotes lung adenocarcinoma progression through the TGF-β1/Smad3 pathway (51). It was also reported that downregulation of AHNAK2 expression inhibited the phosphorylation of ERK, which inactivated the MAPK signaling pathway and led to the proliferation and migration of lung adenocarcinoma cells (52). A study showed that AHNAK2 expression in lung adenocarcinoma was negatively correlated with activated B cells, activated CD8+ T cells and immature B cell infiltration, while it was positively correlated with central memory CD4+ T cells, tumor-associated macrophages, M1 macrophages and M2 macrophage infiltration, promising a new target for immunotherapy (53).
3.1.2. Lung squamous carcinoma
Radiation sensitivity index (RSI) may predict the sensitivity of radiotherapy. A study collected and analyzed lung squamous cancer cell carcinoma datasets from both TCGA and GEO (GSE73403 and GSE37745) databases and found significant differences in AHNAK2 expression between high and low RSI groups, suggesting that AHNAK2 may be related to the molecular mechanisms of regulating radiotherapy sensitivity in lung squamous carcinoma (54).
3.1.3. Mutation of AHNAK2 in non-small cell lung cancer
A study reported that The Cancer Genome Atlas (TCGA) database revealed a high mutation rate of AHNAK2 in lung cancer, which reached 18.8%. And a collection of 12 specimens from patients with brain metastases from lung cancer was sequenced, and the mutation rate of AHNAK2 in lung cancer patients with brain metastases was found to be as high as 26.9% (55). Cui et al. found that approximately 11% of NSCLC patients carry AHNAK2 mutations and defined those with a score <-2.5 as deleterious mutations by the PROVEAN tool. Del-AHNAK2-mut was further found to be strongly associated with tumor mutational burden (TMB), neoantigen load (NAL) levels and tumor-infiltrating immune cell (TIIC) to better predict patient PFS and OS (56). These results suggested that mutations in AHNAK2 may potentially be involved in the regulation of non-small cell lung cancer progression.
3.2. Melanoma
HACAT cells, human immortalized keratin-forming cells, were derived from normal skin at the periphery of the lesion of a 62-year-old male melanoma patient, which showed reduced migratory and invasive capacity after AHNAK knockdown. Treatment of the highly metastatic and tumorigenic melanoma cell line B16F10 with TGF-β and shAHNAK further confirmed that AHNAK knockdown resulted in a significant reduction in N-calmodulin expression and Smad3 phosphorylation and that TGF-β did not induce metastasis and invasion in AHNAK knockdown B16F10 cells (57). A study by Suh et al. also confirmed that AHNAK -/- mice were more resistant to lung metastasis by B16F10 cells than wild type mice (58). However, two other studies presented opposite findings. Huang et al. reported that AHNAK expression was downregulated in primary malignant melanoma PMM in ureteral tissue compared to paracancerous ureteral tissue (59). Another study noted that AHNAK expression was significantly downregulated in melanoma and correlated with poor prognosis, and that knockdown of AHNAK in primary melanocytes resulted in reduced expression of E-calcineurin (60). The role of AHNAK in melanoma development still appears to be controversial and needs to be further investigated.
Analysis of the data provided by TCGA and Hodis et al. showed that mutations in AHNAK2 were more common in melanoma patients of older age (over 40 years old). However, the significance of this phenomenon is unclear and has not been further explained or investigated by the authors (61). Li et al. reported that AHNAK2 was highly expressed in uveal melanoma (UM), which is closely associated with high expression and shorter survival in UM, and that the expression of AHNAK2 was higher in primary tumor tissues with metastatic UM than that in primary tumor tissues without metastatic UM. Compared to corneal epithelial cell line D78, the expression of AHNAK2 was significantly upregulated in UM cell lines M17 and SP6.5. And knockdown of AHNAK2 in UM cell lines inhibited the activation of PI3K signaling and reduced cell proliferation, migration and invasion (62).
3.3. Glioma
Zhao et al. examined the expression of AHNAK in six glioma cell lines. Compared to normal glial cell lines (HEB), AHNAK mRNA levels were downregulated in four cell types (SHG-44, A172, U87, U251), especially in the A172, U87 and U251 cell lines (p<0.001). In addition, 30 normal brain tissue and 73 glioma tissue specimens were collected in this study, which confirmed that the low expression of AHNAK in glioma was correlated with poor prognosis (63). Overexpression of AHNAK in U87 and U251 cells inhibited the proliferation and invasion of glioma cells and induced apoptosis (63). Another study using TCGA database analysis showed that AHNAK transcript levels were significantly reduced in glioma stem cells with greater proliferation and migration capacity compared to differentiated glioma cells, although there was no significant difference of AHNAK expression in glioblastoma (GBM) and normal brain tissue (64). These results seem to indicate that AHNAK plays an inhibition role in gliomas. Moreover, the prognosis of glioblastoma patients with AHNAK mutations is worse (median survival of 4.16 months for AHNAK mutated patients compared to 13.53 months for wild-type AHNAK patients), which has been confirmed to act as an independent factor for poor prognosis in GBM (64).
Research in the relationship between AHNAK2 and glioma is still lacking. The Glioma Centre of Southern Medical University reported a rare case of isocitrate dehydrogenase (IDH) wild-type epithelioid glioblastoma with a BRAF V600E mutation and good results with virofenib treatment, in which the AHNAK2 mutation was identified in the whole exome sequencing results, promising a new target for the treatment of GBM (65).
3.4. Laryngeal cancer
A study showed that in normal tissues AHNAK showed only weak or moderate staining with no strong staining present, whereas in laryngeal cancer tissue specimens AHNAK strong staining accounted for 25% (21/83), in which tumor tissue specimens were collected from 83 laryngeal cancer patients and 12 normal tissue specimens (9 tonsils and 3 healthy epithelial tissues of the palatal suspensor) for immunohistochemical analysis. And the patients with high AHNAK expression had a worse prognosis (66). In another study, hanfungin was found to inhibit the growth of laryngeal cancer Hep-2 cells by upregulating AHNAK expression (67). The role of AHNAK in the two reports appears to be inconsistent and needs further investigation. Studies on the relationship between AHNAK2 and laryngeal cancer are lacking. The role of AHNAK2 is not clear.
3.5. Thyroid cancer
In a study of papillary thyroid carcinoma (PTC) in China, tumor and normal tissue specimens from 16 PTC patients were collected and high-throughput sequencing was performed. The analysis showed that AHNAK may be a driver gene for the development of PTC (68).
Compared with AHNAK, AHNAK2 in thyroid cancer has been relatively well studied and been considered to influence thyroid cancer progression as a pro-carcinogenic factor. Several studies using bioinformatics analysis have shown that AHNAK2 expression is upregulated in PTC, indicating poor prognosis. Additional functional analysis has shown that AHNAK2 is closely associated with immune cell infiltration and may contribute to thyroid cancer progression by regulating cell adhesion, cell junctions and immune-related pathways (69, 70). Ye et al. confirmed through various experiments that AHNAK2 is upregulated in thyroid cancer tissues, especially in metastatic thyroid cancer, where its expression is higher, and that high expression of AHNAK2 suggests poor prognosis (71). It was also found that AHNAK2 may achieve regulation of thyroid cancer migration, invasion and lymph node metastasis through the NF-κB signaling pathway (71).
3.6. Breast cancer
MMTV-PyVT (Mouse mammary tumor virus, Polyoma Virus middle T antigen) mice carry murine mammary tumor virus and females develop significant mammary tumors as early as 5 weeks, which are often used in human breast cancer research. The study successfully screened MMTV mice with AHNAK-/- and AHNAK+/+ phenotypes and found that AHNAK-/- mice developed more mammary tumors. It was also found that AHNAK expression was decreased in breast cancer tissues compared to normal tissues, and it was hypothesized that AHNAK may be a tumor suppressor and that its deficiency promoted mammary cell proliferation and tumor development in mice (72). Using the TCGA and METABRIC databases, J. Cimas et al. found that AHNAK is mutated in approximately 5% of basal cell-like breast cancers and that AHNAK mutations are associated with a good prognosis (73). Studies on AHNAK2 and breast cancer are lacking. The biological role of AHNAK2 needs to be further investigated.
3.6.1. Triple-negative breast cancer
The production of vesicles by cancer cells, which promote the migration of recipient fibroblasts, plays an important part in promoting fibroblast migration and regulating the tumor microenvironment. One report showed that AHNAK is more highly expressed in vesicles produced by MDA-MB-231 cells from highly invasive breast cancers compared to the less invasive MC7 cells, and appears to be an essential element for vesicle formation. Treatment of non-transformed fibroblasts with vesicles from MDA-MB-231 cells improved their migratory capacity. The study also found that AHNAK expression was upregulated in invasive ductal carcinoma and metastatic carcinoma compared to normal breast epithelium, with higher expression in metastatic carcinoma in particular (74). These data seem to suggest that AHNAK is abnormally highly expressed in breast cancer and that it can promote tumor progression by inducing fibroblast migration and extracellular matrix destruction around Triple-negative breast cancer (TNBC) cells. However, some studies have suggested the opposite that AHNAK is a suppressor in breast cancer. Chen et al. reported that AHNAK mRNA levels were significantly downregulated in human breast cancer cell lines, particularly in triple-negative breast cancer cell lines, and that downregulation of AHNAK expression was associated with poor prognosis in TNBC. In vivo and in vitro experiments confirmed that AHNAK can regulate signaling pathways such as AKT/MAPK and Wnt/βcatenin to inhibit proliferation and metastasis of TNBC cells (75). In doxorubicin-resistant triple-negative breast cancer cells, AHNAK knockdown was found to prevent a decrease in doxorubicin-regulated activated caspase 7 expression as well as an increase in S-phase, whereas AHNAK overexpression decreased activated caspase 7 expression and induced an increase in S-phase, suggesting that AHNAK may be involved in the regulation of chemoresistance in TNBC (76). The roles of AHNAK and AHNAK2 in lung and breast cancer subtypes are compared in Table S1 .
3.7. Hepatocellular carcinoma
A study examined AHNAK mRNA levels in 60 liver cancer tissues and paracancerous tissues and found that AHNAK expression was significantly elevated in liver cancer tissues, in which the methylation level of AHNAK promoter in peripheral blood mononuclear cells (PBMC) from 260 patients with varying degrees of liver disease was also measured (77). Interestingly, the level of AHNAK methylation was negatively correlated with disease severity, with 44.44% methylation in the normal control group, 38.23% in the chronic hepatitis B group, 34.38% in the compensated cirrhosis group, 31.43% in the decompensated cirrhosis group, and the lowest in the liver cancer group, about 27.7%. It was demonstrated by receiver operating characteristic (ROC) curves that AHNAK methylation can serve as a marker for the diagnosis of hepatocellular carcinoma (Area Under Curve, AUC=0.98). Methylation of CpG islands in the promoter region is known to cause gene silencing, resulting in reduced or absent gene expression. In this study, AHNAK showed hypomethylation and increased expression in HCC, suggesting that AHNAK may act as a cancer promoter. Another study found that GATA binding protein 4 (GATA4) was highly expressed in hepatoblastoma and correlated with the mesenchymal migration phenotype of hepatoblastoma cells, and that GATA4 gene silencing inhibited the migration of HUH6 cells (78). While AHNAK expression decreased after GATA4 knockdown, overexpression of GATA4 caused AHNAK elevation, suggesting that AHNAK upregulation may promote hepatoblastoma progression. Li et al. found that AHNAK can co-localize and interact with insulin-like growth factor 1 (IGF-1R) and promote the growth of hepatocellular carcinoma (HCC) (79). Contrary to the findings reported above, Rui et al. found that upregulated ring finger protein 38 (RNF38) promoted TGF-β signaling through ubiquitinated degradation of AHNAK and induced epithelial-mesenchymal transition in HCC cells. Furthermore, AHNAK knockdown restored the reduced migratory and invasive capacity of HCC cells due to RNF38 downregulation, which appears to indicate the presence of AHNAK as an oncogenic factor in hepatocellular carcinoma (80).
AHNAK2 has rarely been studied in HCC. A study collected 39 specimens of HCC patients from China, 22 of which were subjected to whole exome sequencing, and revealed a high mutation rate in AHNAK2 (22.7%, 5/22), although the high mutation rate in AHNAK2 was not explored in more detail (81).
3.8. Pancreatic cancer
Studies have confirmed that AHNAK is aberrantly highly expressed in pancreatic ductal adenocarcinoma (PDAC) and is associated with shorter disease-free survival and poorer overall survival. Knockdown of AHNAK induced P53 downregulation and inhibited EMT, proliferation and migration of PDAC cells, while addition of P53 protein reversed the effect of AHNAK downregulation on PDAC cells, which suggests that AHNAK may be a novel biomarker for PDAC (82).
Bioinformatic analysis has shown that AHNAK2 expression is upregulated in PDAC. Several studies have demonstrated high diagnostic and predictive prognostic value of clinical prediction models for PDAC constructed with AHNAK2 as a factor (83–85). Specific molecular mechanism studies of AHNAK2 in PDAC are still lacking and need to be further explored and verified.
3.9. Esophageal cancer
One study indicated that SMYD2 can directly methylate AHNAK as well as multiple sites in the CRU region of AHNAK2, which may be involved in regulating cell adhesion, cell signaling, and tumor cell migration and invasion (86). Hou et al. reported that AHNAK2 may be involved in the regulation of radioresistance in esophageal cancer. Knockdown of AHNAK2 resulted in increased radioresistance in esophageal cancer KYSE-150 cells. Whole-exome sequencing analysis of esophageal cancer cells with AHNAK2 knockdown revealed that AHNAK2 may act through NF-κB and TNF signaling pathways to regulate the expression of interleukins, interleukin receptors and chemokines (87).
3.10. Gastric cancer
Studies have shown that upregulation of MicroRNA-93-5p suppressed AHNAK expression, which activated the Wnt signaling pathway and promoted EMT, proliferation, and migration of gastric cancer cells. In contrast, overexpression of AHNAK inhibited the migration, invasion and EMT of gastric cancer HGC-27 cells, which indicates that AHNAK may act as a suppressor to regulate the progression of gastric cancer (88, 89).
Epstein-Barr virus (EBV)-associated gastric cancer (GC) is characterized by high DNA methylation and is more sensitive to 5-fluorouracil and cisplatin chemotherapy. It was reported that AHNAK2 methylation was increased in EBVGC cells compared to normal GC cells. And decreased expression of AHNAK2 in EBVGC metastatic sites was confirmed using IHC. It has also been reported that AHNAK2 may be involved in the regulation of 5-fluorouracil and cisplatin resistance. In summary, Ohmura et al. speculated that gene silencing due to increased AHNAK2 methylation may mediate the regulation of EBVGC sensitivity to chemotherapy, although detailed molecular mechanistic studies are still required to confirm their speculation (90). One study found that gastric cancer patients with PIK3CA, LRP1B and AHNAK2 mutations had a better prognosis, and that investigating the molecular mechanisms of the three-gene interaction has potential value in improving the prognosis of gastric cancer (91). Another study constructed a clinical prediction model for gastric cancer using AHNAK2 as a factor and validated that the model has some predictive value for recurrence and prognosis (92).
3.11. Colorectal cancer
In colorectal cancer, SORBS1 could co-localize with AHNAK and induce ERK phosphorylation and up-regulation of ROCK1 expression through inhibition of AHNAK expression, which promoted the proliferation and migration of colorectal cancer cells, suggesting that AHNAK may act as a suppressor in colorectal cancer (93).
AHNAK2 was identified as a differentially expressed protein in colorectal cancer RKO cells possessing KRAS mutations, without Ras pathway mutations. In addition, AHNAK2 is also a differentially expressed protein in KRAS G13D mutations and BRAF V600E mutations, although the specific roles of these differentially expressed proteins, including AHNAK2, were not further explored (94).
3.12. Renal cancer
AHNAK is lacking in renal cancer studies while AHNAK2 has been more intensively explored and confirmed to be a pro-oncogenic factor. Wang et al. reported that AHNAK2 expression was elevated in renal clear cell carcinoma and correlated with poorer overall survival (95). Downregulation of AHNAK2 inhibits lipid synthesis and thus affects tumor cell metabolism. According to chromatin immunoprecipitation and dual luciferase reporter gene analysis, HIF1α and the hre1 promoter of AHANK2 bind, and hypoxia-induced upregulation of AHNAK2 expression is dependent on HIF1α. Knockdown of AHNAK2 impairs hypoxia-induced epithelial mesenchymal transition and stem cell properties of tumor cells.
3.13. Bladder cancer
In one study, 10 urine samples were collected from patients with bladder urothelial carcinoma (BLCA) and benign urothelial lesion (BUL), and 3 tissue specimens were collected for proteomic analysis. Four candidate BLCA diagnostic proteins including AHNAK, EPPK1, MYH14 and OLFM4 were screened using public databases such as TCGA for joint comparisons. It was further confirmed by cellular immunohistochemistry that only AHNAK2 could discriminate well between BLCA and BUL samples. The expression of AHNAK was also found to be significantly decreased in BLCA compared to BUL, which is consistent with the results in the TCGA database, suggesting that AHNAK may be a suppressor gene and diagnostic marker for bladder cancer (96). However, Okusa et al. reported that AHNAK was more highly expressed in the cytoplasmic membrane of uroepithelial cancer tissues compared to adjacent normal tissues (97). The two studies reached inconsistent conclusions. Lee et al. speculated that the inconsistency might be due to the two different processing performed on samples by immunocytochemistry and immunohistochemistry (96). In another study, a clinical prediction model was constructed for bladder cancer using AHNAK as a factor, which verified that the model is of some predictive value for diagnosis and prognosis (98, 99).
Immunohistochemical analysis was performed in a study and it was found that the sensitivity and specificity of AHNAK2 for identifying severe cystitis with reactive urothelial atypia (RUA) and carcinoma in situ (CIS) were 97% and 69% respectively, 97% and 55% between CIS and low-grade invasive bladder cancer, and 80% and 86% between low-grade and high-grade invasive bladder cancer, suggesting that AHNAK2 may be a valuable pathological diagnostic marker for IHC pathology in bladder lesions (100). Koguchi et al. collected pathological specimens from 120 patients who underwent radical resection for bladder cancer for IHC analysis and divided the cases into two groups of high and low AHNAK2 expression based on staining. They found that patients with high AHNAK2 expression had significantly lower recurrence-free survival (RFS) and cancer-specific survival (CSS). Multifactorial analysis showed that high AHNAK2 expression could be an independent risk factor for worse RFS and CSS (101). A similar conclusion was reached in another study in which Komina et al. collected urine samples from 67 cancer patients with bladder occupancy (bladder cancer group N=37, benign bladder tumors N=30) and found that the mean urinary AHNAK2 protein level was almost 10 times higher in bladder cancer patients than that in controls (49.08 pg/mL vs. 5.28 pg/mL) and that AHNAK2 expression level in invasive bladder cancer was significantly higher than that in non-invasive bladder cancer (117.99 pg/mL vs. 7.14 pg/mL). Similarly, AHNAK2 level was significantly higher in muscle-invasive bladder cancer than that in non-muscle-invasive bladder cancer (160.05 pg/mL vs. 23.19 pg/mL) (102). A large sample analysis using the TCGA and GEO databases has been reported to confirm that AHNAK2 is overexpressed in bladder cancer and is associated with poorer overall survival (103).
3.14. Prostate cancer
In castration-resistant prostate cancer (CRPC), AHNAK is a downstream target molecule of BRD4, and knockdown of AHNAK or BRD4 inhibits the migration of prostate cancer cells. A decrease in AHNAK expression and cell migration can be seen in treatment of CRPC cells with MZ1, which degrades BRD4. Analysis using the TCGA database revealed a significant correlation between BRD4 and AHNAK mRNA expression with high expression of both being associated with poorer recurrence-free survival (104). Another study reported that AHNAK mutation rate was approximately 4.82% in prostate cancer, ranking it fourth, although it was not studied in depth (105). Studies on the relationship between AHNAK2 and prostate cancer are still lacking.
3.15. Tumors in female reproductive system
It was found that AHNAK expression was downregulated in ovarian cancer. Knockdown of AHNAK in ovarian cancer cells was seen to activate the Wnt signaling pathway, and overexpression of AHNAK inhibited cell proliferation, migration, and EMT. The authors hypothesized that AHNAK might regulate ovarian cancer progression through the Wnt pathway (106). The interesting thing is that another report indicated AHNAK overexpression in ovarian cancer was associated with poor prognosis, yet in vitro, knockdown of AHNAK inhibited the proliferation, migration, and invasion of CAOV3 and SKOV3 cells in ovarian cancer (107). The authors speculated that this “paradoxical” phenomenon may be related to epigenetic modifications and multiple protein interactions, indicating a complex role of AHNAK proteins in tumor regulation.
A study found by immunohistochemistry and bioinformatic analysis that AHNAK2 was highly expressed in cervical adenocarcinoma and was associated with a poorer prognosis. Moreover, knockdown of AHNAK2 inhibited the value-added and metastasis of cervical cancer Hela cells (108). Using mouse embryonic fibroblasts, Lee et al. confirmed that AHNAK interacts with the receptor-activated Smad (R-Smad, including Smad1, Smad2, and Smad3) and found that it can bind to endogenous Smad3 and regulate the nuclear translocation of Smad3, playing a key role in transforming growth factor-β (TGF-β) induced R-Smad activation (72). Furetermore, AHNAK can induce c-Myc and cell cycle protein D1 downregulation through TGFβ/R-Smad, leading to cervical squamous cell carcinoma cell (siHa) arrest in the G0/G1 phase and inhibition of cell growth (72).
3.16. Other malignant tumors
Sudo et al. found that AHNAK was overexpressed in mesothelioma tissues. In the study they examined AHNAK expression in eight mesothelioma cell lines and found that seven mesothelioma cell lines (211H, H28, H226, H2052, H2452, MESO1, and MESO4) had high AHNAK expression, and only MeT-5A had no detectable AHNAK expression. Migration and invasion of the other seven mesothelioma cells were significantly elevated compared to the AHNAK-deficient MeT-5A cell line, although knockdown of AHNAK significantly inhibited the migration and invasion of the seven cells (109).
AHNAK2 mutations in cutaneous squamous cell carcinoma was identified in a study, in which its role was not further explored (110). Saito et al. collected 10 surgically resected specimens of thymic carcinoma and its paraneoplastic tissues for genomic and epigenomic aberration studies. The sequencing revealed an amplification of AHNAK2 located in the region of chromosome 14q32.33, which may be the oncogene of thymic carcinoma, although more in-depth studies are needed to elucidate the specific mechanism (111).
3.17. Briefly summarize the signal pathways related to AHNAK family in malignant tumors
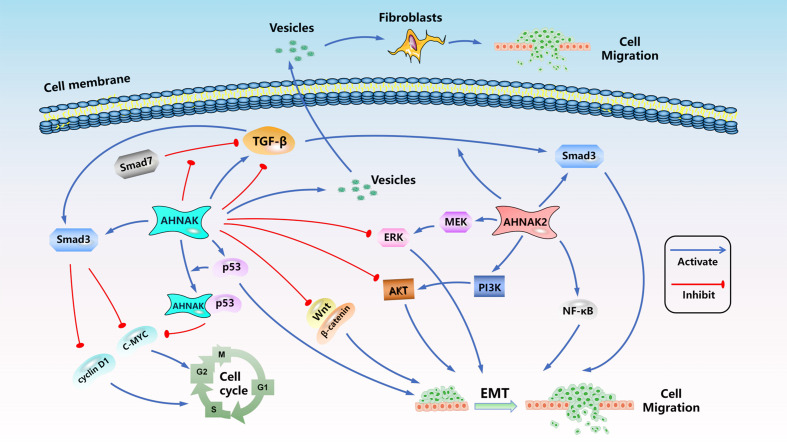
Hitherto, the exhaustive molecular mechanisms and signaling pathways regulation of AHNAK and AHNAK2 are still in the exploratory stage. One research had pointed out that AHNAK can relieve the inhibitory effect of samd7 on TGF-β, and could inhibit the expression of c-MYC and cyclin D1 by activating the TGF-β/smad3 pathway, resulting in cell cycle arrest (72). AHNAK can also inhibit EMT and migration of tumor cells by inducing inactivation of TGF-β signaling (80). Nevertheless, another study reported that AHNAK is an indispensable factor in tumor cell migration induced by TGF-β (57). TGF-β has two distinct roles in malignancy: tumor suppression based on the induction of growth arrest and apoptosis, and tumor suppression based on the induction of angiogenic capacity and epithelial mesenchymal transition (EMT) (112). AHNAK is closely related to TGF-β. It is speculated that the different functions it exhibits in cancer cells may be related to TGF-β. The results suggested that the regulation of TGF-β signaling pathway by AHNAK seems to be bidirectional. It has been reported that AHNAK2 can promote tumor cell invasion, migration and EMT by activating TGF-β/smad3 signaling (51). At present, there are no reports about bidirectional regulation of TGF-β signal by AHNAK2. P53 plays crucial roles in the regulation of tumor progression, studies have shown that AHNAK could form a complex with P53 and inhibit cell cycle (47). Knockdown AHNAK induced down-regulation of p53 and inhibited tumor cell migration and EMT (82). Silva et al. reported that AHNAK could be transported to extracellular through vesicles and act on fibroblasts, induce fibroblast migration and ECM destruction, and promote tumor progression (74). AHNAK seems to have more inhibitory effect on the regulation of signal pathway. AHNAK could inhibit the signals of Wnt/β-catenin, ERK and AKT, resulting in the decrease of EMT, migration and invasion of tumor cells (75, 88, 93, 106). However, AHNAK2 seems to exist more as an activator of signal pathways, which could promote the activation of PI3K/AKT, MEK/ERK and NF-κB signals and induce EMT, invasion and migration of tumor cells (52, 62, 71). We briefly summarized the AHNAKs-related signal pathways in Figure 3 . As the AHNAK family, which has been paid more attention recently, its regulatory mechanism in tumor process urgently needs more in-depth research.
Figure 3.
The signal pathways related to AHNAK family in malignant tumors. ( : Activation effect,
: Activation effect,  : Inhibition effect).
: Inhibition effect).
4. AHNAKs in cancers in bioinformatics database
4.1. The expression of AHNAKs in cancers
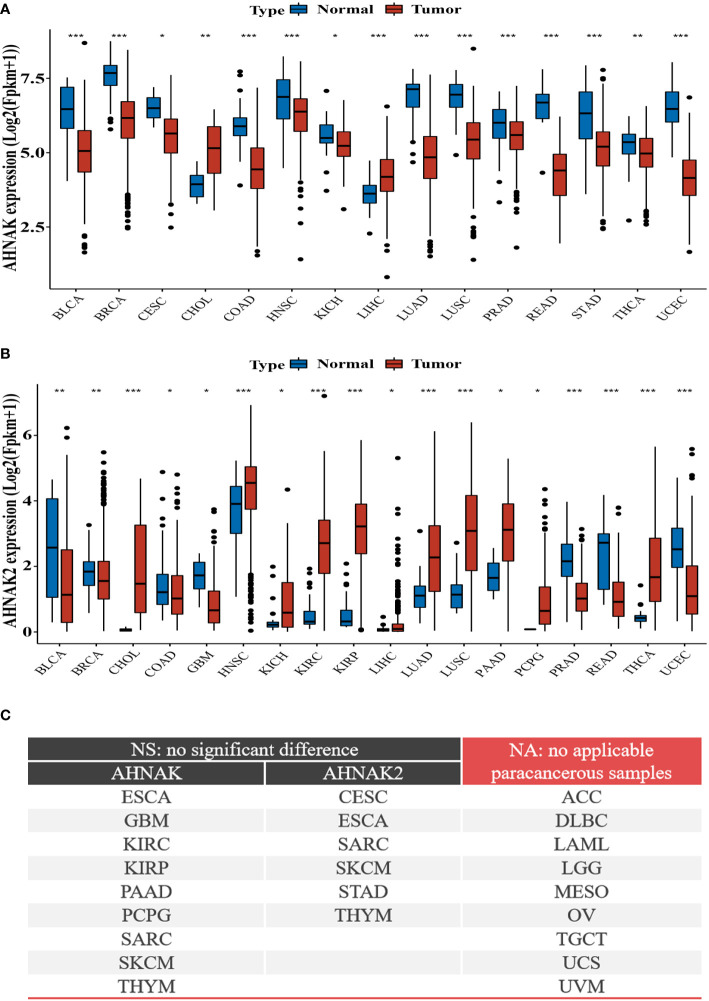
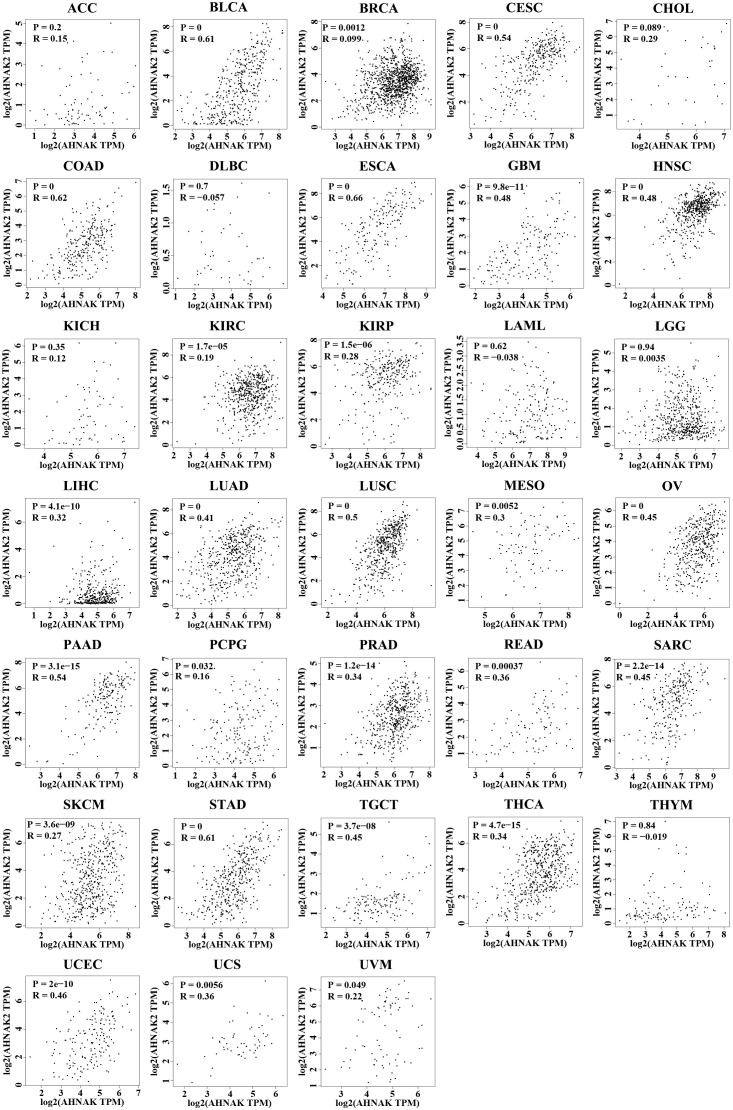
To further analyze the relationship between AHNAKs and cancers, the mRNA expression and clinical data of 33 cancers in TCGA were collected from UCSC Xena database (https://xenabrowser.net/datapages/). We used R language (4.1.0) to analysis the data. It turned out that AHNAK and AHNAK2 were significantly differentially expressed in cancer and paraneoplastic tissues of some tumors. Among them, AHNAK expression was downregulated in BLCA, BRCA, CESC, COAD, HNSC, KICH, LUAD, LUSC, PRAD, READ, STAD, THCA, UCEC, while upregulated in CHOL and LIHC ( Figure 4A ). For AHNAK2, its expression was downregulated in BLCA, BRCA, COAD, GBM, PRAD, READ and UCEC while up-regulated in CHOL, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PAAD, PCPG and THCA ( Figure 4B ). Figure 4C illustrated cancer types with no significant differences or no applicable paracancerous samples. Detailed data on differences in expression between tumor and normal tissues were shown in Table ( Table 1 ). Moreover, because of AHNAK and AHNAK2 exhibit roughly 90% homologous sequences, with certain resemblances in their molecular structures and biological functions, we examined the correlation in mRNA expression between AHNAK and AHNAK2 from different tumor types in the TCGA datasets in GEPIA (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/) database. The mRNA expression of AHNAK and AHNAK2 showed significant postive correlation in 27 types of tumors, only 6 types of tumors (ACC, DLBC KICH, LAML, LGG, THYM) showed no significant correlation ( Figure 5 ). Interestingly, there was no significant negative correlation in any type of tumor. Detailed correlation data were shown in Table ( Table 2 ).
Figure 4.
Differential expression of AHNAKs mRNA in tumor and corresponding non-tumor tissues. (A) AHNAK downregulation in BLCA, BRCA, CESC, COAD, HNSC, KICH, LUAD, LUSC, PRAD, READ, STAD, THCA, UCEC, and upregulation in CHOL, LIHC. (B) AHNAK2 downregulation in BLCA, BRCA, COAD, GBM, PRAD, READ, UCEC and upregulation in CHOL, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PAAD, PCPG, THCA. (C) Cancer types with no significant differences or no applicable paracancerous samples. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. (NA, no applicable paracancerous samples; NS, no significance; *, p<0.05; **, p<0.01; ***, p<0.001).
Table 1.
Differential expression of AHNAKs mRNA in tumor and corresponding non-tumor tissues.
| Cancer Type |
Number of Cases | AHNAK (Exp: Mean ± SD) | AHNAK2 (Exp: Mean ± SD) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor | Normal | Tumor Exp | Normal Exp | logFC | P-Value | Tumor | Normal | logFC | P-Value | |
| ACC | 79 | – | 3.131 ± 0.990 | – | – | – | 0.457 ± 0.605 | – | – | – |
| BLCA | 408 | 19 | 4.983 ± 1.047 | 6.399 ± 0.965 | -1.416 | <0.001* | 1.530 ± 1.420 | 2.565 ± 1.562 | -1.036 | 0.003* |
| BRCA | 1098 | 113 | 6.012 ± 0.948 | 7.603 ± 0.599 | -1.591 | <0.001* | 1.662 ± 0.902 | 1.800 ± 0.530 | -0.137 | 0.002* |
| CESC | 306 | 3 | 5.500 ± 0.879 | 6.517 ± 0.678 | -1.017 | 0.046* | 2.744 ± 1.288 | 2.961 ± 1.037 | -0.217 | 0.830 |
| CHOL | 36 | 9 | 5.058 ± 0.975 | 3.911 ± 0.486 | 1.147 | 0.002* | 1.853 ± 1.448 | 0.069 ± 0.048 | 1.784 | <0.001* |
| COAD | 458 | 41 | 4.419 ± 0.953 | 5.905 ± 0.731 | -1.486 | <0.001* | 1.208 ± 0.870 | 1.558 ± 1.078 | -0.349 | 0.038* |
| DLBC | 48 | – | 3.468 ± 1.196 | – | – | – | 0.139 ± 0.141 | – | – | – |
| ESCA | 162 | 11 | 5.745 ± 0.838 | 6.151 ± 1.049 | -0.406 | 0.185 | 3.326 ± 1.599 | 2.574 ± 2.209 | 0.752 | 0.183 |
| GBM | 167 | 5 | 3.239 ± 0.921 | 3.567 ± 0.508 | -0.329 | 0.441 | 0.878 ± 0.759 | 1.663 ± 0.654 | -0.785 | 0.017* |
| HNSC | 502 | 44 | 6.233 ± 0.878 | 6.751 ± 0.893 | -0.518 | <0.001* | 4.265 ± 1.226 | 3.583 ± 1.185 | 0.683 | <0.001* |
| KICH | 65 | 24 | 5.229 ± 0.718 | 5.543 ± 0.698 | -0.314 | 0.045* | 0.946 ± 0.971 | 0.389 ± 0.497 | 0.557 | 0.027* |
| KIRC | 531 | 72 | 6.066 ± 0.770 | 6.133 ± 0.567 | -0.067 | 0.517 | 2.643 ± 1.151 | 0.474 ± 0.390 | 2.168 | <0.001* |
| KIRP | 289 | 32 | 5.588 ± 0.916 | 5.682 ± 0.470 | -0.094 | 0.861 | 3.020 ± 1.247 | 0.545 ± 0.508 | 2.475 | <0.001* |
| LAML | 151 | – | 6.385 ± 0.991 | – | – | – | 0.627 ± 0.522 | – | – | – |
| LGG | 525 | – | 4.479 ± 0.937 | – | – | – | 0.550 ± 0.537 | – | – | – |
| LIHC | 373 | 50 | 4.171 ± 0.811 | 3.622 ± 0.509 | 0.549 | <0.001* | 0.275 ± 0.558 | 0.075 ± 0.076 | 0.200 | |
| LUAD | 515 | 59 | 4.764 ± 1.115 | 6.835 ± 0.706 | -2.071 | <0.001* | 2.291 ± 1.270 | 1.102 ± 0.509 | 1.189 | <0.001* |
| LUSC | 501 | 49 | 5.327 ± 0.953 | 6.833 ± 0.621 | -1.506 | <0.001* | 2.994 ± 1.441 | 1.202 ± 0.519 | 1.792 | <0.001* |
| MESO | 86 | – | 6.029 ± 0.703 | – | – | – | 2.544 ± 1.177 | – | – | – |
| OV | 379 | – | 4.815 ± 0.877 | – | – | – | 2.106 ± 1.014 | – | – | – |
| PAAD | 178 | 4 | 5.716 ± 0.796 | 5.414 ± 0.306 | 0.302 | 0.132 | 2.973 ± 1.245 | 1.715 ± 0.688 | 1.258 | 0.0300* |
| PCPG | 183 | 3 | 3.610 ± 0.964 | 3.665 ± 0.479 | -0.054 | 0.910 | 0.982 ± 0.946 | 0.080 ± 0.015 | 0.902 | 0.023* |
| PRAD | 496 | 52 | 5.516 ± 0.765 | 5.908 ± 0.748 | -0.392 | <0.001* | 1.095 ± 0.613 | 2.169 ± 0.857 | -1.073 | <0.001* |
| READ | 167 | 10 | 4.251 ± 0.952 | 6.505 ± 0.935 | -2.254 | <0.001* | 1.108 ± 0.802 | 2.401 ± 1.168 | -1.293 | <0.001* |
| SARC | 263 | 2 | 5.642 ± 0.882 | 5.322 ± 0.866 | 0.319 | 0.598 | 2.557 ± 1.373 | 1.956 ± 2.402 | 0.600 | 0.611 |
| SKCM | 471 | 1 | 4.794 ± 1.071 | 5.133 | – | – | 1.836 ± 1.388 | 1.020 | – | – |
| STAD | 375 | 32 | 5.127 ± 0.930 | 6.2 ± 1.052 | -1.073 | <0.001* | 2.100 ± 1.321 | 2.828 ± 1.910 | -0.728 | 0.059 |
| TGCT | 156 | – | 4.212 ± 0.970 | – | – | – | 0.651 ± 0.490 | – | – | – |
| THCA | 510 | 58 | 4.929 ± 0.756 | 5.236 ± 0.617 | -0.307 | 0.001* | 1.901 ± 1.209 | 0.439 ± 0.194 | 1.462 | <0.001* |
| THYM | 119 | 2 | 3.901 ± 1.326 | 5.612 ± 2.351 | -1.711 | 0.173 | 0.447 ± 0.675 | 0.778 ± 0.812 | -0.330 | 0.309 |
| UCEC | 544 | 35 | 4.157 ± 0.868 | 6.439 ± 0.797 | -2.282 | <0.001* | 1.386 ± 1.062 | 2.507 ± 1.011 | -1.121 | <0.001* |
| UCS | 56 | – | 4.041 ± 0.850 | – | – | – | 1.251 ± 0.630 | – | – | – |
| UVM | 80 | – | 4.321 ± 0.709 | – | – | – | 2.673 ± 1.444 | – | – | – |
Exp, expression; SD, Standard Deviation; LogFC, Log2 fold change; -, no applicable datas; *, P-value < 0.05 was considered statistically significant.
Figure 5.
The correlation in mRNA expression between AHNAK and AHNAK2 from different tumor types in the TCGA. (R: Pearson correlation coefficient, p < 0.05 was considered statistically significant).
Table 2.
The correlation in mRNA expression between AHNAK and AHNAK2 from different tumor types in the TCGA.
| Positive | Negative | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | R | P | Type | R | P | Type | R | P | None | ||
| BLCA | 0.610 | <0.001** | LIHC | 0.320 | <0.001* | SARC | 0.450 | <0.001** | Non-correlation | ||
| BRCA | 0.099 | 0.001* | LUAD | 0.410 | <0.001* | SKCM | 0.270 | <0.001* | Type | R | P |
| CESC | 0.540 | <0.001* | LUSC | 0.500 | <0.001* | STAD | 0.610 | <0.001* | ACC | 0.15 | 0.2000 |
| COAD | 0.620 | <0.001* | MESO | 0.300 | 0.005* | TGCT | 0.450 | <0.001* | CHOL | 0.29 | 0.089 |
| ESCA | 0.620 | <0.001* | OV | 0.450 | <0.001* | THCA | 0.340 | <0.001* | DLBC | -0.057 | 0.700 |
| GBM | 0.480 | <0.001* | PAAD | 0.540 | <0.001** | UCEC | 0.460 | <0.001* | KICH | 0.12 | 0.350 |
| HNSC | 0.480 | <0.001* | PCPG | 0.160 | 0.032* | UCS | 0.360 | 0.006* | LAML | -0.038 | 0.620 |
| KIRC | 0.190 | <0.001* | PRAD | 0.340 | <0.001** | UVM | 0.220 | 0.049* | LGG | 0.004 | 0.940 |
| KIRP | 0.280 | <0.001* | READ | 0.360 | <0.001* | THYM | -0.019 | 0.840 | |||
R: Pearson correlation coefficient, *: p < 0.05 was considered statistically significant.
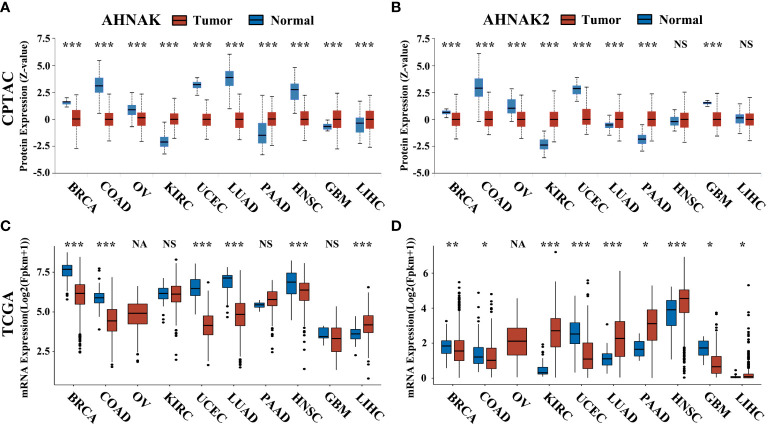
In addition, we analyezed the protein expression of AHNAK and AHNAK2 in CPTAC through UALCAN database (https://ualcan.path.uab.edu/). In the CPTAC database of UALCAN, 8 types of tumors have relatively complete comparative data of protein expression in tumor and normal samples. Among them, AHNAK expression was downregulated in BRCA, COAD, OV, UCEC, LUAD and HNSC, while upregulated in KIRC, PAAD, GBM and LIHC ( Figure 6A ). For AHNAK2, its expression was downregulated in BRCA, COAD, OV and GBM, while upregulated in KIRC, PAAD, LUAD and PAAD ( Figure 6B ). Understanding the correlation between protein abundance and RNA level is crucial, so we performed simultaneous comparisons. The protein and mRNA expression of AHNAK had different trends in KIRC and GBM, while AHNAK2 was not consistent only in LIHC ( Figure 6 ). The results revealed that the protein and mRNA levels of AHNAK and AHNAK2 were highly consistent.
Figure 6.
The protein expression of AHNAK and AHNAK2 in CPTAC and the corresponding mRNA expression in TCGA. (A, B) The AHNAK or AHNAK2 protein expression of eight cancers in CPTAC. (C, D) The AHNAK or AHNAK2 mRNA expression of the eight cancers in TCGA. (NA, no applicable paracancerous samples; NS, no significance; *, p<0.05; **, p<0.01; ***, p<0.001).
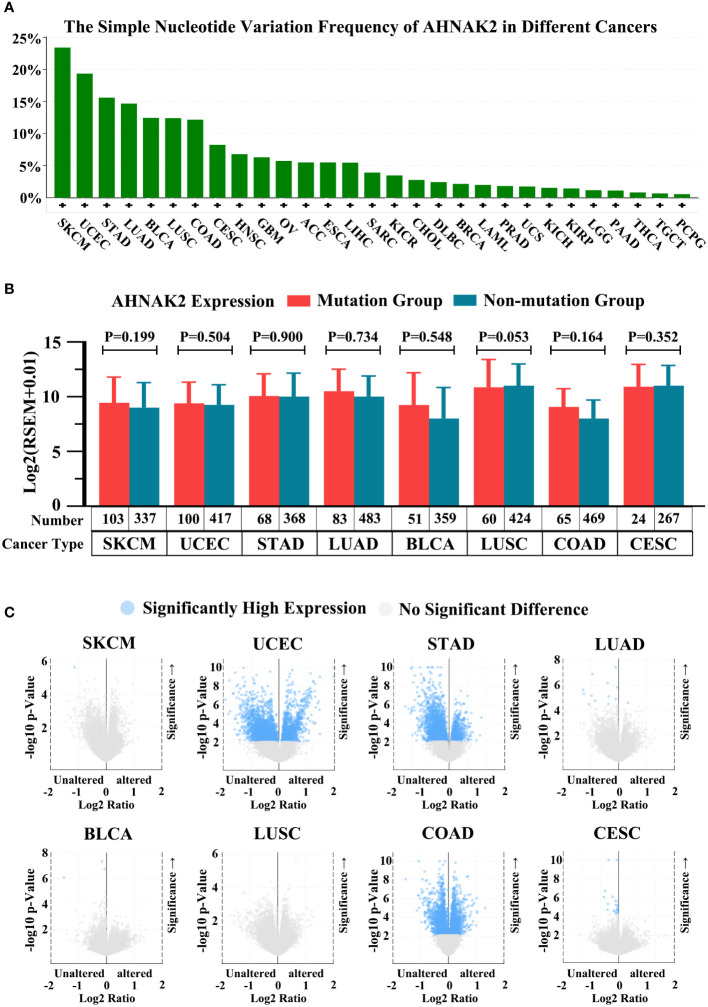
In present studies reported AHNAK2 is among the most frequently mutated genes in several tumor types. It is common knowledge that gene mutations may affect its expression levels, consequently, we analyzed the simple nucleotide variation frequency in different cancers and the mRNA expression of AHNAK2 between mutation and not-mutation samples in TCGA in cBioPortal database (https://www.cbioportal.org/). As shown in Figure 7 , AHNAK2 gene mutations were found in a variety of tumors, and the mutation frequency in SKCM was as high as 24% ( Figure 7A ). We selected the top eight cancers for subsequent analyses, including SKCM, UCEC, STAD, LUAD, BLCA, LUSC, COAD and CESC in turn. Confusingly, AHNAK2 mutation did not seem to have a significant effect on mRNA expression ( Figure 7B ). Of course, due to the limitations of the sample and other reasons, more research is needed in the future. Moreover, We also analyzed the expression of other genes after the mutation of AHNAK2, the brief results are shown in Figure 7 in the form of a volcano map and the detailed results can be seen in the Supplementary Material ( Figure 7C , Supplementary Excel ). Due to the lack of in-depth researches, the upstream and downstream genes of AHNAK2 were not yet fully determined, so we have not further analyzed and discussed the results.
Figure 7.
Analysis of AHNAK2 mutations in cancers. (A) The simple nucleotide variation frequency of AHNAK2 in different cancers in TCGA. (B) The mRNA expression of AHNAK2 in mutation groups and non-mutation groups. (C) Comparison of mRNA expression of other genes in AHNAK2 mutation groups and non-mutation groups. (p < 0.05 was considered statistically significant).
4.2. AHNAKs and prognosis
Using univariate regression analysis, we examined the associations of AHNAK and AHNAK2 with the prognosis of 33 cancers by R language ( Tables 3 , 4 ). It is worth proposing that AHNAK2 was significantly associated with all four indicators including OS, PFS, DFS and DSS in lung adenocarcinoma patients, which is also consistent with our previous findings (50).
Table 3.
Contribution of the expression of AHNAK to survival by univariate Cox regression analysis in cancers.
| Cancer Type | OS | PFS | DFS | DSS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | HR | 95%CI | P-value | HR | 95%CI | P | |
| ACC | 1.129 | 0.750-1.698 | 0.561 | 1.170 | 0.847-1.617 | 0.341 | 1.210 | 0.636-2.300 | 0.561 | 1.091 | 0.715-1.665 | 0.686 |
| BLCA | 1.380 | 1.180-1.615 | 0.000* | 1.290 | 1.103-1.508 | 0.001* | 0.956 | 0.667-1.371 | 0.807 | 1.442 | 1.188-1.750 | 0.000* |
| BRCA | 0.980 | 0.827-1.160 | 0.812 | 0.941 | 0.793-1.117 | 0.489 | 0.857 | 0.690-1.064 | 0.162 | 0.870 | 0.699-1.082 | 0.211 |
| CESC | 1.335 | 0.997-1.787 | 0.053 | 1.068 | 0.810-1.408 | 0.642 | 0.886 | 0.567-1.383 | 0.593 | 1.278 | 0.918-1.780 | 0.146 |
| CHOL | 0.874 | 0.541-1.411 | 0.582 | 0.953 | 0.601-1.511 | 0.837 | 1.411 | 0.672-2.960 | 0.363 | 0.947 | 0.567-1.583 | 0.836 |
| COAD | 1.049 | 0.844-1.304 | 0.667 | 1.064 | 0.885-1.279 | 0.508 | 1.095 | 0.736-1.630 | 0.653 | 1.058 | 0.819-1.366 | 0.667 |
| DLBC | 1.236 | 0.672-2.275 | 0.495 | 1.280 | 0.763-2.148 | 0.350 | 1.278 | 0.512-3.192 | 0.599 | 1.232 | 0.487-3.115 | 0.659 |
| ESCA | 0.720 | 0.542-0.957 | 0.024* | 0.794 | 0.614-1.027 | 0.079 | 0.940 | 0.569-1.551 | 0.807 | 0.742 | 0.534-1.032 | 0.076 |
| GBM | 1.121 | 0.949-1.325 | 0.179 | 1.113 | 0.930-1.331 | 0.243 | – | – | – | 1.143 | 0.954-1.370 | 0.147 |
| HNSC | 1.036 | 0.879-1.221 | 0.676 | 0.911 | 0.771-1.075 | 0.270 | 0.918 | 0.580-1.453 | 0.716 | 1.011 | 0.817-1.251 | 0.921 |
| KICH | 0.774 | 0.320-1.871 | 0.569 | 0.665 | 0.303-1.458 | 0.309 | 0.697 | 0.151-3.215 | 0.643 | 0.859 | 0.312-2.367 | 0.769 |
| KIRC | 0.757 | 0.641-0.894 | 0.001* | 0.837 | 0.698-1.003 | 0.054 | 1.117 | 0.634-1.969 | 0.702 | 0.708 | 0.579-0.867 | 0.001* |
| KIRP | 0.930 | 0.664-1.302 | 0.672 | 0.690 | 0.530-0.898 | 0.006* | 0.743 | 0.500-1.104 | 0.141 | 0.685 | 0.469-1.001 | 0.051 |
| LAML | 1.142 | 0.926-1.409 | 0.214 | – | – | – | – | – | – | – | – | – |
| LGG | 0.880 | 0.733-1.057 | 0.172 | 0.929 | 0.801-1.079 | 0.335 | 0.978 | 0.618-1.546 | 0.923 | 0.937 | 0.771-1.139 | 0.512 |
| LIHC | 0.966 | 0.777-1.202 | 0.758 | 0.989 | 0.816-1.198 | 0.910 | 0.860 | 0.693-1.068 | 0.172 | 0.939 | 0.707-1.248 | 0.665 |
| LUAD | 1.091 | 0.958-1.242 | 0.190 | 1.042 | 0.922-1.176 | 0.511 | 0.957 | 0.798-1.147 | 0.632 | 1.103 | 0.936-1.301 | 0.241 |
| LUSC | 1.069 | 0.919-1.244 | 0.387 | 1.188 | 0.982-1.437 | 0.077 | 0.946 | 0.714-1.254 | 0.699 | 1.141 | 0.894-1.456 | 0.290 |
| MESO | 1.156 | 0.803-1.665 | 0.436 | 1.097 | 0.755-1.594 | 0.628 | 0.791 | 0.342-1.827 | 0.583 | 1.068 | 0.670-1.701 | 0.783 |
| OV | 1.142 | 0.985-1.324 | 0.079 | 0.957 | 0.837-1.093 | 0.514 | 0.912 | 0.751-1.106 | 0.349 | 1.166 | 0.994-1.367 | 0.059 |
| PAAD | 1.743 | 1.287-2.36 | 0.000* | 1.701 | 1.284-2.252 | 0.000* | 2.870 | 1.409-5.846 | 0.004* | 1.775 | 1.261-2.499 | 0.001* |
| PCPG | 1.006 | 0.489-2.069 | 0.988 | 0.951 | 0.618-1.464 | 0.820 | 1.523 | 0.554-4.186 | 0.414 | 1.178 | 0.504-2.754 | 0.706 |
| PRAD | 0.669 | 0.333-1.341 | 0.257 | 0.779 | 0.602-1.008 | 0.057 | 0.779 | 0.488-1.242 | 0.294 | 0.438 | 0.157-1.220 | 0.114 |
| READ | 0.769 | 0.490-1.209 | 0.255 | 1.004 | 0.685-1.471 | 0.985 | 1.290 | 0.425-3.911 | 0.653 | 0.528 | 0.293-0.951 | 0.034* |
| SARC | 0.758 | 0.594-0.967 | 0.026* | 0.777 | 0.636-0.949 | 0.014* | 0.865 | 0.651-1.148 | 0.316 | 0.654 | 0.499-0.858 | 0.002* |
| SKCM | 0.981 | 0.863-1.114 | 0.763 | 0.955 | 0.858-1.063 | 0.401 | – | – | – | 0.937 | 0.818-1.073 | 0.346 |
| STAD | 1.113 | 0.929-1.333 | 0.244 | 1.105 | 0.910-1.343 | 0.313 | 1.149 | 0.813-1.623 | 0.431 | 1.122 | 0.893-1.411 | 0.324 |
| TGCT | 1.619 | 0.528-4.959 | 0.399 | 1.074 | 0.787-1.464 | 0.654 | 0.971 | 0.695-1.358 | 0.865 | 1.792 | 0.524-6.127 | 0.352 |
| THCA | 1.224 | 0.619-2.421 | 0.561 | 0.908 | 0.639-1.291 | 0.591 | 1.076 | 0.658-1.759 | 0.770 | 0.707 | 0.275-1.817 | 0.472 |
| THYM | 1.019 | 0.617-1.683 | 0.941 | 0.912 | 0.652-1.276 | 0.591 | – | – | – | 0.644 | 0.262-1.583 | 0.338 |
| UCEC | 0.984 | 0.777-1.247 | 0.895 | 0.917 | 0.749-1.124 | 0.405 | 0.846 | 0.628-1.141 | 0.274 | 1.039 | 0.779-1.386 | 0.793 |
| UCS | 0.823 | 0.574-1.180 | 0.289 | 0.773 | 0.542-1.102 | 0.155 | 0.572 | 0.281-1.166 | 0.124 | 0.708 | 0.481-1.043 | 0.081 |
| UVM | 0.686 | 0.344-1.365 | 0.283 | 0.922 | 0.509-1.670 | 0.789 | – | – | – | 0.652 | 0.314-1.353 | 0.251 |
OS, Overall survival, PFS, Progression free survival, DFS, Disease free survival, DSS, Disease specific survival. *, P-value < 0.05 was considered statistically significant.
Table 4.
Contribution of the expression of AHNAK2 to survival by univariate Cox regression analysis in cancers.
| Cancer Type | OS | PFS | DFS | DSS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | HR | 95%CI | P-value | HR | 95%CI | P | |
| ACC | 0.932 | 0.475-1.826 | 0.837 | 0.825 | 0.463-1.470 | 0.514 | 1.108 | 0.400-3.068 | 0.844 | 0.725 | 0.319-1.647 | 0.443 |
| BLCA | 1.210 | 1.096-1.336 | 0.000*8** | 1.193 | 1.079-1.319 | 0.001* | 1.151 | 0.908-1.459 | 0.244 | 1.281 | 1.138-1.442 | 0.000* |
| BRCA | 1.070 | 0.889-1.289 | 0.473 | 1.282 | 1.078-1.525 | 0.005* | 1.361 | 1.096-1.690 | 0.005* | 1.206 | 0.950-1.530 | 0.124 |
| CESC | 1.110 | 0.924-1.334 | 0.264 | 0.977 | 0.815-1.171 | 0.799 | 0.806 | 0.598-1.087 | 0.158 | 1.123 | 0.908-1.390 | 0.285 |
| CHOL | 1.115 | 0.811-1.532 | 0.504 | 1.102 | 0.816-1.489 | 0.527 | 1.239 | 0.759-2.021 | 0.391 | 1.142 | 0.816-1.598 | 0.440 |
| COAD | 1.165 | 0.940-1.445 | 0.163 | 1.344 | 1.119-1.614 | 0.002* | 1.435 | 0.949-2.169 | 0.087 | 1.356 | 1.047-1.756 | 0.021* |
| DLBC | 2.853 | 0.019-418.4 | 0.680 | 0.001 | 0.000-3.901 | 0.097 | 0.014 | 0-30614.376 | 0.565 | 0.005 | 0.0-2540.95 | 0.426 |
| ESCA | 0.887 | 0.761-1.035 | 0.128 | 0.963 | 0.842-1.102 | 0.585 | 1.111 | 0.853-1.447 | 0.433 | 0.865 | 0.721-1.038 | 0.119 |
| GBM | 1.147 | 0.948-1.388 | 0.160 | 1.277 | 1.060-1.539 | 0.010* | – | – | – | 1.173 | 0.962-1.430 | 0.114 |
| HNSC | 1.071 | 0.952-1.204 | 0.256 | 0.989 | 0.880-1.112 | 0.857 | 0.871 | 0.639-1.187 | 0.381 | 1.054 | 0.908-1.225 | 0.488 |
| KICH | 1.166 | 0.574-2.370 | 0.671 | 1.200 | 0.625-2.304 | 0.584 | 1.423 | 0.527-3.844 | 0.487 | 1.086 | 0.473-2.491 | 0.846 |
| KIRC | 1.096 | 0.961-1.250 | 0.172 | 0.981 | 0.857-1.124 | 0.786 | 0.977 | 0.633-1.507 | 0.915 | 1.074 | 0.910-1.267 | 0.397 |
| KIRP | 1.032 | 0.802-1.328 | 0.806 | 0.873 | 0.709-1.074 | 0.199 | 0.842 | 0.630-1.127 | 0.248 | 0.931 | 0.691-1.253 | 0.636 |
| LAML | 1.342 | 0.925-1.947 | 0.121 | – | – | – | – | – | – | – | – | – |
| LGG | 1.251 | 0.968-1.617 | 0.087 | 1.217 | 0.965-1.535 | 0.097 | 0.429 | 0.136-1.356 | 0.149 | 1.237 | 0.940-1.626 | 0.128 |
| LIHC | 1.061 | 0.790-1.425 | 0.695 | 1.016 | 0.785-1.315 | 0.902 | 0.948 | 0.690-1.303 | 0.743 | 1.114 | 0.783-1.585 | 0.550 |
| LUAD | 1.305 | 1.156-1.472 | 0.000* | 1.222 | 1.094-1.365 | 0.000* | 1.260 | 1.068-1.485 | 0.006* | 1.347 | 1.156-1.571 | 0.000* |
| LUSC | 0.997 | 0.903-1.101 | 0.955 | 1.051 | 0.933-1.185 | 0.410 | 0.911 | 0.763-1.089 | 0.308 | 1.040 | 0.892-1.212 | 0.618 |
| MESO | 1.244 | 1.027-1.506 | 0.025* | 1.179 | 0.961-1.446 | 0.115 | 1.556 | 0.806-3.003 | 0.188 | 1.308 | 1.022-1.674 | 0.033* |
| OV | 1.180 | 1.042-1.335 | 0.009* | 0.996 | 0.886-1.119 | 0.944 | 0.984 | 0.827-1.171 | 0.855 | 1.185 | 1.035-1.356 | 0.014* |
| PAAD | 1.479 | 1.236-1.772 | 0.000* | 1.401 | 1.187-1.654 | 0.000* | 1.904 | 1.295-2.799 | 0.001* | 1.473 | 1.205-1.800 | 0.000* |
| PCPG | 0.381 | 0.102-1.427 | 0.152 | 0.795 | 0.490-1.289 | 0.352 | 0.958 | 0.356-2.583 | 0.933 | 0.218 | 0.027-1.764 | 0.153 |
| PRAD | 0.884 | 0.326-2.398 | 0.808 | 0.658 | 0.463-0.935 | 0.019* | 0.450 | 0.235-0.861 | 0.016* | 0.399 | 0.063-2.537 | 0.330 |
| READ | 1.296 | 0.832-2.018 | 0.251 | 1.318 | 0.898-1.934 | 0.158 | 1.268 | 0.465-3.456 | 0.643 | 0.963 | 0.487-1.903 | 0.913 |
| SARC | 0.835 | 0.720-0.969 | 0.017* | 0.868 | 0.766-0.983 | 0.026* | 0.857 | 0.716-1.026 | 0.094 | 0.818 | 0.695-0.963 | 0.016* |
| SKCM | 1.115 | 1.017-1.221 | 0.020* | 1.035 | 0.956-1.120 | 0.393 | – | – | – | 1.070 | 0.967-1.184 | 0.188 |
| STAD | 1.097 | 0.975-1.235 | 0.125 | 1.152 | 1.016-1.306 | 0.028* | 1.158 | 0.931-1.440 | 0.186 | 1.139 | 0.980-1.323 | 0.090 |
| TGCT | 0.067 | 0.000-15.07 | 0.328 | 1.168 | 0.625-2.183 | 0.626 | 0.964 | 0.486-1.912 | 0.917 | 0.008 | 0.000-13.35 | 0.203 |
| THCA | 1.139 | 0.762-1.703 | 0.526 | 1.316 | 1.059-1.636 | 0.013* | 1.258 | 0.930-1.702 | 0.137 | 1.000 | 0.538-1.858 | 0.999 |
| THYM | 1.308 | 0.577-2.965 | 0.520 | 1.427 | 0.889-2.291 | 0.141 | – | – | – | 0.501 | 0.033-7.715 | 0.620 |
| UCEC | 1.264 | 1.056-1.513 | 0.011* | 1.054 | 0.892-1.245 | 0.539 | 0.933 | 0.717-1.214 | 0.606 | 1.378 | 1.116-1.700 | 0.003* |
| UCS | 0.821 | 0.464-1.452 | 0.497 | 0.775 | 0.456-1.318 | 0.347 | 0.774 | 0.283-2.115 | 0.617 | 0.819 | 0.462-1.452 | 0.495 |
| UVM | 1.937 | 1.344-2.791 | 0.000* | 1.969 | 1.410-2.751 | 0.000** | – | – | – | 2.091 | 1.404-3.113 | 0.000* |
OS, Overall survival; PFS, Progression free survival; DFS, Disease free survival; DSS, Disease specific survival. *, P-value < 0.05 was considered statistically significant.
4.3. Functional Enrichment of AHNAKs in TCGA database
We also respectively screened the 50 genes most closely associated with AHNAK or AHNAK2 expression in GEPIA using data from tumor tissue specimens of 33 cancers in TCGA, in order to explore the function of AHNAKs ( Table 5 ). Gene Ontology analysis was performed by virtue of DAVID Bioinformatics Resources (containing three parts: Biological Process, BP; Cellular Component, CC; Molecular Function, MF, with the top 10 enriched functions for each section, https://david.ncifcrf.gov/). We found that AHNAK has a relatively complex functional enrichment, including Wnt, MAPK/AKT signaling pathway, cell adhesion, membrane repair, calcium channels and other related ( Table 6 ). In contrast, AHNAK2 function was relatively concentrated, mainly related to cell adhesion, cytoskeleton structure and calcium channel regulation ( Table 7 ).
Table 5.
AHNAKs expression related genes.
| AHNAK expression related genes (Top 50) | AHNAK2 expression related genes (Top 50) | ||||
|---|---|---|---|---|---|
| Gene Symbol | Gene ID | PCC | Gene Symbol | Gene ID | PCC |
| ATL3 | ENSG00000184743.12 | 0.6 | KRT6A | ENSG00000205420.10 | 0.58 |
| LRRFIP1 | ENSG00000124831.18 | 0.6 | GJB3 | ENSG00000188910.7 | 0.58 |
| TMOD3 | ENSG00000138594.12 | 0.6 | PKP1 | ENSG00000081277.11 | 0.58 |
| HIPK3 | ENSG00000110422.11 | 0.58 | GJB5 | ENSG00000189280.3 | 0.58 |
| SLAIN2 | ENSG00000109171.14 | 0.58 | KRT5 | ENSG00000186081.11 | 0.58 |
| SP1 | ENSG00000185591.9 | 0.58 | BICD2 | ENSG00000185963.13 | 0.57 |
| ELMSAN1 | ENSG00000156030.12 | 0.56 | PERP | ENSG00000112378.11 | 0.57 |
| MAP3K2 | ENSG00000169967.16 | 0.56 | TUBA4A | ENSG00000127824.13 | 0.56 |
| MYOF | ENSG00000138119.16 | 0.56 | MICALL1 | ENSG00000100139.13 | 0.56 |
| SLK | ENSG00000065613.13 | 0.56 | CLCA2 | ENSG00000137975.7 | 0.56 |
| TRIP11 | ENSG00000100815.12 | 0.56 | LAMB3 | ENSG00000196878.12 | 0.56 |
| AP4E1 | ENSG00000081014.10 | 0.55 | DSC3 | ENSG00000134762.16 | 0.56 |
| C16orf72 | ENSG00000182831.11 | 0.55 | FAM83G | ENSG00000188522.14 | 0.56 |
| FBXW2 | ENSG00000119402.16 | 0.55 | SERPINB5 | ENSG00000206075.13 | 0.55 |
| CSNK1G1 | ENSG00000169118.15 | 0.54 | DSG3 | ENSG00000134757.4 | 0.55 |
| MFAP3 | ENSG00000037749.11 | 0.54 | SFN | ENSG00000175793.11 | 0.55 |
| SEC24B | ENSG00000138802.11 | 0.54 | DSP | ENSG00000096696.13 | 0.54 |
| CSNK1A1 | ENSG00000113712.16 | 0.53 | TRIM29 | ENSG00000137699.16 | 0.54 |
| SPOPL | ENSG00000144228.8 | 0.53 | KCTD11 | ENSG00000213859.4 | 0.54 |
| SUSD6 | ENSG00000100647.7 | 0.53 | KRT14 | ENSG00000186847.5 | 0.54 |
| WASF2 | ENSG00000158195.10 | 0.53 | TLDC1 | ENSG00000140950.15 | 0.53 |
| BTBD7 | ENSG00000011114.14 | 0.52 | ANXA2 | ENSG00000182718.16 | 0.53 |
| CNOT6L | ENSG00000138767.12 | 0.52 | FAT2 | ENSG00000086570.12 | 0.53 |
| KLF7 | ENSG00000118263.14 | 0.52 | BNC1 | ENSG00000169594.12 | 0.53 |
| LUZP1 | ENSG00000169641.13 | 0.52 | COL17A1 | ENSG00000065618.16 | 0.52 |
| MARK2 | ENSG00000072518.20 | 0.52 | PKP3 | ENSG00000184363.9 | 0.52 |
| MTF1 | ENSG00000188786.9 | 0.52 | TUBB6 | ENSG00000176014.12 | 0.52 |
| SPTY2D1 | ENSG00000179119.14 | 0.52 | ANXA2P2 | ENSG00000231991.4 | 0.52 |
| TRIP12 | ENSG00000153827.13 | 0.52 | DUSP7 | ENSG00000164086.9 | 0.52 |
| VPS4B | ENSG00000119541.9 | 0.52 | LAMC2 | ENSG00000058085.14 | 0.52 |
| EFCAB14 | ENSG00000159658.10 | 0.51 | KRT17 | ENSG00000128422.15 | 0.51 |
| JAK1 | ENSG00000162434.11 | 0.51 | CDH3 | ENSG00000062038.13 | 0.51 |
| MYH9 | ENSG00000100345.20 | 0.51 | ADGRF4 | ENSG00000153294.11 | 0.51 |
| STRN | ENSG00000115808.11 | 0.51 | KLC3 | ENSG00000104892.16 | 0.5 |
| TMEM127 | ENSG00000135956.8 | 0.51 | FAM83B | ENSG00000168143.8 | 0.5 |
| UBN1 | ENSG00000118900.14 | 0.51 | CERS3 | ENSG00000154227.13 | 0.5 |
| ZFP91 | ENSG00000186660.14 | 0.51 | IL20RB | ENSG00000174564.12 | 0.5 |
| AFF4 | ENSG00000072364.12 | 0.5 | RP3-523K23.2 | ENSG00000261116.1 | 0.5 |
| ASXL2 | ENSG00000143970.16 | 0.5 | TENM2 | ENSG00000145934.15 | 0.5 |
| CAB39 | ENSG00000135932.10 | 0.5 | SLC2A1 | ENSG00000117394.19 | 0.49 |
| CACUL1 | ENSG00000151893.14 | 0.5 | DSC2 | ENSG00000134755.14 | 0.49 |
| FKBP15 | ENSG00000119321.8 | 0.5 | GJB2 | ENSG00000165474.5 | 0.49 |
| GAPVD1 | ENSG00000165219.21 | 0.5 | ITGA6 | ENSG00000091409.14 | 0.49 |
| MYO9A | ENSG00000066933.15 | 0.5 | GSDMC | ENSG00000147697.8 | 0.48 |
| NUMB | ENSG00000133961.19 | 0.5 | LAD1 | ENSG00000159166.13 | 0.48 |
| RIC1 | ENSG00000107036.11 | 0.5 | LAMA3 | ENSG00000053747.15 | 0.48 |
| ZDHHC5 | ENSG00000156599.10 | 0.5 | FSCN1 | ENSG00000075618.17 | 0.48 |
| ADAM10 | ENSG00000137845.14 | 0.49 | FAM83A | ENSG00000147689.16 | 0.48 |
| CAST | ENSG00000153113.23 | 0.49 | IFFO2 | ENSG00000169991.10 | 0.48 |
| CTNND1 | ENSG00000198561.12 | 0.49 | S100A2 | ENSG00000196754.10 | 0.48 |
PCC, Pearson Correlation Coefficient.
Table 6.
Gene ontology analysis of AHNAK by DAVID Bioinformatics Resources.
| Category | Term | Count | P-Value | FDR |
|---|---|---|---|---|
| BP | peptidyl-serine phosphorylation | 5 | 0.001 | 0.150 |
| BP | Wnt signaling pathway | 5 | 0.001 | 0.150 |
| BP | protein phosphorylation | 7 | 0.002 | 0.150 |
| BP | plasma membrane repair | 3 | 0.002 | 0.150 |
| BP | endocytosis | 4 | 0.015 | 0.940 |
| BP | intracellular signal transduction | 5 | 0.023 | 1.000 |
| BP | interleukin-15-mediated signaling pathway | 2 | 0.026 | 1.000 |
| BP | actin filament-based movement | 2 | 0.040 | 1.000 |
| BP | Golgi organization | 3 | 0.040 | 1.000 |
| BP | RPSMNRL | 2 | 0.049 | 1.000 |
| CC | cytosol | 23 | 0.002 | 0.200 |
| CC | cytoplasm | 23 | 0.003 | 0.200 |
| CC | adherens junction | 4 | 0.008 | 0.380 |
| CC | membrane | 16 | 0.011 | 0.390 |
| CC | nucleus | 22 | 0.013 | 0.390 |
| CC | early endosome | 4 | 0.032 | 0.700 |
| CC | cytoskeleton | 5 | 0.034 | 0.700 |
| CC | growth cone | 3 | 0.038 | 0.700 |
| CC | glutamatergic synapse | 4 | 0.064 | 0.88 |
| CC | macromolecular complex | 5 | 0.066 | 0.88 |
| MF | cadherin binding | 9 | 0.000 | 0.000 |
| MF | protein serine/threonine/tyrosine kinase activity | 7 | 0.001 | 0.028 |
| MF | protein serine/threonine kinase activity | 6 | 0.002 | 0.079 |
| MF | actin binding | 5 | 0.009 | 0.180 |
| MF | ATP binding | 10 | 0.009 | 0.180 |
| MF | protein kinase activity | 5 | 0.012 | 0.200 |
| MF | protein binding | 38 | 0.017 | 0.230 |
| MF | protein homodimerization activity | 6 | 0.030 | 0.340 |
| MF | small GTPase binding | 4 | 0.032 | 0.340 |
| MF | histone acetyltransferase binding | 2 | 0.053 | 0.520 |
BP, Biological Process; CC, Cellular Component; MF, Molecular Function. RPSMNRL, regulation of postsynaptic specialization membrane neurotransmitter receptor levels.
Table 7.
Gene ontology analysis of AHNAK2 by DAVID Bioinformatics Resources.
| Category | Term | Count | P-Value | FDR |
|---|---|---|---|---|
| BP | cell-cell adhesion | 14 | 0.000 | 0.000 |
| BP | epidermis development | 10 | 0.000 | 0.000 |
| BP | intermediate filament organization | 5 | 0.000 | 0.002 |
| BP | cell adhesion | 9 | 0.000 | 0.003 |
| BP | keratinization | 5 | 0.000 | 0.003 |
| BP | system development | 3 | 0.000 | 0.006 |
| BP | cell-cell junction assembly | 4 | 0.000 | 0.006 |
| BP | HCAVPMAM | 5 | 0.001 | 0.024 |
| BP | cornification | 3 | 0.001 | 0.024 |
| BP | tissue development | 3 | 0.003 | 0.080 |
| CC | cornified envelope | 10 | 0.000 | 0.000 |
| CC | desmosome | 7 | 0.000 | 0.000 |
| CC | adherens junction | 9 | 0.000 | 0.000 |
| CC | cell-cell junction | 8 | 0.000 | 0.000 |
| CC | intermediate filament | 7 | 0.000 | 0.000 |
| CC | basement membrane | 6 | 0.000 | 0.000 |
| CC | cell junction | 7 | 0.000 | 0.000 |
| CC | extracellular exosome | 16 | 0.000 | 0.001 |
| CC | connexin complex | 3 | 0.001 | 0.013 |
| CC | keratin filament | 4 | 0.002 | 0.019 |
| MF | structural constituent of cytoskeleton | 6 | 0.000 | 0.000 |
| MF | cadherin binding | 8 | 0.000 | 0.000 |
| MF | calcium ion binding | 10 | 0.000 | 0.002 |
| MF | structural molecule activity | 6 | 0.000 | 0.002 |
| MF | cadherin binding involved in cell-cell adhesion | 3 | 0.001 | 0.015 |
| MF | gap junction channel activity | 3 | 0.001 | 0.019 |
| MF | structural constituent of epidermis | 3 | 0.003 | 0.042 |
| MF | extracellular matrix structural constituent | 4 | 0.005 | 0.049 |
| MF | phosphatidylserine binding | 3 | 0.010 | 0.095 |
| MF | CAPBIBHCPMC | 2 | 0.012 | 0.095 |
BP, Biological Process; CC, Cellular Component; MF, Molecular Function. HCAVPMAM, homophilic cell adhesion via plasma membrane adhesion molecules; CAPBIBHCPMC, cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication.
5. Conclusion and prospect
Given all that, the AHNAK family has a variety of important biological functions, such as calcium channel regulation, barrier formation, and cytoskeleton and cell adhesion regulation, and plays an important part in the development of malignant tumors. The AHNAK family mediates the regulation of tumor cell migration and invasion through various pathways. From the current studies, it appears that AHNAK may play different roles in different tumors, even though within the same tumor its role is controversial, such as TNBC, melanoma and bladder cancer. Unfortunately, the role of AHNAK in all cancer types reported in three or more studies has been somewhat controversial. However, AHNAK2 seems more likely to act as an oncogenic gene. The aberrant overexpression of AHNAK2 has been consistently recognized to promote the progression of lung adenocarcinoma, thyroid cancer, and bladder cancer (with at least three reports for each type). However, as a tumor-related huge protein family that has only been highlighted in recent years, in-depth molecular mechanistic studies on this family, especially regarding the regulation of EMT and downstream signaling pathways such as P53, MAPK/AKT, TGF-β by the AHNAK family are lacking and need to be further explored in the future with a view to providing new targets for tumor diagnosis and treatment.
Author contributions
SZ: Conceptualization, Data curation, Funding acquisition, Writing – original draft. ZC: Conceptualization, Supervision, Writing – review & editing. HL: Conceptualization, Supervision, Writing – review & editing.
Acknowledgments
We acknowledge the TCGA, UCSC, GEPIA, UALCAN and cBioPortal databases for free use.
Glossary
| ACC | adrenocortical carcinoma |
| AF | atrial fibrillation |
| AKT | protein kinase B |
| AR-CMT | autosomal recessive CMT |
| AUC | area under curve |
| BLCA | bladder urothelial carcinoma |
| BLCA | bladder urothelial carcinoma |
| BMP2 | human hone morphogenetic protein 2 |
| BMP4 | human hone morphogenetic protein 4 |
| BP | biological Process |
| BRCA | breast invasive carcinoma |
| BRD4 | bromodomain containing 4 |
| BUL | benign urothelial lesion |
| CAD | coronary artery disease |
| Cavβ2 | the β2 subunit of cardiomyocyte L type calcium channels |
| Cavβ4 | the β4 subunit of cardiomyocyte L-type calcium channels |
| CC | cellular component |
| CESC | cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | cholangiocarcinoma |
| CIS | carcinoma in situ |
| CMT | charcot marie tooth |
| COAD | colon adenocarcinoma |
| CRPC | castration-resistant prostate cancer |
| CRU | center recall unit |
| CSS | cancer-specific survival |
| DFS | disease free survival |
| DLBC | lymphoid neoplasm diffuse large B-cell lymphoma |
| DSS | disease specific survival |
| EBV | epstein barr virus |
| EMT | epithelial mesenchymal transition |
| EPPK1 | Human Epiplakin 1 |
| ERK | extracellular regulated protein kinases |
| ESCA | esophageal carcinoma |
| FGF1 | fibroblast growth factor 1 |
| FGF21 | fibroblast growth factor 21 |
| GATA4 | GATA binding protein 4 |
| GBM | glioblastoma |
| GBM | glioblastoma multiforme |
| GC | gastric cancer |
| GEO | gene omnibus |
| GO | gene ontology |
| HCC | hepatocellular carcinoma |
| HIF1α | hypoxia inducible factor-1α |
| HNSC | head and neck squamous cell carcinoma |
| IDH | isocitrate dehydrogenase |
| IGF-1R | interact with insulin-like growth factor 1 receptor |
| IHC | immunohistochemistry |
| KICH | kidney chromophobe |
| KIRC | kidney renal clear cell carcinoma |
| KIRP | kidney renal papillary cell carcinoma |
| LAML | acute myeloid leukemia |
| LGG | brain lower grade glioma |
| LIHC | liver hepatocellular carcinoma |
| LRP1B | low density lipoprotein receptor-related protein 1B |
| LUAD | lung adenocarcinoma |
| LUSC | lung squamous cell carcinoma |
| LVGCC | type voltage gated calcium channels |
| MAPK | mitogen-activated protein kinase |
| MESO | mesothelioma |
| MF | molecular function |
| MMTV | mouse mammary tumor virus |
| MYH14 | myosin heavy chain 14 |
| NAL | neoantigen load levels |
| NF-κB | nuclear factor kappa B |
| OLFM4 | Olfactomedin-4 |
| OS | overall survival |
| OV | ovarian serous cystadenocarcinoma |
| PAAD | pancreatic adenocarcinoma |
| PBMC | peripheral blood mononuclear cells |
| PCPG | pheochromocytoma and paraganglioma |
| PDAC | pancreatic ductal adenocarcinoma |
| PFS | progression free survival |
| PI3K | Phosphatidylinositol 3 hydroxykinase |
| PIK3CA | phosphatidylinositol 3-kinase catalytic alpha |
| PKA | protein kinase A |
| PRAD | prostate adenocarcinoma |
| PRX | Periaxin |
| PTC | papillary thyroid carcinoma |
| PyVT | Polyoma Virus middle T antigen |
| READ | rectum adenocarcinoma |
| RFS | recurrence-free survival |
| RNF38 | ring finger protein 38 |
| RPL | recurrent pregnancy loss |
| RSI | radiation sensitivity index |
| RUA | reactive urothelial atypia |
| SARC | sarcoma |
| SKCM | skin cutaneous melanoma |
| ST2 | tumorigenicity 2 |
| STAD | stomach adenocarcinoma |
| TCGA | The Cancer Genome Atlas |
| TGCT | testicular germ cell tumors |
| TGF-β | transforming growth factor β |
| THCA | thyroid carcinoma |
| THYM | thymoma |
| TIIC | tumor-infiltrating immune cell |
| TMB | tumor mutational burden |
| TNBC | triple-negative breast cancer |
| UBE3C | ubiquitin protein ligase E3C |
| UCEC | uterine corpus endometrial carcinoma |
| UCS | uterine carcinosarcoma |
| UM | uveal melanoma |
| UVM | uveal melanoma |
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Hebei Province (No.H2021108003)
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1258951/full#supplementary-material
References
- 1. Sundararaj S, Ravindran A, Casarotto MG. AHNAK: The quiet giant in calcium homeostasis. Cell Calcium. (2021) 96:102403. doi: 10.1016/j.ceca.2021.102403 [DOI] [PubMed] [Google Scholar]
- 2. Hieda Y, Tsukita S, Tsukita S. A new high molecular mass protein showing unique localization in desmosomal plaque. J Cell Biol (1989) 109:1511–8. doi: 10.1083/jcb.109.4.1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shtivelman E, Cohen FE, Bishop JM. A human gene (AHNAK) encoding an unusually large protein with a 1.2-microns polyionic rod structure. Proc Natl Acad Sci USA (1992) 89:5472–6. doi: 10.1073/pnas.89.12.5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hashimoto T, Amagai M, Parry DA, Dixon TW, Tsukita S, Tsukita S, et al. Desmoyokin, a 680 kDa keratinocyte plasma membrane-associated protein, is homologous to the protein encoded by human gene AHNAK. J Cell Sci (1993) 105(Pt 2):275–86. doi: 10.1242/jcs.105.2.275 [DOI] [PubMed] [Google Scholar]
- 5. Kudoh J, Wang Y, Minoshima S, Hashimoto T, Amagai M, Nishikawa T, et al. Localization of the human AHNAK/desmoyokin gene (AHNAK) to chromosome band 11q12 by somatic cell hybrid analysis and fluorescence in situ hybridization. Cytogenet Cell Genet (1995) 70:218–20. doi: 10.1159/000134037 [DOI] [PubMed] [Google Scholar]
- 6. de Morree A, Droog M, Grand Moursel L, Bisschop IJ, Impagliazzo A, Frants RR, et al. Self-regulated alternative splicing at the AHNAK locus. FASEB J (2012) 26:93–103. doi: 10.1096/fj.11-187971 [DOI] [PubMed] [Google Scholar]
- 7. Kouno M, Kondoh G, Horie K, Komazawa N, Ishii N, Takahashi Y, et al. AHNAK/Desmoyokin is dispensable for proliferation, differentiation, and maintenance of integrity in mouse epidermis. J Invest Dermatol (2004) 123:700–7. doi: 10.1111/j.0022-202X.2004.23412.x [DOI] [PubMed] [Google Scholar]
- 8. Davis TA, Loos B, Engelbrecht AM. AHNAK: the giant jack of all trades. Cell Signal (2014) 26:2683–93. doi: 10.1016/j.cellsig.2014.08.017 [DOI] [PubMed] [Google Scholar]
- 9. Komuro A, Masuda Y, Kobayashi K, Babbitt R, Gunel M, Flavell RA, et al. The AHNAKs are a class of giant propeller-like proteins that associate with calcium channel proteins of cardiomyocytes and other cells. Proc Natl Acad Sci USA (2004) 101:4053–8. doi: 10.1073/pnas.0308619101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zardab M, Stasinos K, Grose RP, Kocher HM. The obscure potential of AHNAK2. Cancers (Basel) (2022) 14:528. doi: 10.3390/cancers14030528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sundararaj S, Shishmarev D, Lin Y, Aditya S, Casarotto MG. (1)H, (13)C and (15)N backbone NMR chemical shift assignments of the C-terminal P4 domain of AHNAK. Biomol NMR Assign (2018) 12:253–7. doi: 10.1007/s12104-018-9818-3 [DOI] [PubMed] [Google Scholar]
- 12. Haase H, Podzuweit T, Lutsch G, Hohaus A, Kostka S, Lindschau C, et al. Signaling from beta-adrenoceptor to L-type calcium channel: identification of a novel cardiac protein kinase A target possessing similarities to AHNAK. FASEB J (1999) 13:2161–72. doi: 10.1096/fasebj.13.15.2161 [DOI] [PubMed] [Google Scholar]
- 13. Haase H. AHNAK, a new player in beta-adrenergic regulation of the cardiac L-type Ca2+ channel. Cardiovasc Res (2007) 73:19–25. doi: 10.1016/j.cardiores.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 14. Alvarez J, Hamplova J, Hohaus A, Morano I, Haase H, Vassort G. Calcium current in rat cardiomyocytes is modulated by the carboxyl-terminal AHNAK domain. J Biol Chem (2004) 279:12456–61. doi: 10.1074/jbc.M312177200 [DOI] [PubMed] [Google Scholar]
- 15. Cho H, Shen GQ, Wang X, Wang F, Archacki S, Li Y, et al. Long noncoding RNA ANRIL regulates endothelial cell activities associated with coronary artery disease by up-regulating CLIP1, EZR, and LYVE1 genes. J Biol Chem (2019) 294:3881–98. doi: 10.1074/jbc.RA118.005050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu L, Huang J, Wei B, Mo J, Wei Q, Chen C, et al. Multiomics analysis of genetics and epigenetics reveals pathogenesis and therapeutic targets for atrial fibrillation. BioMed Res Int (2021) 2021:6644827. doi: 10.1155/2021/6644827 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Liu L, Huang J, Liu Y, Pan X, Li Z, Zhou L, et al. Multiomics analysis of transcriptome, epigenome, and genome uncovers putative mechanisms for dilated cardiomyopathy. BioMed Res Int (2021) 2021:6653802. doi: 10.1155/2021/6653802 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Marg A, Haase H, Neumann T, Kouno M, Morano I. AHNAK1 and AHNAK2 are costameric proteins: AHNAK1 affects transverse skeletal muscle fiber stiffness. Biochem Biophys Res Commun (2010) 401:143–8. doi: 10.1016/j.bbrc.2010.09.030 [DOI] [PubMed] [Google Scholar]
- 19. Jin J, Bhatti DL, Lee KW, Medrihan L, Cheng J, Wei J, et al. AHNAK scaffolds p11/Anxa2 complex and L-type voltage-gated calcium channel and modulates depressive behavior. Mol Psychiatry (2020) 25:1035–49. doi: 10.1038/s41380-019-0371-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benedetti B, Benedetti A, Flucher BE. Loss of the calcium channel beta4 subunit impairs parallel fibre volley and Purkinje cell firing in cerebellum of adult ataxic mice. Eur J Neurosci (2016) 43:1486–98. doi: 10.1111/ejn.13241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sekiya F, Bae YS, Jhon DY, Hwang SC, Rhee SG. AHNAK, a protein that binds and activates phospholipase C-gamma1 in the presence of arachidonic acid. J Biol Chem (1999) 274:13900–7. doi: 10.1074/jbc.274.20.13900 [DOI] [PubMed] [Google Scholar]
- 22. Haase H, Alvarez J, Petzhold D, Doller A, Behlke J, Erdmann J, et al. AHNAK is critical for cardiac Ca(V)1.2 calcium channel function and its beta-adrenergic regulation. . FASEB J (2005) 19:1969–77. doi: 10.1096/fj.05-3997com [DOI] [PubMed] [Google Scholar]
- 23. Obermeier B, Verma A, Ransohoff RM. The blood-brain barrier. Handb Clin Neurol (2016) 133:39–59. doi: 10.1016/B978-0-444-63432-0.00003-7 [DOI] [PubMed] [Google Scholar]
- 24. Gentil BJ, Benaud C, Delphin C, Remy C, Berezowski V, Cecchelli R, et al. Specific AHNAK expression in brain endothelial cells with barrier properties. J Cell Physiol (2005) 203:362–71. doi: 10.1002/jcp.20232 [DOI] [PubMed] [Google Scholar]
- 25. von Boxberg Y, Salim C, Soares S, Baloui H, Alterio J, Ravaille-Veron M, et al. Spinal cord injury-induced up-regulation of AHNAK, expressed in cells delineating cystic cavities, and associated with neoangiogenesis. Eur J Neurosci (2006) 24:1031–41. doi: 10.1111/j.1460-9568.2006.04994.x [DOI] [PubMed] [Google Scholar]
- 26. Downs KM, McHugh J, Copp AJ, Shtivelman E. Multiple developmental roles of AHNAK are suggested by localization to sites of placentation and neural plate fusion in the mouse conceptus. Gene Expr Patterns (2002) 2:27–34. doi: 10.1016/S0925-4773(02)00349-0 [DOI] [PubMed] [Google Scholar]
- 27. Lim HJ, Kim J, Park CH, Lee SA, Lee MR, Kim KS, et al. Regulation of c-myc expression by AHNAK promotes induced pluripotent stem cell generation. J Biol Chem (2016) 291:752–61. doi: 10.1074/jbc.M115.659276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y, Lv X, Chen H, Zhi Z, Wei Z, Wang B, et al. Peptide derived from AHNAK inhibits cell migration and proliferation in hirschsprung's disease by targeting the ERK1/2 pathway. J Proteome Res (2021) 20:2308–18. doi: 10.1021/acs.jproteome.0c00811 [DOI] [PubMed] [Google Scholar]
- 29. Huang Y, Laval SH, van Remoortere A, Baudier J, Benaud C, Anderson LV, et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J (2007) 21:732–42. doi: 10.1096/fj.06-6628com [DOI] [PubMed] [Google Scholar]
- 30. Yan X, Noel F, Marcotte I, DeWolf CE, Warschawski DE, Boisselier E. AHNAK C-terminal peptide membrane binding-interactions between the residues 5654-5673 of AHNAK and phospholipid monolayers and bilayers. Langmuir (2020) 36:362–9. doi: 10.1021/acs.langmuir.9b02973 [DOI] [PubMed] [Google Scholar]
- 31. Shin JH, Lee SH, Kim YN, Kim IY, Kim YJ, Kyeong DS, et al. AHNAK deficiency promotes browning and lipolysis in mice via increased responsiveness to beta-adrenergic signalling. Sci Rep (2016) 6:23426. doi: 10.1038/srep23426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim YN, Shin JH, Kyeong DS, Cho SY, Kim MY, Lim HJ, et al. AHNAK deficiency attenuates high-fat diet-induced fatty liver in mice through FGF21 induction. Exp Mol Med (2021) 53:468–82. doi: 10.1038/s12276-021-00573-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woo JK, Shin JH, Lee SH, Park HM, Cho SY, Sung YM, et al. Essential role of AHNAK in adipocyte differentiation leading to the transcriptional regulation of Bmpr1alpha expression. Cell Death Dis (2018) 9:864. doi: 10.1038/s41419-018-0873-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin S, Seong JK, Bae YS. AHNAK stimulates BMP2-mediated adipocyte differentiation through Smad1 activation. Obes (Silver Spring) (2016) 24:398–407. doi: 10.1002/oby.21367 [DOI] [PubMed] [Google Scholar]
- 35. Song X, Zhang L, Du X, Zheng Y, Jia T, Zhou T, et al. Neuroblast differentiation-associated protein derived polypeptides: AHNAK(5758-5775) induces inflammation by activating mast cells via ST2. Immunol Invest (2023) 52:178–93. doi: 10.1080/08820139.2022.2151368 [DOI] [PubMed] [Google Scholar]
- 36. Li L, Liu Y, Feng T, Zhou W, Wang Y, Li H. The AHNAK induces increased IL-6 production in CD4+ T cells and serves as a potential diagnostic biomarker for recurrent pregnancy loss. Clin Exp Immunol (2022) 209:291–304. doi: 10.1093/cei/uxac067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salim C, Boxberg YV, Alterio J, Fereol S, Nothias F. The giant protein AHNAK involved in morphogenesis and laminin substrate adhesion of myelinating Schwann cells. Glia (2009) 57:535–49. doi: 10.1002/glia.20782 [DOI] [PubMed] [Google Scholar]
- 38. Gillespie CS, Sherman DL, Blair GE, Brophy PJ. Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment. Neuron (1994) 12:497–508. doi: 10.1016/0896-6273(94)90208-9 [DOI] [PubMed] [Google Scholar]
- 39. Han H, Kursula P. Periaxin and AHNAK nucleoprotein 2 form intertwined homodimers through domain swapping. J Biol Chem (2014) 289:14121–31. doi: 10.1074/jbc.M114.554816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tey S, Shahrizaila N, Drew AP, Samulong S, Goh KJ, Battaloglu E, et al. Linkage analysis and whole exome sequencing reveals AHNAK2 as a novel genetic cause for autosomal recessive CMT in a Malaysian family. Neurogenetics (2019) 20:117–27. doi: 10.1007/s10048-019-00576-3 [DOI] [PubMed] [Google Scholar]
- 41. Pascal A, Gallaud E, Giet R, Benaud C. Annexin A2 and AHNAK control cortical NuMA-dynein localization and mitotic spindle orientation. J Cell Sci (2022) 135:jcs259344. doi: 10.1242/jcs.259344 [DOI] [PubMed] [Google Scholar]
- 42. Kim IY, Yi SS, Shin JH, Kim YN, Ko CY, Kim HS, et al. Intensive morphometric analysis of enormous alterations in skeletal bone system with micro-CT for AHNAK(-/-) mice. Anat Sci Int (2020) 95:323–33. doi: 10.1007/s12565-020-00525-3 [DOI] [PubMed] [Google Scholar]
- 43. Huang Y, de Morree A, van Remoortere A, Bushby K, Frants RR, den Dunnen JT, et al. Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle. Hum Mol Genet (2008) 17:1855–66. doi: 10.1093/hmg/ddn081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skoldberg F, Ronnblom L, Thornemo M, Lindahl A, Bird PI, Rorsman F, et al. Identification of AHNAK as a novel autoantigen in systemic lupus erythematosus. Biochem Biophys Res Commun (2002) 291:951–8. doi: 10.1006/bbrc.2002.6534 [DOI] [PubMed] [Google Scholar]
- 45. Wen L, Zhu C, Zhu Z, Yang C, Zheng X, Liu L, et al. Exome-wide association study identifies four novel loci for systemic lupus erythematosus in Han Chinese population. Ann Rheum Dis (2018) 77:417. doi: 10.1136/annrheumdis-2017-211823 [DOI] [PubMed] [Google Scholar]
- 46. Kirov A, Kacer D, Conley BA, Vary CP, Prudovsky I. AHNAK2 participates in the stress-induced nonclassical FGF1 secretion pathway. J Cell Biochem (2015) 116:1522–31. doi: 10.1002/jcb.25047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gu J, Mao W, Ren W, Xu F, Zhu Q, Lu C, et al. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett (2019) 443:125–34. doi: 10.1016/j.canlet.2018.11.029 [DOI] [PubMed] [Google Scholar]
- 48. Park JW, Kim IY, Choi JW, Lim HJ, Shin JH, Kim YN, et al. AHNAK loss in mice promotes type II pneumocyte hyperplasia and lung tumor development. Mol Cancer Res (2018) 16:1287–98. doi: 10.1158/1541-7786.MCR-17-0726 [DOI] [PubMed] [Google Scholar]
- 49. Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J Proteome Res (2006) 5:1143–54. doi: 10.1021/pr050455t [DOI] [PubMed] [Google Scholar]
- 50. Zhang S, Lu Y, Qi L, Wang H, Wang Z, Cai Z. AHNAK2 is associated with poor prognosis and cell migration in lung adenocarcinoma. BioMed Res Int (2020) 2020:8571932. doi: 10.1155/2020/8571932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu G, Guo Z, Zhang Q, Liu Z, Zhu D. AHNAK2 promotes migration, invasion, and epithelial-mesenchymal transition in lung adenocarcinoma cells via the TGF-beta/smad3 pathway. Onco Targets Ther (2020) 13:12893–903. doi: 10.2147/OTT.S281517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang DW, Zheng HZ, Cha N, Zhang XJ, Zheng M, Chen MM, et al. Down-regulation of AHNAK2 inhibits cell proliferation, migration and invasion through inactivating the MAPK pathway in lung adenocarcinoma. Technol Cancer Res Treat (2020) 19:1533033820957006. doi: 10.1177/1533033820957006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zheng M, Liu J, Bian T, Liu L, Sun H, Zhou H, et al. Correlation between prognostic indicator AHNAK2 and immune infiltrates in lung adenocarcinoma. Int Immunopharmacol (2021) 90:107134. doi: 10.1016/j.intimp.2020.107134 [DOI] [PubMed] [Google Scholar]
- 54. Du Y, Yuan S, Zhuang X, Zhang Q, Qiao T. Multiomics differences in lung squamous cell carcinoma patients with high radiosensitivity index compared with those with low radiosensitivity index. Dis Markers (2021) 2021:3766659. doi: 10.1155/2021/3766659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu Z, Zheng M, Lei B, Zhou Z, Huang Y, Li W, et al. Whole-exome sequencing identifies somatic mutations associated with lung cancer metastasis to the brain. Ann Transl Med (2021) 9:694. doi: 10.21037/atm-21-1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cui Y, Liu X, Wu Y, Liang X, Dai J, Zhang Z, et al. Deleterious AHNAK2 mutation as a novel biomarker for immune checkpoint inhibitors in non-small cell lung cancer. Front Oncol (2022) 12:798401. doi: 10.3389/fonc.2022.798401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sohn M, Shin S, Yoo JY, Goh Y, Lee IH, Bae YS. AHNAK promotes tumor metastasis through transforming growth factor-beta-mediated epithelial-mesenchymal transition. Sci Rep (2018) 8:14379. doi: 10.1038/s41598-018-32796-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suh JM, Son Y, Yoo JY, Goh Y, Seidah NG, Lee S, et al. Proprotein convertase subtilisin/kexin Type 9 is required for AHNAK-mediated metastasis of melanoma into lung epithelial cells. Neoplasia (2021) 23:993–1001. doi: 10.1016/j.neo.2021.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang Y, Wei L, Huang Y, Wen S, Liu T, Duan X, et al. Identification of distinct genomic features reveals frequent somatic AHNAK and PTEN mutations predominantly in primary Malignant melanoma presenting in the ureter. Jpn J Clin Oncol (2022) 52:930–43. doi: 10.1093/jjco/hyac061 [DOI] [PubMed] [Google Scholar]
- 60. Sheppard HM, Feisst V, Chen J, Print C, Dunbar PR. AHNAK is downregulated in melanoma, predicts poor outcome, and may be required for the expression of functional cadherin-1. Melanoma Res (2016) 26:108–16. doi: 10.1097/CMR.0000000000000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Conic RZ, Arbesman J. Melanoma tumor characteristics: an analysis of mutational burden and copy number alterations by patient age and stage. J Invest Dermatol (2018) 138:1218–21. doi: 10.1016/j.jid.2017.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li M, Liu Y, Meng Y, Zhu Y. AHNAK nucleoprotein 2 performs a promoting role in the proliferation and migration of uveal melanoma cells. Cancer Biother Radiopharm (2019) 34:626–33. doi: 10.1089/cbr.2019.2778 [DOI] [PubMed] [Google Scholar]
- 63. Zhao Z, Xiao S, Yuan X, Yuan J, Zhang C, Li H, et al. AHNAK as a prognosis factor suppresses the tumor progression in glioma. J Cancer (2017) 8:2924–32. doi: 10.7150/jca.20277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bhargava S, Patil V, Mahalingam K, Somasundaram K. Elucidation of the genetic and epigenetic landscape alterations in RNA binding proteins in glioblastoma. Oncotarget (2017) 8:16650–68. doi: 10.18632/oncotarget.14287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin Z, Xu H, Yang R, Li Z, Zheng H, Zhang Z, et al. Effective treatment of a BRAF V600E-mutant epithelioid glioblastoma patient by vemurafenib: a case report. Anticancer Drugs (2022) 33:100–4. doi: 10.1097/CAD.0000000000001130 [DOI] [PubMed] [Google Scholar]
- 66. Dumitru CA, Bankfalvi A, Gu X, Zeidler R, Brandau S, Lang S. AHNAK and inflammatory markers predict poor survival in laryngeal carcinoma. PloS One (2013) 8:e56420. doi: 10.1371/journal.pone.0056420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cui X, Zhu W, Wang P, Wang X. Tetrandrine inhibits the intracellular calcium ion level and upregulates the expression of brg1 and AHNAK in hep-2 cells. Clin Lab (2015) 61:1569–76. doi: 10.7754/Clin.Lab.2015.141242 [DOI] [PubMed] [Google Scholar]
- 68. Yang C, Xu W, Gong J, Liu Z, Cui D. Novel somatic alterations underlie Chinese papillary thyroid carcinoma. Cancer biomark (2020) 27:445–60. doi: 10.3233/CBM-191200 [DOI] [PubMed] [Google Scholar]
- 69. Zheng L, Li S, Zheng X, Guo R, Qu W. AHNAK2 is a novel prognostic marker and correlates with immune infiltration in papillary thyroid cancer: Evidence from integrated analysis. Int Immunopharmacol (2021) 90:107185. doi: 10.1016/j.intimp.2020.107185 [DOI] [PubMed] [Google Scholar]
- 70. Xie Z, Lun Y, Li X, He Y, Wu S, Wang S, et al. Bioinformatics analysis of the clinical value and potential mechanisms of AHNAK2 in papillary thyroid carcinoma. Aging (Albany NY) (2020) 12:18163–80. doi: 10.18632/aging.103645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ye R, Liu D, Guan H, AiErken N, Fang Z, Shi Y, et al. AHNAK2 promotes thyroid carcinoma progression by activating the NF-kappaB pathway. Life Sci (2021) 286:120032. doi: 10.1016/j.lfs.2021.120032 [DOI] [PubMed] [Google Scholar]
- 72. Lee IH, Sohn M, Lim HJ, Yoon S, Oh H, Shin S, et al. AHNAK functions as a tumor suppressor via modulation of TGFbeta/Smad signaling pathway. Oncogene (2014) 33:4675–84. doi: 10.1038/onc.2014.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cimas FJ, Manzano A, Baliu-Pique M, Garcia-Gil E, Perez-Segura P, Nagy A, et al. Genomic mapping identifies mutations in RYR2 and AHNAK as associated with favorable outcome in basal-like breast tumors expressing PD1/PD-L1. Cancers (Basel) (2020) 12:2243. doi: 10.3390/cancers12082243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Silva TA, Smuczek B, Valadao IC, Dzik LM, Iglesia RP, Cruz MC, et al. AHNAK enables mammary carcinoma cells to produce extracellular vesicles that increase neighboring fibroblast cell motility. Oncotarget (2016) 7:49998–50016. doi: 10.18632/oncotarget.10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen B, Wang J, Dai D, Zhou Q, Guo X, Tian Z, et al. AHNAK suppresses tumor proliferation and invasion by targeting multiple pathways in triple-negative breast cancer. J Exp Clin Cancer Res (2017) 36:65. doi: 10.1186/s13046-017-0522-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Davis T, van Niekerk G, Peres J, Prince S, Loos B, Engelbrecht AM. Doxorubicin resistance in breast cancer: A novel role for the human protein AHNAK. Biochem Pharmacol (2018) 148:174–83. doi: 10.1016/j.bcp.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 77. Sun L, Li K, Liu G, Xu Y, Zhang A, Lin D, et al. Distinctive pattern of AHNAK methylation level in peripheral blood mononuclear cells and the association with HBV-related liver diseases. Cancer Med (2018) 7:5178–86. doi: 10.1002/cam4.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Soini T, Eloranta K, Pihlajoki M, Kyronlahti A, Akinrinade O, Andersson N, et al. Transcription factor GATA4 associates with mesenchymal-like gene expression in human hepatoblastoma cells. Tumour Biol (2018) 40:1010428318785498. doi: 10.1177/1010428318785498 [DOI] [PubMed] [Google Scholar]
- 79. Li K, Song K, Hou Y, Tian Y, Wang H, Sun L, et al. AHNAK contributes to hepatocellular carcinoma growth by interacting with IGF-1R. . Molecules (2022) 27:8680. doi: 10.3390/molecules27248680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Peng R, Zhang PF, Yang X, Wei CY, Huang XY, Cai JB, et al. Overexpression of RNF38 facilitates TGF-beta signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res (2019) 38:113. doi: 10.1186/s13046-019-1113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ke L, Shen J, Feng J, Chen J, Shen S, Li S, et al. Somatic mutation profiles revealed by next generation sequencing (NGS) in 39 chinese hepatocellular carcinoma patients. Front Mol Biosci (2021) 8:800679. doi: 10.3389/fmolb.2021.800679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang Z, Liu X, Huang R, Liu X, Liang Z, Liu T. Upregulation of nucleoprotein AHNAK is associated with poor outcome of pancreatic ductal adenocarcinoma prognosis via mediating epithelial-mesenchymal transition. J Cancer (2019) 10:3860–70. doi: 10.7150/jca.31291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yang ZQ, Liu YJ, Zhou XL. An integrated microarray analysis reveals significant diagnostic and prognostic biomarkers in pancreatic cancer. Med Sci Monit (2020) 26:e921769. doi: 10.12659/MSM.921769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Klett H, Fuellgraf H, Levit-Zerdoun E, Hussung S, Kowar S, Kusters S, et al. Identification and validation of a diagnostic and prognostic multi-gene biomarker panel for pancreatic ductal adenocarcinoma. Front Genet (2018) 9:108. doi: 10.3389/fgene.2018.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lu D, Wang J, Shi X, Yue B, Hao J. AHNAK2 is a potential prognostic biomarker in patients with PDAC. Oncotarget (2017) 8:31775–84. doi: 10.18632/oncotarget.15990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Olsen JB, Cao XJ, Han B, Chen LH, Horvath A, Richardson TI, et al. Quantitative profiling of the activity of protein lysine methyltransferase SMYD2 using SILAC-based proteomics. Mol Cell Proteomics (2016) 15:892–905. doi: 10.1074/mcp.M115.053280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hou Q, Jiang Z, Li Z, Jiang M. Identification and functional validation of radioresistance-related genes AHNAK2 and EVPL in esophageal squamous cell carcinoma by exome and transcriptome sequencing analyses. Onco Targets Ther (2021) 14:1131–45. doi: 10.2147/OTT.S291007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu ZM, Yang XL, Jiang F, Pan YC, Zhang L. Matrine involves in the progression of gastric cancer through inhibiting miR-93-5p and upregulating the expression of target gene AHNAK. J Cell Biochem (2020) 121:2467–77. doi: 10.1002/jcb.29469 [DOI] [PubMed] [Google Scholar]
- 89. Shen E, Wang X, Liu X, Lv M, Zhang L, Zhu G, et al. MicroRNA-93-5p promotes epithelial-mesenchymal transition in gastric cancer by repressing tumor suppressor AHNAK expression. Cancer Cell Int (2020) 20:76. doi: 10.1186/s12935-019-1092-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90. Ohmura H, Ito M, Uchino K, Okada C, Tanishima S, Yamada Y, et al. Methylation of drug resistance-related genes in chemotherapy-sensitive Epstein-Barr virus-associated gastric cancer. FEBS Open Bio. (2020) 10:147–57. doi: 10.1002/2211-5463.12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yu Y, Xie Z, Zhao M, Lian X. Identification of PIK3CA multigene mutation patterns associated with superior prognosis in stomach cancer. BMC Cancer (2021) 21:368. doi: 10.1186/s12885-021-08115-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhou YY, Kang YT, Chen C, Xu FF, Wang HN, Jin R. Combination of TNM staging and pathway based risk score models in patients with gastric cancer. J Cell Biochem (2018) 119:3608–17. doi: 10.1002/jcb.26563 [DOI] [PubMed] [Google Scholar]
- 93. Cho WC, Jang JE, Kim KH, Yoo BC, Ku JL. SORBS1 serves a metastatic role via suppression of AHNAK in colorectal cancer cell lines. Int J Oncol (2020) 56:1140–51. doi: 10.3892/ijo.2020.5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kundu S, Ali MA, Handin N, Conway LP, Rendo V, Artursson P, et al. Common and mutation specific phenotypes of KRAS and BRAF mutations in colorectal cancer cells revealed by integrative -omics analysis. J Exp Clin Cancer Res (2021) 40:225. doi: 10.1186/s13046-021-02025-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang M, Li X, Zhang J, Yang Q, Chen W, Jin W, et al. AHNAK2 is a novel prognostic marker and oncogenic protein for clear cell renal cell carcinoma. Theranostics (2017) 7:1100–13. doi: 10.7150/thno.18198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lee H, Kim K, Woo J, Park J, Kim H, Lee KE, et al. Quantitative proteomic analysis identifies AHNAK (Neuroblast differentiation-associated protein AHNAK) as a novel candidate biomarker for bladder urothelial carcinoma diagnosis by liquid-based cytology. Mol Cell Proteomics (2018) 17:1788–802. doi: 10.1074/mcp.RA118.000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Okusa H, Kodera Y, Oh-Ishi M, Minamida Y, Tsuchida M, Kavoussi N, et al. Searching for new biomarkers of bladder cancer based on proteomic analysis. J Electrophoresis (2008) 52(1):19–24. doi: 10.2198/jelectroph.52.19 [DOI] [Google Scholar]
- 98. Chu J, Li N, Li F. A risk score staging system based on the expression of seven genes predicts the outcome of bladder cancer. Oncol Lett (2018) 16:2091–6. doi: 10.3892/ol.2018.8904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Qu G, Liu Z, Yang G, Xu Y, Xiang M, Tang C. Development of a prognostic index and screening of prognosis related genes based on an immunogenomic landscape analysis of bladder cancer. Aging (Albany NY) (2021) 13:12099–112. doi: 10.18632/aging.202917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Witzke KE, Grosserueschkamp F, Jutte H, Horn M, Roghmann F, von Landenberg N, et al. Integrated fourier transform infrared imaging and proteomics for identification of a candidate histochemical biomarker in bladder cancer. Am J Pathol (2019) 189:619–31. doi: 10.1016/j.ajpath.2018.11.018 [DOI] [PubMed] [Google Scholar]
- 101. Koguchi D, Matsumoto K, Shimizu Y, Kobayashi M, Hirano S, Ikeda M, et al. Prognostic impact of AHNAK2 expression in patients treated with radical cystectomy. Cancers (Basel) (2021) 13:1748. doi: 10.3390/cancers13081748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Komina S, Petrusevska G, Jovanovic R, Kunovska SK, Stavridis S, Dohcev S, et al. AHNAK2 urinary protein expression as potential biomarker for bladder cancer detection: A pilot study. Turk J Urol (2022) 48:423–30. doi: 10.5152/tud.2022.22132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Luo Y, Zeng G, Wu S. Identification of microenvironment-related prognostic genes in bladder cancer based on gene expression profile. Front Genet (2019) 10:1187. doi: 10.3389/fgene.2019.01187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shafran JS, Andrieu GP, Gyorffy B, Denis GV. BRD4 regulates metastatic potential of castration-resistant prostate cancer through AHNAK. Mol Cancer Res (2019) 17:1627–38. doi: 10.1158/1541-7786.MCR-18-1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhao X, Lei Y, Li G, Cheng Y, Yang H, Xie L, et al. Integrative analysis of cancer driver genes in prostate adenocarcinoma. Mol Med Rep (2019) 19:2707–15. doi: 10.3892/mmr.2019.9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cai Y, Hu Y, Yu F, Tong W, Wang S, Sheng S, et al. AHNAK suppresses ovarian cancer progression through the Wnt/beta-catenin signaling pathway. Aging (Albany NY) (2021) 13:23579–87. doi: 10.18632/aging.203473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nie X, Zheng M, Gao L, Hu Y, Zhuang Y, Li X, et al. Interaction between TMEFF1 and AHNAK proteins in ovarian cancer cells: Implications for clinical prognosis. Int Immunopharmacol (2022) 107:108726. doi: 10.1016/j.intimp.2022.108726 [DOI] [PubMed] [Google Scholar]
- 108. Xu M, Cheng A, Yu L, Wei W, Li J, Cai C. AHNAK2 is a biomarker and a potential therapeutic target of adenocarcinomas. Acta Biochim Biophys Sin (Shanghai) (2022) 54:1708–19. doi: 10.3724/abbs.2022112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sudo H, Tsuji AB, Sugyo A, Abe M, Hino O, Saga T. AHNAK is highly expressed and plays a key role in cell migration and invasion in mesothelioma. Int J Oncol (2014) 44:530–8. doi: 10.3892/ijo.2013.2183 [DOI] [PubMed] [Google Scholar]
- 110. Lobl MB, Clarey D, Schmidt C, Wichman C, Wysong A. Analysis of mutations in cutaneous squamous cell carcinoma reveals novel genes and mutations associated with patient-specific characteristics and metastasis: a systematic review. Arch Dermatol Res (2021) 314:711–8. doi: 10.1007/s00403-021-02213-2 [DOI] [PubMed] [Google Scholar]
- 111. Saito M, Fujiwara Y, Asao T, Honda T, Shimada Y, Kanai Y, et al. The genomic and epigenomic landscape in thymic carcinoma. Carcinogenesis (2017) 38:1084–91. doi: 10.1093/carcin/bgx094 [DOI] [PubMed] [Google Scholar]
- 112. Massague J. TGFbeta in cancer. Cell (2008) 134:215–30. doi: 10.1016/j.cell.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.