Highlights
-
•
Cilevirus infectious clones.
-
•
CiLV-C reverse genetic system.
-
•
Construction and validation of a Kitavirus infectious cDNA clone.
-
•
Gibson assembly and golden gate systems.
Keywords: Cilevirus, Citrus leprosis virus C, CiLV-C infectious clone, Reverse genetics
Abstract
Reverse genetics systems represent an important tool for studying the molecular and functional processes of viral infection. Citrus leprosis virus C (CiLV-C) (genus Cilevirus, family Kitaviridae) is the main pathogen responsible for the citrus leprosis (CL) disease in Latin America, one of the most economically important diseases of the citrus industry. Molecular studies of this pathosystem are limited due to the lack of infectious clones. Here, we report the construction and validation of a CiLV-C infectious cDNA clone based on an agroinfection system. The two viral RNA segments (RNA1 and RNA2) were assembled into two binary vectors (pJL89 and pLXAS). Agroinfiltrated Nicotiana benthamiana plants showed a response similar to that observed in the natural infection process with the formation of localized lesions restricted to the inoculated leaves. The virus recovered from the plant tissue infected with the infectious clones can be mechanically transmitted between N. benthamiana plants. Detection of CiLV-C subgenomic RNAs (sgRNAs) from agroinfiltrated and mechanically inoculated leaves further confirmed the infectivity of the clones. Finally, partial particle-purification preparations or sections of CiLV-C-infected tissue followed by transmission electron microscopy (TEM) analysis showed the formation of CiLV-C virions rescued by the infectious clone. The CiLV-C reverse genetic system now provides a powerful molecular tool to unravel the peculiarities of the CL pathosystem.
1. Introduction
Citrus leprosis (CL) is considered one of the main viral diseases affecting citrus orchards in America. This disease is caused by leprosis-associated viruses from the genera Cilevirus, Dichorhavirus, and perhaps Higrevirus. Currently, citrus leprosis virus C (CiLV-C), genus Cilevirus (family Kitaviridae) is considered the most devastating virus infecting citrus in Brazil (Bastianel et al., 2010). It was described in more than 50 natural and experimental host species belonging to at least 28 plant families (Garita et al., 2014). The interaction between CiLV-C and its vector Brevipalpus yothersi (Acari: Tenuipalpidae) occurs in a persistent manner (Roy et al., 2015; Tassi et al., 2017). Brevipalpus-transmitted viruses (BTV) can accumulate either in the cytoplasm or the nucleus (Freitas-Astua et al., 2018). CiLV-C infection is characterized by the formation of chlorotic and necrotic circular lesions in citrus leaves, fruits, and stems (Bastianel et al., 2010). So far, no systemically infected plants have been reported by cytoplasmic type species responsible for the CL disease in natural infections.
In the virus infection process, vesicles in the endoplasmic reticulum (ER) system comprising bacilliform (40–70 nm x 100–120 nm) viral particles are extensively visualized (Kitajima et al., 1974; Leastro et al., 2018). The genome of cileviruses consists of two segments of positive-sense single-stranded RNA (+ssRNA) carrying 5′-cap and 3′-poly(A) tail structures (Freitas-Astua et al., 2018). For the type species CiLV-C, RNA1 (∼ 8.7 kb) encodes the polymerase precursor (RdRp), containing conserved domains of methyl transferase, helicase, protease, and polymerase (Pascon et al., 2006), and the capsid protein (CP p29) (Leastro et al., 2018; Ortega-Rivera et al., 2023). On the other hand, RNA 2 (∼ 5 kb) encodes a component of the viral RNA silencing suppression (RSS) machinery (p15) (Leastro et al., 2020a), the movement protein (MP p32) (Leastro et al., 2021a, 2021b) and the putative glycoprotein (p61) and matrix protein (p24) (Kuchibhatla et al., 2014; Leastro et al., 2018). Four subgenomic RNAs (sgRNA) are generated during the process of CiLV-C infection. RNA1 produces a sgRNA1 fragment of 0.7 kb in size that transcribe the p29, while the RNA2 generates the sgRNA 2 (3 kb, correspondent to p61, MP, and p24), the sgRNA 3 (1.5 kb that transcribes the MP and p24), and the sgRNA 4 (0.6 kb that corresponds to p24 only) (Pascon et al., 2006).
The development of reverse genetics systems of RNA viruses has expanded the understanding of the functions of viral and host genes and proteins. It provides broad insights about the pathosystem of interaction virus-host-vector, which facilitates the development of anti-viral treatments (Cordero et al., 2017; Peng et al., 2021). More recently, plant virus infectious clones have been used for plant genome editing based on viral vectors engineered for CRISPR-Cas-mediated genome editing in plants (Ali et al., 2015; Liu et al., 2023; Uranga et al., 2023). In this context, the development of infectious clones can facilitate the engineering of molecular tools with the potential to be applied in different areas of knowledge.
In this study, we describe the construction of a genetic reverse system for the CiLV-C. This system relies on the agroinfiltration of plant leaves for transient co-expression of the CiLV-C RNA 1 and RNA 2.
2. Materials and methods
2.1. Plant material and virus source
Two to four-week-old plants of Nicotiana benthamiana were used in agroinfiltration and mechanical transmission assays. The CiLV-C linage CRD (GenBank accession numbers DQ352194.1 and DQ352195.1) from citrus-infected leaves was used in this study. The N. benthamiana were kept under a two-step cycle of 24 °C day/18 °C night and 16 h light/8 h dark. The presence of symptoms was monitored for 30 days-post infiltration or inoculation (days p.i).
2.2. Construction of CiLV-C full-length cDNA infectious clones
For pJL89 constructs, the full-length cDNA of CiLV-C RNA1 and RNA2 were obtained using the Superscript™ III (Thermo Fisher Scientific, Waltham, MA, USA) with oligo(dT) primer according to the manufacturer's instruction. For amplification and assembly of viral full-length genomes, primers were designed containing overlapping fragments with the aid of the NEBuilder assembly tool (https://nebuilder.neb.com/). The vector fragments from the binary vector pJL89 (Lindbo, 2007) were prepared by inverse vector amplification with the primers apJL89 vector fwd and apJL89 vector rev, which anneal to the ribozyme HDV-Rz region, and the promoter region, respectively (Supplementary Table 1 and Fig. 1A). Given the size of ∼ 8.7 kb of RNA1, the strategy used for overlap extension PCR for assembly of this fragment into binary vector pJL89 was performed in 2 steps: i) amplification of the first half (F1) of RNA1 (4.411 bp) with the primers (bRNA1 F1 fwd/ bRNA1 F1 rev; Supplementary Table 1 and Fig. 1A) containing an overlapping region of 25 bp with the 5′end of the vector pJL89 and the second half of RNA1; ii) amplification of the second half (F2) of RNA1 (remaining 4.410 pb) with the primers (cRNA1 F2 fwd/ cRNA1 F2 rev; Supplementary Table 1 and Fig. 1A) containing the overlap of 25 bp with the 3′end of the first half of RNA1 and with the 3′of the binary vector (Fig. 1). The RNA2 (F3) (∼ 5 kb) was amplified with the primers (dRNA2 F3 fwd/ dRNA2 F3 rev; Supplementary Table 1 and Fig. 1A) in a single step with 25 bp overlaps with 5′and 3′ends of the binary vector pJL89 (Fig. 1A).
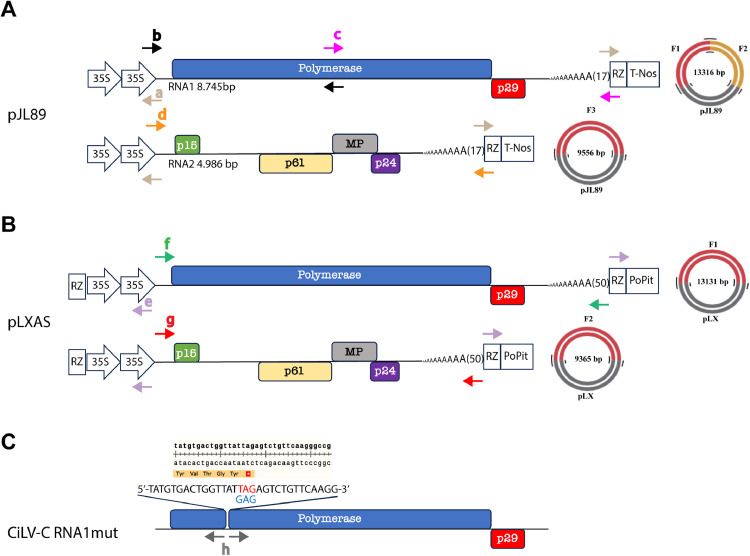
Fig. 1.
Construction of CiLV-C infectious cDNA clones using pJL89 and pLXAS binary vectors. (A) Schematic representation of CiLV-C infectious cDNA clone in binary plasmid. The pJL89 amplification strategy consists of plasmid amplification by inverse PCR with a set of primers that anneal to the HDV Rz region and the 35S CaMV promoter region. The arrows show the direction of amplification. This amplification strategy ensures that the viral fragments are assembled between the double 35S promoter (double arrow) and the HDV Rz (rectangular box), followed by the NOS terminator (square box). It is shown the genomic organization of CiLV-C RNA1 and RNA2. The boxes highlight the virus ORFs: polymerase and p29 (RNA1), and p15, p61, MP, and p24 (RNA2). The black, hot pink, and orange arrows indicate the direction and the primers used for the amplification of the two RNA1 (first -F1- and second -F2-) or the RNA2 (F3) fragments, respectively. On the right, a schematic representation of the pJL89 RNA1 CiLV-C and pJL89 RNA2 CiLV-C constructs assembled. F1 and F2 correspond to the first and second fragments of the RNA1, and F3 corresponds to the RNA2 fragment. The sizes in base pairs (bp) of the viral genome, amplified fragments, and assembled constructs are presented. (B) The pLXAS amplification strategy consists of plasmid amplification by inverse PCR with a set of primers that anneals to the HDV Rz region and the 35S CaMV promoter region. The arrows show the direction of amplification. This amplification strategy ensures that the viral fragments are assembled between the double 35S promoter (double arrow) and the HDV Rz (rectangular box), followed by the PoPit terminator (square box). The green arrows indicate the direction of amplification of the RNA 1 (F1) fragment for the assembly using BsmBI restrict enzyme with nts compatibles to the 3-end of the 35S promoter and 5-end of HDV Rz and the red arrows indicate the direction of amplification of the RNA2 (F2). (C) The stop codon insertion into the viral gene polymerase was performed by replacing a G-T at position 1.405 using a pair primer (Hpolymutstop fwd/Hpolymutstop rev) carrying the point mutation. “a, b, c, d, e, f, g, and h” correspond to the primers named in Supplementary Table 1.
The backbone clone vector and the virus genome fragments (F1, F2, and F3) were amplified with the Phusion High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA, USA), and all fragments were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). Subsequently, the viral fragments were assembled in the vector pJL89, flanked by double cauliflower mosaic virus (CaMV) 35S promoter and the NOS terminator (Fig. 1A), in an isothermal reaction (Gibson Assembly Cloning master mix, New England BioLabs, Ipswich, MA, USA), according to the manufacturer's instructions. The assembled products (named pJL89/2 × 35S-cDNA1:HDVRz-TNOS and pJL89/2 × 35S-cDNA2:HDVRz-TNOS) were transformed in electrocompetent Escherichia coli DH5-alfa cells, and correct clones were selected by colony PCR with primers designed to confirm the correct orientation of the fragments inserted into the vector. In addition, extracted plasmids from positive PCR colonies (4 for each construction) were also confirmed by enzyme digestion (PstI for pJL89 cDNA1 and EcoRI for pJL89 cDNA2), which showed an expected digestion profile (data not shown). The integrity of the recombination regions was confirmed by plasmid DNA sequencing. Finally, the Agrobacterium tumefaciens (strain C58) were separately transformed with the corresponding plasmids containing the full-length CiLV-C cDNA.
For pLXAS (Pasin et al., 2017) construction, the CiLV-C RNA1 and RNA2 were amplified from the cDNA above mentioned using the Phusion High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA, USA). The vector fragments from the binary vector pLXAS were prepared by inverse vector amplification with the primers epLXAS fwd/epLXAS rev, which anneals to the ribozyme HDV-Rz region, and to the 35S promoter region, respectively (Supplementary Table 1 and Fig. 1B). In contrast to the assembly strategy used for insertion of CiLV-C fragments into the pJL89 binary vector, for pLXAS CiLV-C infectious constructs, we proceeded to the golden gate strategy with primers carrying the BsmBI type II restriction site containing sequences complementary to the binary vector. Virus genome fragments (RNA1 and RNA2) were integrally amplified using the primers fRNA1 fwd/fRNA1 rev and gRNA2 fwd/gRNA2 rev, respectively (Supplementary Table 1 and Fig. 1B). After amplification, the respective fragments were digested with BsmbI according to the manufacturer's instruction, then ligated into the pLXAS construct using T4 DNA Ligase (Promega, Madison, WI, USA). The assembled constructs named pLXAS/Rz-2 × 35S-cDNA1:HDVRz-PoPit and pLXAS/Rz-2 × 35S-cDNA2:HDVRz-PoPit were also transformed in DH5-alfa cells and selected by colony PCR as above described. Plasmids from five independent colonies were obtained and analyzed by the Sanger sequencing method. Finally, the selected constructs were used to transform the agrobacterium C58 strain.
To obtain the CiLV-C cDNA1mut control construct carrying a stop codon at the N-terminus of the viral polymerase, we performed an inverse PCR point mutation to replace ‘G’ with ‘T’ at position 1.405. This point mutation impairs the complete translation of the viral polymerase, thus impairing the replication and transcription of the viral genome. The pJL89/2 × 35S-cDNA1:HDVRz-TNOS and pLXAS/Rz-2 × 35S-cDNA1:HDVRz-PoPit were used as a template for inverse PCR amplification with the primers hpolymutstop fwd/hpolymutstop rev (Supplementary Table 1 and Fig. 1C). Next, the amplification product, previously treated with DpnI, was ligated using the T4 DNA ligase (Promega, Madison, WI, USA), generating the constructs named pJL89/2 × 35S-cDNA1mut:HDVRz-PoPit and pLXAS/Rz-2 × 35S-cDNA1mut:HDVRz-PoPit. The introduced point mutation was confirmed by the Sanger sequencing method in five independent constructs.
2.3. Agrobacterium infiltration
At the four-leaf growth stage, N. benthamiana leaves (five plants per construct in three independent experiments) were co-infiltrated (OD600 = 0.5) with A. tumefaciens harboring the full-length infectious clone vector(s) at 1:1 ratio in agroinfiltration buffer (10 mM of MES pH 5,6 and 10 mM of MgCl2). Individual agrobacterium cultures grew at 28 °C in Luria-Bertani (LB) medium containing rifampicin (100 mg mL−1) and kanamycin (50 mg mL−1) antibiotics for plasmid and agrobacterium selection. As mock control, N. benthamiana plants were infiltrated with A. tumefaciens harboring the cDNA1mut plus cDNA2. To increase the stability of the viral genome transcripts, plants were co-agroinfiltrated with a binary vector carrying the silencing suppressor HCPro from the tobacco etch virus (OD600 = 0.1).
2.4. Virus mechanical transmission
At 6 days p.i, agroinfiltrated leaves from several plants were crushed in phosphate buffer (30 mM, pH 8.0) and the leaf extract was used as virus particle inoculum for mechanical inoculation of new N. benthamiana (three-four leaves per plant and five plants per infectious clone) using Carborundum (VWR prolabo, France) as an abrasive.
2.5. RNA extraction and RT-PCR assay
Total RNAs from agroinfiltrated (at 8 days p.i) or mechanical inoculated (at 4 days p.i) N. benthamiana leaves were extracted using VWR Life Science AMRESCO RiboZol RNA extraction Reagent. 1 μg of these RNA extracts was treated with DNase I, RNase free 1 U/μL (Thermo Fisher Scientific, Waltham, MA, USA), and then reverse transcribed with CiLV-C p29 and p24 primers for virus detection (Supplementary Table 1) using the One-Step RT-PCR system (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer's instructions. A blank (non-template control) was used in all RT-PCR assays.
2.6. Northern blot
For CiLV-C genomic (g) and subgenomic (sg) RNA detection, all procedures of RNA electrophoresis, membrane transference, hybridization, and detection were conducted as previously described by Leastro et al. (2020a) and Pallas et al. (1998). For CiLV-C RNA1 g and sg detections, we generated a DIG-riboprobe (Roche, Mannheim, Germany) complementary to the p29 gene. For the detection of gRNA2 and its respective subgenomic RNAs, a DIG-probe complementary to the complete sequence of the p24 was used. Northern blot analysis from agroinfiltrated leaves expressing the RNA1mut control construct, the functional full-length cDNA infectious clone, or the mechanically inoculated plants was performed using 6 μg, 6 μg or 5 μg of total RNA, respectively. The quantification of the RNA bands was performed using ImageJ version 2.0cr software with the ISAC plugin.
2.7. Partial purification of CiLV-C particles, SDS-PAGE, and transmission electron microscope (TEM)
Partial purification of virion particles was performed by PEG precipitation. Briefly, 0.5 g of agroinfiltrated tissue was crushed in 1 mL of extraction buffer (100 mM K2HPO4, 100 mM Ascorbic acid, and 20 mM EDTA, pH 7.1) at room temperature. Then, mixed with 0.5 mL of chloroform/butanol (1:1) solution. The supernatant phase was recovered after 14.000 rpm for 10 min, then mixed with 1/5 vol of PEG 20.000 (30%) and kept on ice for 20 min. After low centrifugation (7.500 rpm for 15 min.), the pellet was resuspended in PEN buffer (10 mM NaH2PO4 [pH 7.0], 1 mM EDTA) for 30 min. The supernatant and pellet fractions were formed after centrifugation at 6.500 rpm for five minutes and analyzed in 1% agarose gel. Part of the supernatant fraction was used for protein and RNA extraction. Before RT-PCR, the RNA was treated with DNase I as aforementioned. SDS-PAGE was performed as previously described by Leastro et al. (2015).
A portion of the partially purified virions was negatively stained using 2% phosphotungstic acid and 4% uranyl acetate. The samples were adsorbed in formvar/carbon-coated grids. The preparations were visualized with a JOEL-JEM 1010 (DEBEN AMT) transmission electron microscope operated at 100 kV. The images were obtained in magnification between X50k-X150k. Particle size (length and diameter) was estimated from the measurement of 25 virions using the ImageJ program.
Small pieces of Nicotiana benthamiana leaves inoculated with CiLV-C infectious clone were fixed in a mixture of 2% glutaraldehyde, 2.5% paraformaldehyde in 0.05 M cacodylate buffer pH 7.2. They were then post-fixed in 1% osmium tetroxide in the same buffer, dehydrated in acetone, infiltrated and embedded in the low viscosity Spurr's epoxy resin (Kitajima and Alberti, 2014). The blocks were sectioned using a Leica EMUC7 ultramicrotome equipped with Diatome diamond knife. Thin sections were collected on 300 mesh copper grids, stained with 3% uranyl acetate and Reynold's lead citrate, and examined using a Jeol JEM 1011 transmission electron microscope. Images were registered digitally with a Gatan camera.
3. Results
3.1. Construction of the full-length cDNA clone of CiLV-C
The full-length cDNAs of two CiLV-C RNA fragments were individually cloned into the pJL89 and pLXAS vectors, between a double CaMV 35S promoter and hepatitis delta virus (HDV) ribozyme (Rz) followed by the NOS (pJL89) or the potato proteinase inhibitor (PoPit) terminator sequences (pLXAS) (Fig. 1A and B), to generate pJL89/35S-cDNA1, pJL89/35S-cDNA2, pLXAS/35S-cDNA1 and pLXAS/35S-cDNA2 constructs. The resultant viral constructs contained at the 3′ termini of the viral RNA, a poly (A) tail of 17 nt (pJL89) or 50 nt (pLX) (Fig. 1A and B). Together with the infectious CiLV-C clones, a mutated version of the RNA1 carrying a premature stop codon in the CiLV-C polymerase (Fig. 1C), was generated by replacing the nucleotide ‘G’ with a ‘T’ at position 1.405, generating the pJL89/35S-cDNA1mut and pLXAS/35S-cDNA1mut constructs.
3.2. Infectivity of CiLV-C full-length cDNA clones in N. benthamiana
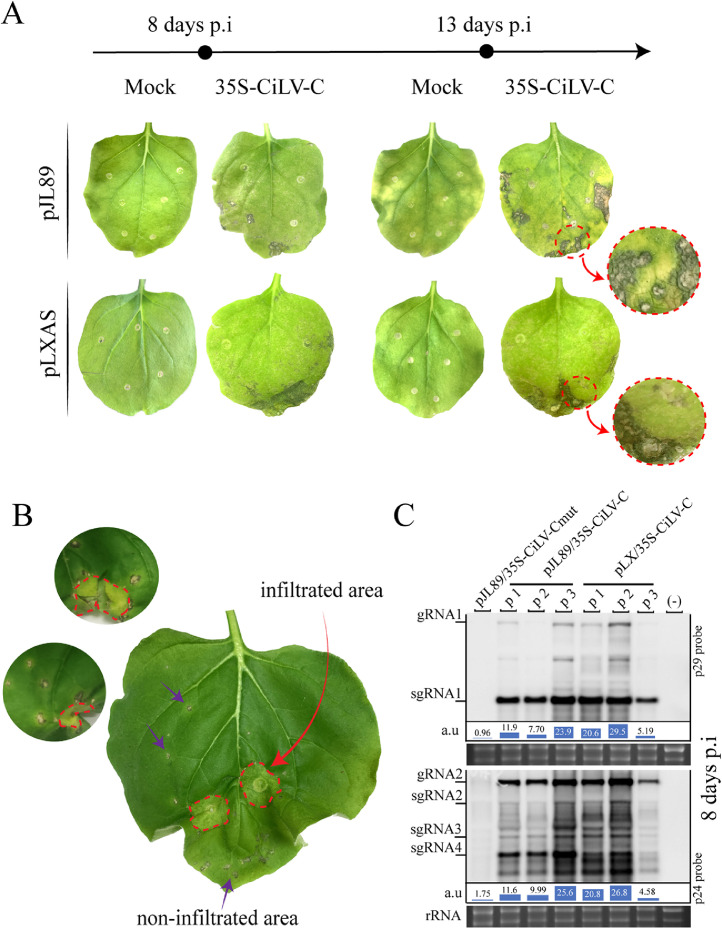
The infectivity of the CiLV-C full-length cDNA clones was tested by agroinfiltration of N. benthamiana leaves with pJL89/35S-cDNA1 + pJL89/35S-cDNA2 (pJL89/35S-CiLV-C) and pLXAS/35S-cDNA1 + pLXAS/35S-cDNA2 (pLXAS/35S-CiLV-C) constructs. As a negative control and due to the incapacity of the CiLV-C to move systemically, the N. benthamiana plants were agroinfiltrated with a construct carrying a stop codon at the RNA polymerase gene. To this end, pJL89/35S-cDNA1mut + pJL89/35S-cDNA2 (pJL89/35S-CiLV-Cmut) and pLXAS/35S-cDNA1mut + pLXAS/35S-cDNA2 (pLXAS/35S-CiLV-Cmut) were agroinfiltrated into N. benthamiana plants at the four-leaf growth stage. At 8- and 13-days p.i, non-symptoms were visualized in the agroinfiltrated leaves of the negative controls (Fig. 2A). In contrast, at 8 days p.i, N. benthamiana leaves agroinfiltrated with pJL89/35S-CiLV-C and pLXAS/35S-CiLV-C showed necrosis formation that increased over the course of the infection, resulting in completely necrotic leaf areas and necrotic spots (Fig. 2A, at 13 days p.i). The necrotic spots remind the symptoms previously reported in N. benthamiana experimentally-mite inoculated with CiLV-C (Garita et al., 2014). To investigate if the necrosis could be formed outside the infiltrated areas, which would suggest that the rescued virus could spread locally (cell-to-cell), we infiltrate small regions of the leaves and observed the formation of necrosis in areas around and far from the infiltrated regions. At 10–15 days p.i, we observed typical CiLV-C necrotic spots around infiltrated zones and in non-infiltrated regions of the leaf (Fig. 2B). The CiLV-C infectious clone showed a high efficiency of infection in the agroinfiltration assay with the 100% of infiltrated plants (five plants per construct in three independent experiments) showing necrotic symptoms in the infiltrated leaves. No plant showed systemic infection.
Fig. 2.
Infectivity analysis of the full-length cDNA citrus leprosis virus C clone. (A) Necrotic symptoms (necrotic leaf areas and necrotic spots) are visualized in infiltrated N. benthamiana leaves at 8- and 13-days post infiltration, co-infiltrated with pJL89/35S-cDNA1 + pJL89/35S-cDNA2 (pJL89/35S-CiLV-C) and pLXAS/35S-cDNA1 + pLXAS/35S-cDNA2 (pLXAS/35S-CiLV-C) constructs. Mock corresponds to plants infiltrated with the pJL89 or pLXAS constructs carrying the CiLV-C RNA1 mutant, containing a premature stop codon in the CiLV-C polymerase, co-infiltrated with the respective CiLV-C RNA2 construct. (B) Cell-to-cell spread of CiLV-C infectious clone. Small regions of the leaf were infiltrated to evaluate the necrotic symptoms outside the infiltrated region. The red arrow indicates a small leaf area infiltrated and the purple arrows indicate necrotic lesions outside the infiltrated area. (C) Northern blot showing the profile of acid nucleic detection from N. benthamina leaves agroinfiltrated with the CiLV-C pJL89 or pLXAS constructs at 8 days post-infiltration, using a DIG-riboprobe complementary to the CiLV-C p29 and p24 genes. (-) corresponds to a non-infiltrated plant. Each lane corresponds to a different N. benthamiana plant. p1, p2, and p3 = plant 1, plant 2, and plant 3. rRNA stained with ethidium bromide indicates equal loading of samples. The localization of CiLV-C gRNA1 and 2 and sgRNA1, 2, 3, and 4 are indicated. The graphs represent the relative accumulation (a.u) of total CiLV-C RNAs.
Northern blot analysis using total RNA extracted from leaves expressing the pJL89/35S-CiLV-Cmut and pJL89 or pLX/35S-CiLV-C constructs were loaded at the same concentration (6 μg) in an RNA denaturing gel. The results confirmed the virus replication by the formation of subgenomics RNAs from CiLV-C RNA 1 and 2 when compared with the control. A very abundant band that corresponds to p29 (sgRNA1) and p24 (sgRNA4) was detected (Fig. 2C). Furthermore sgRNAs 2 and 3 from gRNA2 were visualized, in contrast to the control expressing the truncated polymerase, which showed a soft background (Fig. 2C) corresponding to the agrobacterium transcribed viral RNAs. The quantification of genomic and subgenomic CiLV-C RNAs revealed that the accumulation of viral RNAs increased more in those plants agroinfiltrated with CiLV-C expressing the complete polymerase when compared with the control (Fig. 2C). Quantification of the accumulation of CiLV-C RNAs revealed that leaves expressing the non-mutated clones generated approximately 16.9 (RNA1, 16.25 vs. 0.96) and 9.25 (RNA2, 16.20 vs. 1.75) times more hybridization signal than the control (Fig. 2C), thus indicating that the signal detected in the samples expressing the complete polymerase comes from the CiLV-C RNA replication. This result together with the abundant detection of p29 and p24 sgRNAs, allows us to conclude that the CiLV-C full-length cDNA clones generate replicative CiLV-C vRNAs in N. benthamiana cells.
3.3. The infectivity of the full-length cDNA clone of CiLV-C is confirmed by mechanical inoculation
The infectivity of the selected clones was also confirmed by mechanical inoculation. Since the two sets of constructs derived from the pJL89 and pLXAS plasmids do not exhibit significant differences in virus accumulation in the agroinfiltrated N. benthamiana leaves, this assay was performed using plants agroinfiltrated with pJL89 derivatives.
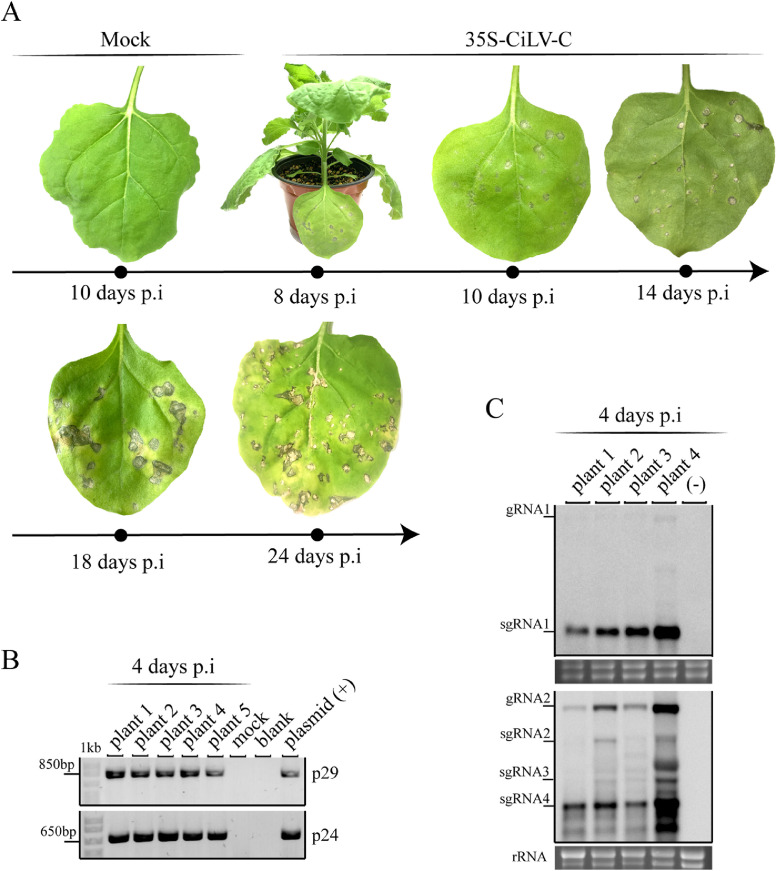
Extracts of the agroinfiltrated leaves from several plants were used as sources of inocula. Leaf extracts from plants infiltrated with pJL89/35S-CiLV-Cmut were also used as control inoculum (mock control). The infiltrated leaves were ground in 30 mM phosphate buffer (pH 8.0) and the extracts obtained were used for the inoculation of new N. benthamiana plants. The typical CiLV-C symptoms corresponding to a localized necrotic lesion became evident 8 days after mechanical inoculation and progressed in the course of the viral infection (Fig. 3A). On the other hand, extracts from plants expressing the mock control did not present symptoms (Fig. 3A). Several inoculated leaves, in three independent experiments (five plants per experiment) showed necrotic symptoms in the inoculated leaves indicating a high efficiency in the mechanical transmission. Total RNA from inoculated leaves was extracted at 4 days p.i., treated with DNase I and the presence of the virus was evaluated by RT-PCR. Specific amplicons corresponding to the CiLV-C p29 and p24 genes were detected in all inoculated plants except the mock control (Fig. 3B). To further confirm the CiLV-C infectivity, we performed a northern blot from some of the positive RT-PCR samples. Northern blot analysis confirmed the virus replications by the detection of subgenomics RNAs from CiLV-C RNA 1 and 2 (Fig. 3C). Variation in the concentration of viral RNA in the samples was noticed.
Fig. 3.
CiLV-C derived from the cDNA clone is mechanically transmitted. (A) CiLV-C symptoms are clearly seen as necrotic spots at 8 days and progress in the course of the viral infection. Mock corresponds to the plants inoculated with extract of plants agroinfiltrated with pJL89/35S-cDNA1mut + pJL89/35S-cDNA2 (pJL89/35S-CiLV-Cmut). (B) RT-PCR analysis from RNA extracted (DNase I treated) of the leaves mechanically inoculated with CiLV-C inoculum derived from the cDNA clone or the CiLV-C mutated version (Mock). Blank, no template added. (+), positive control (plasmid) to amplify the CiLV-C p29 and p24 genes. p29 and p24 have 792 bp and 645 bp, respectively. The marker band sizes of 850 bp and 650 bp are indicated. 1 kb corresponds to GeneRuler 1 kb ladder marker. (C) Northern blot at 4 days post-mechanical inoculation, using a DIG-riboprobe complementary to the CiLV-C p29 and p24 genes. rRNA stained with ethidium bromide indicates equal loading of samples. The localization of gRNA1 and 2 and sgRNA1, 2, 3, and 4 are indicated.
3.4. Partial particle-purification assay and TEM shows the presence of CiLV-C virions
To demonstrate the rescue of virus particles from the CiLV-C infectious clone, we subjected samples from agroinfiltrated tissue to partial purification using PEG for virion precipitation. After purification, a portion of the pellet generated from leaf extracts of plants expressing pJL89/35S-CiLV-Cmut (mock), pJL89/35S-CiLV-C, and pLXAS/35S-CiLV-C were loaded onto a 1% agarose gel. For the samples expressing the functional infectious clones (pJL89/35S-CiLV-C and pLXAS/35S-CiLV-C), a clear virion band was visualized (Fig. 4A), while the same band was not noticed at the control (pJL89/35S-CiLV-Cmut). To confirm that the band corresponds to CiLV-C virus particles, a part of the supernatant obtained after the purification was subjected to RNA extraction, treated with DNase I, and used for an RT-PCR reaction with specific primer for CiLV-C p29 and p24 genes. Positive p29 and p24 amplicons were visualized (Fig. 4B) from leaves expressing pJL89 or pLXAS/35S-CiLV-C constructs, while no amplification was obtained in the sample containing the control pJL89/35S-CiLV-Cmut construct. To exclude that the positive amplification was generated by the presence of plasmid from the agrobacterium - even after the RNA has been treated with DNase - we performed a PCR directly using the RNA as a template and, as expected, positive amplicons for CiLV-C p29 and p24 genes were not noticed. We also evaluate the possible presence of viral proteins by SDS-PAGE analysis. A portion of the supernatant obtained was mixed with Laemmli sample buffer (0.125 M This HCl pH 6.8, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, and 0.004% bromophenol blue) next, the samples were denatured and loaded in a 12% SDS-PAGE gel electrophoresis. Bands similar in size to p29 and p24 (that sgRNAs are transcribed abundantly during CiLV-C infection (Pascon et al., 2006)) (Fig. 4C, red and black arrows, respectively) were visualized in the gel lane loaded with pJL89/35S-CiLV-C sample. Bands of similar size were not observed in the lane loaded with the mock sample (pJL89/35S-CiLV-Cmut). To provide further evidence of the particle formation from the CiLV-C infectious clone, the supernatant fraction of partially purified virions preparation from leaves expressing the pJL89/35S-CiLV-C constructs and small tissue pieces of N. benthamiana leaves experimentally inoculated with the CiLV-C infectious clone were negatively stained and examined under TEM. Electro-dense short-bacilliform structures (likely virions) were observed (Fig. 4D), exhibiting sizes ranging from 20 to 40 nm in width and 40–130 nm in length (Fig. 4E) in samples from the partially purified virus. In samples obtained from ultra-thin sections of tissue infected with CiLV-C infectious clones, a large number of membrane-bounded particles were observed. These particles exhibited circular or bacilliform profiles (40–50 nm in width and 80–100 nm in length) in several leaf parenchymal cells, and were consistently found within the cisternae of the endoplasmic reticulum (Fig. 4F). These observed particles were essentially similar to those described and identified as CiLV-C particles in cells from leprotic lesions in orange (Citrus sinensis Osbeck) leaf, stem, or fruit cells infected by CiLV-C (Calegario et al., 2013; Rodrigues et al., 2003), and also in the intercellular space (celoma) of viruliferous Brevipalpus yothersi mite vector (Kitajima and Alberti, 2014). Taken together, these findings strongly indicate the formation of viral particles rescued from the CiLV-C infectious clone.
Fig. 4.
Partial purification of CiLV-C virions from agroinfiltrated Nicotiana benthamiana leaves. (A) Agarose gel electrophoresis analysis of CiLV-C virions PEG precipitated from leaves infiltrated with pJL89/35S-CiLV-Cmut (mock), pJL89/35S-CiLV-C, and pLX/35S-CiLV-C clone. Arrow indicates the putative CiLV-C virion. (B) RT-PCR or PCR reactions using total RNA (DNase I treated) extracted from samples analyzed in A. (+), positive control in which the amplifications were performed using a plasmid containing the CiLV-C p29 and p24 genes, as templates. p29 and p24 genes have 792 bp and 645 bp, respectively. Blank, no template added. The marker band sizes of 850 bp and 650 bp are indicated. 1 kb corresponds to GeneRuler 1 kb ladder marker. (C) SDS-PAGE of partial purification of CiLV-C virus particles. The gel was stained by Coomassie Brilliant Blue. The gel image shows the PageRuler Plus Prestained protein ladder ranging from 10 to 250 kDa. Red and black arrows indicate bands similar in size to CiLV-C p29 and p24 proteins, respectively. The asterisk (*) highlights equivalent bands, demonstrating that the samples were loaded in similar concentrations. (D) Electro-dense short-bacilliform structures (putative CiLV-C virions) were negatively stained with 2% phosphotungstic acid and visualized with the aid of a TEM. Bars correspond to 50 nm (right image) or 100 nm (left image). (E) Schematic representation of short-bacilliform virion showing the particle size range (width and length) obtained from analyzing 25 putative virions using the ImageJ software. (F) Transmission electron micrographs of leaf parenchymal cells from N. benthamiana experimentally infected with CiLV-C infectious clone at 15 days post-inoculation. Many putative CiLV-C virions (40–50 nm in width and 80–100 nm in length) are present within membrane-bounded cisternae of the endoplasmic reticulum in the cytoplasm. Bars correspond to 2 μm, 1 μm, and 200 nm (from left to right). The red-dotted circle highlights the increased area observed in the images on the right. The red arrows indicate individual viral particles.
4. Discussion
CiLV-C, the type species of the genus Cilevirus, is an important viral pathogen that greatly impacts citrus orchard production. Despite its detrimental effects on citrus production, particularly in the Americas, CiLV-C and the citrus leprosis pathosystem remain poorly studied. This knowledge gap can be attributed, in part, to the lack of an available CiLV-C infectious clone. Over the years many infectious clones for plant viruses have been constructed, providing an excellent tool for gene expression systems, the investigation of virus-host interactions, and the study of viral gene and protein functions (Shakir et al., 2023). Additionally, these clones have been used to confer virus resistance to host plants through the silencing of host genes or gene overexpression (Boyer and Haenni, 1994; Gleba et al., 2004; Jiang et al., 2018; Nagyová and Subr, 2007).
In this study, we report a functional infectious cDNA clone for a member of the genus Cilevirus. The infectivity of the virus was confirmed in N. benthamiana infiltrated plants, and further validated through viral transmission to mechanically inoculated plants. Symptoms consisting of necrotic circular lesions, usually associated with CL, were observed after agroinfiltration and mechanical inoculation with the infectious clone, although only in the inoculated leaves, reproducing what is observed in the natural citrus infection and in the experimental host, which shows local necrotic lesions restricted to leaves inoculated with the virus (Bastianel et al., 2010; Garita et al., 2014). In this sense, the introduction of a premature stop codon in the CiLV-C polymerase gene, which significantly reduced the accumulation of viral RNA, will be a very attractive tool to study the viral cycle, allowing the discrimination between the viral progeny and the 35S-transcribed viral RNAs.
Members of the family Kitaviridae generate an unusual infection phenotype consisting of locally restricted infection (Ramos-Gonzalez et al., 2023). The molecular factors associated with this restriction in systemic infection are still unknown. The lack of systemic movement in the natural context of cilevirus infection has led to questioning the efficiency and functionality of its encoded movement protein (MP). However, recent studies demonstrate the capacity of cileviruses MP to rescue the local or systemic movement of defective-infectious clones belonging to the genera Alfamovirus, Tobamovirus, and Betacarmovirus (Leastro et al., 2020a, 2020b, 2021a), indicating that other viral proteins or genome regions should trigger the restricted local infection of cileviruses. In this sense, viral necrotic lesions associated with the plant hypersensitive response (HR) should play a role in restricting virus movement (Pallas and Garcia, 2011; Paudel and Sanfaçon, 2018). Many viral proteins could trigger HR, including RSS (Aguilar et al., 2019; Inaba et al., 2011). We have recently reported RSS activity for the p29, p15, and p61 CiLV-C proteins (Leastro et al., 2020a) opening the possibility that the restricted local transport of Cilevirus could be associated with the strong necrotic lesions observed in the inoculated leaves triggered by one or some of the encoded RSS. Further analysis will be addressed to this hypothesis by using the infectious CiLV-C clones described herein.
Several electro-dense short-bacilliform virions were observed, measuring a range of 20–40 nm in width and 40–130 nm in length for samples from partial particle-purification assay. Previous studies speculated that CiLV-C has a bacilliform particle measuring 40–70 nm in width and 100–120 nm in length (Kitajima et al., 1974; Ramos-Gonzalez et al., 2023). Nevertheless, ultra-thin sections from CiLV-C infected tissue showed virions similar to those previously described and identified as CiLV-C particles in cells from Citrus sinensis Osbeck. These virions have a circular or bacilliform particle profile, measuring 40–50 nm in width and 80–100 nm in length (Calegario et al., 2013; Rodrigues et al., 2003). The variation in particle size can be attributed to the preservation state of the tissue, which can affect virions morphology, tending to become spheroidal when they are not well preserved, and the method used for particle observation (observations made by EWK).
On the other hand, all analyzes performed in the present study have been based on N. benthamiana species since this plant has been previously tested as an experimental host for the virus (Garita et al., 2014) and has long been used as a host model to study plant virus infection. The region of Valencia is the largest producer of citrus in all of Europe, and CiLV-C, along with other CL viruses, is considered as a Union quarantine pest (Lázaro et al., 2022). Consequently, no bioassays using CiLV-C infectious clones will be addressed on its natural host upon receiving authorization, representing one of our future steps. However, these infectivity tests could be done in countries where the virus has been present for a long time.
In conclusion, we present here, for the first time in the genus Cilevirus, a CiLV-C infectious clone, which will be an excellent tool to analyze crucial aspects of the viral cycle including the peculiar local restriction of the viral infection attributed to members of the family Kitaviridae.
Ethical approval
This article does not contain any studies with human participants or animals requiring ethical approval.
Funding
This work was supported by grant PID2020-115571RB-100 from the Spanish MCIN/AEI/10.13039/501100011033 granting agency.
CRediT authorship contribution statement
Mikhail Oliveira Leastro: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Elliot Watanabe Kitajima: Methodology, Investigation. Vicente Pallás: Writing – review & editing, Funding acquisition. Jesús Ángel Sánchez-Navarro: Supervision, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Isabel Font for her assistance in preparing the samples analyzed in TEM. We thank Lorena Corachán for her excellent technical support. We Thank Rodolfo De La Torre to provide total RNA citrus samples. The biological tests were carried out in Brazil.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199264.
Contributor Information
Mikhail Oliveira Leastro, Email: molilea@ibmcp.upv.es.
Jesús Ángel Sánchez-Navarro, Email: jesanche@ibmcp.upv.es.
Appendix. Supplementary materials
Data availability
No data was used for the research described in the article.
References
- Aguilar E., Del Toro F.J., Brosseau C., Moffett P., Canto T., Tenllado F. Cell death triggered by the P25 protein in Potato virus X-associated synergisms results from endoplasmic reticulum stress in Nicotiana benthamiana. Mol. Plant Pathol. 2019;20(2):194–210. doi: 10.1111/mpp.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z., Abul-faraj A., Li L., Ghosh N., Piatek M., Mahjoub A., Aouida M., Piatek A., Baltes N.J., Voytas D.F., Dinesh-Kumar S., Mahfouz M.M. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant. 2015;8(8):1288–1291. doi: 10.1016/j.molp.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Bastianel M., Novelli V., Kitajima E., Kubo K. Citrus leprosis: centennial of an unusual mite virus pathosystem. Plant Dis. 2010;94:284–292. doi: 10.1094/PDIS-94-3-0284. [DOI] [PubMed] [Google Scholar]
- Boyer J.C., Haenni A.L. Infectious transcripts and cDNA Clones of RNA viruses. Virology. 1994;198(2):415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- Calegario R.F., Locali E.C., Stach-Machado D.R., Peroni L.A., Caserta R., Salaroli R.B., Freitas-Astúa J., Machado M.A., Kitajima E.W. Polyclonal antibodies to the putative coat protein of Citrus leprosis virus C expressed in Escherichia coli: production and use in immunodiagnosis. Trop. Plant Pathol. 2013;38 [Google Scholar]
- Cordero T., Mohamed M.A., López-Moya J.J., Daròs J.A. A recombinant potato virus Y infectious clone tagged with the rosea1 visual marker (PVY-Ros1) facilitates the analysis of viral infectivity and allows the production of large amounts of anthocyanins in plants. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Astua J., Ramos-Gonzalez P.L., Arena G.D., Tassi A.D., Kitajima E.W. Brevipalpus-transmitted viruses: parallelism beyond a common vector or convergent evolution of distantly related pathogens? Curr. Opin. Virol. 2018;33:66–73. doi: 10.1016/j.coviro.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Garita L.C., Tassi A.D., Calegario R.F., Freitas-Astúa J., Salaroli R.B., Romão G.O., Kitajima E.W. Experimental host range of Citrus leprosis virus C (CiLV-C) Trop Plant Pathol. 2014;39:43–55. [Google Scholar]
- Gleba Y., Marillonnet S., Klimyuk V. Engineering viral expression vectors for plants: the 'full virus' and the 'deconstructed virus' strategies. Curr. Opin. Plant Biol. 2004;7(2):182–188. doi: 10.1016/j.pbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Inaba J., Kim B.M., Shimura H., Masuta C. Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol. 2011;156(4):2026–2036. doi: 10.1104/pp.111.180042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Jiang L., Yang J., Peng J., Lu Y., Zheng H., Lin L., Chen J., Yan F. Over-expression of Oryza sativa Xrn4 confers plant resistance to virus infection. Gene. 2018;639:44–51. doi: 10.1016/j.gene.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Kitajima E.W., Alberti G. Anatomy and fine structure of Brevipalpus mites (Tenuipalpidae, Prostigmata, Acrinotrichida) Part 7. Ultrastructural detection of cytoplasmic and nuclear types of Brevipalpus transmitted viruses. Zoologica. 2014;160:173–192. [Google Scholar]
- Kitajima, E.W., Rosillo, M.A., Portillo, M.M., Müller, G.W., and Costa, A.S., 1974. Microscopia eletrônica de tecidos foliares de laranjeiras infectadas pela lepra explosiva da Argentina. Fitopatologia Brasileira, Brasília. 9, 55–56.
- Kuchibhatla D.B., Sherman W.A., Chung B.Y., Cook S., Schneider G., Eisenhaber B., Karlin D.G. Powerful sequence similarity search methods and in-depth manual analyses can identify remote homologs in many apparently "orphan" viral proteins. J. Virol. 2014;88(1):10–20. doi: 10.1128/JVI.02595-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro, E., Vanaclocha, P., Vicent, A., Vives, M.A., Delbianco, A., 2022. Pest survey card on Citrus leprosis viruses. European Food Safety Authority, 1–6.
- Leastro M.O., Castro D.Y.O., Freitas-Astua J., Kitajima E.W., Pallas V., Sanchez-Navarro J.A. Citrus leprosis virus C encodes three proteins with gene silencing suppression activity. Front. Microbiol. 2020;11:1231. doi: 10.3389/fmicb.2020.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leastro M.O., Freitas-Astua J., Kitajima E.W., Pallas V., Sanchez-Navarro J.A. Dichorhaviruses movement protein and nucleoprotein form a protein complex that may be required for virus spread and interacts in vivo with viral movement-related Cilevirus proteins. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.571807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leastro M.O., Freitas-Astua J., Kitajima E.W., Pallas V., Sanchez-Navarro J.A. Unravelling the involvement of Cilevirus p32 protein in the viral transport. Sci. Rep. 2021;11(1):2943. doi: 10.1038/s41598-021-82453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leastro M.O., Kitajima E.W., Silva M.S., Resende R.O., Freitas-Astua J. Dissecting the subcellular localization, intracellular trafficking, interactions, membrane association, and topology of citrus leprosis virus C proteins. Front. Plant Sci. 2018;9:1299. doi: 10.3389/fpls.2018.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leastro M.O., Pallas V., Resende R.O., Sanchez-Navarro J.A. The movement proteins (NSm) of distinct tospoviruses peripherally associate with cellular membranes and interact with homologous and heterologous NSm and nucleocapsid proteins. Virology. 2015;478:39–49. doi: 10.1016/j.virol.2015.01.031. [DOI] [PubMed] [Google Scholar]
- Leastro M.O., Villar-Alvarez D., Freitas-Astua J., Kitajima E.W., Pallas V., Sanchez-Navarro J.A. Spontaneous mutation in the movement protein of citrus leprosis virus C2, in a heterologous virus infection context, increases cell-to-cell transport and generates fitness advantage. Viruses. 2021;13(12) doi: 10.3390/v13122498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo J.A. TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 2007;145(4):1232–1240. doi: 10.1104/pp.107.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhao C., Sun K., Deng Y., Li Z. Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes. Mol. Plant. 2023;16(3):616–631. doi: 10.1016/j.molp.2023.02.003. [DOI] [PubMed] [Google Scholar]
- Nagyová A., Subr Z. Infectious full-length clones of plant viruses and their use for construction of viral vectors. Acta Virol. 2007;51(4):223–237. [PubMed] [Google Scholar]
- Ortega-Rivera O.A., Beiss V., Osota E.O., Chan S.K., Karan S., Steinmetz N.F. Production of cytoplasmic type citrus leprosis virus-like particles by plant molecular farming. Virology. 2023;578:7–12. doi: 10.1016/j.virol.2022.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas V., Garcia J.A. How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 2011;92(Pt 12):2691–2705. doi: 10.1099/vir.0.034603-0. [DOI] [PubMed] [Google Scholar]
- Pallas V., Mas P., Sanchez-Navarro J.A. Detection of plant RNA viruses by nonisotopic dot-blot hybridization. Methods Mol. Biol. 1998;81:461–468. doi: 10.1385/0-89603-385-6:461. [DOI] [PubMed] [Google Scholar]
- Pascon R.C., Kitajima J.P., Breton M.C., Assumpcao L., Greggio C., Zanca A.S., Okura V.K., Alegria M.C., Camargo M.E., Silva G.G., Cardozo J.C., Vallim M.A., Franco S.F., Silva V.H., Jordao H., Oliveira F., Giachetto P.F., Ferrari F., Aguilar-Vildoso C.I., Franchiscini F.J., Silva J.M., Arruda P., Ferro J.A., Reinach F., da Silva A.C. The complete nucleotide sequence and genomic organization of Citrus Leprosis associated Virus, Cytoplasmatic type (CiLV-C) Virus Genes. 2006;32(3):289–298. doi: 10.1007/s11262-005-6913-1. [DOI] [PubMed] [Google Scholar]
- Pasin F., Bedoya L.C., Bernabe-Orts J.M., Gallo A., Simon-Mateo C., Orzaez D., Garcia J.A. Multiple T-DNA delivery to plants using novel mini binary vectors with compatible replication origins. ACS Synth. Biol. 2017;6(10):1962–1968. doi: 10.1021/acssynbio.6b00354. [DOI] [PubMed] [Google Scholar]
- Paudel D.B., Sanfaçon H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018;9:1575. doi: 10.3389/fpls.2018.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Ma X., Lu S., Li Z. A versatile plant rhabdovirus-based vector for gene silencing, miRNA expression and depletion, and antibody production. Front. Plant Sci. 2021;11 doi: 10.3389/fpls.2020.627880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gonzalez P.L., Arena G.D., Tassi A.D., Chabi-Jesus C., Kitajima E.W., Freitas-Astua J. Kitaviruses: a window to atypical plant viruses causing nonsystemic diseases. Annu. Rev. Phytopathol. 2023;61:97–118. doi: 10.1146/annurev-phyto-021622-121351. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.C.V., Kitajima E.W., Childers C.C., Chagas C.M. Citrus leprosis virus vectored by brevipalpus phoenicis (Acari: tenuipalpidae) on citrus in Brazil. Exp. Appl. Acarol. 2003;30(1):161–179. doi: 10.1023/b:appa.0000006547.76802.6e. [DOI] [PubMed] [Google Scholar]
- Roy A., Hartung J.S., Schneider W.L., Shao J., Leon G., Melzer M.J., Beard J.J., Otero-Colina G., Bauchan G.R., Ochoa R., Brlansky R.H. Role bending: complex relationships between viruses, hosts, and vectors related to citrus leprosis, an emerging disease. Phytopathology. 2015;105(7):1013–1025. doi: 10.1094/PHYTO-12-14-0375-FI. [DOI] [PubMed] [Google Scholar]
- Shakir S., Zaidi S.S.A., Hashemi F.S.G., Nyirakanani C., Vanderschuren H. Harnessing plant viruses in the metagenomics era: from the development of infectious clones to applications. Trends Plant Sci. 2023;28(3):297–311. doi: 10.1016/j.tplants.2022.10.005. [DOI] [PubMed] [Google Scholar]
- Tassi A.D., Garita-Salazar L.C., Amorim L., Novelli V.M., Freitas-Astua J., Childers C.C., Kitajima E.W. Virus-vector relationship in the Citrus leprosis pathosystem. Exp. Appl. Acarol. 2017;71(3):227–241. doi: 10.1007/s10493-017-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uranga M., Aragonés V., Daròs J.A., Pasin F. Heritable CRISPR-Cas9 editing of plant genomes using RNA virus vectors. STAR Protoc. 2023;4(1) doi: 10.1016/j.xpro.2023.102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.