ABSTRACT
Periodic pattern formation on the cellular and tissue scale is an important process and has been extensively studied. However, periodic pattern formation at the subcellular level still remains poorly understood. The C. elegans epidermis displays a highly ordered parallel stripe pattern as part of its subcellular structure, making it an ideal model to study the formation and reorganization of periodic patterns within cells. Here, we show that the initial formation of periodic striped patterns in the C. elegans epidermis is dependent on actin and spectrin, and requires the apical membrane attachment structures for maintenance. The periodic subcellular structures do not accommodate cell growth by continuously making new stripes. Instead, they increase the number of stripes by going through one round of uniform duplication, which is independent of the increasing epidermal length or the developmental cycles. This long-range synchronized reorganization of subcellular structures is achieved by physical links established by extracellular collagens together with extension forces generated from epidermal cell growth. Our studies uncover a novel strategy employed by evenly spaced and interlinked subcellular structures to maintain their integrity and equidistribution during cell growth and tissue development.
Keywords: C. elegans, Epidermis, Periodic pattern, Hemidesmosome, Collagen
Highlighted Article: By using C. elegans epidermis as a model, we find that periodically organized subcellular structures perform synchronized duplication to increase their numbers and accommodate epidermal cell growth.
INTRODUCTION
Regularly spaced and periodically arranged patterns, such as evenly distributed dots and stripes, are a common sight in the biological world. Numerous studies have already revealed the complex mechanisms underlying periodic pattern formation on the cellular and tissue scales (Schroeder et al., 2011; Economou et al., 2012; Dalle Nogare and Chitnis, 2017; Schweisguth and Corson, 2019). Intracellular pattern formation, on the other hand, is relatively less well studied. Thanks to recent development of super-resolution imaging techniques, such as stimulated emission depletion and stochastic optical reconstruction microscopy, investigations regarding intracellular periodic patterns have been greatly accelerated. In particular, the Zhuang group reported the existence of a strictly periodic cytoskeletal structure composed of actin rings and spectrin tetramers in neuronal cells across species (Xu et al., 2013; He et al., 2016). Periodically arranged membrane skeletons are also found in the cilia and seam cells of C. elegans, indicating the prevalence of intracellular periodic patterns in multiple cell types (Jia et al., 2019). Subsequent studies further revealed that periodic subcellular patterns are vitally important for cellular morphogenesis and functioning. They provide stable and elastic mechanical support, organize membrane protein distribution, compartmentalize intracellular molecular motions, provide signaling platforms and drive nanoscale alignment of neighboring cells (Albrecht et al., 2016; Qu et al., 2017; Huang et al., 2017; Hauser et al., 2018; Wang et al., 2019; Zhou et al., 2019). However, as cells constantly change their shape and volume in various biological processes, how the periodic subcellular structures manage to maintain their functions and organization during these changes still remains unknown.
The epidermis of Caenorhabditis elegans, which possesses a highly ordered periodic subcellular framework, is an ideal model to address this question. The transparent body and the enormous cell size of the C. elegans epidermis ensure that its subcellular patterns can be easily observed with conventional confocal microscopes inside live animals. The adult C. elegans epidermis is a single epithelial layer formed mostly by the giant hyp7 (the cell number 7 of the hypodermis) syncytium of ∼1 mm in length. It is apically anchored to the cuticle exoskeleton and basally supported by body-wall muscles (Chisholm and Xu, 2012). The trans-epidermal attachment structures known as the C. elegans hemidesmosomes (CeHDs) connect the cuticle, epidermis and basement membrane together through intermediate filaments (IFs) (Zhang and Labouesse, 2010). Interestingly, multiple subcellular structures in the C. elegans epidermis, including actin, microtubule, intermediate filaments, spectrins and CeHDs, all display neatly paralleled periodic patterns and share a common periodic framework (Fig. S1) (Francis and Waterson, 1991; Costa et al., 1997; Hresko et al., 1999; Hong et al., 2001; Bosher et al., 2003; Liégeois et al., 2007; Zhang et al., 2015). In addition, certain extracellular matrix proteins and non-structural signaling proteins are also periodically arranged (Fig. S1) (McMahon et al., 2003; Hao et al., 2006; Soloviev et al., 2011). This periodic framework maintains the epidermal structure and compartmentalizes membrane receptors and extracellular matrix (ECM) proteins. In this work, we employ C. elegans as the model and systemically investigate the formation and maturation of the periodically arranged subcellular structures during epidermal morphogenesis. Our discoveries may help us understand how interlinked subcellular structures manage to maintain stability and even distribution during cell growth and tissue morphogenesis.
RESULTS
The initiation of the periodic-patterned frame in the C. elegans epidermis is dependent on actin and spectrin
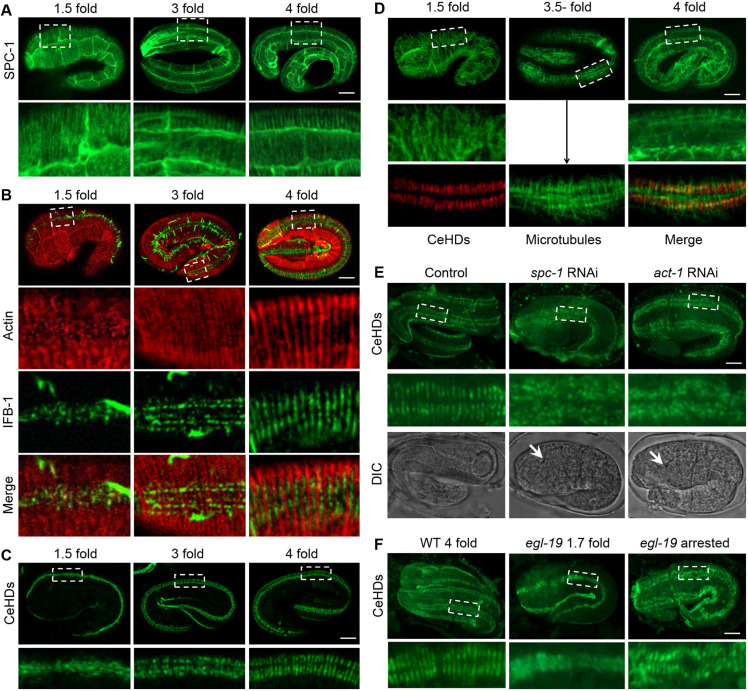
To determine the sequence of events that leads to the initial establishment of the periodic framework, we examined the formation of several major periodic structures in early embryos. We found that the first structure that assumed the striped pattern is the spectrin network. As shown in Fig. 1A, at as early as the 1.5-fold stage, the SPC-1 (the C. elegans α-spectrin protein) had already formed parallel stripes in the epidermis (Norman and Moerman, 2002). This striped pattern of SPC-1 persisted throughout embryogenesis and became increasingly prominent in older embryos (Fig. 1A). The actin filaments display organization of parallel bundles at about the 2-fold stage (Fig. 1B) (Priess and Hirsh, 1986; Gally et al., 2009). Double-labeling revealed that the stripe-formation process for the IFs followed that of the actin bundles. As the cells elongate, the IFs appeared to align according to the position of the actin bundles and filled up the spaces between adjacent bundles, then gradually fused together to form continuous stripes (Fig. 1B). Being in the same anchoring complex, the reorganization process of the CeHDs essentially mimics that of the IFs (Hresko et al., 1994; Bosher et al., 2003). The IF and CeHD units did not fuse into continuous stripes until 4-fold embryonic stage (Fig. 1C). The organization of the microtubule bundles was comparatively irregular and only became temporarily semi-parallel at around the 3-fold stage. Double-labeling experiments showed that the positions of the microtubule bundles do not strictly correlate with those of the CeHD stripes (Fig. 1D).
Fig. 1.
The initialization of epidermal intracellular periodic patterns is actin and spectrin dependent. (A) SPC-1::GFP localization in the epidermis of embryos at early (1.5-fold), mid (3-fold) and late (4-fold) elongation stages. (B) Double-labeling of actin (red) and intermediate filaments (green) by ABD::mCherry and IFB-1::GFP in embryos at 1.5-fold, 3-fold and 4-fold elongation stages. (C) Epidermal distribution of CeHDs visualized by VAB-10A::GFP in embryos at 1.5-fold, 3-fold and 4-fold elongation stages. (D) Epidermal distribution of microtubules (green) labeled by anti-tubulin in embryos at 1.5-fold, 3.5-fold and 4-fold elongation stages. Double-labeling of microtubules and CeHDs (red, anti-VAB-10A) at the 3.5-fold embryonic stage is also shown (arrow). (E) Organization of CeHDs visualized by anti-VAB-10A in NR222 embryos with or without epidermal-specific RNAi against spc-1 and act-1. White arrows point to developed pharynx structures in late stage embryos. (F) Epidermal organization of CeHDs visualized by anti-VAB-10A in late stage wild-type (WT 4-fold), and early stage egl-19(n2368) mutant embryos before muscle defects occur (egl-19 1.7 fold), or late stage egl-19(n2368), which are arrested at 1.5-fold elongation (egl-19 arrested). Boxed areas in A–F are enlarged below. Scale bars: 5 μm.
Previous reports claim that the formation of intracellular periodic structures relies on the actin and spectrin membrane skeleton (Xu et al., 2013; Zhong et al., 2014). To test whether this process is conserved in C. elegans, we disrupted the embryonic actin and spectrin network and examined the effects on stripe formation. As shown in Fig. 1E, upon inactivation of spc-1 or act-1 expression in the epidermis, the CeHD units failed to organize into parallel stripes and remained irregularly distributed even in late stage embryos (Fig. 1E). The intact periodic patterns in egl-19 (encoding a voltage-gated Ca2+ channel) mutants, which display elongation arrest caused by lack of muscle contraction but not epidermal structural defects (Zhang et al., 2011; Lardennois et al., 2019), clarified that abolished periodic patterns in spc-1 and act-1-inactivated embryos is not due to shortened embryonic body length (Fig. 1F). Taken together, the data suggest that the formation of the striped patterns in the C. elegans epidermis starts from the alignment of spectrin molecules and actin bundles, which in turn provides the framework for the reorganization of IFs and CeHD stripes without the participation of microtubules.
The periodic structures reorganize through stripe duplication during post-embryonic epidermal growth
We next documented the distribution and reorganization of the major periodic structures during post-embryonic development, a process in which the epidermal hyp7 syncytium elongates 6-fold from hatching till adulthood (Chisholm and Hsiao, 2012). We first visualized the localization of CeHDs by immunostaining for the apical and basal CeHD receptors MUP-4 and LET-805, respectively. Confocal imaging showed that both apical and basal CeHDs maintained their periodic striped patterns throughout development, and that no disorganization occurred (Fig. 2A). Specifically, the CeHD stripes gradually thickened from the L2 stage, until fissures appeared in the middle of the thickened stripes at around the L3 stage (Fig. 2A, arrow). As the fissure widened, one stripe was subsequently divided into two new, and thinner, parallel stripes. The newly divided stripes thickened again when the nematode reached adulthood (Fig. 2A). MUP-4 distribution in live animals, visualized by means of a GFP fusion reporter, confirmed the above observation (Fig. S2). Interestingly, the number of total CeHD stripes was essentially the same before and after hatching, and did not progressively increase according to the length of the epidermis, cell fusion events or the development cycles. Instead, the stripes underwent only one round of duplication at about the L3 stage, multiplying from ∼300 to ∼600 (Fig. 2B). Analysis of mutants with longer (lon-2) or shorter (sma-6) body lengths also confirmed that the total number of CeHD stripes did not fluctuate according to changes in epidermal lengths (Fig. 2C).
Fig. 2.
The epidermal periodic patterns reorganize through synchronized stripe duplication. (A) Immunostaining of apical (green, anti-MUP-4) and basal (red, anti-LET-805) CeHDs in L1–L4 larvae and adults. Arrows point to dividing stripes. See also Fig. S2. (B) Quantification of the total numbers of CeHD stripes and body lengths in the embryo, L1–L4 larvae and adults stages (mean±s.e.m., n=20 biological samples/stage). (C) Quantification of the total numbers of CeHD stripes and body lengths of wild-type (WT), sma-6(wk7) and lon-2(e678) mutants at the adult stage (mean±s.e.m., n=20 biological samples/genotype). (D) Double-labeling of microtubules (green, anti-tubulin) and CeHDs (red, anti-MUP-4) in L1–L4 larvae and adults. Arrows point to dividing stripes and nascent microtubule bundles. (E) Double-labeling of α-spectrin (green, SPC-1::GFP) and CeHDs (red, LET-805::RFP) in L1–L4 larvae and adults. Boxed areas in A, D, E are enlarged on the right. ***P<0.001; ns, not significant [Tukey's multiple comparison test (B), unpaired t-test (C)]. Scale bars: 5 μm.
Unlike the CeHD stripes, which duplicate by stripe thickening and splitting, the microtubule bundles were positioned within the gaps of the CeHD stripes throughout development (Fig. 2D). In the L3 stage right after the duplication of the CeHD stripes, we observed newly formed microtubule strands invading the fissure of a dividing CeHD stripe (Fig. 2D, arrow). It appeared that the microtubules utilize the newly generated CeHD gaps as a guide for the positioning of their own nascent bundles. SPC-1 (α-spectrin), on the other hand, remained co-localized with the CeHDs throughout development (Fig. 2E). After completion of the reorganization process at the adult stage, the stripes formed by SPC-1 and those formed by CeHDs switched their relative positions from overlapping to alternating each other (Fig. 2E). These observations suggest that both microtubules and spectrins employ the CeHD stripes as the template for the reorganization of their own periodic patterns. Because the actin filaments remain mostly scattered in post-embryonic C. elegans epidermis, and do not form long-lasting periodic patterns (Costa et al., 1997), the reorganization of actin filaments is not included in this study. To summarize, the apical and basal CeHD attachment structures reorganize through stripe duplication during post-embryonic epidermal growth, accompanied by formation of new microtubule bundles and rearrangement of periodic spectrin skeletons.
The CeHD stripe duplication process relies on physical links between adjacent stripes and extension force generated from epidermal cell growth
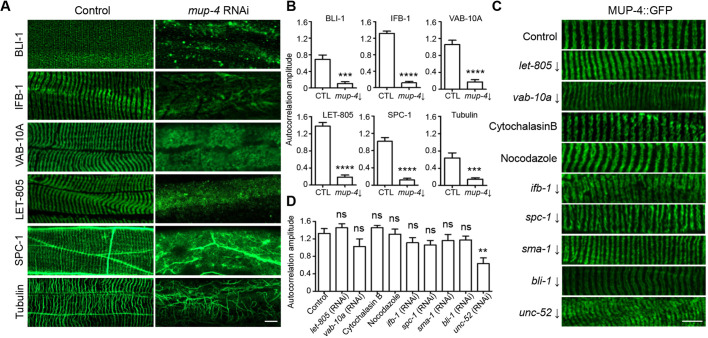
The fact that the duplication of CeHD stripes precedes the doubling of the microtubule bundles suggests that the reorganization of the CeHDs may happen upstream of other periodic structures. To test this hypothesis, we disrupted the apical CeHDs by inactivating mup-4, the EGF-like repeat-containing transmembrane receptor, and examined the changes in the periodic patterns formed by apical ECM (BLI-1), intermediate filaments (IFB-1), spectraplakin family cytolinker (VAB-10A), basal CeHDs (LET-805), spectrins (SPC-1) and microtubules. The results showed that disintegrated apical CeHDs caused a complete loss of the striped patterns of all the other subcellular structures tested (Fig. 3A,B; Fig. S3A). Conversely, we examined whether damage to other periodic structures could affect the striped pattern of apical CeHDs. We employed RNAi or drug treatment to disrupt periodic structures, including the basal CeHDs (let-805), the actin (cytochalasin B), microtubules (nocodazole) and IFs (ifb-1) cytoskeletons, the α- and β-spectrins (spc-1 and sma-1), the cytolinker VAB-10A, and the neighboring tissues, including apical ECM (bli-1) and basement membrane (unc-52) (Zhang et al., 2015). The results showed that none of the disrupted periodic structures could abolish the striped patterns of the apical CeHDs (Fig. 3C,D, Fig. S3B–H). Disruption of the basement membrane structure by unc-52 RNAi resulted in lateral distortion of the MUP-4::GFP stripes, thereby lowered the degree of periodicity, but the striped pattern was still visible (Fig. 3C,D). Taken together, these results suggest that the apical CeHD stripes are responsible for guiding and maintaining the patterns of other subcellular structures.
Fig. 3.
The apical CeHDs are required to maintain the periodic patterns of other subcellular structures. (A,B) Localization patterns (A) and average amplitude of autocorrelation analysis (B) of BLI-1::GFP, IFB-1::GFP, VAB-10A::GFP, LET-805::GFP, SPC-1::GFP and microtubules (anti-tubulin) in wild-type young adult epidermis treated with or without an mup-4 RNAi clone targeting 3857–4958 bp of the mup-4 transcript. (mean±s.e.m., n=10 biological samples/group). (C,D) Localization patterns (C) and average amplitude of autocorrelation analysis (D) of MUP-4::GFP in young adult epidermis intact or with disrupted basal CeHDs (let-805 RNAi), cytolinker (vab-10a RNAi), cytoskeletons (cytochalasin B, nocodazole and ifb-1 RNAi), spectrins (spc-1 and sma-1 RNAi), apical and basal ECM (bli-1 and unc-52 RNAi) (mean±s.e.m., n=10 biological samples/group). RNA target is indicated with a downwards arrow in C. **P<0.01; ***P<0.001; ****P<0.0001; ns, not significant (unpaired t-test). Scale bars: 5 μm.
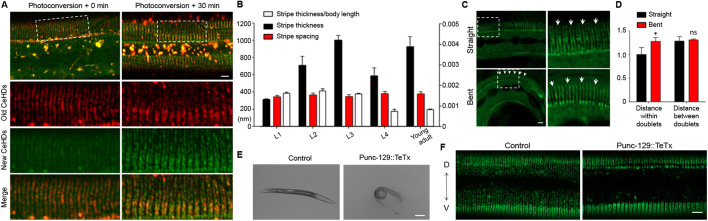
Having established that the apical CeHDs are the key structures controlling the reorganization of periodic patterns, we next wanted to dissect the process of CeHD stripe duplication and elucidate its underlying mechanisms. Our above results showed that CeHD stripe reorganization mainly consists of two steps: thickening and splitting (Fig. 2A). Regarding the first step, there are potentially two ways to achieve stripe thickening – the new CeHD units could either use the old stripes as a template to align into a brand new stripe, or they could randomly insert into the old stripes (Fig. S4A). In order to know which scenario is more likely, we fused MUP-4 with the photoconvertible mEosFP molecule (Wiedenmann et al., 2004). The resulting MUP-4::mEosFP strain was subjected to whole-body photoconversion treatment with the result of labeling all MUP-4 molecules at time zero with red fluorescence. After recovery for 30 min, newly synthesized MUP-4 foci labeled by unconverted green mEosFP were observed in the CeHD stripes, inserting in between the old CeHD units and aligning into two single rows of units (Fig. 4A). This observation is further confirmed by labeling of newly assembled CeHD units with heat shock promoter-driven MUP-4::GFP. As shown in Fig. S4B, the newly synthesized MUP-4 units induced by a 30-min heat shock treatment were inserted randomly into the old stripes, as visualized by anti-MUP-4 immunostaining. We hence conclude that the CeHD stripe thickening is achieved by randomly inserting new CeHD units into the old stripes without any side preference.
Fig. 4.
The apical CeHDs utilize physical links and extension force to drive stripe duplication process. (A) Relative positions of old CeHD units labeled by MUP-4::mEosFP (red) through whole-body photoconversion, and newly formed CeHD units labeled by unconverted MUP-4::mEosFP (green) during stripe separation. Boxed areas are enlarged below. (B) Measurements of the CeHD stripe thickness and the distance between adjacent stripes at the stages of L1–L4 larvae and young adults (mean±s.e.m., n=6 biological samples/stage). (C) MUP-4::GFP labeling showing the different positions of the CeHD stripes within the same L3 larva at regions of straight versus bent body posture. Boxed areas are enlarged on the right. Arrows point to fissures within the same stripe doublet. (D) Measurements of the distance within stripe doublets and between adjacent stripe doublets at regions of straight versus bent body posture (mean±s.e.m., n=6 biological samples/stage). (E) Bright-field images showing the ventral coiler phenotype caused by inactivation of dorsal body wall muscles in transgenic animals expressing Tetanus toxin under the control of the unc-129 promoter. (F) Desynchronized stripe duplication between dorsal and ventral (D↔V) CeHDs (MUP-4::GFP) caused by excessive extension of the dorsal epidermis in transgenic animals expressing Tetanus toxin under the control of the unc-129 promoter. *P<0.05; ns, not significant (unpaired t-test). Scale bars: 5 μm (A,C,F), 100 μm (E).
Our next goal was to understand how one thickened stripe separates into two at the L3 stage. In order to find the driving force for stripe separation, we measured the thickness of the CeHD stripes and the distance between neighboring CeHD stripes at all development stages. The data revealed that changes of the stripe thickness are in accordance with the extension of the epidermis and the stripe duplication process. Specifically, the ratio of stripe thickness to animal length remained relatively steady throughout development, except for halving during stripe duplication (Fig. 4B). By contrast, the distance between neighboring stripes maintained a constant value that was independent of epidermal growth or the number of stripes (Fig. 4B). These results suggest that that the distance between neighboring CeHD stripes is fixed by certain molecules and that the thickened stripes are pulled apart when the epidermal cell elongates (Fig. S4C). This fixed-gap hypothesis is supported by the fact that when the epidermis was temporarily stretched owing to body bends during normal sinusoidal movement, the distance between neighboring CeHD doublets remained constant whereas the fissures within the CeHD doublets were greatly widened (Fig. 4C,D). To test whether indeed the extension of the epidermis is the driving force for stripe separation, we caused persistent extension of the dorsal epidermis by inactivating dorsal muscle contraction with unc-129 promoter-driven Tetanus toxin, thereby confining muscle contraction to the ventral body-wall only (Fig. 4E). In this situation, we found that the CeHD stripes on the dorsal side divided earlier than those on the ventral side, abolishing the synchronized dorsal-ventral stripe reorganization observed in the control group (Fig. 4F). Together, these results suggest that the CeHD stripe duplication is achieved by fixed physical links between adjacent stripes and extension forces generated from epidermal cell growth.
The duplication of CeHD stripes requires the function of apical extracellular collagens
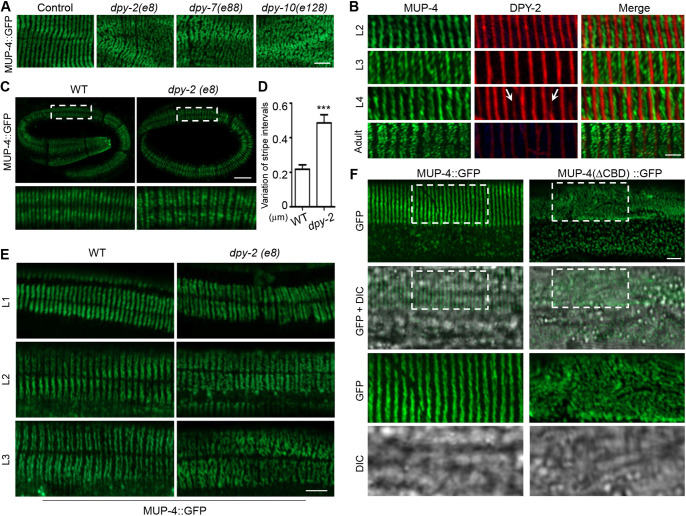
We next sought to find which molecules are responsible for fixing the gaps between adjacent periodic stripes. We discovered by testing all the molecules known to be periodically organized in the epidermis that only a few apically secreted collagens could significantly affect the striped patterning of the apical CeHDs (Figs 3C and 5A) (Francis and Waterston, 1991; Hresko et al., 1994, 1999; Rogalski et al., 1995; Costa et al., 1997; Hong et al., 2001; Bosher et al., 2003; McMahon et al., 2003; Woo et al., 2004; Zhang et al., 2011, 2015). Particularly, mutations of collagen-encoding dpy-2, dpy-7 and dpy-10 genes caused a complete loss of the striped CeHD pattern in the epidermis (Fig. 5A). This result suggests that these collagens located at the apical ECM are promising candidates for acting as the ‘gap-holders’ between adjacent CeHD stripes. Double-labeling experiments showed that these collagens were indeed constantly positioned at the gaps between CeHD stripes at all stages (Fig. 5B). Moreover, new collagen stripes formed and filled the new gaps only after the CeHD stripes were completely separated into two, suggesting that the thickened CeHD stripes were torn apart from the outside instead of cleaved through new collagen molecules becoming inserted into the middle of the stripes (Fig. 5B, arrows). The fact that the expression of the collagens greatly diminished at the adult stage also explains why the CeHD stripes do not duplicate again in older animals in response to further epidermal growth (Figs 2B and 5B). Examination of the collagen mutant embryos revealed that loss of collagens did not affect the initial formation of the periodic stripes, but greatly disturbed the regularity of the intervals between CeHD stripes (Fig. 5C,D). Analysis of collagen mutants at the L1–L3 larval stages subsequently showed that loss of epidermal periodicity occurred during the processes of CeHD stripe thickening and separation (Fig. 5E). Altogether, these results support the notion that certain collagens at the apical ECM are responsible for binding and fixing the distance between adjacent MUP-4 stripes. The fact that duplication of CeHDs is not concomitant with the molting cycles, and that cuticle blistering does not affect CeHD stripe patterning, suggests that it is the collagens at the basal zone, but not the cuticular zone, of the apical ECM that participate in CeHD stripe organization (Figs 2B and 3C). To further confirm that the CeHDs require collagen binding to duplicate their stripes, we deleted the conserved von Willebrand factor type A (vWFA) domain, a well-known collagen-binding domain across species, in the extracellular region of MUP-4 (Hong et al., 2001). The results showed that without the putative collagen-binding ability of MUP-4, the distribution of apical CeHDs became entirely random and no periodic stripes were found (Fig. 5F). Taken together, we conclude that certain apical extracellular collagens are responsible for holding the gaps between adjacent CeHD stripes and tearing the thickened stripes into two to achieve synchronized duplication during epidermal growth.
Fig. 5.
CeHD stripe duplication requires the functions of apical extracellular collagens. (A) MUP-4::GFP localization in young adult epidermis of wild-type and dpy-2(e8), dpy-7(e88), dpy-10(e128) mutants. (B) Double-labeling of MUP-4::GFP and DPY-2::mCherry in L2–L4 larvae and adults. Arrows point to nascent DPY-2 stripes assembling in the already established CeHD gaps. (C) MUP-4::GFP localization in wild-type and dpy-2(e8) mutant embryos. (D) Variation of the stripe positions in wild-type and dpy-2(e8) embryos represented by the difference between the maximum and minimum distance between adjacent stripes in each embryo (mean±s.e.m., n=10 biological samples/genotype). (E) MUP-4::GFP localization in wild-type and dpy-2(e8) mutant at the L1, L2 and L3 stage. (F) Localization patterns of full-length MUP-4 (MUP-4::GFP) and MUP-4 without the collagen-binding domain (MUP-4::△CBD::GFP) in young adult epidermis in a mup-4(mg36) mutant background. Boxed areas are enlarged below. ***P<0.001 (unpaired t-test). Scale bars: 5 μm (A,C,E,F), 2 μm (B).
DISCUSSION
In summary, our work describes a novel strategy employed to maintain the periodic-patterned subcellular structures during cell growth and tissue development (Fig. S5). The evolutionarily conserved actin and spectrin network is only responsible for the initialization but not the reorganization process of the periodic patterns. The periodic stripes are fixed and maintained by stable membrane attachment structures, such as CeHDs. Other periodically arranged structures could then utilize the CeHD stripes as a template to guide their own reorganization process. Most interestingly, the intracellular periodic structures do not accommodate cell growth by continuously making new stripes to fill the expanded space. Instead, they manage to maintain constant spacing by performing synchronized stripe duplication over a long distance. Because the first cell fusion event of hyp7 syncytium occurs in embryos before comma stage, followed by more cell fusions that progress throughout embryogenesis and during the entire post-embryonic development, the initiation/development of the periodic structures appears to not be correlated with hyp7 cell fusion (Podbilewicz and White, 1994) (Figs 1 and 2). During the stripe reorganization process, the epidermal cells utilize a double-anchor system composed of transmembrane CeHD receptors and their extracellular ligands to further stabilize the intracellular periodic patterns. The old CeHD stripes determine the extracellular positions of the newly secreted collagens, and selected collagens in turn immobilize the CeHD units and provide tearing forces for the splitting and creation of new stripe doublets (Fig. S5). It has been proposed that trimerization of collagen molecules could likely happen between members of cuticle collagens with the same temporal expression patterns (Johnstone, 2000). The fact that multiple collagens are involved in stripe organization suggests that a small cross-linked collagen network could be responsible for holding the gaps between adjacent CeHD stripes (Fig. 5A). In conclusion, the stripe duplication process exhibits a series of elegant choices made by the epidermal cells to bring into full play the distinct characteristics of each type of molecules. Together, they manage to maintain the stability and constant spacing of subcellular structures as the epidermal layer elongates 6-fold in length. Because intracellular periodicity is tightly linked to cellular structure and signaling (Albrecht et al., 2016; Qu et al., 2017; Huang et al., 2017; Wang et al., 2019; Zhou et al., 2019), this strategy of synchronized reorganization could ensure that the cells do not suffer from local dysfunction or deformation during cell growth and tissue development. Furthermore, our observations also indicate that the reorganization of subcellular structures could happen in a highly uniform and collective manner, rather than as isolated disassembly and reassembly events. This implies that besides diffusible molecules and chemical signals, intracellular structures could also utilize physical links and mechanical forces to achieve long-distance communication and coordination during development. The findings in this work may provide insights on the mechanisms of other similarly repetitive subcellular structures in different cell types.
MATERIALS AND METHODS
C. elegans strains
C. elegans strains were maintained at 20°C on nematode growth medium (NGM) as described previously (Brenner, 1974). The CRISPR knock-in strain carrying cas815(spc-1::GFP KI) was a gift from the Guangshuo Ou laboratory (Tsinghua-Peking Center for Life Sciences, Tsinghua University, China; Jia et al., 2019). The strain carrying mcIs43[Plin-26::ABDvab-10-mCherry+Pmyo-2-GFP] is from the Michel Labouesse laboratory (Laboratoire de Biologie du Développement, Institut de Biologie Paris Seine, France; Gally et al., 2009). The epidermal-specific RNAi strain NR222[rde-1(ne219)V;kzIs9[pKK1260(lin-26p::nls::GFP)+pKK1253(lin-26p::rde-1)+pRF6(rol-6(su1006)] was a gift from Dr Hiroshi Qadota (Department of Pathology, Emory University, USA. Qadota et al., 2007). The wild-type N2 strain and strains carrying sma-6(wk7), lon-2(e678), egl-19(n2368), dpy-2(e8), dpy-7(e88), dpy-10(e128), mup-4(mg36), juls176[ IFB-1A::GFP+pRF4], ieSi57[eft-3p::TIR1::mRuby::unc-54-3′UTR+Cbr-unc-119(+)], vab-10a(cas602[vab-10a::GFP]), chEx1682[pLH070(qua-1(full-length)::GFP+rol-6(su1006)], cgEx198[(pJC14)bli-1::GFP+unc-119(+)], let-805(mc73[let-805::GFP+unc-119(+)])unc-119(ed3)III and upIs1[mup-4::GFP+pRF4(rol-6(su1006)] were obtained from Caenorhabditis Genetics Center (Woo et al., 2004; Zhang et al., 2015; Yang et al., 2017; Hao et al., 2006; Hong et al., 2001). Strains carrying sdaEx183[Pmup-4::mup-4(ΔCBD)::GFP+Pmyo-2::GFP], sdaEx64[dpy-2::mcherry+Pmyo-2::GFP], sdaEx127[Phsp-16.2::mup-4::GFP+Pmyo-2::GFP], let-805(sda14{let-805::RFP]) and mup-4(sda18[mup-4::mcherry]) were generated for this study.
Molecular cloning and transgenesis
The Pmup-4::mup-4::GFP construct was generated by PCR, amplifying a 4933 bp mup-4 promoter plus the entire mup-4 coding sequence and inserting it upstream of GFP into pPD95,75 vector. Pmup-4::mup-4 (ΔCBD)::GFP was generated by deleting the genomic sequence encoding amino acids (aa) 460–565 (corresponding to the vWFA domain of MUP-4 protein) by enzyme digestion. The photoconvertible Pmup-4::mup-4::mEosFP construct was generated by replacing the GFP in Pmup-4::mup-4::GFP with mEosFP (Wiedenmann et al., 2004). To construct Pdpy-2::dpy-2::mCherry transgene, 1995 bp dpy-2 promoter plus the entire dpy-2 coding sequence were PCR amplified from N2 genomic DNA, and then inserted upstream of mCherry into pPD49.78 vector. Constructs used in this study were generated by infusion recombination using ClonExpress TMOne Step Cloning Kit (Vazyme biotech, Nanjing, CHINA). All PCR amplifications were performed by using PrimeSTAR® GXL DNA Polymerase kit (TaKaRa, Shiga, Japan) and PCR fragments were confirmed by sequencing. Punc-129::TeTx::mCherry plasmid (2894 bp unc-129 promoter-driven Tetanus toxin light chain and mCherry) was a gift from Yingchuan Qi and Yishi Jin (Division of Biological Sciences, University of California, USA). Pmup-4-mup-4::mEosFP and Pdpy-2::dpy-2::mCherry was injected into N2 at a concentration of 10 ng/µl. Punc-129::TeTx::mCherry was injected into EE86 at a concentration of 10 ng/µl. Pmup-4::mup-4 (ΔCBD)::GFP was injected into mup-4(mg36)/+ heterozygous at a concentration of 20 ng/µl. Pmyo-2::GFP was used as co-injection marker
Generation of CRISPR knock-in strains
To generate the let-805::rfp knock-in strain, the let-805 sgRNA (5′-GCACTCATCCCGTTAATCTCTGG-3′) was selected by the optimizing CAS9 gRNA site (http://crispr.mit.edu/) and inserted into the single-guide RNA Cas9 vector (Addgene #47549) using Q5® Site-Directed Mutagenesis Kit as previously described (Dickinson et al., 2015). To insert RFP into the C-terminus of let-805, 1 kb sequences upstream and downstream of let-805 stop codon was selected as 5′ and 3′ homologous arms, respectively. The let-805 donor, let-805 sgRNA and dpy-10 sgRNA were injected into N2 at the concentration of 100 ng/µl, 50 ng/µl and 50 ng/µl, respectively. dpy-10 sgRNA was used as positive selective marker according to a previously established method (Dickinson et al., 2015). The mup-4(sda18[mup-4::mcherry]) strain was constructed by inserting the mCherry sequence before the termination codon of mup-4 using the same CRISPR/Cas9-mediated editing method as let-805(sda14[let-805::RFP]).
C. elegans RNAi and drug treatment
RNAi experiments were performed by bacteria feeding as previously described (Kamath et al., 2003). RNAi clones were recovered from the MRC RNAi library, and the inserts verified by sequencing. In ifb-1 RNAi, unc-52 RNAi, spc-1 RNAi, bli-1 RNAi and sma-1 RNAi experiments, worms were fed on RNAi plates against target genes from the L1 stage. In let-805 RNAi, mup-4 RNAi and vab-10a RNAi experiments, worms were fed on the HT115 plates until the L2 stage and transferred to RNAi plates against target genes. act-1 and spc-1 knockdown in embryos were carried out by injecting dsRNA into the gonad of young adult NR222 mothers, and embryos were collected within the next 24 h for immunostaining. Feeding of HT115 bacteria carrying L4440 empty vector was used as negative control for all RNAi experiments. For disruption of microtubule bundles, L4-stage worms were soaked in 60 µg/ml nocodazole (Sangon Biotech, Shanghai, China) for 2 h in M9 buffer. For disruption of actin filaments, L4-stage worms were soaked in 10 µg/ml Cytochalasin B (Solarbio Science & Technology, Beijing, China) for 1 h in M9 buffer. All treatments were repeated at least three times and the representative data are shown.
C. elegans embryonic and post-embryonic immunostaining
Immunostaining was performed based on previously established protocols (Costa et al., 1997; Zahreddin et al., 2010). The following primary antibodies were used in this work: anti-GFP (DSHB-GFP-12A6), 1:500; anti-tubulin (AA4.3), 1:100; anti-LET-805/myotactin (MH46), 1:100; anti-VAB-10A (MH5), 1:50; anti-UNC-52 (MH3), 1:50; anti-MUP-4, 1:500. 12A6, AA4.3, MH3, MH5 and MH46 were obtained from DSHB (University of Iowa, USA). The MUP-4 polyclonal antibody was custom generated as previously described (Zhang et al., 2015). AlexaFluor® 488- and AlexaFluor® 568-labeled secondary antibodies were obtained from Molecular Probes (Eugene, OR) and used at 1:800 dilution.
Microscopy imaging and measurements
Fluorescence and DIC images were captured by using a Zeiss LSM800 confocal microscope equipped with a 63× oil immersion objective. 488-nm excitation and a 400–570 nm emission filter was used for GFP and AlexaFluor® 488, 568-nm excitation and a 570–700 nm emission filter for RFP, mCherry and AlexaFluor® 568. For single-plane confocal imaging of larvae and adult C. elegans, worms were mounted on 2% agarose pads on a microscopic slide and immobilized with 0.5 mM levamisole. For confocal imaging of C. elegans embryos, eggs were collected onto 2% agarose pads on a microscopic slide and immobilized by oxygen deprivation using a small amount of OP50 bacteria. Stacks of images were captured with 1-µm interval spanning from the top to the mid-body of the embryos. Bright-field images of worms on plates were acquired using a OLYMPUS SZX16 stereomicroscope equipped with a LEICA DFC310 camera.
Photoconversion of Pmup-4-mup-4::mEosFP was achieved by using a LEICA DM2500 upright epifluorescence microscope with a 40× objective lens (Wiedenmann et al., 2004; Mathur et al., 2010). L3-stage transgenic animals were mounted onto 2% agarose pads and exposed to the epifluorescent lighting through the DAPI filter cube with maximum power for 2.5 min. Images were captured with a ZEISS LSM800 confocal microscope immediately after photoconversion or after 30 min recovery.
For measuring C. elegans body lengths, bright-field images of worms cultured on the plates at different stages and different genotypes were captured by means of a stereomicroscope. The body length of each individual was measured from the nose to the tail tip using Image J software (http://rsb.info.nih.gov/ij/). For measuring the thickness and gap distance of the CeHD stripes at different stages, confocal images were taken at the mid-bodies of the MUP-4::GFP transgenic worms, using six animals for each group. For every image, the widths of at least 20 stripes and 20 gaps were measured using Image J, and the mean values were calculated. To count the total number of CeHD stripes in each individual, a series of confocal images were taken from the nose to the tail of anti-MUP-4 immunostained animals with a 63× oil immersion objective. The images were then pieced together to reconstruct the entire worm body and the total number of CeHD stripes was counted by a researcher who was blind to the experimental conditions. To generate the autocorrelation curves, intensity profiles were extracted from confocal images of periodic patterns using ImageJ software and then analyzed using the built-in routine autocorrection function of MATLAB (https://www.mathworks.com/) as previously described (Zhong et al., 2014; Han et al., 2017). The autocorrelation amplitude is defined as the difference between the first peak and the average of the two first valleys of the autocorrelation curve.
Statistical analysis
Statistical analyses were performed using GraphPad Prism7.0 software (www.graphpad.com). No statistical method was used to predetermine sample size. Statistical analysis was performed with an unpaired two-tailed Student's t-test. Data normality was verified by using the D'Agostino and Pearson normality test and the Shapiro-Wilk normality test. One-way ANOVA followed by Tukey's multiple comparison test was performed on data in Fig. 2B. Significance was accepted for P<0.05.
Supplementary Material
Acknowledgements
We thank Guangshuo Ou, Yingchuan Qi, Yishi Jin, Hiroshi Qadota and Michel Labouesse for reagents. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Author contributions
Conceptualization: H.Z.; Validation: C.W., Y.Y.; Formal analysis: C.W.; Investigation: C.W., Y.Y., R.F.; Resources: Y.Z.; Data curation: R.F., Y.Z.; Writing - original draft: C.W., H.Z.; Writing - review & editing: H.Z.; Visualization: C.W., R.F.; Supervision: H.Z.; Funding acquisition: H.Z.
Funding
This work was supported by National Natural Science Foundation of China (31670912, 31871384 and 31900545), National Key R&D Program of China (2019YFA0802400), Jiangsu Provincial Distinguished Young Scholars (BK20160009), Jiangsu Provincial Innovative Research Team, the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT-IRT1075) and the Priority Academic Development Program of Jiangsu Province Higher Education Institutions (PAPD).
Contributor Information
Chunxia Wang, Jiangsu Key Laboratory of Infection and Immunity, Institutes of Biology and Medical Sciences, Soochow University, Suzhou 215123, China.
Yuyan Yang, Jiangsu Key Laboratory of Infection and Immunity, Institutes of Biology and Medical Sciences, Soochow University, Suzhou 215123, China.
Rong Fu, Jiangsu Key Laboratory of Infection and Immunity, Institutes of Biology and Medical Sciences, Soochow University, Suzhou 215123, China.
Yi Zhu, Jiangsu Key Laboratory of Infection and Immunity, Institutes of Biology and Medical Sciences, Soochow University, Suzhou 215123, China.
Huimin Zhang, Jiangsu Key Laboratory of Infection and Immunity, Institutes of Biology and Medical Sciences, Soochow University, Suzhou 215123, China.
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.246793.reviewer-comments.pdf
References
- Albrecht, D., Winterflood, C. M., Sadeghi, M., Tschager, T., Noé, F. and Ewers, H. (2016). Nanoscopic compartmentalization of membrane protein motion at the axon initial segment. J. Cell Biol. 215, 37-46. 10.1083/jcb.201603108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher, J. M., Hahn, B.-S., Legouis, R., Sookhareea, S., Weimer, R. M., Gansmuller, A., Chisholm, A. D., Rose, A. M., Bessereau, J.-L. and Labouesse, M. (2003). The Caenorhabditis elegans vab-10 spectraplakin isoforms protect the epidermis against internal and external forces. J. Cell Biol. 161, 757-768. 10.1083/jcb.200302151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, A. D. and Hsiao, T. I. (2012). The Caenorhabditis elegans epidermis as a model skin. I: development, patterning, and growth. Wiley Interdiscip. Rev. Dev. Biol. 1, 861-878. 10.1002/wdev.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, A. D. and Xu, S. (2012). The Caenorhabditis elegans epidermis as a model skin. II: differentiation and physiological roles. Wiley Interdiscip. Rev. Dev. Biol. 1, 879-902. 10.1002/wdev.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M., Draper, B. W. and Priess, J. R. (1997). The role of actin filaments in patterning the Caenorhabditis eleganscuticle. Dev. Biol. 184, 373-384. 10.1006/dbio.1997.8530 [DOI] [PubMed] [Google Scholar]
- Dalle Nogare, D. and Chitnis, A. B. (2017). Self-organizing spots get under your skin. PLoS Biol. 15, e2004412. 10.1371/journal.pbio.2004412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D. J., Pani, A. M., Heppert, J. K., Higgins, C. D. and Goldstein, B. (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035-1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou, A. D., Ohazama, A., Porntaveetus, T., Sharpe, P. T., Kondo, S., Basson, M. A., Gritli-Linde, A., Cobourne, M. T. and Green, J. B. A. (2012). Periodic stripe formation by a turing mechanism operating at growth zones in the mammalian palate. Nat. Genet. 44, 348-351. 10.1038/ng.1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, R. and Waterston, R. H. (1991). Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 114, 465-479. 10.1083/jcb.114.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally, C., Wissler, F., Zahreddine, H., Quintin, S., Landmann, F. and Labouesse, M. (2009). Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development 136, 3109-3119. 10.1242/dev.039412 [DOI] [PubMed] [Google Scholar]
- Han, B., Zhou, R., Xia, C. and Zhuang, X. (2017). Structural organization of the actin-spectrin-based membrane skeleton in dendrites and soma of neurons. Proc. Natl. Acad. Sci. USA 114, E6678-E6685. 10.1073/pnas.1705043114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, L., Mukherjee, K., Liegeois, S., Baillie, D., Labouesse, M. and Bürglin, T. R. (2006). The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev. Dyn. 235, 1469-1481. 10.1002/dvdy.20721 [DOI] [PubMed] [Google Scholar]
- Hauser, M., Yan, R., Li, W., Repina, N. A., Schaffer, D. V. and Xu, K. (2018). The spectrin-actin-based periodic cytoskeleton as a conserved nanoscale scaffold and ruler of the neural stem cell lineage. Cell Rep. 24, 1512-1522. 10.1016/j.celrep.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J., Zhou, R., Wu, Z., Carrasco, M. A., Kurshan, P. T., Farley, J. E., Simon, D. J., Wang, G., Han, B., Hao, J., et al. (2016). Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc. Natl. Acad. Sci. USA 113, 6029-6034. 10.1073/pnas.1605707113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Elbl, T., Ward, J., Franzini-Armstrong, C., Rybicka, K. K., Gatewood, B. K., Baillie, D. L. and Bucher, E. A. (2001). MUP-4 is a novel transmembrane protein with functions in epithelial cell adhesion in Caenorhabditis elegans. J. Cell Biol. 154, 403-414. 10.1083/jcb.200007075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko, M. C., Williams, B. D. and Waterston, R. H. (1994). Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J. Cell Biol. 124, 491-506. 10.1083/jcb.124.4.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko, M. C., Schriefer, L. A., Shrimankar, P. and Waterston, R. H. (1999). Myotactin, a novel hypodermal protein involved in muscle–cell adhesion in Caenorhabditis elegans. J. Cell Biol. 146, 659-672. 10.1083/jcb.146.3.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. Y.-M., Zhang, C., Zollinger, D. R., Leterrier, C. and Rasband, M. N. (2017). An αII Spectrin-Based Cytoskeleton Protects Large-Diameter Myelinated Axons from Degeneration. J. Neurosci. 37, 11323-11334. 10.1523/JNEUROSCI.2113-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, R., Li, D., Li, M., Chai, Y., Liu, Y., Xie, Z., Shao, W., Xie, C., Li, L., Huang, X., et al. (2019). Spectrin-based membrane skeleton supports ciliogenesis. PLoS Biol. 17, e3000369. 10.1371/journal.pbio.3000369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone, I. L. (2000). Cuticle collagen genes expression in Caenorhabditis elegans. Trends Genet. 16, 21-27. 10.1016/S0168-9525(99)01857-0 [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Lardennois, A., Pásti, G., Ferraro, T., Llense, F., Mahou, P., Pontabry, J., Rodriguez, D., Kim, S., Ono, S., Beaurepaire, E., et al. (2019). An actin-based viscoplastic lock ensures progressive body-axis elongation. Nature 573, 266-270. 10.1038/s41586-019-1509-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liégeois, S., Benedetto, A., Michaux, G., Belliard, G. and Labouesse, M. (2007). Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics 175, 709-724. 10.1534/genetics.106.066035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Radhamony, R., Sinclair, A. M., Donoso, A., Dunn, N., Roach, E., Radford, D., Mohammad Mohaghegh, P. S., Logan, D. C., Kokolic, K., et al. (2010). mEosFP-based green-to-red photoconvertible subcellular probes for plants. Plant Physiol. 154, 1573-1587. 10.1104/pp.110.165431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, L., Muriel, J. M., Roberts, B., Quinn, M. and Johnstone, I. L. (2003). Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol. Biol. Cell 14, 1366-1378. 10.1091/mbc.e02-08-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, K. R. and Moerman, D. G. (2002). α spectrin is essential for morphogenesis and body wall muscle formation in Caenorhabditis elegans. J. Cell Biol. 157, 665-677. 10.1083/jcb.200111051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz, B. and White, J. G. (1994). Cell fusions in the developing epithelial of C. elegans. Dev. Biol. 161, 408-424. 10.1006/dbio.1994.1041 [DOI] [PubMed] [Google Scholar]
- Priess, J. R. and Hirsh, D. I. (1986). Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev. Biol. 117, 156-173. 10.1016/0012-1606(86)90358-1 [DOI] [PubMed] [Google Scholar]
- Qadota, H., Inoue, M., Hikita, T., Köppen, M., Hardin, J. D., Amano, M., Moerman, D. G. and Kaibuchi, K. (2007). Establishment of a tissue-specific RNAi system in C. elegans. Gene 400, 166-173. 10.1016/j.gene.2007.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Y., Hahn, I., Webb, S. E. D., Pearce, S. P. and Prokop, A. (2017). Periodic actin structures in neuronal axons are required to maintain microtubules. Mol. Biol. Cell 28, 296-308. 10.1091/mbc.e16-10-0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski, T. M., Gilchrist, E. J., Mullen, G. P. and Moerman, D. G. (1995). Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics 139, 159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, M. D., Greer, C. and Gaul, U. (2011). How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. Development 138, 3067-3078. 10.1242/dev.062141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth, F. and Corson, F. (2019). Self-organization in pattern formation. Dev. Cell 49, 659-677. 10.1016/j.devcel.2019.05.019 [DOI] [PubMed] [Google Scholar]
- Soloviev, A., Gallagher, J., Marnef, A. and Kuwabara, P. E. (2011). C. elegans patched-3 is an essential gene implicated in osmoregulation and requiring an intact permease transporter domain. Dev. Biol. 351, 242-253. 10.1016/j.ydbio.2010.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Simon, D. J., Wu, Z., Belsky, D. M., Heller, E., O'Rourke, M. K., Hertz, N. T., Molina, H., Zhong, G., Tessier-Lavigne, M., et al. (2019). Structural plasticity of actin-spectrin membrane skeleton and functional role of actin and spectrin in axon degeneration. eLife 8, e38730. 10.7554/eLife.38730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann, J., Ivanchenko, S., Oswald, F., Schmitt, F., Röcker, C., Salih, A., Spindler, K.-D. and Nienhaus, G. U. (2004). EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 101, 15905-15910. 10.1073/pnas.0403668101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, W.-M., Goncharov, A., Jin, Y. and Chisholm, A. D. (2004). Intermediate filaments are required for C. elegans epidermal elongation. Dev. Biol. 267, 216-229. 10.1016/j.ydbio.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Xu, K., Zhong, G. and Zhuang, X. (2013). Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452-456. 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Zhang, Y., Li, W.-J., Jiang, Y., Zhu, Z., Hu, H., Li, W., Wu, J.-W., Wang, Z.-X., Dong, M.-Q., et al. (2017). Spectraplakin Induces Positive Feedback between Fusogens and the Actin Cytoskeleton to Promote Cell-Cell Fusion. Dev. Cell. 41, 107-120.e4. 10.1016/j.devcel.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Zahreddine, H., Zhang, H., Diogon, M., Nagamatsu, Y. and Labouesse, M. (2010). CRT-1/calreticulin and the E3 ligase EEL-1/HUWE1 control hemidesmosome maturation in C. elegans development. Curr. Biol. 20, 322-327. 10.1016/j.cub.2009.12.061 [DOI] [PubMed] [Google Scholar]
- Zhang, H. and Labouesse, M. (2010). The making of hemidesmosome structures in vivo. Dev. Dyn. 239, 1465-1476. 10.1002/dvdy.22255 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Landmann, F., Zahreddine, H., Rodriguez, D., Koch, M. and Labouesse, M. (2011). A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature 471, 99-103. 10.1038/nature09765 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Li, W., Li, L., Li, Y., Fu, R., Zhu, Y., Li, J., Zhou, Y., Xiong, S. and Zhang, H. (2015). Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity 42, 309-320. 10.1016/j.immuni.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Zhong, G., He, J., Zhou, R., Lorenzo, D., Babcock, H. P., Bennett, V. and Zhuang, X. (2014). Developmental mechanism of the periodic membrane skeleton in axons. eLife 3, e04581. 10.7554/eLife.04581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, R., Han, B., Xia, C. and Zhuang, X. (2019). Membrane-associated periodic skeleton is a signaling platform for RTK transactivation in neurons. Science. 365, 929-934. 10.1126/science.aaw5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Fig. 2. The epidermal periodic patterns reorganize through synchronized stripe duplication. (A) Immunostaining of apical (green, anti-MUP-4) and basal (red, anti-LET-805) CeHDs in L1–L4 larvae and adults. Arrows point to dividing stripes. See also Fig. S2. (B) Quantification of the total numbers of CeHD stripes and body lengths in the embryo, L1–L4 larvae and adults stages (mean±s.e.m., n=20 biological samples/stage). (C) Quantification of the total numbers of CeHD stripes and body lengths of wild-type (WT), sma-6(wk7) and lon-2(e678) mutants at the adult stage (mean±s.e.m., n=20 biological samples/genotype). (D) Double-labeling of microtubules (green, anti-tubulin) and CeHDs (red, anti-MUP-4) in L1–L4 larvae and adults. Arrows point to dividing stripes and nascent microtubule bundles. (E) Double-labeling of α-spectrin (green, SPC-1::GFP) and CeHDs (red, LET-805::RFP) in L1–L4 larvae and adults. Boxed areas in A, D, E are enlarged on the right. ***P<0.001; ns, not significant [Tukey's multiple comparison test (B), unpaired t-test (C)]. Scale bars: 5 μm.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/66e8/10682509/d5d12ff0aacb/joces-133-246793-g2.jpg)